Abstract
The development of tumor vaccines represents a significant focus within cancer therapeutics research. Nonetheless, the efficiency of antigen presentation in tumor vaccine remains suboptimal. We introduce an innovative mRNA-lipid nanoparticle platform designed to express tumor antigenic epitopes fused with the transmembrane domain and cytoplasmic tail of the neonatal Fc receptor (FcRn). This novel design exploits FcRn trafficking signals to direct the epitope-FcRn fusion toward endolysosomal degradation, thereby generating epitopes capable of eliciting targeted T cell responses and establishing immune memory. The FcRn-directed presentation of epitopes enhances MHC class I and II antigen presentation, thereby robustly inducing CD4+ and CD8+ T cell responses, which translates to the inhibition of tumor growth and extension of survival in preclinical mouse models. In summary, the deliberate incorporation of FcRn trafficking signals into vaccine design markedly boosts T cell responses, underscoring the promise of this novel strategy in advancing the efficacy of tumor vaccines.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-024-03888-y.
Keywords: FcRn, mRNA vaccine, Antigen presentation, Tumor, T cell epitope
Introduction
The immune system is crucial for maintaining health and combating disease. Genetic and evolutionary studies have underscored the pivotal role of the major histocompatibility complex (MHC) in immune recognition, defense, and surveillance [1]. MHC molecules on antigen-presenting cells (APCs) bind to peptides resulting from proteasomal degradation, presenting cellular status cues on the cell surface for T cell recognition and activation. This enables the immune system to recognize the state of the cell in a timely manner and to clear the abnormal cell efficiently and specifically [2]. Nevertheless, in certain pathological contexts, MHC presentation can be compromised. For example, tumor cells may diminish the display of MHC molecules or essential antigen-processing proteins, thereby reducing epitope presentation and facilitating immune evasion [3, 4].
Identifying functional T cell epitopes poses significant challenges. Advances in algorithms for predicting MHC-binding epitopes and identifying T cell epitopes have facilitated the creation of epitope vaccines for disease treatment and prevention [5–12]. However, the practical implementation of epitope-based vaccines continues to face unresolved challenges. For example, the presentation of epitopes by MHC class I to activate the immune response has been reported to be an inefficient process. In contrast, MHC class II presents not only exogenous proteins but also intracellular antigens [13–16]. In recent years, various strategies have been employed to optimize MHC II presentation, primarily by annexing antigenic sequences to localization motifs within the endosomal-lysosomal system. This integration allows for the co-presentation of antigens with the MHC II pathway, enhancing antigen presentation [17–21].
FcRn, a non-classical Fc receptor, predominantly binds to IgG, shielding it from endosomal-lysosomal degradation and extending its half-life [22–24]. Studies have shown that the cytoplasmic tail of FcRn is the principal domain containing endosomal-lysosomal sorting signals [25, 26]. Moreover, the excision of Fc-binding and transmembrane domains does not impede the lysosomal routing of FcRn-linked moieties [25, 27].
In this study, we engineered a novel mRNA construct leveraging FcRn-mediated transport, expressing FcRn's transmembrane and cytoplasmic domains fused to antigenic epitopes. The objective was to enhance degradation and presentation efficiency by steering mRNA-encoded products toward lysosomal degradation, yielding the desired epitopes. Our findings revealed that this vaccine could increase the proportion of T cells activated in tumor models, thereby delaying tumor growth. Additionally, the vaccine also could be used to preventing tumorigenesis.
Results
Trafficking of FcRn-tagged fusion proteins into endolysosomal compartments
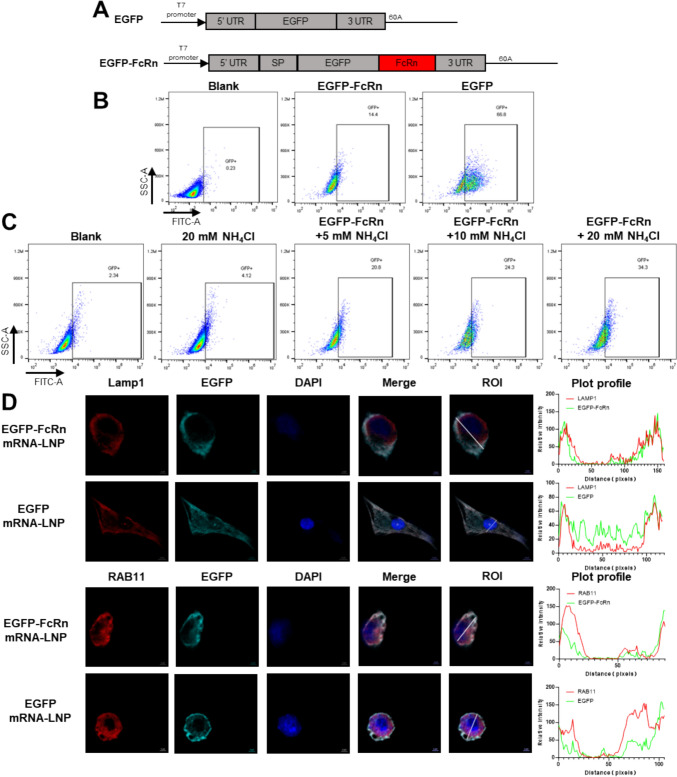
To elicit an adaptive immune response against the encoded antigens, the mRNA vaccine must be functionally delivered to the cytosol of the cell and undergo the process of transcription and translation, thus the products could enter the antigen presentation pathway, which presents the antigenic message to the T cells [28, 29]. To ascertain whether the mRNA encoding the FcRn protein could be translated and transported to the lysosome, we utilized EGFP as a tracer in DCs (DC2.4 cells). To enhance transfection efficiency, the mRNAs were encapsulated within LNPs [30, 31]. Post-transfection, a higher frequency of GFP+ cells was observed in those transfected with EGFP mRNA-LNP (Fig. 1B). The addition of NH₄Cl, which directly neutralises the lysosomal pH, to the culture medium [32], led to an elevation in the frequency of GFP+ in EGFP-FcRn mRNA-LNP-transfected cells. And the elevation was found to be dose-dependent with respect to NH₄Cl (Fig. 1C). To investigate cytoplasmic trafficking, transfected cells were co-stained with a polyclonal rabbit anti-EGFP antibody and monoclonal antibodies against LAMP1 (a lysosomal marker) and RAB11 (endosome maker protein). The fluorescence intensity trends observed for EGFP-FcRn within cells were largely consistent with those of LAMP1 and RAB11. However, this consistency was relatively less pronounced for EGFP (Fig. 1D). Consequently, EGFP-FcRn colocalization with endosomes and lysosomes was observed, indicating that FcRn directs the linked EGFP into the endosomal-lysosomal system. In summary, these findings demonstrate that FcRn facilitates the entry of associated proteins into endosomal-lysosomal compartments for timely lysosomal degradation.
Fig. 1.
Trafficking of FcRn-tagged fusion proteins into endolysosomal compartments. A Schematic representation of EGFP and EGFP-FcRn mRNA constructs. B DC2.4 cells were transfected with EGFP, EGFP-FcRn, or a blank control, GFP+ cell frequencies were assessed using flow cytometry. C DC2.4 cells transfected with EGFP-FcRn were treated with NH4Cl at 6 h post-transfection. GFP-positive cell frequencies were then evaluated by flow cytometry. D Cellular localization of EGFP and EGFP-FcRn in DC2.4 cells was analyzed 48 h post-transfection with 5 μg of mRNA-LNP. Cells were stained with antibodies targeting Lamp1 and RAB11 (red), and EGFP signal was amplified using a polyclonal anti-EGFP antibody (cyan). Colocalization was analyzed using the Plot Profile tool in ImageJ, with ROI indicating the region of interest
Improved MHC class I and II epitope presentation mediated by FcRn trafficking signals
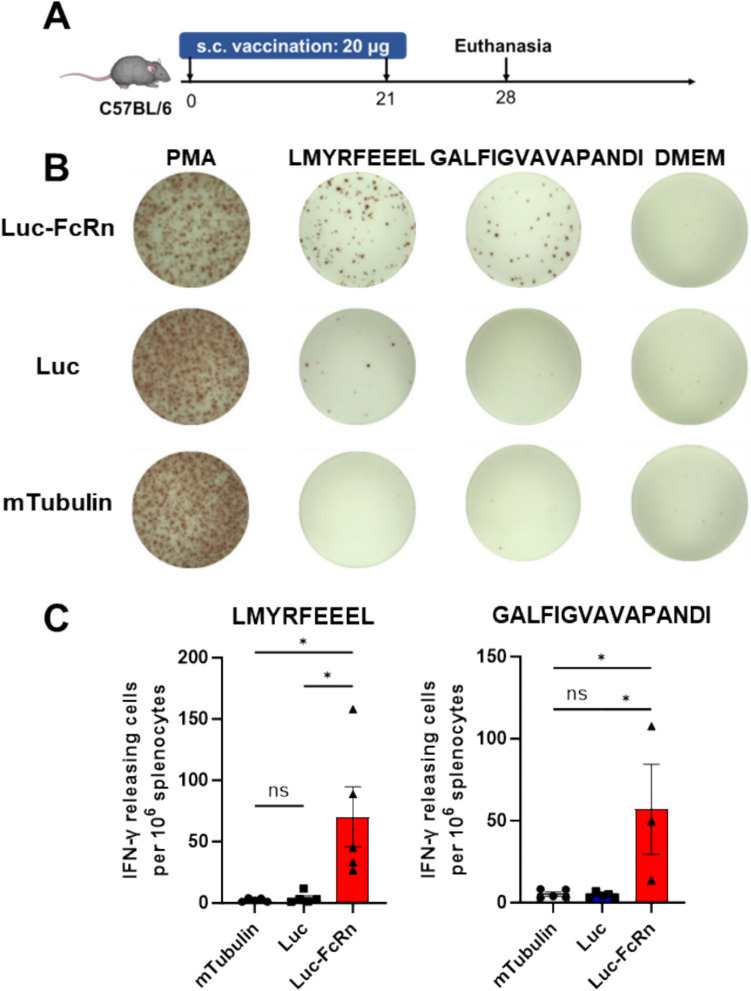
Epitope vaccines are classified as pathogen-derived peptide vaccines recognized by T cells via presentation by MHC molecules [33]. Short peptides are the end products of epitope vaccines, which may originate from DNA, RNA, synthetic long peptides, and other sources. Unlike other vaccine types, mRNA-based epitope vaccines can rapidly translate within the cytoplasmic ribosomes into numerous long peptides containing epitopes upon cell entry. Subsequently, these long peptides are processed and cleaved by intracellular proteasomes or lysosomes into short peptides that bind to MHC class I or II molecules, presented on the cell surface for T cell recognition, initiating immune responses and memory formation. To assess the impact of FcRn addition on immune response enhancement, mice were immunized using mRNAs encoding luciferase antigenic fragments (48–147, 238–315), either with or without FcRn. ELISPOT assays were then conducted to compare T cell responses to H-2-Kb-restricted epitope LMYRFEEEL or H-2-IAb-restricted epitope GALFIGVAVAPANDI, both derived from luciferase. Spot counts revealed that mice immunized with FcRn-containing mRNA exhibited a significantly higher response than treated with PBS (as blank control) and mTubulin (as a control of LNP) (Fig. 2). This, in conjunction with intracellular trafficking, demonstrated that FcRn directs associated proteins to the endosomal-lysosomal system, enhancing degradation and epitope generation, which augments antigen presentation efficiency and T cell activation.
Fig. 2.
Epitopes fusion with FcRn could lead to greater T cell response after immunization. Mice were allocated into three groups and immunized twice with mRNA-LNPs encoding Fluc (luciferase), Fluc-FcRn, and mTubulin (tubulin from Mus musculus). The specific T cell responses were then assessed using ELISPOT. A Schematic of the immunization schedule with mRNA-LNPs. B Splenocytes were stimulated with single peptides from luciferase. C Statistical analysis of the spot counts. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Elicitation of antitumoral immunity in a therapeutic mouse model
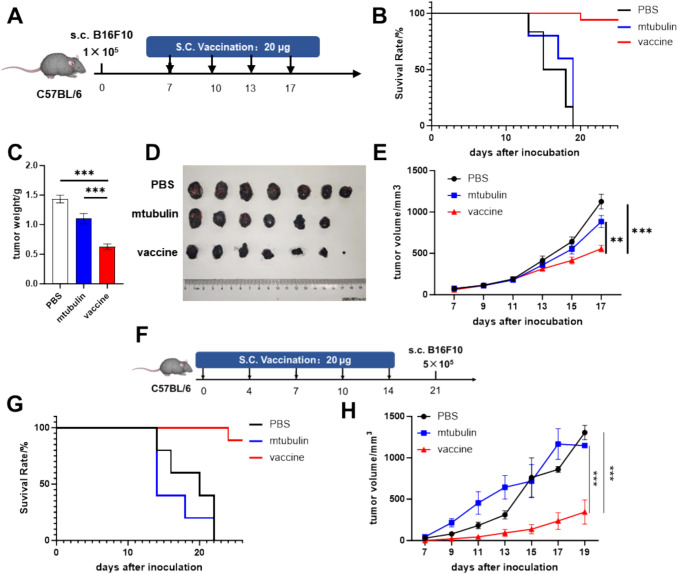
We further validated the efficacy of mRNA-encoded FcRn-antigen fragment fusions in a tumor model designed as a vaccine. We utilized a B16F10 melanoma mouse model, known for expressing classical melanoma-associated antigens and epitopes, including the highly expressed tyrosinase-related proteins 1 and 2 (TRP1 and TRP2) in melanocytes [34, 35]. We designed mRNAs encoding antigenic fragments of TRP1 (108–201, 354–469) and TRP2 (64–198, 358–464), which enriched with epitopes predicted to have high binding affinity to H-2-Kb, H-2-Db, and H-2-IAb according to IEDB, and fused these with FcRn before LNP encapsulation. Mice received four vaccine treatments post-tumor development (Fig. 3A). Treatment with vaccine resulted in partial tumor inhibition and reduced tumor growth rates (Fig. 3E), along with prolonged survival compared to PBS (blank control) and mTubulin (LNP control) (Fig. 3B). Moreover, tumor profiles significantly improved with vaccination, as indicated by reduced weights and volumes (Fig. 3C, D). This indicates that the vaccine exerts a degree of efficacy in inhibiting tumor progression.
Fig. 3.
The epitope-FcRn vaccine could be used in both treatment and prevention for tumor. The A-E depict the tumor treatment models. A The establishment of the melanoma model and the treatment regimen. Mice were divided into three groups and injected with 200 μL PBS (n = 10), 200 μL mTubulin mRNA-LNP (n = 10), or 200 μL vaccine ( a combination of 100 μL TRP1 mRNA and 100 μL TRP2 mRNA) (n = 10); B the survival curves for the three groups; C, D the mice were enthusiasm on day18 post- tumor inoculation. Tumor tissues were isolated, and tumor weights were measured; E the tumor volumes were measured throughout the treatment period. The F–H describe the tumor prevention models. F the vaccination and tumor challenge schedule, with mice divided into three groups receiving 200 μL PBS (n = 5), 200 μL mTubulin mRNA-LNP (n = 5), or 200 μL vaccine (a combination of 100 μL TRP1 mRNA and 100 μL TRP2 mRNA) (n = 5); G the survival curves for the three groups; H tumor volume post-tumor cell inoculation. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Furthermore, we assessed the vaccine's potential for tumor prevention. Following four prophylactic vaccinations, mice were challenged with tumor cells (Fig. 3F). We observed a slowdown in tumor growth and enhancement in survival rates among immunized mice (Fig. 3G, H). These findings suggest that the antigen fragments-FcRn vaccine can confer both therapeutic and preventive effects.
Robust induction of CD4+ and CD8+ T cell response by stimulation with FcRn fusion Ags
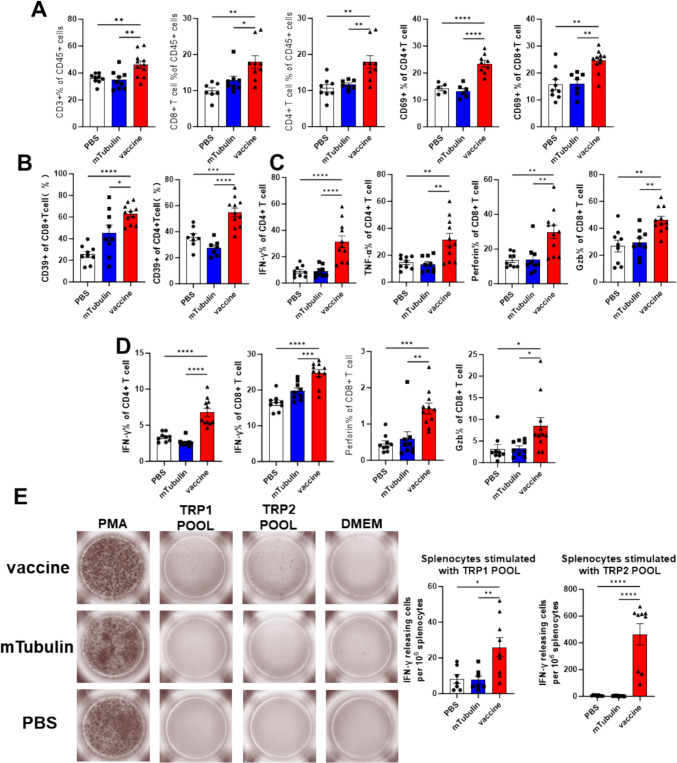
Immune profiles were assessed using flow cytometry following tumor treatment at day18. Firstly, the T cells isolated from tumor were changed notably, and vaccination significantly increased the proportion of lymphocytes (CD3+ cells) from 38 to 50%. CD8+ and CD4+ T cell populations were augmented in vaccine-treated mice, exhibiting heightened activation as evidenced by increased CD69 expression (Fig. 4A). Given that CD39 is recognized as a marker for tumor-reactive T cells [36–38], we assessed CD39 expression and the frequency of these cells. The vaccine increased the frequency of both CD8+ and CD4+ T cells with tumor reactivity, with a basal response observed in the control group (Fig. 4B). Tumor-infiltrating immune cells and splenocytes were isolated, and cytokine release, including TNF-α, IFN-γ, perforin, and Granzyme B, was quantified (Fig. 4C, D). A significant enhancement in T cell activity and cytotoxicity was observed in both tumor-infiltrating lymphocytes (TILs) and splenocytes.
Fig. 4.
The T cell responses were detected after treatment and prevention. A–C the T cells isolated from tumor were analyzed by flow cytometry, D the T cells from splenocytes were analyzed by flow cytometry. E Splenocytes from mice vaccinated with PBS, mTubulin mRNA-LNP, or vaccine were stimulated with peptide pool derived from TRP1 and TRP2. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Furthermore, splenocytes from prophylactically vaccinated mice were analyzed for T cell responses to vaccine-included peptides using ELISPOT. The appearance of spots in the vaccinated group post-peptide stimulation (Fig. 4E) indicated the successful establishment of immune memory.
Vaccination with FcRn fusion Ags remodels tumor microenvironment
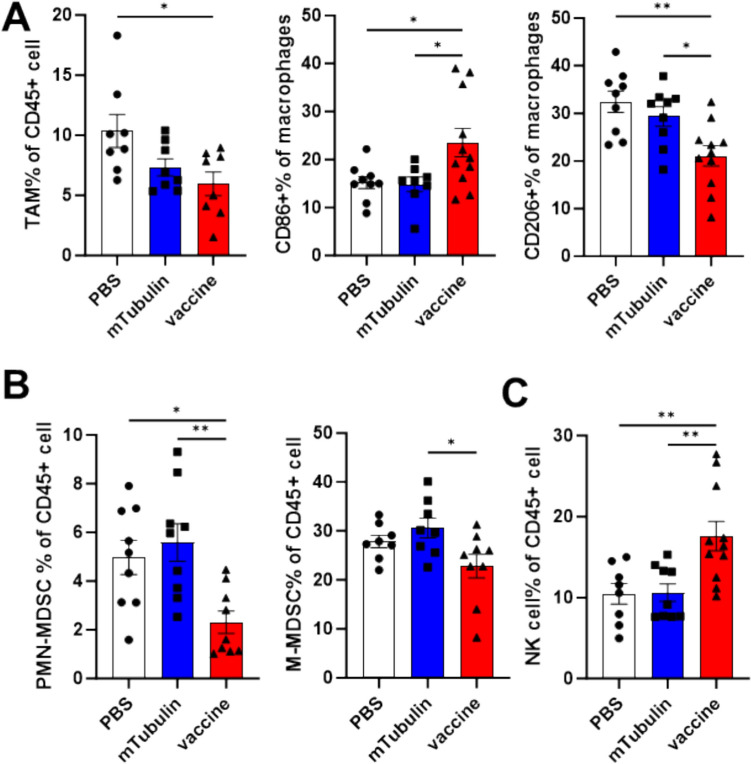
In addition to assessing the impact of mRNA vaccination on T-cell populations, we aimed to determine if there was a detectable effect on other leukocytes within the tumor microenvironment (TME). In the group treated with the vaccine, the proportion of tumor-infiltrating tumor-associated macrophages (TAMs) was modestly reduced, with a notable increase in the percentage of M1-type macrophages, while the other groups displayed predominantly M2-type macrophages (Fig. 5A). Moreover, both M-MDSCs and PMN-MDSCs showed reduced infiltration in the tumor microenvironment of vaccine-treated mice (Fig. 5B). Additionally, the frequency of NK cells increased by approximately 10% (Fig. 5C). This suggests that the vaccine may improve the immunosuppressive nature of the TME, although the underlying mechanisms remain unexplored in this study.
Fig. 5.
Vaccination with FcRn-fusion Ags remodels tumor microenvironment. Tumor-infiltrating immune cells were isolated from tumor tissues on day 18 post-tumor inoculation and analyzed by flow cytometry. A The tumor associated macrophages and the ratio of M1 and M2 phenotypes. B The ratio of PMN-MDSCs and M-MDSCs. C The frequency of NK cells. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Safety profile of mRNA-LNPs
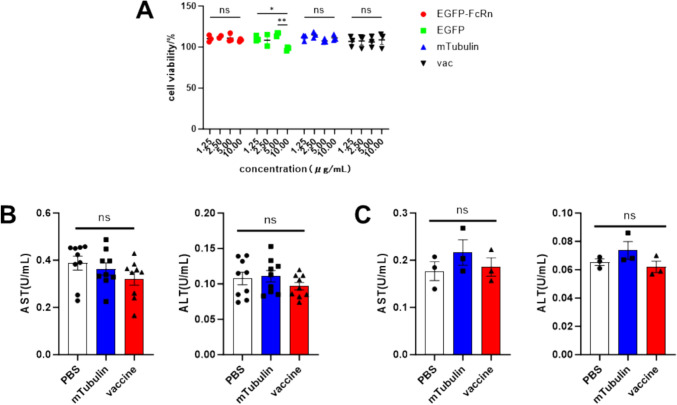
We assessed the impact of the mRNA-LNP on cell viability using the CCK8 assay, and our findings indicated that none of the transfection reagents utilized in this study exerted any detrimental effects on cell viability. Furthermore, serum was collected during the assessment of therapeutic and immunogenic effects. Our analysis revealed that the vaccine did not induce hepatotoxicity, as evidenced by the measurement of AST and ALT levels, thereby confirming that the mRNA-LNP did not compromise the health of the mice in vivo. These findings suggested that the vaccine is safe (Fig. 6).
Fig. 6.
Safety profiles. A The cytotoxicity of the four mRNA-LNPs used in the study was detected using CCK-8 kit. B AST and ALT activities in peripheral blood were measured in tumor-bearing mice treated with PBS, mTubulin mRNA-LNP, and vaccine. C AST and ALT activities in peripheral blood were evaluated in mice immunized with PBS, mTubulin mRNA-LNP, and vaccine. Data are shown as mean values ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001
Discussion
In this study, we developed a novel mRNA-lipid nanoparticle (mRNALNP) vaccine platform that harnesses the trafficking signals of the neonatal Fc receptor (FcRn) to augment the presentation of tumor-specific antigenic epitopes and elicit robust antitumor immune responses. By fusing tumor epitopes with the cytoplasmic tail of FcRn, we guided the antigenic cargo to the lysosomal compartment, facilitating efficient processing and presentation on MHC class I and II molecules. This strategic harnessing of FcRn trafficking signals resulted in a robust induction of both CD4+ and CD8+ T cell responses, ultimately leading to significant tumor growth inhibition and enhanced survival in a mouse model.
The observed enhancement in antigen presentation facilitated by FcRn trafficking signals represents a key mechanistic insight from this work. In contrast to methods that target antigens to the endoplasmic reticulum (ER) or cytosol [39, 40], such as the MITD domain in the BioNTech mRNA vaccine platform [17], our system's FcRn-directed lysosomal trafficking may offer several potential advantages. By directing the antigens into the endocytic pathway and lysosomal compartment, we facilitate efficient processing and loading onto MHC class II molecules, thereby inducing potent CD4+ T cell help essential for the establishment and maintenance of durable antitumor immunity. Furthermore, the FcRn-guided lysosomal degradation of the antigen, as indicated by our use of predicted MHC-I and MHC-II epitopes, enhances the generation of a diverse array of MHC class I and II-presented epitopes, broadening the repertoire of both CD8+ and CD4+ T cell responses, as confirmed by ELISPOT and FACS analysis. This comprehensive activation of both CD4+ and CD8+ T cells likely contribute to the enhanced therapeutic efficacy observed in our preclinical tumor model. This FcRn-based approach underscores the value of harnessing specific trafficking mechanisms to augment the immunogenicity of tumor antigens. Rather than relying on passive or nontargeted antigen delivery, our strategic harnessing of the FcRn recycling pathway enables targeted transport of tumor epitopes to the optimal subcellular compartments for efficient antigen processing and presentation. This targeted strategy ensures that the critical steps in the antigen presentation cascade are optimized, resulting in the robust induction of tumor-specific T cell responses. Furthermore, the FcRn-mediated trafficking signals could potentially be applied to other vaccine platforms or immunotherapeutic strategies, expanding the applicability of this innovative approach.
In summary, our study explored the potential of FcRn trafficking signals in enhancing antigen presentation and immune response within mRNA vaccines. These findings highlight the efficacy of rational vaccine design that incorporates specific trafficking mechanisms to unleash the full potential of tumor antigen-based immunotherapy.
Method
Cells
The murine melanoma cell line B16F10 and the dendritic cell line DC2.4 were cultured under standard conditions. The B16F10 cell line was used to establish a tumor model. The DC2.4 cell line was used to assess the intracellular distribution and localization of mRNA product.
Animals
C57BL/6 mice were obtained from Gempharmatech and were 7–8 weeks old at the initiation of the experiments. The mice were group-housed in ventilated cages at an ambient temperature of 21–23 °C, with an automated 12:12 h light–dark cycle, and provided with water and commercial rodent food. Animal care was conducted in compliance with the animal welfare guidelines of the University of Science and Technology of China. All animal experiments were approved by the Animal Care and Use Committee of the University of Science and Technology of China (USTCACUC24080124056).
Construction of clone and mRNA in vitro transcription
All mRNAs used in this study were synthesized via in vitro transcription using the modified pUC57 plasmid as a vector. mRNA synthesis was carried out by in vitro transcription from linearized plasmids using the MEGAscript® T7 Transcription Kit, following an optimized protocol. During this process, mRNA was co-transcriptionally capped at the 5' end with the CleanCap cap1 analog (Cat# N-7413, TriLink). After incubating for 6 h, the transcription products were isolated by selective precipitation with lithium chloride. The resulting mRNA pellet was reconstituted in nuclease- and endotoxin-free water. Concentration was measured with a Nanodrop, and purity was assessed through capillary electrophoresis. The final mRNA solution was aliquoted and stored at − 80 °C.
The non-coding region, T7 promoter, and polyA tail transcription element were incorporated at both ends of the mRNA coding sequence to form a complete T7 promoter-5'UTR-minigene-3'UTR-polyA transcribed region, replacing the lactose manipulator region in the pUC57 plasmid. EGFP mRNA and mTubulin mRNA encoded the full-length versions of EGFP and mTubulin, respectively. The EGFP-FcRn mRNA encoded the transmembrane region and intracellular segment of FcRn (referred to as FcRn throughout the text) at the 3'-end of the EGFP nucleic acid.
The Fluc mRNA encoded a multiepitope fragment (48–147, 238–315) of luciferase (BAA93575.1), which was ligated at the GS linker. The Fluc-FcRn mRNA encoded FcRn at the 3'-end of Fluc.The vaccine comprised two mRNAs. TRP1 mRNA encoded a multiepitope fragment of the antigen TRP1 (108–201, 354–469 for TRP1) and FcRn, and TRP2 encoded a multiepitope fragment of TRP2 (64–198, 358–464 for TRP2) and FcRn. The antigenic fragments were linked to one another, and to FcRn, by the GS linker.
Formulation of mRNA into LNP
The mRNA was solubilized in 100 mM pH 4.0 citrate buffer. The LNPs were composed of an ionizable lipid (SM102), helper lipids (DSPC and cholesterol), and a PEG-modified lipid (DMG-PEG2000), dissolved at a molar ratio of 50:10:38.5:1.5 (SM102:DSPC:cholesterol:DMG-PEG2000). The N/P ratio was 6:1. Subsequently, the lipids dissolved in ethanol were mixed with mRNA at a 3:1 volume ratio. Subsequently, the mixture was dialyzed against pH 7.4 buffer for 18 h, filtered through a 0.22 μm filter membrane to eliminate bacteria, and stored at 4 °C for future use.
Immunofluorescence
Cells were seeded at the count of 3 × 104, after 24 h for adherence, the cells were transfected with 5 μg of EGFP mRNA-LNP or EGFP-FcRn mRNA-LNP. After 48 h, wash the plates with pre-warmed PBS. Fix cells with 4% paraformaldehyde for 15 min and wash the cells 3 times with pre-chilled deionized water. Add permeabilization reagent 0.1% PBST to the wells and kept at room temperature for 15 min. Subsequently, after discarding the liquid, the blocking solution was added to the wells and blocked for 1 h at room temperature. Discard the liquid and add PBS-diluted Anti-EGFP Polyclonal Antibody (Solarbio, K009757P), Purified Mouse Anti-RAB11 (BD, 610656), and purified anti-CD107a(LAMP-1) (Biolegend, 328601) to the transfection wells and incubate overnight at 4 °C. After washing with PBST, add PBS-diluted Cy3 Conjugated Goat anti-mouse IgG (Solarbio, K1031G-Cy3) and FITC Conjugated Goat anti-rabbit IgG (Solarbio, SF134) to the wells, incubate for 1 h at room temperature, and wash 3 times with PBST. Drop 20 μL of antifluorescence quenching medium containing DAPI on the glass slide, remove the crawler to absorb the water, cover the slide, seal the edge with nail polish, and observe using ZEISS LSM880 with Airyscan confocal fluorescence microscopy.
Enzyme-linked immunospot assay
Antigen-specific T cell responses were assessed using the Mouse IFN-γ Pre-coated ELISPOT Kit (Dakewe Biotech, 2210006) following the manual instructions. Briefly, splenocytes from immunized mice with mRNA-LNP were isolated and resuspended in complete cell medium before being seeded onto plates pre-coated with mouse IFN-γ capture antibody at a density of 5 × 105 cells.The cells were stimulated with PMA, DMEM, single peptides, or peptide pools for 20 h. Subsequently, the supernatant was discarded, and the cells were lysed; clones with IFN-γ secreting ability became visible upon incubation with HRP-conjugated antibodies and chromogenic substrates.
Assessment of theraputic efficacy of vaccine
In order to evaluate the in vivo therapeutic efficacy of melanoma, we established mouse models of experimental subcutaneous tumors by inoculating B16F10 cells. For the experimental melanoma model, 1 × 105 B16F10 cells in 100 µL of PBS were implanted via s.c. injection into C57BL/6 male mice. Following implantation, mice were monitored for tumor growth every 3 days using a digital caliper. Treatment was administered via s.c. injection at a dose of 100 µg per mouse. The initial treatment was given on day 7 post-tumor inoculation, followed by three additional injections on days 10, 13, and 17 (in total, four injections).
Flow cytometry analysis
To evaluate mRNA-LNP expression in DC2.4 cells, the cells were plated in 12-well plates at a density of 100,000 cells per well and incubated for 24 h. Subsequently, the cells were transfected with mRNA-LNP. After a 48 h post-transfection, the cells were harvested and washed three times with PBS before analysis.
To identify immune cells in mice post-tumor treatment, tumor-infiltrating immune cells were isolated using density gradient centrifugation in the 40%-80% Percoll system. Then, splenocytes were isolated by grinding the spleen and lysing erythrocytes, followed by resuspension in PBS. The cells were incubated with fluorescently labelled antibodies for one hour, after which flow cytometry analysis was conducted.
For assessing T cell cytokine expression, splenocytes and tumor-infiltrating immune cells from treated mice were collected and resuspended in RPMI-1640 full medium. Then, phorbol 12-myristate 13-acetate (PMA), ionomycin, and monensin were added to stimulate the cells for four hours. After this, the cells were washed and stained with fluorescently labelled anti-CD3, CD8, and CD4 antibodies (Biolegend). Then, the cells were fixed and permeabilized with a Biolegend fixation and permeabilization kit according to the manufacturer's instructions. The cells were then intracellularly stained with fluorescently labelled cytokine antibodies and analyzed by flow cytometry (Beckman, CytoFLEX).
Cell viability assay in vitro.
DC2.4 cells were seeded in 96-well plates until reaching approximately 80% confluence confluence and subsequently treated with EGFP-mRNA-LNPs, EGFP-FcRn mRNA-LNPs, mTubulin-mRNA-LNPs, and a combination of TRP1-mRNA-LNPs and TRP2-mRNA-LNPs.Two days post-transfection, the mRNA-LNPs were removed and fresh medium was introduced. The CCK-8 reagent (YEASEN, 40203ES76) was added to each well at a volume of 10 μL per well and the plates were incubated at 37 °C for one hour prior to detection. The OD450 of each well was measured using a microplate reader, and cell viability was determined.
In vivo toxicity evaluation
To assess in vivo toxicity, serum was collected from vaccinated mice 7 days post the final injection (day 21) and from tumor-bearing mice at the end of the efficacy study (day 22).AST and ALT levels were measured using assay kits for AST (Solarbio) and ALT (Solarbio) following the manufacturers’ instructions.
Statistical analysis
All graphs were generated with GraphPad Prism 10 software, and statistical analyses were conducted using the same software, including one-way analysis of variance (ANOVA) and two-sided t tests. A P value less than 0.05 was considered statistically significant. All significant values are indicated in various figures as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study is supported by the National Key Research and Development Program of China (Grant Nos. 2022YFC2304102 and 2022YFC2303300), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB0490000), the National Natural Science Foundation of China (Grant Nos. 82272301 and 32100745), the Anhui Provincial Key Research and Development Project (Grant No. 2022i01020025) and the Fundamental Research Funds for the Central Universities (WK9100000001).
Author contributions
TJ, SZ and SX provided funding, designed the study, participated in data analysis, and revised the manuscript. MH designed the study, performed experiments, analyzed the data, and drafted the manuscript. Other authors participated in the experiments and/or revised the manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declaration
Conflict of interest
The authors declare that they have no conflict of interest relevant to this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shoubing Zhou, Email: zhoushoubingseu@126.com.
Shi Xu, Email: xushi@therarna.cn.
Tengchuan Jin, Email: jint@ustc.edu.cn.
References
- 1.Bentkowski P, Radwan J (2019) Evolution of major histocompatibility complex gene copy number. PLoS Comput Biol 15(5):e1007015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pishesha N, Harmand TJ, Ploegh HL (2022) A guide to antigen processing and presentation. Nat Rev Immunol 22(12):751–764 [DOI] [PubMed] [Google Scholar]
- 3.Dersh D, Hollý J, Yewdell JW (2021) A few good peptides: MHC class I- based cancer immunosurveillance and immunoevasion. Nat Rev Immunol 21:116–128 [DOI] [PubMed] [Google Scholar]
- 4.Garrido F (2019) MHC/HLA class I loss in cancer cells. Springer, Cham, pp 15–78 [Google Scholar]
- 5.Stoeckius M et al (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14(9):865–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreiter S et al (2015) Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520(7549):692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleri W et al (2017) The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol 8:278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dey J et al (2022) Designing a novel multi-epitope vaccine to evoke a robust immune response against pathogenic multidrug-resistant Enterococcus faecium bacterium. Gut Pathogens 14(1):21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dacon C, Tucker C, Peng L, Lee CCD, Lin TH, Yuan M, Cong Y, Wang L, Purser L, Williams JK, Pyo CW (2022) Broadly neutralizing antibodies target the coronavirus fusion peptide. Science 377:728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grifoni A, Sidney J, Vita R, Peters B, Crotty S, Weiskopf D, Sette A (2021) SARS-CoV-2 human T cell epitopes: adaptive immune response against COVID-19. Cell Host Microbe 29(7):1076–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skwarczynski M, Toth I (2016) Peptide-based synthetic vaccines. Chem Sci 7(2):842–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keskin DB et al (2019) Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565(7738):234–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Princiotta MF et al (2003) Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity 18(3):343–354 [DOI] [PubMed] [Google Scholar]
- 14.Yewdell JW, Reits E, Neefjes J (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol 3(12):952–961 [DOI] [PubMed] [Google Scholar]
- 15.Calin-Laurens V et al (1992) High efficiency of endogenous antigen presentation by MHC class II molecules. Int Immunol 4(10):1113–1121 [DOI] [PubMed] [Google Scholar]
- 16.Storni T, Bachmann MF (2004) Loading of MHC class I and II presentation pathways by exogenous antigens: a quantitative in vivo comparison. J Immunol 172(10):6129–6135 [DOI] [PubMed] [Google Scholar]
- 17.Kreiter S et al (2008) Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J Immunol 180(1):309–318 [DOI] [PubMed] [Google Scholar]
- 18.Alspach E et al (2019) MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 574(7780):696–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto K et al (2020) Selective autophagy of MHC-I promotes immune evasion of pancreatic cancer. Autophagy 16(8):1524–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu JQ et al (2020) IL-2 enhanced MHC class I expression in papillary thyroid cancer with Hashimoto’s thyroiditis overcomes immune escape in vitro. J Cancer 11(14):4250–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stifter K et al (2020) IFN-γ treatment protocol for MHC-Ilo/PD-L1+ pancreatic tumor cells selectively restores their TAP-mediated presentation competence and CD8 T-cell priming potential. J Immunother Cancer 8(2):e000692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt EGW et al (2017) Direct demonstration of a neonatal Fc receptor (FcRn)-driven endosomal sorting pathway for cellular recycling of albumin. J Biol Chem 292(32):13312–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin WL et al (2001) Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell 7(4):867–877 [DOI] [PubMed] [Google Scholar]
- 24.Gan Z et al (2009) Analyses of the recycling receptor, FcRn, in live cells reveal novel pathways for lysosomal delivery. Traffic 10(5):600–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y et al (2018) Truncation of the murine neonatal fc receptor cytoplasmic tail does not alter IgG metabolism or transport in vivo. J Immunol 200(4):1413–1424 [DOI] [PubMed] [Google Scholar]
- 26.Tesar DB, Björkman PJ (2010) An intracellular traffic jam: Fc receptor-mediated transport of immunoglobulin G. Curr Opin Struct Biol 20(2):226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmoud IS et al (2017) Signal dependent transport of a membrane cargo from early endosomes to recycling endosomes. Eur J Cell Biol 96(5):418–431 [DOI] [PubMed] [Google Scholar]
- 28.Girard J, Moussion C, Förster R (2012) HEVs, lymphatics and homeostatic immune cell trafficking in lymph nodes. Nat Rev Immunol 12(11):762–773 [DOI] [PubMed] [Google Scholar]
- 29.Miao L, Zhang Y, Huang L (2021) mRNA vaccine for cancer immunotherapy. Mol Cancer 20(1):41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zong Y et al (2023) Lipid nanoparticle (LNP) enables mRNA delivery for cancer therapy. Adv Mater 35(51):2303261 [DOI] [PubMed] [Google Scholar]
- 31.Schoenmaker L et al (2021) mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int J Pharm 601:120586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lahusen TJ, Deng C (2015) SRT1720 induces lysosomal-dependent cell death of breast cancer cells. Mol Cancer Ther 14(1):183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W et al (2009) Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine 27(36):5013–5019 [DOI] [PubMed] [Google Scholar]
- 34.Mansour M et al (2007) Therapy of established B16–F10 melanoma tumors by a single vaccination of CTL/T helper peptides in VacciMax®. J Transl Med 5(1):1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fässler M et al (2019) Antibodies as biomarker candidates for response and survival to checkpoint inhibitors in melanoma patients. J ImmunoTherapy Cancer 7(1):1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kortekaas KE et al (2020) CD39 identifies the CD4+ tumor-specific T-cell population in human cancer. Cancer Immunol Res 8(10):1311–1321 [DOI] [PubMed] [Google Scholar]
- 37.Shen Y et al (2024) CD39hi identifies an exhausted tumor-reactive CD8+ T cell population associated with tumor progression in human gastric cancer. Pharmacol Res 202:107122 [DOI] [PubMed] [Google Scholar]
- 38.Zhu W et al (2021) CD8+CD39+ T cells mediate anti-tumor cytotoxicity in bladder cancer. Onco Targets Ther 14:2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosas-Taraco A et al (2010) Targeting and retention of antigen to the endoplasmic reticulum enhances immune tumor protection. Hum Gene Ther 21(10):1421–1421 [Google Scholar]
- 40.Martínez-Puente DH et al (2021) Targeting E7 antigen to the endoplasmic reticulum degradation pathway promotes a potent therapeutic antitumor effect. J Drug Target 29(10):1102–1110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.