Abstract
Hepatic cystic echinococcosis (HCE), a life-threatening liver disease, has 5 subtypes, i.e., single-cystic, polycystic, internal capsule collapse, solid mass, and calcified subtypes. And each subtype has different treatment methods. An accurate diagnosis is the prerequisite for effective HCE treatment. However, clinicians with less diagnostic experience often make misdiagnoses of HCE and confuse its 5 subtypes in clinical practice. Computer-aided diagnosis (CAD) techniques can help clinicians to improve their diagnostic performance. This paper aims to propose an efficient CAD system that automatically differentiates 5 subtypes of HCE from the ultrasound images. The proposed CAD system adopts the concept of deep transfer learning and uses a pre-trained convolutional neural network (CNN) named VGG19 to extract deep CNN features from the ultrasound images. The proven classifier models, k - nearest neighbor (KNN) and support vecter machine (SVM) models, are integrated to classify the extracted deep CNN features. 3 distinct experiments with the same deep CNN features but different classifier models (softmax, KNN, SVM) are performed. The experiments followed 10 runs of the five-fold cross-validation process on a total of 1820 ultrasound images and the results were compared using Wilcoxon signed-rank test. The overall classification accuracy from low to high was 90.46 ± 1.59% for KNN classifier, 90.92 ± 2.49% for transfer learned VGG19, and 92.01 ± 1.48% for SVM, indicating SVM classifiers with deep CNN features achieved the best performance (P < 0.05). Other performance measures used in the study are specificity, sensitivity, precision, F1-score, and area under the curve (AUC). In addition, the paper addresses a practical aspect by evaluating the system with smaller training data to demonstrate the capability of the proposed classification system. The observations of the study imply that transfer learning is a useful technique when the availability of medical images is limited. The proposed classification system by using deep CNN features and SVM classifier is potentially helpful for clinicians to improve their HCE diagnostic performance in clinical practice.
Keywords: Hepatic cystic echinococcosis, Multi-classification, Convolutional neural networks, Ultrasound images, Transfer learning, Support vector machine
Subject terms: Computational biology and bioinformatics, Diseases
Introduction
Echinococcosis, also known as hydatid disease, is an infection caused by a larval-stage tapeworm, echinococcus1. Human echinococcosis is a zoonotic disease caused by parasites of the genus echinococcus transmitted by dogs in livestock-raising areas2. According to the World Health Organization (WHO), echinococcus granulosus is endemic in South America, Eastern Europe, Russia, the Middle East, and China, where human incidence rates are as high as 50 per 100,000 person-years3. The two most important forms in humans are cystic echinococcosis (CE) and alveolar echinococcosis (AE) caused by infection with the metacestode stages of echinococcus granulosus and echinococcus multilocularis, respectively4. A study from Philip S. Craig5 indicated that in China human CE cases are responsible nationally for more than 98% of echinococcosis infections, with AE in the remainder of infections (2%). The liver is the most common site of disease in humans accounting for 50–70% of cases, followed by the lungs (20–30%), and less frequently the spleen, kidneys, heart, bones, central nervous system, and other organs6. The signs and symptoms of hepatic echinococcosis (HE) can include hepatic enlargement (with or without a palpable mass in the right upper quadrant), right epigastric pain, nausea, and vomiting. If a cyst ruptures, the sudden release of its contents can precipitate allergic reactions ranging from mild to fatal anaphylaxis7. Although some progress has been made in the control of echinococcosis, this zoonosis continues to be a major public health issue in several countries8. Take China as an example, it has been estimated that 600,000–1.3 million existing cases of human echinococcosis occur in China9. The disease is very highly endemic in Xinjiang Uygur Autonomous Region; from 2004 to 2010, a total of 4,768 human echinococcosis cases were reported in Xinjiang Uygur Autonomous Region, accounting for 32.6% of total cases in China and most of them were hepatic cystic echinococcosis (HCE)10.
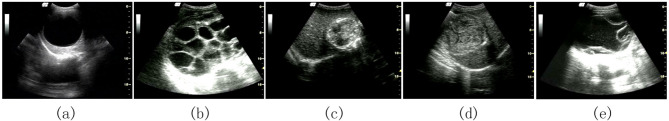
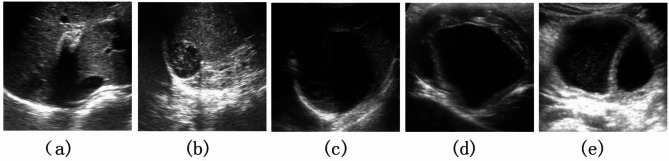
Currently, the diagnosis of HE is based on clinical findings, imaging results, and serology11, among which imaging results play a key role in diagnosing and staging CE. Ultrasound remains the cornerstone of diagnosis, staging, and follow-up of CE cysts12. According to the World Health Organization Working Group of Experts on Hydatidosis(WHO/IWGE) based on Gharbi ultrasound typing, HCE consists of 5 different subtypes shown in Fig. 1, i.e. single-cystic type (SC or CE1), polycystic type (PC or CE2), internal capsule collapse type (ICC or CE3), solid mass type (SM or CE4), and calcified type (CA or CE5)3,13, which need different clinical management and therapeutic methods. However, due to the complexity of ultrasound images of various HCE subtypes, the manual diagnosis of HCE subtypes requires a high degree of skill and concentration, which is time-consuming, subjective, and prone to operator bias.
Fig. 1.
Ultrasound image samples of each HCE subtype (a) single-cystic type (b) polycystic type (c) calcified type (d) solid mass type (e) internal capsule collapse type.
Computer-aided diagnosis (CAD), a technology based on the machine learning algorithm, has been wildly used in various diseases in recent years to help clinicians improving their diagnostic performance. In recent years, Yadav Niranjan and his research team focused on thyroid tumor CAD systems by using machine learning algorithms and ultrasound images and achieved great success. These works included thyroid tumor ultrasound image classification work14,15, image segmentation work16, image preprocessing work17–19, and systematic review work of thyroid tumor CAD system20. Kriti et al.21 developed a CAD system for breast cancer based on deep learning models and ultrasound images, which yield an accuracy of 98.0% with individual class accuracy values of 100% and 96% for benign and malignant classes respectively. Wang et al.22developed a multibranch transformer network for the automated classification of multiple ophthalmic diseases using B-mode ultrasound images, which got an accuracy of 82.21% (95% CI 78.45–85.44%) in the temporal external testing set. To the best of our knowledge, few studies on echinococcosis CAD systems have been carried out. An existing study from Xin Shenghai, et al.23 designed an echinococcosis CAD system for binary classification from computed tomography (CT) images, which achieved a classification accuracy of 80.32% and 82.45% for cystic vs. alveolar echinococcosis and calcified vs. noncalcified lesion, respectively. However, these studies have limited clinical significance as they inadequately considered multi-types of echinococcosis in real clinical practice. Therefore, we mainly worked on classifying HCE into 5 different subtypes including SC, PC, ICC, SM, and CA. Our previous study24 has demonstrated that CNN has a potential ability for this multi-classification task. To further improve the classification performance and build a more efficient HCE CAD system, we extended the findings from the previous study by exploring the deep CNN features via transfer learning and using the proven classifiers for the multi-classification in this paper.
The major novelties of this paper are listed below.
The concept of deep transfer learning is applied to the specific 5-class HCE classification problem.
A significant improvement in the classification performance is observed when the SVM classifier is applied with features extracted by the transfer-learned deep CNN model.
The proposed method performs well even with a smaller number of training samples.
The phenomenon of overfitting with smaller training data and the impact on classification performance are studied.
The remaining part of this paper is arranged as follows. In Sect. 2, we describe the dataset used in the study and its preprocessing. Section 3 presents an overview concept of transfer learning and the complete framework for the proposed classification algorithm. Section 4 gives details of the experiments performed, presents evaluation results, and the discussion. Section 5 gives the conclusion of this paper.
Dataset and preprocessing
This study was approved by the Institutional Review Board of The First Affiliated Hospital of Xinjiang Medical University in China, and informed consent was obtained from the research subjects. A total of 1820 ultrasound images from 967 HCE patients were collected from the First Affiliated Hospital of Xinjiang Medical University between the years 2008 and 2020 and clinically confirmed by at least two radiologists with more than 8 years of working experience in ultrasound, including 358 images of 174 SC patients, 383 images of 194 PC patients, 350 images of 156 ICC patients, 351 images of 213 CA patients, 378 images of 230 SM patients, which were shown in Table 1. Controversies have been solved through the discussion. The personal private information has been manually removed from the images.
Table 1.
The ultrasound images and patients’ number of five subtypes of HCE involved in the study.
| Subtypes of HCE | Number of ultrasound images | Number of patients |
|---|---|---|
| Single-cystic (SC) | 358 | 174 |
| Polycystic (PC) | 383 | 194 |
| Internal Capsule Collapse (ICC) | 350 | 156 |
| Calcified (CA) | 351 | 213 |
| Solid Mass (SM) | 378 | 230 |
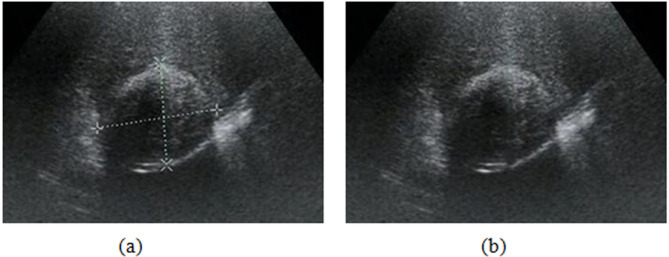
Before inputting ultrasound images into the proposed model, preprocessing is needed. The preprocessing phase mainly includes ROI extraction and artificial marker repair. A small portion of these ultrasound images was artificially marked by radiologists to record the location of the cystic capsule, cortical thickness, etc. As shown in Fig. 2(a)24, artificial markers occlude the texture region and break the integrity of an image, affecting adversely image analysis. Hence, we tried to use the Spot Healing Brush in Adobe Photoshop (version 19.1.1) to remove the markers and repair the image. Especially, our Spot Healing Brush is a rounded brush with a diameter of 15 pixels that slightly larger than the width of the graticules, a hardness of 0% to avoid harsh and unnatural edges, a spacing of 25%, an angle of 0%, and a roundness of 100%. Then, we carefully painted along the graticules with the brush to remove the makers (graticules). As shown in Fig. 2(b)24, the makers are entirely repaired with good image quality, and the training images are maximally restored as the original ultrasound images with no artificially labeled information that may affect the classifier’s performance, enhancing the models’ reliability.
Fig. 2.
Removal and reparation of artificial markers in an HCE ultrasound image. (a) image with markers. (b) repaired image.
The original images show a large echo region, including cystic lesions and other peripheral organ parts. To eliminate the interference of irrelevant areas and reduce the computational burden of the models, we manually cropped the lesion area from each image and resized them to an uninformed size of 224 × 224 pixels.
Methods
Since our previous study24 indicated that the CNN model named VGG1925 has an outstanding performance on this HCE classification task, in this study, we still use VGG19 to further improve the classification performance based on the deep CNN features via transfer learning and proven classifiers. And we confirmed that all methods were performed in accordance with relevant guidelines and regulations.
Transfer learning in an inductive setting
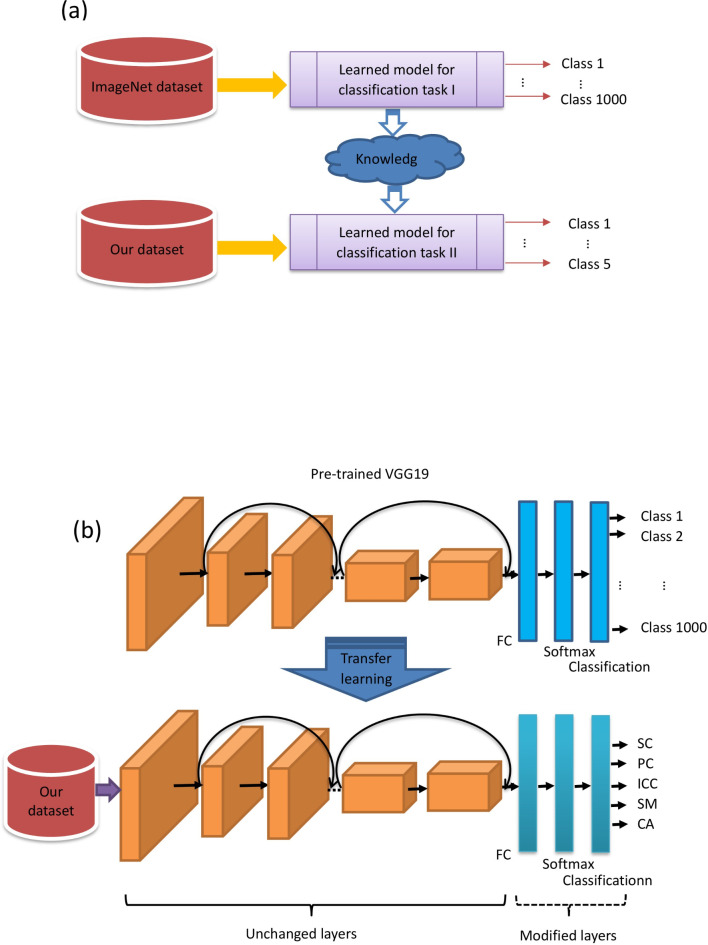
Transfer learning refers to the process of using the knowledge that has been acquired in a previously learned task, the source task, in order to enhance the learning procedure in a new task, the target task26. Given a source domain (Ds) and a learning task (Ts) in the source task, a target domain (Dt), and the corresponding learning task (Tt), transfer learning aims to improve the learning in Dt, using the knowledge gained in Ds and Ts. Based on the type of task and the nature of the data available at the Ds and Ts, different settings are specified for transfer learning27. It is known as inductive transfer learning when labeled data is available at Ds and Dt, for a classification task28. In such a scenario, the domain is represented as D=(Fi Ci) ∀ i, where Fi is the feature vector for the i-th training sample and Ci is the corresponding class label. The setting for transfer learning with respect to our study is introduced as follows.
VGG19 is a convolutional neural network that is 19 layers deep, including 16 convolutional layers,5 pooling layers, 3 fully-connected (FC) layers, and 2 dropout layers with dropout probabilities of 0.5. VGG19 is an integration of an image feature extractor and a classifier. The extractor captures image features that can be connected to various classifiers29. VGG19 is a very accurate and efficient model that has an outstanding performance in the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC) 2014. A pre-trained VGG19 was trained by using 1.2 million natural images (cat, dog, car, Tree, etc. ) in the ImageNet which consisted of 1000 defined classes. Training samples in the ImageNet define the source domain and the source task is a 1000-class classification problem. We define our task as the classification of HCE into 5 subtypes. The HCE ultrasound images formed our dataset and defined the target domain. Figure 3(a) demonstrates the basic framework of the transfer learning approach applied in our study. Since our target domain has 5 different classes, we modified the last FC layers of VGG19 to fit our target domain. In particular, the original fully connected (FC) layer with an output size of 1000 in VGG19 was replaced by a new FC layer with an output size of 5. The softmax layer, following the FC layer, and the cross-entropy-based classification layer at the output were replaced with new ones. The modified deep learning model is shown in Fig. 3(b). Then, we fine-tune the modified VGG19 by training it with our images. The learning factor for weights and bias at the FC layer was set to a high value of 10 to make the network learn abstract high-level features specific to the target domain. The low-level features were expected to be learned by the pre-trained layers from the original VGG19. The transfer learned and fine-tuned CNN model could then be used for the experiments by feeding the ultrasound images.
Fig. 3.
Deep learning framework. (a) A transfer learning framework (b) Modifying VGG19 for the application.
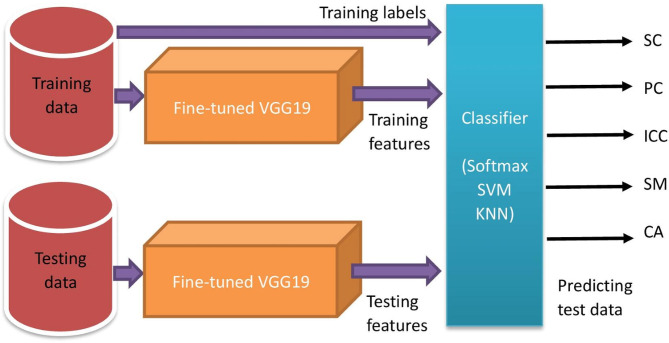
Proposed classification Framework
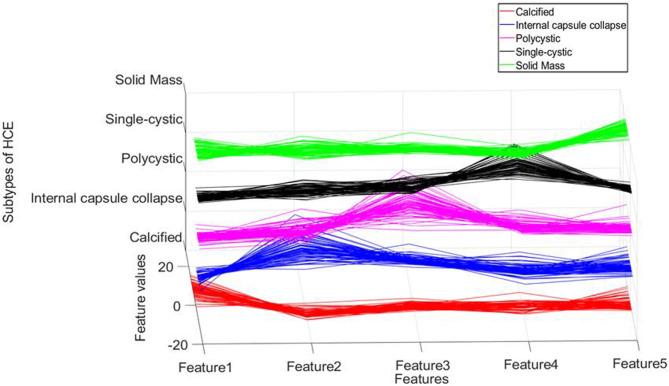
The proposed classification framework makes use of the modified and fine-tuned VGG19 to learn the features of HCE ultrasound images. (The features extracted by the modified and fine-tuned VGG19 are referred to as deep CNN features in the rest of this paper.) Features extracted from layers near the FC layer give good discriminative representation for trained images. Figure 4 shows the deep features of our dataset at the FC layer.
Fig. 4.
Features distribution of cystic echinococcosis at FC layer of VGG19 (red lines denote features of calcified type, blue lines denote features of internal capsule collapse, purple lines denote features of polycystic type, black lines denote features of single-cystic, green lines denote features of solid mass type).
The transfer learned model then uses its softmax classifier layer to classify the images into 5 HCE subtypes. However, improved performance is expected when deep CNN features are provided to well-established pattern classifiers30. Thus, in the proposed work, the deep CNN features were further classified by two proven classifier models other than the softmax classifier belonging to deep CNN. These two proven classifiers are support vector machine (SVM) and k-nearest neighbors (KNN). SVM is a discriminative classifier defined by a separating hyperplane which is the best decision boundary that helps classify data points in n-dimensional space31. The reason for choosing Multiclass-SVM as the classifier in our experiments was its outstanding performance in classifying CNN features in many previous works32–34. However, in cases when data points can not be easily separated by SVM hyperplane, a density estimation based on the classifier may be helpful. This is the reason for considering KNN in our experiments. Multiclass-SVM with polynomial kernel function maps inputs into high-dimensional feature spaces for nonlinear classification of our 5-class problem whereas KNN calculates the Euclidean distance to build the training model for class-membership prediction using majority voting from K-neighbors. Figure 5 shows the overall framework for the proposed classification system. In particular, the deep CNN features of the training data and their class labels were used to train the classifier. Then, we use this trained classifier to predict the output class labels for the test data with their deep CNN features.
Fig. 5.
Overall framework for HCE classification.
Experiments and results
The proposed classification system was implemented in MATLAB 2020b on a computer having specifications of 16GB RAM and Intel® Core™ i5-4590 @ 3.3 GHz CPU.
Classifier settings
In addition to the image quality that can affect the performance of an image classification system, the image features and the classifier model also play very important roles in the system. In this study, we performed 3 distinct experiments with different classifier models used in the final stage of the proposed CE image classification system, which are described as follows.
Transfer learned deep CNN model with its softmax classifier, as a stand-alone system
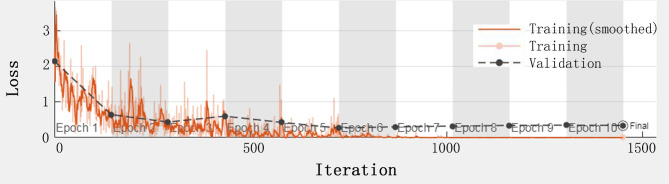
The pre-trained VGG19 was further fine-tuned by the training set. The hyperparameters of the CNN were heuristically adjusted so as to facilitate the convergence of the loss function during the training process (Fig. 6). The stochastic gradient descent with momentum (SGDM) was the chosen optimizer, considering its good learning rate and the parameter-specific adaptive nature of the learning rates. For SGDM, the initial learning rate was set to 0.0003. The setting of a high value might prevent the loss function from converging and could cause overshoots, whereas a very small value of the learning rate increases the computational burden and training time. The mini-bach size was set to 10. This setting is a compromise between the training speed (a larger batch size means faster training) and hardware limitation. Besides, a very large batch size adversely affects the model performance. The maximum epochs were set to 10, considering the occurrence of overfitting. The hyperparameter settings of the experiments are listed in Table 2.
Fig. 6.
Loss function of the transfer learned VGG19 during the training process.
Table 2.
Experimental parameters and settings.
| Model | Parameters | Settings(values) |
|---|---|---|
| Transfer learned VGG19 | Learning algorithm | SGDM |
| Initial learning rate | 0.0003 | |
| Mini-bach size | 10 | |
| Maximum epochs | 10 | |
| SVM | Model sub-type | ECOC hinge |
| Loss function | One-vs-all | |
| solver | BFGS | |
| Coding | SVM linear | |
| KNN | Number of neighbors | 38 |
| Distance metric | Euclidean |
Deep CNN features with SVM classifier
We extract deep CNN features from the pooling layer of the VGG19 which is next to the FC layer. These features were further classified using SVM. A multi-class SVM with an error-correcting output code (ECOC) model was used as the classifier in this study. A one-vs-all strategy was set in the SVM for multi-class classification. There were three binary SVM learners, each of them with a linear kernel. Other parameters of SVM are listed in Table 2.
Deep CNN features with KNN classifier
A classification experiment was then performed by the KNN classifier with the deep CNN features. The key parameters of KNN include k, the number of nearest neighbors, and the distance metric. We set the value of k to 38, the square root of the number of training samples as per cross-validation settings. A lower value of k can make the system susceptible to noise and overfitting, whereas a higher value means more computations. Furthermore, data imbalances with respect to classes may dominate the final classification results if the value of k is set too high. Euclidean distance was used as the distance metric in the experiment.
Performance evaluation
The systems’ performance was validated using 5-fold cross-validation. To avoid the possible bias during data set partitioning for cross-validation, the experiment was repeated 10 times for each classification system forementioned, and each followed a five-fold cross-validation process. The average results of the 50 trials were presented in a mean ± standard deviation format. The overall classification accuracy from low to high was 90.46 ± 1.59% for the KNN classifier, 90.92 ± 2.49% for the transfer learned VGG19, and 92.01 ± 1.48% for SVM. The Wilcoxon signed-rank test indicated that there is a statistically significant difference between the mean classification accuracy of SVM and VGG19 (P = 0.0129, < 0.05), SVM and KNN (P = 0.000, < 0.05), indicating that superior performance is achieved when the SVM classifier is used to classify the deep CNN features. Thus, we choose the SVM classifier with deep CNN features as our classification system. Compared with previous work24, this system has significantly improved the classification accuracy by 1.40%, indicating SVM classifier has a superior discriminative ability than KNN and Softmax classifier. In fact, the same algorithms using deep features extracted by CNN and other proven classifiers to the classification work, such as SVM and KNN, have been applied to many other disease diagnoses and achieved great success35–37. In particular, Kumar, et al.35 proposed brain tumor detection and classification framework deep feature extraction from Resnet 18 pre-trained network in conjunction with feature fusion and optimized Machine Learning classifiers that can efficiently improve the system performance with a high accuracy, sensitivity, specificity, precision, F1 score, of 99.11%, 98.99%, 99.22%, 99.09%, 99.09%. Jeba, et al.36 also used the same approach as we did in this paper and obtained a high accuracy of 94.65% for the brain tumor classification work, while other stand-alone machine learning methods got a low accuracy, such as AlexNet, KNN, etc. Khagi et al.37 also applied the same algorithm to the Alzheimer’s disease classification and got a high accuracy around 98-99%.
A confusion matrix summarizes correct and incorrect classifications in a tabular form. Table 3 shows the accumulated confusion matrix for the SVM classifier on deep CNN features, obtained from the 10 runs of 5-fold cross-validation experiments.
Table 3.
Accumulated confusion matrix from 10 runs of 5-fold cross-validation of SVM classifier on the deep CNN features (where CA, ICC, PC, SC, and SM refer to the calcified type, internal capsule collapse type, polycystic, respectively).
| Predicted | ||||||
|---|---|---|---|---|---|---|
| CA | ICC | PC | SC | SM | ||
| Actual | CA | 3302 | 4 | 6 | 9 | 189 |
| ICC | 13 | 3048 | 171 | 134 | 134 | |
| PC | 32 | 211 | 3425 | 101 | 61 | |
| SC | 45 | 40 | 76 | 3414 | 5 | |
| SM | 125 | 64 | 23 | 10 | 3558 |
From the confusion matrix, different metrics can be derived to indicate the classifier’s performance, specific to each class. Essential metrics are sensitivity, specificity, precision, and F1-score calculated by the following Eqs. (1–4).
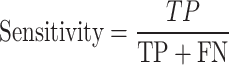 |
1 |
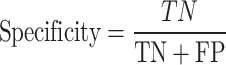 |
2 |
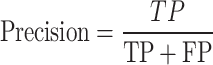 |
3 |
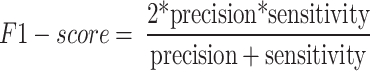 |
4 |
Where TP, FP, TN, and FN represent the numbers of true positives, false positives, true negatives, and false negatives, respectively. Table 4 presents the class-specific performance of the proposed system when the SVM classifier was used with deep CNN features, which achieved the best performance in our experiment.
Table 4.
Class-specific evaluation of SVM classifier with deep CNN features.
| CE type | Sensitivity (%) | Specificity (%) | Precision (%) | F1-score(%) |
|---|---|---|---|---|
| CA | 94.08 ± 3.06 | 98.54 ± 0.78 | 93.99 ± 2.94 | 93.98 ± 2.09 |
| ICC | 87.09 ± 4.56 | 97.83 ± 0.90 | 90.66 ± 3.48 | 88.74 ± 3.00 |
| PC | 89.43 ± 3.06 | 98.08 ± 0.95 | 92.65 ± 3.28 | 90.96 ± 2.32 |
| SC | 95.36 ± 2.27 | 98.26 ± 0.82 | 93.17 ± 3.06 | 94.22 ± 2.01 |
| SM | 94.13 ± 2.51 | 97.30 ± 1.01 | 90.27 ± 3.22 | 92.11 ± 2.01 |
Smaller training dataset
Since we performed five-fold cross-validation to evaluate the performance of our models, the original training set consists of 80% of the total images, and the rest 20% is the testing set. Transfer learning is a technique that can help in overcoming the scarcity of images when training a deep CNN38. To test the capability of our classification system and the efficiency of the transfer learning technique, we performed the experiments under the condition when the availability of training data is limited:
By randomly selecting 75% of the original training set (i.e. 60% of the total images).
By randomly selecting 50% of the original training set (i.e. 40% of the total images).
By randomly selecting 25% of the original training set (i.e. 20% of the total images).
For each condition above, we use the deep CNN features extracted from the smaller training set to train the SVM classifier, and then 20% of the total images not included in the training set were provided as the testing set to test the model’s performance. Table 5 presents the corresponding performance measures.
Table 5.
The performance of SVM classifier with reduced training data.
| Training data | Accuracy(%) | AUC(%) | ||||
|---|---|---|---|---|---|---|
| CA | ICC | PC | SC | SM | ||
| 100% | 92.01 ± 1.48 | 99.52 ± 0.26 | 98.37 ± 0.66 | 98.08 ± 0.86 | 99.37 ± 0.36 | 99.03 ± 0.45 |
| 75% | 90.89 ± 1.56 | 99.37 ± 0.37 | 97.99 ± 0.87 | 97.74 ± 1.01 | 99.16 ± 0.53 | 98.82 ± 0.64 |
| 50% | 89.48 ± 1.91 | 99.27 ± 0.40 | 97.69 ± 0.87 | 97.46 ± 1.40 | 98.95 ± 0.46 | 98.49 ± 0.72 |
| 25% | 86.40 ± 2.27 | 98.70 ± 0.61 | 96.62 ± 1.68 | 96.70 ± 1.52 | 98.17 ± 1.13 | 97.68 ± 0.80 |
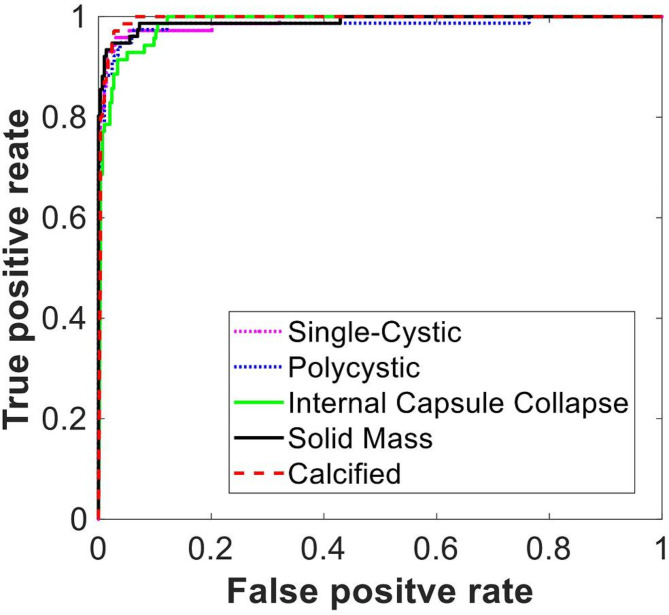
In addition to the overall classification accuracy, we also analyzed the receiver operating characteristic (ROC) curves of the SVM classifiers on each subtype of HCE. Figure 7 represents the ROC curves under the condition that 50% of the training data is used. The average values of the area under the curve (AUC) listed in Table 5 indicate that the accuracy of the SVM classifier decreases with the size of the training data. It is notable that the reduction in size of the training data has not impaired the system performance significantly. Specially, the classification accuracy only decreased by 2.53% when the training data was reduced from 100 to 50%. This observation suggests that our system has a decent capability even with a limited number of training data, which is helpful in practical scenarios because the number of training samples available could be limited due to the unavailability of more medical data. Another obvious advantage of the smaller training set is a shorter training time. In our experiment, the average training time of our system for 100%, 75%, 50%, and 25% training data was approximately 184 min, 144 min, 101 min, and 61 min, respectively.
Fig. 7.
ROC curves of the SVM classifier.
Regarding misclassifications
Based on the performance evaluation and detailed analysis, we give the following reference about the proposed classification systems. The classification accuracy of the system improved when SVM was used instead of the classification layers within the transfer learned mode, meaning that some of the classification samples, i.e. Figure 9(a) and (b), which went wrong with the softmax-based classifier of the VGG19, were corrected by the SVM-based classifier. Since the features used for classification are the same (i.e. deep CNN features) for different classifiers, the different classification results must be caused by the classifiers themselves, indicating SVM has a superior classification capability. However, there are still some samples that can not be correctly predicted for both two classifiers. As shown in Fig. 9 (c-e), these samples were all misclassified by the 2 classifiers. It may be because these misclassified images have more complex heterogeneity compared with other samples in the same group and parts of the image features are similar to other types. Besides, the number of such special images is also very limited in the training set, which directly leads the insufficiently trained classifiers to misclassifications.
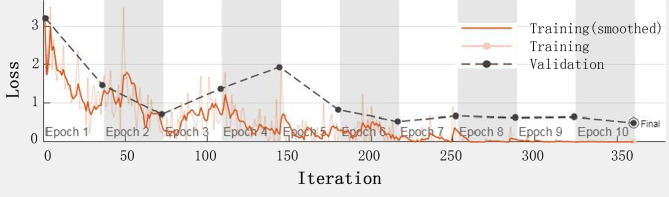
Fig. 8.
Training and validation loss on training the model with 25% of training data.
Another aspect of this study we concentrated on was the handling of the smaller training data. We noticed that there was a decline in the performance of the classification system (Table 5) with the reduction in the training data, which suggests that the discriminative power of deep CNN features is affected. This effect is further analyzed by studying the variation of training and validation losses against iterations. Figure 8 presents a sample instance of training progress when 25% of the training data was used. In this case, we find that the training loss decreases, whereas the validation loss increases approximately between 110 and 150 iterations. This reaction indicates the phenomenon of overfitting took place. This is the reason for lower classification as the model has learned specific training images without gaining a good generalization capability. In other words, the model complexity is greater compared to the available training samples. Properly tuning the learning and regularization rate parameters can prevent the model from overfitting, as well as the data augmentation39. Since this is a pilot study of the HCE CAD system, we focused on validating the feasibility of the proposed system and improving the system’s performance, a limitation in the current study is that our data were all collected from one hospital, therefore, the generalizability of the model was unknown. Additional studies, ideally covering a large number of cases from multiple centers are necessary to validate the models’ generalizability.
Fig. 9.
Misclassified samples. (a) CA type misclassified as ICC by softmax-based classifier, but correctly classified by SVM. (b) SM type misclassified as polycystic by softmax-based classifier, but correctly classified by SVM. (c) SC type misclassified as ICC for both softmax-based classifier and SVM. (d) PC type misclassified as ICC for both softmax-based classifier and SVM. (e) ICC type missclassified as PC for both softmax-based classifier and SVM.
In future studies, we also plan to conduct a more in-depth study by using other state-of-the-art CNN, i.e., Transformer40, Yolov841, etc. machine-learning algorithms, and optimization techniques in our scenario, to realize a high-efficiency and fully automatic HCE classification system without manually extracting the lesion area.
Conclusion
In this paper, we demonstrate automatic multi-classification systems for HCE by using transfer-learned VGG19, KNN, and SVM based on the deep CNN features of the ultrasound images. The experiments were conducted under a five-fold cross-validation process. The best performance was achieved by the SVM with an accuracy of 92.01 ± 1.48%, followed by the VGG19 and KNN with an accuracy of 90.92 ± 2.49% and 90.46 ± 1.59%, respectively, indicating the hybrid approach of CNN with SVM, CNN is used for feature extraction and SoftMax layer is removed and replaced with SVM, could effectively improve the performance in terms of accuracy. The proposed approach also showed a decent performance even with smaller training data. In sum, the proposed HCE classification system is efficient and potentially applicable to clinical practice, assisting clinicians in enhancing diagnostic accuracy and increasing the probability of successful treatment.
Acknowledgements
This work was supported by the Project of Top-notch Talents of Technological Youth of Xinjiang (2023TSYCCX0067), and Natural Science Foundation Xinjiang Uygur Autonomous Region (2022D01C436, 2022D01C184).
Author contributions
Miao Wu performed the computations , analyse the results and wrote the paper. Chuanbo Yan contributed the data. Gan Sen gave useful advice.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available as they contain identifiable and personal information but are available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethical approval was obtained from the Research Ethics Committee of the Xinjiang Medical University. Both written consent and verbal consent were allowed according to the Ethics Committee.
Consent for publication
Written informed consent for publication of their clinical details and clinical images was obtained from the patients. A copy of the consent form is available for review by the editor of the journal.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Czermak, B. V. et al. Echinococcosis of the liver. Abdom. Imaging. 33 (2), 133–143 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Eckert, J., Conraths, F. J. & Tackmann, K. Echinococcosis: an emerging or re-emerging zoonosis? Int. J. Parasitol.30 (12–13), 1283–1294 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Brunetti, E. et al. Expert consensus for the diagnosis and treatment of cystic and alveolar echinococcosis in humans. Acta Trop.114 (1), 1–16 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Feng, X. et al. Human cystic and alveolar echinococcosis in the Tibet Autonomous Region (TAR), China. J. Helminthol.89 (6), 671–679 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig, P. S. C. The Echinococcosis Working Group in, and C. Echinococcosis Working Group in, Epidemiology of human alveolar echinococcosis in China. Parasitology international, 55: pp. S221-S225. (2006). [DOI] [PubMed]
- 6.Kammerer, W. S. & Schantz, P. M. Echinococcal disease. Infect. Dis. Clin. N. Am.7 (3), 605–618 (1993). [PubMed] [Google Scholar]
- 7.Bhutani, N. & Kajal, P. Hepatic echinococcosis: a review. Annals Med. Surg.36, 99–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dakkak, A. J. V. P. Echinococcosis/hydatidosis: a severe threat in Mediterranean countries. 174(1–2): pp. 2–11. (2010). [DOI] [PubMed]
- 9.Ito, A. et al. Control of Echinococcosis and Cysticercosis: A Public Health Challenge to International Cooperation in Chinap. 3–17 (AMSTERDAM, 2003). [DOI] [PubMed]
- 10.Wang, N. et al. The complete mitochondrial genome of G3 genotype of Echinococcus granulosus (Cestoda: Taeniidae). Mitochondrial DNA Part. DNA Mapp. Sequencing Anal.27 (3), 1701–1702 (2016). [DOI] [PubMed] [Google Scholar]
- 11.McManus, D. P. et al. Echinococcosis Lancet (British Edition), 362(9392): 1295–1304. (2003). [DOI] [PubMed] [Google Scholar]
- 12.Stojkovic, M. et al. Diagnosing and staging of cystic echinococcosis: How do CT and MRI perform in comparison to Ultrasound? PLoS Negl. Trop. Dis.6 (10), e1880–e1880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharbi, H. A. et al. Ultrasound examination of the hydatic liver. Radiology139 (2), 459–463 (1981). [DOI] [PubMed] [Google Scholar]
- 14.Yadav, N., Dass, R. & Virmani, J. Deep learning-based CAD System Design for Thyroid Tumor Characterization Using Ultrasound Images (Multimedia Tools and Applications, 2023).
- 15.Yadav, N., Dass, R. & Virmani, J. Machine learning-based CAD system for thyroid tumour characterisation using ultrasound images. Int. J. Med. Eng. Inf.16 (6), 547–559 (2024). [Google Scholar]
- 16.Yadav, N., Dass, R. & Virmani, J. Objective assessment of segmentation models for thyroid ultrasound images. J. Ultrasound. 26 (3), 673–685 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav, N., Dass, R. & Virmani, J. Despeckling filters applied to thyroid ultrasound images: a comparative analysis. Multimedia Tools Appl.81 (6), 8905–8937 (2022). [Google Scholar]
- 18.Kriti, J., Virmani & Agarwal, R. Assessment of despeckle filtering algorithms for segmentation of breast tumours from ultrasound images. Biocybernetics Biomedical Eng.39 (1), 100–121 (2019). [Google Scholar]
- 19.A, R. D. Image Quality Assessment parameters for Despeckling filters - ScienceDirect. Procedia Comput. Sci.167, 2382–2392 (2020). [Google Scholar]
- 20.Yadav, N., Dass, R. & Virmani, J. A systematic review of machine learning based thyroid tumor characterisation using ultrasonographic images. J. Ultrasound. 27 (2), 209–224 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kriti, J., Virmani & Agarwal, R. Deep feature extraction and classification of breast ultrasound images. Multimedia Tools Appl.79 (37–38), 27257–27292 (2020). [Google Scholar]
- 22.Wang, Y. J. et al. Automated classification of multiple ophthalmic diseases using ultrasound images by deep learning. Br. J. Ophthalmol.108 (7), 999–1004 (2024). [DOI] [PubMed] [Google Scholar]
- 23.Xin, S. et al. Automatic Lesion Segmentation and Classification of Hepatic Echinococcosis Using a multiscale-feature Convolutional Neural Network58p. 659–668 (Medical & biological engineering & computing, 2020). 3. [DOI] [PubMed]
- 24.Wu, M. et al. Automatic Classification of Hepatic Cystic Echinococcosis Using Ultrasound Images and Deep Learning. Journal of Ultrasound in Medicine (official journal of the American Institute of Ultrasound in Medicine, 2021). [DOI] [PubMed]
- 25.Simonyan, K. & Zisserman, A. J. C. S. Very Deep Convolutional Networks for Large-Scale Image Recognition. (2014).
- 26.Fachantidis, A. et al. Transferring task models in reinforcement learning agents. Neurocomputing (Amsterdam). 107, 23–32 (2013). [Google Scholar]
- 27.Shao, L., Zhu, F. & Li, X. Transfer learning for visual categorization: a Survey. Ieee Trans. Neural Networks Learn. Syst.26 (5), 1019–1034 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Weiss, K., Khoshgoftaar, T. M. & Wang, D. A survey of transfer learning. J. big data. 3 (1), 1–40 (2016). [Google Scholar]
- 29.Nasir, I. M. et al. Deep learning-based classification of Fruit diseases: an application for Precision Agriculture. Cmc-Computers Mater. Continua. 66 (2), 1949–1962 (2021). [Google Scholar]
- 30.Xue, D. X. et al. CNN-SVM for Microvascular Morphological Type Recognition with Data Augmentation. J. Med. Biol. Eng.36 (6), 755–764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lian, L. Intelligent optimization algorithm for support Vector Machine: Research and Analysis of Prediction ability. Int. J. Artif. Intell. Tools, 33(01). (2024).
- 32.Sethy, P. K. et al. Deep Feature Based rice leaf Disease Identification Using Support Vector Machine175 (Computers and Electronics in Agriculture, 2020).
- 33.Sun, Z., Li, F. & Huang, H. Large Scale Image Classification Based on CNN and Parallel SVMp. 545–555 (Springer International Publishing, 2017).
- 34.Sahoo, J. P., Ari, S. & Patra, S. K. Hand Gesture Recognition Using PCA Based Deep CNN Reduced Features and SVM Classifier. in 2019 IEEE International Symposium on Smart Electronic Systems (iSES) (Formerly iNiS). (2019).
- 35.Kumar, S. A. & Sasikala, S. Automated Brain Tumour Detection and Classification Using Deep Features and Bayesian Optimised Classifiers20 (Current Medical Imaging, 2023). [DOI] [PubMed]
- 36.Jeba, J. A., Devi, S. N. & Meena, M. Modified CNN Architecture for efficient classification of Glioma Brain Tumour. Iete J. Res.69 (12), 9310–9323 (2023). [Google Scholar]
- 37.Khagi, B., Kwon, G. R. & Lama, R. Comparative analysis of Alzheimer’s disease classification by CDR level using CNN, feature selection, and machine-learning techniques. Int. J. Imaging Syst. Technol.29 (3), 297–310 (2019). [Google Scholar]
- 38.Kandel, I. & Castelli, M. Transfer Learning with Convolutional Neural Networks for Diabetic Retinopathy Image Classification. A Review. Applied sciences, 10(6): p. 2021. (2020).
- 39.Gavrilov, A. D. et al. Preventing Model Overfitting and Underfitting in Convolutional neural networks. Int. J. Softw. Sci. Comput. Intell. (IJSSCI). 10 (4), 19–28 (2018). [Google Scholar]
- 40.Chen, Y. L. et al. Transformer-CNN for small image object detection signal. Processing-Image Communication, 129. (2024).
- 41.Yuan, Z. J. et al. YOLOv8-ACU: improved YOLOv8-pose for facial acupoint detection. Front. Neurorobotics., 18. (2024). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are not publicly available as they contain identifiable and personal information but are available from the corresponding author on reasonable request.