Abstract
Our previous study found that Dunaliella parva WRINKLED1-like (DpWRI1-like) was a key regulatory factor of lipid biosynthesis in D. parva. DpWRI1-like gene and target genes of DpWRI1-like have been obtained in our previous study, but the interacting proteins of DpWRI1-like are unclear now, which has limited a deep understanding of the function of DpWRI1-like. Yeast two-hybrid was widely used to identify protein-protein interaction. In this study, the interacting proteins of DpWRI1-like were obtained using yeast two-hybrid technique to further realize the role of DpWRI1-like. Three important interacting proteins have the following predicted activities: acyl-CoA-binding domain-containing protein 6 (interacting protein 1, ACBD6), duplicated carbonic anhydrase (interacting protein 2, DCA) and DNA-binding transcription factor (interacting protein 3, TF). Bimolecular fluorescence complementation assay further validated the interaction between DpWRI1-like and interacting proteins ACBD6 and DCA. The further bioinformatics analyses of interacting proteins were conducted. Protein-protein docking indicated the strong affinity between DpWRI1-like and three interacting proteins. Since interacting proteins have been found to be related to lipid biosynthesis in other organisms, this study contributes to a deeper understanding of the role of DpWRI1-like in lipid synthesis. In conclusion, this study firstly reported three interacting proteins (ACBD6, DCA and TF) of DpWRI1-like related to lipid biosynthesis, and conducted their bioinformatics analyses, which would be conducive to a deep understanding of the function of DpWRI1-like in lipid biosynthesis.
Keywords: Dunaliella parva, DpWRI1-like, Yeast two-hybrid system, Interacting proteins, Lipid biosynthesis
Highlights
-
•
Three interacting proteins (ACBD6, DCA and TF) of DpWRI1-like were obtained.
-
•
DCA significantly enhanced organic compound content.
-
•
TF significantly reduced organic compound content.
1. Introduction
As a kind of clean renewable energy, biodiesel has received the widespread attention. Compared with oil plants, as raw material of biodiesel, microalgae have the advantage of faster growth, shorter life cycle and no occupation of farmland [1]. Microalgal lipid can be used to efficiently produce biodiesel. The commercial production of microalgal biodiesel has become a research hotspot in the field of new energy. However, low biomass, low lipid yield and energy-intensive harvest affected the commercial production of microalgal biodiesel [2]. Therefore, how to increase lipid content in algae becomes research focus.
Many reports found that nitrogen limitation enhanced lipid content in microalgae. Nitrogen limitation enhanced lipid content by upregulating expression of some lipid-related genes in Chlorella vulgaris [3]. Nevertheless, nitrogen limitation usually decreases biomass, which limits its utilization. Our previous research also found that nitrogen limitation increased lipid content (from 25 % to 40 %) in Dunaliella parva (D. parva). Nevertheless, under nitrogen limitation condition, biomass reduced by 39 % compared with biomass under nitrogen sufficient condition in D. parva [4]. Therefore, increasing lipid content by biochemical way has great limitation. Recently, it is a new strategy to regulate lipid metabolism by regulating transcription factor genes in microalgae [5]. Transcription factors are proteins that specifically bind to the specific DNA sequences and can activate or inhibit gene transcription. Unlike the conventional genetic engineering with single gene, transcription factor strategy affects many genes related to several metabolic pathways. Therefore, these metabolic pathways can be simultaneously regulated.
D. parva can normally grow and reproduce in 0.05–5 M NaCl [6]. Its outstanding advantages include photosynthetic autotrophy, strong stress resistance and simple cultivation conditions. It is a lipid-producing microalga without cell wall, which is conducive to genetic transformation. D. parva can produce functional proteins with high activity, β-carotene and glycerin with high nutritional value, which can be directly used as natural health food [7]. Compared with model microalgae, D. parva is more suitable for producing biofuels and β-carotene, which can reduce the cost of biodiesel preparation. However, lipid content of D. parva is relatively low, therefore increasing lipid content has become an important way to reduce the cost of biodiesel preparation.
WRINKLED1 (WRI1) was originally discovered in Arabidopsis wri1-1 mutant. AtWRI1 overexpression recovered the normal seed surface in wri1-1 mutant. The wri1-1 mutant was unable to convert glucose/sucrose into substrates of fatty acid biosynthesis during seed development, and the activities of many enzymes such as hexokinase/phosphofructokinase reduced, resulting in 80 % reduction of lipid content [8]. WRI1s are important AP2-type transcription factors associated with lipid yield [9]. Scientists extensively studied WRI1 in plants. WRI1 and its target genes mainly expressed in endosperm [10]. WRI1 could regulate the level of genes related to the synthesis of fatty acid and triacylglycerol in Arabidopsis, converting carbon flow to lipid synthesis [11]. The expression of WRI1 was closely associated with lipid yield in Siberian apricot kernel [12]. About WRI1 in microalgae, there were a few reports. In our previous work, DpWRI1-like gene (GenBank no., KR185335) was also related to lipid accumulation [4]. We also found that DpWRI1-like regulated many target genes involved in carbohydrate metabolism, lipid metabolism, photosynthesis and transcription factor. It was proposed that DpWRI1-like participated in a regulatory network controlling lipid biosynthesis [13]. In a word, WRI1 gene has been extensively studied in plants but not in D. parva, which is closely related to lipid accumulation as a transcription factor.
We conducted transcriptome sequencing of D. parva under the conditions of nitrogen deficiency and sufficient nitrogen source, analyzed and constructed metabolic pathways of fatty acid, triacylglycerol, starch, and identified transcription factor gene DpWRI1-like that responded to nitrogen deficiency and its promoter in our previous study [4]. Our previous study indicated that there was a positive correlation between DpWRI1-like level and lipid content under nitrogen limitation condition in D. parva [8]. In addition, through chromatin immunoprecipitation sequencing, we identified many target genes of DpWRI1-like related to carbohydrate and lipid metabolism [13]. The regulatory effects of transcription factors on metabolism are closely related to their target genes and interacting proteins. Therefore, interacting proteins of transcription factor DpWRI1-like also need further research. Some studies identified proteins that interacted with WRI1 in plants. In Arabidopsis, BLISTER regulated chromatin dynamics, and promoted seed maturation and fatty acid biosynthesis by interacting with WRI1 [14]. Proteins interacting with Arabidopsis thaliana WRI1 (AtWRI1) were identified by yeast two-hybrid (Y2H) assay, including TCP family transcription factors (TCP4, TCP10 and TCP24) [15]. Zhai et al. found that KIN10 kinase physically interacted with AtWRI1 and triggered phosphorylation of AtWRI1 [16]. However, the interacting proteins of DpWIR1-like have not been identified. In order to illustrate the function of DpWRI1-like more fully, this article identified the interacting proteins of DpWIR1-like through Y2H technology, and conducted bioinformatic analysis and further discussion. In addition, bimolecular fluorescence complementation (BiFC) assay validated the interacting proteins of DpWIR1-like. This study contributed to a deeper understanding of the function of DpWRI1-like in lipid synthesis.
2. Materials and methods
2.1. Microalgal cultivation
D. parva (No. FACHB-815) was preserved on Dunaliella medium in glass-bottom dish (NEST Biotechnology, Wuxi, China) in our laboratory. Our previous paper provided a detailed description of the cultivation of D. parva [4]. The bottle containing D. parva cells was gently swirled, which was conducive to the growth.
2.2. RNA extraction and first-strand cDNA synthesis
Total RNA was isolated using Trizol reagent (Takara, Dalian, China) for D. parva based on manufacturer's instruction. The specific procedures were as follows. Before isolation of RNA, 1 mL D. parva culture was centrifuged to obtain cell pellet, then cell pellet was washed with PBS solution. Add 1 mL Trizol to completely resuspend cell pellet. Leave the mixture at room temperature for 5 min. Add 0.2 mL chloroform to the above mixture, and mix until the mixture becomes milky. Keep the mixture at room temperature for 5 min. Centrifuge at 12,000×g for 15 min at 4 °C. Transfer the top liquid layer to new centrifuge tube. Add 0.5 mL isopropanol and mix well. Keep the mixture at room temperature for 10 min. Centrifuge at 12,000×g for 10 min at 4 °C to precipitate RNA. Carefully remove the supernatant, and dry the precipitate for several minutes. After the precipitate is dry, dissolve it with RNase-free water. The OD260/OD280 values of RNA samples were in range of 1.8–2.0 using NanoPhotometer NP80 (Implen, Munich, Germany). Using 1 % agarose gel electrophoresis, RNA's purity was investigated, and protein impurity and RNA degradation were not found. Then using RNA as template, first-strand cDNA was obtained using PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara, Dalian, China) and stored at −20 °C. The specific procedures were as follows. Prepare the following mixture in a microtube (total 10 μL): 1 μL Oligo dT Primer (50 μM), 1 μL dNTP mixture (10 mM each), total RNA (<5 μg) x μL, RNase free ddH2O (8-x) μL. Incubate mixture for 5 min at 65 °C, then cool immediately on ice. Prepare reaction mixture in a total volume of 20 μL and mix gently: 10 μL the above mixture, 4 μL 5× PrimeScript buffer, 0.5 μL RNase inhibitor (40 U/μL), 1.0 μL PrimeScript RTase (200 U/μL), 4.5 μL RNase free ddH2O. Incubate reaction mixture at 42 °C for 60 min. Inactivate enzyme at 95 °C for 5 min, then cool on ice to obtain cDNA.
2.3. Cloning of DpWRI1-like gene from D. parva
According to Y2H manual, primers (Wri1-F and Wri1-R) were obtained according to DpWRI1-like cDNA (GenBank no., KR185335) (Table 1). Used first-strand cDNA as a template, PCR was prepared with PrimeSTAR HS DNA Polymerase (Takara, Dalian, China) using the procedure: 94 °C 3 min, 35 cycles (98 °C10 s, 42 °C 15 s, 72 °C 1 min), 72 °C 10 min, 4 °C 1 min. The dA was added to 3′ end of PCR product by TaKaRa Taq (Takara, Dalian, China). Then PCR band was recovered, and ligated into pMD19-T vector with dT at 3′ end by TA-cloning strategy for sequencing.
Table 1.
Primers used in this study.
| Primer name | Primer sequence (5′–3′) |
|---|---|
| Wri1-F | CATGGAGGCCGAATTCATGGACTTCCGCAGCACTCCTCCC |
| Wri1-R | GCAGGTCGACGGATCCCTAGTTACTCTCTCCGTGGATCCA |
| Wri1-N | CATGGAGGCCGAATTCTCCAAATTCCGTGGCGTGCACCAGAC |
| Wri1-C | GCAGGTCGACGGATCCATCGTAGTTTGTGATGCCGCCCTT |
| BiFN-SeqF(wri1) | CTAGTCTAGAATGGACTTCCGCAGCACTC |
| BiFN-SeqR(wri1) | CCGCTCGAGGTTACTCTCCGTGGATCCA |
| BiFC-SeqF(Protein 1) | CTAGTCTAGAATGGACTCTAGCGCAGATG |
| BiFC-SeqR(Protein 1) | CGCGGATCCTCGTTTAAGTAGGTGGGCC |
| BiFC-SeqF(Protein 2) | CTAGTCTAGAATGGCCACCACCGTCCGCG |
| BiFC-SeqR(Protein 2) | CGCGGATCCGTAGCTAGAGACTGGCCTG |
2.4. Preparation of cDNA library and yeast bait strain
The cDNA library of D. parva (cell density>2 × 107/mL) has been obtained in our previous work [17]. Y2H test were finished using Matchmaker Gold Yeast Two-Hybrid System Kit (Takara, Dalian, China). The cDNA fragment of DpWRI1-like (nucleotide positions: 613–804, bait sequence DpWri1N) corresponding to C-terminus (the binding region) was amplified using primers Wri1-N/Wri1-C. Bait sequence was inserted into pGBKT7 vector at BamHI/EcoRI sites. To determine whether the recombinant vector was successfully constructed, vector was sequenced by T7 Primer. Before screening the library, autoactivation and toxicity of bait sequence need to be excluded in Y2HGold strain [18]. The method transforming strains Y2HGold and Y187 was shown in our previous paper [17].
2.5. Two-hybrid library screening by yeast mating
Mate 1 mL Y187 cDNA library with 5 mL Y2HGold culture (OD600 ≥ 0.8) transformed with pGBKT7-Wri1N plasmid for library screening [19]. The specific procedure was based on our previous paper [17]. All positive clones on Synthetic Defined Medium (SD)/-Ade/-His/-Trp/-Leu/X-α-Gal/AbA (QDO/X/A) plate need to exclude duplicate and confirm the authenticity.
2.6. Identification of positive clones
The selected positive clones were cultured in QDO/X/A liquid medium. After 24 h, yeast plasmid was extracted using MiniBEST plasmid purification kit (Takara). The extracted yeast plasmid and primers (T7 and 3′AD) were used for PCR amplification. The positive plasmid was transformed into DH5α strain using ampicillin for screening. The positive DH5α colonies were subjected to sequencing and similarity comparison.
2.7. Bimolecular fluorescence complementation (BiFC) assay
DpWRI1-like gene was cloned into pUC-SPYNE vector containing YFPN sequence. The encoding sequences of interacting proteins (acyl-CoA-binding domain-containing protein 6, ACBD6 and duplicated carbonic anhydrase, DCA) identified through Y2H assay were cloned into pUC-SPYCE vector containing YFPC sequence, respectively. Primers for BiFC assay were listed in Table 1. For transient transformation, Agrobacterium tumefaciens strain GV3101 carrying empty pUC-SPYNE/pUC-SPYCE plasmids or recombinant pUC-SPYNE/pUC-SPYCE plasmids was infiltrated into Nicotiana benthamiana leaves. Nicotiana benthamiana was always cultured in soil at 70 % humidity with a 24 °C, 16 h, photon flux density of 100 μmol/(m2 × s)/20 °C, 8 h, dark cycle. After injection, Nicotiana benthamiana leaves were cultured for three days, and the TCS SP8 confocal laser scanning microscope (Leica, Wetzlar, Germany) was utilized to investigate fluorescent signal.
2.8. Bioinformatics analysis
Based on the encoding sequences of interacting proteins (ACBD6 and DCA), their properties were analyzed by various bioinformatics software. The physicochemical properties, hydrophilicity, signal peptide and transmembrane region were analyzed by ProtParam, ProtScale, SignalP-4.1 and TMHMM-2.0, respectively. SOPMATF and SWISS-MODEL predicted the secondary and tertiary structure, respectively. HDOCK was used to conduct protein-protein docking. The specific software and website were shown in Supplementary materials: Table S1.
3. Results
3.1. Production of Y2HGold bait strain
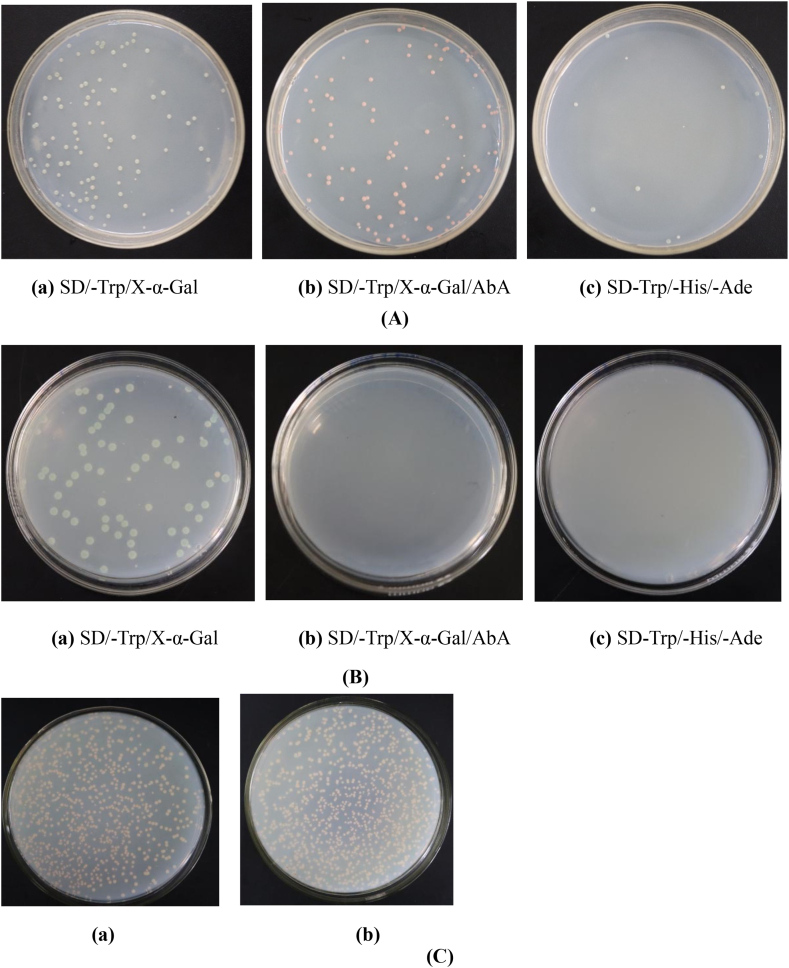
Y2HGold bait strain was constructed for Y2H assay according to the following steps. Full-length DpWRI1-like cDNA (1023 bp) amplified by primers Wri1-F and Wri1-R were shown in Supplementary materials: Fig. S1A. As a first step for Y2H assay, it is very important to confirm that bait does not autonomously activate reporter genes in Y2HGold without prey protein. Our result showed that full-length DpWRI1-like protein autonomously activated reporter genes (Fig. 1A). Therefore, to determine binding region of DpWRI1-like, cDNA fragment (DpWri1N, nucleotide positions: 613–804) encoding C-terminus was amplified by primers Wri1-N/Wri1-C based on prediction of conserved domains in DpWRI1-like (Supplementary materials: Fig. S1B). The blue Y2HGold-pGBKT7-DpWri1N colonies were detected on SD/-Trp/X-α-Gal plate. However, Y2HGold-pGBKT7-DpWri1N blue colony was not observed on SD/-Trp/X-α-Gal/AbA and SD/-Trp/-His/-Ade plates (Fig. 1B). It was proved that DpWri1N protein had not the ability of autoactivation. In the absence of reporter genes, pGBKT7-DpWri1N was nontoxic for the growth of Y2HGold strain. The size and quantity of colonies were similar on SD/-Trp medium between Y2HGold-pGBKT7-DpWri1N and Y2HGold-pGBKT7 strains (Fig. 1C). The above results indicated that Y2HGold bait strain was successfully constructed. Then bait strain was used for yeast mating assay.
Fig. 1.
Production of Y2HGold bait strain. (A) Detection of autoactivation of full-length DpWRI1-like protein in yeast strain Y2HGold. Yeast strain Y2HGold grew on SD/-Trp/X-α-Gal/AbA and SD/-Trp/-His/-Ade plates, which indicated that pGBKT7-DpWRI1-like had the ability of autoactivation. (B) Detection of the autoactivation of DpWri1N protein in yeast strain Y2HGold. Yeast strain Y2HGold didn't grow on SD/-Trp/X-α-Gal/AbA and SD/-Trp/-His/-Ade plates, which indicated that pGBKT7-Wri1N hadn't the ability of autoactivation. (C) Toxicity assay of DpWri1N in yeast strain Y2HGold on SD/-Trp plate. (a) Yeast strain Y2HGold containing PGBKT7-DpWri1N. (b) Yeast strain Y2HGold containing PGBKT7.
3.2. Identification of interacting proteins
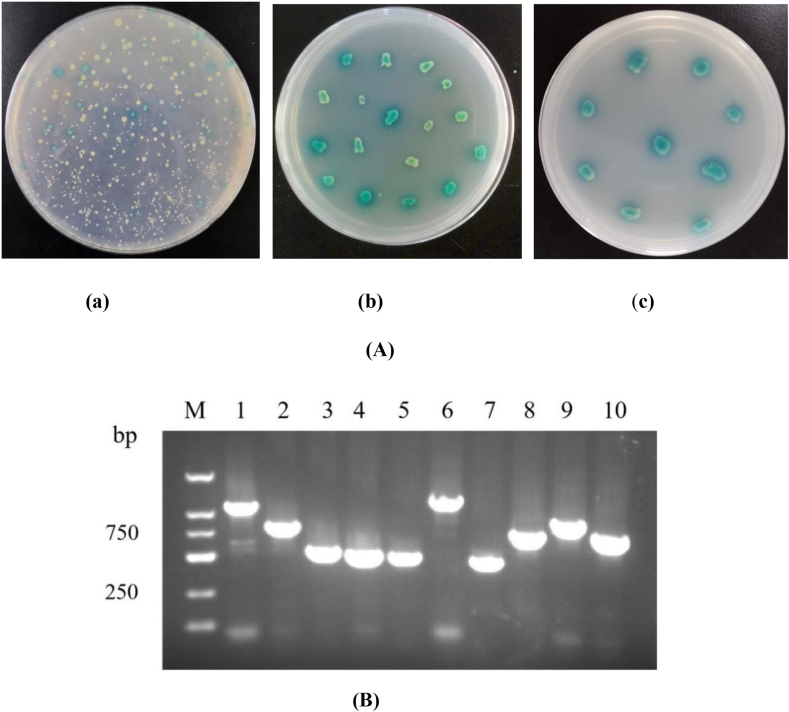
Yeast mating assay was conducted for Y2H assay according to the following steps. After 24 h of co-culture, there was successful yeast mating (Supplementary materials: Fig. S2). After centrifugation, yeast zygotes were cultured on SD/-Leu/-Trp/X-α-Gal/AbA medium. By counting colony number, the mating efficiency reached 4 %. Many positive colonies were clearly observed on SD/-Trp/-Leu/X-α-Gal/AbA plate. To reduce false positives, colonies were re-cultured for 3 times on SD/-Trp/-Leu/-His/-Ade/X-α-Gal/AbA plate (Fig. 2A). Finally, 86 positive yeast colonies were obtained.
Fig. 2.
(A) Screening of positive colonies. (a) The first screening. (b) The second screening. (c) The third screening. (B) Plasmid PCR of positive colonies. M: DL2000 DNA Marker. 1–10: positive colonies. Positive colonies were amplified with primers T7/3′AD.
Plasmids were extracted and purified from 86 yeast colonies, and 27 cDNA bands were confirmed using primers T7/3′AD (Fig. 2B). Then 27 yeast plasmids with genes encoding interacting proteins were transformed into E. coli strain DH5α using ampicillin for screening.
After sequencing, Blastx search showed three important proteins interacting with DpWRI1-like. They were protein 1 (acyl-CoA-binding domain-containing protein 6, ACBD6), protein 2 (duplicated carbonic anhydrase, DCA) and protein 3 (DNA-binding transcription factor, TF) (Table 2). The encoding sequences for three interacting proteins were shown in Fig. S3. The above results indicated that interacting proteins of DpWRI1-like were successfully identified through yeast mating assay, plasmid extraction and sequencing.
Table 2.
Proteins interacting with DpWRI1-like.
| Proteins No. | Annotation |
|---|---|
| Protein 1 | Acyl-CoA-binding domain-containing protein 6, ACBD6 |
| Protein 2 | Duplicated carbonic anhydrase, DCA |
| Protein 3 | DNA-binding transcription factor, TF |
3.3. Results of BiFC assay
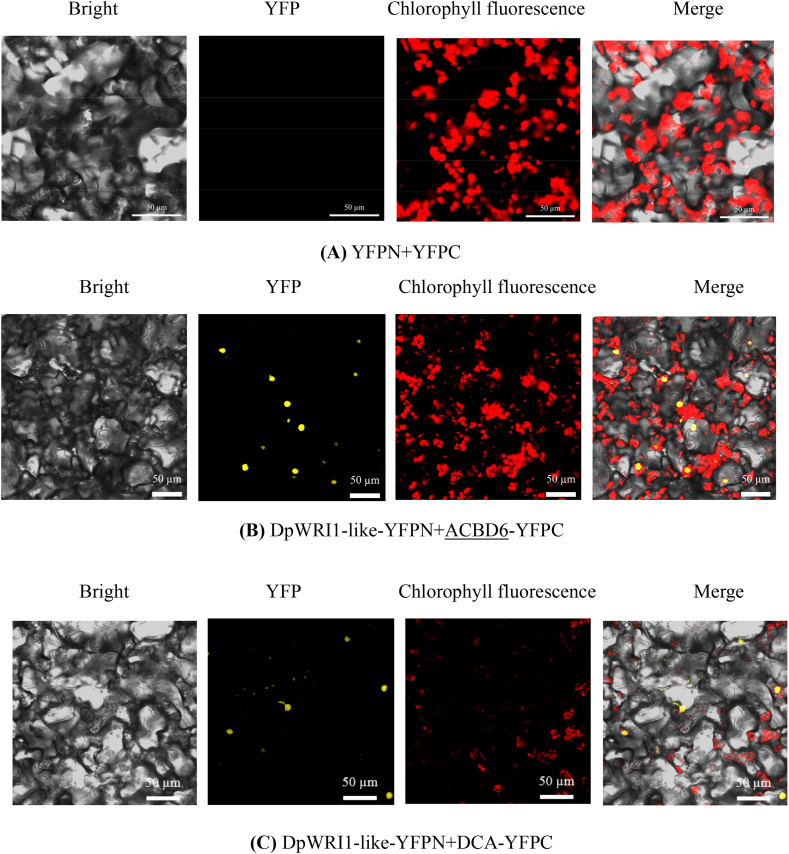
BiFC assay was extensively used to verify the result of Y2H assay. The results of BiFC assay were shown in Fig. 3. BiFC assay was performed using Agrobacterium tumefaciens strain GV3101, which provided additional evidence for the interaction between DpWRI1-like and ACBD6/DCA proteins in Y2HGold cell. Fig. 3 showed that YFP signals of DpWRI1-like-YFPN/ACBD6-YFPC and DpWri1-like-YFPN/DCA-YFPC groups were clearly visible in nucleus. Thus, BiFC assay further demonstrated that DpWRI1-like protein authentically interacted with ACBD6 and DCA proteins.
Fig. 3.
Results of bimolecular fluorescence complementary assay. BiFC analysis was performed using Agrobacterium tumefaciens strain GV3101. Bars = 50 μm.
3.4. Bioinformatics analysis of interacting proteins
Bioinformatics analysis of interacting proteins including hydrophobicity, signal peptide prediction, transmembrane prediction and prediction of secondary structure and three-dimensional structure, can lay a solid foundation for better studying the functions of these proteins in the future. Protein sequences encoded by ACBD6, DCA and TF genes were analyzed by ExPasy-ProtParam/ExPasy-ProtScale. The results showed that ACBD6 contained 247 aa, the molecular formula was C1127H1737N323O385S12, the theoretical isoelectric point was 4.39, the relative molecular weight was 26355.84 Da, and total number of atoms was 3584. The number of acidic amino acid residues (Asp + Glu) was 38, the number of alkaline amino acid residues (Arg + Lys) was 16. The aliphatic index was 63.77 and the instability coefficient was 43.99. Grand average hydropathicity (GRAVY) was −0.539. Therefore, ACBD6 protein was a hydrophilic and unstable protein (Supplementary materials: Fig. S4A).
The results showed that DCA contained 485 aa, the molecular formula was C2376H3644N654O742S15, the theoretical isoelectric point was 5.11, the relative molecular weight was 53723.91 Da, and total number of atoms was 7431. The number of acidic amino acid residues (Asp + Glu) was 59, the number of alkaline amino acid residues (Arg + Lys) was 35. The aliphatic index was 77.61 and the instability coefficient was 33.18. GRAVY for DCA was −0.361. So DCA protein was a hydrophilic and stable protein (Supplementary materials: Fig. S4A).
TF contained 161 aa, the molecular formula was C732H1230N224O239S7, the theoretical isoelectric point was 8.32, the relative molecular weight was 17217.60 Da, and total number of atoms was 2432. The number of acidic amino acid residues (Asp + Glu) was 20, the number of alkaline amino acid residues (Arg + Lys) was 22. The aliphatic index was 89.07 and the instability coefficient was 38.12. GRAVY for DCA was −0.294. Therefore, TF protein was a hydrophilic and stable protein (Supplementary materials: Fig. S4A).
SignalP 4.1 was utilized to predict signal peptides of ACBD6, DCA and TE proteins (Supplementary materials: Fig. S4B). C value (0.157) at the 47th amino acid of ACBD6 was the highest. Signal peptide value (S value, 0.173) at the 37th amino acid of ACBD6 was the highest. The highest Y value (0.137) was at the 47th amino acid. The average S value and D value of amino acids (from 1st to 46th) were 0.106 and 0.119, respectively. According to the criteria, the average S value (>0.5) is a signal peptide, and the average D value (>0.5) is a secreted protein. Fig. S4B suggests that ACBD6 does not contain a signal peptide and is not a secreted protein.
For DCA protein, the highest C value (0.205) was at the 38th amino acid. The S value (0.155) at the 37th amino acid was the highest. The highest Y value (0.154) was at the 38th amino acid. The average S value and D value of amino acids (from 1st to 37th) were 0.110 and 0.130, respectively. Fig. S4B suggests that DCA does not contain signal peptide and is not a secretory protein.
For TF protein, the highest C value (0.110) was at the 61st amino acid. The S value (0.115) at the 54th amino acid was the highest. The highest Y value (0.103) was at the 41st amino acid. The average S value and D value of amino acids (from 1st to 40th) were 0.094 and 0.098, respectively. Fig. S4B suggests that TF does not contain signal peptide and is not a secretory protein. According to the prediction of transmembrane domain (Supplementary materials: Fig. S4C), ACBD6, DCA and TF have not transmembrane segment.
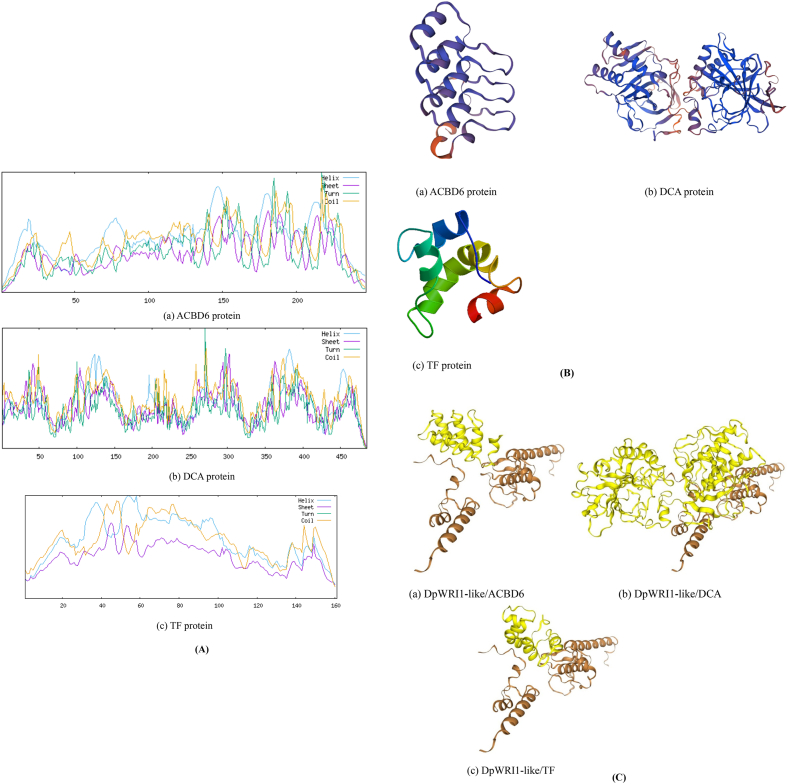
SOPMA software was utilized to predict secondary structure of ACBD6, DCA and TF (Fig. 4A). ACBD6 protein includes 47.77 % α helix, 5.26 % extended strand, 8.50 % β turn and 38.46 % random coil. The secondary structure of ACBD6 protein was dominated by α-helix and random coil. DCA protein includes 16.49 % α helix, 26.39 % extended strand, 2.27 % β turn and 54.85 % random coil. The secondary structure of DCA was dominated by extended strand and random coil. TF protein includes 39.13 % α helix and 60.87 % random coil. The secondary structure of TF was dominated by random coil. The predicted tertiary structure of ACBD6, DCA and TF was shown in Fig. 4B. As shown in Fig. 4C scores of protein-protein docking were −262.89, −245.36 and −198.41 between DpWRI1-like/ACBD6, DpWRI1-like/DCA and DpWRI1-like/TF, which indicated the strong affinity.
Fig. 4.
Bioinformatics analysis of interacting proteins. (A) Prediction of secondary structure. (B) Prediction of tertiary structure. (C) Protein-protein docking between DpWRI1-like/ACBD6, DpWRI1-like/DCA and DpWRI1-like/TF.
4. Discussion
Transcription factors regulate metabolism through their target genes and interacting proteins. Many target genes of transcription factor DpWRI1-like related to lipid biosynthesis was identified from microalga Dunaliella parva in our previous study. These target genes were related to carbohydrate and lipid metabolism. The interacting proteins of transcription factor DpWRI1-like also need further research. Y2H assay identified three interacting proteins including acyl-CoA-binding domain-containing protein 6 (interacting protein 1, ACBD6), duplicated carbonic anhydrase (interacting protein 2, DCA) and DNA-binding transcription factor (interacting protein 3, TF). BiFC assay verified the result of Y2H assay. Bioinformatics analysis of interacting proteins can lay a good foundation for the research on the functions of these proteins in the future. This study helps to better understand the function of DpWRI1-like and its interacting proteins.
ACBD family played important roles in the upkeep of various functions [[20], [21], [22]]. They are crucial to various functions, possibly through the interaction with a variety of proteins related to neural stem cell self-renewal, stress resistance, neurodegeneration, intracellular vesicle trafficking, lipid homeostasis, viral replication, organelle formation and apoptotic response [[21], [22], [23], [24], [25]]. ACBD6 controls the composition of lipid and protein in cell membrane of human. The transfer of acyl chain from acyl-CoA to protein and lipid is achieved through the interaction of various enzyme and protein, including ACBD6 [26]. ACBD6 interacts with N-myristoyltransferase to produce a dimer enzyme complex that regulates the specificity of myristoylation [[27], [28], [29]]. N-myristoylation is a necessary modification that regulates the function, stability and membrane binding of various cytoplasmic proteins in cells [30,31]. In our previous study, DpWRI1-like gene was strongly associated with lipid synthesis, which further confirmed the reliability of this experiment [4]. Whereas, the function of ACBD6 has not been further studied in D. parva.
DCA is a member of carbonic anhydrases (CAs) family. CAs are zinc metalloenzymes, which exist widely [32]. CAs can catalyze the reversible hydration/dehydration reaction: CO2+H2O ⇔ H2CO3⇔HCO3−+H+, which is the foundation of many biological processes besides acid-base regulation, such as photosynthesis, respiration, osmoregulation, bone resorption and biomineralization [[33], [34], [35], [36], [37]]. In higher plants, CAs include three independent families (α/β/γ type) [38]. Carbonic anhydrase is highly salt-resistant. Dunaliella salina carbonic anhydrase could optimize the utilization of inorganic carbon under high salinity condition [39]. In brief, carbonic anhydrase is the foundation of many biological processes, which catalyzes the reversible hydration/dehydration reaction to produces HCO3−. It is speculated that duplicated carbonic anhydrase can promote the synthesis of lipid in D. parva, which has aroused our great interest. In our unpublished study (submitted to Journal of Applied Phycology), D. parva DCA gene was transformed into D. parva by genetic engineering technique. The carotenoid, total carbohydrate, starch, protein, and oil contents of transgenic D. parva increased by 16.31 %, 31.68 %, 43.97 %, 52.91 %, and 12.32 %, compared to control. Perhaps D. parva DCA affected photosynthesis through cis-element. Therefore, our unpublished study about D. parva DCA gene lays a foundation for studying the function of DCA gene in D. parva. Further experiments will be conducted to demonstrate its function in D. parva.
The analysis indicated that protein 3 had DNA-binding transcription factor activity. Transcription factor can bind to the nearby DNA to activate or inhibit gene transcription. In our previous study, transcription factor DpAP2 related to carotenoid biosynthesis also interacted with this protein [17]. It is indicated that protein 3 might be related to the synthesis of carotenoid and lipid. Whereas, the function and interacting genes of protein 3 in D. parva are unclear. In our unpublished study, D. parva TF gene was cloned into pET-30a vector, then transformed into E. coli Rosetta (DE3) strain to produce recombinant TF protein. After purification by Ni2+-NTA column, the purified TF protein was used to immunize rabbit to produce polyclonal antibody. Then the polyclonal antibody will be used in chromatin immunoprecipitation assay to detect the binding genes of D. parva TF protein in the future. In another unpublished study (submitted to Journal of Applied Phycology), we cloned D. parva TF gene into pBI221-GFP-UbiΩ-CAT vector and overexpressed it in D. parva to investigate its effects on organic content and antioxidant activity in D. parva. The results showed that D. parva TF gene had inhibitory effects on the contents of carbohydrate, protein, chlorophyll, carotenoid and oil, and antioxidant activities. In the future, target genes of protein 3 and its function will be further demonstrated by ChIP-Seq and CRISPR/Cas9 technology.
The previous studies identified various interacting proteins of WRI1 in different higher plants through Y2H. However, the progress of the research about the interacting proteins of WRI1 is very slow in microalgae. CRISPR/Cas9 technology will be used to investigate the effect of DpWRI1-like gene knockout on three important interacting proteins (ACBD6, DCA and TF) and the effect of gene knockout of genes encoding three interacting proteins on DpWRI1-like in D. parva.
5. Conclusion
DpWRI1-like is a key regulatory factor of lipid biosynthesis. DpWRI1-like gene and target genes of DpWRI1-like have been obtained in our previous study. Whereas, target proteins of DpWRI1-like remain unclear. In this study, the interacting proteins of DpWRI1-like (ACBD6, DCA and TF) were found by Y2H to further understand the function of DpWRI1-like. This study laid a good foundation for further understanding the regulatory mechanism of DpWRI1-like in lipid synthesis.
CRediT authorship contribution statement
Lingru Ruan: Writing – original draft, Data curation. Limei Huang: Writing – original draft, Data curation. Lina Wu: Writing – original draft, Data curation. Jinghui Gu: Writing – original draft, Data curation. Yanyan Liang: Writing – original draft, Data curation. Xiuli Liang: Writing – original draft, Data curation. Changhua Shang: Writing – review & editing, Writing – original draft, Data curation, Conceptualization.
Ethical approval statement
Not applicable.
Data and code availability statement
Data generated during this study can be obtained from the corresponding author upon reasonable request. mRNA data have been deposited at NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers (Acyl-CoA-binding domain-containing protein 6, ACBD6, GenBank no.: PQ321315.1; Duplicated carbonic anhydrase, DCA, GenBank no.: PQ321311.1 and DNA-binding transcription factor, TF, GenBank no.: ON548534.1).
Funding
This study was financially supported by Natural Science Foundation of Guangxi Zhuang Autonomous Region (No. 2024JJA130020), Guangxi Key Research and Development Program (No. 2023AB01132) and 2024 Joint Fund for Guangxi Normal University and National Natural Science Foundation of China (No. 2024PY020).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e41165.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Han X.T., Zheng L., Sun S., Zou J.Z. An application prospect of biodiesel from marine microalgae. Mar. Sci. 2008;8:76–81. [Google Scholar]
- 2.Shahid A., Rehman A.U., Usman M., Ashraf M.U.F., Javed M.R., Khan A.Z., Gill S.S., Mehmood M.A. Engineering the metabolic pathways of lipid biosynthesis to develop robust microalgal strains for biodiesel production. Biotechnol. Appl. Biochem. 2020;67:41–51. doi: 10.1002/bab.1812. [DOI] [PubMed] [Google Scholar]
- 3.Kumari K., Samantaray S., Sahoo D., Tripathy B.C. Nitrogen, phosphorus and high CO2 modulate photosynthesis, biomass and lipid production in the green alga Chlorella vulgaris. Photosynth. Res. 2021;148:17–32. doi: 10.1007/s11120-021-00828-0. [DOI] [PubMed] [Google Scholar]
- 4.Shang C., Bi G., Yuan Z., Wang Z., Alam M.A., Xie J. Discovery of genes for production of biofuels through transcriptome sequencing of Dunaliella parva. Algal Res. 2016;13:318–326. doi: 10.1016/j.algal.2015.12.012. [DOI] [Google Scholar]
- 5.Courchesne N.M., Parisien A., Wang B., Lan C.Q. Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J. Biotechnol. 2009;141:31–41. doi: 10.1016/j.jbiotec.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Chai Y.R., Tian F., Liu H.T., Li J., Xue L.X. Isolation and functional analyse of promoter of the rbcS gene from Dunaliella salina. China Biotechnol. 2008;28:47–52. doi: 10.13523/j.cb.20080410. [DOI] [Google Scholar]
- 7.Walker T.L., Becker D.K., Collet C. Characterisation of the Dunaliella tertiolecta RbcS genes and their promoter activity in Chlamydomonas reinhardtii. Plant Cell Rep. 2005;23:727–735. doi: 10.1007/s00299-004-0884-x. [DOI] [PubMed] [Google Scholar]
- 8.Focks N., Benning C. wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118:91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma W., Kong Q., Arondel V., Kilaru A., Bates P.D., Thrower N.A., Benning C., Ohlrogge J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Z., Liu X., Li N., Du C., Wang K., Zhao C., Wang Z., Hu Y., Zhang M. WRINKLED1 homologs highly and functionally express in oil-rich endosperms of oat and castor. Plant Sci. 2019;287 doi: 10.1016/j.plantsci.2019.110193. [DOI] [PubMed] [Google Scholar]
- 11.Maeo K., Tokuda T., Ayame A., Mitsui N., Kawai T., Tsukagoshi H., Ishiguro S., Nakamura K. An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009;60:476–487. doi: 10.1111/j.1365-313X.2009.03967.x. [DOI] [PubMed] [Google Scholar]
- 12.Deng S., Mai Y., Shui L., Niu J. WRINKLED1 transcription factor orchestrates the regulation of carbon partitioning for C18:1 (oleic acid) accumulation in Siberian apricot kernel. Sci. Rep. 2019;9:2693. doi: 10.1038/s41598-019-39236-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang C., Pang B., Yu H., Gan S., Li Y. Identification of targets of transcription factor WRINKLED1-Like related to lipid biosynthesis from marine microalga Dunaliella parva. Front. Mar. Sci. 2022;8 doi: 10.3389/fmars.2021.807493. [DOI] [Google Scholar]
- 14.Huang R., Liu M., Gong G., Wu P., Bai M., Qin H., Wang G., Liao H., Wang X., Li Y., Wu H., Wang X., Yang C., Schubert D., Zhang S. BLISTER promotes seed maturation and fatty acid biosynthesis by interacting with WRINKLED1 to regulate chromatin dynamics in Arabidopsis. Plant Cell. 2022;34:2242–2265. doi: 10.1093/plcell/koac083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong Q., Singh S.K., Mantyla J.J., Pattanaik S., Guo L., Yuan L., Benning C., Ma W. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiol. 2020;184:658–665. doi: 10.1104/pp.20.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai Z., Liu H., Shanklin J. Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell. 2017;29:871–889. doi: 10.1105/tpc.17.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang C., Pang B., Zhang J., Yu L., Gan S., Li Y., Wu H. Identification of interacting proteins of transcription factor DpAP2 related to carotenoid biosynthesis from marine microalga Dunaliella parva. Front. Mar. Sci. 2022;9 doi: 10.3389/fmars.2024.1448420. Corrigendum:Front.Mar.Sci.11 (2024) 1448420. [DOI] [Google Scholar]
- 18.Chung I.Y.W., Li L., Tyurin O., Gagarinova A., Wibawa R., Li P., Hartland E.L., Cygler M. Structural and functional study of Legionella pneumophila effector RavA. Protein Sci. 2021;30:940–955. doi: 10.1002/pro.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z.M., Zhuang M., Wang B.T., Jin L., Jin F.J. Identification and characterization of a DevR-interacting protein in Aspergillus oryzae. Fungal Biol. 2020;124:155–163. doi: 10.1016/j.funbio.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Burton M., Rose T.M., Faergeman N.J., Knudsen J. Evolution of the acyl-CoA binding protein (ACBP) Biochem. J. 2005;392:299–307. doi: 10.1042/BJ20050664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faergeman N.J., Wadum M., Feddersen S., Burton M., Kragelund B.B., Knudsen J. Acyl-CoA binding proteins; structural and functional conservation over 2000 MYA. Mol. Cell. Biochem. 2007;299:55–65. doi: 10.1007/s11010-005-9040-3. [DOI] [PubMed] [Google Scholar]
- 22.Fan J., Liu J., Culty M., Papadopoulos V. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog. Lipid Res. 2010;49:218–234. doi: 10.1016/j.plipres.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao S., Chye M.L. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol. Biochem. 2009;47:479–484. doi: 10.1016/j.plaphy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Xiao S., Chye M.L. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog. Lipid Res. 2011;50:141–151. doi: 10.1016/j.plipres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Soupene E., Kuypers F.A. Ligand binding to the ACBD6 protein regulates the acyl-CoA transferase reactions in membranes. J. Lipid Res. 2015;56:1961–1971. doi: 10.1194/jlr.M061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soupene E., Kuypers F.A. Dual role of ACBD6 in the acylation remodeling of lipids and proteins. Biomolecules. 2022;12:1726. doi: 10.3390/biom12121726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soupene E., Kao J., Cheng D.H., Wang D., Greninger A.L., Knudsen G.M., DeRisi J.L., Kuypers F.A. Association of NMT2 with the acyl-CoA carrier ACBD6 protects the N-myristoyltransferase reaction from palmitoyl-CoA. J. Lipid Res. 2016;57:288–298. doi: 10.1194/jlr.M065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soupene E., Kuypers F.A. ACBD6 protein controls acyl chain availability and specificity of the N-myristoylation modification of proteins. J. Lipid Res. 2019;60:624–635. doi: 10.1194/jlr.M091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soupene E., Schatz U.A., Rudnik-Schöneborn S., Kuypers F.A. Requirement of the acyl-CoA carrier ACBD6 in myristoylation of proteins: activation by ligand binding and protein interaction. PLoS One. 2020;15 doi: 10.1371/journal.pone.0229718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castrec B., Dian C., Ciccone S., Ebert C.L., Bienvenut W.V., Le Caer J.P., Steyaert J.M., Giglione C., Meinnel T. Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern. Nat. Chem. Biol. 2018;14:671–679. doi: 10.1038/s41589-018-0077-5. [DOI] [PubMed] [Google Scholar]
- 31.Giglione C., Meinnel T. Mapping the myristoylome through a complete understanding of protein myristoylation biochemistry. Prog. Lipid Res. 2022;85 doi: 10.1016/j.plipres.2021.101139. [DOI] [PubMed] [Google Scholar]
- 32.Hewett-Emmett D., Tashian R.E. Functional diversity, conservation, and convergence in the evolution of the alpha-, beta-, and gamma-carbonic anhydrase gene families. Mol. Phylogenet. Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- 33.Badger M.R., Price G.D. The role of carbonic anhydrase in photosynthesis. Annu. Rev. Plant Biol. 1994;45:369–392. doi: 10.1146/annurev.pp.45.060194.002101. [DOI] [Google Scholar]
- 34.Henry R.P., Cameron J.N. The role of carbonic anhydrase in respiration, ion regulation and acid-base balance in the aquatic crab Callinectes sapidus and the terrestrial crab Gecarcinus lateralis. J. Exp. Biol. 1983;103:205–223. doi: 10.1242/jeb.103.1.205. [DOI] [Google Scholar]
- 35.Weber A.K., Pirow R. Physiological responses of Daphnia pulex to acid stress. BMC Physiol. 2009;9:9. doi: 10.1186/1472-6793-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teitelbaum S.L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 37.Le Roy N., Jackson D.J., Marie B., Ramos-Silva P., Marin F. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front. Zool. 2014;11:75. doi: 10.1186/s12983-014-0075-8. [DOI] [Google Scholar]
- 38.Moroney J.V., Ma Y., Frey W.D., Fusilier K.A., Pham T.T., Simms T.A., DiMario R.J., Yang J., Mukherjee B. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: intracellular location, expression, and physiological roles. Photosynth. Res. 2011;109:133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- 39.Fisher M., Gokhman I., Pick U., Zamir A. A salt-resistant plasma membrane carbonic anhydrase is induced by salt in Dunaliella salina. J. Biol. Chem. 1996;271:17718–17723. doi: 10.1074/jbc.271.30.17718. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated during this study can be obtained from the corresponding author upon reasonable request. mRNA data have been deposited at NCBI (https://www.ncbi.nlm.nih.gov/) with accession numbers (Acyl-CoA-binding domain-containing protein 6, ACBD6, GenBank no.: PQ321315.1; Duplicated carbonic anhydrase, DCA, GenBank no.: PQ321311.1 and DNA-binding transcription factor, TF, GenBank no.: ON548534.1).