Abstract
The intricate nature of microbiota sequencing data—high dimensionality and sparsity—presents a challenge in identifying informative and reproducible microbial features for both research and clinical applications. Addressing this, we introduce PreLect, an innovative feature selection framework that harnesses microbes’ prevalence to facilitate consistent selection in sparse microbiota data. Upon rigorous benchmarking against established feature selection methodologies across 42 microbiome datasets, PreLect demonstrated superior classification capabilities compared to statistical methods and outperformed machine learning-based methods by selecting features with greater prevalence and abundance. A significant strength of PreLect lies in its ability to reliably identify reproducible microbial features across varied cohorts. Applied to colorectal cancer, PreLect identifies key microbes and highlights crucial pathways, such as lipopolysaccharide and glycerophospholipid biosynthesis, in cancer progression. This case study exemplifies PreLect’s utility in discerning clinically relevant microbial signatures. In summary, PreLect’s accuracy and robustness make it a significant advancement in the analysis of complex microbiota data.
Subject terms: Next-generation sequencing, Clinical microbiology
Introduction
The microbiota, which comprises diverse microbial communities inhabiting the human body, plays a crucial role in maintaining host health and influencing disease1. Advanced sequencing techniques, like 16S rRNA gene sequencing2, have revolutionized our ability to profile the microbiome comprehensively and investigate the associations between microbial taxa and host health. Despite these advances, the inherent complexity of microbiota data, characterized by high dimensionality and sparsity, presents significant challenges for accurate modeling and interpretation. Therefore, in microbiota research, feature selection becomes essential for identifying potential biomarkers for diagnosis or prognosis3. Such identification may guide the development of targeted therapeutic interventions.
Previous studies have employed differential abundance analysis for identifying features that significantly differ in abundance between groups of samples. Methods such as DEseq24, edgeR5, and LEfSe6 have been used widely. However, these methods have come under scrutiny for their propensity to yield an inordinately high number of false positives7. In addition, univariate models such as statistical testing may result in false results if complex interactions are ignored. Efforts to overcome these limitations have seen the adoption of machine learning (ML) algorithms with multivariate models for feature engineering of microbiota data. Models such as LASSO8, random forest (RF)9, and eXtreme Gradient Boosting (XGBoost)10 adeptly capture intricate variable interactions, leading to more precise predictions. However, microbiota data are typically sparse; many taxa present in only a small subset of the samples, often containing 70–90% zeros11. This sparsity makes it challenging to determine whether a taxon is truly informative or merely a result of noise12. Furthermore, sparsity can lead to instability in feature selection, resulting in discrepancies in data interpretation13.
To tackle the challenges arising from the sparsity, researchers have looked towards alternative strategies. For instance, dictionary learning has been proposed to achieve a compact and informative data representation by linearly combining a set of essential functions or atoms in a sparse signal14. Nardone et al. introduced a method of sparse dictionary learning, SMBA-CSFS15, showing promising results in biological data applications. Mutual information16 is another popular approach in information theory, which has been implemented in various feature selection methods. For instance, Peng, Long, and Ding introduced a mutual information-based feature selection method called max-relevance and min-redundancy (mRMR)17. This method aims to minimize redundancy among features and maximize the dependency between a feature subset and a class label, which can lead to improved discriminative power of the selected features. Another noteworthy feature selection method is ReliefF18, which can handle sparse data. This approach randomly selects instances and evaluates the differences in feature values between the current instance and its nearest neighbor, both from the same class and different classes. The feature importance scores are then calculated as the average differences over all instances.
Sparse learning primarily employs L1-regularization, as seen in the LASSO method that sets regression coefficients of non-informative features to zero for facilitating automatic feature selection. Several LASSO variants have since been proposed, such as LAD-Lasso19, resistant to heavy-tailed errors or outliers in response, and sparse group Lasso20, capable of identifying sparse sets by adopting the L1 and L2 norm penalty. The sparse group Lasso has been introduced to generate group-wise and within-group sparsity, facilitating the identification of important groups and essential variables within groups21. However, a challenge in microbiota research is the varying feature selection across different cohorts, influenced by diverse microbial compositions and the inherent high prevalence observed in the original cohort.
To address these gaps, our study introduces PreLect, which incorporates a prevalence penalty to eliminate irrelevant features and enhance the reliability and reproducibility of feature selection in microbiome research. Our approach offers a more robust and accurate feature selection process, leading to a deeper understanding of microbial interactions and potential disease biomarkers. We benchmarked PreLect across multiple microbiome datasets and compared it to other statistics and ML-based methods. Our study emphasizes PreLect’s potential to enhance our grasp on microbiota’s role in health and disease, opening pathways for developing microbial markers in clinical settings.
Results
Assessing PreLect’s efficacy in an ultra-sparse dataset
In this study, we developed PreLect, an embedded feature selection framework that utilizes the force of regularization rate (lambda, λ) to select informative features (Fig. 1). More specifically, PreLect incorporates a prevalence penalty to discourage the selection of low-prevalence (local) features effectively. Therefore, it is particularly beneficial when dealing with sparse data. To demonstrate its effectiveness, we compared PreLect with other popular feature engineering methods, including LASSO, SVMLASSO, elastic net (EN), RF, XGBoost, and mutual information (MI), using an ultra-sparse dataset (“real-sim” in LIBSVM), which contains only 0.24% non-zero values.
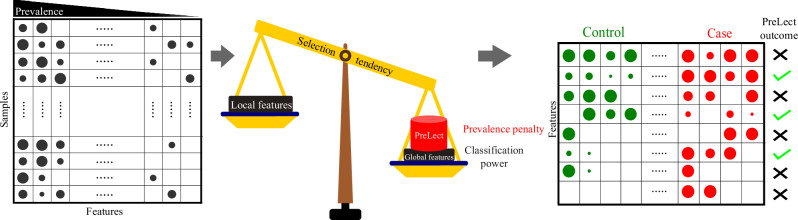
Fig. 1. PreLect framework for microbial data analysis.
This diagram illustrates the PreLect framework, a process for identifying significant microbial features in sparse, high-dimensional datasets. The workflow begins with inputting a microbial data table, which details the microbial characteristics of each sample. PreLect employs an inverse prevalence penalty to ensure the universality of selected features. The final output is a selection of microbes with significant classification ability to distinguish between patient groups and consistency across different cohorts, which are then used in further analyses.
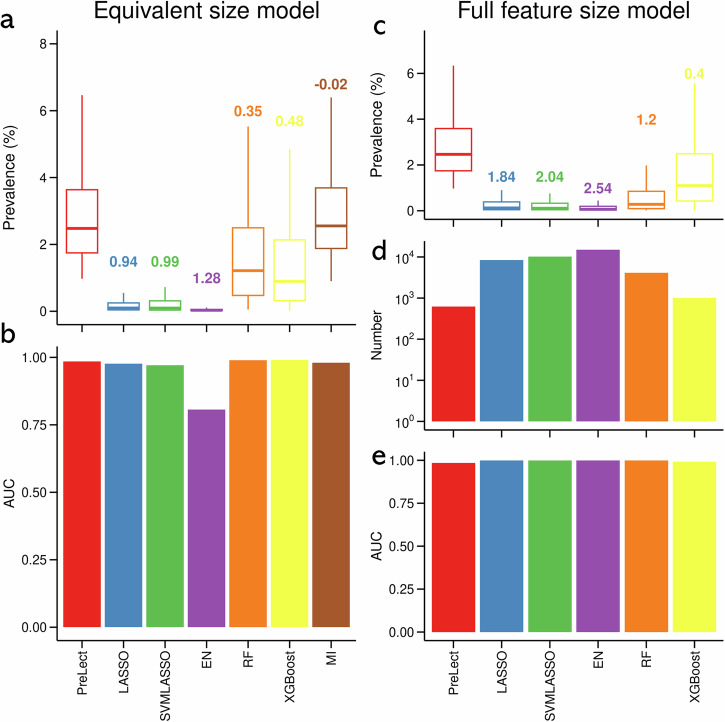
Initially, we applied the six methods, limiting the number of selected features in each to match that of PreLect to evaluate the universality and performance of the features selected by PreLect. Our results revealed that PreLect had a slightly lower but comparable prevalence than MI (median prevalence: 2.584% vs. 2.667%; Fig. 2a) but a higher prevalence than the other five methods. Among the six methods evaluated, five demonstrated similarly high areas under the receiver operating characteristic curve (AUC), as shown in Fig. 2b (LASSO: 0.976, SVMLASSO: 0.971, random forest (RF): 0.989, XGBoost: 0.991, mutual information (MI): 0.98), all comparable to PreLect’s AUC of 0.985. Elastic net (EN), however, displayed the lowest performance with an AUC of 0.806. Furthermore, when using the full feature set, which is the feature set selected by each method with parameters that achieved the best performance, PreLect outperformed the other approaches by demonstrating both the highest prevalence (Fig. 2c) and the smallest feature set size (Fig. 2d; 618 features). Despite the L1-based methods having feature sets approximately ten times larger than PreLect, they achieved only marginally better AUC scores compared to PreLect, as shown in Fig. 2e (PreLect: 0.985, LASSO: 1.0, SVMLASSO: 1.0). In summary, PreLect offers substantial advantages over the other methods by effectively extracting global features (high prevalence) and providing sufficient discrimination in sparse non-microbiome data.
Fig. 2. A comprehensive evaluation of feature prevalence and classifier efficacy across different feature selection algorithms on an ultra-sparse dataset.
a Feature prevalence in ‘equivalent size model.’ This panel showcases the distributions of feature prevalence when each algorithm is restricted to an equal number of features. Embedded values represent Cohen’s d, accentuating the differential prominence of PreLect compared to other methods. A positive Cohen’s d value indicates that the features selected by PreLect have a higher prevalence compared to those chosen by the corresponding benchmarking method. b Classifier performance in ‘equivalent size model.’ The AUC scores are displayed here, indicating the classification capability for each algorithm under the model that holds feature counts consistent across methods. c Feature prevalence in ‘full feature size model.’ In contrast to panel a, this representation of the non-zero importance features selected by each algorithm. d Feature counts in ‘full feature size model.’ A logarithmic scale captures the number of features chosen by each algorithm, providing insights into their inherent feature selection tendencies. e Classifier efficacy in ‘full feature size model.’ Presented here are the AUC scores that quantify the discriminatory power of each algorithm, this time under the setting where no limitations are imposed on feature count. The ‘equivalent size model’ limits feature assessment to PreLect’s selection count, while the ‘full feature size model’ uses each method’s typical feature count. The prediction performance of all benchmarking methods was assessed using logistic regression, employing a 7/3 train-test split ratio. The evaluation was conducted on the testing set to ensure the accuracy and reliability of the results. Detailed descriptions of the computational issues encountered are provided in Supplementary Note 3.
PreLect captures the universal and critical features across 42 microbiome datasets
In our detailed exploration of microbiota research, we utilized PreLect on a broad collection of 42 microbiome datasets. Our analysis compared PreLect’s efficiency against fifteen well-known approaches: six based on statistics (Fig. 3) and nine specialized in feature selection (Fig. 4). Our results were revealing: PreLect-selected features consistently demonstrated higher mean relative abundance across samples when compared to other methods in the majority of the datasets (Figs. 3b and 4b). Furthermore, PreLect’s feature set distribution differed notably from benchmarked statistics and ML methods (Supplementary Figs. 1 and 2). The features selected by PreLect in the balanced dataset (crc_zeller) are equally represented across case and control samples, showing no bias toward either group, as detailed in Supplementary Figs. 1 and 2. Furthermore, we investigated the sw_sed_detender dataset, which is imbalanced and shows the greatest difference in feature prevalence between the two conditions (seawater vs. sediment; Cohen’s d = 0.724). The findings suggest that PreLect consistently focuses on selecting features with high prevalence in both conditions (Supplementary Fig. 3). However, we observed a slight preference for PreLect to select features more frequently in the predominant case, in this instance, seawater. This observation indicates that the PreLect algorithm might be slightly influenced by the case-to-control ratio of the dataset, yet it remains robust in handling imbalanced datasets.
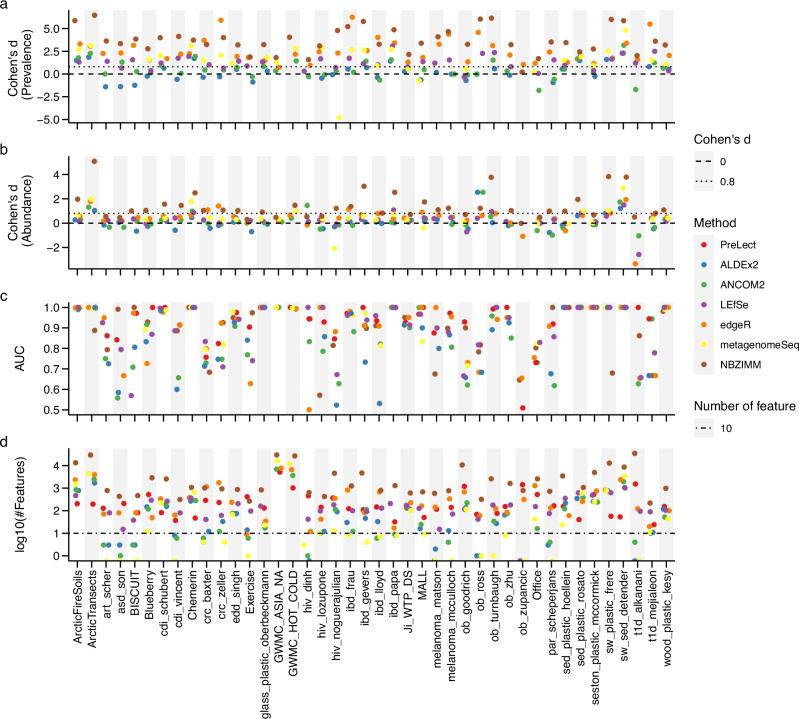
Fig. 3. Comparative analysis of PreLect with statistical methods across 42 microbiome datasets.
a Effect size of prevalence difference. Utilizing Cohen’s d, this panel represents the effect size of the prevalence difference between PreLect and the benchmarked methods. A positive Cohen’s d value indicates that the features selected by PreLect have a higher prevalence compared to those chosen by the corresponding benchmarking method. b Effect size of feature abundance difference. This depicts the relative abundance of selected features between PreLect and the other methods. A Cohen’s d value surpassing 0.8 (indicated by the dotted line) suggests that PreLect selects features with markedly higher prevalence or abundance than other techniques. c Classification performance. Here, the AUC score is extracted from a basic logistic regression model that utilizes the chosen features to differentiate between case and control samples. d Number of features selected by each method in the corresponding dataset. The horizontal dashed line serves as a reference, marking the selection of 10 features. Each point’s color corresponds to a specific method, as the inset legend details. The prediction performance of all benchmarking methods was assessed using logistic regression, employing a 7/3 train-test split ratio. The evaluation was conducted on the testing set to ensure the accuracy and reliability of the results. Detailed descriptions of the computational issues encountered are provided in Supplementary Note 3.
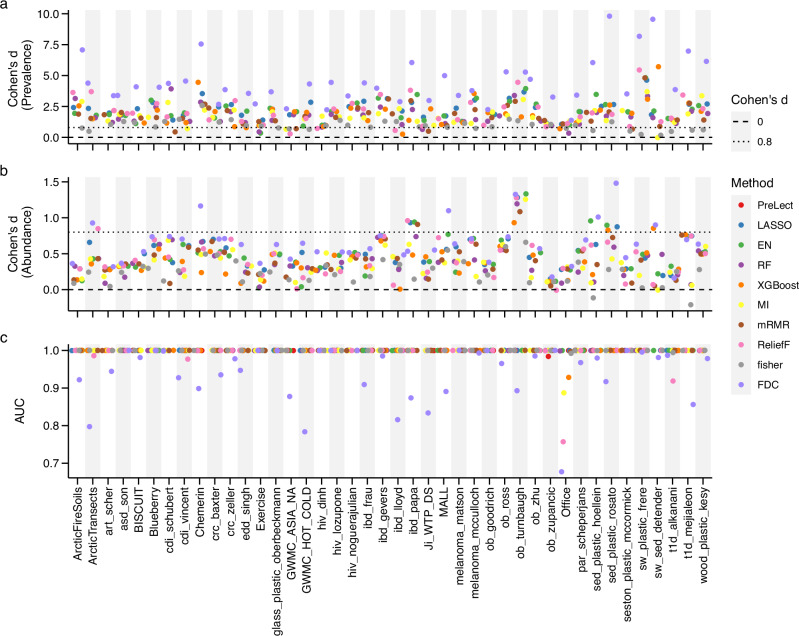
Fig. 4. Comparative analysis of PreLect with ML-based methods across 42 microbiome datasets in the ‘equivalent size model’.
a and b Effect size of prevalence and abundance difference. Cohen’s d measures the effect size of the prevalence difference between PreLect and other benchmarked machine learning methods. Values above 0.8 (dotted line) indicate a notable higher feature prevalence or abundance of PreLect. c Classification performance. The AUC values are derived from a naïve logistic regression model to classify case and control samples. Herein, all the ML-based methods use the top n weighted features. n equals the number of features selected by PreLect. The prediction performance of all benchmarking methods was assessed using logistic regression, employing a 7/3 train-test split ratio. The evaluation was conducted on the testing set to ensure the accuracy and reliability of the results.
Looking closely at the statistics-based methods, edgeR, LEfSe, and NBZIMM22 were found to choose feature sets with fewer occurrences compared to PreLect (Fig. 3a). This pattern was consistent across 41 datasets for edgeR, 29 for LEfSe, and 41 for NBZIMM. A high Cohen’s d value, above 0.8, indicated PreLect’s superior performance (as seen in Fig. 3a). This finding aligns with previous research7, suggesting that benchmarked methods might produce more false positives. In contrast, ALDEx2 and ANCOM2 focused on fewer features of higher prevalence (Fig. 3a and d). However, their broader distribution of feature prevalence fell short compared to PreLect. Notably, 14 datasets for ALDEx2 and 11 for ANCOM2 had fewer than ten features, which might have impacted their ability to detect significant differences. Our analysis shows that PreLect provides a balance: it selects enough features of high prevalence while ensuring accurate predictions, especially when compared to edgeR, LEfSe, and NBZIMM. By minimizing false positives, PreLect presents a dependable method for microbiome data analysis.
In our side-by-side comparison with other ML methods, we used a detailed grid search (explained in the “Methods” section) for the nine ML techniques to find the optimal parameters. Importantly, to maintain fairness, we matched the feature size to what was selected by PreLect. Our findings showcased PreLect’s dominance in feature prevalence over most ML methods across the 42 datasets. Except for MI, only one dataset exhibited a Cohen’s d above 0 (Fig. 4a). PreLect also consistently outperformed other methods in prediction accuracy across the datasets (Fig. 4c). Interestingly, the feature dispersion criterion (FDC)23, which is an unsupervised feature selection technique developed for text classification, selected features with an exceptionally low prevalence in all datasets. This observation suggests that the variance filter approach used by FDC may not be suitable for microbiota data, as low prevalence features can exhibit small fluctuations. We also evaluated the performance of PreLect against benchmarked methods using their respective default full feature sets to discern variations in feature selection across methods further. The features selected by PreLect still outperformed other methods in almost all 42 datasets (Supplementary Fig. 5).
To evaluate PreLect’s capability in selecting high-prevalence and informative feature sets, we synthesized five datasets by designating the top 100 most prevalent features as true positive features, with the remaining classified as negative features (detailed in the “Methods” section and Supplementary Fig. 6). The results demonstrate that ALDEx2, ANCOM2, metagenomeSeq, and MI are particularly effective at accurately identifying true positive features. This effectiveness is likely attributed to univariate models being highly sensitive to case-control signals. However, in real-world data, prevalent features often do not show strong case-control signals, as illustrated in Fig. 3. Despite this, PreLect consistently outperforms other ML-based methods in our comparisons.
In conclusion, PreLect empowers researchers to pinpoint critical features in intricate microbiome datasets, enriching our grasp on microbial community behaviors and their roles in various health conditions.
PreLect promotes consistent selection of microbe features in cross-cohort analyses
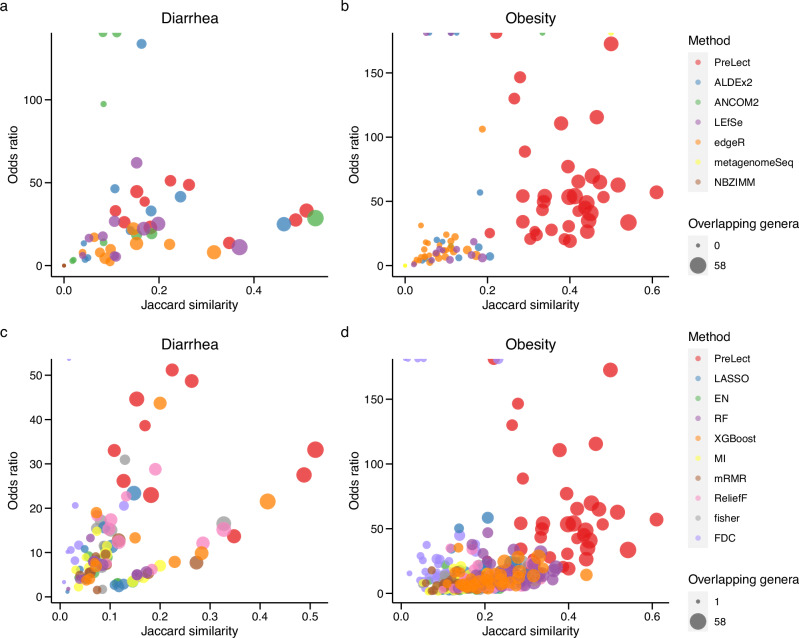
Consistency in identifying influential microbes from different datasets for the same health condition is vital for microbiology research24. Nevertheless, ensuring reproducibility across diverse studies remains a significant challenge. We addressed this issue by evaluating the consistency of microbe features selected by PreLect. We specifically targeted two prevalent diseases: diarrhea (with five datasets) and obesity (with nine datasets). We analyzed the overlapping genera across cohorts to assess the degree of consistency between different studies.
Using the well-known statistics methods, PreLect exhibited the highest overlapping number and odds ratio in the obesity datasets (Fig. 5a and b), suggesting that PreLect identified the most common results and the best match across the obesity datasets. For diarrhea, ALDEx2 and ANCOM2 displayed high odds ratios but low overlapping numbers in some comparisons. This could be because they only chose a small number of features they selected to start with. In addition, the Jaccard similarity revealed that the intersection ratio of ALDEx2 remained lower than that of PreLect. On the other hand, edgeR and LEfSe demonstrated large overlapping genera numbers but low odds ratios in diarrhea, suggesting a bias derived from a larger number of selected features by edgeR and LEfSe. Looking at machine learning methods that used the same number of features as PreLect (Fig. 5c and d), PreLect maintained the highest overlapping number, Jaccard similarity, and odds ratio in diarrhea and was almost dominant in obesity. The decision-tree-based models, including RF and XGBoost, exhibited a high Jaccard similarity due to their mechanisms but lower odds ratios. We observed that features present in multiple datasets tend to have higher prevalence within those datasets (Supplementary Fig. 7; diarrhea: r = 0.458, p-value < 2.2e−16; obesity: r = 0.528, p-value < 2.2e−16). This finding suggests that features with higher prevalence in one dataset are more likely to be universal across different datasets. Consequently, this supports the ability of PreLect to select features that are universally applicable, by leveraging the prevalence distribution within individual datasets. To conclude, our consistency analysis revealed that PreLect outperformed other benchmarked methods. This can be attributed to PreLect’s ability to select universally relevant features.
Fig. 5. Genera selection consistency across cohorts for diarrhea and obesity.
a and b Comparison with statistical methods. c and d Comparison with ML-based methods. The results derived from ML-based methods are in the ‘equivalent size model.’ The x-axis shows Jaccard similarity, which measures the similarity of shared genera; the y-axis shows the odds ratio that indicates the strength of association between selected genera and the condition (diarrhea or obesity). The size of data points reflects the number of shared genera, and color represents the feature selection method.
Decoding the microbial landscape of colorectal cancer with PreLect
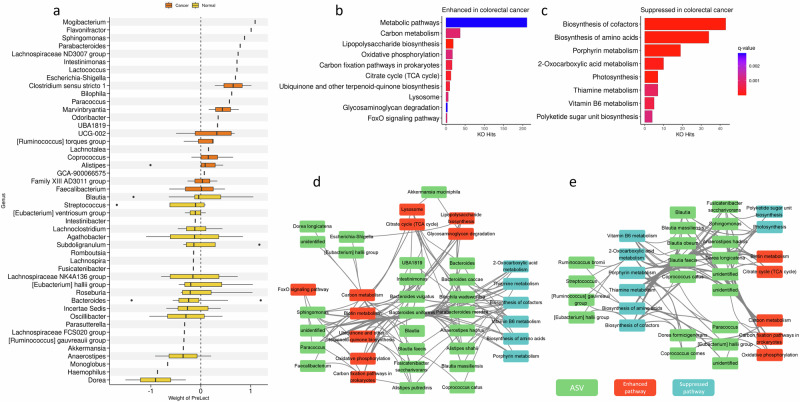
To further elucidate the biological significance of the features selected by PreLect, we investigated the microbes featured by PreLect in a colorectal cancer dataset25, which collected 50 health control, 63 adenoma, and 41 CRC patients. To detect microbial community shifts during cancer progression, we categorized adenoma patients as normal to maintain a clear distinction between healthy and cancer-affected cases. In the dataset, PreLect selected 231 ASV features, which can achieve an impressive AUC of 0.907 for distinguishing normal subjects from cancer patients (accuracy: 0.795, sensitivity: 0.952, specificity: 0.917, and F1 score: 0.934). Among the 94 genera represented by these ASVs, 46 genera have a prevalence higher than 0.5 in both normal and cancer samples, and their weights in the predictive model for tumor samples are shown in Fig. 6a. We observed that Flavonifractor, Bilophila, and Escherichia-shigella genera positively contribute to predicting tumor samples. This observation suggests that the patients with a high abundance of these three genera could potentially be predicted as having colorectal cancer. Indeed, Flavonifractor and Bilophila have been reported to be more abundant in CRC patients than in healthy controls26, and Escherichia-shigella has been identified as a pathogen of CRC27. On the other hand, the Akkermansia genus, which has a negative contribution in predicting tumor samples, has been reported as an anticancer probiotic with anti-inflammatory properties28; and the Ruminococcus gauvreauii group, which also negatively contributed to predicting tumor samples, has been reported to promote the activation of CD8+ T cells, reducing colon tumor growth29.
Fig. 6. Analysis of colorectal cancer dataset (Zeller).
a The weight distribution of PreLect-selected ASVs with prevalence higher than 0.5 in both normal and tumor samples. The genera with a median weight of selected ASV larger and smaller than zero are marked as normal (yellow) and tumor (orange), respectively. The enhanced (b) and suppressed (c) pathways in which PreLect-featured ASVs are involved. The x-axis shows the number of KOs identified as significantly enriched (b) and suppressed (c) in colorectal cancer by GSEA analysis. The color indicates the BH-corrected q-value for each pathway. d and e The bacteria-pathway correlation network, where orange nodes represent ASV labeled with genus or species names, and green nodes represent KO terms. The blue represents significant pathways according to (b and c, see the “Methods” section). The edges between the nodes represent correlations, with red indicating positive correlations, blue indicating negative correlations, and green indicating KOs belonging to the same pathway.
To understand the biological implications of these selected features, we examined the microbiome-based functional alterations between normal and colorectal cancer using gene set enrichment analysis (GSEA). We found that carbon metabolism had the highest number of hits in the enhanced pathway of CRC, with carbon fixation and the citrate cycle also enriched in CRC, suggesting that gut bacteria might have adapted to the abnormal energy metabolism environment in CRC30 (Fig. 6b). We also observed that lipopolysaccharide (LPS) biosynthesis is the second highest enhanced pathway in CRC. The LPS is a major component of the outer membrane of Gram-negative bacteria and a classic inflammatory activator known to trigger the Toll-like receptor 4 (TLR4)-mediated signal transduction31. In addition, oxidative phosphorylation emerged as the third highest enhanced pathway in CRC. Previous studies have reported that enteric bacteria, such as enterotoxigenic Bacteroides fragilis (ETBF) and Helicobacter pylori32, can induce reactive oxygen species (ROS) production in colonic epithelium, causing DNA damage that is considered to initiate CRC progression. Furthermore, our analysis revealed that relevant KOs in Vitamin B1, B6, and B12 were inhibited in CRC, corresponding to the Thiamine metabolism, vitamin B6 metabolism, and porphyrin metabolism pathways, respectively (Fig. 6c), suggesting that the bacteria involved in these pathways may help maintain gut microbiome environment to prevent CRC progression.
Our results indicated that oxidative phosphorylation was enriched in cancer and had a higher association with, Sphingomonas, and Paracoccus (Fig. 6d). In addition, our analysis revealed enrichment of catalase (K03781) and superoxide dismutase (K04564) KOs in the FoxO signaling pathway of CRC (Supplementary Fig. 8), which exhibited a strong correlation with the taxa mentioned above, indicating that these three taxa may be involved in ROS production. Overall, PreLect highlights the potential roles of specific bacterial taxa, providing insights into potential therapeutic targets and diagnostic markers.
Expanding the horizons of PreLect: beyond microbiome to microRNA transcriptome, binary to multi-class domains and regression task
To elucidate the PreLect algorithm’s adaptability, we explored its potential in a broader spectrum of omics data types. We collected shotgun data from six CRC studies and identified differential features between cancer and normal samples, comparing these with ML-based benchmark methods as shown in Supplementary Fig. 9. Under the “Equivalent Size Model” features selected by PreLect demonstrated the highest consistency across the six datasets. Conversely, in the “full feature size model”, due to the large number of features selected by elastic net (EN), it exhibited the highest consistency in cross-cohort analysis. Additionally, our pursuit led us to integrate the PreLect model with microRNA (miRNA) data. This data, inherently sparse, was derived from a comprehensive collection of 14 distinct cancer varieties archived in the cancer genome atlas (TCGA) database (Supplementary Fig. 10). In a comparative analysis with MI and mRMR—both of which recognize more abundant features—PreLect stood out by manifesting paramount prevalence and an unparalleled classification prowess. This was particularly evident when juxtaposed against selections made via other machine learning paradigms (highlighted in Supplementary Fig. 11). Such outcomes accentuate PreLect’s adeptness in proficiently navigating diverse sparse data realms, which it achieves by leveraging the prevalence penalty.
Diving deeper into the realm of classification, we endeavored to extend PreLect’s versatility to accommodate multi-class scenarios. Our methodology incorporated a ‘one-vs-rest’ stratagem (detailed in Supplementary Note 1). In our quest for feature selection, two distinct paradigms were adopted: the ‘intersection,’ wherein features were unanimously selected by all classifiers, and the ‘union,’ where a feature’s selection by even a single classifier sufficed. Post this selection, rigorous validation was ensured using multiclass logistic regression on both CRC and IBD datasets (as depicted in Supplementary Fig. 12). A noteworthy observation was the superior efficacy of the union sets over their intersection counterparts, a phenomenon potentially attributed to the richer informative content of the union feature set. The union set’s mean AUC in IBD notably surpassed the 0.8 threshold (see Supplementary Fig. 12). A caveat, however, emerged in the form of inconsistent performance metrics across CRC datasets. While PreLect showcased commendable proficiency on the CRC dataset sourced from Zeller, it faltered in the Baxter-derived dataset. Such disparities underscore both the promise and challenges for PreLect in multi-class terrains, emphasizing the imperative for continued refinement.
Finally, we expanded PreLect to include regression capabilities, as detailed in Supplementary Note 2. To test this, we gathered data from four studies on overweight individuals and used human gut 16S data to regress BMI. We also benchmarked the regression version of PreLect against other methods and compared it to PreLect’s performance in binary classification tasks. The results indicate that the penalty term strategy of PreLect’s regression version is effective, and the features it selects share similar characteristics with those identified in classification tasks (Supplementary Fig. 13).
Discussion
We have meticulously engineered the PreLect framework, seamlessly integrating a prevalence penalty. This innovation addresses the dual challenges of feature sparsity and high dimensionality intrinsic to microbiota data, significantly elevating the feature selection process in the domain of microbiome research. Comprehensive assessments of PreLect across various microbiota datasets showcased its superiority over traditional statistical paradigms and ML-driven feature selection methodologies. Our investigations shed light on the inherent shortcomings of conventional methods, notably the propensity to yield an elevated count of false-positive identifications. Furthermore, methods grounded in univariate statistics, particularly those hinging on “differential expression,” often grapple with delineating the complex interplay among variables, culminating in data interpretation disparities. When examining ML-centric approaches, the specter of overfitting looms large, gravitating towards the selection of low-prevalence, localized features. This predisposition accentuates the reproducibility quandary across diverse cohorts. The PreLect architecture deftly sidesteps this pitfall by assimilating a prevalence penalty, positioning it as a nimble, empirically informed alternative to prevalent-filtering paradigms. Additionally, it is common practice to filter out features with low prevalence prior to applying conventional feature selection methods, such as LASSO and the Wilcoxon test, with the aim of enhancing their performance. Notably, even with this preprocessing step, the performance of these two conventional methods does not improve (Supplementary Fig. 14). On the other hand, using prevalence alone to select features does not yield satisfactory classification performance (Supplementary Fig. 15). Consequently, PreLect consistently outperforms these two preprocessing strategies, further underscoring its effectiveness in selecting informative features.
Nevertheless, like all frameworks, PreLect’s efficacy is occasionally modulated by the unique prevalence distribution intrinsic to specific datasets. An illustrative case in point is the ‘ob_zupancic’ dataset, which manifested a comparatively subdued AUC value (Supplementary Fig. 16). Intriguingly, the prevalence spectrum of ‘ob_zupancic’ is punctuated by two prominent peaks, deviating from the typical distribution observed in other datasets. This observation hints at a potential Achilles’ heel in PreLect’s design: its performance might wane in settings dominated by a multimodal prevalence distribution.
In summary, our primary contribution is the introduction of PreLect, an L1-based model adept at identifying features with significant biological relevance. While our model places importance on prevalence, potentially affecting its predictive performance, it still yields results comparable to other leading methods. A key strength of our approach is its consistency across various cohorts, ensuring reproducibility in microbiome studies and the analysis of other types of sparse data. Our efforts have enhanced the understanding of microbial interactions and the identification of potential disease biomarkers. We hope that our research serves as a foundation for further studies, promoting a deeper understanding of the role of microbiota in health and disease and supporting the development of practical microbial markers for clinical applications.
Methods
Dataset collection and preprocessing
To evaluate the effectiveness of PreLect in analyzing sparse data, we employed the binary dataset, called real-sim, from LIBSVM33. This dataset is derived from a text categorization problem and consists of 20,958 features and 72,309 observations (samples). Notably, only about 0.24% of the values in the dataset are non-zero, making it an excellent example of sparse data. Each feature represents a term’s occurrence frequency within a document, and each observation corresponds to a specific document. The objective of the binary classification is to categorize the documents into two distinct classes based on their content.
We further incorporated 38 microbiome datasets from Nearing’s study34. These datasets were obtained from various sources, including humans, mice, soil, marine environments, wastewater, and buildings. The majority of these datasets were processed using the QIIME2 version 2019.7 standard operating procedure. Primers were removed using cutadapt and stitched by QIIME2 VSEARCH. The initial dataset comprised 38 entries, but two colorectal cancer datasets were excluded due to duplication with other datasets we independently collected. The complete list of included datasets is presented in Supplementary Data 1.
Additionally, we gathered 16S rRNA sequencing data from published studies focusing on Colorectal Cancer (CRC), Inflammatory Bowel Disease (IBD), and Melanoma. These datasets (crc_baxter, crc_zeller, ibd_frau, ibd_lloyd, melanoma_matson, and melanoma_mcculloch) were processed using the dada2 pipeline (v1.16.0), which has included stitching into its standard pipeline. For CRC analysis, we used two cohorts: Zeller et al. (PRJEB6070)25 and Baxter et al. (SRP062005)35, comprising French and international samples, respectively. Both datasets include samples from healthy individuals and colorectal cancer patients. To maintain a clear distinction between healthy and cancer-affected cases, we treated adenoma (precancerous) samples as normal.
We obtained IBD data from two sources: the Human Microbiome Project (PRJNA398089)36 and Frau et al. (PRJEB38969)37. The Human Microbiome Project includes a diverse collection of samples from individuals with IBD, specifically Crohn’s disease (CD) and ulcerative colitis (UC), as well as healthy controls. Frau et al.‘s dataset comprises samples from a European cohort featuring CD and UC patients alongside healthy individuals. In our analysis, we collectively classified CD and UC samples as IBD, comparing them against the healthy control group.
For Melanoma analysis, we collected two distinct cohorts from McCulloch et al. (PRJNA762360)38 and Matson et al. (PRJNA399742)39, respectively. The McCulloch et al. dataset consists of samples from a US cohort, encompassing both immunotherapy responders and non-responders. The Matson et al. dataset provides data on a diverse population, classifying melanoma patients into case and control groups according to their response to PD-1 therapy. Matson et al. contribute another dataset, focusing on the association between the gut microbiome and melanoma in a separate cohort.
We excluded samples with sequencing depths lower than 50,000 reads in these six independent datasets. Subsequently, we processed raw sequences using the dada2 pipeline (v1.16.0)40 with default settings in R (version 3.6.3) to generate amplicon sequence variants (ASVs) and assign taxonomy using the SILVA (v138.1) short-subunit reference database41. Following this, we normalized the abundance tables of ASVs for each dataset using variance stabilizing transformation (VST) from the DESeq2 package4, VST is specifically applied to normalize samples within a dataset instead of its features. The primary goal of VST is to stabilize the variance across samples by transforming the data so that the relationship between the mean and variance becomes approximately constant. We then performed z-score standardization to normalize the features. This step ensures that features with different scales or units are comparable42 and do not disproportionately influence the model performance (Supplementary Fig. 17).
PreLect algorithm
In the context of high-sparsity settings, we consider the feature selection problem for a dataset (, ) with , where represents the sample with features and denotes binary labels. We introduce an innovative feature selection framework—PreLect—that integrates feature prevalence with LASSO in logistic loss (binary cross-entropy, BCE) by minimizing the following objective function:
| 1 |
where denotes the prevalence of feature , which is the proportion of non-zero sample numbers to the total sample numbers for each feature . denotes the weight of feature . The term represents a modified L1-norm regularization that uses feature prevalence to address the high sparsity problem. The user-defined parameter, lambda (λ), represents the intensity of the regularization term intensity and is used as the cut-off in the feature selection procedure. A detailed process for determining an appropriate λ value to select informative features in the real-sim dataset is depicted in Supplementary Fig. 18.
To solve the non-differentiable convex optimization problem arising from the L1-norm, we employ the proximal gradient descent (PGD) algorithm. PGD is particularly suitable for solving optimization problems involving non-differentiable convex functions. Using the proximal operator, PGD can effectively optimize the objective function while handling non-differentiability. PGD calculates intermediate weights sequentially and updates the new weights at each iteration using the soft-threshold function:
| 2 |
where is the intermediate weight during the kth iteration for feature . As weight updates also involve determining the learning rate, we introduced the root mean square propagation (RMSprop), combined with prevalence, to adjust each weight’s learning rate. RMSprop is a method that adaptively optimizes learning rate the learning rate for each parameter based on the magnitude of its gradients. The RMSprop function in PreLect is described as
| 3.1 |
| 3.2 |
where is the basic learning rate (default: 0.001), is the discounting factor for the history and present gradient (default: 0.9), and is a small constant for numerical stability (default: 10−8). The PreLect training terminates after a maximum of 100,000 iterations or when convergence is achieved with an error <10−4.
Lambda optimization strategy
PreLect leverages the regularization parameter, λ, to identify informative features. Therefore, we proposed a two-layer scanning procedure to select an appropriate λ. For each dataset, we initially conducted a preliminary scan of λ values ranging from 10−10 to 1, with intervals of 0.1, using the entire dataset. Based on the initial scan, we established a lower bound, defined as the λ value retaining fewer than 90% of the features, and an upper bound, defined as the λ value retaining fewer than ten features. Subsequently, we divided the region between the lower and upper bounds into 50 equally sized segments and performed a 5-fold cross-validation (CV) for each λ value within this refined range. The PreLect algorithm offers several evaluation metrics, including the area under the precision-recall curve (AUCPR), the Matthews correlation coefficient (MCC), convergence, and minimal BCE loss, to facilitate the selection of a suitable lambda that captures informative features.
This study employed the BCE loss to ascertain the optimal λ value. Specifically, we pinpointed the inflection point on the BCE loss curve, where it transitions into a gentle slope towards the pull-up region on the curve. PreLect used the segmented regression algorithm to identify this inflection point. The segmented regression algorithm applied a decision tree regression to detect the segmented signal at the k-point, which users can define according to the gradient of the BCE loss curve. In this study, we defined k, which varies from 5 to 7 for different datasets (see Supplementary Data 1), and the first break point was denoted as the inflection point. Consequently, PreLect identified the λ corresponding to the inflection point as optimal for selecting informative features. This inflection point balances regularization strength and model loss, ensuring the selection of meaningful features while avoiding overfitting.
Benchmarked methods
To assess the effectiveness of PreLect, we compared it with six statistics-based methods: ALDEx243, ANCOM244, edgeR5, LEfSe6, metagenomeSeq11 and NBZIMM22; and nine ML-based methods: LASSO8, EN45, RF9, XGBoost10, MI16, mRMR17, Relief-F46, Fisher Score47, and FDC23. The details of these benchmarked methods are described in Supplementary Note 3. All the statistics-based methods use raw count table as input except edgeR and AMCOM2, which use pseudo count. The input data for ML-based benchmarked methods were processed through z-standardization and variance stabilizing transformation (VST). Prior to performing VST, we add a pseudo count to aid in data smoothing, ensuring stable transformation. Furthermore, in comparing PreLect with ML-based methods, we employed two criteria to determine the number of features selected by each method. (1) Equivalent size model: only the top n weighted features of each method were used for comparison, where n is the number of features selected by PreLect. (2) Full feature size model: the default number (non-zero importance features) of features for each method was applied.
The parameters for ML-based methods were tuned using a “grid search” strategy through a five-fold CV procedure. We adopted evaluation metrics such as AUC, AUCPR, and accuracy. We determined the final parameter set via a voting procedure, focusing on the combination boasting the highest mean metric value. In cases where multiple parameter combinations received equal votes, priority was assigned to the combination with the highest prevalence among selected features.
In summary, our benchmarking of PreLect against a diverse array of statistical and ML-based feature selection methodologies offers an extensive evaluation of its capabilities. Through this meticulous comparison, we elucidated the potential merits of PreLect, emphasizing its prowess in feature identification and subsequent enhancement of model performance for downstream analyses.
Cross-cohort consistency analysis
In this study, we conducted a cross-cohort consistency analysis based on data from Nearing’s research, which includes five independent datasets related to diarrhea and nine cohorts of obesity (Supplementary Data 1). In each dataset, ASVs were classified taxonomically utilizing the SILVA (v138.1) short-subunit reference database41, subsequently aggregating their abundance at the genus level. We used benchmarked methods and PreLect to select features from all fourteen datasets. We compared the selected genera within each disease category, employing metrics like overlap count, Jaccard similarity, and odds ratio via pairwise comparisons. The primary objective of this analysis was to evaluate the robustness and reliability of the microbial features identified.
Synthetic data evaluation
To assess PreLect’s capacity in identifying prevalent and informative features, we analyzed five diverse datasets ArcticFireSoils, crc_zeller, ibd_lloyd, melanoma_mcculloch, and sw_sed_detender representing various diseases or ecological niches. We selected the top 100 prevalent features as true positives, enhancing case-control distinctions by sorting their non-zero values. The other features are considered as true negatives, with their non-zero values shuffled randomly to diminish biological signals. We compared PreLect with 15 benchmarking methods quantitatively using Precision and F1-score metrics (Supplementary Fig. 6).
Functional annotation and enrichment analysis of PreLect-selected ASVs
We conducted a series of functional enrichment analyses to elucidate the enriched and suppressed functions associated with the featured microbes in colorectal cancer. We first utilized PICRUSt2 (phylogenetic investigation of communities by reconstruction of unobserved states)48 to predict the Kyoto encyclopedia of genes and genomes (KEGG) orthologs (KO) for each ASV. Next, we performed gene set enrichment analysis (GSEA)49 to assess the activity of the KOs associated with PreLect-selected ASVs in cancer. During the GSEA, we arranged the ASVs in descending order based on their log-fold changes (logFC) when comparing cancer to normal samples. The enrichment score was then calculated by hitting the PreLect-selected ASVs associated with the tested KO. KOs with a z-score > 2, as determined by GSEA organized by descending and ascending fold change values, were classified as being enriched in cancer and normal conditions, respectively.
Subsequently, using the KEGGREST package50, we interrogated the KEGG database to identify pathways associated with the predicted KOs. Subsequently, we performed Fisher’s exact test to assess the enrichment of KOs enriched in cancer or normal for each pathway. Pathways with significant enrichment of cancer-enriched KOs were termed ‘enhanced in cancer.’ Those enriched with normal-enriched KOs were labeled ‘suppressed in cancer. P-values obtained from Fisher’s exact test underwent correction using the Benjamini and Hochberg (BH) method, with a set q-value threshold of <0.05.
Construction of bacteria-pathway association network
We utilized significant KO terms derived from CRC’s enriched and suppressed pathways to build the bacteria-pathway association network, as identified through GSEA. We measured the correlation of normalized abundances between these KO terms and the selected ASVs using the Pearson correlation coefficient (PCC) after centered log-ratio transformation. The network was generated by applying a threshold of an absolute PCC value > 0.5 and a significance level of BH-adjusted q-value < 0.001.
miRNA dataset preprocessing
The miRNA profiling data were obtained from TCGA encompassing 14 distinct cancer types. We specifically opted for the ‘Isoform Expression Quantification’ data type, processed using the BCGSC pipeline51. To discern the 5’ end isoforms, we relied upon miRBase (v21)52. Isoform expression was quantified by calculating reads per million miRNA mapped (RPM). To evaluate PreLect’s performance, we gathered ‘primary tumor’ and ‘normal solid tissue’ samples for the binary classification task, and we employed z-standardization before PreLect and all ML-based benchmarked methods. In PreLect, a 5-fold cross-validation was performed during the lambda scanning process, using segmented regression with k = 5 to determine the optimal lambda across all cancer types. Simultaneously, a grid-search approach, as described earlier, was used for the ML-based methods.
Shotgun dataset preprocessing
The six benchmarking datasets30,53–56 consisted of raw sequencing FASTQ files obtained from the NCBI SRA Archive. We processed these files by removing low-quality or host-contaminated reads using KneadData (version 0.10.0) with the hg37 human reference genome and default settings. Taxonomic profiling and quantification were then performed using MetaPhlAn 4.0.257 (with the database updated as of October 2022). In this study, our analysis focused on the class, order, family, genus, and species levels. Relative abundance data was standardized using the z-score method before being input into PreLect and other benchmarking methods for comparison.
BMI dataset preprocessing and analysis
In the four datasets58–61, with the exception of Kennedy (KM_2023), where the ASV count table was directly sourced from the author’s GitHub repository, the remaining datasets were processed using the dada2 pipeline to generate ASV tables from raw sequence files. Taxonomic assignments were carried out using the SILVA database. For consistency across cohorts, raw ASV counts were aggregated at the genus level and underwent variance-stabilizing transformations. In the classification analysis using PreLect, we categorized samples with a BMI > 30 as obese and the rest as normal. We then compared the feature selection patterns of PreLect with those of 15 other benchmarking methods.
Limitations
In this version of PreLect, we have adopted the sub-gradient method of proximal gradient descent (PGD) to address the non-differentiability issue associated with L1 regularization. However, the PGD algorithm is not the most efficient approach. To enhance optimization speed, we incorporated RMSprop, yet thousands of iterations are still required to achieve convergence for some larger datasets, such as real-sim and gwmc_hot_cold. Additionally, to determine the optimal lambda value, PreLect employs k-fold cross-validation (CV) scanning across multiple lambda values, which significantly extends the computational time. Although GPU acceleration can speed up PreLect, larger datasets like gwmc_hot_cold, which includes 1021 samples and 92,126 taxa, still require several days to process. Moving forward, we plan to incorporate a more efficient L1 regularization solver to improve runtime efficiency. Moreover, while PreLect has shown the capability to handle bias derived from imbalanced datasets, it still struggles with datasets that have an extremely skewed case-control ratio. To enhance PreLect’s generalizability, developing a solution to effectively address dataset imbalance is a necessary future step.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This research was funded by the National Science and Technology Council in Taiwan (NSTC 109-2221-E-010-014-MY3 and NSTC 112-2221-E-A49-106-MY3) and Ministry of Health and Welfare in Taiwan (MOHW112-TDU-B-222-124013 and MOHW111-TDU-B-221-114007). We thank Dr. Hsuan-Cheng Huang at the Institute of Biomedical Informatics, National Yang Ming Chiao Tung University, for giving us valuable comments to improve this work. The results of miRNA section data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Author contributions
Conceptualization: C.-C.L. Methodology: C.-C.L., Y.-C.C., Y.-Y.S., T.-Y.C., M.-F.W., C.-C.H. Implementation: Y.-C.C. Supervision: C.-C.L. Writing—original draft: Y.-C.C. Writing—review & editing: C.-C.L. and Y.-C.C.
Data availability
The accessions of 42 microbiota datasets, shotgun and BMI study are listed in (Supplementary Data 1). The processed datasets, which include microbiota and miRNA, are available at zenodo [10.5281/zenodo.10062236].
Code availability
The data processing and analysis code is available in this GitHub repository at https://github.com/YinchengChen23/PreLect_manuscript. The PreLect toolkit is written in Python using the Pytorch library. The code with the User Manual is available on GitHub at https://github.com/YinchengChen23/PreLect under the MIT license. Additionally, we have provided an R package for the PreLect toolkit, which implements multi-class classification and time-to-event analysis, available at https://github.com/YinchengChen23/PreLectR/tree/main.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-024-00598-2.
References
- 1.Alhinai, E. A., Walton, G. E. & Commane, D. M. The role of the gut microbiota in colorectal cancer causation. Int. J. Mol. Sci.20, 5295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim, H., Kim, S. & Jung, S. Instruction of microbiome taxonomic profiling based on 16S rRNA sequencing. J. Microbiol.58, 193–205 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Chen, Z.-J. et al. Association of Parkinson’s disease with microbes and microbiological therapy. Front. Cell. Infect. Microbiol.11, 619354 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 1–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol.11, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, 1–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calgaro, M., Romualdi, C., Waldron, L., Risso, D. & Vitulo, N. Assessment of statistical methods from single cell, bulk RNA-seq, and metagenomics applied to microbiome data. Genome Biol.21, 1–31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc.: Ser. B (Methodol.)58, 267–288 (1996). [Google Scholar]
- 9.Breiman, L. Random forests. Mach. Learn.45, 5–32 (2001). [Google Scholar]
- 10.Chen, T. Q. & Guestrin, C. Xgboost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 785–794 (Association for Computing Machinery, 2016).
- 11.Paulson, J. N., Stine, O. C., Bravo, H. C. & Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods10, 1200–1202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee, K., Chen, J. & Zhan, X. Adaptive and powerful microbiome multivariate association analysis via feature selection. NAR Genom. Bioinform.4, lqab120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, L. et al. Utilizing stability criteria in choosing feature selection methods yields reproducible results in microbiome data. Biometrics78, 1155–1167 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aharon, M., Elad, M. & Bruckstein, A. K-SVD: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans. Signal Process.54, 4311–4322 (2006). [Google Scholar]
- 15.Nardone, D., Ciaramella, A. & Staiano, A. A sparse-modeling based approach for class specific feature selection. PeerJ Comput. Sci.5, e237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraskov, A., Stögbauer, H. & Grassberger, P. Estimating mutual information. Phys. Rev. E69, 066138 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Peng, H., Long, F. & Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell.27, 1226–1238 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Kira, K. & Rendell, L. A. A practical approach to feature selection. In Machine Learning Proceedings, 249–256 (Morgan Kaufmann, 1992).
- 19.Lambert-Lacroix, S. & Zwald, L. Robust regression through the Huber’s criterion and adaptive lasso penalty. Electron. J. Stat.5, 1015–1053 (2011). [Google Scholar]
- 20.Yuan, M. & Lin, Y. Model selection and estimation in regression with grouped variables. J. R. Stat. Soc.: Ser. B (Stat. Methodol.)68, 49–67 (2006). [Google Scholar]
- 21.Simon, N., Friedman, J., Hastie, T. & Tibshirani, R. A sparse-group lasso. J. Comput. Graph. Stat.22, 231–245 (2013). [Google Scholar]
- 22.Zhang, X. & Yi, N. NBZIMM: negative binomial and zero-inflated mixed models, with application to microbiome/metagenomics data analysis. BMC Bioinform.21, 1–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira, A. and Figueiredo, M. Efficient unsupervised feature selection for sparse data. 2011 IEEE EUROCON - International Conference on Computer as a Tool, 1–4 (IEEE, 2011).
- 24.Schloss, P. D. Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. MBio9, e00525–00518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeller, G. et al. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol. Syst. Biol.10, 766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai, D. et al. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front. Microbiol.10, 826 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stecher, B. The roles of inflammation, nutrient availability and the commensal microbiota in enteric pathogen infection. Metab. Bact. Pathog.3, 297–320 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Derrien, M., Belzer, C. & de Vos, W. M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog.106, 171–181 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Zhang, X. et al. Tissue-resident Lachnospiraceae family bacteria protect against colorectal carcinogenesis by promoting tumor immune surveillance. Cell Host Microbe31, 418–432.e418 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Yachida, S. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med.25, 968–976 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Wu, X. et al. Lipopolysaccharide promotes metastasis via acceleration of glycolysis by the nuclear factor-κB/snail/hexokinase3 signaling axis in colorectal cancer. Cancer Metab.9, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodwin, A. C. et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA108, 15354–15359 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, C.-C. & Lin, C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol.2, 1–27 (2011). [Google Scholar]
- 34.Nearing, J. T. et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun.13, 342 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter, N. T., Ruffin, M. T., Rogers, M. A. & Schloss, P. D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med.8, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frau, A. et al. Inter-kingdom relationships in Crohn’s disease explored using a multi-omics approach. Gut Microbes13, 1930871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCulloch, J. A. et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med.28, 545–556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res.41, D590–D596 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey, Y. N. et al. Overview of machine learning and deep learning concepts. In Machine Learning in the Oil and Gas Industry: Including Geosciences, Reservoir Engineering, and Production Engineering with Python 75–152 (2020).
- 43.Fernandes, A. D. et al. Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome2, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaul, A., Mandal, S., Davidov, O. & Peddada, S. D. Analysis of microbiome data in the presence of excess zeros. Front. Microbiol.8, 2114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc.: Ser. B (Stat. Methodol.)67, 301–320 (2005). [Google Scholar]
- 46.Kononenko, I., Šimec, E. & Robnik-Šikonja, M. Overcoming the myopia of inductive learning algorithms with RELIEFF. Appl. Intell.7, 39–55 (1997). [Google Scholar]
- 47.Gu, Q., Li, Z. & Han, J. Generalized Fisher Score for Feature Selection. In Proceedings of the 27th Conference on Uncertainty in Artificial Intelligence, 266–273 (Uncertainty in Artificial Intelligence, 2011).
- 48.Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol.38, 685–688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenenbaum, D. et al. Package ‘keggrest’ (R Foundation for Statistical Computing, Vienna, Austria, 2019).
- 51.Chu, A. et al. Large-scale profiling of microRNAs for the cancer genome atlas. Nucleic Acids Res.44, e3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic acids Res.47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng, Q. et al. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat. Commun.6, 6528 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Gupta, A. et al. Association of Flavonifractor plautii, a flavonoid-degrading bacterium, with the gut microbiome of colorectal cancer patients in India. MSystems4, 00438–00419 (2019). [DOI] [PMC free article] [PubMed]
- 55.Hannigan, G. D., Duhaime, M. B., Ruffin IV, M. T., Koumpouras, C. C. & Schloss, P. D. Diagnostic potential and interactive dynamics of the colorectal cancer virome. MBio9, 02248–02218 (2018). [DOI] [PMC free article] [PubMed]
- 56.Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med.25, 667–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol.41, 1633–1644 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Escobar, J. S., Klotz, B., Valdes, B. E. & Agudelo, G. M. The gut microbiota of Colombians differs from that of Americans, Europeans and Asians. BMC Microbiol.14, 1–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kennedy, K. M. et al. Parity modulates impact of BMI and gestational weight gain on gut microbiota in human pregnancy. Gut Microbes15, 2259316 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lippert, K. et al. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef. Microbes8, 545–556 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Somnuk, S. et al. Metabolic and inflammatory profiles, gut microbiota and lifestyle factors in overweight and normal weight young Thai adults. PLoS ONE18, e0288286 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accessions of 42 microbiota datasets, shotgun and BMI study are listed in (Supplementary Data 1). The processed datasets, which include microbiota and miRNA, are available at zenodo [10.5281/zenodo.10062236].
The data processing and analysis code is available in this GitHub repository at https://github.com/YinchengChen23/PreLect_manuscript. The PreLect toolkit is written in Python using the Pytorch library. The code with the User Manual is available on GitHub at https://github.com/YinchengChen23/PreLect under the MIT license. Additionally, we have provided an R package for the PreLect toolkit, which implements multi-class classification and time-to-event analysis, available at https://github.com/YinchengChen23/PreLectR/tree/main.