Abstract
WNT/β-catenin signaling plays key roles in development and cancer1,2. ZNRF3/RNF43 modulates Frizzleds through ubiquitination, dampening WNT/β-catenin signaling. Conversely, RSPO1-4 binding to LGR4-6 and ZNRF3/RNF43 enhances WNT/β-catenin signaling3–5. Here, we elucidate the overall landscape of architectures in multiple LGR4, RSPO2, and ZNRF3 assemblies, showcasing varying stoichiometries and arrangements. These structures reveal that LGR4 and RSPO2 capture distinct states of ZNRF3. The intrinsic heterogeneity of the LGR4-RSPO2-ZNRF3 assembly is influenced by LGR4 content. Particularly, in the assembly complex with a 2:2:2 ratio, two LGR4 protomers induce and stabilize the inactive state of ZNRF3, characterized by a wide inward-open conformation of two transmembrane helices (TM helices). This specific assembly promotes a stable complex, facilitating LGR4-induced endocytosis of ZNRF3. In contrast, the active dimeric ZNRF3, bound by a single LGR4, adopts a coiled-coil TM helices conformation and dimerization of RING domains. Our findings unveil how LGR4 content mediates diverse assemblies, leading to conformational rearrangements in ZNRF3 to regulate WNT/β-catenin signaling, and provide a structural foundation for drug development targeting Wnt-driven cancers.
Subject terms: Ligases, Cryoelectron microscopy
The Wnt signalling pathway is essential for development and the maintenance of stem cells. Here, the authors reveal how LGR4 and RSPO2 modulate ZNRF3 conformations and assembly heterogeneity, stabilising ZNRF3 in its inactive state to enhance Wnt signalling. These findings offer insights for the development of targeted therapies for Wnt-driven cancers.
Introduction
WNT/β-catenin signaling plays central roles in embryonic development and adult tissue homeostasis1,2. Two cell-surface single transmembrane E3 ubiquitin ligases, Zinc ring finger 3 (ZNRF3) and ring finger protein 43 (RNF43), are integral components acting as negative feedback modulators within this intricate pathway3,4. The two E3 ubiquitin ligases play a crucial role by recognizing and ubiquitinating Frizzleds, leading to their endocytosis and degradation, effectively reducing WNT/β-catenin signaling. In contrast, leucine-rich-repeat G protein-coupled receptors 4-6 (LGR4-6) and their ligands R-spondin (RSPO1-4) serve as well-established positive modulators. They inhibit ZNRF3/RNF43 activity, increasing the abundance of Frizzleds on the cell surface, thereby amplifying WNT/β-catenin signaling5–12. Additionally, dysfunctional ZNRF3/RNF43 mutants, aberrant expression of LGR4-6, and RSPOs translocation contribute to oncogenic Wnt/β-catenin signaling in various cancers13–20. Understanding the structural principles governing these regulatory modules is essential for developing strategies to precisely control Wnt signaling output.
LGR4, classified as a class A orphan GPCR, features a relatively expansive horseshoe-shaped extracellular domain (ECD) containing 17 leucine-rich-repeats (LRRs), followed by a seven-helix transmembrane domain of the rhodopsin type21,22. In contrast, ZNRF3 is a transmembrane E3 ubiquitin ligase with an extracellular N-terminal protease-associated domain (PAD) and a single transmembrane helix, followed by an intracellular C-terminal RING domain housing the E3 ligase activity20–23. Numerous previously reported crystal structures of their ectodomains have elucidated the interaction of RSPOs with both the LGR4/5 ECD and ZNRF3/RNF43 ECD21,22,24–30. However, these structures reveal diverse quaternary arrangements, potentially influenced by the absence of the cell membrane and the 7TM region. The assembly mechanism of the full-length LGR4, ZNRF3, and RSPOs complex warrants further exploration. Importantly, LGR4 does not depend on a G protein for WNT/β-catenin signaling3,7, prompting questions about signal transduction through its transmembrane domains and the subsequent impact on WNT/β-catenin signaling.
While cytosolic E3 ubiquitin ligases have been extensively studied, limited research has been conducted on the structure and regulation of cell-surface single transmembrane E3 ubiquitin ligases like ZNRF3 and RNF4320,23. These ligases play a pivotal role in remodeling the cell surface proteome and controlling cellular responses to extracellular ligands in diverse biological processes. ZNRF3, in particular, has shown promise as a potent degrader of disease-causing cell-surface proteins through anti-ZNRF3-antibody-based PROTABs (proteolysis-targeting antibodies)31–33. Comprehending the mechanisms of WNT/β-catenin signaling regulation through E3 ubiquitin ligases and the structural rearrangements involved will shed light on the intricate control of WNT/β-catenin signaling and pave the way for novel therapeutic strategies in various diseases.
In this work, we observe the diverse structures of the assembly of LGR4, RSPO2, and ZNRF3 subunits at different stoichiometric ratios using cryo-electron microscopy (cryo-EM). The intrinsic heterogeneity of the LGR4-RSPO2-ZNRF3 assembly is influenced by the content of LGR4. A higher proportion of LGR4 induces and stabilizes the inactive state of ZNRF3, potentially forming a stable assembly that facilitates internalization and, consequently, promotes WNT/β-catenin signaling. Moreover, ZNRF3 undergoes conformational changes in response to different assemblies. These findings uncover the mechanism by which LGR4 and RSPO2 mediate the heterogeneity of the assembly, regulate the association with ZNRF3 for the appropriate WNT/β-catenin signaling output, and provide a structural platform for drug development targeting Wnt-driven cancers.
Results
Structure determination of LGR4-RSPO2-ZNRF3 complexes
We developed a high-affinity nanobody (NB52) targeting the ectodomain of LGR4 (Supplementary Fig. 1a), which was further expanded into a larger “megabody” (MB52) to optimize the orientation of the particle (Supplementary Fig. 1b, c)34. To stabilize the interaction between ZNRF3 and LGR4 for structural determination, we employed NanoBiT technology35,36. Specifically, fragments of LgBiT and HiBiT were genetically fused to the C-terminus of LGR4 and the RING domain of ZNRF3, respectively (Supplementary Fig. 2a). LGR4 and ZNRF3 were expressed separately, then mixed with RSPO2(Fu) (specifically the Furin domain, which includes two adjacent Furin-like cysteine-rich domains responsible for its function) during extraction, and subsequently purified to form the LGR4-RSPO2-ZNRF3 complex (Supplementary Fig. 2b). Different assemblies were observed in 1:1:1 and 2:2:2 stoichiometric ratios. The cryo-EM analysis revealed that the predominant complex observed was the LGR4-RSPO2-ZNRF3 complex with a 1:1:1 stoichiometry, in comparison to other assemblies. The 1:1:1 particle formation was used for 3D reconstruction, homogeneous refinement, global CTF refinement, and non-uniform refinement, resulting in a reconstruction map at 2.70 Å resolution. Although the RING domain’s density was not visible in the high-resolution maps, it was observed extending into the cytoplasm in low-pass-filtered maps (Supplementary Fig. 2c–e). The 1:1:1 LGR4-RSPO2-ZNRF3 complex was classified as a heterotrimer. In contrast, the 2:2:2 particles accounted for 3.1% of the good particles, and a final map resolution of approximately 6.78 Å was obtained after 3D reconstruction, which was inadequate to achieve a high-resolution model. This 2:2:2 complex is referred to as the di-heterotrimer (Supplementary Fig. 2d).
To improve the resolution of the 2:2:2 LGR4-RSPO2-ZNRF3 complexes, we optimized the expression system, and purification protocol (Supplementary Fig. 3a, b). These enhancements resulted in the predominant formation of the LGR4-RSPO2-ZNRF3 complex, which facilitated cryo-EM data collection and processing. Co-expression and purification of LGR4 and ZNRF3(containing RING domain) in the presence of RSPO2-Fu primarily yielded pentamer B, with a final map resolution of approximately 3.20 Å (Supplementary Fig. 3c–e). Notably, the RING domain was observed extending into the cytoplasm in low-pass-filtered maps (Supplementary Fig. 3f). Cryo-EM analysis also identified a few particles of the di-heterotrimer, achieving a map resolution of approximately 7.66 Å (Supplementary Fig. 3d). Even after merging the two datasets of the di-heterotrimer, the map resolution improved only slightly, reaching approximately 6.4 Å. 3D variability analysis in cluster mode revealed that the 2:2:2 LGR4-RSPO2-ZNRF3 complexes exhibit significant dynamics.
Obtaining sufficient particles of the LGR4-RSPO2-ZNRF3 (2:2:2) complex using the ZNRF3 construct with the RING domain proved challenging. To address this, GFP and anti-GFP nanobody were genetically fused to the C-terminus of LGR4 and ZNRF3ΔRING (RING domain truncated), respectively, with suitable length and flexible linkers (Supplementary Fig. 4a). The sample preparation involved co-expressing and purifying LGR4 and ZNRF3ΔRING in the presence of RSPO2. During 2D analysis, two distinct assemblies of LGR4, RSPO2, and ZNRF3 subunits were observed (Supplementary Fig. 4c). Rigorous 3D classification successfully resolved the Di-heterotrimer structure (2:2:2) with an overall resolution of 3.38 Å and another assembly map with a 1:2:2 stoichiometry, designated as pentamer A, with approximately 3.21 Å resolution (Supplementary Fig. 4b–g). These structures collectively provide evidence for the heterogeneity in the assembly of RSPO2, ZNRF3, and LGR4.
Overall architecture of LGR4-RSPO2-ZNRF3 complex with a 2:2:2 stoichiometry
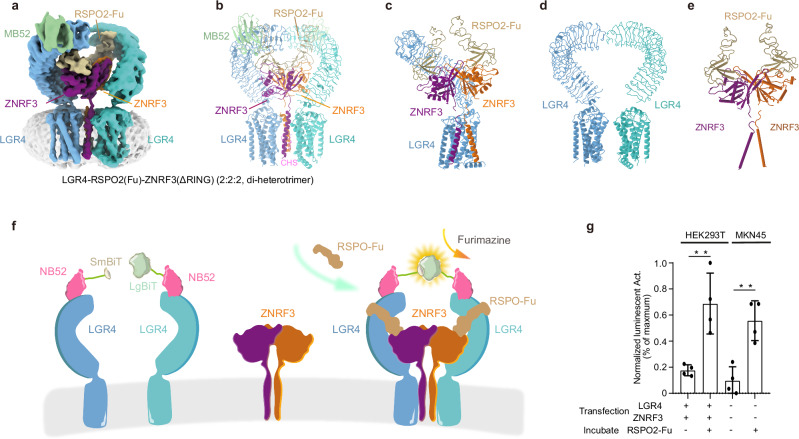
The structure of the LGR4-RSPO2-ZNRF3 complex in a 2:2:2 stoichiometry (Di-heterotrimer, specifically referring to ZNRF3 without the RING domain) exhibits a rough twofold symmetry in both the transmembrane and extracellular domain (ECD) regions (Fig. 1a–c, Supplementary Fig. 5a). A pair of horseshoe-shaped LGR4 protomers is located at the periphery of the complex, and their ectodomains face opposite directions without interacting (Fig. 1d), wrapping around the ZNRF3 ectodomain and RSPO2(Fu) (Fig. 1a, b). Meanwhile, the ZNRF3 dimer resides at the center of the architecture (Fig. 1c). The pair of RSPO2 separate from each other without contact, and nests on the top of each ZNRF3 through its N-terminus, looking like two goat horns standing on the head of a ZNRF3 dimer (Fig. 1e). While their arrangement closely aligns with a previously reported crystal structure of ZNRF3 ECD complexed with RSPO2, the C-terminus of RSPO2 rotates upward about 19.1°27 (Supplementary Fig. 5b). RSPO2 attaches to the concave surface near the tip of the LGR4 ectodomain using its C-terminus (Fig. 1c), resembling the crystal structure of LGR4 ECD-RSPO1 complex21,22. In the extracellular region of the structure, aside from the bridging by RSPO2, ZNRF3, and LGR4 do not interact (Fig. 1b, c).
Fig. 1. Cryo-EM structure of LGR4-RSPO2-ZNRF3(ΔRING) (2:2:2, di-heterotrimer) complex.
Structure of LGR4-RSPO2-ZNRF3(ΔRING) (2:2:2, di-heterotrimer) complex, Cryo-EM map (a) and atomic model (b) are shown. c The model of the di-heterotrimer complex (one LGR4 is omitted for clarity). d The arrangements of two LGR4 protomers in di-heterotrimer are shown from the front view. e The arrangements of RSPO2and ZNRF3 subunits are shown from the front view. Subunits LGR4, ZNRF3, and RSPO2 are colored in light blue/cyan, violet/orange, and brown, respectively. The nanobody portion of MB52 is shown in slate green (other segment is omitted for clarity). The same color scheme is used throughout the manuscript unless stated otherwise. f Schematic of the NanoBiT cell-based assay. The structure shows the proximity of two NB52s (pink). LgBiT (large subunit, light green) and SmBiT (small subunit, light brown) fragments are fused to the C-terminus of two NB52, respectively. The assembly of LGR4-RSPO2-ZNRF3(2:2:2) complex in the schematic (colored identically as in panel a) facilitates the luminescence complementation of NB52-LgBiT and NB52-SmBiT. g The results demonstrate a strong luminescent signal when RSPO2, NB52-LgBiT, and NB52-SmBiT, along with furimazine, are added to 293T cells (p = 0.005, n = 4) or MKN45 cancer cells (p = 0.0026, n = 4) expressing LGR4 and ZNRF3. Each value represents the mean ± SEM from four independent experiments. Source data are provided as a Source Data file.
In the transmembrane region, two LGR4 seven-transmembrane domains (7TMD) sandwich a pair of ZNRF3 TM helices (Fig. 1a–c, Supplementary Fig. 5c–e). The pair of TM helices are drawn close together on the extracellular side but diverge on the cytoplasmic side, creating an inverted V-shape configuration, clipping two distinct elongated densities that are attributed to cholesteryl hemisuccinate (CHS) (Fig. 1b, c, e). Additionally, MB52 binds to the convex outer surface near the tip of the LGR4 ECD (Fig. 1a, b).
Additionally, for the identification of the LGR4-RSPO2-ZNRF3 complex (2:2:2) on the cell surface, we developed a NanoBiT sensor utilizing the NB52 binding site. Structure shows that the proximity of two NB52 facilitates the complementarity of LgBiT and SmBiT fragments (Fig. 1f). Upon introducing NB52-LgBiT and NB52-SmBiT, along with a substrate, to HEK293T cells (or MKN45 cancer cells) expressing LGR4 and ZNRF3, a robust luminance signal is generated upon the addition of RSPO2 (Fig. 1g). This outcome signifies the presence of the LGR4-RSPO2(Fu)-ZNRF3 complex (2:2:2) on the cell surface. Our findings align with a recent report indicating that LGR4 and RNF43/ZNRF3 form a 2:2 dimer, allowing for the bivalent binding of RSPO on the cellular membrane37.
Structure of LGR4-RSPO2-ZNRF3 complexes with a 1:1:1 stoichiometry
In the LGR4-RSPO2-ZNRF3 complex (1:1:1), RSPO2 serves as a bridge between LGR4 and ZNRF3, aligning with the aforementioned di-heterotrimer configuration (Fig. 2a). Within the transmembrane region, the N-terminus of the single transmembrane helix of ZNRF3 leaned against the extracellular side of TM1 and TM7 of LGR4, with its C-terminus loosely in contact with the C-terminus of the bent TM7 helix (Supplementary Fig. 5f–h).
Fig. 2. Cryo-EM structures of LGR4-RSPO2ZNRF3(RING) (1:1:1, heterotrimer), LGR4-RSPO2-ZNRF3(RING) (1:2:2, pentamer B), LGR4-RSPO2-ZNRF3 (ΔRING) (1:2:2, pentamer A).
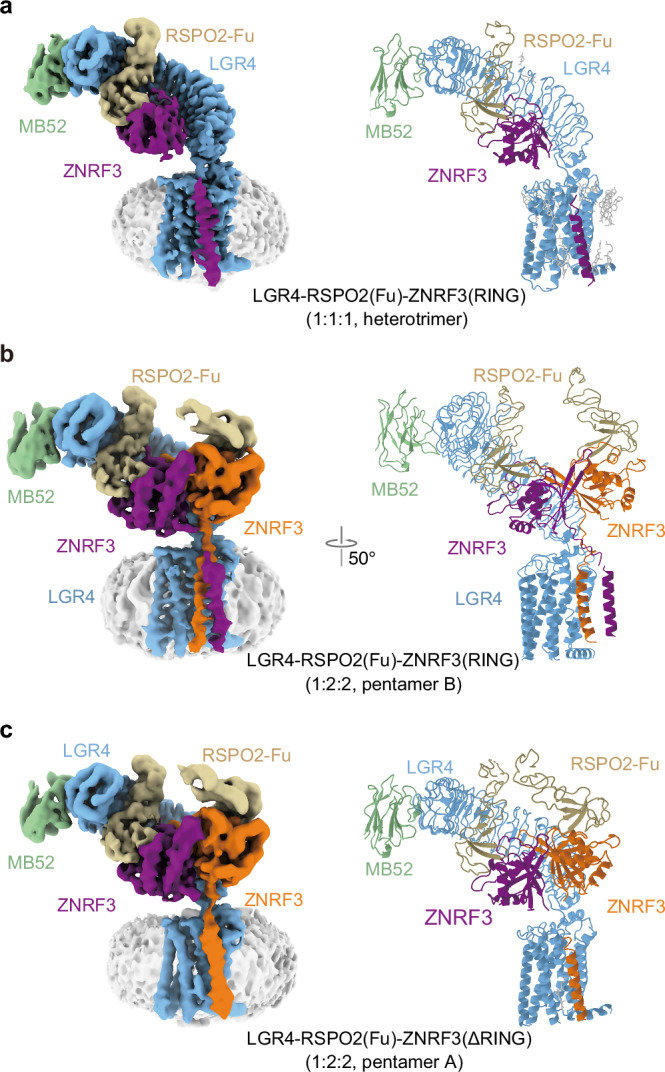
Structure of LGR4-RSPO2-ZNRF3(RING) (1:1:1, heterotrimer) (a), LGR4-RSPO2-ZNRF3(RING) (1:2:2, pentamer B) (b), and LGR4-RSPO2-ZNRF3(ΔRING) (1:2:2, pentamer A) (c), Cryo-EM map (left) and atomic model (right) are shown.
Structures of LGR4-RSPO2-ZNRF3 complexes with a 1:2:2 stoichiometry
The structure of pentamer B unveiled that RSPO2 connected LGR4 and ZNRF3 in a manner analogous to the di-heterotrimer (Fig. 2b). Both RSPO2 and ZNRF3 PAD were symmetrically assembled, aligning with the previously reported arrangement27. The linker between PAD and TM helix of ZNRF3 exhibited high flexibility (Fig. 2b, Supplementary Fig. 5i). The dimeric ZNRF3 assumed a cross-dimeric conformation, with the two transmembrane helices forming a coiled-coil configuration and their C-termini parallel to each other (Fig. 2b and Supplementary Fig. 5i). These coiled-coil helices tilted towards the transmembrane domain of LGR4, establishing interactions between the top of the single transmembrane helix of ZNRF3b and the edge of the outer surface of TM1 and TM7 in LGR4 (in the architecture assembled by the same RSPO2 bridging LGR4 and ZNRF3, the subunits labeled as “a”, otherwise as “b”) (Supplementary Fig. 5i–k).
Furthermore, as previously mentioned, another LGR4-RSPO2-ZNRF3(ΔRING) complex with a stoichiometry of 1:2:2 (pentamer A) was derived from the same sample where the structure was obtained with di-heterotrimer (Fig. 2c). In this complex, both LGR4 and the PAD of ZNRF3 remained unchanged. However, the TM helix(a) of dimeric ZNRF3 lacked discernible density, indicating its conformational flexibility (Fig. 2c). The TM helix(b) of ZNRF3 exhibited interactions with the TMD of LGR4 at its N-terminus (Supplementary Fig. 5l–n)
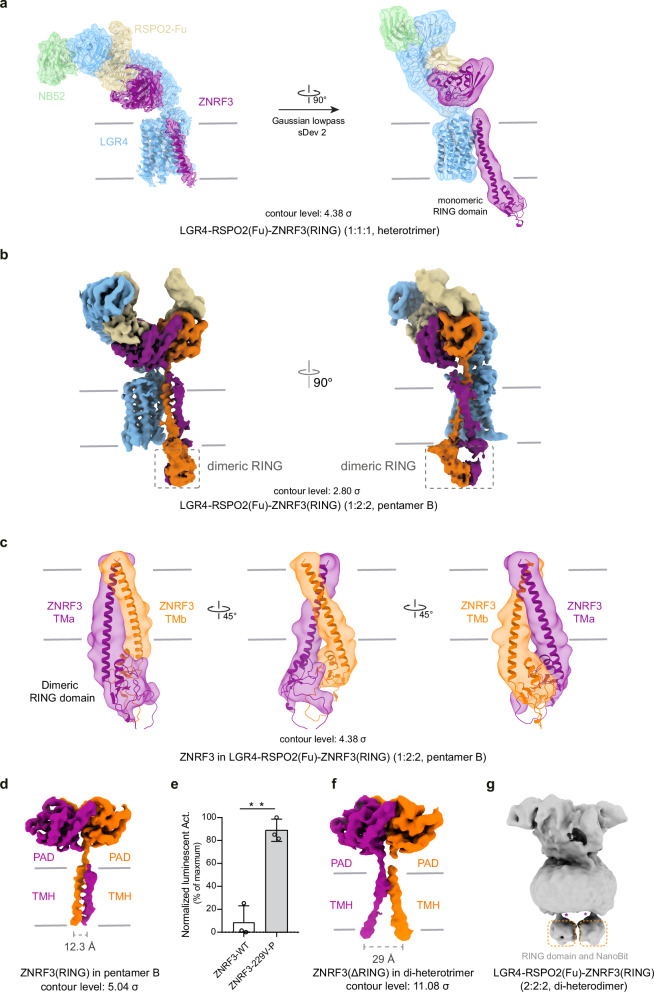
Active conformations of dimeric ZNRF3 in LGR4-RSPO2-ZNRF3 complex (1:2:2)
We used AlphaFold predictions to gain structural insights. Notably, there is a stable cytoplasmic helix connecting the TM helix to the RING domain, which supports our assumption of a relatively rigid connection (Supplementary Fig. 6a). Using these AlphaFold results, we fit the predicted structure of ZNRF3(RING) into the low-pass-filtered map of both the 1:1:1 (heterotrimer) and the 1:2:2 (pentamer B) complexes. The dimerization of the PA domain (PAD), TM helix, and RING domains in the ZNRF3 dimer is clearly observed, and despite the lower resolution of the RING domain, the fit is reasonable within the available map (Fig. 3a–c).
Fig. 3. The rearrangement RING domain in different complexes.
a Superposition of the map and model of the LGR4-RSPO2-ZNRF3(RING) (1:1:1, heterotrimer, contour level: 2.8σ). b Map of the LGR4-RSPO2-ZNRF3(RING) (1:2:2, pentamer B, contour level: 2.8σ). c Superposition of the map and model of the TM helix and RING domain in pentamer B, shown in the low-pass map (contour level: 4.38σ) from different perspectives. d Map of ZNRF3(RING) in pentamer B (contour level: 5.4σ). e Wnt3a-stimulated-TOPFlash activity regulated by WT or V229-P-S230 mutant of ZNRF3, n = 4, p value = 0.0356. Source data are provided as a Source Data file. f Map of ZNRF3 (ΔRING) in di-heterotrimer (contour level: 11.08σ). g The map of the LGR4-RSPO2-ZNRF3 complex in its di-heterodimer form (2:2:2), with ZNRF3 containing the RING domain. Each value represents the mean ± SEM from four independent experiments.
In pentamer B, the “finger-crossed” arrangement of the two TM helices of ZNRF3 is evident (Fig. 3b). The upper portions of these two TM helices cross over each other, swapping positions, while their lower segments form a coiled-coil arrangement (Fig. 3b, d). This coiled-coil configuration leads to the dimerization of the subsequent RING domains, as supported by the observed density of the two RING domains integrated on the cytoplasmic side in the low-pass-filtered map (Fig. 3c). This observation implies that the dimeric ZNRF3 might signify an active conformation, given that the dimerization of RING domains is a critical prerequisite for the enzymatic activity of E3 ligase38–46. We engineered a mutant in which a proline residue was inserted between V229 and S230 to disrupt the helix structure. This disruptive mutant (V229-P-S230), which targets the coiled-coil helices, significantly enhances Wnt3a-stimulated TOPFlash activity (Fig. 3e), indicating that the dimeric ZNRF3 assumes an active conformation, the mutant impairs ZNRF3 ubiquitination for Frizzleds, resulting in the enhancement of WNT/β-catenin signaling. While, in pentamer A, the two ZNRF3 helices apparently lacked a stable direct interaction, unlike the compact coiled-coil configuration observed in pentamer B. This discrepancy results in the non-visibility of TM helix (a) in the dimeric ZNRF3 (Fig. 2c).
Inactive conformation of dimeric ZNRF3 in the LGR4-RSPO2-ZNRF3(ΔRING) complex (2:2:2)
The structure of LGR4-RSPO2-ZNRF3(ΔRING) complex (2:2:2) illustrates that the dimeric ZNRF3 resembles an “opening scissors” shape, and its dimerization is primarily mediated by PADs (Fig. 3f). The most prominent feature of dimeric ZNRF3 in the di-heterotrimer is that two TM helices juxtapose at the N-terminal end of the two TM helices, but splay apart at the C-terminus of the TM helix on the cytoplasmic side, with a separation distance of 29 Å (Fig. 3f). In contrast, the distance between the two C-termini of the TM helices in the dimeric ZNRF3 in the LGR4-RSPO2-ZNRF3(RING) complex (1:2:2) is reduced to 12.3 Å (Fig. 3d). The map of the LGR4-RSPO2(Fu)-ZNRF3 complex (2:2:2, including the ZNRF3 RING domain) reveals that the intracellular RING domains of ZNRF3 are separated from each other in low-pass-filtered maps (Fig. 3g). This observation provides evidence that in the inverted V-shaped TM helices configuration, the RING domains are unable to dimerize. Therefore, it is inferred that the inverted V-shape configuration of the two TM helices extends to the two RING domains, resulting in their separation to prevent catalysis (Supplementary Fig. 6b, c). Hence, within the LGR4-RSPO2-ZNRF3 complex (2:2:2), the dimeric ZNRF3 is considered to be in an inactive state, in contrast to the active state characterized by the dimerization of their two RING domains in the LGR4-RSPO2-ZNRF3(RING) complex (1:2:2).
LGR4 deactivates ZNRF3
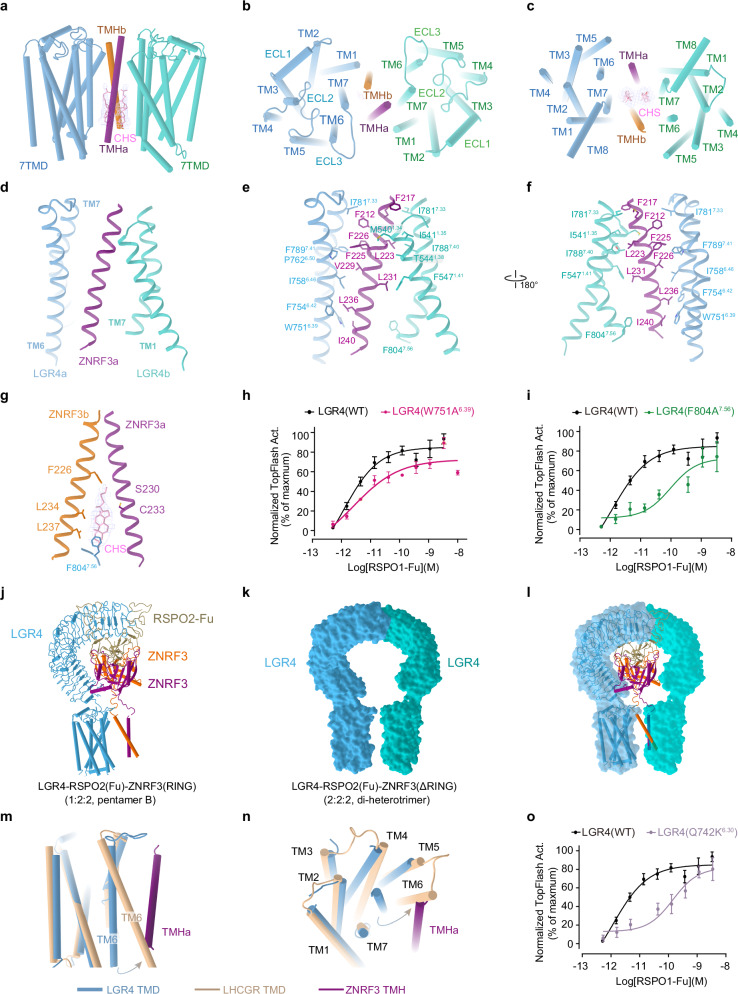
The configurations of LGR4-RSPO2-ZNRF3 complexes, each incorporating a single LGR4, indicate that LGR4 and ZNRF3 adopt loosely packed conformations during the assembly process (Fig. 2). However, the incorporation of a second LGR4 leads to a substantial compaction of the overall structure of the LGR4-RSPO2-ZNRF3 complex (2:2:2), tightly holding ZNRF3 (Fig. 1a, b). In the transmembrane region of the LGR4-RSPO2-ZNRF3 complex (2:2:2), a pair of ZNRF3 TM helices are squeezed between two LGR4 TMDs, forming two completely symmetrical interfaces composed of TM6 and TM7 of LGR4 as well as the two TM helices from ZNRF3. Each single TM helix of ZNRF3 is framed by TM6 and TM7 of one LGR4 and TM7 of the other LGR4 (Fig. 4a–f). Near the extracellular top of the transmembrane region, one ZNRF3 TM helix (a) primarily interacts with TM7 and TM1 of an LGR4 (LGR4b) (Fig. 4b). At the intracellular bottom of the transmembrane region, this ZNRF3 TM helix(a) engages with TM6 of the other LGR4 (LGR4a) (Fig. 4c). Interestingly, the F8047.56 residue on LGR4a’s TM6 interacts with a cholesteryl hemisuccinate molecule positioned between the two ZNRF3 TM helices, which in turn interacts with the other ZNRF3 TM helix(b) (Fig. 4g). This suggests that the cholesteryl hemisuccinate molecule regulates both the interaction between the two ZNRF3 TM helices and the transmembrane interaction between LGR4 and ZNRF3, thereby modulating ZNRF3’s catalytic activity. Mutations (W751A6.39 or F804A7.56) affecting the LGR4-ZNRF3 interface impair RSPO1-dependent TOPFlash activity, highlighting the importance of this interaction in the WNT/β-catenin signaling pathway (Fig. 4h, i, and Supplementary Fig. 7a, b).
Fig. 4. LGR4 induces ZNRF3 into an inactive state.
The transmembrane interface between LGR4 and ZNRF3 in the LGR4-RSPO2-ZNRF3(ΔRING) complex (2:2:2, di-heterotrimer) is shown from the front view (a), top view (b) and bottom view (c). d The interface between the transmembrane domains of LGR4 and ZNRF3. e, f The side chain interactions between LGR4 and ZNRF3 within the transmembrane region of the LGR4-RSPO2(Fu)-ZNRF3(ΔRING) complex (2:2:2) are shown in detail. g The specific interactions between the transmembrane domain of LGR4, ZNRF3 and cholesteryl hemisuccinate within the LGR4-RSPO2(Fu)-ZNRF3(ΔRING) complex (2:2:2) are highlighted. h, i Dose-dependent TOPFlash activity induced by WT (black) or W751A6.39 mutant (red, e) and F804A7.56 mutant (green, f) of LGR4 after stimulation with RSPO1, n = 3. Source data are provided as a Source Data file. j–l Comparing the LGR4 in pentamer B and di-heterotrimer. j The model of LGR4-RSPO2ZNRF3(RING) (1:2:2, pentamer B). k The surface of two LGR4 in LGR4-RSPO2ZNRF3(ΔRING) (2:2:2, di-heterotrimer). l Superposition of the LGR4 in pentamer B and di-heterotrimer (ZNRF3 and RSPO2 are deleted for clarity). m, n Conformational comparison of the TMD of LGR4 (light blue) in the di-heterotrimer with that of active LHCGR (wheat, PDB:7FII, RMSD = 1.524, 182 to 182 atoms) from the front view (m), and bottom view (n). The potential steric clash between TM6 of the active LGR4 and the single TM helix (violet) of ZNRF3 in the di-heterotrimer complex is shown. o TOPFlash plot illustrating the effect of breaking ionic lock (Q742K6.30 mutant) (purple) in the transmembrane domain of LGR4 on the activity of RSPO1, compared to that of WT (black), n = 4. Source data are provided as a Source Data file. Each value represents the mean ± SEM from independent experiments. The LGR4(WT) datasets in all three panels are identical.
Even without RSPOs, LGR4 and ZNRF3 may directly interact through their TMDs, although this interaction might be too weak to form a stable complex. Nevertheless, LGR4 can still internalize some ZNRF3 from the cell surface, reducing its abundance on the membrane. RSPOs bridge the extracellular domains of LGR4 and ZNRF3, strengthening their TMD-TMD interactions. These interactions, in turn, facilitate the association of their extracellular domains. Both the bridging by RSPOs and the TMD-TMD interactions contribute to the assembly of the LGR4-RSPOs-ZNRF3 complex. Therefore, LGR4 F804A7.56 and W751A6.39 display an impaired response to RSPO1 at lower ligand concentrations, which diminishes the potency of RSPOs. However, increased RSPOs molecules can effectively bridge the extracellular domains of LGR4 and ZNRF3, compensating for the impaired TMD-TMD interactions. This results in a maximal response of RSPOs for the endocytosis of ZNRF3 with LGR4 mutants similar to that of wild-type LGR4.
Overlaying LGR4 in the di-heterotrimer and pentamer B reveals that the coiled-coil helices of ZNRF3 hinder the approach of a second LGR4 (Fig. 4j–l). This suggests that the transmembrane interplay between LGR4 and ZNRF3 within the di-heterotrimer could facilitate the transition of ZNRF3’s TM helix from a coiled-coil configuration to an inverted V-shape, leading to the separation of ZNRF3’s RING domain from its dimerization partner. Consequently, the transmembrane engagements between ZNRF3 and LGR4 induce and stabilize the inactive conformation of ZNRF3, thereby suppressing its enzymatic activity responsible for Frizzled ubiquitination. Additionally, within the di-heterotrimer, where two LGR4 transmembrane domains compress ZNRF3 helices, this interaction fosters a stronger association between LGR4 and ZNRF3 (Fig. 4a–f), thereby potentially promoting the removal of ZNRF3 from the membrane due to their joint endocytosis.
Furthermore, LGR4 TMDs align very well in these complexes (Supplementary Fig. 8a), and bear resemblance to the classical inactive state observed in class A GPCRs, such as rhodopsin and the β2 adrenergic receptor47,48 (Supplementary Fig. 8b–d). This implies that RSPO2 binding does not influence the conformation of LGR4’s 7TMD, and RSPO2 does not act as an agonist of LGR4. When comparing the 7TMD of LGR4 in the complex with those of the active LHCGR structuree49–51 (Fig. 4m, n), we noted that the outward movement and rotation of TM6 on the cytosolic side—a common feature for G protein coupling activation across the GPCR superfamily—were restricted. Our analysis revealed that the TM6 of LGR4 in the LGR4-RSPO2-ZNRF3 complex (2:2:2) is unable to move outward due to steric clashes caused by the single transmembrane domain of ZNRF3.
The “ionic lock” acts as a molecular switch within rhodopsin-family GPCRs, essential for maintaining these receptors in their resting state (Supplementary Fig. 8e)52–55. Interestingly, we observed that mutating the ionic lock (Q742K6.30) in the transmembrane domain of the classical inactive LGR4 resulted in decreased TOPFlash activity (Fig. 4o, Supplementary Fig. 7c). This mutation may increase the propensity for TM6 to move outward, thereby releasing the restraint imposed by ZNRF3. These findings suggest that LGR4 in the complex retains its classical inactive conformation and cannot adopt an active conformation. Unlike other class A GPCRs, LGR4 directly regulates assembly in a context-dependent manner, rather than through conformational changes induced by signal molecules.
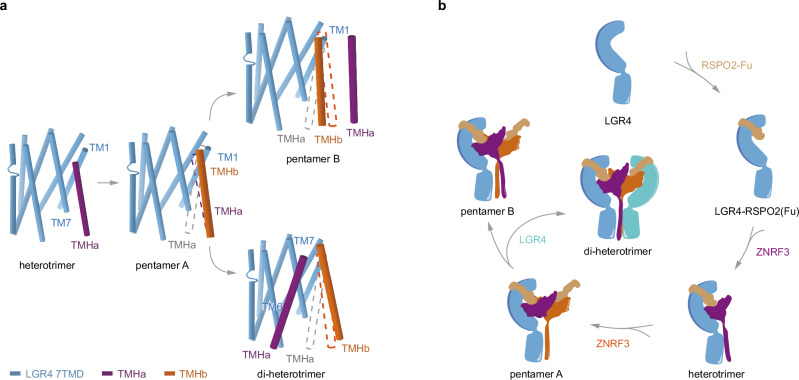
Pathway for the assembly of different complexes
Upon analyzing the structures of LGR4 in all complexes, it becomes apparent that LGR4 consistently adopts a classical inactive conformation. In the context of conventional class A GPCRs, activation typically involves the outward swing of TM6, which facilitates G protein coupling or binding to other transducers. Our analysis indicates that LGR4 does not exhibit this outward movement of TM6, even in the presence of RSPO2 and ZNRF3 (Supplementary Fig. 9). The orientation of ZNRF3 PAD is stabilized by RSPO2 attaching to LGR4 ECD, while the proline-rich peptide linker between ECD and TM helix of ZNRF3 exhibits flexibility, allowing for conformational transitions of TM helix during the assembly of the LGR4-RSPO2-ZNRF3 complex (Fig. 5).
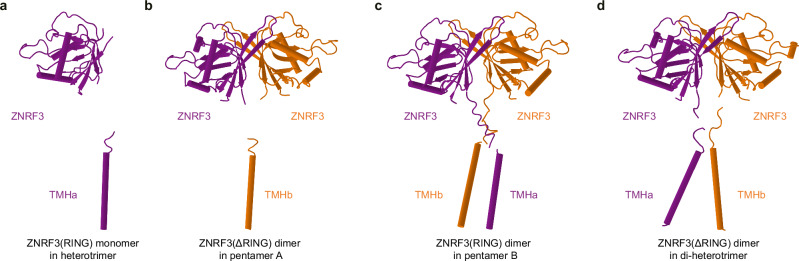
Fig. 5. Different conformational states of ZNRF3 in various assemblies of the LGR4-RSPO2(Fu)-ZNRF3 complexes.
a ZNRF3 in the heterotrimer: the linker between the extracellular domain of ZNRF3 and TM helix is absent. b ZNRF3 in pentamer A: one of the TM helices in the ZNRF3 dimer is completely missing, and the linker between the extracellular domain and the TM helix is also absent. c ZNRF3 in pentamer B: the two TM helices exhibit a “finger-crossed” arrangement. d ZNRF3 in the di-heterotrimer: the two TM helices adopt an inverted V-shaped configuration. ZNRF3 is depicted in purple or orange.
RSPO2 initiates the assembly of the LGR4-RSPO2-ZNRF3 (1:1:1) complex by bridging LGR4 and ZNRF3 (Figs. 2a and 6). The binding of RSPO2 to ZNRF3 promotes the dimerization propensity of ZNRF327, enabling the formation of the LGR4-RSPO2-ZNRF3 (1:2:2) complex (pentamer A) (Figs. 2c and 6). Pentamer A can differentiate into two distinct states based on the presence or absence of another LGR4 induction (Fig. 6). In the absence of another LGR4, the two TM helices form a coiled-coil configuration, and the two RING domains dimerize to adopt the conformation as observed in pentamer B (Figs. 2b and 6). However, in the presence of another LGR4, pentamer A transitions to the final assembly state (Figs. 1a, b, and 6). The LGR4-RSPO2-ZNRF3 complex (2:2:2) exhibits a stronger interaction network in the membrane region due to the participation of a second LGR4 (Fig. 4a–f), resulting in a more compact conformation.
Fig. 6. Proposed model for the LGR4-RSPO2-ZNRF3 complex assembly pathway.
a Front view of the transmembrane region of the LGR4-RSPO2-ZNRF3 in the different states aligned by the LGR4 TMD. The dashed line (violet/orange) indicates the position of ZNRF3 TM helices in the previous state. TM helix(a) is not visible in pentamer A, and the dashed line is colored in gray. For clarity, one LGR4 is omitted from the di-heterotrimer. b Cartoon illustrates the assembly pathway from the LGR4-apo state, and RSPO2-bound state, through LGR4-RSPO2-ZNRF3 (1:1:1, heterotrimer) state to LGR4-RSPO2-ZNRF3 (1:2:2, pentamer A) state, and then differentiates into the LGR4-RSPO2-ZNRF3 (1:2:2, pentamer B) state in the absence of LGR4, or into the LGR4-RSPO2-ZNRF3 (2:2:2, di-heterotrimer) state in the presence of LGR4.
In summary, the assembly pathway of the LGR4-RSPO2-ZNRF3 complex involves transitioning from heterotrimer to pentamer A and then differentiating into either pentamer B conformation in the absence of sufficient LGR4 or a completely assembly state (di-heterotrimer) when LGR4 is present in abundant amounts (Fig. 6b, Supplementary Movie 1).
Discussion
The Wnt/β-catenin signal transduction cascade is a critical regulator controlling developmental gene expression programs and maintaining the balance of stem cell proliferation, differentiation, and adult tissue homeostasis1,2. Within this complex pathway, a critical regulatory module is formed by LGR4-6, RSPO1-4, and ZNRF3/RNF433–12. The diverse composition and arrangement contribute to the complexity of the regulatory system. Understanding the assembly mechanism and structural basis of this module’s regulation of WNT/β-catenin signaling is crucial for advancing regenerative therapeutics and cancer treatments.
Multiple crystal structures of their ectodomains have been documented, delineating the interactions between RSPOs and the LGR4/5 ECD, as well as RSPOs and ZNRF3/RNF43 ECD. These structures have shed light on their stoichiometries and configurations in both solution and crystalline states. However, the LGR4/5 ECD-RSPO1/2-RNF43/ZNRF3 complex manifests a 1:1 or 1:1:1 stoichiometry in solution, while the crystal-packing interactions yield diverse other arrangements with a 2:2 or 2:2:2 stoichiometry21,22,24–29. Nonetheless, these models were established devoid of the cell membrane and the 7TM region. The varied stoichiometry and arrangement of the LGR4/5 ECD-RSPO1/2-RNF43/ZNRF3 ECD complexes pose challenges in unifying theoretical understanding of their roles in RSPOs signal transmission.
In our investigation, we determined the structures of multiple assemblies of full-length LGR4-RSPO2-ZNRF3 with varied stoichiometries and arrangements, encompassing structures with 1:1:1, 1:2:2, and 2:2:2 stoichiometric ratios (Figs. 1 and 2). These structural insights highlight that ZNRF3 adopts distinct conformations within the different assemblies (Figs. 3 and 5). A higher proportion of LGR4 induces and stabilizes the inactive conformation of ZNRF3 to promote WNT/β-catenin signaling (Fig. 1). Moreover, LGR4 strengthens its association with ZNRF3 in the assembly with a proposed 2:2:2 stoichiometric ratio (Fig. 4a–f), which we hypothesize may facilitate their joint endocytosis and promote WNT/β-catenin signaling. While co-internalization is highlighted as an important mechanism, further experiments are required to assess the precise role of stoichiometry in the endocytosis efficiency of individual components.
The structures of various assemblies unveil the mechanism through which LGR4 inhibits the E3 activity of ZNRF3. Firstly, the involvement of a second LGR4 results in a more compact conformation, enhancing its association with ZNRF3 and intensifying the impact on endocytosis. Secondly, the final assembly reveals that LGR4 envelops ZNRF3 (Fig. 1a, b), indicating spatial inhibition and preventing ZNRF3 from recruiting other proteins for ubiquitination. Furthermore, within the LGR4-RSPO2-ZNRF3 (1:2:2) complex, ZNRF3 assumes a coiled-coil configuration, prompting the dimerization of two consecutive RING domains (Fig. 3b, c). This finding suggests that the dimeric ZNRF3 may denote an active conformation, as the dimerization of RING domains is a pivotal prerequisite for the enzymatic activity of E3 ligase38-46. Conversely, in the LGR4-RSPO2(Fu)-ZNRF3 (2:2:2) complex, the inverted V-shape configuration of the two transmembrane helices extends to the two RING domains, potentially causing their separation preventing catalysis (Fig. 3f, g). This configuration implies that the dimeric ZNRF3 may be in an inactive conformation. Additionally, within their transmembrane region, LGR4 and ZNRF3 exhibit strong interactions that serve to stabilize the V-shape conformation (Fig. 4a–g). We postulate that the presence of additional LGR4 induces a conformational rearrangement of ZNRF3, stabilizing its inactive dimeric state.
Our sequence alignments of LGR4 and LGR5, as well as ZNRF3 and RNF43, reveal a significant conservation of key residues at the TMD-TMD interface (Supplementary Fig. 10a). In LGR4, residues W7516.39 and F8047.56 are essential for interactions with ZNRF3, while the corresponding residues in LGR5, L7706.39 and F8237.56, suggest a shared interaction mechanism across both receptors. The transmembrane coiled-coil helices of ZNRF3 are crucial for regulating Frizzled ubiquitination, and the AlphaFold-predicted structure of RNF43 displays a similarly conserved helix, supporting functional parallels (Supplementary Fig. 10b). However, RNF43 may operate through a slightly different mechanism, as the flexible loop between its transmembrane helix and RING domain forms a cytoplasmic helix, potentially impacting catalytic activity (Supplementary Fig. 10c).
Interestingly, our studies suggest that LGR4 remains in a conventional inactive state during assembly, and RSPO2 cannot activate LGR4, contrary to previous speculations (Supplementary Fig. 8a–d)4,5,9,10. This distinct mechanism of LGR4-mediated signal transduction via ZNRF3 is different from the classic G protein signaling and offers a unique perspective on GPCR system transduction.
Moreover, we investigated cell-surface single transmembrane E3 ubiquitin ligases, specifically ZNRF3, which play a significant role in regulating signaling receptor abundance and cellular responses to extracellular ligands in WNT/β-catenin signaling. ZNRF3 adopts different conformations during the regulation of LGR4 (Fig. 5), leading to dynamic control of Frizzleds homeostasis. Understanding these regulatory mechanisms could pave the way for future studies on substrate recognition and ubiquitination in this class of E3 ligases. Our findings have implications beyond basic research, as they provide valuable insights for the development of PROTABs, based on anti-ZNRF3 antibodies31–33. By optimizing PROTABs according to the structure and regulatory mechanism of ZNRF3, the field of targeted protein degradation is now better equipped to address challenging protein targets that were previously considered “undruggable”.
In conclusion, our study uncovers the assembly mechanism of LGR4, RSPO2, and ZNRF3 and provides deep mechanistic insights into the RSPO-mediated regulation of Wnt/β-catenin signaling. These structural insights lay a foundation for future regenerative therapeutics and cancer treatments.
Methods
Animal experiment statement
The camel immunization and blood collection were conducted by Shanghai Kailuo Biotechnology Co., Ltd, Shanghai, 201600, China, in compliance with the requirements of the Shanghai Animal Ethics Association.
Construct design
Human LGR4 (residues 25–833) and human ZNRF3 (residues 56–334, including the RING domain) were fused with C-terminal LgBiT and HiBiT, respectively. Alternatively, human LGR4 (residues 25–833) and human ZNRF3 (56–241, with the RING domain truncated) were fused with C-terminal GFP and anti-GFP nanobody, respectively. A 15-residue linker was inserted between the components. These four fragments were then cloned into a modified pEG BacMam vector, incorporating an N-terminal influenza hemagglutinin (HA) signal peptide and a C-terminal Flag tag. Additionally, the Furin domain of RSPO1 or RSPO2 was cloned into the pEG BacMam vector, featuring an N-terminal Gaussia signal peptide and a C-terminal HRV-3C protease site followed by a Fc domain of human IgG. Furthermore, the full-length sequences of human LGR4 and ZNRF3, along with their respective mutants, were cloned into the pcDNA3.1(+) vector for subsequent use in cell-based assays (see Supplementary Table 2).
MB52 generation, expression, and purification
In brief, two camels were immunized subcutaneously with approximately 1 mg human LGR4 protein combined with equal volume of Gerbu FAMA adjuvant once a week for seven consecutive weeks. Three days after the final boost, peripheral blood lymphocytes (PBLs) were isolated from the whole blood using Ficoll-Paque Plus according to the manufacturer’s instructions. Total RNA from the PBLs was extracted and reverse transcribed into cDNA using a Super-Script III FIRST-Strand SUPERMIX Kit (Invitrogen). The VHH encoding sequences were amplified with two-step enriched-nested PCR using VHH-specific primers and cloned between PstI and BsteII sites of the pMECS vector. Electro-competent E. coli TG1 cells (Lucigen) were transformed.
E. coli strain TG1 cells containing the VHH library were superinfected with M13KO7 helper phages to obtain a library of VHH-presenting phages. Phages presenting LGR4-specific VHHs were enriched after two rounds of bio-panning. Periplasmic extracts were made and analyzed using ELISA screens. NB52 was cloned into a pMECS vector (NTCC) that contains a PelB signal peptide and a hemagglutinin (HA) tag followed by a His6 tag at the C-terminus. It was expressed in the periplasm of E. coli strain TOP10F’ cells.
The VHH gene of NB52 was expanded by fusion to the circular permutated extracellular adhesin domain of Helicobacter pylori (HopQ, 45 kDa) to generate the megabody referred to as MB52. MB52 was expressed as a periplasmic protein in E. coli strain TOP10F’ cells and purified56,57.
The MB52 was expressed in TOP10F’ cells. After induction, the culture supernatant was collected by centrifugation at 18,000 × g for 30 min to remove cells and debris. The clarified supernatant was loaded onto a HisTrap HP column (Cytiva) pre-equilibrated with buffer A (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 10 mM imidazole).
To eliminate nonspecific proteins, the column was washed with 10 column volumes of buffer A supplemented with 20 mM imidazole. The nanobody was then eluted using buffer A containing 250 mM imidazole.
The eluted fractions were concentrated and subjected to size-exclusion chromatography using a Superdex 75 Increase 10/300 GL column (Cytiva) equilibrated in gel filtration buffer (20 mM HEPES, pH 7.5, 150 mM NaCl). Purity and monodispersity of the nanobody were confirmed by SDS-PAGE and analytical size-exclusion chromatography.
Expression and purification of RSPO1 and RSPO2 Fu domain
The plasmid of the RSPO2-Fu domain protein was transiently transfected into the HEK293F cells using Polyethyleneimine (PEI) at a cell density of 2 × 106 cells/ml. 24 h post-transfection, sodium butyrate was added to a final concentration of 10 mM. Cells were incubated at 30 °C for 72 h, and then cell supernatant was harvested by centrifugation at 5000 × g for 30 min. The RSPO2 or RSPO1 Fu domain protein was isolated from cell supernatant by protein A affinity chromatography. The elution was digested with HRV-3C protease and subjected to HiLoad 16/600 Superdex 75 (20 mM HEPES, 150 mM NaCl) to remove the fused Fc domain.
Expression and purification of complexes
Human LGR4 and ZNRF3 were expressed together or separately in HEK293 GnTI- cells that cultured in the FreeStyleTM 293 medium (Gibco). The plasmid of LGR4 and/or ZNRF3 were transiently transfected into the HEK293 GnTI- cells using Polyethyleneimine (PEI) at a cell density of 2 × 106 cells/ml. 24 h post-transfection, the cells were supplemented with 10 mM sodium butyrate to improve the protein expression level and incubated at 30 °C for 72 h before harvest. The cells were lysed using the homogenizer in the buffer containing 20 mM HEPES, pH 7.4, 10 mM NaCl, 5% glycerol supplement with of protease inhibitor cocktail, EDTA-free. The complex was formed by the addition of 1 μM RSPO2 and 1 μM MB52. After centrifugation, the membrane was solubilized in the same buffer with the addition of 1% Lauryl maltose neopentyl glycol (LMNG) and 0.1% cholesteryl hemisuccinate (CHS) at 4 °C for 2 h. The supernatant was collected by centrifugation at 15,000 × g for 1 h and applied to an anti-Flag M2 antibody affinity chromatography column, and then washed with the same buffer with the detergent concentration gradually decreased to 0.02% LMNG and 0.002% CHS, and finally eluted with 20 mM HEPES, pH 7.4, 10 mM NaCl, 5% glycerol, 0.02% LMNG, 0.002% CHS and 0.2 mg/ml Flag-peptide. The complex protein was concentrated for final purification of size exclusion in 20 mM HEPES, pH 7.4, 50 mM NaCl, 0.002% LMNG, and 0.0002% CHS using Superose™ 6 Increase 10/300 GL.
Genetic fusions of LgBiT and HiBiT were attached to the C-terminus of LGR4 and the RING domain of ZNRF3 (inclusive of the RING domain), respectively. LGR4, and ZNRF3 were expressed separately, and the expressing cells were mixed and purified with RSPO2. The LGR4-RSPO2-ZNFR3 complex with a stoichiometric ratio of 1:1:1 was obtained. Co-expression and purification of LGR4 and ZNRF3 in the presence of RSPO2 led to a predominant assembly with a 1:2:2 stoichiometry.
Furthermore, GFP and anti-GFP nanobody were fused to the C-terminus of LGR4 and the single TM helix of ZNRF3 (the RING domain of ZNRF3 was truncated), respectively. Subsequently, LGR4 and ZNRF3 were co-expressed and purified in the presence of RSPO2, resulting in a predominant assembly with a 2:2:2 stoichiometry.
Cryo-EM data collection
For the preparation of cryo-EM grids, 3μl of purified complexes at a concentration of 2-3 mg/ml was applied onto the freshly glow-discharged 300 mesh R1.2/R1.3 UltrAufoil holey gold grids (Quantifoil) or ANTcryoTM Au300-1.2/1.3 (Nanodim) under 100% humidity at 4°C. The grid was blotted with a wait time of 5 s, and a blot time for 3 s, and plunged-frozen into liquid ethane using the Vitrobot Mark IV (Thermo Fisher Scientific, FEI), and cooled by liquid nitrogen.
Final datasets were collected on a Titan Krios, with a K3 detector. All micrographs were acquired at a calibrated pixel size of 0.5355 Å with a dose rate of 23.3 electrons per pixel per second, and defocus values range from −1.5 μm to −2.5 μm. The micrograph has 36 frames each and was collected over a 3 s exposure and resulting in a total dose of 70 electrons per Å2. At these settings, a total of 6801 movies for the heterotrimer complex, 7042 movies for the pentamer B complex, and 5208 movies for the di-heterotrimer and the pentamer A complex were collected.
Cryo-EM image processing
All datasets were imported within the cryoSPARC58. The image stack was aligned using the patch motion correction module. The final 2x binning was done in relation to the data collection with a pixel size of 0.5355. Contrast transfer function (CTF) parameters for each no does-weight micrograph were determined by patch CTF estimation. According to CTF estimation result and relative ice thickness, the high-quality micrographs were selected to ensure that estimated CTF fit resolution is higher than 4 Å for further processing. Manual-picker was used to pick the particles and subjected to several rounds of 2D classification to generate a particular template for the next round auto-pick. Auto-picked particles were extracted for two rounds of 2D classification, then good particles were selected for 3D Ab-initio reconstruction and several rounds of 3D heterogeneous refinement to remove the poor density classes. The good particles were further subjected to 3D Ab-initio reconstruction, homogenous refinement, and non-uniform refinement to produce model and high-quality particles. The particles were subsequently subjected to global CTF refinement and local CTF refinement. Reference-based motion correction was then applied, and the particles were stacked using UCSF PyEM59 and RELION60 before being imported into CryoSieve61 for additional processing. The final screened particles were re-imported into CryoSPARC for ab initio reconstruction, homogeneous refinement, and non-uniform refinement to generate the final map.
For the LGR4-RSPO2-ZNRF3(RING) heterotrimer complex, 1,896,148 particles were extracted and subjected to reference-free 2D classification, resulting in good particles with two distinct structural features. These particles were processed through heterogeneous refinement to produce an ab initio model with two classes. Particles with a stoichiometric ratio of 1:1:1 were used for 3D reconstruction and heterogeneous refinement. After multiple rounds of heterogeneous refinement, 789,902 particles underwent homogeneous refinement, global CTF refinement, and non-uniform refinement, yielding a reconstruction map with a resolution of 2.76 Å. Reference-based motion correction was applied, and the particles were stacked using UCSF PyEM and RELION before being processed in CryoSieve. The final 106,020 screened particles were re-imported into CryoSPARC for ab initio reconstruction, homogeneous refinement, and non-uniform refinement. These results were then imported into RELION for Post-processing, culminating in a final reconstruction map at 2.70 Å resolution. Another class of ab initio models with a stoichiometric ratio of 2:2:2, constituting 3.10% of the good particles, resulted in a final map resolution of approximately 6.78 Å. Due to the low resolution, an atomic model could not be built for the 2:2:2 complex (ZNRF3 containing the RING domain), referred to as the Di-heterotrimer.
For the LGR4-RSPO2-ZNRF3(RING) pentamer B complex, 1,547,111 particles were picked and subjected to reference-free 2D classification, yielding good particles with two distinct structural features. These particles were processed through heterogeneous refinement to produce an ab initio model with two classes. The 186,955 particles with a stoichiometric ratio of 1:2:2 were used for 3D reconstruction and heterogeneous refinement. Through multiple rounds of heterogeneous refinement, 263,077 high-quality particles underwent homogeneous refinement and non-uniform refinement, producing a reconstruction map at 3.29 Å resolution. These particles were stacked using UCSF PyEM and RELION and then processed in CryoSieve. The final set of 61,066 screened particles was re-imported into CryoSPARC for ab initio reconstruction, homogeneous refinement, and non-uniform refinement, resulting in a reconstruction map at 3.20 Å resolution. In this sample, a 2:2:2 complex assembly (ZNRF3 containing the RING domain) was also observed, consisting of 39,995 particles. After homogeneous refinement and non-uniform refinement with C2 symmetry, the map resolution reached 7.66 Å.
For the LGR4-RSPO2ZNRF3(ΔRING) pentamer A complex and Di-heterotrimer complex (ZNRF3 truncated RING domain), a total of 689,122 particles were picked, and then subjected to reference-free 2D classification, revealing two different classes. For the LGR4-RSPO2-ZNRF3(ΔRING) di-heterotrimer complex, 180,960 particles underwent homogeneous refinement with C2 symmetry, global CTF refinement, and non-uniform refinement with C2 symmetry, resulting in a reconstruction map with a resolution of 3.44 Å. Reference-based motion correction was applied, and the particles were stacked using UCSF PyEM and RELION. Finally, 47,438 particles were imported into CryoSieve for further processing, re-imported into CryoSPARC, and subjected to ab initio reconstruction, homogeneous refinement with C2 symmetry, local CTF refinement, and non-uniform refinement with C2 symmetry, yielding a reconstruction map with a resolution of 3.38 Å. For the low-resolution transmembrane domain, a mask was generated, and local refinements were performed, significantly improving the density map. The map was then optimized using EMReady62 to further enhance the quality and interpretability of the cryo-EM images.
For the LGR4-RSPO2-ZNRF3(ΔRING) pentamer A complex, 142,915 particles were used for 3D reconstruction, homogeneous refinement, global CTF refinement, and non-uniform refinement, resulting in a reconstruction map at 3.36 Å resolution. After reference-based motion correction and particle stacking using UCSF PyEM and RELION, the final 73,170 particles were processed in CryoSieve, re-imported into CryoSPARC, and subjected to ab initio reconstruction, homogeneous refinement, and non-uniform refinement, resulting in a 3.32 Å resolution reconstruction map. The final local CTF refinement produced a reconstruction map with a resolution of 3.21 Å.
Model building and refinement
For the LGR4-RSPO2-ZNRF3 complexes cryo-EM reconstruction, the structures of LGR4 and ZNRF3 single TM were predicted by the AlphaFold2 and the initial template of MB52 was generated using SWISS-MODEL. These structures, RSPO2 domain (PDB code: 4UFR), and ZNRF3 PAD (PDB code: 4UFS) were fit into the composite cryo-EM map of the LGR4-RSPO2-ZNRF3 complexes as starting model in ChimeraX63. These models were then iteratively refined with manual adjustments in Coot64, followed by real-space refinements in Phenix65. All models were validated using MolProbity66. Structural figures were prepared in PyMOL (PyMOL | pymol.org).
Luciferase reporter assay
In our TOPFlash experiments, we utilize the Wnt3a-conditioned medium (Wnt3a-CM) to ensure consistent activation of the Wnt/β-catenin signaling pathway. The preparation of Wnt3a-CM involves culturing L cells stably transfected with Wnt3a (L-Wnt3a cells), which express and secrete the Wnt3a protein. For the Wnt/β-catenin signaling TOPFlash reporter assay, HEK293T cells are transiently transfected with plasmids encoding either wild-type or mutant forms of LGR4 or ZNRF3, along with the super 8×TOPFlash firefly luciferase and pRL-SV40-renilla luciferase reporter (Beyotime) at a mass ratio of 1:1:0.1. Following a 12-hour transfection period, the cells are serum-starved overnight to synchronize their signaling response. To assess the RSPO1 effect, the Fu domain of recombinant RSPO1 is serially diluted (3-fold) in either DMEM or Wnt3a-CM. These mixtures are then added to a half-well white 96-well plate containing 5000 cells/well and incubated at 37 °C for 10 h. The TOPFlash signal is subsequently measured using the Dual-Glo luciferase assay kit, following the manufacturer’s protocol. All experiments were performed for three times. Data was analyzed by non-linear regression in Prism (GraphPad Software).
Flow cytometric analysis
HEK293T cells were transiently transfected with the indicated plasmids. 12 h after transfection, cells were serum-starved overnight and then treated with the Fu domain of recombinant RSPO1 or RSPO2 at 37 °C for 2 h. Cells were washed three times with ice-cold PBS and collected using a trypsin-free dissociation buffer. After blocking for 30 min, primary staining of cells was performed with anti-Flag-Tag (DYKDDDDK), Mouse mAb (YEASEN, Cat: 30505ES20) at a dilution of 1:5000 for 1 h at 4 °C. Cells were then washed three times with ice-cold 0.2% BSA in PBS and incubated with YSFluor™ 488 Goat Anti-Mouse IgG (H + L) (YEASEN, Cat: 33206ES60) at a dilution of 1:400. All reagents were used according to the manufacturer’s instructions. After extensive washes using PBS, cells were subjected to analysis using a BD Fortessa™ flow cytometer and data were analyzed with FlowJo Software (FlowJo, LLC). FACS sequential gating listed in Supplementary Table 3 and Supplementary Data 1.
Supplementary information
Source data
Acknowledgements
The cryo-EM data were collected at the Cryo-Electron Microscopy Research Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences. Camel immunization, nanobody (Nb) library generation, screening, and identification of nanobody were performed at Shanghai Kailuo Biotechnology Co., Ltd. This work is supported by National Key Research and Development Program of China (NKPs, No. SQ2023YFB3200066, H.L.), Science and Technology Commission of Shanghai Municipality (No. 18JC1415400, Y.G.), and National Natural Science Foundation of China (31670743 Y.G., U24A20675 J.Q.W., 62273329 Z.N.W.), and partially funded by Shanghai Kailuo Biotechnology Co., Ltd.
Author contributions
Y.G. conceived and designed the study. L.W. purified the LGR4-RSPO2-ZNRF3 complex, the LGR4-RSPO2 and LGR4. L.W. performed cryo-EM data collection, map calculations and model building, structural analysis, and figure preparation, wrote methods, and participated in manuscript preparation. Y.Y.L. initiated the project, designed the expression constructs of LGR4, and discovered nanobody. Q.Q.C. performed the TOPFlash functional studies and NanoBit experiment. H.R.Q and F.Z.H designed NanoBit and purified the fragment NB52-LgBiT and NB52-SmBiT and participated in figure preparation. T.J.G., and Z.C.S. cultured and purified the MB52. T.J.G., Z.C.S. and S.J.G. purified RSPO2 and RSPO1. L.Y.L. designed the construct of MB52 and participated in manuscript preparation. T.J.G. and S.Y.Z. participated in constructs of mutants of LGR4 and ZNRF3. Z.Y.L performed Flow cytometric analysis. J.Q.W., Y.Y.D., Z.N.W., Q.Y., and Y.H., give important suggestions about the manuscripts. Y.G. supervised the project, analyzed the structures, and wrote the manuscript.
Peer review
Peer review information
Nature Communications thanks Gunnar Schulte, who co-reviewed with Julien Bous, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support this study. Source data are provided as a Source Data file. The PDB data generated in this study have been deposited in the RCSB PDB database under accession code 8XFT, 8XFS, 8XFP and 8Y69, Cryo-EM maps generated in this study have been deposited with EMDB under accession codes EMD-38309, EMD-38308, EMD-38307 and EMD-38982 (https:// www.ebi.ac.uk/emdb/EMD-38982). Accession numbers are also listed in Supplementary Table 1. The TopFlash, NanoBit assay and FACS data generated in this study are provided in the Source Data file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lu Wang, Fangzheng Hu, Qianqian Cui, Huarui Qiao, Lingyun Li, Tengjie Geng, Yuying Li.
Contributor Information
Zenan Wang, Email: zn.wang1@siat.ac.cn.
Yuanyuan Dai, Email: daiyuanyuan@cicams.ac.cn.
Yong Geng, Email: gengyong@simm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55431-3.
References
- 1.Nusse, R. & Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Steinhart, Z. & Angers, S. Wnt signaling in development and tissue homeostasis. Development145, 10.1242/dev.146589 (2018). [DOI] [PubMed]
- 3.Hao, H. X. et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature485, 195–200 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Koo, B. K. et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature488, 665–669 (2012). [DOI] [PubMed] [Google Scholar]
- 5.de Lau, W. et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature476, 293–297 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Carmon, K. S., Lin, Q., Gong, X., Thomas, A. & Liu, Q. LGR5 interacts and cointernalizes with Wnt receptors to modulate Wnt/β-catenin signaling. Mol. Cell Biol.32, 2054–2064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmon, K. S., Gong, X., Yi, J., Thomas, A. & Liu, Q. RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc. Natl Acad. Sci. USA111, E1221–E1229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binnerts, M. E. et al. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl Acad. Sci. USA104, 14700–14705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glinka, A. et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep.12, 1055–1061 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmon, K. S., Gong, X., Lin, Q. S., Thomas, A. & Liu, Q. Y. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. P Natl Acad. Sci. USA108, 11452–11457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huch, M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature494, 247–250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park, S. et al. Unlike LGR4, LGR5 potentiates Wnt-beta-catenin signaling without sequestering E3 ligases. Sci. Signal13, eaaz4051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seshagiri, S. et al. Recurrent R-spondin fusions in colon cancer. Nature488, 660–664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Styrkarsdottir, U. et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature497, 517–520 (2013). [DOI] [PubMed] [Google Scholar]
- 15.van Andel, H. et al. Aberrantly expressed LGR4 empowers Wnt signaling in multiple myeloma by hijacking osteoblast-derived R-spondins. Proc. Natl Acad. Sci. USA114, 376–381 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakis, M. et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat. Genet46, 1264–1266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salik, B. et al. Targeting RSPO3-LGR4 Signaling for Leukemia Stem Cell Eradication in Acute Myeloid Leukemia. Cancer Cell38, 263–278 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X. et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl Acad. Sci. USA110, 12649–12654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, C. et al. RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nat. Commun.5, 3149 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Lebensohn, A. M., Bazan, J. F. & Rohatgi, R. Receptor control by membrane-tethered ubiquitin ligases in development and tissue homeostasis. Curr. Top. Dev. Biol.150, 25–89 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu, K., Xu, Y., Rajashankar, K. R., Robev, D. & Nikolov, D. B. Crystal structures of Lgr4 and its complex with R-spondin1. Structure21, 1683–1689 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu, J. G. et al. Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G protein-coupled receptors (LGRs). J. Biol. Chem.290, 2455–2465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng, N. & Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Chen, P. H., Chen, X., Lin, Z., Fang, D. & He, X. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev.27, 1345–1350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felce, J. H. et al. Receptor Quaternary Organization Explains G Protein-Coupled Receptor Family Structure. Cell Rep.20, 2654–2665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng, W. C. et al. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep.3, 1885–1892 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Zebisch, M. et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat. Commun.4, 2787 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zebisch, M. & Jones, E. Y. Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J. Struct. Biol.191, 149–155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, D. et al. Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev.27, 1339–1344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng, W. C. et al. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. Plos One8, e83110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marei, H. et al. Antibody targeting of E3 ubiquitin ligases for receptor degradation. Nature610, 182–189 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crunkhorn, S. Developing antibody-based PROTACs. Nat. Rev. Drug Discov.21, 795 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Kargbo, R. B. Emerging Proteolysis Targeting Antibodies (PROTABs) for Application in Cancer Therapy. ACS Med Chem. Lett.13, 1833–1834 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uchański, T. et al. Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM. Nat. Methods18, 60–68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan, J. et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun.11, 4121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia, R. et al. Cryo-EM structure of the human histamine H(1) receptor/G(q) complex. Nat. Commun.12, 2086 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toh, Y. et al. LGR4 and LGR5 form distinct homodimers that only LGR4 complexes with RNF43/ZNRF3 to provide high affinity binding of R-spondin ligands. Sci. Rep.13, 10796 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nikolay, R. et al. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem.279, 2673–2678 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Peschard, P. et al. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol. Cell27, 474–485 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Feltham, R. et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J. Biol. Chem.286, 17015–17028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plechanovová, A. et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol.18, 1052–1059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liew, C. W., Sun, H., Hunter, T. & Day, C. L. RING domain dimerization is essential for RNF4 function. Biochem J.431, 23–29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yudina, Z. et al. RING Dimerization Links Higher-Order Assembly of TRIM5α to Synthesis of K63-Linked Polyubiquitin. Cell Rep.12, 788–797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plechanovová, A., Jaffray, E. G., Tatham, M. H., Naismith, J. H. & Hay, R. T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature489, 115–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiorentini, F., Esposito, D. & Rittinger, K. Does it take two to tango? RING domain self-association and activity in TRIM E3 ubiquitin ligases. Biochem Soc. Trans.48, 2615–2624 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balaji, V. & Hoppe, T. Regulation of E3 ubiquitin ligases by homotypic and heterotypic assembly. F1000Res9, Faculty Rev–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherezov, V. et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science318, 1258–1265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen, S. G. et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature450, 383–387 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Faust, B. et al. Autoantibody mimicry of hormone action at the thyrotropin receptor. Nature609, 846–853 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duan, J. et al. Hormone- and antibody-mediated activation of the thyrotropin receptor. Nature609, 854–859 (2022). [DOI] [PubMed] [Google Scholar]
- 51.Duan, J. et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature598, 688–692 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Ballesteros, J. A. et al. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem.276, 29171–29177 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Greasley, P. J., Fanelli, F., Rossier, O., Abuin, L. & Cotecchia, S. Mutagenesis and modelling of the alpha(1b)-adrenergic receptor highlight the role of the helix 3/helix 6 interface in receptor activation. Mol. Pharm.61, 1025–1032 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Shapiro, D. A., Kristiansen, K., Weiner, D. M., Kroeze, W. K. & Roth, B. L. Evidence for a model of agonist-induced activation of 5-hydroxytryptamine 2A serotonin receptors that involves the disruption of a strong ionic interaction between helices 3 and 6. J. Biol. Chem.277, 11441–11449 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Xie, X. Q. & Chowdhury, A. Advances in methods to characterize ligand-induced ionic lock and rotamer toggle molecular switch in G protein-coupled receptors. Methods Enzymol.520, 153–174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardon, E. et al. A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc.9, 674–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, X. et al. Structural insights into the activation of human calcium-sensing receptor. Elife10, 10.7554/eLife.68578 (2021). [DOI] [PMC free article] [PubMed]
- 58.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Asarnow, D., Palovcak, E. & Cheng, Y. UCSF pyem v0.5. Zenodo,10.5281/zenodo.3576630 (2019).
- 60.Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, J. et al. A minority of final stacks yields superior amplitude in single-particle cryo-EM. Nat. Commun.14, 7822 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He, J., Li, T. & Huang, S. Y. Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun.14, 3217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr.66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study. Source data are provided as a Source Data file. The PDB data generated in this study have been deposited in the RCSB PDB database under accession code 8XFT, 8XFS, 8XFP and 8Y69, Cryo-EM maps generated in this study have been deposited with EMDB under accession codes EMD-38309, EMD-38308, EMD-38307 and EMD-38982 (https:// www.ebi.ac.uk/emdb/EMD-38982). Accession numbers are also listed in Supplementary Table 1. The TopFlash, NanoBit assay and FACS data generated in this study are provided in the Source Data file. Source data are provided with this paper.