Abstract
Cutaneous squamous cell carcinoma (cSCC) is the second most common skin cancer, originating from the malignant proliferation of squamous epithelial cells. However, its pathogenesis remains unclear. To further explore the mechanisms underlying cSCC, we analyzed the data from one single-cell RNA sequencing study and discovered a significant upregulation of tryptophan 2,3-dioxygenase (TDO2) in the cancer-associated fibroblasts (CAFs). Nonetheless, the specific expression and potential biological significance of TDO2 in cSCC have not yet been reported. In this study, we confirmed that TDO2 is highly expressed in CAFs of cSCC. Clinical correlation analysis indicated that high TDO2 expression was significantly associated with poor tumor differentiation. Furthermore, increased TDO2 expression in cSCC correlated with reduced CD8 + T cell infiltration, suggesting its role in modulating immune responses. TDO2 inhibitors significantly reduced the size and number of tumors in mice and effectively increased CD8 + T cell infiltration. RNA sequencing analysis revealed that TDO2 inhibitors modulate immune cell activity and downregulate the PI3K-Akt signaling pathway. In summary, our study demonstrates that TDO2 + CAFs induce immune evasion by inhibiting CD8 + T cell infiltration in cSCC. Inhibiting TDO2 could enhance antitumor immune responses, providing a promising strategy to improve treatment outcomes in cSCC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-024-03921-0.
Keywords: Cutaneous squamous cell carcinoma; Tryptophan 2,3-dioxygenase; Cancer-associated fibroblasts; Immune escape
Introduction
Cutaneous squamous cell carcinoma (cSCC) ranks as the second most prevalent type of non-melanoma skin cancer (NMSC), constituting about 20% of all NMSC cases [1]. There has been a remarkable increase in the burden of SCC [2]. The disease predominantly affects the elderly, with a lifetime incidence rate ranging from 6 %to 11% [3], and the annual disease-specific mortality is estimated to be 1.5% to 2% [4]. Factors contributing to the increased incidence of cSCC include aging populations and enhanced skin cancer screening protocols [5]. Primary risk factors for cSCC include prolonged exposure to ultraviolet radiation, older age, male gender, and HPV infection [6]. Notably, high-risk cSCC patients experience a metastasis rate of up to 37% [7], with a median survival of less than two years for those with distant metastases [8], underscoring the critical need for early detection and intervention.
The development and progression of cSCC are driven by intricate interactions within the tumor microenvironment (TME), which comprises a diverse array of tumor and non-tumor cells, including immune cells, cancer-associated fibroblasts (CAFs), endothelial cells, and adipocytes [9]. These cells collaborate to promote tumor growth, dissemination, and treatment resistance through the secretion of cytokines, chemokines, extracellular matrix (ECM) components, and signaling molecules [10]. Mutations in keratinocytes, alterations in stromal cells, and changes in the dermal stromal support matrix all play pivotal roles in the pathogenesis of cSCC [11]. The dermal matrix provides structural support and facilitates the secretion of growth factors by infiltrating immune cells and CAFs, promoting neovascularization through endothelial proliferation. CAFs, as key components of the ECM, exert a profound influence on the TME by modulating tumor cell behavior and fostering an inflammatory, immune-suppressive, and pro-angiogenic milieu [12].
TDO2 serves as a principal rate-limiting enzyme in the tryptophan (Trp) metabolic pathway and is chiefly expressed in the human liver, where it plays a critical role in the breakdown of Trp [13]. TDO2 exhibits pan-cancerous properties, with high expression levels observed across various cancer types, establishing it as a potential therapeutic target. Numerous studies have associated elevated TDO2 expression with poorer patient prognosis [14]. Research has demonstrated that TDO2 predominantly drives the progression of several cancers, including liver, colorectal, glioma, and breast cancers, primarily through the Kynurenine (Kyn)/Aryl hydrocarbon Receptor (AHR) pathway [14–17]. Additionally, TDO2 contributes to tumor advancement by promoting immune escape [18], highlighting its significant role in molding the TME and facilitating cancer immune evasion. However, the exact functions and mechanisms of TDO2 in cSCC remain undefined.
In this study, we employed single-cell transcriptome data analysis and discovered high expression levels of TDO2 in CAFs. We further confirmed the co-expression of TDO2 with the CAF-specific marker alpha-smooth muscle actin (α-SMA) in cSCC patient tissues using in situ hybridization and immunofluorescence staining. This finding underscores the crucial role of TDO2 in regulating the tumor stroma. Subsequent investigations into the mechanism of TDO2 in cSCC and its correlations with clinical pathological features bolster the evaluation of TDO2 as a viable therapeutic target. Our results indicate that elevated TDO2 expression is associated with reduced immune cell infiltration in cSCC, suggesting that TDO2 may modulate tumor immune escape and progression by impacting immune cell functions, particularly the activity of CD8 + T cells. These insights pave the way for developing novel immunotherapeutic strategies against cSCC, potentially offering new therapeutic avenues for cSCC patients.
Material and methods
Single-cell sequencing analysis
A publicly accessible single-cell RNA sequencing (scRNA-seq) dataset [19] from cSCC patients was used, providing comprehensive transcriptomic profiles for exploring cell-specific expression patterns of TDO2. During the data preprocessing phase, stringent quality controls were applied, including removing low-quality reads and eliminating contaminated cells and doublets, thereby ensuring data integrity. An expression matrix was constructed using specialized sequencing analysis software, facilitating the quantification of gene expression levels across individual cells. Various cell types, such as fibroblasts and immune cells, were identified using established marker genes or single-cell clustering analysis techniques. The expression of TDO2 in these differentiated cell types is closely examined to reveal its distinctive expression patterns within cSCC.
RNA in situ hybridization
Tissues obtained from surgically resected cSCC specimens were fixed in 10% neutral-buffered formalin at room temperature for 16 to 24 h. After fixation, the tissues were dehydrated through a series of graded ethanol solutions, cleared in xylene, and then embedded in paraffin. The tissues were sectioned into 5 μm slices using a microtome (Leica Biosystems), transferred to microscope slides, and dried at 60 °C for 30 min on a slide warmer. Dewaxing was followed by a 10-min incubation in pretreatment solution A at room temperature to remove endogenous enzymes, followed by boiling in citrate-containing pre-reaction solution B for 15 min. To expose mRNA, sections were digested with protease at 40 °C for 15 min, then incubated with a gene-specific probe in hybridization solution at 40 °C for two hours. After hybridization, unhybridized probes were washed off, and signal amplification was performed using a cascade signal amplification technique based on nucleic acid-protein hybridization. A fast red substrate was added for an alkaline phosphatase-based color reaction, showing target RNA as red or green dots or patches. The mRNA in situ detection for each gene was performed manually using the PinpoRNA™ RNA in situ hybridization detection kit (PIF2000, GD Pinpoease). The probes targeted the 132-1177 base region of TDO2 (Cat. No.69991-A2, GD Pinpoease) and the 45-1242 base region of Actin alpha 2 (ACTA2) (Cat. No.591-A1, GD Pinpoease).
Multiplex immunohistochemical (mIHC) staining
mIHC staining was performed on formalin-fixed, paraffin-embedded (FFPE) samples from cSCC patients using the Opal IHC Kit (Akoya Biosciences) in accordance with the manufacturer’s instructions. Antibodies targeting TDO2, α-SMA, and CD8 were obtained from Abcam. The staining procedure began with peroxidase blocking, followed by sequential application of primary antibodies. HRP-conjugated secondary antibodies were used for detection, and fluorescent signals were developed with corresponding Opal dyes, selected to minimize spectral overlap. To prevent cross-reactivity, each antibody was applied in separate staining cycles, with microwave treatment used to strip bound antibodies between rounds. Antigen retrieval was performed using optimized buffers provided by Akoya Biosciences to ensure optimal epitope exposure. After staining, the slides were counterstained with DAPI, ensuring fluorescence signals were adjusted to avoid interference with Opal dyes. The samples were then mounted in antifade solution for imaging. Imaging was conducted on the Vectra multispectral platform, using optimized settings to maximize the signal-to-noise ratio and enable accurate quantification of fluorescence signals.
Isolation and culture of primary CAFs
Collected cSCC surgical resection tissues from consenting patients were immediately immersed in phosphate-buffered saline (PBS, Meilunbio) containing 10% antibiotics (penicillin, 100 units/mL; streptomycin, 100 mg/mL; Gibco). Under sterile conditions, the tissues were gently rinsed to remove excess blood, fat, and necrotic tissue. The tissues were then minced into approximately 1 mm3 fragments and transferred to a 15 mL centrifuge tube containing Type IV collagenase. The tube was incubated in a 37 °C water bath for 1 h with periodic agitation. After digestion, the supernatant was transferred to a new centrifuge tube containing complete culture medium and centrifuged at 1000 rpm for 5 min. The supernatant was discarded, and the cells and tissue fragments were resuspended in a small volume of complete culture medium and subsequently transferred to a culture flask for further cultivation. When cells reached 70% confluence, fibroblasts were separated from keratinocytes using 0.25% Trypsin–EDTA. Cell growth was regularly monitored, and medium changes or subculturing were performed as needed. Primary fibroblasts were grown in Dulbecco’s Modified Eagle Medium (DMEM, EpiZyme) supplemented with 20% fetal bovine serum (FBS, OriCell) and 1% antibiotics. Cell cultures were maintained at 37 °C with 5% CO₂ and 95% humidity.
Immunofluorescence
For indirect immunofluorescence (IF), fibroblasts were cultured on glass coverslips until they reached approximately 50% confluence. The cells were then fixed with 4% formaldehyde at room temperature for 15 min, washed three times with PBS, and permeabilized with 0.3% Triton X-100 in PBS at room temperature for 5 min. The cells were incubated overnight at 4 °C with primary antibodies against TDO2 and α-SMA (both from Abcam), which were diluted in 3% bovine serum albumin (BSA, Sigma-Aldrich) in PBS. After incubation, the cells were washed three times with PBS and then incubated with secondary antibodies, AF594 goat anti-rabbit IgG or AF488 goat anti-mouse IgG (both from Thermo Fisher Scientific), for 1 h at 37 °C. After another round of washing, the cells were incubated with DAPI (Sigma-Aldrich) for 5 min at 37 °C to counterstain the nuclei. Finally, the coverslips were mounted.
Immunohistochemistry staining
Sections were processed in a thermostatic oven at 60 to 65 °C for 30 min, sequentially deparaffinized with xylene and ethanol, and hydrated. Antigen retrieval was performed by heating the sections in antigen retrieval solution to boiling for 10 min, followed by natural cooling to room temperature. Three percent hydrogen peroxide was added dropwise and incubated in the dark for 15 min. Blocking solution was then added dropwise and incubated for 30 min. The sections were incubated overnight at 4 °C with primary antibodies targeting TDO2, CD4 (Abcam), CD8 (Abcam), FOXP3 (Abcam), CD206 (Cell Signaling Technology), and Ki67 (Cell Signaling Technology). After rinsing with PBS, the sections were incubated with horseradish peroxidase (HRP)-coupled secondary antibodies diluted in blocking buffer for 1 h. The slides were then stained with DAB chromogen and counterstained with hematoxylin. To distinguish between high and low expression groups of TDO2 + cells or CD8 + T cells, the median number of positive cells was determined from random fields across 30 samples. For correlation analysis between TDO2 + cells and other markers (CD8, CD4, CD206, and FOXP3), consecutive sections from the same patient samples were stained for each marker. Correlations were analyzed by selecting consistent fields of view across the sections.
Mouse model construction and treatment procedures
Seven-week-old SKH-1 mice were acquired from the Shanghai Public Health Clinical Center. An AUV-induced cSCC mouse model was established in this study. Mice in the ultraviolet radiation (UVR) group were irradiated using a daylight ultraviolet simulator (SUV1000, Sigma) equipped with UVA and UVB filters. The initial minimal erythema dose (MED) was set at 160 mJ/cm2 for UVB and 2520 mJ/cm2 for UVA. UV radiation was administered five times weekly for 24 weeks. As the mice developed increased tolerance to UVR, the radiation dose was incrementally increased to 1.625 MED over 20 weeks. At the conclusion of the experiment, cumulative doses reached 242.91 J/cm2 for UVA and 26.99 J/cm2 for UVB. Tumor diameters in the UVR group were measured three times weekly. Fifteen mice were divided into the untreated group (n = 5), TDO2 inhibitor (TDO2i) group (n = 5), and AHR inhibitor (AHRi) group (n = 5). For mice in the TDO2i group, the TDO2 inhibitor LM10 (AbMole) was dissolved in 2% dimethyl sulfoxide (DMSO) in sterile water and administered at a dose of 160 mg/kg per day by oral gavage, while the mice in the untreated group were fed the same concentration of DMSO (2%) in water from week 30[19]. For the AHRi group, the AHR inhibitor CH22319 (MedChemExpress) was intraperitoneally injected three times a week at a dose of 100 mg/mouse in a 200 mL total injection volume[20].
Flow cytometry analysis
Each sample was processed to generate single-cell suspensions, primarily achieved through enzymatic digestion. Tissues were incubated in DMEM containing Type IV collagenase and DNase (both from Sigma-Aldrich) at 37 °C for one hour. After dissociation, any remaining erythrocytes were eliminated using a red blood cell lysis buffer (Sigma-Aldrich), thereby purifying the sample. The purified cells were then washed twice with PBS to remove residual enzymes and debris. Following the washes, the cells were stained with specific antibodies (CD45, CD3, CD4, and CD8) according to the manufacturers’ protocols. These antibodies target unique cell surface markers, allowing for the identification and quantification of various cell populations using flow cytometry.
RNA isolation and library preparation
Total RNA was extracted using the Trizol reagent (Invitrogen) following the manufacturer’s guidelines. RNA purity and concentration were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), and RNA integrity was evaluated with an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA libraries were then synthesized using the VAHTS Universal V6 RNA sequencing (RNA-seq) Library Prep Kit, following the specified protocols. Transcriptome sequencing and analysis were conducted by OE Biotech Co., Ltd. (Shanghai, China).
RNA sequencing and differentially expressed genes analysis
For RNA-seq and the identification of differentially expressed genes, libraries were sequenced on an Illumina NovaSeq 6000 platform, producing 150 bp paired-end reads. An average of approximately 48.23 million raw reads per sample were generated. The raw reads in fastq format were processed with the fastp tool to remove reads of inferior quality, resulting in approximately 46.74 million clean reads per sample. Clean reads were aligned to the reference genome using HISAT2 for both the untreated and TDO2i groups. FPKM values of each gene were calculated, and read counts were obtained using HTSeq-count. Principal Component Analysis (PCA) was performed in R (v 3.2.0) to assess sample consistency. Differential expression analysis was conducted using DESeq2, with a q-value (adjusted p-value) threshold of < 0.05 and a |log2FoldChange|> 1 indicating significantly differentially expressed genes (DEGs). Hierarchical clustering analysis of DEGs was carried out in R (v 3.2.0) to elucidate gene expression patterns across groups and samples. Enrichment analyses, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses, were performed to identify significantly enriched terms based on the hypergeometric distribution using R (v 3.2.0). Column diagrams depicting the enrichment results were generated in R. Gene Set Enrichment Analysis (GSEA) was conducted using GSEA software to determine whether a predefined gene set was significantly enriched at either the top or bottom of the ranked list based on differential expression.
Western blot analysis
Total protein extracted from tissue samples was subjected to separation by SDS-PAGE and transferred to a PVDF membrane (Millipore). Antibodies against PI3K (Cell Signaling Technology), phosphorylated PI3K (Cell Signaling Technology), AKT (Cell Signaling Technology), phosphorylated AKT (Cell Signaling Technology), and β-actin (Cell Signaling Technology) were used. The PVDF membranes were incubated with the primary antibody overnight at 4 °C, and subsequently visualized using a Phototope-Horseradish Peroxidase Western Blot Detection Kit.
Statistical analysis
Statistical tests were conducted using GraphPad Prism 8.3.0 and SPSS 27.0. One-way or two-way ANOVA was employed to determine significant differences among multiple groups. Continuous data were presented as mean ± Standard Error of the Mean (SEM), while categorical data are shown as frequency (percentage). A p-value of less than 0.05 was deemed statistically significant for all analyses.
Results
High expression of TDO2 in CAFs of cSCC
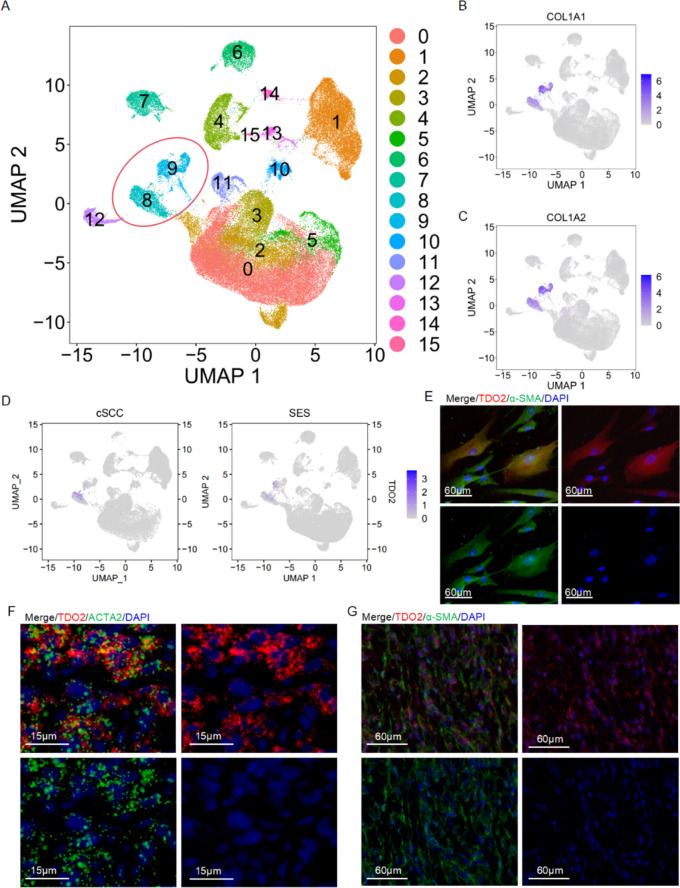
To elucidate TDO2 expression across different cellular populations in cSCC, we analyzed the public scRNA-seq dataset GSE193304 from the Chinese population, which encompasses data from tumor and sun-exposed skin (SES) tissues in three cSCC sample pairs. Rigorous data quality control yielded 69,970 high-quality cells, divided into 32,329 cells from cSCC tissues and 37,641 cells from SES tissues. We employed the “mnn” algorithm to mitigate batch effects and normalize the data, facilitating accurate cell type identification through dimensionality reduction via Uniform Manifold Approximation and Projection (UMAP). This analysis distinguished 15 cellular subgroups (Fig. 1A), with Clusters 8 and 9 identified as fibroblast subgroups, characterized by elevated COL1A1 and COL1A2 expression (Fig. 1B-C). The subsequent analysis highlighted that TDO2 was predominantly expressed in the CAFs, where its expression notably surpassed that in the fibroblasts from SES tissues (Fig. 1D). Following the enzymatic isolation of CAFs from cSCC patient tumor tissues, immunofluorescence assays further demonstrated that these primary isolated CAFs not only expressed α-SMA but also prominently displayed TDO2 (Fig. 1E). In situ hybridization and immunofluorescence staining further validated TDO2’s cellular localization within cSCC tissues, revealing its co-localization with ACTA2, also recognized as α-SMA, which is a definitive marker of CAFs (Fig. 1F-G). Collectively, these findings underscore the pronounced expression of TDO2 in CAFs associated with cSCC.
Fig. 1.
Analysis of TDO2 expression in CAFs of cSCC. A UMAP projection visualizes the clustering of cell populations in cSCC, with each color representing a distinct cell subgroup, identifying a total of 15 subgroups. B UMAP plots illustrate the expression of the gene COL1A1 within cells from cSCC, with color intensity reflecting expression levels. C Similar to B, these UMAP plots display the expression of the gene COL1A2. D UMAP visualization of TDO2 expression across the cell populations, showing localized upregulation. E Immunofluorescence images demonstrating the co-localization of TDO2 (red) with α-SMA (green) in isolated CAFs, confirming their concurrent overexpression; the scale bar denotes 60 μm. F-G In situ hybridization and immunofluorescence staining of cSCC tissues showing the colocalization of TDO2 (red) with ACTA2/α-SMA (green). DAPI (blue) marks the nuclei. Scale bars represent 15 μm (Fig. 1F) and 60 μm (Fig. 1G)
Expression of TDO2 in cSCC and its clinical correlation analysis
We conducted a statistical analysis of TDO2 expression levels in tumor tissues from 30 cSCC patients, dividing the samples into a low-expression group (n = 15) and a high-expression group (n = 15). We further analyzed the correlation between TDO2 expression and various clinical pathological parameters. The analysis showed that there was no statistically significant difference in TDO2 expression with respect to gender, age, maximum tumor diameter, tumor location, and depth (p > 0.05). However, a significant statistical correlation was found between TDO2 expression and the degree of differentiation of cSCC (p < 0.05); tumors with moderate to low differentiation were significantly more prevalent in the high-expression group than in the low-expression group. This finding suggests that the level of TDO2 expression may be related to the degree of differentiation of cSCC tumors and might serve as a potential biomarker for the differentiation status of cSCC (Table 1).
Table 1.
The correlation between TDO2 expression and clinical pathological characteristics in cutaneous squamous cell carcinoma
| Characteristics | TDO2 Expression Levels | p value | ||
|---|---|---|---|---|
| Low (n = 15) | High (n = 15) | |||
| Gender | Male | 6 | 8 | 0.464 |
| Female | 9 | 7 | ||
| Age | < 65 | 2 | 3 | 0.624 |
| ≥ 65 | 13 | 12 | ||
| Maximum Tumor Diameter | < 2 cm | 10 | 9 | 0.705 |
| ≥ 2 cm | 5 | 6 | ||
| Tumor Location | Head and Neck | 10 | 11 | 0.690 |
| Trunk and Limbs | 5 | 4 | ||
| Depth | < 6 mm | 9 | S | 0.713 |
| ≥ 6 mm | 6 | 7 | ||
| Degree of Differentiation | High Differentiation | 13 | 4 | < 0.05* |
|
Medium–low Differentiation |
2 | 11 | ||
*p < 0.05
Correlation analysis of TDO2 upregulation and immune cell infiltration in cSCC
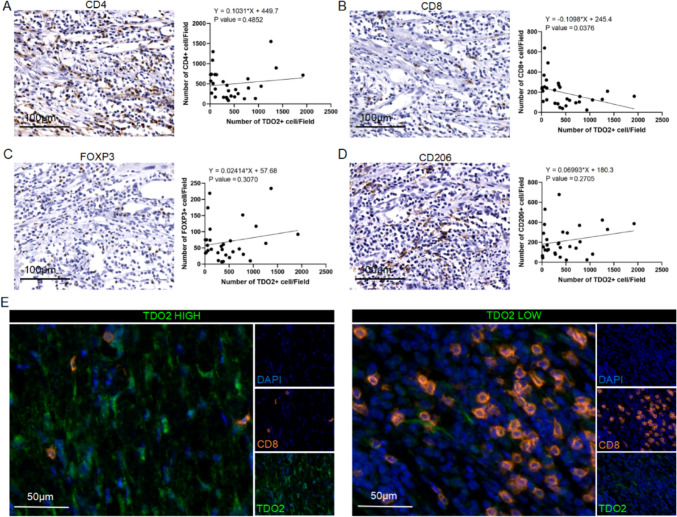
After refining the clinical dataset by removing patients with incomplete clinical information, we performed serial sectioning and immunohistochemical staining on samples obtained from 30 cSCC patients. We specifically targeted markers such as TDO2, CD4 (indicative of helper T cells), CD8 (representing cytotoxic T lymphocytes [CTL]), FOXP3 (a marker for regulatory T cells [Tregs]), and CD206 (associated with M2 macrophages), to investigate the relationship between TDO2 expression and the infiltration of various immune cell types. Quantitative analysis of the immunohistochemistry data revealed a significant negative correlation between the presence of TDO2 + cells and CD8 + T cells, as illustrated in Fig. 2B. However, no significant correlation was observed between TDO2 expression and other immune cell types, such as CD4 + helper T cells, FOXP3 + Tregs, and CD206 + M2 macrophages (Fig. 2A, C–D). Moreover, mIHC staining of the clinical specimens showed that regions with high TDO2 expression had a reduced number of CD8 + CTLs. Conversely, regions with lower TDO2 expression exhibited a higher density of these cells, as evidenced in Fig. 2E.
Fig. 2.
Correlation between TDO2 expression and CD8 + T cell infiltration in cSCC patients. A-D Immunohistochemical analysis of the correlation between TDO2 + cells and CD4 + T cells, CD8 + T cells, FOXP3 + cells, and CD206 + cells. The figure displays scatter plots and regression analysis demonstrating the correlation between the quantity of TDO2 + cells and the numbers of CD4 + T cells, CD8 + T cells, FOXP3 + cells, and CD206 + cells. The linear regression equation and P-value adjacent to the scatter plots indicate a significant negative correlation between the number of TDO2 + cells and CD8 + T cells, while the correlation with other immune cell markers is not significant. Scale bar = 100 µm. E Representative mIHC images of TDO2 + cells and CD8 + T cells. The image shows the expression of TDO2 (green) and CD8 (orange) via immunofluorescence staining in cSCC patient tissue samples. The left image represents an area of high TDO2 expression, while the right image represents an area of low TDO2 expression. DAPI (blue) is used for nuclear staining. Scale bar = 50 µm. Statistical significance is indicated as *p < 0.05
Next, we conducted a statistical assessment of the relationship between CD8 + T cell infiltration and tumor differentiation grades in the tumor samples of 30 cSCC patients. The results indicated a statistically significant difference in CD8 + T cell infiltration between highly differentiated and medium–low differentiated tumors (p < 0.05). Highly differentiated tumors were more frequently associated with elevated CD8 + T cell levels, while medium–low differentiated tumors were predominantly found in the low CD8 + T cell group. This suggests a correlation between CD8 + T cell infiltration and tumor differentiation in cSCC, with poorly differentiated tumors exhibiting lower CD8 + T cell infiltration (Table 2).
Table 2.
The correlation between CD8 + T cell expression and tumor differentiation grade in cutaneous squamous cell carcinoma
| Tumor Differentiation Grade | CD8 + T cell expression | p value | |
|---|---|---|---|
| Low (n = 15) | High (n = 15) | ||
| High Differentiation | 5 | 12 | < 0.05* |
| Medium–Low Differentiation | 10 | 3 | |
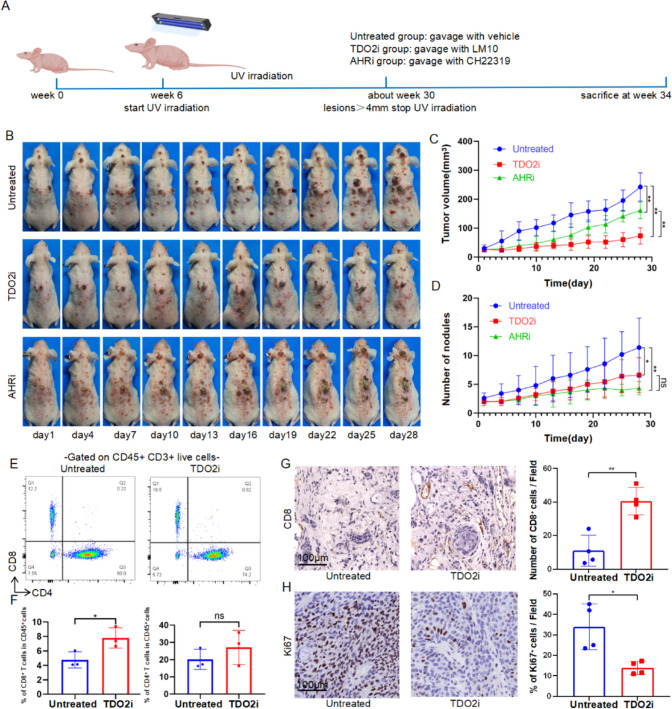
Intervening of TDO2 inhibition to suppress tumor growth in mice
Considering the shared mechanisms between actinic keratosis (AK) and cSCC, most lesions smaller than 3 mm are classified as AK, while those larger than 4 mm are typically classified as cSCC, with lesions between 3 and 4 mm potentially representing a transition from AK to cSCC[21]. Therefore, medical intervention was initiated for the tumors when they reached 4 mm, and the number of tumors larger than 3 mm was recorded (Fig. 3A). In vivo experiments were conducted using a UV-induced cSCC mouse model to evaluate the antitumor efficacy of TDO2 inhibitors. The results shown in Fig. 3B-D indicate that TDO2 inhibitors significantly suppressed tumor volume growth and reduced the number of tumors, exhibiting stronger antitumor effects compared to those of AHR inhibitors. Under the influence of TDO2 inhibitors, flow cytometric analysis revealed an increased proportion of CD8 + T cells within CD45 + cells, while the proportion of CD4 + T cells remained unchanged (Fig. 3E-F). Immunohistochemical analysis also showed that the number of CD8 + T cells increased in tumor tissues treated with TDO2 inhibitors, while the number of Ki67 + cells decreased (Fig. 3G-H), suggesting that TDO2 inhibitors may suppress tumor progression by enhancing the infiltration of CD8 + T cells.
Fig. 3.
Intervention of TDO2 inhibits tumor growth in mice. A Timeline for mouse model establishment and drug intervention. B Representative schematic of tumor development in mice from the untreated group, the TDO2i group, and the AHRi group. C Curves showing changes in tumor volume over time in different treatment groups (n = 5/group). D Statistical analysis of the number of tumors over time in different treatment groups (n = 5/group). E Flow cytometry analysis of immune cell infiltration. Representative flow cytometry scatter plots showing CD4 + T cells and CD8 + T cells expression in CD45 + CD3 + live cells from mice in the untreated group and the TDO2i group. F The left graph shows the percentage of CD8 + T cells within CD45 + cells, indicating a significant increase in the proportion of CD8 + T cells in the TDO2i group; the right graph shows the percentage of CD4 + T cells within CD45 + cells, with no significant difference observed (n = 3/group). G Immunohistochemical staining for CD8 + T cells, showing the comparison of the number of CD8 + T cells in tumor tissues between the untreated group and the TDO2i group (n = 4/group). H Immunohistochemical staining for Ki67, showing the comparison of the proportion of Ki67 + cells in tumor tissues between the untreated group and the TDO2i group (n = 4/group). Scale bar = 100 μm. The data are presented as mean ± SEM. Statistical significance is indicated as ∗ p < 0.05, ∗ ∗ p < 0.01
TDO2 inhibitors suppress tumor progression by modulating immune cell function
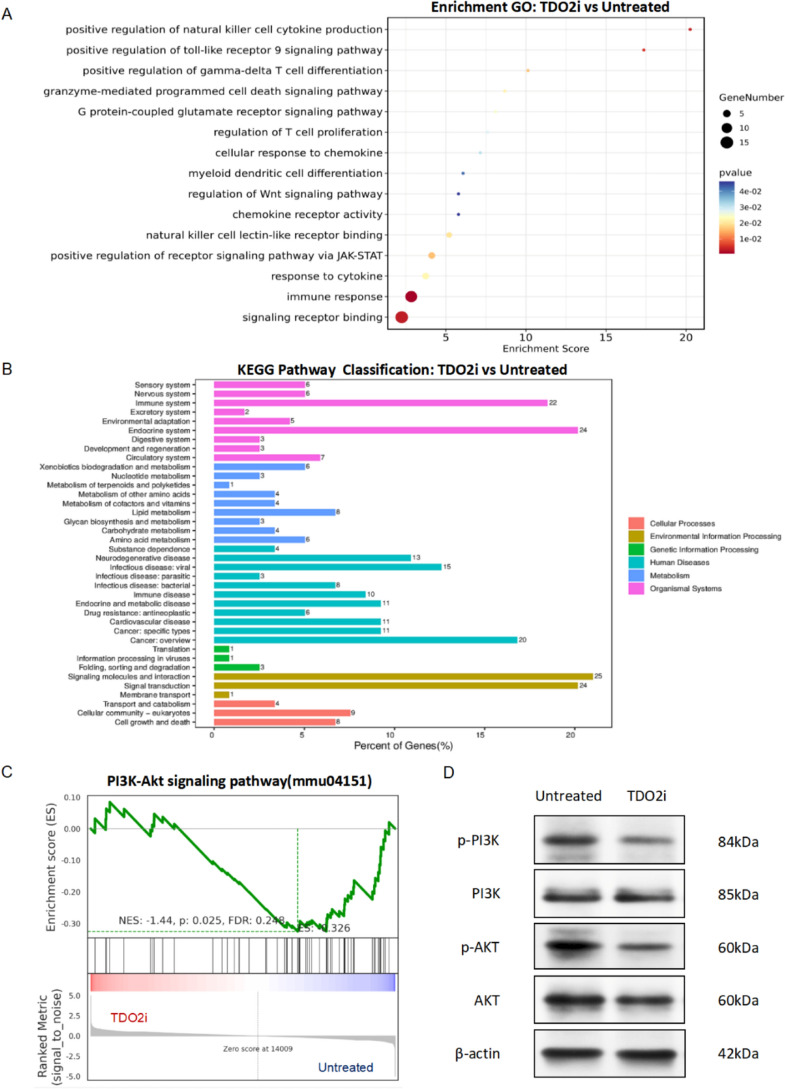
To further investigate the molecular mechanisms by which TDO2 inhibitors suppress tumor progression through modulation of immune cell functions, we utilized TDO2 inhibitors in a mouse cSCC model. Using RNA-seq technology, we conducted GO enrichment analysis, KEGG enrichment analysis, and GSEA analysis on cSCC tissues from the untreated group (n = 4/group) and the TDO2i group (n = 4/group), revealing the key mechanisms by which TDO2 inhibitors regulate immune cell activity.
As shown in Fig. 4A, the results of the GO enrichment analysis results indicate that TDO2 inhibitors primarily affect several immune cell-related biological processes, including signal receptor binding, immune response, cytokine response, and chemokine response. Notably, significant enrichment was observed in the regulation of T cell proliferation, positive regulation of gamma-delta T cell differentiation, and granzyme-mediated programmed cell death signaling pathways, suggesting that TDO2 inhibitors suppress tumor progression by modulating these critical immune cell functions. The results of the KEGG enrichment analysis, as shown in Fig. 4B, were primarily enriched in signaling molecules and interactions, signal transduction, the endocrine system, the immune system, and cancer-related pathways. The GSEA analysis in Fig. 4C highlighted significant enrichment in the PI3K-Akt signaling pathway, further elucidating how TDO2 inhibitors alter the phenotype and function of immune cells through specific immunoregulatory pathways. We elaborated on the effect of TDO2 inhibitors on the PI3K-Akt signaling pathway. As shown in Fig. 4D, the levels of phosphorylated PI3K and AKT were significantly decreased in the TDO2i group compared to the untreated group.
Fig. 4.
TDO2 inhibitors suppress tumor progression by modulating immune cell functions. A GO functional annotation chart for the untreated group and the TDO2i group. The figure displays the results of GO functional enrichment analysis, highlighting the key biological processes affected by TDO2 inhibitors, the analysis compares the untreated group and the TDO2i group. The size of the dots represents the number of genes involved in each process, while the gradient of color indicates the significance of the P-values, ranging from blue (least significant) to red (most significant). B KEGG pathway annotation chart for untreated group and TDO2i group. The bar graph displays the pathways most significantly affected according to KEGG pathway enrichment analysis. This analysis also compares the untreated group and the TDO2i groups, illustrating pathways predominantly influenced by the treatment with TDO2 inhibitors. The length of each bar represents the percentage of genes involved, while the color denotes different categories of biological processes. C GSEA functional enrichment analysis chart for the untreated group and the TDO2i group. These line graphs represent the enrichment of the PI3K-Akt signaling pathway based on GSEA. n = 4/group. The Normalized Enrichment Score (NES), p-value, and False Discovery Rate (FDR) provide statistical significance of the enrichment for each set. D Western blot analysis of the expression of p-PI3K, PI3K, p-AKT, and AKT in untreated and TDO2i groups. β-actin served as an internal control
In summary, TDO2 inhibitors regulate immune cell activity within the TME through multiple molecular mechanisms, particularly by enhancing the cytotoxic function of CD8 + T cells. This finding provides a new strategic direction for the treatment of cSCC. These findings not only provide deeper insights into the role of TDO2 in tumor immunoregulation but also offer important scientific evidence for the development of new therapeutic drugs targeting these pathways.
Discussion
cSCC, as a common malignant skin tumor on the head and face in the elderly, can be effectively treated with surgery at its early stages [22]. However, patients often ignore early symptoms, leading to a more aggressive or metastatic stage by the time of diagnosis [23]. Additionally, factors such as patient age, tumor location, and overall health status limit treatment options. Currently, even with PD-1 monoclonal antibody treatment, the efficacy rate is only 50% [24], which significantly impacts the survival and quality of life of patients. Thus, finding effective treatments for advanced cSCC is a pressing issue.
The progression of cSCC is not solely driven by the uncontrolled growth of tumor cells but is closely related to interactions among diverse cells within the TME. In this complex microenvironment, various cell types are present surrounding the tumor cells, including Langerhans cells, dendritic cells, macrophages, myeloid-derived suppressor cells, T cells, B cells, and cancer-associated fibroblasts [25]. CAFs, in particular, play a central role in the TME, significantly influencing tumor growth and spread [26]. They release specific chemical signals that affect immune cells, such as limiting the accumulation of immune effector cells like CD8 + T cells in the tumor area [27]. Additionally, CAF activity promotes the accumulation of immunosuppressive cells such as M2 macrophages, Tregs, and myeloid-derived suppressor cells (MDSCs) in the TME, enhancing tumor immune suppression [28]. Moreover, cytokines like IL-1β can induce the transformation of normal fibroblasts into CAFs, further increasing the accumulation of suppressive immune cells and exacerbating the immunosuppressive environment [29]. Clearly, understanding the intricate network of interactions between CAFs and immune cells in the TME is crucial for elucidating the mechanisms of tumor immune evasion.
In our study, single-cell transcriptome analysis revealed that the TDO2 in the TME of cSCC is expressed by CAFs. In situ hybridization and immunofluorescence staining further confirmed the co-expression of TDO2 and α-SMA in CAFs, highlighting the potential role of TDO2 in extracellular matrix synthesis and remodeling. In other reported tumors, TDO2 is highly expressed in tumor tissues, but the cells expressing it vary across different tumor types. In liver cancer, renal cell carcinoma, bladder cancer, and ovarian cancer [30–33], TDO2 is highly expressed in tumor cells, directly affecting their proliferation, metastasis, and invasion. In contrast, in oral squamous cell carcinoma, breast cancer, and lung cancer [19, 34, 35], TDO2 is highly expressed in stromal cells, influencing tumor progression through the immune microenvironment.
Our study demonstrates that TDO2 is highly expressed in CAFs in cSCC, underscoring its potential role in cSCC and its interactions with other signaling pathways. This finding is significant for assessing its potential as a therapeutic target. Firstly, as a key enzyme in tryptophan metabolism, TDO2 expression in tumors may alter the tryptophan metabolic balance in the TME, thereby affecting tumor cell proliferation [36]. Secondly, TDO2 expression in CAFs may be involved in regulating tumor immune evasion, impacting immune cell activity [37]. This effect is not limited to cSCC but is also observed in other tumor types, such as lung cancer [35] and esophageal squamous cell carcinoma [38]. This indicates that TDO2 facilitates tumor immune evasion by establishing a local immunosuppressive environment, representing a general mechanism of tumor cell escape from immune surveillance.
In studies involving cSCC patients, we observed an association between TDO2 expression levels and the extent of immune cell infiltration within tumors. Specifically, regions with higher TDO2 expression had fewer CD8 + T cells, while regions with lower TDO2 expression had more CD8 + T cells. This suggests that TDO2 expression levels might be related to the suppression of the tumor immune environment, influencing tumor progression. Further analysis indicated that TDO2 expression correlated with the clinicopathological features of cSCC and might serve as a potential indicator of tumor differentiation, helping to evaluate its value as a therapeutic target. Similarly, in triple-negative breast cancer, reduced TDO2 expression decreased CD8 + T cell activity and IFN-γ production [39]. In colorectal cancer, TDO2 overexpression reduced the proportions of major T lymphocyte subsets (CD3, CD4, CD8, Treg, and IFN-γ CD8), promoting liver metastasis through the TDO2-Kyn-AhR pathway and stimulating PD-L1-mediated antitumor immunosuppression and Wnt signaling-related cancer stemness [40]. These findings suggest a close association between TDO2 and the shaping of the TME and tumor immune evasion.
Through transcriptome sequencing and GO and KEGG enrichment analyses of cSCC samples, we revealed that TDO2 inhibitors are primarily enriched in pathways affecting immune-related processes, such as T cell proliferation regulation, positive regulation of gamma-delta T cell differentiation, and granzyme-mediated apoptotic signaling pathways. Furthermore, GSEA analysis emphasized the role of TDO2 inhibitors in the PI3K-AKT signaling pathway, inhibition of TDO2 affects the phosphorylation levels of PI3K and AKT. It has been reported that the overexpression of TDO2 promotes the activation of the AKT signaling pathway, resulting in enhanced proliferative properties and tumorigenic potential in glioma cells [15]. In esophageal squamous cell carcinoma, TDO2 was found to significantly increase phosphorylated AKT/GSK3 levels and promote M2 macrophage polarization by upregulating IL-8 expression, thereby accelerating tumor progression [38].
Despite the significant findings, our study has several limitations that need to be addressed. First, the sample size for animal experiments was relatively small. Larger sample sizes could provide more reliable and generalizable results. Although we obtained consistent results with human data in the mouse cSCC model, which showed upregulation of TDO2 in CAFs (Fig. S1), it may not fully replicate the tumor microenvironment. For instance, the immune response in mice may differ from that in humans, which could affect the translatability of our findings. Lastly, although the study highlighted the involvement of the PI3K-AKT signaling pathway, functional assays to confirm these effects were not included. Future research should incorporate functional assays, such as flow cytometry and cytokine profiling, to validate the effects of TDO2 inhibitors on immune function. In summary, while our study provides important insights into the role of TDO2 inhibitors in regulating immune cell functions and tumor progression, addressing these limitations in future research will be crucial for validating and expanding our findings.
In conclusion, studying TDO2 expression and its regulatory mechanisms in cSCC and its tumor immune microenvironment not only aids in understanding the molecular basis of tumor development but also provides scientific evidence and new therapeutic perspectives for the development of novel immunotherapy strategies targeting TDO2. These findings highlight the importance of TDO2 as a potential therapeutic target and offer promising new approaches for the treatment of advanced cSCC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Declared none.
Author contributions
FL and GY contributed to this study equally. FL performed the experiments and wrote this original draft. GY collected all public datasets and was responsible for the main analysis. ZZ, ZZ, and YW was responsible for the collection of cSCC tissues. LW analyzed data. YL conducted the pathological diagnoses. QZ reviewed and edited this original draft. GZ was responsible for funding acquisition and supervision of the study. All authors read and approved the final manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82073016) and the National Key Research and Development Program of China (Grant Nos. 2022YFC2504700 and 2022YFC2504703).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
The study was approved by the Ethics Committee of Shanghai Skin Disease Hospital, and written informed consent was obtained from all patients (Grant No. 2020–12). Additionally, all animal experiments were approved by the Animal Ethics Committee of Shanghai Skin Disease Hospital (Grant No. 2020–038(Animal)). All methods were carried out in accordance with the relevant guidelines under the ethical approval and consent to participate section.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fangqi Lu and Guorong Yan have contributed equally to this work and shared the first authorship.
Contributor Information
Qingyu Zeng, Email: zengqingyu2011@tongji.edu.cn.
Guolong Zhang, Email: glzhangtj@tongji.edu.cn.
References
- 1.Heppt MV, Leiter U (2023) Cutaneous squamous cell carcinoma: state of the art, perspectives and unmet needs. J Dtsch Dermatol Ges 21(4):421–424. 10.1111/ddg.15052 [DOI] [PubMed] [Google Scholar]
- 2.Zhang W, Zeng W, Jiang A, He Z, Shen X, Dong X, Feng J, Lu H (2021) Global, regional and national incidence, mortality and disability-adjusted life-years of skin cancers and trend analysis from 1990 to 2019: an analysis of the global burden of disease study 2019. Cancer Med 10(14):4905–4922. 10.1002/cam4.4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dessinioti C, Pitoulias M, Stratigos AJ (2022) Epidemiology of advanced cutaneous squamous cell carcinoma. J Eur Acad Dermatol Venereol 36(1):39–50. 10.1111/jdv.17709 [DOI] [PubMed] [Google Scholar]
- 4.Waldman A, Schmults C (2019) Cutaneous squamous cell carcinoma. Hematol Oncol Clin North Am 33(1):1–12. 10.1016/j.hoc.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 5.Kosmadaki MG, Gilchrest BA (2002) The demographics of aging in the United States: implications for dermatology. Arch Dermatol 138(11):1427–1428. 10.1001/archderm.138.11.1427-a [DOI] [PubMed] [Google Scholar]
- 6.Que SKT, Zwald FO, Schmults CD (2018) Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol 78(2):237–247. 10.1016/j.jaad.2017.08.059 [DOI] [PubMed] [Google Scholar]
- 7.De Jong E, Lammerts MUPA, Genders RE, Bouwes Bavinck JN (2022) Update of advanced cutaneous squamous cell carcinoma. J Europ Acad Dermatol Venereol 36:6–10. 10.1111/jdv.17728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzic JG, Schmitt AR, Wright AC, Alniemi DT, Zubair AS, Olazagasti Lourido JM, Sosa Seda IM, Weaver AL, Baum CL (2017) Incidence and trends of basal cell carcinoma and cutaneous squamous cell carcinoma: a population-based study in Olmsted county Minnesota 2000 to 2010. Mayo Clin Proc 92(6):890–898. 10.1016/j.mayocp.2017.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, Zhang B, Meng Q, Yu X, Shi S (2021) Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 20(1):131. 10.1186/s12943-021-01428-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Visser KE, Joyce JA (2023) The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell 41(3):374–403. 10.1016/j.ccell.2023.02.016 [DOI] [PubMed] [Google Scholar]
- 11.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, Hynes RO, Jain RK, Janowitz T, Jorgensen C, Kimmelman AC, Kolonin MG, Maki RG, Powers RS, Puré E, Ramirez DC, Scherz-Shouval R, Sherman MH, Stewart S, Tlsty TD, Tuveson DA, Watt FM, Weaver V, Weeraratna AT, Werb Z (2020) A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 20(3):174–186. 10.1038/s41568-019-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21(3):309–322. 10.1016/j.ccr.2012.02.022 [DOI] [PubMed] [Google Scholar]
- 13.Cheong JE, Sun L (2018) Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy - Challenges and opportunities. Trends Pharmacol Sci 39(3):307–325. 10.1016/j.tips.2017.11.007 [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Zhai J, Kong X, Wang X, Wang Z, Fang Y, Wang J (2020) Comprehensive analysis of the expression and prognosis for TDO2 in breast cancer. Mol Ther Oncolytics 17:153–168. 10.1016/j.omto.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong C, Peng L, Tao B, Yin S, Lyu L, Ding H, Yang X, Peng T, He H, Zhou P (2022) TDO2 and tryptophan metabolites promote kynurenine/AhR signals to facilitate glioma progression and immunosuppression. Am J Cancer Res 12(6):2558–2575 [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H, Xiang Y, Zong QB, Dai ZT, Wu H, Zhang HM, Huang Y, Shen C, Wang J, Lu ZX, Ponnambalam S, Chen K, Wu Y, Zhang TC, Liao XH (2022) TDO2 modulates liver cancer cell migration and invasion via the Wnt5a pathway. Int J Oncol 60(6):5362. 10.3892/ijo.2022.5362 [DOI] [PubMed] [Google Scholar]
- 17.Lee R, Li J, Li J, Wu CJ, Jiang S, Hsu WH, Chakravarti D, Chen P, LaBella KA, Li J, Spring DJ, Zhao D, Wang YA, DePinho RA (2022) Synthetic essentiality of tryptophan 2,3-dioxygenase 2 in APC-mutated colorectal cancer. Cancer Discov 12(7):1702–1717. 10.1158/2159-8290.Cd-21-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H (2015) Cancer-associated fibroblasts: their characteristics and their roles in tumor growth. Cancers (Basel) 7(4):2443–2458. 10.3390/cancers7040902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Lu H, Xie W, Wang D, Shan Z, Xing X, Wang XM, Fang J, Dong W, Dai W, Guo J, Zhang Y, Wen S, Guo XY, Chen Q, Bai F, Wang Z (2022) TDO2+ myofibroblasts mediate immune suppression in malignant transformation of squamous cell carcinoma. J Clin Invest 132(19):e157649. 10.1172/jci157649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, Quevedo R, Neufeld L, Liu ZQ, Jin R, Grünwald BT, Foerster EG, Chaharlangi D, Guo M, Makhijani P, Zhang X, Pugh TJ, Pinto DM, Co IL, McGuigan AP, Jang GH, Khokha R, Ohashi PS, O’Kane GM, Gallinger S, Navarre WW, Maughan H, Philpott DJ, Brooks DG, McGaha TL (2022) Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 55(2):324-340.e8. 10.1016/j.immuni.2022.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J, West CE, Roh YS, Sutaria N, Kwatra SG, Kwatra MM (2021) Mouse models for actinic keratoses. J Pharmacol Toxicol Methods 110:107071. 10.1016/j.vascn.2021.107071 [DOI] [PubMed] [Google Scholar]
- 22.Corchado-Cobos R, García-Sancha N, González-Sarmiento R, Pérez-Losada J, Cañueto J (2020) Cutaneous squamous cell carcinoma: from biology to therapy. Int J Mol Sci 21(8):2956. 10.3390/ijms21082956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dessinioti C, Stratigos AJ (2022) Recent advances in the diagnosis and management of high-risk cutaneous squamous cell carcinoma. Cancers 14(14):3556. 10.3390/cancers14143556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claveau J, Archambault J, Ernst DS, Giacomantonio C, Limacher JJ, Murray C, Parent F, Zloty D (2020) Multidisciplinary management of locally advanced and metastatic cutaneous squamous cell carcinoma. Curr Oncol 27(4):e399–e407. 10.3747/co.27.6015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saeidi V, Doudican N, Carucci JA (2023) Understanding the squamous cell carcinoma immune microenvironment. Front Immunol 14:1084873. 10.3389/fimmu.2023.1084873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Nieto GA, Teppo HR, Petrelius N, Izzi V, Devarajan R, Petäistö T, Liu H, Kim KS, Karppinen SM, Ruotsalainen H, Koivunen J, Mäki JM, Walker GC, Pihlajaniemi T, Gullberg D, Heljasvaara R (2022) Upregulated integrin α11 in the stroma of cutaneous squamous cell carcinoma promotes skin carcinogenesis. Front Oncol 12:981009. 10.3389/fonc.2022.981009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ene-Obong A, Clear AJ, Watt J, Wang J, Fatah R, Riches JC, Marshall JF, Chin-Aleong J, Chelala C, Gribben JG, Ramsay AG, Kocher HM (2013) Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology 145(5):1121–1132. 10.1053/j.gastro.2013.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang A, Qian Y, Ye Z, Chen H, Xie H, Zhou L, Shen Y, Zheng S (2017) Cancer-associated fibroblasts promote M2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med 6(2):463–470. 10.1002/cam4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erez N, Truitt M, Olson P, Arron ST, Hanahan D (2010) Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-kappaB-dependent manner. Cancer Cell 17(2):135–147. 10.1016/j.ccr.2009.12.041 [DOI] [PubMed] [Google Scholar]
- 30.Ai Y, Luo S, Wang B, Xiao S, Wang Y (2022) MiR-126-5p promotes tumor cell proliferation, metastasis and invasion by targeting TDO2 in hepatocellular carcinoma. Molecules 27(2):443. 10.3390/molecules27020443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham QT, Taniyama D, Sekino Y, Akabane S, Babasaki T, Kobayashi G, Sakamoto N, Sentani K, Oue N, Yasui W (2021) Clinicopathologic features of TDO2 overexpression in renal cell carcinoma. BMC Cancer 21(1):737. 10.1186/s12885-021-08477-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham QT, Taniyama D, Akabane S, Harada K, Babasaki T, Sekino Y, Hayashi T, Sakamoto N, Sentani K, Oue N, Yasui W (2021) TDO2 overexpression correlates with poor prognosis cancer stemness and resistance to cetuximab in bladder cancer. Cancer Rep Hoboken 4(6):e1417. 10.1002/cnr2.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Tao F, Jiang J, Chen L, Du J, Cheng X, He Q, Zhong S, Chen W, Wu X, Ou R, Xu Y, Tang KF (2021) Tryptophan 2, 3-dioxygenase promotes proliferation, migration and invasion of ovarian cancer cells. Mol Med Rep 23(6):12084. 10.3892/mmr.2021.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du R, Zhang X, Lu X, Ma X, Guo X, Shi C, Ren X, Ma X, He Y, Gao Y, Liu Y (2023) PDPN positive CAFs contribute to HER2 positive breast cancer resistance to trastuzumab by inhibiting antibody-dependent NK cell-mediated cytotoxicity. Drug Resist Updat 68:100947. 10.1016/j.drup.2023.100947 [DOI] [PubMed] [Google Scholar]
- 35.Hsu YL, Hung JY, Chiang SY, Jian SF, Wu CY, Lin YS, Tsai YM, Chou SH, Tsai MJ, Kuo PL (2016) Lung cancer-derived galectin-1 contributes to cancer associated fibroblast-mediated cancer progression and immune suppression through TDO2/kynurenine axis. Oncotarget 7(19):27584–27598. 10.18632/oncotarget.8488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding X, Jin Y, Shi X, Wang Y, Jin Z, Yin L, Gao S, Lei Y, Yang J (2024) TDO2 promotes bladder cancer progression via AhR-mediated SPARC/FILIP1L signaling. Biochem Pharmacol 223:116172. 10.1016/j.bcp.2024.116172 [DOI] [PubMed] [Google Scholar]
- 37.Perez-Castro L, Garcia R, Venkateswaran N, Barnes S, Conacci-Sorrell M (2023) Tryptophan and its metabolites in normal physiology and cancer etiology. Febs j 290(1):7–27. 10.1111/febs.16245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, Sun J, Li Y, Zhou X, Zhai W, Wu Y, Chen G, Gou S, Sui X, Zhao W, Qiu L, Yao Y, Sun Y, Chen C, Qi Y, Gao Y (2021) Tryptophan 2,3-dioxygenase 2 controls M2 macrophages polarization to promote esophageal squamous cell carcinoma progression via AKT/GSK3β/IL-8 signaling pathway. Acta Pharm Sin B 11(9):2835–2849. 10.1016/j.apsb.2021.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene LI, Bruno TC, Christenson JL, D’Alessandro A, Culp-Hill R, Torkko K, Borges VF, Slansky JE, Richer JK (2019) A role for tryptophan-2,3-dioxygenase in CD8 T-cell suppression and evidence of tryptophan catabolism in breast cancer patient plasma. Mol Cancer Res 17(1):131–139. 10.1158/1541-7786.Mcr-18-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki T, Chung S, Sakai H, Ohata H, Obata Y, Shiokawa D, Mizoguchi Y, Kubo T, Ichikawa H, Taniguchi H, Aoki K, Soga T, Nakagama H, Okamoto K (2022) Stemness and immune evasion conferred by the TDO2-AHR pathway are associated with liver metastasis of colon cancer. Cancer Sci 113(1):170–181. 10.1111/cas.15182 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.