Abstract
Traditionally, medical research is based on randomized controlled trials (RCTs) for interventions such as drugs and operative procedures. However, increasingly, there is a need for health research to evolve. RCTs are expensive to run, are generally formulated with a single research question in mind, and analyze a limited dataset for a restricted period. Progressively, health decision makers are focusing on real-world data (RWD) to deliver large-scale longitudinal insights that are actionable. RWD are collected as part of routine care in real time using digital health infrastructure. For example, understanding the effectiveness of an intervention could be enhanced by combining evidence from RCTs with RWD, providing insights into long-term outcomes in real-life situations. Clinicians and researchers struggle in the digital era to harness RWD for digital health research in an efficient and ethically and morally appropriate manner. This struggle encompasses challenges such as ensuring data quality, integrating diverse sources, establishing governance policies, ensuring regulatory compliance, developing analytical capabilities, and translating insights into actionable strategies. The same way that drug trials require infrastructure to support their conduct, digital health also necessitates new and disruptive research data infrastructure. Novel methods such as common data models, federated learning, and synthetic data generation are emerging to enhance the utility of research using RWD, which are often siloed across health systems. A continued focus on data privacy and ethical compliance remains. The past 25 years have seen a notable shift from an emphasis on RCTs as the only source of practice-guiding clinical evidence to the inclusion of modern-day methods harnessing RWD. This paper describes the evolution of synthetic data, common data models, and federated learning supported by strong cross-sector collaboration to support digital health research. Lessons learned are offered as a model for other jurisdictions with similar RWD infrastructure requirements.
Keywords: real-world data, digital health research, synthetic data, common data models, federated learning, university-industry collaboration
Background
While randomized controlled trials (RCTs) have long been accepted as the gold standard in evidence-based medicine, increasingly, there is a need to evolve this practice [1]. Well-designed RCTs are ideal for investigating the safety and efficacy of an intervention in a highly controlled setting, for example, treatment effects in drug development [2]. RCTs can fail to demonstrate the effectiveness of the intervention under complex, “real-world,” dynamic conditions [3]. This can have serious cost implications for health systems when the outcomes promised under RCT conditions fail to deliver during postmarket surveillance [4]. Increasingly, health decision makers are focusing on real-world data (RWD) to deliver large-scale longitudinal insights that are actionable. RWD are collected as part of routine care in real time using digital health infrastructure [3,5]. Modern-day health research can capitalize on the benefits of RWD with a focus on translating the findings into clinical practice. Together, the findings generated through RCTs and RWD can bridge evidence gaps to support regulatory decision-making [6]. RWD “can provide valuable complementary evidence by answering important questions on treatment effects in clinical practice that are not answered by RCTs” [7]. Perspectives in medical research regarding RCTs as the only source of practice-guiding clinical evidence need to evolve. Certainly, the use of RWD for regulatory decision-making must address key considerations to ensure that the evidence generated is fit for purpose. This includes evaluation of data relevancy and quality, including accuracy, completeness, provenance, and transparency of RWD processing [8]. Steps to address these considerations are evident in the frameworks and policies emerging over the past decade, for example, to support the Food and Drug Administration (FDA) with harnessing RWD for postmarket safety surveillance [9]. Both data obtained through RCTs and RWD have their strengths and weaknesses (Textbox 1), further emphasizing a complementary approach to both methods in modern-day health research.
Comparing data capture methods for randomized controlled trials (RCTs) versus real-world data (RWD).
Data capture for RCTs
Demonstrate efficacy under controlled conditions (internal validity)
Describe effect and causal relationships between an intervention and an outcome
Data collected in a controlled and scheduled manner in accordance with the clinical trial
Collected specifically to answer a small number of questions
Other data regarding comorbidities may be incomplete or contain recall bias
Intervention compared to either placebo or selected alternative
Quality assessment tools used to review risk of bias resulting from imperfect RCT methodology
Data elements centered on a specific research question with limited longitudinal insights
RWD
Demonstrate effectiveness under real-world conditions (external validity)
Describe the association or correlation between an intervention and an outcome
Can be used to derive causal relationships but entail strong assumptions and rigorous methods, including evaluation of the RWD relevancy and quality
Data often offer the advantage of being available in real time or near real time (recency of data capture)
Provide a comprehensive picture of the patient (including details of the illness and social determinants)
The same data used for clinical care are used for research purposes, noting that RWD can be subject to other forms of bias; for example, the care received may be a function of socioeconomic resources
No control arm or intervention compared to standard treatment or care
Evaluation of data quality is necessary to ensure accuracy, completeness, provenance, and transparency of processing
Data assets may offer fragmented real-world trajectories across health systems
The interest in RWD for medical research has coincided with the rapid expansion of health IT (HIT), generating vast volumes of digital data through a myriad of sources. These include electronic medical records (EMRs), personal health records, wearable devices, mobile health, registries, and administrative data (such as claims and billing activities) [10]. However, the massive amounts of data now generated across various health care systems and platforms pose challenges in data integration and interoperability. The European Commission’s funding initiatives, such as Horizon Europe and the Innovative Health Initiative, emphasize the importance of cross-sector collaboration and data integration to foster improved interoperability and advance health care research [11,12]. Other challenges faced by RWD capture for research include privacy and confidentiality concerns [13]. Using RWD for research requires the secondary use of the data for purposes other than those for which they were originally collected [14]. Ethical and governance considerations must reflect both social license and privacy-protecting regulations. However, a difficulty faced by researchers in the digital era is conforming to regulatory frameworks established before digitization. While efforts are underway to integrate access to RWD for secondary use into updated legislation, novel methods are necessary to harness “big data” for digital health research. The same way that drug trials require infrastructure such as research nurses to support their conduct, digital health and the use of RWD also have research infrastructure needs [15]. These are not yet present in most academic institutions.
Health care research urgently requires the transformative power of data and HIT. Solutions are emerging to capture RWD siloed across HIT systems while addressing critical challenges such as interoperability, privacy, security, and effectiveness. This paper describes the rapid evolution of the medical research landscape and the ongoing development of modern-day research infrastructure. Such methods include common data models (CDMs) [16], federated learning (FL) [17], and synthetic data generation [18] supported by strong cross-sector collaboration. These novel methods are explored and, in turn, lessons learned are offered as a model for other jurisdictions with similar RWD infrastructure requirements.
Methodology
Health data collection methods have undergone significant evolution alongside the widespread adoption of HIT systems, EMRs, and other digital health technologies. To comprehensively understand this evolution, we conducted a review and perspective study, tracing the progression from traditional data capture methods such as RCTs to the integration of RWD into medical research. Our objective was to provide both a retrospective examination and a forward-looking perspective on the evolution of research infrastructure for digital health over the past 25 years. In our methodology, we outlined the trends and strategies identified through the rapid review to overcome barriers to using RWD and enhance health research infrastructure. We emphasized the incorporation of all available health data resources to ensure a comprehensive analysis, with continued attention to data privacy, ethical compliance in digital health, and mitigation of disclosure risk.
The Right Data for the Right Problem
Overview
To support the evolution of modern-day digital health research, a multifaceted approach, including synthetic data generation, mapping to CDMs, FL, and enablers to promote RWD extraction for research, is proposed. Figure 1 conceptualizes such an approach using CDM frameworks to support access to routinely collected health data, synthetic data generation, and FL infrastructure. Such an approach provides flexibility, offering the right data for the right problem at hand. Scenarios will always exist in research that require the extraction of identifiable or potentially reidentifiable patient information from data repositories for research purposes. In such circumstances, while the clinical validity of the data is high, so, too, can be the disclosure risk. Strict adherence to ethics and governance research protocols is essential. However, in recent years, there has been growing interest in alternative methods to harness RWD while minimizing disclosure risk. Methods to support RWD access in a deidentified manner, standardizing terminologies and mitigating the need for data sharing outside of enterprise structures are of particular focus. In doing so, the need to access identifiable or potentially reidentifiable patient health care data is minimized. The strategies identified to deliver each alternative method, balancing privacy concerns against clinical usefulness, are outlined in Figure 1.
Figure 1.
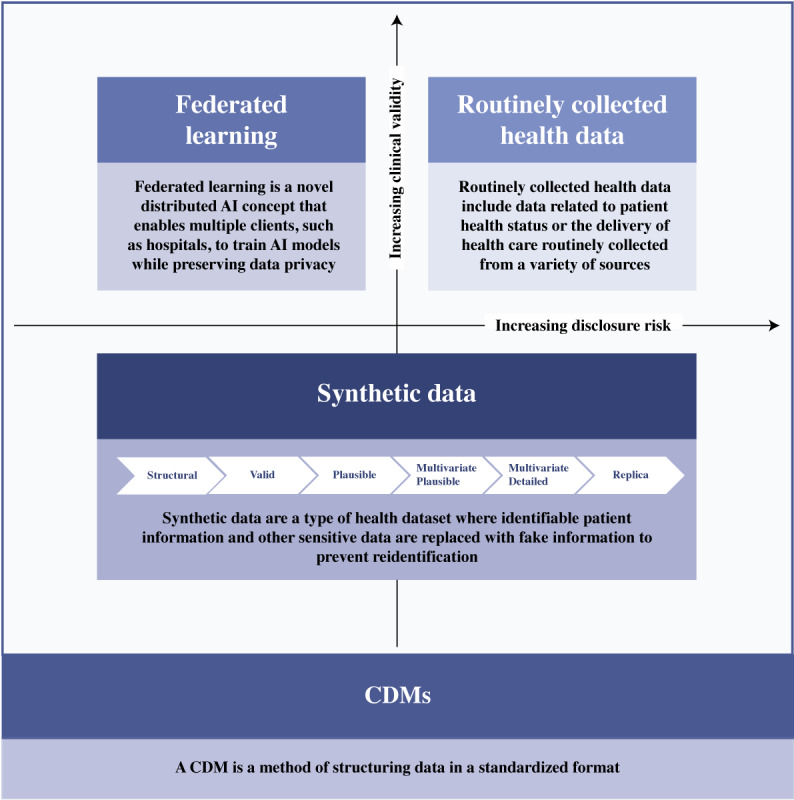
Approaches to accessing data for modern-day health research. AI: artificial intelligence; CDM: common data model.
Goal 1: Synthetic Data Generation
Historically, accessing RWD has been associated with many challenges, such as laborious data access and consent procedures [19], particularly in environments in which privacy protection is prioritized and public scrutiny of digital privacy is rising [20]. Synthetic datasets, generated by a model to represent essential aspects of RWD [21], have been proposed to offer a solution for both privacy concerns and the need for widespread data access for analysis [22].
Synthetic datasets are generally classified into 3 broad categories: fully synthetic, partially synthetic, and hybrid [23]. Fully synthetic datasets entirely synthesize data without original values, ensuring privacy but compromising data validity [24-26]. In contrast, partially synthetic datasets replace selected attributes with synthetic values to preserve privacy while retaining original data, which is beneficial for imputing missing values [24-26]. Hybrid synthetic datasets combine original and synthetic data for strong privacy preservation, increasing data validity to help achieve a balance between privacy and fidelity [24-26]. However, there is a more detailed classification by the UK Office for National Statistics, which describes synthetic data in 6 levels [27], as shown in Figure 1. On the basis of this classification, a synthetic structural dataset (lowest level), developed solely from metadata, lacks clinical value and disclosure risk but is suitable only for basic code testing [27]. Conversely, a replica-level synthetically augmented dataset (highest level), which preserves format, structure, and patterns, offers high analytical value but increases disclosure risks due to its similarity to the original data [27]. The selection of synthetic data would depend on the nature of the application.
The use of synthetic data has a long-standing history dating back to the early stages of computing [28]. The early foundational work of Stanislaw Ulam and John von Neumann in the 1940s, particularly focusing on the Monte Carlo simulation technique [29], is one such example. However, the notion of fabricating synthetic data to ensure valid statistical inferences and uphold disclosure control was first suggested by Rubin (as cited in the work by Raghunathan [22]) as a discussion of the work by Jabine (as cited in the work by Raghunathan [22]). Over time, the generation of synthetic data has moved from the use of statistical methods (eg, multiple data imputation and Bayesian bootstrap) [23] to more robust algorithms [30] due to the rise of several novel tools and services [23]. An early example is the synthetic minority oversampling technique algorithm, where synthetic data points are generated by selecting a predetermined number of neighbors for each underrepresented instance, randomly choosing some minority class instances, and creating artificial observations along the line between the selected minority instance and its closest neighbors [31]. This algorithm underwent maturation over time, leading to the emergence of several variants [32-35], which predominately focused on continuous variables but failed to identify nominal features when applied to datasets with categorical features, necessitating the creation of new labels for these attributes [36].
The introduction of deep learning methodologies, exemplified by the inception of variational autoencoders in 2013 and generative adversarial networks (GANs) in 2014, catalyzed the evolution of more promising paradigms in the domain of synthetic data generation [37]. GANs, most importantly [37], had the potential to generate synthetic data without direct engagement with the original dataset, a feature with potential implications for reducing disclosure risk [38]. The GAN model first proposed by Goodfellow et al [38] considers simultaneously training two neural network models: (1) a generative model that captures the data distribution and (2) a discriminative model that determines where the sample is generated from the model or data distribution (Figure 2) [39]. Initially, the generative model commences with noise inputs, devoid of access to the training or original dataset, relying on feedback from the discriminative model to generate a data sample [39]. Currently, GANs have gained a lot of interest due to their capability to produce high-quality synthetic data that closely match real data, especially in health care applications [40], including (1) forecasting and planning, (2) design and evaluation of new health technology and algorithms, (3) data augmentation, (4) testing and benchmarking, and (5) education [41].
Figure 2.
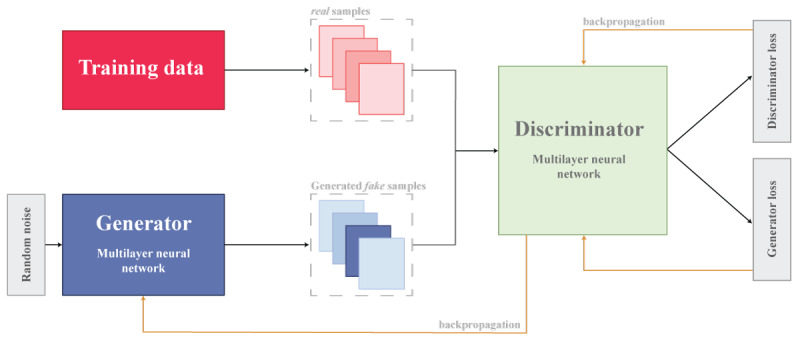
Generative adversarial network model.
In the domain of published literature, GAN models are frequently discussed for their role in generating synthetic data [42-46]. However, various applications and services are now accessible for creating synthetic data tailored specifically for health care applications [23]. Among these tools are Synthea, implemented in Java; DataSynthesizer and SynSys, which are Python packages; and synthpop and simPop, both packages based on R [30,47,48]. Synthea uses the PADARSER (publicly available data approach to the realistic synthetic electronic health record) framework for synthetic data generation, relying on publicly available datasets instead of real electronic health records (EHRs) [49]. The framework emphasizes (1) using health statistics, (2) assuming no access to real EHRs, (3) integrating clinical guidelines, and (4) ensuring realistic properties in synthetic EHRs, as shown in Figure 3 [49].
Figure 3.
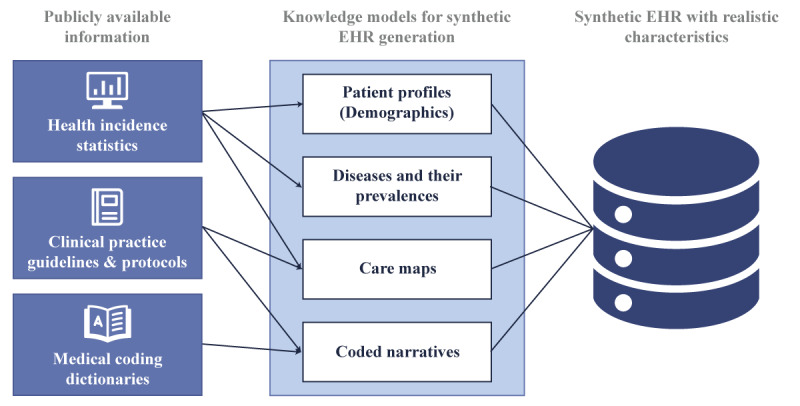
PADARSER (publicly available data approach to the realistic synthetic electronic health record) framework reproduced from Walonoski J et al [49], which is published under Creative Commons Attribution 4.0 International License [52]. EHR: electronic health record.
synthpop uses regression trees for generating variables in a synthetic population but cannot handle complex data structures such as sophisticated sampling designs or hierarchical clusters (eg, individuals within households) [50], whereas simPop focuses on a modular object-oriented concept that uses various approaches, such as calibration through iterative proportional fitting and simulated annealing and modeling or data fusion through logistic regression, to generate a synthetic population [50]. In contrast, DataSynthesizer and SynSys use real patient data for the generation of synthetic datasets. For example, the DataSynthesizer includes 3 key modules for the generation of synthetic data: DataDescriber, which analyzes attribute types and distributions while preserving privacy; DataGenerator, which uses this analysis to create synthetic data; and Model Inspector, which provides an intuitive summary for evaluation and adjustment of parameters [51]. SynSys uses real data to train Markov and regression models to generate more realistic synthetic data, as shown in Figure 4 [30].
Figure 4.
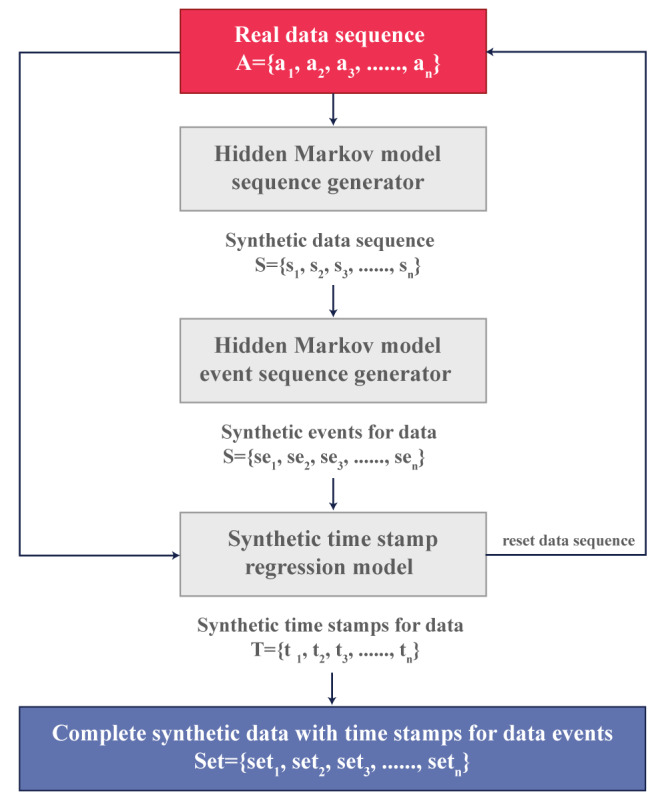
SynSys model adapted from Dahmen J et al [30], which is published under Creative Commons Attribution 4.0 International License [53].
Goal 2: CDMs
Sharing clinical data, including clinical trial data, for research is increasingly recognized as an efficient way to advance scientific knowledge [54]. However, the sharing of clinical data in health care is not without its challenges, with research highlighting concerns related to privacy, security, and interoperability [55]. While literature exists with regard to mitigating privacy and security issues in clinical data sharing for research purposes, interoperability issues persist [56]. One potential solution that has been touted to limit issues related to interoperability are CDMs [55].
CDMs are commonly used in research to enable the exchange or sharing of datasets for specific purposes [57]. The objective of a CDM is to streamline the conversion of data from diverse databases into a consistent format with standardized terminology, thereby enabling systematic analysis [58]. Over the past decade, several CDMs have been collaboratively developed and risen to the level of de facto standards for clinical research data. These include the Health Care Systems Research Network (formerly known as the HMO Research Network) Virtual Data Warehouse, the National Patient-Centered Clinical Research Network CDM, the Observational Medical Outcomes Partnership (OMOP) CDM, the Clinical Data Interchange Standards Consortium (CDISC) Study Data Tabulation Model, and the Sentinel CDM [59].
The CDISC was one of the oldest known CDMs, established in 1998, and has been pivotal in streamlining clinical data acquisition, interchange, and submission processes. With its 12 domains (Textbox 2) and unique variable naming conventions, the CDISC ensures clarity and consistency in data representation. However, it mainly aims to provide guidelines rather than imposing strict data collection requirements, allowing for flexibility for different study designs and objectives [60].
Clinical Data Interchange Standards Consortium domains and their data structures [60].
Demographics: 1 record per subject
Disposition: 1 record per subject
Exposure: 1 record per subject per phase or dose
Adverse events: 1 record per subject per adverse event
Concomitant medications: 1 record per subject per medication
Serum chemistry: 1 record per subject per visit per measurement
Hematology: 1 record per subject per visit per measurement
Urinalysis: 1 record per subject per visit per measurement
Electrocardiogram: 1 record per subject per visit
Vital signs: 1 record per subject per visit (per position)
Physical examination: 1 record per subject per examination, body system, or finding
Medical history: 1 record per subject per examination, body system, or condition
Another significant CDM, Sentinel, initiated as part of the FDA’s Sentinel Initiative to monitor FDA-regulated medical products on a national scale [61]. It uses standardized concept codes with 19 tables (Textbox 3) [62], although users may need to map data due to variations in coding systems [63]. On the other hand, the Health Care Systems Research Network Virtual Data Warehouse aims to centralize data extraction and loading processes across 17 health care systems in the United States [64]. Its comprehensive structure comprises 7 content areas and >450 variables spread across 18 tables, as illustrated in Figure 5 [64], enhancing research efficiency by consolidating data management efforts [64].
Sentinel Common Data Model [62].
Administrative data
Enrollment
Demographic
Dispensing
Encounter
Diagnosis
Procedure
Prescribing
Mother-infant linkage data
Mother-infant linkage
Auxiliary data
Facility
Provider
Feature engineering data
Feature engineering
Registry data
Death
Cause of death
State vaccine
Inpatient data
Inpatient pharmacy
Inpatient transfusion
Clinical data
Laboratory test results
Vital signs
Patient-reported measure (PRM) data
PRM survey
PRM survey response
Figure 5.
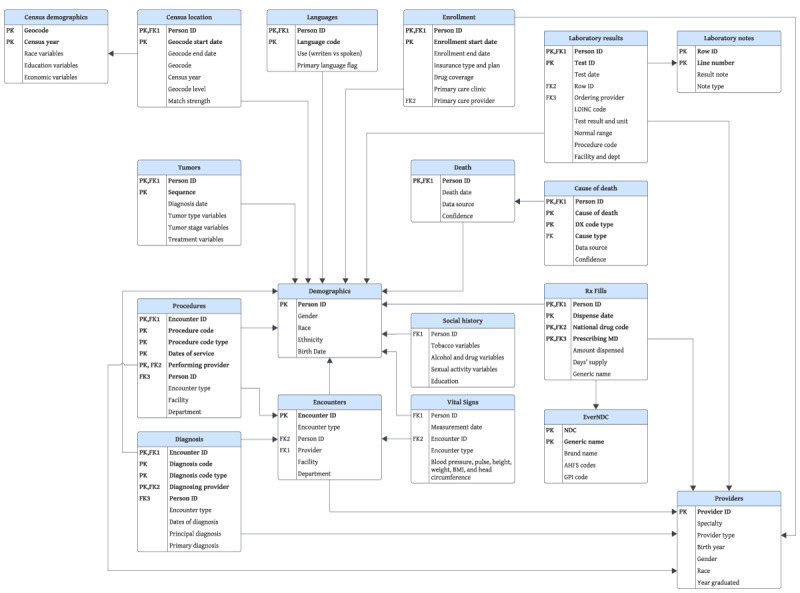
Health Care Systems Research Network Virtual Data Warehouse common data model, modified from Ross TR et al [62], which is published under a Creative Commons Attribution 4.0 International License [65]. AHFS: American hospital formulary service; DX: diagnostic; EverNDC: Ever National Drug Code; GPI: generic product identifier; LOINC: Logical Observation Identifiers Names and Codes; MD: medical doctor; NDC: National Drug Code; Rx: prescription.
The National Patient-Centered Clinical Research Network was implemented to support patient-centered studies and stands out for its expansive data coverage, storing information from >100 million individuals [66] in a common format across its 23 interconnected tables [67]. It incorporates actual dates and a unique patient identifier for efficient data navigation, ensuring data integrity and facilitating comprehensive analysis [68]. In addition, the Observational Health Data Sciences and Informatics program focused on standardizing medical data representation across diverse source systems [69]. With its OMOP CDM comprising 18 tables [70], the Observational Health Data Sciences and Informatics program integrates data from >100 databases worldwide [71], addressing the need for standardized EHR data and consistent patient-level information in observational databases [69], as shown in Figure 6 [72].
Figure 6.
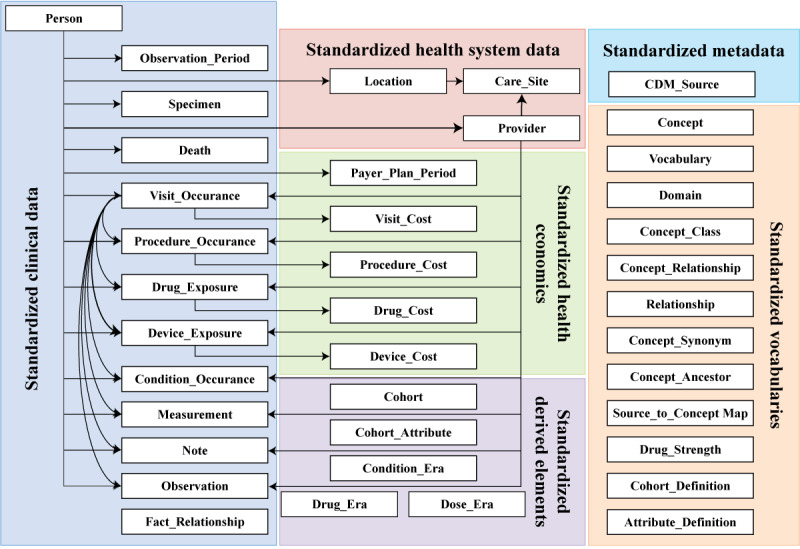
Observational Medical Outcomes Partnership Common Data Model, reproduced from Jiang G et al [72], which is published under Creative Commons Attribution 4.0 International License [52].
While most approaches follow a different structure for storing their data, most of these models use an extract, transform, and load (ETL) process to map the data from the source database to the target structure, as shown in Figure 7 [73]. The source database may come from hospital-wide systems (such as Epic EMRs, Oracle Health, and Meditech [75]) or departmental systems (such as MOSAIQ by Elekta [76], ARIA by Varian [76], picture archiving and communication systems [77], pathology or laboratory information systems [78], and others).
Figure 7.
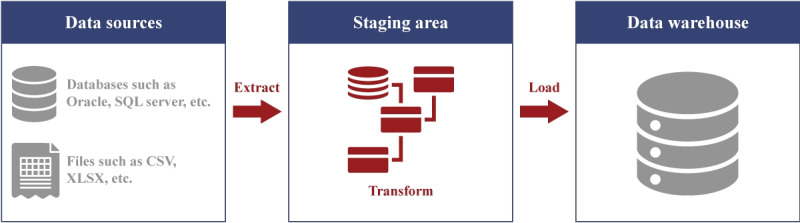
Extract, transform, and load process adapted from the work published by Abd Al-Rahman SQ et al [73], under the CC-BY-SA license [74].
The ETL process operates via 3 principal stages: extraction, transformation, and loading [79]. Extraction refers to retrieving data from relevant sources, often in file formats such as CSV [79], relational databases such as MySQL [79], nonrelational databases such as NoSQL [80], graph databases such as Neo4j [81], or accessed through Representational State Transfer clients [79]. Transformation entails the refinement and adaptation of the data to conform to the prescribed schema, encompassing tasks such as normalization, deduplication, and quality validation procedures [79]. This stage may also involve aligning the data with standardized terminologies such as the Systemized Nomenclature of Medicine–Clinical Terms or the International Classification of Diseases to ensure semantic consistency and interoperability across systems [82] or understanding preexisting standards (such as Digital Imaging and Communications in Medicine [83], the National Council for Prescription Drug Programs SCRIPT standard [84], and so on) toward mapping relevant information. Loading involves the transfer of the refined data into operational databases, data marts, or data warehouses for subsequent use [79].
Goal 3: FL
Traditional centralized machine learning (ML) approaches face privacy and security risks [85] and limited predictive accuracy due to single-source data constraints [86]. To limit these challenges, FL has emerged as a solution by facilitating distributed model training on local devices. Google introduced FL in 2016, which uses distributed learning platforms to leverage enhanced computational abilities of devices, connect devices executing local training models, and facilitate cooperation among devices to build consensus global models of learning [87]. FL offers a secure and efficient approach to analyzing fragmented health care data [88]. This decentralized approach reduces the risk of data exposure and vulnerability to cyberattacks [89].
Over the past years, there has been a notable trend regarding how medical data are processed and used. EMRs play an important role in health care data collection and retrieval. However, strict regulations on data sharing necessitate the anonymization of sensitive patient attributes [90]. Health care organizations face challenges in aggregating clinical records for deep learning models due to privacy, data ownership, and legal concerns. Balancing data protection with leveraging collective knowledge is challenging [88]. In health care, FL initiatives are emerging as a privacy-enhancing approach to artificial intelligence and ML. These initiatives aim to collaboratively train predictive models across various institutions without centralizing sensitive personal data [91]. Recently, FL has been applied to the health care domain and life science industry, addressing the need for high-quality models in ML applications [92,93]. The FL paradigm has gained popularity for its scalable and privacy-preserving approach to joint training across federated health data repositories [85,93,94]. FL develops ML models over distributed datasets in locations such as hospitals, laboratories, and mobile devices, ensuring data privacy [88]. FL aims to overcome barriers associated with transferring sensitive clinical data to a central repository in conventional centralized artificial intelligence and ML models [85]. This approach allows for training of ML models on distributed client nodes, preserving the privacy and integrity of patient data [85]. The core concept of FL involves sharing only the parameters of the ML model being trained rather than sharing the actual data [94-96].
The FL methodology involves a network of nodes, each sharing models instead of raw training data with the central server. FL is conducted iteratively as follows. Initially, the server distributes the current global ML model parameters to all participating edge nodes. Each node then uses its locally stored data samples to update its own model based on the received parameters. Subsequently, each node transmits its updated model parameters back to the server. The server performs a global aggregation operation, combining and weighting the model parameters received from each node to generate a new set of global model parameters. This process is iterated multiple times until convergence. Importantly, at no stage do the nodes share their training data with each other or the central server, enhancing privacy and reducing bandwidth use [87,97,98].
Goal 4: Cross-Sector Collaboration (Enablers to Promote RWD Access for Research)
The methods outlined previously provide novel approaches to RWD (or simulated RWD) access to promote digital health research. While these methods may meet most digital health research requests, access to ethically approved identifiable RWD cannot be dismissed. However, a conundrum in the digital era, with EMRs now generating vast volumes of health care data, is the limited skilled informaticians trained in data extraction and analysis. The Joint Science Academies Statement on Global Issues specific to “Digital Health and the Learning Health System” noted the basic requirement of developing and cultivating a digital health workforce, stating that “the training challenge for leveraging digital health is vast—in health care, public health and biomedical science” [99]. Those trained in data extraction are often focused on the operational activities of the health care organization. Support is needed to streamline RWD extraction for digital health research. Assigning domain experts to handle the manual data extraction steps to support researchers with access to medical RWD is necessary [100]. Academia-industry digital health collaborations can leverage uniquely skilled resources and networks to benefit both sectors [101]. Embedding staff with affiliations to both the university and health care sectors is one potential method. To overcome barriers related to university-industry collaboration, an environment fostering the missions of both sectors is necessary [102]. Being cognizant of the notable differences between the primary cross-sector objectives is necessary, for example, feasible timelines and balancing competing demands [103]. This approach is explored further in the use case below.
Use Case
In reviewing the evolution of digital techniques used to harness RWD, consideration must be given to the application of such methods to support modern-day research. An illustrative use case is provided in this section to offer a forward-looking perspective on where such techniques may be headed.
A center dedicated to digital health research was established in Queensland, Australia. The center spanned 6 university faculties, collaborating with external government and industry partners. To overcome the challenges of harnessing RWD for research, the center established a service offering a multifaceted approach to RWD access (Figure 1). The needs and current and future state of each research infrastructure goal have been summarized in Table 1.
Table 1.
The needs and current and future state of the research infrastructure goals of a center dedicated to digital health research established in Queensland, Australia.
|
|
Synthetic data | CDMsa | FLb | Routinely collected health data (EMRc) |
| Needs |
|
|
|
|
| Current state |
|
|
|
|
| Future state |
|
|
|
|
aCDM: common data model.
bFL: federated learning.
cEMR: electronic medical record.
dRWD: real-world data.
eAI: artificial intelligence.
fOMOP: Observational Medical Outcomes Partnership.
The infrastructure goals highlighted in Table 1 draw upon techniques emerging in recent decades through the maturation of digital health technologies and strong cross-sector collaborations. The use case signifies how organizations are joining forces to advance modern-day research through RWD capture. No individual goal was deemed superior, yet through commitment to drive each approach to RWD access (Figure 1), this dedicated service is a method for providing researchers with the right data for the right problem.
Discussion
Overview
The evolution of digital health has seen many health care organizations shifting beyond the foundational levels of implementation to established methods of harnessing RWD to promote a learning health system [105]. A learning health system needs academic inquiry brought close to the routinely generated health care data, yet data security and privacy must remain paramount. While the clinical validity of the data is always greatest via direct access and extraction from the data source, so, too, is the disclosure risk. Novel methods have emerged and evolved to support access to RWD for modern-day health care research. Application of these techniques over time has provided an opportunity to reflect on the emerging needs, including the strengths and weaknesses of each goal and the future directions. In addition, the lessons learned for the described digital health research center case in point (Table 1) are included for each goal in the following sections.
Synthetic Data Strengths, Weaknesses, and Lessons Learned
Synthetic data generation has made significant advancements in recent decades, from statistical methods to robust algorithms and established applications and services tailored to synthetic data generation for health care needs. The synthetic data created by the various models have the potential to reduce costs and accelerate data generation [106]. As such, synthetic data can have numerous applications in health care, such as estimating the impact of policies, augmenting ML algorithms, and improving predictive public health models [29]. Although synthetic data hold promise, significant work needs to be done to make them a clear option to replace RWD [107]. The reason for this conundrum is the lack of a clear understanding as to whether such a dataset can be used for decision-making or whether the final analysis would require original data [108]. Locally, the use of semirepresentative synthetic datasets (Table 1) has been effective in supporting researchers with projects less reliant on accurate representations within the synthetic data to enable research to progress while awaiting the necessary approvals to access production data. Example projects include the support of qualitative focus group sessions to co-design clinical analytics tools or development of the infrastructure for future FL projects. Work continues to explore whether similar results and accurate conclusions can be drawn from representative synthetic data when compared to RWD, with some demonstrating promising results [109,110].
Synthetic data are not free from bias [111], privacy [112], and data quality assessment [41] issues. Bias, inherent in human society, especially affects marginalized groups and is reflected in data access and generation [113]. This poses a risk with ML algorithm adoption, potentially perpetuating or amplifying societal biases [111]. Regarding privacy, while synthetic data have been claimed to be a potential solution for mitigating privacy concerns, Stadler et al [112] highlight that synthetic datasets often contain residual information from their training data, making them vulnerable to ML-based attacks that can reveal features preserved by the generative model. However, it is challenging to predict the type of information retained in synthetic data or the specific features targeted by adversaries, thereby complicating the assessment of the privacy benefits provided by synthetic data generation [112]. In addition, Stadler et al [112] explain that differential privacy, a technique used in synthetic data generation to inject noise into the original statistical information for enhanced privacy [114], provides limited defense against ML-based inference attacks, particularly for high-dimensional datasets [112]. The evaluation of data quality is another such issue, which remains an open challenge [115]. The problem arises from the absence of a standardized quality metric, which impedes fair and definitive comparisons between methods, consequently affecting the selection of an appropriate approach [41]. As a consequence of these issues, there is a crucial need for tailored regulations on synthetic data use in medicine and health care to ensure quality and minimize potential risks [116].
Synthetic data frequently reside in a regulatory gray zone concerning their use [117], and existing data protection laws such as the General Data Protection Regulation and Health Insurance Portability and Accountability Act (HIPAA) have constraints in adequately addressing all potential risks linked to synthetic data [29]. For instance, HIPAA’s privacy rule considers the creation of deidentified data as a health care operation, thus exempting them from the need for patient consent, a principle similarly applied in the General Data Protection Regulation [117]. However, synthetic health data, while not deidentified, closely replicate real data, raising questions about whether they should be classified as protected health information and require informed consent and research ethics review [117]. Some studies have demonstrated the use of synthetic data in research, eliminating the need for an ethics review [118]. Whether this is a scalable future direction for synthetic data use in research remains to be seen.
CDM Strengths, Weaknesses, and Lessons Learned
The past 2 decades have seen the emergence of numerous CDMs to support collaborative health care research through data standardization. For example, the use of the OMOP CDM to conduct observational studies has grown extensively in recent years (from 14 publications in 2016 to 57 publications in 2020) [119], and its utility has been demonstrated in numerous, large-scale, multinational studies, such as estimating comparative drug safety and effectiveness [120-122]. The benefits are obvious for observational research in the digital era, when research questions can be addressed through combining databases with different underlying models, different information types, and different coding systems. What must not be overlooked is the potential for different biases to exist within different datasets and these nuances to be lost during translation to the CDM. Due to the complex transformations between sources and targets with varying schemas, databases, and technologies, the ETL implementations are considered prone to faults or issues [123].
According to Nwokeji and Matovu [124], these issues include complexity, cost, data heterogeneity, lack of automation, maintenance, standardization, and time. First, the growing complexity of data structures presents formidable obstacles to devising streamlined strategies [124]. In addition, the cost-intensive nature of ETL solution development imposes significant financial burdens [120]. Data heterogeneity, stemming from diverse sources and formats, further complicates the integration process [124]. Many existing ETL solutions continue to rely on manual procedures or necessitate human intervention, indicating an incomplete transition toward automation [124]. A lesson learned through the local mapping of the statewide EMR to the OMOP CDM within a nonproduction environment [104] (Table 1) highlighted the requirement for a joint clinical and technical venture. Establishing appropriate governance structures with input from clinical and technical staff is necessary to clearly articulate and endorse CDM implementation and ongoing maintenance decisions. Maintenance of ETL solutions is rendered demanding by the variety of data schemas and the dynamic nature of application requirements [124]. Furthermore, the absence of standardized methodologies for modeling ETL processes and executing workflows exacerbates these challenges [124]. Finally, the protracted process of designing, developing, implementing, and executing ETL solutions entails considerable time investments [124]. Despite these challenges, a multitude of commercial tools, including Microsoft SQL Server Integration Services, Oracle Warehouse Builder, IBM InfoSphere, and Informatica PowerCenter, alongside open-source alternatives such as Talend Open Studio and Pentaho Kettle, serve to facilitate and simplify these processes [79].
To address interoperability issues, the use of CDMs continues to expand within the health domain. Areas of future focus include the ongoing development of CDMs, their vocabularies, and tools to support their use. Further work is warranted to establish guidelines for CDM development [125] and achieving consensus on governance practices across institutions using RWD for secondary purposes [104].
FL Strengths, Weaknesses, and Lessons Learned
Of the goals discussed, FL is the most recent technique emerging in the field of RWD access. This technology allows for learnings to be obtained from health data across organizations and locations without attempting traditional integration [87,97]. The adoption of FL in the health care domain addresses the challenges of data privacy, confidentiality, and security while still enabling efficient model training [126]. Existing works on FL in the health sector reveal a diverse range of applications categorized into prognosis, diagnosis, and clinical workflow. Prognosis-related applications encompass endeavors such as stroke prediction and prevention, brain data meta-analysis, and brain tumor segmentation [88,127,128]. Diagnosis-related applications include COVID-19 diagnosis, morphometry for Alzheimer disease, and heart disease predictions from EHRs [88,129,130]. In addition to prognosis and diagnosis, FL holds significant potential in optimizing clinical workflows within the health care sector. These applications encompass various aspects, such as drug sensitivity prediction, integration of medical data, and clinical decision support systems [88,131,132]. These advancements highlight FL in streamlining clinical workflow efficiencies, enhancing patient care, and fostering innovation in health care delivery [88]. The application of FL demonstrates its potential to enhance health care outcomes while preserving data privacy and security, highlighting the significance of interdisciplinary research and innovative solutions in advancing FL across scientific domains.
Despite the numerous advantages of FL, this methodology presents several challenges that must be addressed for its effective implementation in scientific settings. The challenges facing FL can be categorized into several critical domains. First, privacy and security concerns arise from compromised servers or clients, potentially jeopardizing data integrity and confidentiality, with active and passive attacks posing threats to overall data security [87,88,91,133,134]. The distributed nature of FL gives rise to potential new privacy and security issues that must be avoided, including the leakage of sensitive patient information (privacy) and poisoning of data (security) [135]. Second, communication bottlenecks exacerbate these challenges, hindering seamless data exchange between clients and servers and raising issues regarding network state and protocol efficacy [87,88,91]. Third, addressing the heterogeneity in data distribution poses significant challenges, particularly in handling nonindependent and non–identically distributed data [88,91,136]. Fourth, the rising computing costs, especially considering the varied capabilities of devices, highlight the critical need to address challenges related to asymmetric computing and mitigate concerns regarding energy consumption in scenarios involving on-device training [85,88,91]. Moreover, the reliability of central servers responsible for managing local training and updates is also uncertain, increasing the likelihood of data leakage and security breaches [87,88,137]. Finally, the development of new FL computing frameworks, which include redundant servers, hardware accelerators, and decentralized training models, necessitates a comprehensive and thorough investigation [87,138]. These multifaceted challenges highlight the urgent need for interdisciplinary research and innovative solutions to facilitate the successful implementation and advancement of FL across scientific domains.
FL offers a novel approach to collaborative training across health care data repositories, bypassing the need for data sharing and safeguarding sensitive medical information [139]. In the use case provided in this paper (Table 1), the process involved a combination of approaches. Standardizing the data from health databases such as EMRs and health registries via a CDM was necessary, including provision to the FL client to then test the FL model using a synthetically generated dataset. This innovative method has the potential to address various health care issues by using distributed datasets across health care facilities. By doing so, it creates opportunities for pioneering research and business opportunities in the future of health care. Researchers will focus on integrating FL into upcoming medical devices such as intelligent implants and wearables. This will lead to the development of new eHealth services, improving patient well-being.
Personalization is key in preventive health care and chronic disease management through tailored interventions. It is expected that FL will drive precision medicine and elevate health care standards in the coming years. FL also stands to transform health care delivery, offering improved precision, accessibility, and patient-centered care [85,88,97,139].
Looking ahead, challenges such as ensuring data quality and incorporating expert knowledge into FL models need attention. Designing effective incentive mechanisms is crucial to encourage users of mobile and wearable devices to participate in the FL process. This participation involves these devices collecting high-quality data locally, training local models, and sharing model updates with a central server.
Cross-Sector Collaboration: Enablers to Promote RWD Access for Research
Digitization can accelerate RWD access through the novel technical methods emerging in recent decades. However, a holistic approach is necessary to support modern-day research in a system as multifaceted as that of digital health. The types of collaboration between the university and industry or health care sectors to drive digital transformation are varied [140]. Human factors are as important as the technologies themselves. Rybnicek and Königsgruber [141] identified 4 categories to drive the success of these cross-sector collaborations: institutional factors, relationship factors, output factors, and framework factors. The illustrative use case (Table 1) supports this approach. Embedding staff members across both types of organizations with access to both academic and health care networks and governed by the policies and procedures of the health care sector was key to supporting RWD access for research. Contractual agreements were critical to outline the key roles and responsibilities of conjoint staff, the governance frameworks by which they must abide, and clear reporting lines across both organizations. Colocation was deemed essential to build the relationship and trust. This takes both time and commitment from both sectors. As organizations continue to strive for advancements in HITs, it is the interpersonal relations that are fostering this growth. “As much as we talk about technology, at the end of the day collaboration is about people” [140].
Conclusions
The past 25 years have seen a maturation in digital health at large. HITs are opening new and efficient ways to deliver patient care. This evolution of patient care delivery and its ability to digitally capture data through routine care has underpinned the progression of medical research techniques. A shift in perspective is necessary, moving away from the emphasis on RCTs as the only source of practice-guiding clinical evidence to include the use of RWD. Novel methods are necessary to harness the vast volumes of RWD now generated through these digital platforms. Techniques such as synthetic data generation, CDMs, FL, and collaborations between the health care and university sectors all support this common goal. Appropriate policies and frameworks are essential to address the challenges of using RWD for research. We demonstrated how, by mapping health care data to a CDM and generating a synthetic dataset, these approaches facilitate the establishment of FL infrastructure, highlighting the interoperability of these methodologies across various research environments. To achieve a learning health system, a new and disruptive research infrastructure must be established, maintained, and enhanced to expedite the translation of research findings into clinical practice. This infrastructure, equipped with emerging digital health techniques and supported by strong cross-sector collaborations, advances research by enabling more effective RWD capture, providing researchers with “the right data for the right problem.”
Abbreviations
- CDISC
Clinical Data Interchange Standards Consortium
- CDM
common data model
- EHR
electronic health record
- EMR
electronic medical record
- ETL
extract, transform, and load
- FDA
Food and Drug Administration
- FL
federated learning
- GAN
generative adversarial network
- HIPAA
Health Insurance Portability and Accountability Act
- HIT
health IT
- ML
machine learning
- OMOP
Observational Medical Outcomes Partnership
- PADARSER
publicly available data approach to the realistic synthetic electronic health record
- RCT
randomized controlled trial
- RWD
real-world data
Footnotes
Conflicts of Interest: None declared.
References
- 1.Bondemark L, Ruf S. Randomized controlled trial: the gold standard or an unobtainable fallacy? Eur J Orthod. 2015 Oct 01;37(5):457–61. doi: 10.1093/ejo/cjv046.cjv046 [DOI] [PubMed] [Google Scholar]
- 2.Wieseler B, Neyt M, Kaiser T, Hulstaert F, Windeler J. Replacing RCTs with real world data for regulatory decision making: a self-fulfilling prophecy? BMJ. 2023 Mar 02;380:e073100. doi: 10.1136/bmj-2022-073100. https://doi.org/10.1136/bmj-2022-073100 . [DOI] [PubMed] [Google Scholar]
- 3.Chodankar D. Introduction to real-world evidence studies. Perspect Clin Res. 2021;12(3):171–4. doi: 10.4103/picr.picr_62_21. https://europepmc.org/abstract/MED/34386383 .PCR-12-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pihlstrom BL, Curran AE, Voelker HT, Kingman A. Randomized controlled trials: what are they and who needs them? Periodontol 2000. 2012 Jun;59(1):14–31. doi: 10.1111/j.1600-0757.2011.00439.x. [DOI] [PubMed] [Google Scholar]
- 5.Sherman RE, Anderson SA, Dal Pan GJ, Gray GW, Gross T, Hunter NL, LaVange L, Marinac-Dabic D, Marks PW, Robb MA, Shuren J, Temple R, Woodcock J, Yue LQ, Califf RM. Real-world evidence - what is it and what can it tell us? N Engl J Med. 2016 Dec 08;375(23):2293–7. doi: 10.1056/NEJMsb1609216. [DOI] [PubMed] [Google Scholar]
- 6.Morales DR, Arlett P. RCTs and real world evidence are complementary, not alternatives. BMJ. 2023 Apr 03;381:736. doi: 10.1136/bmj.p736. [DOI] [PubMed] [Google Scholar]
- 7.Wang SV, Schneeweiss S, RCT-DUPLICATE Initiative. Franklin JM, Desai RJ, Feldman W, Garry EM, Glynn RJ, Lin KJ, Paik J, Patorno E, Suissa S, D'Andrea E, Jawaid D, Lee H, Pawar A, Sreedhara SK, Tesfaye H, Bessette LG, Zabotka L, Lee SB, Gautam N, York C, Zakoul H, Concato J, Martin D, Paraoan D, Quinto K. Emulation of randomized clinical trials with nonrandomized database analyses: results of 32 clinical trials. JAMA. 2023 Apr 25;329(16):1376–85. doi: 10.1001/jama.2023.4221. https://europepmc.org/abstract/MED/37097356 .2804067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel GS, Bryan J, McClellan M, Romine M, Frank K, Silcox C. Characterizing RWD quality and relevancy for regulatory purposes. Duke-Margolis Center. 2018. Oct 01, [2024-09-20]. https://healthpolicy.duke.edu/sites/default/files/2020-03/characterizing_rwd.pdf .
- 9.Berger M, Daniel G, Frank K, Hernandez A, McClellan M, Okun S, Overhage M, Platt R, Romine M, Tunis S, Wilson M. A framework for regulatory use of real-world evidence. Duke-Margolis Center. 2017. Sep 13, [2024-09-20]. https://healthpolicy.duke.edu/sites/default/files/2020-08/rwe_white_paper_2017.09.06.pdf .
- 10.Liu F, Panagiotakos D. Real-world data: a brief review of the methods, applications, challenges and opportunities. BMC Med Res Methodol. 2022 Nov 05;22(1):287. doi: 10.1186/s12874-022-01768-6. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-022-01768-6 .10.1186/s12874-022-01768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Innovative health initiative. European Commission. [2024-09-25]. https://research-and-innovation.ec.europa.eu/research-area/health/innovative-health-initiative_en .
- 12.Innovative Health Initiative launches first five projects. Innovative Health Initiative. [2024-09-25]. https://www.ihi.europa.eu/news-events/newsroom/innovative-health-initiative-launches-first-five-projects .
- 13.Verma A, Bhattacharya P, Patel Y, Shah K, Tanwar S, Khan B. Data localization and privacy-preserving healthcare for big data applications: architecture and future directions. Proceedings of Emerging Technologies for Computing, Communication and Smart Cities; ETCCS 2021; August 21-22, 2021; Punjab, India. 2021. [DOI] [Google Scholar]
- 14.Näher AF, Vorisek CN, Klopfenstein SA, Lehne M, Thun S, Alsalamah S, Pujari S, Heider D, Ahrens W, Pigeot I, Marckmann G, Jenny MA, Renard BY, von Kleist M, Wieler LH, Balzer F, Grabenhenrich L. Secondary data for global health digitalisation. Lancet Digit Health. 2023 Feb;5(2):e93–101. doi: 10.1016/S2589-7500(22)00195-9. https://linkinghub.elsevier.com/retrieve/pii/S2589-7500(22)00195-9 .S2589-7500(22)00195-9 [DOI] [PubMed] [Google Scholar]
- 15.Togo K, Yonemoto N. Real world data and data science in medical research: present and future. Jpn J Stat Data Sci. 2022 Apr 13;5(2):769–81. doi: 10.1007/s42081-022-00156-0. https://europepmc.org/abstract/MED/35437515 .156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent S, Burn E, Dawoud D, Jonsson P, Østby JT, Hughes N, Rijnbeek P, Bouvy JC. Common problems, common data model solutions: evidence generation for health technology assessment. Pharmacoeconomics. 2021 Mar;39(3):275–85. doi: 10.1007/s40273-020-00981-9. https://europepmc.org/abstract/MED/33336320 .10.1007/s40273-020-00981-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savage N. Synthetic data could be better than real data. Nature. 2023 Apr 27; doi: 10.1038/d41586-023-01445-8.10.1038/d41586-023-01445-8 [DOI] [PubMed] [Google Scholar]
- 18.Nikolentzos G, Vazirgiannis M, Xypolopoulos C, Lingman M, Brandt EG. Synthetic electronic health records generated with variational graph autoencoders. NPJ Digit Med. 2023 Apr 29;6(1):83. doi: 10.1038/s41746-023-00822-x. https://doi.org/10.1038/s41746-023-00822-x .10.1038/s41746-023-00822-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bietz MJ, Bloss CS, Calvert S, Godino JG, Gregory J, Claffey MP, Sheehan J, Patrick K. Opportunities and challenges in the use of personal health data for health research. J Am Med Inform Assoc. 2016 Apr;23(e1):e42–8. doi: 10.1093/jamia/ocv118. https://europepmc.org/abstract/MED/26335984 .ocv118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudrapatna VA, Butte AJ. Opportunities and challenges in using real-world data for health care. J Clin Invest. 2020 Feb 03;130(2):565–74. doi: 10.1172/JCI129197. https://doi.org/10.1172/JCI129197 .129197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordon J, Szpruch L, Houssiau F, Bottarelli M, Cherubin G, Maple C, Cohen SN, Weller A. Synthetic data -- what, why and how? arXiv. Preprint posted online on May 6, 2022. 2024 doi: 10.48550/arXiv.2205.03257. [DOI] [Google Scholar]
- 22.Raghunathan TE. Synthetic data. Annu Rev Stat Appl. 2021 Mar 07;8(1):129–40. doi: 10.1146/annurev-statistics-040720-031848. [DOI] [Google Scholar]
- 23.Gonzales A, Guruswamy G, Smith SR. Synthetic data in health care: a narrative review. PLOS Digit Health. 2023 Jan;2(1):e0000082. doi: 10.1371/journal.pdig.0000082. https://europepmc.org/abstract/MED/36812604 .PDIG-D-22-00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domingo-Ferrer J, Montes F. Privacy in Statistical Databases: International Conference, PSD 2022, Paris, France, September 21–23, 2022, Proceedings. Cham, Switzerland: Springer; 2022. [Google Scholar]
- 25.Hernandez-Matamoros A, Fujita H, Perez-Meana H. A novel approach to create synthetic biomedical signals using BiRNN. Inf Sci. 2020 Dec;541:218–41. doi: 10.1016/j.ins.2020.06.019. [DOI] [Google Scholar]
- 26.Sano N. Synthetic data by principal component analysis. 2020 International Conference on Data Mining Workshops; ICDMW; November 17-20, 2020; Sorrento, Italy. 2020. [DOI] [Google Scholar]
- 27.ONS methodology working paper series number 16 - synthetic data pilot. Office for National Statistics. [2024-12-02]. https://www.ons.gov.uk/methodology/methodologicalpublications/generalmethodology/onsworkingpaperseries/onsmethodologyworkingpaperseriesnumber16syntheticdatapilot .
- 28.Nikolenko SI. Synthetic Data for Deep Learning. Cham, Switzerland: Springer; 2021. [Google Scholar]
- 29.Giuffrè Mauro, Shung DL. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. NPJ Digit Med. 2023 Oct 09;6(1):186. doi: 10.1038/s41746-023-00927-3. https://doi.org/10.1038/s41746-023-00927-3 .10.1038/s41746-023-00927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahmen J, Cook D. SynSys: a synthetic data generation system for healthcare applications. Sensors (Basel) 2019 Mar 08;19(5):1181. doi: 10.3390/s19051181. https://www.mdpi.com/resolver?pii=s19051181 .s19051181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002 Jun 01;16:321–57. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 32.Han H, Wang WY, Mao BH. Borderline-SMOTE: a new over-sampling method in imbalanced data sets learning. Proceedings of the International Conference on Intelligent Computing; ICIC 2005; August 23-26, 2005; Hefei, China. 2005. [DOI] [Google Scholar]
- 33.Batista GE, Prati RC, Monard MC. A study of the behavior of several methods for balancing machine learning training data. SIGKDD Explor Newsl. 2004 Jun;6(1):20–9. doi: 10.1145/1007730.1007735. [DOI] [Google Scholar]
- 34.He H, Bai Y, Garcia EA, Li S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. Proceedings of the IEEE International Joint Conference on Neural Networks; IJCNN 2008; June 1-8, 2008; Hong Kong, China. 2008. [DOI] [Google Scholar]
- 35.Torres FR, Carrasco-Ochoa JA, Martínez-Trinidad JF. SMOTE-D a deterministic version of SMOTE. Proceedings of the 8th Mexican Conference on Pattern Recognition; MCPR 2016; June 22-25, 2016; Guanajuato, Mexico. 2016. [DOI] [Google Scholar]
- 36.Mukherjee M, Khushi M. SMOTE-ENC: a novel SMOTE-based method to generate synthetic data for nominal and continuous features. Appl Syst Innov. 2021 Mar 02;4(1):18. doi: 10.3390/asi4010018. [DOI] [Google Scholar]
- 37.Figueira A, Vaz B. Survey on synthetic data generation, evaluation methods and GANs. Mathematics. 2022 Aug 02;10(15):2733. doi: 10.3390/math10152733. [DOI] [Google Scholar]
- 38.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial networks. Commun ACM. 2020 Oct 22;63(11):139–44. doi: 10.1145/3422622. [DOI] [Google Scholar]
- 39.Little RJ. Statistical analysis of masked data. J Off Stat. 1993;9(2):407–26. https://www.proquest.com/openview/970596f2406469cc1d5edae5d4d0d890 . [Google Scholar]
- 40.Ghosheh G, Li J, Zhu T. A review of Generative Adversarial Networks for Electronic Health Records: applications, evaluation measures and data sources. arXiv. Preprint posted online on March 14, 2022. 2024 https://arxiv.org/abs/2203.07018 . [Google Scholar]
- 41.Murtaza H, Ahmed M, Khan NF, Murtaza G, Zafar S, Bano A. Synthetic data generation: state of the art in health care domain. Comput Sci Rev. 2023 May;48:100546. doi: 10.1016/j.cosrev.2023.100546. [DOI] [Google Scholar]
- 42.Rashidian S, Wang F, Moffitt R, Garcia V, Dutt A, Chang W, Pandya V, Hajagos J, Saltz M, Saltz J. SMOOTH-GAN: towards sharp and smooth synthetic EHR data generation. Proceedings of the 18th International Conference on Artificial Intelligence in Medicine; AIME 2020; August 25-28, 2020; Minneapolis, MN. 2020. [DOI] [Google Scholar]
- 43.Imtiaz S, Arsalan M, Vlassov V, Sadre R. Synthetic and private smart health care data generation using GANs. Proceedings of the 2021 International Conference on Computer Communications and Networks; ICCCN 2021; July 19-22, 2021; Athens, Greece. 2021. [DOI] [Google Scholar]
- 44.Abedi M, Hempel L, Sadeghi S, Kirsten T. GAN-based approaches for generating structured data in the medical domain. Appl Sci. 2022 Jul 13;12(14):7075. doi: 10.3390/app12147075. [DOI] [Google Scholar]
- 45.Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. Proceedings of the IEEE 15th International Symposium on Biomedical Imaging; ISBI 2018; April 4-7, 2018; Washington, DC. 2018. [DOI] [Google Scholar]
- 46.Torfi A, Fox EA. CorGAN: correlation-capturing convolutional generative adversarial networks for generating synthetic healthcare records. arXiv. Preprint posted online on January 25, 2020. 2024 doi: 10.48550/arXiv.2001.09346. [DOI] [Google Scholar]
- 47.Goncalves A, Ray P, Soper B, Stevens J, Coyle L, Sales AP. Generation and evaluation of synthetic patient data. BMC Med Res Methodol. 2020 May 07;20(1):108. doi: 10.1186/s12874-020-00977-1. https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/s12874-020-00977-1 .10.1186/s12874-020-00977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dankar FK, Ibrahim M. Fake it till you make it: guidelines for effective synthetic data generation. Appl Sci. 2021 Feb 28;11(5):2158. doi: 10.3390/app11052158. [DOI] [Google Scholar]
- 49.Walonoski J, Kramer M, Nichols J, Quina A, Moesel C, Hall D, Duffett C, Dube K, Gallagher T, McLachlan S. Synthea: an approach, method, and software mechanism for generating synthetic patients and the synthetic electronic health care record. J Am Med Inform Assoc. 2018 Mar 01;25(3):230–8. doi: 10.1093/jamia/ocx079. https://europepmc.org/backend/ptpmcrender.fcgi?accid=PMC7651916&blobtype=pdf .4098271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Templ M, Meindl B, Kowarik A, Dupriez O. Simulation of synthetic complex data: the R package simPop. J Stat Softw. 2017;79(10):1–38. doi: 10.18637/jss.v079.i10. [DOI] [Google Scholar]
- 51.Ping H, Stoyanovich J, Howe B. DataSynthesizer: privacy-preserving synthetic datasets. Proceedings of the 29th International Conference on Scientific and Statistical Database Management; SSDBM '17; June 27-29, 2017; Chicago, IL. 2017. [DOI] [Google Scholar]
- 52.Attribution-Non-Commercial 4.0 International (CC BY-NC 4.0) Creative Commons. [2024-12-19]. https://creativecommons.org/licenses/by-nc/4.0/deed.en .
- 53.Attribution 4.0 International (CC BY4.0) Creative Commons. [2024-12-19]. https://creativecommons.org/licenses/by/4.0/deed.en .
- 54.Kalkman S, Mostert M, Udo-Beauvisage N, van Delden JJ, van Thiel GJ. Responsible data sharing in a big data-driven translational research platform: lessons learned. BMC Med Inform Decis Mak. 2019 Dec 30;19(1):283. doi: 10.1186/s12911-019-1001-y. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-019-1001-y .10.1186/s12911-019-1001-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kush RD, Warzel D, Kush MA, Sherman A, Navarro EA, Fitzmartin R, Pétavy F, Galvez J, Becnel LB, Zhou FL, Harmon N, Jauregui B, Jackson T, Hudson L. FAIR data sharing: the roles of common data elements and harmonization. J Biomed Inform. 2020 Jul;107:103421. doi: 10.1016/j.jbi.2020.103421. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(20)30049-6 .S1532-0464(20)30049-6 [DOI] [PubMed] [Google Scholar]
- 56.Aneja S, Avesta A, Xu H, Machado LO. Clinical informatics approaches to facilitate cancer data sharing. Yearb Med Inform. 2023 Aug;32(1):104–10. doi: 10.1055/s-0043-1768721. http://www.thieme-connect.com/DOI/DOI?10.1055/s-0043-1768721 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, Tsui R. How to improve the reuse of clinical data-- openEHR and OMOP CDM. J Phys Conf Ser. 2020 Oct 01;1624:032041. doi: 10.1088/1742-6596/1624/3/032041. [DOI] [Google Scholar]
- 58.Voss EA, Makadia R, Matcho A, Ma Q, Knoll C, Schuemie M, DeFalco FJ, Londhe A, Zhu V, Ryan PB. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J Am Med Inform Assoc. 2015 May;22(3):553–64. doi: 10.1093/jamia/ocu023. https://europepmc.org/abstract/MED/25670757 .ocu023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garza M, Del Fiol G, Tenenbaum J, Walden A, Zozus MN. Evaluating common data models for use with a longitudinal community registry. J Biomed Inform. 2016 Dec;64:333–41. doi: 10.1016/j.jbi.2016.10.016. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(16)30153-8 .S1532-0464(16)30153-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood FE Jr, Fitzsimmons MJ. Clinical data interchange standards consortium (CDISC) standards and their implementation in a clinical data management system. Drug Inf J. 2001 Dec 30;35:853–62. doi: 10.1177/009286150103500323. [DOI] [Google Scholar]
- 61.Kawai AT, Martin D, Henrickson SE, Goff A, Reidy M, Santiago D, Selvam N, Selvan M, McMahill-Walraven C, Lee GM. Validation of febrile seizures identified in the sentinel post-licensure rapid immunization safety monitoring program. Vaccine. 2019 Jul 09;37(30):4172–6. doi: 10.1016/j.vaccine.2019.05.042.S0264-410X(19)30666-8 [DOI] [PubMed] [Google Scholar]
- 62.Sentinel common data model. Sentinel Initiative. [2024-03-02]. https://www.sentinelinitiative.org/sites/default/files/Sentinel%20Common%20Data%20Model_01102024.PNG .
- 63.Ogunyemi OI, Meeker D, Kim HE, Ashish N, Farzaneh S, Boxwala A. Identifying appropriate reference data models for comparative effectiveness research (CER) studies based on data from clinical information systems. Med Care. 2013 Aug;51(8 Suppl 3):S45–52. doi: 10.1097/MLR.0b013e31829b1e0b. [DOI] [PubMed] [Google Scholar]
- 64.Ross TR, Ng D, Brown JS, Pardee R, Hornbrook MC, Hart G, Steiner JF. The HMO research network virtual data warehouse: a public data model to support collaboration. EGEMS (Wash DC) 2014;2(1):1049. doi: 10.13063/2327-9214.1049. https://europepmc.org/abstract/MED/25848584 .egems1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0) Creative Commons. [2024-12-19]. https://creativecommons.org/licenses/by-nc-nd/4.0/
- 66.Toh S, Rasmussen-Torvik LJ, Harmata EE, Pardee R, Saizan R, Malanga E, Sturtevant JL, Horgan CE, Anau J, Janning CD, Wellman RD, Coley RY, Cook AJ, Courcoulas AP, Coleman KJ, Williams NA, McTigue KM, Arterburn D, McClay J. The national patient-centered clinical research network (PCORnet) bariatric study cohort: rationale, methods, and baseline characteristics. JMIR Res Protoc. 2017 Dec 05;6(12):e222. doi: 10.2196/resprot.8323. https://www.researchprotocols.org/2017/12/e222/ v6i12e222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Y, Zong N, Wen A, Liu S, Stone DJ, Knaack D, Chamberlain AM, Pfaff E, Gabriel D, Chute CG, Shah N, Jiang G. Developing an ETL tool for converting the PCORnet CDM into the OMOP CDM to facilitate the COVID-19 data integration. J Biomed Inform. 2022 Mar;127:104002. doi: 10.1016/j.jbi.2022.104002. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(22)00018-1 .S1532-0464(22)00018-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hossain MS. Design and implementation of serverless architecture for i2b2 on AWS cloud and Snowflake data warehouse. University of Missouri. 2023. [2024-09-20]. https://mospace.umsystem.edu/xmlui/handle/10355/96163 .
- 69.Carus J, Nürnberg S, Ückert F, Schlüter C, Bartels S. Mapping cancer registry data to the episode domain of the observational medical outcomes partnership model (OMOP) Appl Sci. 2022 Apr 15;12(8):4010. doi: 10.3390/app12084010. [DOI] [Google Scholar]
- 70.Makadia R, Ryan PB. Transforming the premier perspective hospital database into the observational medical outcomes partnership (OMOP) common data model. EGEMS (Wash DC) 2014;2(1):1110. doi: 10.13063/2327-9214.1110. https://europepmc.org/abstract/MED/25848597 .egems1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lamer A, Abou-Arab O, Bourgeois A, Parrot A, Popoff B, Beuscart JB, Tavernier B, Moussa MD. Transforming anesthesia data into the observational medical outcomes partnership common data model: development and usability study. J Med Internet Res. 2021 Oct 29;23(10):e29259. doi: 10.2196/29259. https://www.jmir.org/2021/10/e29259/ v23i10e29259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang G, Kiefer RC, Sharma DK, Prud'hommeaux E, Solbrig HR. A consensus-based approach for harmonizing the OHDSI common data model with HL7 FHIR. Stud Health Technol Inform. 2017;245:887–91. https://europepmc.org/abstract/MED/29295227 . [PMC free article] [PubMed] [Google Scholar]
- 73.Abd Al-Rahman SQ, Hasan EH, Sagheer AM. Design and implementation of the web (extract, transform, load) process in data warehouse application. IAES Int J Artif Intell. 2023 Jun 01;12(2):765. doi: 10.11591/ijai.v12.i2.pp765-775. [DOI] [Google Scholar]
- 74.Attribution-ShareAlike 4.0 International (CC BY-SA 4.0) Creative Commons. [2024-12-19]. https://creativecommons.org/licenses/by-sa/4.0/
- 75.Beauvais B, Kruse CS, Fulton L, Shanmugam R, Ramamonjiarivelo Z, Brooks M. Association of electronic health record vendors with hospital financial and quality performance: retrospective data analysis. J Med Internet Res. 2021 Apr 14;23(4):e23961. doi: 10.2196/23961. https://www.jmir.org/2021/4/e23961/ v23i4e23961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kirrmann S, Gainey M, Röhner F, Hall M, Bruggmoser G, Schmucker M, Heinemann FE. Visualization of data in radiotherapy using web services for optimization of workflow. Radiat Oncol. 2015 Jan 20;10(1):22. doi: 10.1186/s13014-014-0322-3. https://ro-journal.biomedcentral.com/articles/10.1186/s13014-014-0322-3 .s13014-014-0322-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang HK, Taira RK. Infrastructure design of a picture archiving and communication system. AJR Am J Roentgenol. 1992 Apr;158(4):743–9. doi: 10.2214/ajr.158.4.1546584. [DOI] [PubMed] [Google Scholar]
- 78.Sinard J. Practical Pathology Informatics. New York, NY: Springer; 2006. Pathology LIS: relationship to institutional systems; pp. 173–206. [Google Scholar]
- 79.Bansal SK, Kagemann S. Integrating big data: a semantic extract-transform-load framework. Computer. 2015 Mar;48(3):42–50. doi: 10.1109/mc.2015.76. [DOI] [Google Scholar]
- 80.Yangui R, Nabli A, Gargouri F. ETL based framework for NoSQL warehousing. Proceedings of the 14th European, Mediterranean, and Middle Eastern Conference; EMCIS 2017; September 7-8, 2017; Coimbra, Portugal. 2017. [DOI] [Google Scholar]
- 81.Baghal A. Leveraging graph models to design acute kidney injury disease research data warehouse. Proceedings of the Sixth International Conference on Social Networks Analysis, Management and Security; SNAMS 2019; October 22-25, 2019; Granada, Spain. 2019. [DOI] [Google Scholar]
- 82.Burrows E, Razzaghi H, Utidjian L, Bailey L. Standardizing clinical diagnoses: evaluating alternate terminology selection. AMIA Jt Summits Transl Sci Proc. 2020 May;2020:71–9. https://europepmc.org/abstract/MED/32477625 . [PMC free article] [PubMed] [Google Scholar]
- 83.Bidgood WD Jr, Horii SC, Prior FW, Van Syckle DE. Understanding and using DICOM, the data interchange standard for biomedical imaging. J Am Med Inform Assoc. 1997 May 01;4(3):199–212. doi: 10.1136/jamia.1997.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dhavle AA, Rupp MT. Towards creating the perfect electronic prescription. J Am Med Inform Assoc. 2015 Apr;22(e1):e7–12. doi: 10.1136/amiajnl-2014-002738.amiajnl-2014-002738 [DOI] [PubMed] [Google Scholar]
- 85.Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, Ourselin S, Sheller M, Summers RM, Trask A, Xu D, Baust M, Cardoso MJ. The future of digital health with federated learning. NPJ Digit Med. 2020 Sep 14;3(1):119. doi: 10.1038/s41746-020-00323-1. https://doi.org/10.1038/s41746-020-00323-1 .10.1038/s41746-020-00323-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu J, Glicksberg BS, Su C, Walker P, Bian J, Wang F. Federated Learning for Healthcare Informatics. J Healthc Inform Res. 2021;5(1):1–19. doi: 10.1007/s41666-020-00082-4. https://europepmc.org/abstract/MED/33204939 .82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krishnan S, Anand AJ, Srinivasan R, Kavitha R, Suresh S. Handbook on Federated Learning: Advances, Applications and Opportunities. Boca Raton, FL: CRC Press; 2023. [Google Scholar]
- 88.Joshi M, Pal A, Sankarasubbu M. Federated learning for healthcare domain - pipeline, applications and challenges. ACM Trans Comput Healthcare. 2022 Nov 03;3(4):1–36. doi: 10.1145/3533708. [DOI] [Google Scholar]
- 89.Pfitzner B, Steckhan N, Arnrich B. Federated learning in a medical context: a systematic literature review. ACM Trans Internet Technol. 2021 Jun 02;21(2):1–31. doi: 10.1145/3412357. [DOI] [Google Scholar]
- 90.Olatunji IE, Rauch J, Katzensteiner M, Khosla M. A review of anonymization for healthcare data. Big Data. 2022 Mar 10; doi: 10.1089/big.2021.0169. [DOI] [PubMed] [Google Scholar]
- 91.Dhade P, Shirke P. Federated learning for healthcare: a comprehensive review. Eng Proc. 2023;59(1):230. doi: 10.3390/engproc2023059230. [DOI] [Google Scholar]
- 92.Antunes RS, André da Costa C, Küderle A, Yari IA, Eskofier B. Federated learning for healthcare: systematic review and architecture proposal. ACM Trans Intell Syst Technol. 2022 May 03;13(4):1–23. doi: 10.1145/3501813. [DOI] [Google Scholar]
- 93.Cremonesi F, Planat V, Kalokyri V, Kondylakis H, Sanavia T, Miguel Mateos Resinas V, Singh B, Uribe S. The need for multimodal health data modeling: a practical approach for a federated-learning healthcare platform. J Biomed Inform. 2023 May;141:104338. doi: 10.1016/j.jbi.2023.104338. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(23)00059-X .S1532-0464(23)00059-X [DOI] [PubMed] [Google Scholar]
- 94.Guendouzi BS, Ouchani S, EL Assaad H, EL Zaher M. A systematic review of federated learning: challenges, aggregation methods, and development tools. J Netw Comput Appl. 2023 Nov;220:103714. doi: 10.1016/j.jnca.2023.103714. [DOI] [Google Scholar]
- 95.Berghout T, Benbouzid M, Bentrcia T, Lim WH, Amirat Y. Federated learning for condition monitoring of industrial processes: a review on fault diagnosis methods, challenges, and prospects. Electronics. 2022 Dec 29;12(1):158. doi: 10.3390/electronics12010158. [DOI] [Google Scholar]
- 96.McMahan HB, Moore E, Ramage D, Hampson S, Arcas BA. Communication-efficient learning of deep networks from decentralized data. arXiv. Preprint posted online on February 17, 2016. 2024 doi: 10.1002/9781119845041.ch10. [DOI] [Google Scholar]
- 97.Yang Q, Fan L, Yu H. Federated Learning: Privacy and Incentive. Cham, Switzerland: Springer; 2020. [Google Scholar]
- 98.Wang Y. Performance enhancement schemes and effective incentives for federated learning [thesis] University of Ottawa. 2021. Nov 16, [2024-12-09]. https://ruor.uottawa.ca/items/fb7bb89a-0b36-4116-99c0-cd03e0c6517e .
- 99.2020 digital health and learning health system. National Academies. 2020. May, [2024-10-10]. https://www.nationalacademies.org/documents/link/LF5F46A0F4D8D3765F6DBFFA9DD3EA606B9CCD57CD98/file/DC3D040E01F6D31C3E3ECB934FB1EF25E407789B8DE2 .
- 100.Gehrmann J, Herczog E, Decker S, Beyan O. What prevents us from reusing medical real-world data in research. Sci Data. 2023 Jul 13;10(1):459. doi: 10.1038/s41597-023-02361-2. https://doi.org/10.1038/s41597-023-02361-2 .10.1038/s41597-023-02361-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu C, Shao S, Liu C, Bennett GG, Prvu Bettger J, Yan LL. Academia-industry digital health collaborations: a cross-cultural analysis of barriers and facilitators. Digit Health. 2019 Sep 26;5:2055207619878627. doi: 10.1177/2055207619878627. https://journals.sagepub.com/doi/abs/10.1177/2055207619878627?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub0pubmed .10.1177_2055207619878627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Awasthy R, Flint S, Sankarnarayana R, Jones RL. A framework to improve university–industry collaboration. J Industry Univ Collab. 2020 Feb 23;2(1):49–62. doi: 10.1108/jiuc-09-2019-0016. [DOI] [Google Scholar]
- 103.Hingle M, Patrick H, Sacher PM, Sweet CC. The intersection of behavioral science and digital health: the case for academic-industry partnerships. Health Educ Behav. 2019 Feb 24;46(1):5–9. doi: 10.1177/1090198118788600. [DOI] [PubMed] [Google Scholar]
- 104.Hallinan CM, Ward R, Hart GK, Sullivan C, Pratt N, Ng AP, Capurro D, Van Der Vegt A, Liaw ST, Daly O, Luxan BG, Bunker D, Boyle D. Seamless EMR data access: integrated governance, digital health and the OMOP-CDM. BMJ Health Care Inform. 2024 Feb 21;31(1):e100953. doi: 10.1136/bmjhci-2023-100953. https://informatics.bmj.com/lookup/pmidlookup?view=long&pmid=38387992 .bmjhci-2023-100953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mandl KD, Kohane IS, McFadden D, Weber GM, Natter M, Mandel J, Schneeweiss S, Weiler S, Klann JG, Bickel J, Adams WG, Ge Y, Zhou X, Perkins J, Marsolo K, Bernstam E, Showalter J, Quarshie A, Ofili E, Hripcsak G, Murphy SN. Scalable collaborative infrastructure for a learning healthcare system (SCILHS): architecture. J Am Med Inform Assoc. 2014;21(4):615–20. doi: 10.1136/amiajnl-2014-002727. https://europepmc.org/abstract/MED/24821734 .amiajnl-2014-002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gal MS, Lynskey O. Synthetic data: legal implications of the data-generation revolution. Iowa Law Rev. 2024;109(3) doi: 10.2139/ssrn.4414385. [DOI] [Google Scholar]
- 107.Alloza C, Knox B, Raad H, Aguilà M, Coakley C, Mohrova Z, Boin É, Bénard M, Davies J, Jacquot E, Lecomte C, Fabre A, Batech M. A case for synthetic data in regulatory decision-making in Europe. Clin Pharmacol Ther. 2023 Oct;114(4):795–801. doi: 10.1002/cpt.3001. [DOI] [PubMed] [Google Scholar]
- 108.Kokosi T, Harron K. Synthetic data in medical research. BMJ Med. 2022;1(1):e000167. doi: 10.1136/bmjmed-2022-000167. https://europepmc.org/abstract/MED/36936569 .bmjmed-2022-000167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Azizi Z, Zheng C, Mosquera L, Pilote L, El Emam K, GOING-FWD Collaborators Can synthetic data be a proxy for real clinical trial data? A validation study. BMJ Open. 2021 Apr 16;11(4):e043497. doi: 10.1136/bmjopen-2020-043497. https://bmjopen.bmj.com/lookup/pmidlookup?view=long&pmid=33863713 .bmjopen-2020-043497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reiner Benaim A, Almog R, Gorelik Y, Hochberg I, Nassar L, Mashiach T, Khamaisi M, Lurie Y, Azzam ZS, Khoury J, Kurnik D, Beyar R. Analyzing medical research results based on synthetic data and their relation to real data results: systematic comparison from five observational studies. JMIR Med Inform. 2020 Feb 20;8(2):e16492. doi: 10.2196/16492. https://medinform.jmir.org/2020/2/e16492/ v8i2e16492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baumann J, Castelnovo A, Crupi R, Inverardi N, Regoli D. Bias on demand: a modelling framework that generates synthetic data with bias. Proceedings of the 2023 ACM Conference on Fairness, Accountability, and Transparency; FAccT '23; June 12-15, 2023; Chicago, IL. 2023. [DOI] [Google Scholar]
- 112.Stadler T, Oprisanu B, Troncoso C. Synthetic data -- anonymisation groundhog day. arXiv. Preprint posted online on November 13, 2020. 2024 doi: 10.1109/msp.2004.9. [DOI] [Google Scholar]
- 113.Draghi B, Wang Z, Myles P, Tucker A. Identifying and handling data bias within primary healthcare data using synthetic data generators. Heliyon. 2024 Jan 30;10(2):e24164. doi: 10.1016/j.heliyon.2024.e24164. https://linkinghub.elsevier.com/retrieve/pii/S2405-8440(24)00195-6 .S2405-8440(24)00195-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosenblatt L, Liu X, Pouyanfar S, de Leon E, Desai A, Allen J. Differentially private synthetic data: applied evaluations and enhancements. arXiv. Preprint posted online on June 12, 2017. 2024 https://arxiv.org/abs/2011.05537 . [Google Scholar]
- 115.Platzer M, Reutterer T. Holdout-based empirical assessment of mixed-type synthetic data. Front Big Data. 2021;4:679939. doi: 10.3389/fdata.2021.679939. https://europepmc.org/abstract/MED/34268491 .679939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lu Y, Shen M, Wang H, Wang X, van Rechem C, Fu T, Wei W. Machine learning for synthetic data generation: a review. arXiv. Preprint posted online on February 8, 2023. 2024 doi: 10.48550/arXiv.2302.04062. [DOI] [Google Scholar]
- 117.Tsao SF, Sharma K, Noor H, Forster A, Chen H. Health synthetic data to enable health learning system and innovation: a scoping review. Stud Health Technol Inform. 2023 May 18;302:53–7. doi: 10.3233/SHTI230063.SHTI230063 [DOI] [PubMed] [Google Scholar]
- 118.Guo A, Foraker RE, MacGregor RM, Masood FM, Cupps BP, Pasque MK. The use of synthetic electronic health record data and deep learning to improve timing of high-risk heart failure surgical intervention by predicting proximity to catastrophic decompensation. Front Digit Health. 2020 Dec 7;2:576945. doi: 10.3389/fdgth.2020.576945. https://europepmc.org/abstract/MED/34713050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reinecke I, Zoch M, Reich C, Sedlmayr M, Bathelt F. The usage of OHDSI OMOP - a scoping review. Stud Health Technol Inform. 2021 Sep 21;283:95–103. doi: 10.3233/SHTI210546.SHTI210546 [DOI] [PubMed] [Google Scholar]
- 120.Burn E, Weaver J, Morales D, Prats-Uribe A, Delmestri A, Strauss VY, He Y, Robinson DE, Pinedo-Villanueva R, Kolovos S, Duarte-Salles T, Sproviero W, Yu D, Van Speybroeck M, Williams R, John LH, Hughes N, Sena AG, Costello R, Birlie B, Culliford D, O'Leary C, Morgan H, Burkard T, Prieto-Alhambra D, Ryan P. Opioid use, postoperative complications, and implant survival after unicompartmental versus total knee replacement: a population-based network study. Lancet Rheumatol. 2019 Dec;1(4):e229–36. doi: 10.1016/S2665-9913(19)30075-X.S2665-9913(19)30075-X [DOI] [PubMed] [Google Scholar]
- 121.Lane JC, Weaver J, Kostka K, Duarte-Salles T, Abrahao MT, Alghoul H, Alser O, Alshammari TM, Biedermann P, Banda JM, Burn E, Casajust P, Conover MM, Culhane AC, Davydov A, DuVall SL, Dymshyts D, Fernandez-Bertolin S, Fišter K, Hardin J, Hester L, Hripcsak G, Kaas-Hansen BS, Kent S, Khosla S, Kolovos S, Lambert CG, van der Lei J, Lynch KE, Makadia R, Margulis AV, Matheny ME, Mehta P, Morales DR, Morgan-Stewart H, Mosseveld M, Newby D, Nyberg F, Ostropolets A, Park RW, Prats-Uribe A, Rao GA, Reich C, Reps J, Rijnbeek P, Sathappan SM, Schuemie M, Seager S, Sena AG, Shoaibi A, Spotnitz M, Suchard MA, Torre CO, Vizcaya D, Wen H, de Wilde M, Xie J, You SC, Zhang L, Zhuk O, Ryan P, Prieto-Alhambra D. Risk of hydroxychloroquine alone and in combination with azithromycin in the treatment of rheumatoid arthritis: a multinational, retrospective study. The Lancet Rheumatology. 2020 Nov;2(11):e698–711. doi: 10.1016/s2665-9913(20)30276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suchard MA, Schuemie MJ, Krumholz HM, You SC, Chen R, Pratt N, Reich CG, Duke J, Madigan D, Hripcsak G, Ryan PB. Comprehensive comparative effectiveness and safety of first-line antihypertensive drug classes: a systematic, multinational, large-scale analysis. Lancet. 2019 Nov 16;394(10211):1816–26. doi: 10.1016/S0140-6736(19)32317-7. https://europepmc.org/abstract/MED/31668726 .S0140-6736(19)32317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Homayouni H, Ghosh S, Ray I. An approach for testing the extract-transform-load process in data warehouse systems. Proceedings of the 22nd International Database Engineering & Applications Symposium; IDEAS '18; June 18-20, 2018; Villa San Giovanni, Italy. 2018. [DOI] [Google Scholar]
- 124.Nwokeji JC, Matovu R. A systematic literature review on big data extraction, transformation and loading (ETL) In: Arai K, editor. Intelligent Computing. Cham, Switzerland: Springer; 2021. [Google Scholar]
- 125.Ahmadi N, Zoch M, Kelbert P, Noll R, Schaaf J, Wolfien M, Sedlmayr M. Methods used in the development of common data models for health data: scoping review. JMIR Med Inform. 2023 Aug 03;11:e45116. doi: 10.2196/45116. https://medinform.jmir.org/2023//e45116/ v11i1e45116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mbunge E, Muchemwa B, Jiyane S, Batani J. Sensors and healthcare 5.0: transformative shift in virtual care through emerging digital health technologies. Global Health J. 2021 Dec;5(4):169–77. doi: 10.1016/j.glohj.2021.11.008. [DOI] [Google Scholar]
- 127.Ju C, Zhao R, Sun J, Wei X, Zhao B, Liu Y, Li H, Chen T, Zhang X, Gao D, Tan B, Yu H, He C, Jin Y. Privacy-preserving technology to help millions of people: federated prediction model for stroke prevention. arXiv. Preprint posted online on June 15, 2020. 2024 doi: 10.48550/arXiv.2006.10517. [DOI] [Google Scholar]
- 128.Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp. Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Raviv TR, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Szekely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K. The multimodal brain tumor image segmentation benchmark (BRATS) IEEE Trans Med Imaging. 2015 Oct;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. https://europepmc.org/abstract/MED/25494501 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brisimi TS, Chen R, Mela T, Olshevsky A, Paschalidis IC, Shi W. Federated learning of predictive models from federated electronic health records. Int J Med Inform. 2018 Apr;112:59–67. doi: 10.1016/j.ijmedinf.2018.01.007. https://europepmc.org/abstract/MED/29500022 .S1386-5056(18)30008-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu X, Liang Z, Wang J. FedMed: a federated learning framework for language modeling. Sensors (Basel) 2020 Jul 21;20(14):4048. doi: 10.3390/s20144048. https://www.mdpi.com/resolver?pii=s20144048 .s20144048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Honkela A, Das M, Nieminen A, Dikmen O, Kaski S. Efficient differentially private learning improves drug sensitivity prediction. Biol Direct. 2018 Feb 06;13(1):1. doi: 10.1186/s13062-017-0203-4. https://biologydirect.biomedcentral.com/articles/10.1186/s13062-017-0203-4 .10.1186/s13062-017-0203-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Valdes G, Simone CB 2nd, Chen J, Lin A, Yom SS, Pattison AJ, Carpenter CM, Solberg TD. Clinical decision support of radiotherapy treatment planning: a data-driven machine learning strategy for patient-specific dosimetric decision making. Radiother Oncol. 2017 Dec;125(3):392–7. doi: 10.1016/j.radonc.2017.10.014.S0167-8140(17)32654-3 [DOI] [PubMed] [Google Scholar]
- 133.Lyu L, Yu H, Ma X, Chen C, Sun L, Zhao J, Yang Q, Yu PS. Privacy and robustness in federated learning: attacks and defenses. IEEE Trans Neural Netw Learn Syst. 2024 Jul;35(7):8726–46. doi: 10.1109/TNNLS.2022.3216981. [DOI] [PubMed] [Google Scholar]
- 134.Yang Q, Huang A, Fan L, Chan CS, Lim JH, Ng KW, Ong DS, Li B. Federated learning with privacy-preserving and model IP-right-protection. Mach Intell Res. 2023 Jan 10;20(1):19–37. doi: 10.1007/s11633-022-1343-2. [DOI] [Google Scholar]
- 135.Kairouz P, McMahan HB, Avent B, Bellet A, Bennis M, Nitin Bhagoji A, Bonawitz K, Charles Z, Cormode G, Cummings R, D’Oliveira RGL, Eichner H, El Rouayheb S, Evans D, Gardner J, Garrett Z, Gascón A, Ghazi B, Gibbons PB, Gruteser M, Harchaoui Z, He C, He L, Huo Z, Hutchinson B, Hsu J, Jaggi M, Javidi T, Joshi G, Khodak M, Konecný J, Korolova A, Koushanfar F, Koyejo S, Lepoint T, Liu Y, Mittal P, Mohri M, Nock R, Özgür A, Pagh R, Qi H, Ramage D, Raskar R, Raykova M, Song D, Song W, Stich SU, Sun Z, Suresh AT, Tramèr F, Vepakomma P, Wang J, Xiong L, Xu Z, Yang Q, Yu FX, Yu H, Zhao S. Advances and open problems in federated learning. FNT Mach Learn. 2021;14(1–2):1–210. doi: 10.1561/2200000083. [DOI] [Google Scholar]
- 136.Abdelmoniem AM, Ho CY, Papageorgiou P, Canini M. A comprehensive empirical study of heterogeneity in federated learning. IEEE Internet Things J. 2023 Aug 15;10(16):14071–83. doi: 10.1109/JIOT.2023.3250275. https://linkinghub.elsevier.com/retrieve/pii/S2666-3899(24)00001-1 .S2666-3899(24)00001-1 [DOI] [Google Scholar]
- 137.Truong N, Sun K, Wang S, Guitton F, Guo Y. Privacy preservation in federated learning: an insightful survey from the GDPR perspective. Comput Secur. 2021 Nov;110:102402. doi: 10.1016/j.cose.2021.102402. [DOI] [Google Scholar]
- 138.Abreha HG, Hayajneh M, Serhani MA. Federated learning in edge computing: a systematic survey. Sensors (Basel) 2022 Jan 07;22(2):450. doi: 10.3390/s22020450. https://www.mdpi.com/resolver?pii=s22020450 .s22020450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kumar Y, Singla R. Federated learning systems for healthcare: perspective and recent progress. In: Rehman MH, Gaber MM, editors. Federated Learning Systems. Cham, Switzerland: Springer; 2021. [Google Scholar]
- 140.Evans N, Miklosik A, Du JT. University-industry collaboration as a driver of digital transformation: types, benefits and enablers. Heliyon. 2023 Oct;9(10):e21017. doi: 10.1016/j.heliyon.2023.e21017. https://doi.org/10.1016/j.heliyon.2023.e21017 .S2405-8440(23)08225-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rybnicek R, Königsgruber R. What makes industry–university collaboration succeed? A systematic review of the literature. J Bus Econ. 2018 Sep 12;89(2):221–50. doi: 10.1007/s11573-018-0916-6. [DOI] [Google Scholar]


