Abstract
Staphylococcus aureus is a relevant pathogen in bloodstream infections (BSI), and the emergency of the COVID-19 pandemic increased its antimicrobial resistance. S. aureus isolates from BSI (September/2019 - March/2021) were analyzed phenotypically and molecularly, in addition to the clinical features of the patients. Of 88 S. aureus isolates recovered from 85 patients, 25 were isolated before the pandemic and 63 during it, and 16 were from patients with COVID-19. A rate of 45.5% of methicillin-resistant isolates (MRSA) were found, and 5% of them were ceftaroline susceptible dose-dependent. Daptomycin non-susceptibility was observed in 9.1% of isolates. The USA800/ST5/SCCmecIV lineage was prevalent among MRSA isolates (41.8%). Besides, 30.2% of the isolates were associated with community-associated MRSA (CA-MRSA) genotypes. There was a significant impact on the resistance rates for cefoxitin, clindamycin and erythromycin among S. aureus isolates from BSI in COVID-19 patients and association with the previous use of azithromycin by them (p < 0.05). A clonal alternation and an increase in the emergence of CA-MRSA lineages were also found, highlighting the importance of constant microbiological surveillance.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84307-1.
Keywords: COVID-19, Bloodstream infection, S. aureus, Antimicrobial resistance, MRSA lineages
Subject terms: Antimicrobial resistance, Pathogens, Policy and public health in microbiology
Introduction
Staphylococcus aureus is one of the main pathogens associated with bloodstream infections (BSI) worldwide1. The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2020, which led to the COVID-19 (coronavirus disease 2019) pandemic has disrupted healthcare systems and practices causing a great impact on the development of secondary bacterial infections2, especially among critically ill patients3. Studies from this period suggest an increase in the incidence of BSI caused by multidrug-resistant bacteria such as methicillin-resistant S. aureus (MRSA)2.
During the pandemic, some studies highlighted an increase in the incidence of BSI by MRSA, especially among COVID-19 patients2,3. A study conducted in Italy, comparing the incidence of BSI before and after the pandemic, revealed that S. aureus was the most frequently isolated microorganism from COVID-positive patients, with MRSA constituting 40.5% of all S. aureus isolates3. Another study conducted in Madrid, Spain, described an increase in the incidence of S. aureus bacteremia during the pandemic, with rates reaching 1.96 episodes per 1000 non-COVID-19 admissions, while among COVID-19 patients it rose to 10.59 episodes per 1000 admissions. Notably, MRSA isolates were more commonly identified among COVID-positive patients4.
The COVID-19 pandemic played an important role in the antimicrobial resistance scenario of S. aureus isolates, by selecting and helping the dissemination of multidrug-resistant isolates among hospital and community environments. Some authors had reported an increased resistance to oxacillin, erythromycin, and clindamycin for S. aureus recovered from BSI and other sources during the pandemic5,6. It is noteworthy to mention that the increased resistance to erythromycin in S. aureus could be associated with high azithromycin use5,7.
In Brazil, in recent years, well-established clones have been giving way to new clones in hospitals in Rio de Janeiro, as observed by our group8,9. Recently we found community-associated MRSA (CA-MRSA) isolates causing BSI among patients in our hospital9. Furthermore, in the pre-pandemic period, one isolate from the LV-USA300/ST8 lineage was detected for the first time as a cause of BSI10. The emergence of CA-MRSA causing infections in the hospital setting is worrying, given their virulent attributes that provide greater adaptability to hospital environments11. Here, we characterized S. aureus isolates from BSI of patients admitted to a University Hospital in Rio de Janeiro before and during the COVID-19 pandemic, comparing the resistance rates, previous use of azithromycin, and the bacterial genetic background in these periods.
Materials and methods
Clinical isolates and setting
We conducted a prospective cohort study to evaluate the clinical aspects of the patients and phenotypic and molecular profiles of S. aureus isolates recovered from episodes of BSI in adult individuals (> 18 years old) admitted to the Clementino Fraga Filho University Hospital, between September of 2019 and March of 2021. This is a tertiary care public hospital in Rio de Janeiro with 300 active beds. During the COVID-19 pandemic, this hospital was a reference center for treating the disease, with 37 intensive care beds allocated for COVID-19 patients and 14 for non-COVID-19 patients, totaling 51 active ICU beds. The study was approved by the Human Research Ethics Committee of Clementino Fraga Filho University Hospital (CAAE 40458520.7.0000.5257), and informed consent was obtained from all subjects or their legal guardian(s). All experiments were performed following relevant guidelines and regulations.
We analyzed the first bacterial isolate recovered from an episode of S. aureus BSI, with subsequent documentation of blood cultures. Besides, for each patient enrolled in the study, demographic data, classification of the BSI episode, treatment, COVID-19 status during the pandemic, length of stay, and outcome were collected. Data on azithromycin use initiated for the treatment of COVID-19 before hospitalization were also obtained.
All blood cultures were processed using the BacT/ALERT system (BioMerieux, Durham, NC, USA). Staphylococcus aureus identification was performed by the automated VITEK2® system (BioMerieux) and confirmed by MALDI-TOF MS® (Matrix Assisted Laser Desorption Ionization/ Time of Flight Mass Spectrometry) (Bruker Daltonics, Billerica, MA, USA).
Antimicrobial susceptibility tests and SCCmec typing
All S. aureus isolates were characterized by their antimicrobial susceptibility profile using the disk-diffusion test method (Oxoid, Cambridge, UK), according to Clinical & Laboratory Standards Institute12. The broth microdilution (BMD) method was performed to determine the Minimum Inhibitory Concentration (MICs) for daptomycin, linezolid, oxacillin, and vancomycin (Sigma-Aldrich Chemical Company, St Louis, MO, USA). For ceftaroline, the gradient diffusion method (Etest®, BioMérieux) was used to access the MIC values for MRSA isolates12. S. aureus presenting a MIC ≥ 2 mg/L for vancomycin were screened for the hVISA (heteroresistant vancomycin-intermediate S. aureus) phenotype13. S. aureus ATCC 25,923 and ATCC 29,213 were used as controls for the disk-diffusion and BMD tests, respectively, and ATCC 29,213 (MSSA) and Mu3 (hVISA) were used as controls for the hVISA screening.
All MRSA isolates characterized as oxacillin-resistant by the disk-diffusion method were characterized as their SCCmec type14 by multiplex polymerase chain reaction (PCR), using bacterial DNA obtained using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany).
Genotypic profile of MRSA isolates
All MRSA isolates were typed by pulsed-field gel electrophoresis (PFGE) after digestion of whole-cell DNA with SmaI in a CHEF-DRIII® system (Bio-Rad, Richmond, CA, USA)15. The PFGE fingerprints were compared by the unweighted pair-group method with arithmetic mean clustering analysis, applying the Dice correlation coefficient. The clonal lineages were defined by comparison with national8 and international clones16. Representative isolates from each genotype, identified in PFGE, underwent Multilocus Sequence Typing (MLST)17.
A PCR for the detection of PVL genes was carried out for all MRSA isolates18 and those belonging to the USA300/ST8/SCCmecIV lineage also underwent ACME typing19.
Statistical analysis
Categorical variables were compared using the chi-square or Fisher exact tests, and continuous variables were compared using the Mann-Whitney U test. A p-value < 0.05 was statistically significant. All analyses were performed using GraphPad Prism software, version 8 (GraphPad*, San Diego, CA, USA).
Results
Patients and S. aureus BSI episodes
A total of 85 patients were enrolled in the present study presenting 88 S. aureus BSI episodes – one patient had two S. aureus BSI episodes, and another patient had three episodes. Among them, 25 had S. aureus BSI before the onset of the pandemic, whereas 60 developed the infection during the SARS-CoV-2 pandemic. Throughout the pandemic period, patients were further classified based on their COVID-19 diagnosis at the time of BSI occurrence. Consequently, out of the 60 patients affected during the pandemic, 16 received a concurrent diagnosis of COVID-19, confirmed through positive SARS-CoV-2 qPCR results.
We observed that 56.5% of the patients in the study were male, and the mean age was 57 years (15 to 84 age range). Most episodes were hospital-acquired (62.5%), clinically characterized as primary bloodstream infection (78.4%), and associated with central venous catheter usage (67%) (Supplementary Table 1). A higher number of S. aureus BSI episodes occurred among patients without COVID-19 (53.4%) during the pandemic period. Of the total, 46 (57.1%) patients died, with most of them (68.8%) being diagnosed with COVID-19. However, no statistically significant differences were observed between pre-pandemic and pandemic periods or between patients with and without COVID-19, except for the previous use of azithromycin, which was significantly associated with patients diagnosed with COVID-19 (p-value < 0.05). Before the pandemic, none of the patients described the prior use of the antimicrobial. However, during the pandemic, 68.8% of patients with COVID-19 reported previous use of the drug in community settings as outpatients.
Antimicrobial susceptibility and previous use of azithromycin
All 88 S. aureus isolates recovered from BSI were susceptible to trimethoprim-sulfamethoxazole, linezolid, and vancomycin. Resistant rates were observed for penicillin G (94.3%), erythromycin (52.3%), clindamycin (34.1%), ciprofloxacin (20.4%), gentamycin (14.8%) and rifampicin (2.3%).
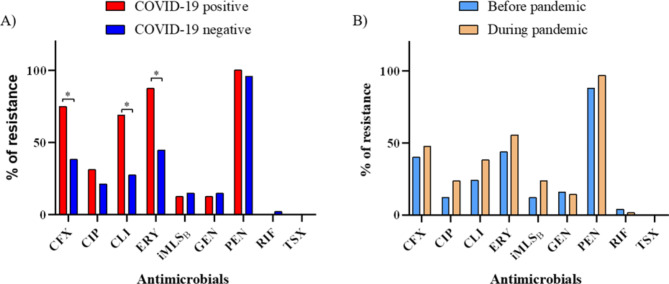
Among 40 (45.5%) S. aureus isolates that were identified as MRSA, 10 were collected before the pandemic, while 30 were from the pandemic period. Although no statistical differences were observed between the two periods, MRSA isolates were more frequent among patients positive for COVID-19 compared to those without COVID-19 (75% vs. 38.3%) (p-value < 0.05). Furthermore, isolates recovered from patients with COVID-19 exhibited higher resistance rates for clindamycin (68.8% vs. 27.7%) and erythromycin (87.5% vs. 44.7%) (p-value < 0.05). About 20% of all S. aureus isolates showed the iMLSB (inducible macrolide-lincosamide-streptogramin B) resistance phenotype, and those isolated from patients during the pandemic showed higher rates of induced resistance (23.8% vs. 12%) (Fig. 1).
Fig. 1.
Antimicrobial resistance rates among S. aureus isolated during (A) and before the COVID-19 pandemic (B). * p-value < 0.05. CFX – Cefoxitin; CIP – Ciprofloxacin; CLI – Clindamycin; ERI – Erythromycin; GEN – Gentamycin; PEN – Penicillin G; RIF – Rifampicin; TSX - Trimethoprim-sulfamethoxazole; iMLSB - inducible macrolide-lincosamide-streptogramin B resistance.
By comparing MRSA and MSSA isolates during both periods and among patients with and without COVID-19, we observed a significantly higher rate of resistance to erythromycin among MRSA isolates, particularly among those recovered from patients with the disease (p-value < 0.05). Moreover, a higher number of MRSA isolates exhibited resistance to ciprofloxacin, regardless of the period analyzed (p-value < 0.05) (Supplementary Table 2).
By analyzing the MIC90 results we noted higher values among isolates recovered during the pandemic period for daptomycin and oxacillin (Table 1). In the pre-pandemic period, a MIC90 of 1 mg/L was observed for daptomycin, while during the pandemic this value reached 2 mg/L. Additionally, we identified that 9.1% (n = 8) of S. aureus isolates presented MIC values > 1 mg/L (non-susceptibility) and seven of them were retrieved during the pandemic period. Regarding oxacillin, the MIC90 values were 64 mg/L during the pre-pandemic period, and > 256 mg/L in the pandemic period, and when we compared the isolates recovered during the pandemic, those recovered from COVID-19 patients presented a higher MIC90 value (> 256 mg/L) in relation to isolates from patients without COVID-19 (128 mg/L) (p-value < 0.05). Additionally, as observed by the disk-diffusion method, MRSA isolates were more prevalent among COVID-19 patients (p-value < 0.05).
Table 1.
MIC90 (mg/L) values and non-susceptibility rates among 88 Staphylococcus aureus isolated from bloodstream infections before and during the COVID-19 pandemic, between September 2019 and March 2021. MIC – Minimum Inhibitory concentration; MIC90 – MIC value capable of inhibit 90% of isolates; N – number; R – resistant; NS – non susceptible; *p-value < 0.05.
| Antimicrobial | Before pandemic (N = 25) |
During pandemic | ||||||
|---|---|---|---|---|---|---|---|---|
| Total (N = 63) |
COVID-19 Positive (N = 16) |
COVID-19 Negative (N = 47) |
||||||
| MIC90 |
N (%) of NS isolates |
MIC90 |
N (%) of NS isolates |
MIC90 |
N (%) of NS isolates |
MIC90 |
N (%) of NS isolates |
|
| Daptomycin | 1 | 1 (4) | 2 | 7 (11.1) | 1 | 1 (6.3) | 2 | 6 (12.8) |
| Linezolid | 1 | 0 (0) | 1 | 0 (0) | 1 | 0 (0) | 1 | 0 (0) |
| Oxacillin | 64 | 10 (40) | > 256 | 30 (47.6) | > 256* | 12 (75)* | 128* | 18 (38.3)* |
| Vancomycin | 2 | 0 (0) | 2 | 0 (0) | 2 | 0 (0) | 1 | 0 (0) |
Although no vancomycin-resistant isolate was detected, we observed a MIC90 of 2 mg/L during both periods (Table 1). Among the 88 S. aureus isolates, ten (6 MRSA and 4 MSSA) presented a MIC of 2 mg/L for vancomycin. However, none of these isolates presented the hVISA phenotype (Supplementary Tables 3 and 4).
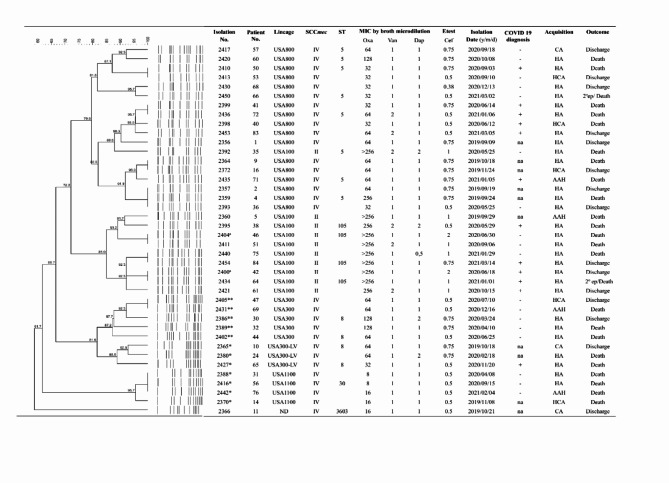
All MRSA isolates underwent ceftaroline MIC determination using the Etest® test (BioMérieux). Only two (5%) (isolates 2400 and 2404) were classified as susceptible dose-dependent (SDD). Notably, both were retrieved during the pandemic period, of which one was collected from a COVID-19-positive patient (Fig. 2; Supplementary Table 3).
Fig. 2.
Dendrogram of the PFGE patterns and characteristics related to the genetic background of 40 MRSA isolates recovered from bloodstream infection. LV: Latin American Variant; SCCmec: Staphylococcal cassette chromosome mec; ST: Sequence Type; MIC: Minimal Inhibitory Concentration in mg/L; Oxa: oxacillin; Van: vancomycin; Dap: daptomycin; Cef: ceftaroline; CA: Community acquisition; HA: hospital acquisition; AAH: Acquisition in another hospital; HCA: Healthcare acquisition; na: not applicable (strain isolated before pandemic); ep: episode; *: pvl gene positive; **: pvl gene and ACME positive; a: susceptible-dose dependent for ceftaroline.
As already mentioned, patients with COVID-19 frequently harbored isolates resistant to clindamycin and erythromycin. Consequently, data regarding the use of azithromycin before the hospitalization, an antibiotic in the same class as erythromycin was collected for all 85 patients. In the period before the pandemic, no patients report the prior use of this drug. However, during the pandemic, its use was reported by 15 patients, representing a significant increase in the previous use of this antimicrobial compared to the pre-pandemic period (p = 0.0047) (Table 2). Furthermore, when comparing BSI episodes that occurred during the pandemic, the prior use of azithromycin was more prevalent among patients with COVID-19 (68.8%; 11/16) than among those without a history of the disease (8.5%; 4/47) (p < 0.0001).
Table 2.
Distribution of the 88 bloodstream infection episodes caused by Staphylococcus aureus according to the previous use of azithromycin and the resistance to erythromycin, before and during the COVID-19 pandemic.
| Characteristics | Total |
N (%) of episodes |
p value |
N (%) of episodes during COVID-19 pandemic |
p value | |||
|---|---|---|---|---|---|---|---|---|
| Before COVID-19 pandemic | During COVID-19 pandemic | Patients with COVID-19 | Patients without COVID-19 |
|||||
| Previous use of Azithromycin |
n = 88 | n = 25 | n = 63 | n = 16 | n = 47 | |||
| 15 (17) | 0 (0) | 15 (23.8) | 0.0047 | 11 (68.8) | 4 (8.5) | < 0.0001 | ||
| Erythromycin resistance | n = 47 | n = 11 | n = 36 | n = 15 | n = 21 | |||
|
Previous use of Azithromycin |
13 (27.7) | 0 (0) | 13 (36.1) | 0.0215 | 10 (66.7) | 3 (14.3) | 0.0019 | |
The analysis of prior azithromycin use was also conducted for erythromycin-resistant isolates (Table 2). Before the pandemic, 23.4% (11/47) of S. aureus isolates were resistant to erythromycin, while during the pandemic, the detected resistance was 76.5% (36/47), revealing a significantly higher incidence of prior azithromycin use during the pandemic (p = 0.0215). Specifically, among patients who used azithromycin, a percentage of 36.1% (13/36) of BSI episodes caused by erythromycin resistant isolates was observed during the pandemic, while no case of BSI caused by erythromycin-resistant isolates were found before the pandemic. Prior use of azithromycin was observed in 66.7% (10/15) of patients with COVID-19 who presented BSI due to erythromycin-resistant S. aureus, a significantly higher percentage compared to cases of patients without the disease, where only 14.3% (3/21) had previously used the antimicrobial (p = 0.0014). Furthermore, the iMLSB phenotype was associated with the use of azithromycin, as isolates from approximately half of the patients who used this drug showed induced resistance (p = 0.0057) (Supplementary Tables 3 and 4).
Genotypic profile of MRSA isolates
Most MRSA isolates were associated with the USA800/SCCmecIV/ST5 lineage (42.5%; 17/40), which was the prevalent lineage found in both periods, pre-pandemic (50%; 5/10) and pandemic (40%; 12/30), as well as among patients with COVID-19 (50%; 6/12) (Fig. 2). Another lineage, the USA100/SCCmecII/ST5 or ST105 were related to 25% (10/40) of all MRSA isolates, and five of them were recovered from patients with COVID-19. Notably, two isolates belonging to this lineage, one from a patient with COVID-19 (isolate number 2400) and another from a patient without COVID-19 (isolate number 2404) were characterized as susceptible dose-dependent (SDD) to ceftaroline (Supplementary Table 3).
Among 40 MRSA isolates, 12 (30%) belonged to the USA300/SCCmecIV/ST8 community lineage (12.5%; 5/40) and were all recovered during the pandemic. Besides these, USA1100/SCCmecIV/ST30 (10%; 4/40) and USA300-LV/SCCmecIV/ST8 (7.5%; 3/40) were also found. All these community-associated MRSA isolates were positive for the pvl gene, while USA300-LV isolates lacked the ACME operon. Classical USA300 isolates showed resistance to erythromycin while USA300-LV isolates were sensitive to this drug (Supplementary Tables 3 and 4). One MRSA isolate (2366) from the pre-pandemic period was characterized as belonging to the ST3603 lineage (Fig. 2).
Discussion
During the COVID-19 pandemic, several authors observed an increase in MRSA rates of isolates from BSI in patients with the disease2–4. In the present study, out of 88 S. aureus isolates, 45.5% were characterized as MRSA. Although no statistical differences were observed between MRSA rates in the periods, this pathogen was isolated more frequently from patients with COVID-19 (75%) than those without the viral infection (38.3%) (p-value < 0.05), as also shown by other authors4,20. Sands and coworkers, in 2023, conducted a cross-sectional retrospective analysis regarding the occurrence of MRSA BSI in patients with and without COVID-19 from hospitals in the United States, between January 2019 and March 2022. The authors described a higher rate of MRSA isolates among COVID-19 patients (11.2%), than among patients without the disease or from pre-pandemic period (3.7% each)20. This is a concerning fact, as morbidity and mortality rates are higher among patients with MRSA infections21. We also found that isolates recovered from patients with COVID-19 exhibited higher resistance rates for clindamycin (68.8% vs. 27.7%) and erythromycin (87.5% vs. 44.7%) (p-value < 0.05). López-Jácome and coworkers, in 2022, compared the antimicrobial susceptibility of different microorganisms from blood, including S. aureus, and described an increase in erythromycin resistance in Mexico during the COVID-19 pandemic5. In 2023, Serra and coworkers found significant resistance to erythromycin among S. aureus isolates from BSI in an Italian hospital. However, the resistance rates to clindamycin (42.2% vs. 50%) and erythromycin (60% vs. 57.1%) were similar for isolates from individuals with and without COVID-1922. Unfortunately, few studies have evaluated and compared the clindamycin and erythromycin resistance rates of S. aureus isolates recovered during the pandemic, making a comparative analysis difficult.
Vancomycin is commonly prescribed as the primary treatment for MRSA BSI, with determination of the minimum inhibitory concentration (MIC) crucial for therapeutic success23. Previous studies of our group recovered VISA and hVISA isolates with vancomycin MIC90 of 2 mg/L causing BSI at the same University Hospital, between 2011 and 201513. But in a study carried out from 2016 to 20189, and in the present study, we observed vancomycin MIC90 values between 1 and 2 mg/L, without isolation of VISA isolates, suggesting that MRSA isolates remain susceptible to this drug in our hospital despite higher values of MIC for vancomycin. Daptomycin is considered as an alternative drug for the treatment of MRSA BSI when vancomycin MIC values > 1.5 mg/L are found24. Despite most isolates being characterized as daptomycin susceptible in the present study, eight (9.1%) isolates showed non-susceptibility to this drug (MIC > 1 mg/L)25. A Chinese study analyzed 472 clinical MRSA isolates collected from 2015 to 2017 and found all isolates sensitive to daptomycin, but 35.2% of them exhibited a higher MIC (> 0.5 mg/L)26. Furthermore, CC5/ST5 isolates were less susceptible to daptomycin. In previous studies at the same hospital, initially between 2011 and 2015, we had found 23.5% of S. aureus isolates from BSI not susceptible to daptomycin8, and in the following years, between 2016 and 2019, the percentage fell to 13.8%9. The decrease in the isolation of VISA isolates and daptomycin non-susceptible isolates may be associated with the change in the molecular epidemiology of S. aureus lineages recovered at our study hospital, as resistance phenotypic profiles were generally associated with USA100/ST5 and ST105 isolates carrying SCCmec II8,9,13, and have been replaced in recent years, but mainly in the present study, by CA-MRSA isolates carrying the SCCmec IV. The efficacy of the combination of daptomycin with ceftaroline for the treatment of MRSA BSI has been shown27,28. However, in Brazil, ceftaroline is only approved for the treatment of MRSA pneumonia or skin and soft tissue infections. Despite this, two (5%) MRSA isolates presented susceptibility dose-dependent (SDD) for this drug in this study, similar to what was found by other authors29,30. The emergency of SDD MRSA isolates for ceftaroline imposes a challenge, as this drug has been proven to be an important therapeutic option for the treatment of recurrent BSI by MRSA27,28.
It is noteworthy that although no significant differences were observed between the two periods for the sociodemographic and clinical characteristics of the patients analyzed, we observed that previous use of azithromycin was reported during the pandemic, mainly among patients with COVID-19. The use of azithromycin as an early therapy for COVID-19 was a usual practice in Brazil31. In a retrospective cohort study conducted in Minas Gerais, Brazil, where the authors evaluated medical record data and microbiological clinical specimens from 79 COVID-19 hospitalized patients with secondary bacterial/fungal infections, it was observed that azithromycin was one of the main drugs previously used by these patients31. Other studies in different countries also described the increased prescription and use of azithromycin during the early stages of the pandemic32,33. Perella and coworkers, in 2023, compared the hospital antibiotic consumption in 2020–2021 with those related to 2019 in Italian hospitals and found an increased consumption of azithromycin in March and April 202033. Therefore, due to the broad use of antimicrobials for the early treatment of COVID-19, especially azithromycin, it was not a surprise that BSI episodes caused by erythromycin, clindamycin, and ciprofloxacin-resistant S. aureus were significantly found among COVID-19 patients, characterizing the general selective pressure caused by previous use of the drug.
Clinicians must be aware of the variable MRSA clones circulating in their hospital to use the most appropriate empiric antimicrobial therapy. Since 2008, our group has reported the complete substitution of the Brazilian endemic clone (ST239/SCCmec III) by USA100/ST5/SCCmecII and ST105/SCCmecII lineages in our hospital8,9,13,34. However, in the present study, these lineages were related to only 25% (10/40) of all MRSA isolates, with a higher number of isolates identified during the pandemic. Gu and coworkers, in 2023, described that ST5/SCCmecII remains the prevalent MRSA lineage in BSI even after COVID-19 pandemic in Wuhan, China35. However, among the MRSA isolates of the present study the majority (42.5%) was associated with the USA800/SCCmecIV/ST5 lineage, found in both periods, as well as among patients with COVID-19. This lineage, also known as the Pediatric clone, has been usually described as a less frequent clone causing BSI36–38. McGuiness and coworkers, in 2021, described a clonal outbreak across Australia caused by CC5/ST5 MRSA isolates that possessed the trimethoprim resistance gene dfrG within SCCmec IVo, but all were sensitive to this drug, suggesting that the presence of dfrG within SCCmec IVo provides a selective advantage to this lineage39. In the present study, all USA800/ST5/SCCmec IV isolates were also sensitive to trimethoprim-sulfamethoxazole. It is interesting to mention that both USA800 and USA100 belong to the same clonal complex (CC) and many isolates also present the sequence type (ST), differing only in the types of SCCmec they carry. Further analyses are needed to better understand why these lineages have become more prevalent.
Additionally, in the present study, we found 30% of CA-MRSA isolates that included eight USA300/SCCmecIV/ST8 (among them three USA300-LV), and four USA1100/SCCmecIV/ST30, the majority (75%) recovered during the pandemic period. More recently, studies from our group described, for the first time in our hospital, the occurrence of BSI caused by isolates related to the MRSA USA300/ST8/SCCmec IV pandemic clone9, as well as its Latin-American variant, USA300-LV, that lacks the ACME operon10. Other studies have also been reporting the emergence of USA300 among MRSA from hospital-acquired BSI40. According to Dyzenhaus and coworkers, in 2023, USA300 strains may optimize their fitness through mutations that alter the regulation of virulence and contribute to their success in a new environment, which may leverage this recent introduction into hospitals41. It is important to mention that all CA-MRSA isolates were positive for the pvl gene, confirming the emergence of virulent MRSA lineages causing BSI and pointing to a change in molecular epidemiology in the hospital during the pandemic.
The present study showed a change in the scenario of BSI caused by S. aureus isolates associated with the COVID-19 pandemic, with an increase in isolates resistant to methicillin, clindamycin, and erythromycin. This may have been driven by the exacerbated use of azithromycin in the community during the pandemic. A clonal alternation and an increase in the emergence of CA-MRSA lineages were also found during the COVID-19 pandemic, highlighting the importance of constant surveillance to better understand the dynamics of this pathogen as the main causative agent of bacteremia worldwide.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
S.A.N. and the Infection Control Group HUCFF/UFRJ took care of the patient. C. O.W., T.L.R.O. and A.L.P.F. analyzed the bacterial samples. S.A.N. and T.L.R.O. gathered the clinical data. R.C.C. and K.R.N.S. drafted the manuscript. R.C.C. prepared Fig. 1. T.L.R.O. and C.O.W. prepared Fig. 2. K.R.N.S., R.C.C. and S.A.N. supervised and critically reviewed the manuscript. All authors approved the final version.
Funding
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil [CAPES, - Finance Code 001]; Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro [FAPERJ, grant numbers: #E- 26/200.419/2023, #E-26/010.000172/2016, #E-26/010.001463/2019 to K.R.N.S., #E- 26/211.554/2019 (to K.R.N.S. and R.C.C.), #E-26/010.101056/2018, #E-26/201.071/2021, #E- 26/211.284/2021 and #E-26/010.001.280/2016 to K.R.N.S. and E-26/010.002497/2019, E-26/211.429/2021 and E-26/201.285/2022 to R.C.C. and E-26/205.939/2022 to T.L.R.O.] and Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq, grant numbers: #307594/2021-1 to K.R.N.S.].
Data availability
The authors declare that the data are available from the corresponding author on request (Availability of data).
Declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the Human Research Ethics Committee of HUCFF (CAAE 40458520.7.0000.5257).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Carolina de Oliveira Whitaker and Tamara Lopes Rocha de Oliveira contributed equally to this work
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Raiane Cardoso Chamon, Email: raianechamon@id.uff.br.
Kátia Regina Netto dos Santos, Email: santoskrn@micro.ufrj.br.
Infection Control Group HUCFF/UFRJ:
Simone Aranha Nouér, Anna Carla Castiñeiras, Christiany Moçali Gonzalez, Joana Freire, Luiz Felipe Guimarães, and Claudia Regina da Costa
References
- 1.Bai, A. D. et al. Staphylococcus aureus bacteraemia mortality: A systematic review and meta-analysis. Clin. Microbiol. Infect.28 (8), 1076–1084. 10.1016/j.cmi.2022.03.015 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Segala, F. V. et al. Impact of SARS-CoV-2 epidemic on antimicrobial resistance: A literature review. Viruses20(11), 2110. 10.3390/v13112110 (2021). [DOI] [PMC free article] [PubMed]
- 3.Segala, F. V. et al. Incidence of bloodstream infections due to multidrug-resistant pathogens in ordinary wards and intensive care units before and during the COVID-19 pandemic: A real-life, retrospective observational study. Infection51 (4), 1061–1069. 10.1007/s15010-023-02000-3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falces-Romero, I., Bloise, I., García-Rodríguez, J., Cendejas-Bueno, E. & SARS-CoV-2 Working Group. Staphylococcus aureus bacteremia in patients with SARS-CoV-2 infection. Med. Clin. (Barc). Jun 9 (11), 495–498. 10.1016/j.medcli.2023.01.012 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Jácome, L. E. et al. Increment antimicrobial resistance suring the COVID-19 pandemic: Results from the Invifar Network. Microb. Drug Resist. 28(3), 338–345 (2022). 10.1089/mdr.2021.0231 [DOI] [PubMed]
- 6.Taleb, M. H., Elmanama, A. A., Taleb, A. H. & Tawfick, M. M. Pre- and post-COVID-19 antimicrobial resistance profile of bacterial pathogens, a comparative study in a tertiary hospital. J. Infect. Dev. Ctries.17 (5), 597–609. 10.3855/jidc.17791 (2023). May 31;. [DOI] [PubMed] [Google Scholar]
- 7.Seabra, G. et al. Azithromycin use in COVID-19 patients: Implications on the antimicrobial resistance. Curr. Top. Med. Chem.21 (8), 677–683. 10.2174/156802662108210319145317 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Damasco, A. P. et al. Daptomycin and vancomycin non-susceptible methicillin-resistant Staphylococcus aureus clonal lineages from bloodstream infection in a Brazilian teaching hospital. Braz J. Infect. Dis.23 (2), 139–142. 10.1016/j.bjid.2019.03.003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Augusto, M. F. et al. Pandemic clone USA300 in a Brazilian hospital: Detection of an emergent lineage among methicillin-resistant Staphylococcus aureus isolates from bloodstream infections. Antimicrob. Resist. Infect. Control. 14 (1), 114. 10.1186/s13756-022-01154-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitaker, C. O., Chamon, R. C., de Oliveira, T. L. R., Nouér, S. A. & Dos Santos, K. R. N. Systemic infection caused by the methicillin-resistant Staphylococcus aureus USA300-LV lineage in a Brazilian child previously colonized. Braz J. Infect. Dis.27 (2), 102737. 10.1016/j.bjid.2022.102737 (2023). Mar-Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyzenhaus, S. et al. MRSA lineage USA300 isolated from bloodstream infections exhibit altered virulence regulation. Cell Host Microbe31 (2), 228–242.e8 (2023). 10.1016/j.chom.2022.12.003 [DOI] [PMC free article] [PubMed]
- 12.CLSI. Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing. 30th Ed. (Clinical and Laboratory Standards Institute, 2020).
- 13.da Costa, T. M. et al. Clinical and microbiological characteristics of heteroresistant and vancomycin-intermediate Staphylococcus aureus from bloodstream infections in a Brazilian teaching hospital. PLoS One11(8), e0160506. 10.1371/journal.pone.0160506 (2016). [DOI] [PMC free article] [PubMed]
- 14.Milheiriço, C., Oliveira, D. C. & de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus.VAntimicrob. Agents Chemother. 51(9), 3374-3377 (2007). 10.1128/AAC.00275-07 (erratum in: Antimicrob Agents Chemother 2007; Dec;51(12):4537). [DOI] [PMC free article] [PubMed]
- 15.Vivoni, A. M. et al. Clonal composition of Staphylococcus aureus isolates at a Brazilian university hospital: Identification of international circulating lineages. J. Clin. Microbiol.44 (5), 1686–1691. 10.1128/JCM.44.5.1686-1691.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDougal, L. K. et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J. Clin. Microbiol.41 (11), 5113–5120. 10.1128/JCM.41.11.5113-5120.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright, M. C., Day, N. P., Davies, C. E., Peacock, S. J. & Spratt, B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol.Mar;38 (3), 1008–1015. 10.1128/JCM.38.3.1008-1015.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuenck, R. P., Cavalcante, F. S., Emery, E., Giambiagi-de Marval, M. & dos Santos, K. R. Staphylococcus aureus isolates belonging to different multilocus sequence types present specific virulence gene profiles. FEMS Immunol. Med. Microbiol.65 (3), 501–504. 10.1111/j.1574-695X.2012.00958.x (2012). [DOI] [PubMed] [Google Scholar]
- 19.Diep, B. A. et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: Convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197(11), 1523-1530 (2008). 10.1086/587907 [DOI] [PubMed]
- 20.Sands, K. E., Blanchard, E. J., Fraker, S., Korwek, K. & Cuffe, M. Health care-associated infections among hospitalized patients with COVID-19, March 2020-March 2022. JAMA Netw Open. 6(4), e238059 (2023). 10.1001/jamanetworkopen.2023.8059 [DOI] [PMC free article] [PubMed]
- 21.Hirabayashi, A. et al. Comparison of disease and economic burden between MRSA infection and MRSA colonization in a university hospital: a retrospective data integration study. Antimicrob. Resist. Infect. Control 13(1), 27. 10.1186/s13756-024-01383-8 [DOI] [PMC free article] [PubMed]
- 22.Serra, N. et al. Staphylococcus aureus and coagulase-negative Staphylococci from bloodstream infections: frequency of occurrence and antimicrobial resistance, 2018–2021. Life (Basel)13(6), 1356 (2023). 10.3390/life13061356 [DOI] [PMC free article] [PubMed]
- 23.Giulieri, S. G. et al. A statistical genomics framework to trace bacterial genomic predictors of clinical outcomes in Staphylococcus aureus bacteremia. Cell. Rep.Sep 26 (9), 113069. 10.1016/j.celrep.2023.113069 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Minter, D. J., Appa, A., Chambers, H. F. & Doernberg, S. B. Contemporary management of Staphylococcus aureus bacteremia-controversies in clinical practice. Clin. Infect. Dis.77 (11), e57–e68. 10.1093/cid/ciad500 (2023). Nov 30;. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Casanova, N., Gutiérrez-Zufiaurre, M. N., Blázquez de Castro, A. M. & Muñoz-Bellido, J. L. Genomic insights into Staphylococcus aureus isolates exhibiting diminished daptomycin susceptibility. Pathogens13(3), 206 (2024). 10.3390/pathogens13030206 [DOI] [PMC free article] [PubMed]
- 26.Jiang, S. et al. Profiling daptomycin resistance among diverse methicillin-resistant Staphylococcus aureus lineages in China. Antimicrob. Agents Chemother.67 (11), e0056323. 10.1128/aac.00563-23 (2023). Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, T. M. et al. Combination ceftaroline and daptomycin salvage therapy for complicated methicillin-resistant Staphylococcus aureus bacteraemia compared with standard of care. Int. J. Antimicrob. Agents. Apr;57 (4), 106310. 10.1016/j.ijantimicag.2021.106310 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Alsowaida, Y. S., Alsolami, A. & Almangour, T. A. Daptomycin and ceftaroline combination for the treatment of persistent methicillin-resistant Staphylococcus aureus bloodstream infections: A case series and literature review. J. Chemother. 12, 1–6. 10.1080/1120009X.2024.2340877 [DOI] [PubMed]
- 29.Chen, C. H. et al. National surveillance of antimicrobial susceptibilities to ceftaroline, dalbavancin, telavancin, tedizolid, eravacycline, omadacycline, and other comparator antibiotics, and genetic characteristics of bacteremic Staphylococcus aureus isolates in adults: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) program in 2020. Int. J. Antimicrob. Agents. 61 (4), 106745. 10.1016/j.ijantimicag.2023.106745 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Sader, H. S., Carvalhaes, C. G. & Mendes, R. E. Ceftaroline activity against Staphylococcus aureus isolated from patients with infective endocarditis, worldwide (2010–2019). Int. J. Infect. Dis.Jan;102, 524–528. 10.1016/j.ijid.2020.11.130 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Singulani, J. L. et al. The impact of COVID-19 on antimicrobial prescription and drug resistance in fungi and bacteria. Braz J. Microbiol.Dec;53 (4), 1925–1935. 10.1007/s42770-022-00818-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdelsalam Elshenawy, R., Umaru, N. & Aslanpour, Z. WHO AWaRe classification for antibiotic stewardship: tackling antimicrobial resistance - A descriptive study from an English NHS Foundation Trust prior to and during the COVID-19 pandemic. Front. Microbiol.14, 1298858. 10.3389/fmicb.2023.1298858 (2023). [DOI] [PMC free article] [PubMed]
- 33.Perrella, A. et al. Hospital antibiotic use during COVID-19 pandemic in Italy. Antibiotics (Basel) 12(1), 168 (2023). 10.3390/antibiotics12010168 [DOI] [PMC free article] [PubMed]
- 34.Chamon, R. C., Ribeiro, S. D., da Costa, T. M., Nouér, S. A. & Dos Santos, K. R. Complete substitution of the Brazilian endemic clone by other methicillin-resistant Staphylococcus aureus lineages in two public hospitals in Rio De Janeiro, Brazil. Braz J. Infect. Dis.21 (2), 185–189. 10.1016/j.bjid.2016.09.015 (2017). Mar-Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu, J. et al. ST7 becomes one of the most common Staphylococcus aureus clones after the COVID-19 epidemic in the city of Wuhan, China. Infect. Drug Resist. 16, 843–852. 10.2147/IDR.S401069 (2023). [DOI] [PMC free article] [PubMed]
- 36.Smith, J. T. et al. Genome evolution of invasive methicillin-resistant Staphylococcus aureus in the Americas. Microbiol. Spectr.10(3), e0020122 (2022). 10.1128/spectrum.00201-22 [DOI] [PMC free article] [PubMed]
- 37.Souza, S. S. R. et al. Demographic fluctuations in bloodstream Staphylococcus aureus lineages configure the mobile gene pool and antimicrobial resistance. NPJ Antimicrob. Resist.2 (1), 14. 10.1038/s44259-024-00032-9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhan, Q., Teng, G., Chen, W. & Yu, X. High prevalence of ST5-SCCmec II-t311 clone of methicillin-resistant Staphylococcus aureus isolated from bloodstream infections in East China. BMC Microbiol.Mar 16 (1), 89. 10.1186/s12866-024-03232-5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGuinness, S. L. et al. Clinical and molecular epidemiology of an emerging panton-valentine leukocidin-positive ST5 methicillin-resistant Staphylococcus aureus clone in Northern Australia. mSphere6(1), e00651-20 (2021). 10.1128/mSphere.00651-20 [DOI] [PMC free article] [PubMed]
- 40.Gulone, L. et al. The changing epidemiology and antimicrobial susceptibility of Staphylococcus aureus isolated from blood xultures in a University Hospital from Argentina. Microb. Drug Resist.Mar;30 (3), 109–117. 10.1089/mdr.2023.0219 (2024). [DOI] [PubMed] [Google Scholar]
- 41.Dyzenhaus, S. et al. MRSA lineage USA300 isolated from bloodstream infections exhibit altered virulence regulation. Cell. Host Microbe. 31 (2), 228–242e8 (2023). Epub 2023 Jan 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data are available from the corresponding author on request (Availability of data).