Abstract
Persistent human papillomavirus (HPV) infection is necessary but insufficient for viral oncogenesis. Additional contributing co-factors, such as immune evasion and viral integration have been implicated in HPV-induced cancer progression. It is widely accepted that HPV + keratinocytes require co-culture with fibroblasts to maintain viral DNA as episomes. How fibroblasts regulate viral episome maintenance is a critical knowledge gap. Here we present comprehensive RNA sequencing and proteomic analysis demonstrating that coculture with fibroblasts is supportive of the viral life cycle, and is confirmatory of previous observations. Novel observations suggest that errors in “cross-talk” between fibroblasts and infected keratinocytes may regulate HPV integration and drive oncogenic progression. Our co-culture models offer new insights into HPV-related transformation mechanisms.
Keywords: Stroma, Human papillomavirus, Oropharyngeal cancer, Microenvironment, Fibroblasts, Transcriptional reprogramming, EMT
Highlights
-
•
Fibroblast coculture supports HPV RNA expression and episomal maintenance in HPV + keratinocytes.
-
•
Fibroblasts reduce EMT related gene expression in HPV + keratinocytes.
-
•
Fibroblasts promote EMT related gene expression in E6E7+ keratinocytes.
1. Introduction
Human papillomaviruses (HPVs) infect the basal keratinocytes of differentiating squamous epithelia [1]. Current estimates suggest there may be more than 400 types of HPV, and there are approximately 12 high-risk HPV types with the capacity to cause cancer in the general population [[2], [3], [4]]. HPV-related cancers (HPV + cancers) continue to contribute to approximately 5 % of the worldwide cancer burden [[5], [6], [7], [8], [9], [10], [11], [12], [13], [14]]. HPV16 is responsible for the majority of HPV + cancers, contributing to 54 % of cervical cancers and ∼90 % of HPV + oropharyngeal squamous cell carcinoma (HPV + OPC) [3,5,[8], [9], [10],[15], [16], [17]]. While these HPV + cancers remain prevalent, the majority of total infections are asymptomatic, self-limiting, and clear before cancer progression [3,[18], [19], [20], [21], [22], [23]].
The stroma is a complex connective tissue comprised of numerous cell types; the main components of the dermal stroma are fibroblasts [15,[24], [25], [26]]. Fibroblasts support tissue homeostasis via the secretion of all components of the extracellular matrix (ECM) and facilitate stromal extracellular signaling; factors produced by fibroblasts are key for angiogenesis, inflammation, wound healing, and are necessary for the proper differentiation of keratinocytes [23,26,27]. Keratinocyte differentiation is critical for the HPV lifecycle [28,29]. Furthermore, the importance of stromal support in the microenvironment is now an emerging field in the context of overall cancer progression, as well as HPV-induced transformation and carcinogenesis [15,[23], [24], [25],[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]]. Precise mechanisms for viral transformation and progression mechanisms remain unclear; however, persistent viral oncogene expression contributes to clear epithelial growth advantages [[43], [44], [45], [46],[46], [46], [47], [48], [49], [50], [51]]. While HPV exclusively infects basal keratinocytes, viral gene products alter the secretion of host factors, indirectly affecting neighboring keratinocytes, fibroblasts, and immune cells in the local microenvironment [22,23,31]. Given the complexity of the tissue infected and the transformation process, the relationship between HPV and epithelial-stromal communication remains at a nascent phase and further investigations are warranted [15,23].
During the HPV lifecycle, the viral genome exists in an episomal form in basal keratinocytes. It is generally accepted that HPV + keratinocyte cell lines must be grown in co-culture with fibroblasts to support viral episome maintenance [[52], [53], [54]]. HPV + keratinocytes maintained in the absence of fibroblasts are noted to quickly integrate or lose viral genome expression [54,55]. From these observations, fibroblasts are influential on the HPV episomal status of adjacent keratinocytes, suggesting their role in controlling this transformative factor. Conversely, HPV genome integration events have been noted as contributing factors in transformation; viral integration correlates with increased viral oncogene expression, loss of functional E2, cellular growth advantages, enhanced tumor progressiveness, cervical cancer progression, and poor clinical prognostics of HPV + OPC [43,[46], [47], [48],52,[56], [57], [58], [59], [60], [61], [62], [63], [64]].
The oncogenes HPV E6 and E7 are considered the major viral oncoproteins that contribute to carcinogenesis following integration; E6 targets and degrades p53, while E7 targets and degrades retinoblastoma protein (pRb) [18,44,[65], [66], [67]]. Oncogene expression alone is considered insufficient for carcinogenesis, and other indeterminate events have been implicated in transformation [68]. While the expression of E6 and E7 extends the proliferative capacity of epithelial cells, fibroblasts have demonstrated a cooperative role in the induction of cell immortalization [15,[69], [70], [71], [72]]. The minor oncoprotein, E5, has demonstrated immune regulatory interactions with the adjacent stroma, and may contribute to viral persistence [22,23]. Of note, the viral DNA binding protein, E2, is not proposed to be oncogenic but has been reported to be involved in the suppression of the innate immune response and is crucial for viral episome persistence [[73], [74], [75], [76], [77], [78]].
We previously reported the value of fibroblast co-culture both in the context of HPV episomal maintenance and as a model for better predicting in vitro to in vivo translational treatment paradigms [55]. In our previous analysis, we demonstrated that mitomycin C (MMC) growth-arrested murine 3T3-J2 fibroblasts (referred to as J2s moving forward) supported HPV16 long control region (LCR) transcriptional regulation [55]. We also found that co-culture supported oncoprotein RNA and viral protein stability in HPV16-genome immortalized keratinocytes. This suggested that the ability of fibroblasts to regulate viral protein expression was at least partially reliant on the expression of the viral LCR [55]. Alterations in the protein levels of p53, pRb, and γH2AX were also demonstrated to be altered in the presence of J2 and further suggested that fibroblasts may alter host protein expression that is supportive of HPV viral genome regulation [55].
Building on these previous data, in this report we utilized RNA sequencing (RNA-seq) and proteomic analysis for a global and comprehensive approach to investigate keratinocyte signaling impacted by fibroblasts. Our investigation confirmed the prior observation, that HPV downregulates portions of innate immune signaling [23,73,74,[79], [80], [81]]. Our analysis of keratinocytes grown in the presence or absence of J2s revealed the novel observation that fibroblasts transcriptionally reprogram keratinocytes. N/Tert-1+HPV16 cells grown with J2s showed a gene regulation pattern similar to that of a suprabasal layer; this epithelium layer is highly supportive of viral replication and amplification [82,83]. While gene ontology (GO) analysis indicated that fibroblasts supported the viral life cycle, removal of fibroblast support promoted viral integration and epithelial to mesenchymal transition (EMT) of the host in N/Tert-1+HPV16 cells. Conversely, N/Tert-1+E6E7 cells grown with J2s showed a greater tendency toward transcriptional reprogramming suggestive of EMT than those grown without J2s, including alterations in cell cycle regulation and oncogenic cytokine expression patterns. Proteomic analysis further supported these observations. Here, we propose that communication with adjacent fibroblasts is critical for viral-host interactions and the viral lifecycle at a transcriptional level. We propose that “cross-talk” errors between fibroblasts and infected keratinocytes promote integration, and are a significant contributing event in the progression of HPV infections to malignancy.
2. Materials and methods
2.1. Cell culture
N/Tert-1 cells and all derived cell lines were cultured in keratinocyte-serum free medium (K-SFM; Invitrogen). Briefly, stable N/Tert-1 lines expressing the complete HPV16 genome were created through a lipid-mediated transfection protocol, utilizing the Cre/LoxP system as previously described and supplemented with 150 μg/ml G418 (Thermo Fisher Scientific) [73,84]. N/Tert-1+E6E7 cell lines were generated via retroviral transduction of HPV16 E6E7 (pLXSNE6E7 [Addgene plasmid 52394; Denise Galloway]) and supplemented with 150 μg/ml G418 (Thermo Fisher Scientific). All derived cell lines have been described previously, STR fingerprinted, and routinely monitored for mycoplasma contamination [43,55,73,74,[85], [86], [87], [88]].
2.2. Culture and mitomycin C (MMC) inactivation of 3T3-J2 mouse embryonic fibroblast feeder cells, and co-culture with keratinocytes
As previously described, 3T3-J2 immortalized mouse embryonic fibroblasts (J2) were grown in DMEM and supplemented with 10 % FBS [55]. 80–90 % confluent plates were supplemented with 4 μg/ml of MMC in DMSO (Cell Signaling Technology) for 4–6 h at 37 °C. MMC-supplemented medium was removed and cells were washed with 1xPBS. Cells were trypsinized, centrifuged at 800 rcf for 5 min, washed once with 1xPBS, centrifuged again, and resuspended at 2 million cells per mL. Quality control of inactivation (lack of proliferation) was monitored for each new batch of mitomycin-C. Unless otherwise stated, 100-mm plate conditions were continually supplemented with 1 × 106 J2 every 2–3 days. Before trypsinization or harvesting, plates were washed to remove residual J2. We have previously confirmed that observations related to residual J2s are null [55]. Please see Supplemental Fig. 5 for workflow related to our analysis.
2.3. RNA isolation
The SV total RNA isolation system kit (Promega) was utilized to isolate RNA from cells, as per the manufacturer's protocol.
2.4. Human sequences RNA-seq bioinformatics pipeline
Library preparation, sequencing, and pre-processing of samples was performed by Novogene. Novogene uses in-house scripts to clean raw reads, filtering out low-quality reads, and reads containing adapter sequences. The genome index was built and cleaned sequences were aligned to the reference human genome using Hisat2 v2.05 [89,90]. Raw gene expression levels were quantified with featureCounts v1.5.0-p3 and then normalized to fragments per kilobase per million (FPKM) [91]. Differential expression analysis was performed using DESeq2 R package v1.20.0 between three experimental groups N/Tert-1, N/Tert-1+E6/E7, and N/Tert-1+HPV16 treated with J2 fibroblasts (n = 3 in each group) and their paired controls respectively (untreated). P-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR), where significance for a differentially expressed gene was determined at FDR <0.05 [92].
2.5. Gene ontology enrichment analysis
GO enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected [93,94]. GO terms with corrected P-value <0.05 were considered significantly enriched by differential expressed genes. Heatmaps were generated with the ‹pheatmap‹ R package using z-score normalized FPKM gene expression averages for each sample condition.
2.6. HPV16 sequences RNA-seq bioinformatics pipeline
Fastq files from Novogene were examined for quality using FastQC and quality control reports were collated by multiQC [95,96]. Reads were filtered to remove low quality sequences and adapter sequences were trimmed using trimmomatic v 0.39 [97]. A genome index was built and all sequences were aligned to the GRCh38.d1.vd1 Reference Sequence, part of the Genomic Data Commons GDC data harmonization pipeline, using STAR aligner v 2.7.9.a [98]. Samtools v1.16.1 was used to index and filter the bam file for reads aligned to HPV16 [99]. The HPV16 filtered bam files were converted back to fastq files using bedtools [100]. The HPV16 fastq sequences were re-aligned to an HPV16 reference genome from NCBI and raw gene expression levels were counted using featureCounts. Raw counts were then normalized using EdgeR's calcNormFactors scaling factor of trimmed mean of M-values (TMM) normalization. EdgeR's quasi-likelihood F-test (QLF) method was then used for differential expression analysis of each gene between three experimental groups N/Tert-1, N/Tert-1+E6/E7, and N/Tert-1+HPV16 treated with J2 fibroblasts (n = 3 in each group) and their paired controls respectively (untreated) [[101], [102], [103]]. The p-value of each QLF test was adjusted using a Benjamini-Hochberg False Discovery Rate (FDR) multiple testing correction using the basic R stats package p.adjust function. Genes passing the FDR cut-off threshold of ≤0.05 for significance were considered statistically significantly different.
2.7. Real-time PCR (qPCR)
A high-capacity cDNA reverse-transcription kit from Invitrogen was used to synthesize cDNA from RNA and processed for qPCR. qPCR was performed on 10 ng of the cDNA isolated. cDNA and relevant primers were mixed with PowerUp SYBR green master mix (Applied Biosystems), and real-time PCR was performed using the 7500 Fast real-time PCR system, using SYBR green reagent. Expression was quantified as relative quantity over GAPDH using the 2−ΔΔCT method. Primer used are as follows. HPV16 E2 F, 5′-ATGGAGACTCTTTGCCAACG-3′; HPV16 E2 R, 5′-TCATATAGACATAAATCCAG-3′; HPV16 E6 F, 5′-TTGAACCGAAACCGGTTAGT-3′; HPV16 E6 R, 5′-GCATAAATCCCGAAAAGCAA-3′; MX1 F, 5′-GGTGGTCCCCAGTAATGTGG-3′; MX1 R, 5′-CGTCAAGATTCCGATGGTCCT-3′; STAT1 F, 5′-CAGCTTGACTCAAAATTCCTGGA-3′; STAT1 R, 5′-TGAAGATTACGCTTGCTTTTCCT-3′; STAT2 F, 5′-CCAGCTTTACTCGCACAGC-3′; STAT2 R, 5′-AGCCTTGGAATCATCACTCCC-3′; STAT3 F, 5′-CAGCAGCTTGACACACGGTA-3′; STAT3 R, 5′-AAACACCAAAGTGGCATGTGA-3′; p53 F, 5′-GAGGTTGGCTCTGACTGTACC-3′; p53 R, 5′-TCCGTCCCAGTAGATTACCAC-3′; Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) F, 5′-GGAGCGAGATCCCTCCAAAAT-3′; GAPDH R, 5′-GGCTGTTGTCATACTTCTCATGG-3′.
2.8. Exo V
PCR based analysis of viral genome status was performed using methods described by Myers et al. [104]. 20 ng of genomic DNA was either treated with exonuclease V (RecBCD, NEB), in a total volume of 30 μl, or left untreated for 1 h at 37 °C followed by heat inactivation at 95 °C for 10 min 2 ng of digested/undigested DNA was then quantified by real time PCR, as noted above, using and 100 nM of primer in a 20 μl reaction. Nuclease free water was used in place of the template for a negative control. The following cycling conditions were used: 50 °C for 2 min, 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and a dissociation stage of 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s, and 60 °C for 15 s. Separate PCR reactions were performed to amplify HPV16 E6 F: 5′- TTGCTTTTCGGGATTTATGC-3′ R: 5′- CAGGACACAGTGGCTTTTGA-3′, HPV16 E2 F: 5′- TGGAAGTGCAGTTTGATGGA-3′ R: 5′- CCGCATGAACTTCCCATACT-3′, human mitochondrial DNA F: 5′-CAGGAGTAGGAGAGAGGGAGGTAAG-3′ R: 5′- TACCCATCATAATCGGAGGCTTTGG -3′, and human GAPDH DNA F: 5′- GGAGCGAGATCCCTCCAAAAT-3′ R: 5′- GGCTGTTGTCATACTTCTCATGG-3′
2.9. Proteomic sample preparation
The samples were digested using commercially available PreOmics iST sample clean up protocol. To the sample containing approximately 100ug of protein, 70ul of lysis buffer was added and mixed, followed by an incubation for 10 min at 950C; 1000 rpm. 50ul of DIGEST solution was added to the mixture, which was then incubated at 370C for 3hrs at 500 rpm. After the digestion, 100ul of STOP solution was added and mixed properly. The digest was then centrifuged at 3800rcf; 3min to ensure complete flow through and washed with 200ul of WASH 1 and 200ul of WASH 2 solution followed by centrifugation after each wash. The cartridge was then placed to the fresh collection tube and 100ul of ELUTE solution was added and centrifuged at 3800rcf; 3min to ensure complete flow through. This step was repeated one more time to ensure maximum recovery. The elutes were then placed in a vacuum evaporator at 450C until completely dried.
2.10. LC-MS/MS
LC-MS/MS analysis were performed using a Q-Exactive HF-X (Thermo) tandem mass spectrometer coupled to an Easy nLC 1200 (Thermo) nanoflow UPLC system. The LC-MS/MS system was fitted with an Easy spray ion source and an Acclaim PepMap 75 μm × 2 cm nanoviper C18 3 μm × 100 Å pre-column in series with an Acclaim PepMap RSLC 75 μm × 50 cm C18 2 μm bead size (Thermo). The mobile phase consists of Buffer A (0.1 % formic acid in water) and Buffer B (80 % acetonitrile in water,0.1 % formic acid). 500 ng of peptides were injected onto the above column assembly and eluted with an acetonitrile/0.1 % formic acid gradient at a flow rate of 300 nL/min over 2 h. The nano-spray ion source was operated at 1.9 kV. The digests were analyzed using a data dependent acquisition (DDA) method acquiring a full scan mass spectrum (MS) followed by 40 tandem mass spectra (MS/MS) in the high energy C- trap Dissociation HCD spectra). This mode of analysis produces approximately 50,000 MS/MS spectra of ions ranging in abundance over several orders of magnitude. Not all MS/MS spectra are derived from peptides.
2.11. Proteomic data analysis
The data were analyzed in Proteome Discoverer (ver 3.0) using the Sequest HT search algorithm and the Human database. Proteins were identified at an FDR <0.01 and quantification used the peptide intensities. Raw protein abundances were normalized in Proteome Discoverer using the “Total Peptide Abundance” method. Differential Enrichment of protein abundance was performed using the ‹DEP‹ package v. 1.26 [105]. First, we filtered for proteins detected in two of three replicates of at least one of the experimental conditions. Variance stabilizing transformation of remaining protein intensity observations was performed using the ‹vsn‹ package v 3.72 via the ‹normalize_vsn‹ function [106]. The quantile regression-based left-censored (QRILC) method was used as the missing value imputation approach. The differential enrichment test was conducted pairwise on each protein using limma v 3.60.4 between three experimental groups N/Tert-1, N/Tert-1+E6/E7, and N/Tert-1+HPV16 treated with J2 fibroblasts (n = 3 in each group) and their paired controls (untreated), respectively [107]. Proteins were identified as significantly differentially expressed between the control and experimental groups with a Benjamini-Hochberg adjusted p-value of <0.05, and a |log2-fold change| > 0.58.
2.12. Immunoblotting
Cells were trypsinized, washed with PBS and resuspended in 2x pellet volume NP40 protein lysis buffer (0.5 % Nonidet P-40, 50 mM Tris [pH 7.8], 150 mM NaCl) supplemented with protease inhibitor (Roche Molecular Biochemicals) and phosphatase inhibitor cocktail (MilliporeSigma). Cell suspension was incubated on ice for 20 min and then centrifuged for 20 min at 184,000 rcf at 4 °C. Protein concentration was determined using the Bio-Rad protein estimation assay according to manufacturer's instructions. 50 μg protein was mixed with 2x Laemmli sample buffer (Bio-Rad) and heated at 95 °C for 5 min. Protein samples were separated on Novex 4–12 % Tris-glycine gel (Invitrogen) and transferred onto a nitrocellulose membrane (Bio-Rad) at 30V overnight using the wet-blot transfer method. Membranes were then blocked with Odyssey (PBS) blocking buffer (diluted 1:1 with PBS) at room temperature for 1 h and probed with indicated primary antibody diluted in Odyssey blocking buffer, overnight. Membranes were washed with PBS supplemented with 0.1 % Tween (PBS-Tween) and probed with the Odyssey secondary antibody (goat anti-mouse IRdye 800CW or goat anti-rabbit IRdye 680CW) (Licor) diluted in Odyssey blocking buffer at 1:10,000. Membranes were washed twice with PBS-Tween and an additional wash with 1X PBS. After the washes, the membrane was imaged using the Odyssey® CLx Imaging System and ImageJ was used for quantification, utilizing GAPDH as internal loading control. Primary antibodies used for western blotting studies are as follows: pRb 1:1000 (Santa Cruz, sc-102), p53 1:1000 (Cell Signaling Technology, CST-2527, and CST-1C12), γH2AX 1:500 (Cell Signaling Technology, CST-80312 and CST-20E3).
2.13. Reproducibility, research integrity, and statistical analysis
All experiments were carried out at least in triplicate in all of the cell lines indicated. Keratinocytes were typed via cell line authentication services. All images shown are representatives from triplicate experiments. Student's t-test or analysis of variance was used to determine significance as appropriate: ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3. Results
3.1. Initial validation of -omics data
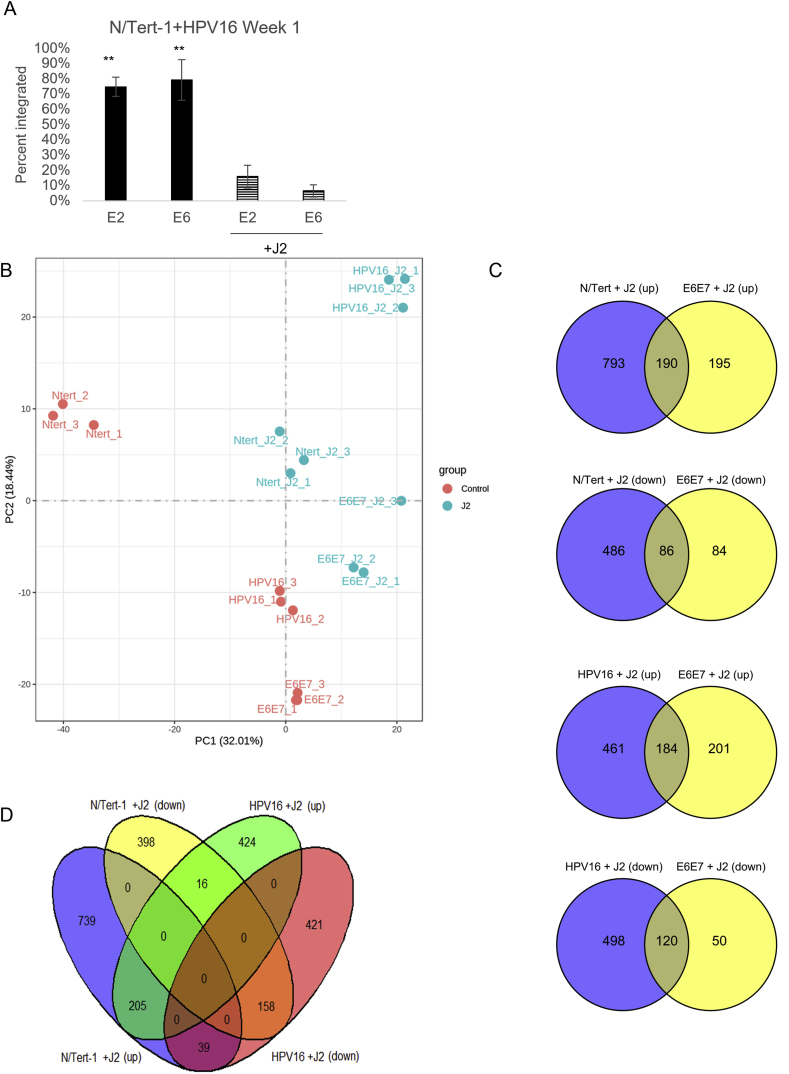
We previously demonstrated that fibroblast co-culture was important for maintaining HPV episomes, influenced HPV16 LCR transcriptional regulation, and supported the expression of HPV16 E7 protein in human foreskin keratinocytes immortalized with HPV16 (HFK + HPV16) [55]. We also observed that fibroblasts altered host protein levels which could affect viral genome regulation, including genes involved in DNA damage repair and innate immune signaling [55]. Our previous analysis also confirmed observations were not due to residual J2s [55]. Taking a more global approach to investigate signaling impacted by fibroblasts, N/Tert-1, N/Tert-1+E6/E7, and N/Tert-1+HPV16 cells were cultured in the presence or absence of J2s for one week. As previously observed, the removal of fibroblasts induces significant viral integration (Fig. 1A) [55]. These matched samples were then subjected to bulk RNA-seq analysis, and label-free liquid chromatography-mass spectrometry-based proteomic analysis (LC-MS/MS). For RNA-seq, triplicate sample data were combined to assess differential gene expression analysis. Evaluations of datasets were compared based on the presence or absence of J2 in N/Tert-1, N/Tert-1+E6E7, or N/Tert-1+HPV16 cell line and cross-compared. Our data revealed numerous genes significantly differentially expressed 1.5 fold or greater when cross-comparing our samples (Principal component analysis (PCA) presented in Fig. 1B, and Venns for the cross-comparison of significant observations are in Fig. 1C and D). A full list of these genes can be found in Supplementary Material S1. The expression level of the HPV16 genes used to generate the gene expression data is given in Supplementary Table S2. Novogene and further bioinformatic analysis identified the most affected canonical pathways, upstream regulators, diseases, and functions predicted to be altered in this data set; significant observations are given in Supplementary Table S3.
Fig. 1.
Global comparison of RNA-seq. 1A. N/Tert-1+HPV16 cells were grown in the presence or absence of J2s for 1 week. Cells were washed to removed J2, then lysed and analyzed for DNA expression of E2 and E6 via the exonuclease V assay, in comparison to GAPDH and mitochondrial DNA controls. Results are presented as percent integration as calculated from the cut ratio of matched GAPDH. ∗∗P < 0.01. 1B. Principal component analysis (PCA) analysis on the gene expression value (FPKM) of all samples. 1C. Venn diagrams of the significantly (up) and (down) regulated RNA expression profile observed in co-culture. 1D. Shared Venn expression profile to cross compare significantly (up) and (down) regulated RNA expression profile observed in co-culture between N/Tert and N/Tert + HPV16.
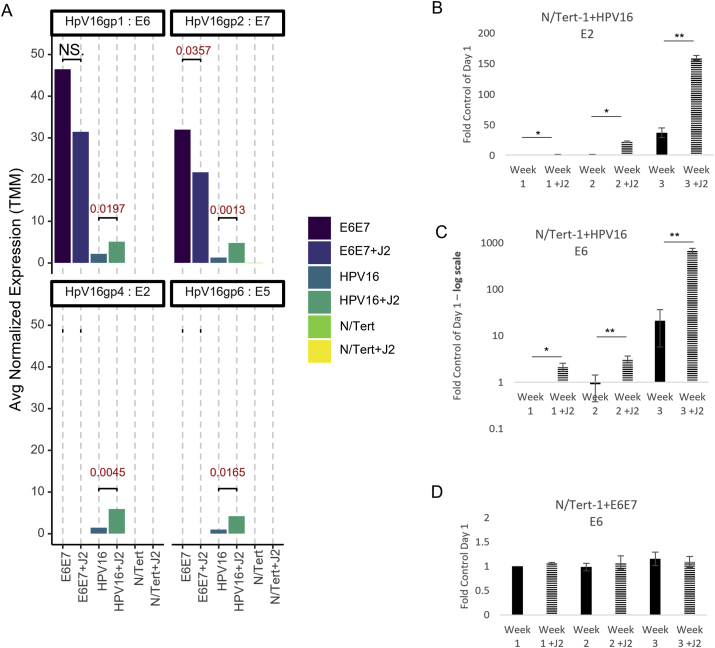
Mining of viral reads from RNA-seq data was performed and interpreted utilizing a technique previously developed [17,108,109]. RNA differential expression analysis demonstrated that N/Tert-1+HPV16 grown in the presence of J2 had significantly higher levels of E2, E5, E6, and E7 transcripts than cells grown in the absence of J2 (RNA-seq reads in Fig. 2A, E2, and E6 qPCR time course validation in Fig. 2B and C, respectively). The reduction of viral RNA expression is consistent with viral integration (shown in Fig. 1A) [55,110]. Alternatively, N/Tert-1+E6E7 grown in the presence of J2 expressed lower RNA transcripts of E7, and similar E6 transcripts in comparison to cells grown in the absence of J2 (RNA-seq reads in Fig. 2A and E6 qPCR time course validation in Fig. 2D). Thus, it appears that stromal support of viral RNA stability is reliant on the episome; this stability may be partially through stromal support of the LCR [55].
Fig. 2.
Fibroblasts support viral RNA expression and episomal maintenance in HPV + keratinocytes. 2A. N/Tert-1+HPV16 cells were grown in the presence or absence of J2s for 1 week. Differential expression data from RNAseq from average normalized reads of E6, E7,E2, and E5 matched to HPV reference genome. Exact significance is presented for each (student's t-test), NS represents no significance. 2B-D. qPCR time course validation of E2 and E6 RNA expression in N/Tert-1+E6E7 and N/Tert-1+HPV16 in the presence or absence of J2 for 3 weeks, 2C is presented in log scale. ∗P < 0.05. ∗∗P < 0.01.
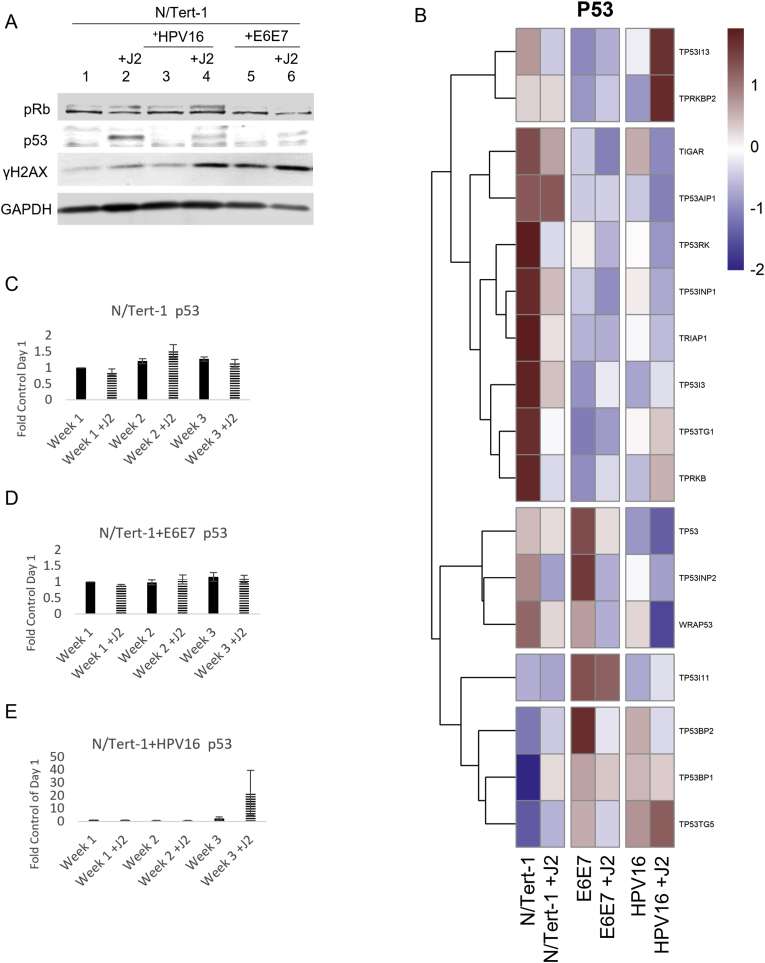
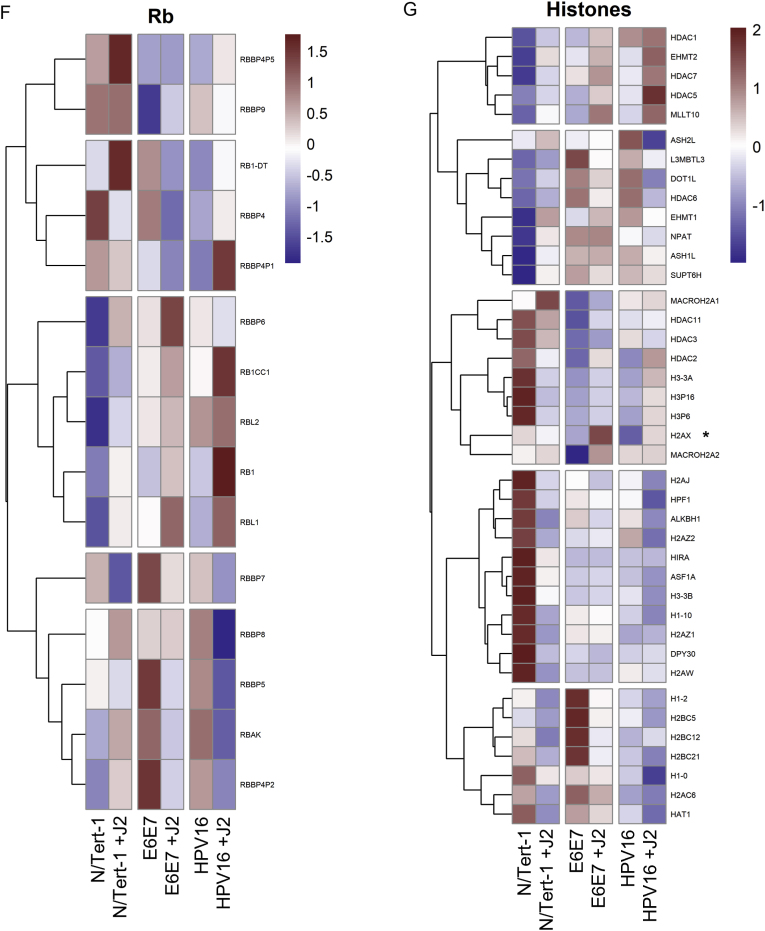
Internal host validation of our RNA-seq analysis was performed on known HPV targets p53, pRb, and γH2AX; this was further confirmed via qRTPCR and Western blot (Fig. 3). Fibroblasts enhanced p53 and γH2AX protein expression in all N/Tert-1 lines, while pRb was enhanced in N/Tert-1 and N/Tert-1+E6E7 (Fig. 3A). Conversely, GO enrichment revealed that TP53 was not enhanced by fibroblasts at the RNA expression level, indicating that the enhancement of p53 protein expression is likely mediated at the level of translation, post-translation, or protein stability (Fig. 3B). Of note, some p53 inducible proteins did appear to be regulated by fibroblasts at the level of RNA (GO enrichment Fig. 3B, p53 qRTPCR time course validation 3C-E) [55]. TP53I13, TP53TG1, and TP53TG5 overexpression have been linked to tumor suppression and the inhibition of cell proliferation [[111], [112], [113]]. Enhancement of these tumor suppressors in N/Tert-1+HPV16 grown in the presence of J2 suggests that fibroblasts promote a genotype of enhanced cell cycle regulation (cell cycle see: section 3.2.3). GO enrichment related to Rb signaling is less clear. The observed RB1, RBL1, RB1CC1, and RBBP4P1 RNA upregulation (Fig. 3F) in N/Tert-1+HPV16 grown in the presence of J2, is suggestive of a genotype that has more cell cycle regulation than in the absence of fibroblasts [[114], [115], [116], [117]]. H2AX RNA upregulation was demonstrated in both N/Tert-1+E6E7 and N/Tert-1+HPV16 grown in the presence of fibroblasts (Fig. 3G, H2AX denoted by ∗), indicating a partial role in the previously observed J2 enhancement of γH2AX protein (the phosphorylated form of the H2AX variant) [55]; additional histone modifications will be elaborated on in section 3.2.4.
Fig. 3.
Fibroblasts differentially regulate p53, pRb, and histone related expression. 3A. N/Tert-1 (lanes 1,2) N/Tert-1+E6E7 (lanes 3,4), N/Tert-1+HPV16 (lanes 5,6) cells were seeded on day 0 and grown in the presence or absence of J2s for 1 week. Cells were washed to remove J2s in noted conditions, trypsinized, lysed, and analyzed via western blotting for pRb, p53, and γH2AX. GAPDH was utilized as a loading control. 3B. Heat map demonstrating significant p53 GO enrichment all groups. 3C. N/Tert-1, 3D. N/Tert-1+E6E7, and 3E. N/Tert-1+HPV16 were grown in the presence or absence of J2s for 3 weeks. Time course of p53 RNA is presented at fold control of day 1. 3F. Heat map demonstrating significant pRb RNA enrichment all groups. 3G. Heat map demonstrating the highested set of significant histone-related RNA enrichment in all groups.
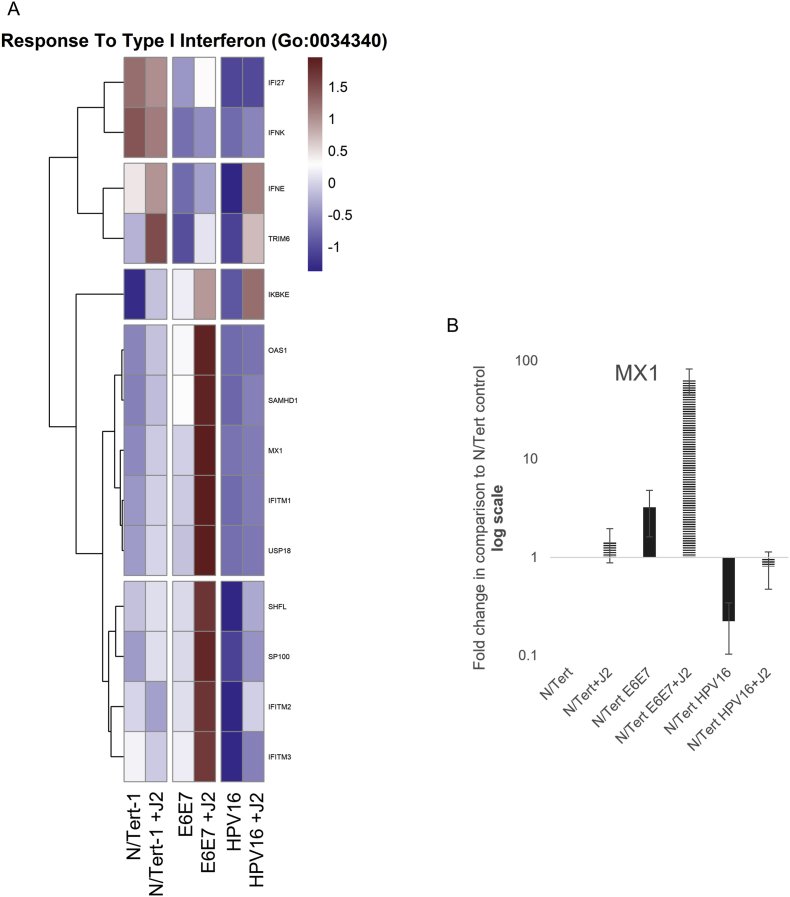
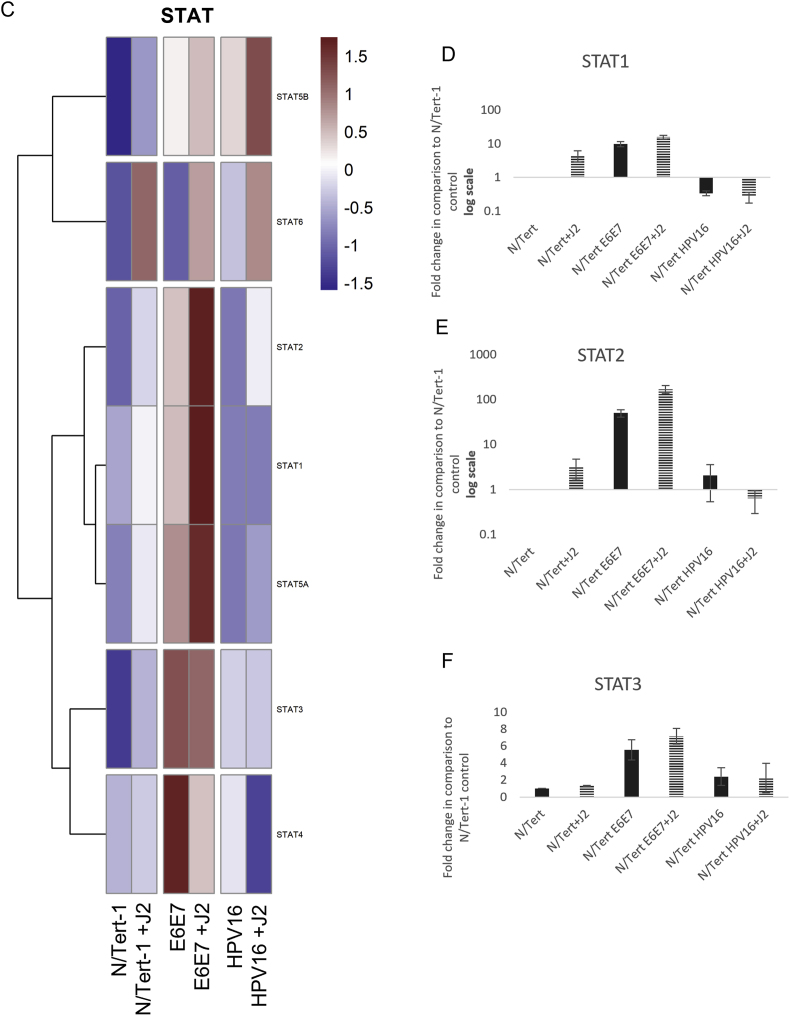
The observation that HPV downregulates innate immune function was also utilized to internally validate our data [23,73,74,[79], [80], [81],118]. In comparison to N/Tert, N/Tert-1+HPV16 keratinocytes have significantly lower RNA expression of interferon (IFN) related response genes, regardless of stromal support (Fig. 4A). GO enrichment of Type I IFN responses is shown in Fig. 4A (numerous immune regulatory events were observed and genes of interest can be found in Supplemental Table 3); HPV suppression of MX1 was validated via qRTPCR (Fig. 4B) [119]. HPV manipulates the JAK/STAT signaling pathway to evade the immune system and encourage cell proliferation; HPV oncoproteins have previously been shown to activate JAK/STAT-related signaling, while suppression of STAT1 is necessary for differentiation-dependent genome amplification and plasmid maintenance [[119], [120], [121]]. E2 and E5 have also been tied to the downregulation of innate immunity [23,73,74]. GO enrichment analysis, and consequent qPCR validation demonstrated that N/Tert-1+E6E7 cells markedly upregulate STAT1,2,3 expression (Fig. 4C–F). N/Tert-1+E6E7 grown in the presence of J2 exhibited the most significant increase in GO enrichment of genes related to IFN (Fig. 4C). Our confirmation of these previous observations with our RNA data established high confidence in proceeding with more extensive analyses to determine novel stromal regulatory pathways.
Fig. 4.
Fibroblasts differentially regulate GO enrichment in relation to innate immune function. 4A. Heat map demonstrating significant GO:0034340 response to type I interferon across all groups. 4B. qPCR validation of MX1 RNA expression, presented in log scale. 4C. Heatmap demonstrating significant STAT RNA expression across all groups. 4D. qPCR validation of STAT1 RNA expression, presented in log scale. 4E. qPCR validation of STAT2 RNA expression, presented in log scale. 4F. qPCR validation of STAT3 RNA expression.
3.2. Differential genomic landscapes altered by fibroblasts in keratinocytes
The utility of a supportive fibroblast feeder layer is broadly accepted as essential for maintaining an episomal HPV genome in primary keratinocyte models; this is a necessary component of 3D models for HPV lifecycle analysis where it is chiefly responsible for proper keratinocyte differentiation [44,54,55,57,69,[122], [123], [124], [125], [126], [127], [128], [129]]. While the coculture of keratinocytes with fibroblast feeders is accepted, the full mechanism of how fibroblasts aid in HPV episomal maintenance has yet to be deciphered. Here we present novel signaling observations that may help elucidate some of the mechanisms behind this unexplained phenomenon.
3.2.1. Cytokine-related regulation
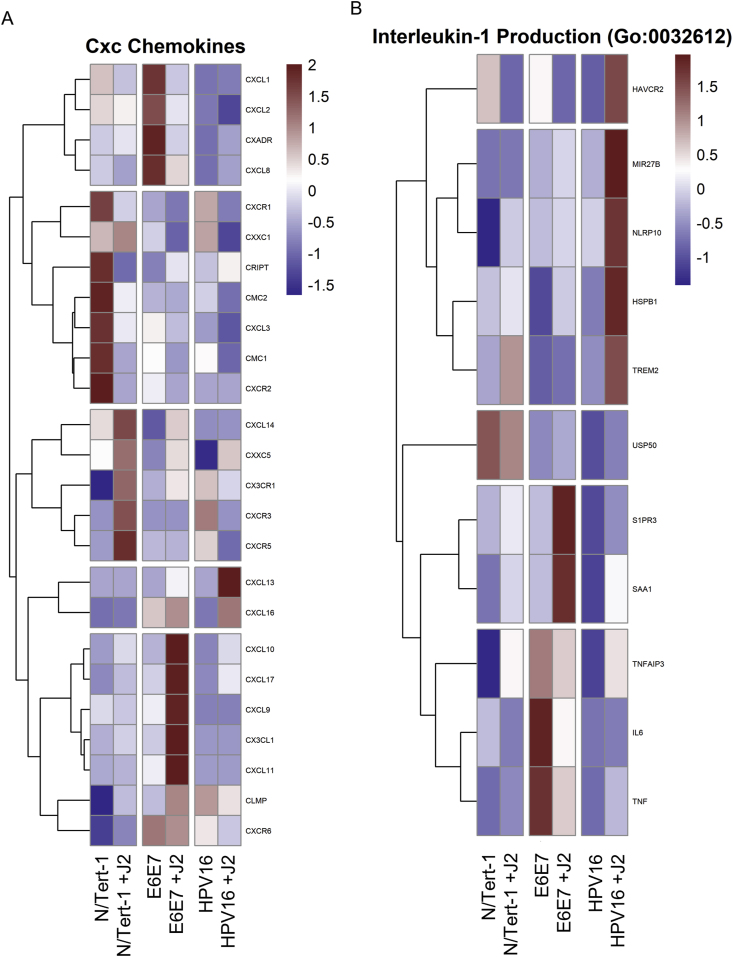
Expanding upon our initial innate immune signaling confirmatory analysis, significant alterations were observed in relation to cytokine and interleukin activity (Fig. 5). Here, we observed significant alterations in CXCL family members via J2 in both N/Tert-1+E6E7 and N/Tert-1+HPV16 when compared to N/Tert-1 (Fig. 5A). These signaling pathways can also have both tumor-promoting and suppressive roles that are cancer-dependent; fibroblasts altered the cytokine transcriptional profile of N/Tert-1+HPV16 in a way that could impede HPV16-driven carcinogenesis. Conversely, N/Tert-1+E6E7 grown in the presence of J2s demonstrated enhanced RNA expression of tumor-promoting CXCL family members (Fig. 5A). N/Tert-1+HPV16 continuously maintained in co-culture with fibroblasts also demonstrated significant upregulation of interleukin antagonist genes related to the repression of EMT progression (Fig. 5B). Reactome enrichment further highlighted the following genes in relation to innate immune and CXCL-related pathways: BST2, CREB5, CSF1, CX3CL1, CXCL1, CXCL2, CXCL3, IFI27, IFI35, IFI6, IFIT1, IFITM1, IFITM3, IL18R1, IL6, IRF7, ISG15, HLA-B, LIF, MMP9, MX1, MX2, OAS1, OAS2, OAS3, PIK3R3, PTAFR, RIPK3, RSAD2, SAMHD1, STAT1, TRIM22, UBE2L6, USP18, XAF1 (Supplemental Table S3).
Fig. 5.
Fibroblasts differentially regulate GO enrichment in relation to Cxc Chemokine and Interleukin-1 Production. 5A. Heat map demonstrating significant CXC chemokines across all groups. 5B. Heatmap demonstrating significant GO:0032612 Interleukin-1 Production across all groups.
3.2.2. Epithelial to mesenchymal transition
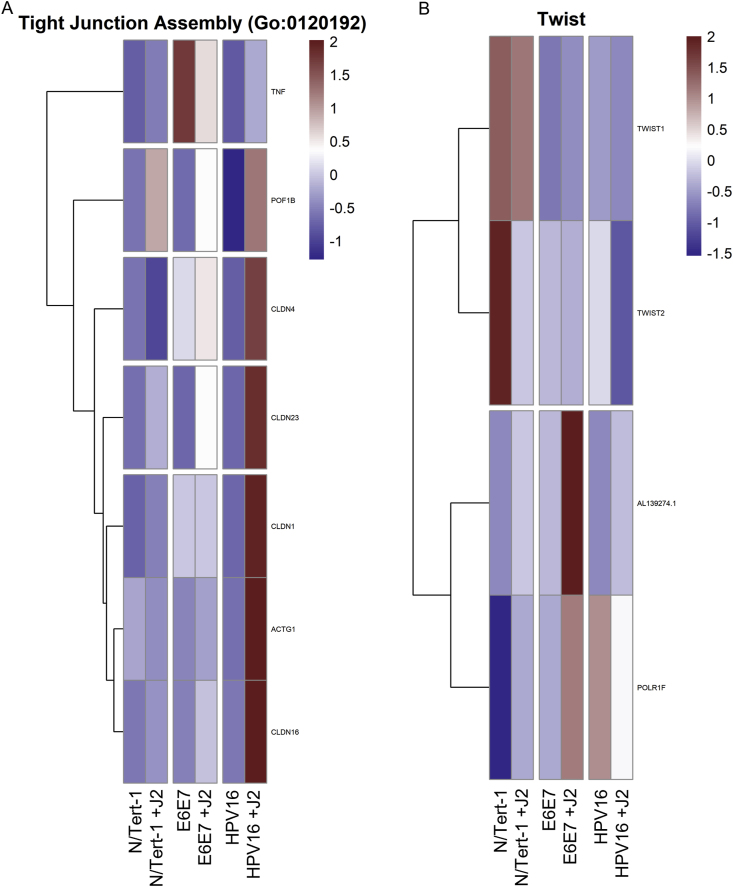
Cytokine and interleukin-related signaling impact EMT and cancer progression via interactions with β-catenin, TNF, Notch/Wnt, and TWIST-related signaling cascades [[130], [131], [132], [133], [134], [135], [136], [137]]. These pathways also regulate cell-cell junction related genes [138]. Another noteworthy observation from the GO enrichment cross-comparison, was the alteration observed in genes related to cell junctions, particularly tight junctions (TJs) (Fig. 6A). In particular, Claudins 1, 4, 16, and 23 are greatly increased in N/Tert-1+HPV16 in the presence of fibroblasts, but unaffected in the other cell lines, regardless of coculture (Fig. 6A). The claudin family of proteins plays a key role in tight junctions (TJs), dysregulation of which is associated with epithelial-to-mesenchymal transition (EMT) and invasive phenotypes, including in HPV + cervical cancer [[139], [140], [141], [142]]. The observed increase in TJ proteins in the presence of fibroblasts in N/Tert-1+HPV16 is a marker of epithelial identity [137,141,143]. GO enrichment of TWIST expression (Fig. 6B) also demonstrated that N/Tert-1+HPV16 grown in the presence of J2 do not demonstrate a progressive EMT-associated genotype [[144], [145], [146]].
Fig. 6.
Fibroblasts differentially regulate Tight Junction Assembly and Twist-related GO enrichment. 6A. Heat map demonstrating significant GO:0120192 Tight Junction Assembly across all groups. 6B. Heat map demonstrating significant Twist-related expression across all groups.
3.2.3. Cell cycle and tissue development
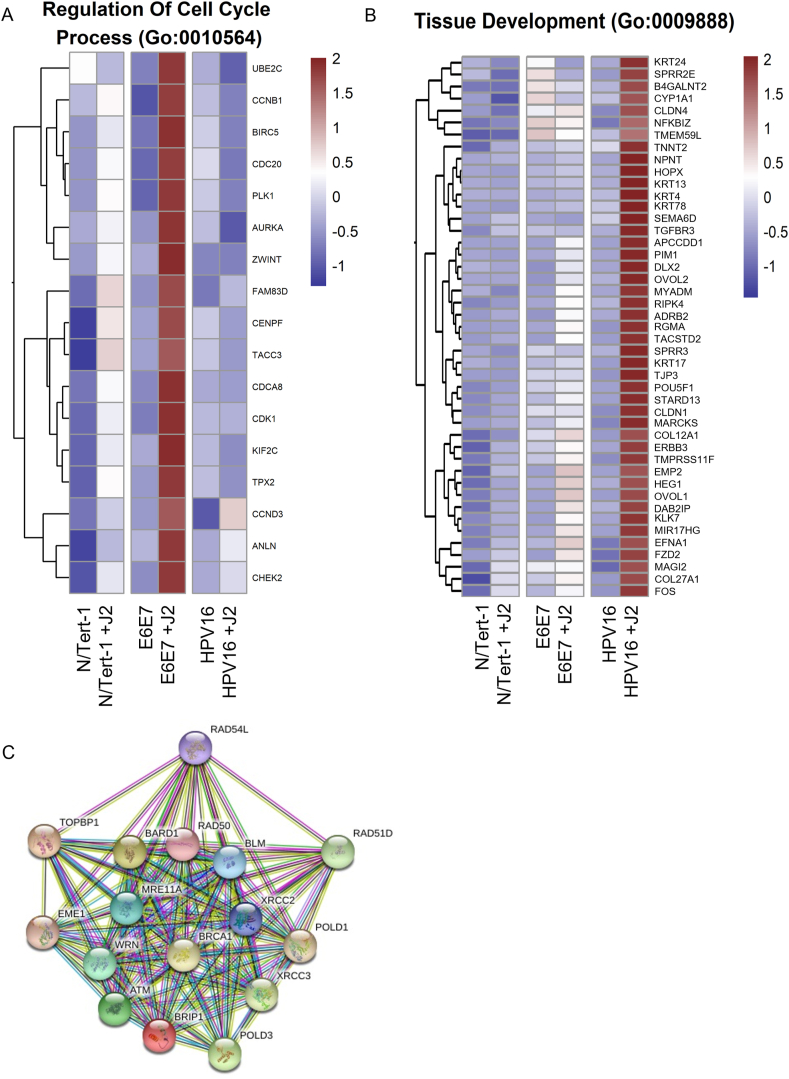
Other significant observations from our GO enrichment cross-comparisons were alterations in genes associated with cell cycle regulation and tissue development (Fig. 7). These pathways are notably altered during EMT, oncogenic transformation, and HPV-related transformation [1,[147], [148], [149]]. N/Tert-1+E6E7 cells cocultured with fibroblasts, markedly upregulated GO enrichment related to cell cycle processes; these alterations were highly suggestive of significant transformation (highest significant gene set highlighted in Fig. 7A) [[150], [151], [152]]. Conversely, N/Tert-1+HPV16 grown in the presence of J2 upregulated GO enrichment in tissue development that was highly suggestive of a more epithelial genotype (Fig. 7B). In particular, the expression of KRT4 and KRT13 decreases in transformed epithelial cells; N/Tert-1+HPV16 grown in the presence of J2 instead showed enhanced KRT13 and KRT4 levels [153].
Fig. 7.
Fibroblasts differentially regulate cell cycle process and tissue development-related GO enrichment. 7A. Condensed heat map demonstrating the most significant GO:0010564 regulation of cell cycle process across all groups. 7B. Condensed heat map demonstrating the most significant GO:0009888 tissue development across all groups. 7C. String network for N/Tert-1+HPV16 which shows the most significantly downregulated gene sets related to homologous recombination (HR) with the removal of stromal support.
3.2.4. Histone modifications and repair mechanisms
When denoting the significance of H2AX, we noted several other significant histone-related alterations via fibroblasts (Fig. 3G). Here, we suggest that chromosomal availability may be a significant contributor to the events observed in this study. Another striking observation was that in N/Tert-1+HPV16, the removal of stromal support significantly downregulated gene sets related to DNA repair factors, homologous recombination (HR) was one of the most significant (Fig. 7C String network of significant factors associated with HR) [154]. Loss of these factors could significantly impair viral replication function. Homologous repair deficits have been suggested to drive integration [155]. We have also demonstrated that viral gene transcripts were significantly reduced in N/Tert-1+HPV16 [55]. These results suggest that the changes induced by fibroblast withdrawal in the keratinocytes could contribute to viral genome integration, irrespective of the transcriptional reprogramming carried out by the virus.
3.3. Differential proteomic landscapes altered by fibroblasts in keratinocytes
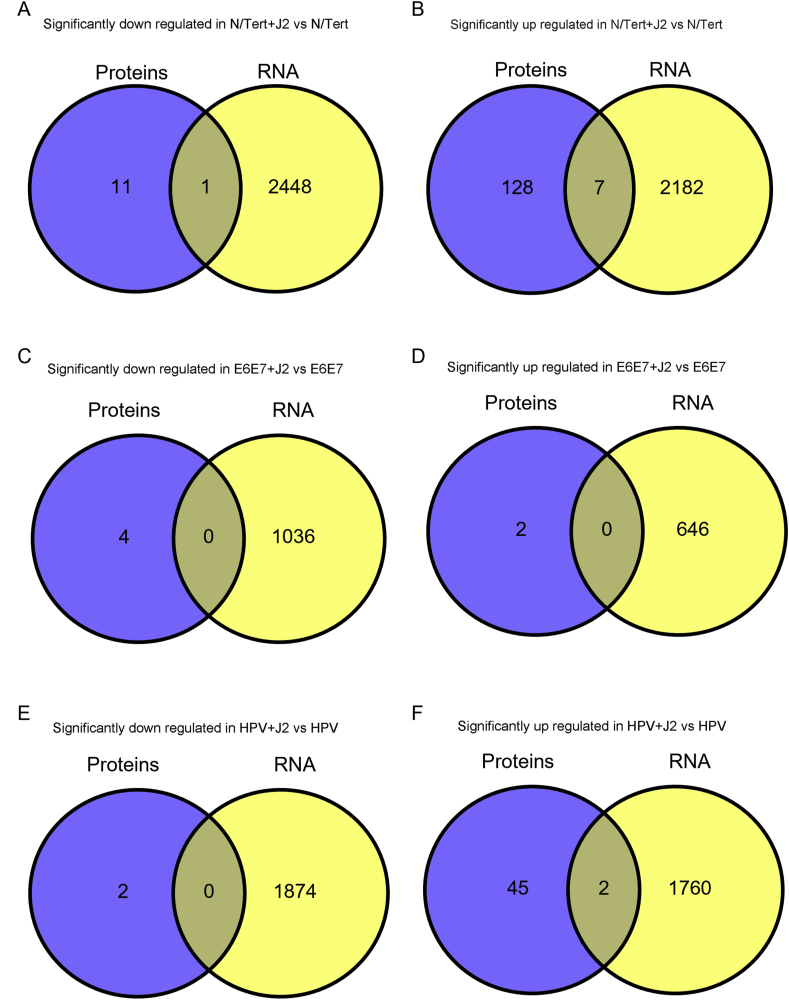
For label-free LC-MS/MS proteomic comparison, matched triplicate samples were harvested at the same time as RNA-seq; differential protein expression and bioinformatic analysis were performed, cross-matched to RNA-seq, and further assessed by bioinformatics. Processed datasets are available in Supplementary Data S4. Cross-comparative analysis is presented as Venn diagrams in Fig. 8. While mRNA expression precedes protein translation, the exact correlation between transcript levels and protein abundance is often poor; correlative assessments can instead be utilized for biomarker trends [[156], [157], [158], [159]]. The Human Protein Atlas was first consulted to assess if comparative analysis supported our RNAseq observations that fibroblasts regulate the transformation potential in HPV + keratinocytes [[160], [161], [162]]. Oncogenic-associated proteins were significantly downregulated in N/Tert-1+HPV16 cells grown in the presence of J2, such as TNFRSF10D; clinical pathology observations have demonstrated that high proteomic expression of many of these proteins correlates with poor prognostics in either cervical cancer and/or head and neck cancer [[160], [161], [162]]. Fibroblast regulation of suprabasal and cell cycle markers, such as cyclins, in N/Tert-1+HPV16 were suggestive of a genotype that would promote the ideal host environment for viral replication and amplification, consistent with our RNA analysis. Global profiling of proteomic trends confirmed differentially regulated subgroups which demonstrated the stromal regulation of EMT-related protein expression. Our overall observations suggest that fibroblasts influence genotypic profiles that support the viral lifecycle while inhibiting EMT progression, and would reduce the oncogenic progression in HPV + keratinocytes. This fibroblast regulation pattern is inversed in E6E7+ keratinocytes, where oncogene expression is outside the control of E2.
Fig. 8.
Differential expression Venn diagrams comparing significant up or down regulation via fibroblasts in RNA-seq and proteomic analysis. The sum of all the numbers in the circle represents the total number in the compared groups, and the overlapping area indicates the number of differential genes shared between the groups, as shown in the following Fig. 8A and B. Cross comparison of N/Tert-1 downregulation, and upregulation, respectively via fibroblasts. 8C,D. Cross comparison of N/Tert-1+E6E7 downregulation, and upregulation, respectively via fibroblasts. 8E,F. Cross comparison of N/Tert-1+HPV16 downregulation, and upregulation, respectively via fibroblasts.
4. Discussion
Decades of research have continued to improve the model systems utilized to mimic HPV infection and progression. Despite the increasing availability of improved models, the progression from primary infected cells to cancer has yet to be fully demonstrated in vitro [[163], [164], [165]]. The addition of fibroblast feeder cells for the generation of epithelial cell lines has improved both the efficiency of immortalization attempts, as well as contributing to viral episomal maintenance, yet the mechanisms behind stromal regulation of the virus has yet to be fully elucidated [70,71,128]. To assess how fibroblasts modulate viral-keratinocyte interactions and episomal maintenance, we sought to evaluated the most effective approach and control for all relevant factors. For this reason, we chose to utilize our well-characterized and matched N/Tert-1 keratinocyte lines that we have previously sequenced for numerous HPV-related signaling studies [55,73,74,77,88,166].
Genomic and proteomic assessments revealed that fibroblasts promoted a suprabasal epithelial state in N/Tert-1+HPV16, whereas N/Tert-1+E6E7 presented a profile that was suggestive of EMT progression in the presence of fibroblasts. The precise mechanisms of viral oncogenic transformation remains largely speculative, although E6E7 expression is vital and a number of host biomarkers are well characterized in this progression [32,44,47,57,67,129,141,[167], [168], [169]]. Our studies confirmed that N/Tert-1+HPV maintained in fibroblasts sustained HPV episomes (Fig. 1A) [52,55,57,59,109]. While we have yet to determine the temporal relationship of fibroblast withdrawal and integration, both fibroblast withdrawal and integration promote a dysregulated host expression pattern that could promote EMT and oncogenic progression.
Cytokine and interleukin signaling are known immune regulatory events in the response to HPV [81,170]. Chemokines are small molecules and secretory peptides are associated with cellular signaling and are broadly divided into subfamilies based on their amino acid motifs: XC, CC, CXC, and CXXXC [130,131]. Chemokine ligands, work jointly with specific chemokine receptors, to control a broad range of biological processes [130,131]. CXC family members are further divided into ELR+ and ELR-members, based on the presence or absence of a Glu-Leu-Arg (ELR) motif in their N-terminus [130]. ELR + CXC chemokines are associated with the progression of cancer, conversely downregulation of these has been found to suppress the motility of cancer [130]. On the other hand, ELR- CXC chemokines are associated with tumor-suppressive effects [130]. Likewise, fibroblasts upregulated the interleukin-related microRNA-27b (miR-27b) in N/Tert-1+HPV16 (Fig. 5B). Upregulation of miR-27b has been shown to alter the transcription factors in the SNAIL, ZEB, and TWIST families and limit EMT progression [136]. The chemokine and interleukin-related GO enrichment observed in N/Tert-1+HPV16 grown in the presence of J2 were highly indicative of an epithelial genotype (Fig. 5). This suggests that fibroblasts may play a regulatory role in the prevention of EMT-related transformation in HPV + keratinocytes.
Additional observations related to stromal regulation of an epithelial-like state in N/Tert-1+HPV16 were the observations related to TJ signaling (Fig. 6A). TJs are comprised of a complex group of molecules and are associated with the suprabasal and intermediate layers of epithelia. While numerous TJ proteins are downregulated in the transformation process, others are overexpressed and mislocalized [143,171]. Dysregulation of TJ proteins is associated with epithelial-to-mesenchymal transition (EMT) and invasive phenotypes [141,143,172]. As large complexes, TJs facilitate signal transduction and are involved in cell proliferation, migration, differentiation, and survival, all of which are also beneficial to the viral lifecycle [173]. In HPV + cervical cancer, HPV16 E7 has been shown to alter the expression and localization of TJ-associated claudins [141,143,172]. Twist1 is also associated with EMT (Fig. 6B); its transcriptional activation of Claudin-4 has been shown to promote cervical cancer migration and invasion [142,144,167]. Our analysis shows partial upregulation of TJ components in N/Tert-1+E6E7 when in coculture with fibroblasts; in N/Tert-1+HPV16, there was a significant upregulation in these molecules (Fig. 6). In particular, there was a marked increase in TJ assembly proteins, including claudins, which are crucial to tight junction integrity (Fig. 6A). Here, we suggest that fibroblast regulation presents a model for stages of transformation. The decreased expression of junctional proteins seen in N/Tert-1+E6E7 is more analogous to later, neoplastic stages of transformation. When the viral genome is integrated, E6E7 is overexpressed, and there is a progression towards EMT. Meanwhile, the increased expression of TJ components in N/Tert-1+HPV16 cultured with fibroblasts is analogous to early viral lifecycle stages. Furthermore, by inducing increased levels of TJ components in infected keratinocytes, the virus induces an environment that mimics a suprabasal phenotype, which is important for the amplification stage of the viral lifecycle [61,125,174]. Differences observed between N/Tert-1+E6E7 and N/Tert-1+HPV16 indicate that the upregulation of junctional proteins seen in full genome containing cells may be driven by other viral factors, possibly E2, and warrants further investigation. It is of interest to further dissect the impact of keratinocyte-fibroblast co-culture upon the subcellular localization of these TJ components and any resulting downstream effects on cell invasive capacity in both E6E7+ and full-genome containing cell lines.
“Fragile sites” are associated with viral integration, and are characterized by faulty chromosome condensation and replication stress [[175], [176], [177], [178], [179]]. The altered histone gene expression we observed following fibroblast removal is consistent with an altered chromatin state, exposing “fragile site” availability, and enabling integration events [54]. We propose that loss of stromal support promotes a chromosomal state that promotes integration; this, in turn, promotes transcriptional reprogramming events that are conducive to EMT and oncogenic progression [57,59,61,75,85,109,110,175]. We also suggest that in the progression of natural infection, errors in the “cross-talk” between fibroblasts and HPV + keratinocytes are likely a factor that contributes to viral integration and epithelial reprogramming.
5. Conclusion
Both our research and that of others have shown that interactions between fibroblasts and keratinocytes in HPV models are critical for maintaining episomal HPV genomes, influencing keratinocyte differentiation, and regulating viral transcription [23,28,29,38,55,128,[180], [181], [182]]. Here we present RNAseq analysis revealing that fibroblasts may regulate the transcriptional signature of HPV + keratinocytes by regulating cytokine activity, cell junction proteins, and EMT-related signatures. Proteomic analysis further supported these findings, highlighting fibroblasts' ability to modulate the expression of signaling events linked to oncogenic transformation. Overall, fibroblasts were found to influence both viral and host cell signaling, promote HPV lifecycle maintenance, and possibly limit EMT and cancer progression of HPV + keratinocytes.
CRediT authorship contribution statement
Claire D. James: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Formal analysis, Data curation. Rachel L. Lewis: Validation, Methodology, Investigation, Data curation. Austin J. Witt: Validation, Methodology, Investigation, Data curation. Christiane Carter: Writing – review & editing, Validation, Software, Methodology, Investigation, Formal analysis. Nabiha M. Rais: Methodology, Investigation, Data curation. Xu Wang: Formal analysis, Data curation. Molly L. Bristol: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Data availability statement
Following the 2023 NIH data management and sharing policy, all data resulting from the development of projects will be available in scientific communications presented at conferences and in manuscripts that will be published in peer-reviewed scientific journals. Data will be deposited in the Open Science Framework (OSF) platform. OSF can be accessed at https://osf.io. VCU is an OSF institutional member, and OSF is an approved generalist repository for the 2023 NIH data management and sharing policy.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the VCU Philips Institute for Oral Health Research, the VCU Quest Fund, the National Institute of Dental and Craniofacial Research/NIH/DHHS R03 DE029548, and the National Cancer Institute-designated Massey Cancer Center grant P30 CA016059. Services in support of the research project were provided by the VCU Massey Comprehensive Cancer Center Bioinformatics Shared Resource. Massey is supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30 CA016059. Services and products in support of the research project were generated by the VCU Massey Comprehensive Cancer Center Proteomics Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30 CA016059.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tvr.2024.200302.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
Data availability
Data will be made available on request.
References
- 1.Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M., Broker T.R., et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 2.Bzhalava D., Eklund C., Dillner J. International standardization and classification of human papillomavirus types. Virology. 2015;476:341–344. doi: 10.1016/j.virol.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Parkin D.M., Bray F. Chapter 2: the burden of HPV-related cancers. Vaccine. 2006;24(Suppl 3):S3/11–25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 4.Burd E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gribb J.P., Wheelock J.H., Park E.S. Human papilloma virus (HPV) and the current state of oropharyngeal cancer prevention and treatment. Del J Public Health. 2023;9:26–28. doi: 10.32481/djph.2023.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogliano V., Baan R., Straif K., Grosse Y., Secretan B., Ghissassi F.E. Carcinogenicity of human papillomaviruses. Lancet Oncol. 2005;6:204. doi: 10.1016/S1470-2045(05)70086-3. [DOI] [PubMed] [Google Scholar]
- 7.Saraiya M., Unger E.R., Thompson T.D., Lynch C.F., Hernandez B.Y., Lyu C.W., et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. JNCI J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brianti P., De Flammineis E., Mercuri S.R. Review of HPV-related diseases and cancers. New Microbiol. 2017;40:80–85. [PubMed] [Google Scholar]
- 9.Marur S., D'Souza G., Westra W.H., Forastiere A.A. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancers Linked With HPV Each Year, Centers for Disease Control and Prevention. (CDC) (Updated 18 September 2024). https://www.cdc.gov/cancer/hpv/cases.html. (Accessed 10 December 2024).
- 11.Liao C.-I., Francoeur A.A., Kapp D.S., Caesar M.A.P., Huh W.K., Chan J.K. Trends in human papillomavirus–associated cancers, demographic characteristics, and vaccinations in the US, 2001-2017. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J., Deng Y., Boakye D., Tin M.S., Lok V., Zhang L., et al. Global distribution, risk factors, and recent trends for cervical cancer: a worldwide country-level analysis. Gynecol. Oncol. 2022;164:85–92. doi: 10.1016/j.ygyno.2021.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 14.Malik S., Sah R., Muhammad K., Waheed Y. Tracking HPV infection, associated cancer development, and recent treatment efforts—a comprehensive review. Vaccines. 2023;11:102. doi: 10.3390/vaccines11010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spurgeon M.E., Lambert P.F. Human papillomavirus and the stroma: bidirectional crosstalk during the virus life cycle and carcinogenesis. Viruses. 2017;9:219. doi: 10.3390/v9080219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins D. A review of cross-protection against oncogenic HPV by an HPV-16/18 AS04-adjuvanted cervical cancer vaccine: importance of virological and clinical endpoints and implications for mass vaccination in cervical cancer prevention. Gynecol. Oncol. 2008;110:S18–S25. doi: 10.1016/j.ygyno.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 17.James C.D., Otoa R., Youssef A.H., Fontan C.T., Sannigrahi M.K., Windle B., et al. Press; 2024. HPV16 Genome Structure Analysis in Oropharyngeal Cancer PDXs Identifies Tumors with Integrated and Episomal Genomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 19.Moscicki A.-B., Schiffman M., Burchell A., Albero G., Giuliano A.R., Goodman M.T., et al. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine. 2012;30(Suppl 5):F24–F33. doi: 10.1016/j.vaccine.2012.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moscicki A.-B., Ma Y., Farhat S., Jay J., Hanson E., Benningfield S., et al. Natural history of anal human papillomavirus infection in heterosexual women and risks associated with persistence. Clin Infect Dis Off Publ Infect Dis Soc Am. 2014;58:804–811. doi: 10.1093/cid/cit947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei F., Goodman M.T., Xia N., Zhang J., Giuliano A.R., D'Souza G., et al. Incidence and clearance of anal human papillomavirus infection in 16 164 individuals, according to human immunodeficiency virus status, sex, and male sexuality: an international pooled analysis of 34 longitudinal studies. Clin Infect Dis Off Publ Infect Dis Soc Am. 2023;76:e692–e701. doi: 10.1093/cid/ciac581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodily J., Laimins L.A. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19:33–39. doi: 10.1016/j.tim.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raikhy G., Woodby B.L., Scott M.L., Shin G., Myers J.E., Scott R.S., et al. Suppression of stromal interferon signaling by human papillomavirus 16. J. Virol. 2019;93 doi: 10.1128/jvi.00458-19. 10.1128/jvi.00458-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathi M., Billet S., Bhowmick N.A. Understanding the role of stromal fibroblasts in cancer progression. Cell Adhes. Migrat. 2012;6:231–235. doi: 10.4161/cam.20419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhowmick N.A., Neilson E.G., Moses H.L. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barcellos-Hoff M.H. In: Encycl. Syst. Biol. Dubitzky W., Wolkenhauer O., Cho K.-H., Yokota H., editors. Springer; New York, NY: 2013. Stroma; pp. 2017–2019. [DOI] [Google Scholar]
- 27.Kendall R.T., Feghali-Bostwick C.A. Fibroblasts in fibrosis: novel roles and mediators. Front. Pharmacol. 2014;5:123. doi: 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert P.F., Ozbun M.A., Collins A., Holmgren S., Lee D., Nakahara T. Using an immortalized cell line to study the HPV life cycle in organotypic “raft” cultures. Methods Mol. Med. 2005;119:141–155. doi: 10.1385/1-59259-982-6:141. [DOI] [PubMed] [Google Scholar]
- 29.Meyers C. Organotypic (raft) epithelial tissue culture system for the differentiation-dependent replication of papillomavirus. Methods Cell Sci. 1996;18:201–210. doi: 10.1007/BF00132885. [DOI] [Google Scholar]
- 30.Barros M.R., de Melo C.M.L., Barros M.L.C.M.G.R., de Cássia Pereira de Lima R., de Freitas A.C., Venuti A. Activities of stromal and immune cells in HPV-related cancers. J. Exp. Clin. Cancer Res. 2018;37:137. doi: 10.1186/s13046-018-0802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahebali S., Van den Eynden G., Murta E.F., Michelin M.A., Cusumano P., Petignat P., et al. Stromal issues in cervical cancer: a review of the role and function of basement membrane, stroma, immune response and angiogenesis in cervical cancer development. Eur. J. Cancer Prev. 2010;19:204–215. doi: 10.1097/CEJ.0b013e32833720de. [DOI] [PubMed] [Google Scholar]
- 32.Chung S.-H., Shin M.K., Korach K.S., Lambert P.F. Requirement for stromal estrogen receptor alpha in cervical neoplasia. Horm Cancer. 2013;4:50–59. doi: 10.1007/s12672-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epithelial–stromal interactions modulating penetration of matrigel membranes by HPV 16-immortalized keratinocytes. J. Invest. Dermatol. 1997;109:619–625. doi: 10.1111/1523-1747.ep12337594. [DOI] [PubMed] [Google Scholar]
- 34.Alkasalias T., Moyano-Galceran L., Arsenian-Henriksson M., Lehti K. Fibroblasts in the tumor microenvironment: shield or spear? Int. J. Mol. Sci. 2018;19:1532. doi: 10.3390/ijms19051532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao X., Xu J., Wang W., Liang C., Hua J., Liu J., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20:131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monteran L., Erez N. The dark side of fibroblasts: cancer-associated fibroblasts as mediators of immunosuppression in the tumor microenvironment. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahrotaban S., Mahdavi N., Abdollahi A., Yazdani F., Kaghazloo A., Derakhshan S. Carcinoma-associated fibroblasts are a common finding in the microenvironment of HPV-positive oropharyngeal squamous cell carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM. 2019;27:683–688. doi: 10.1097/PAI.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 38.Smola H., Stark H.-J., Thiekötter G., Mirancea N., Krieg T., Fusenig N.E. Dynamics of basement membrane formation by keratinocyte–fibroblast interactions in organotypic skin culture. Exp. Cell Res. 1998;239:399–410. doi: 10.1006/excr.1997.3910. [DOI] [PubMed] [Google Scholar]
- 39.Truffi M., Sorrentino L., Corsi F. Fibroblasts in the tumor microenvironment. Adv. Exp. Med. Biol. 2020;1234:15–29. doi: 10.1007/978-3-030-37184-5_2. [DOI] [PubMed] [Google Scholar]
- 40.Almangush A., Jouhi L., Haglund C., Hagström J., Mäkitie A.A., Leivo I. Tumor-stroma ratio is a promising prognostic classifier in oropharyngeal cancer. Hum. Pathol. 2023;136:16–24. doi: 10.1016/j.humpath.2023.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Sharma V., Letson J., Furuta S. Fibrous stroma: driver and passenger in cancer development. Sci. Signal. 2022;15 doi: 10.1126/scisignal.abg3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bremnes R.M., Dønnem T., Al-Saad S., Al-Shibli K., Andersen S., Sirera R., et al. The role of tumor stroma in cancer progression and prognosis: emphasis on carcinoma-associated fibroblasts and non-small cell lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2011;6:209–217. doi: 10.1097/JTO.0b013e3181f8a1bd. [DOI] [PubMed] [Google Scholar]
- 43.James C.D., Fontan C.T., Otoa R., Das D., Prabhakar A.T., Wang X., et al. Human papillomavirus 16 E6 and E7 synergistically repress innate immune gene transcription. mSphere. 2020;5 doi: 10.1128/mSphere.00828-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basukala O., Banks L. The not-so-good, the bad and the ugly: HPV E5, E6 and E7 oncoproteins in the orchestration of carcinogenesis. Viruses. 2021;13:1892. doi: 10.3390/v13101892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeFilippis R.A., Goodwin E.C., Wu L., DiMaio D. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 2003;77:1551–1563. doi: 10.1128/jvi.77.2.1551-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francis D.A., Schmid S.I., Howley P.M. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 2000;74:2679–2686. doi: 10.1128/jvi.74.6.2679-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoppe-Seyler K., Bossler F., Braun J.A., Herrmann A.L., Hoppe-Seyler F. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018;26:158–168. doi: 10.1016/j.tim.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Jeon S., Lambert P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrison M.A., Morreale R.J., Akunuru S., Kofron M., Zheng Y., Wells S.I. Targeting the human papillomavirus E6 and E7 oncogenes through expression of the bovine papillomavirus type 1 E2 protein stimulates cellular motility. J. Virol. 2011;85:10487–10498. doi: 10.1128/JVI.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riley R.R., Duensing S., Brake T., Münger K., Lambert P.F., Arbeit J.M. Dissection of human papillomavirus E6 and E7 function in transgenic mouse models of cervical carcinogenesis. Cancer Res. 2003;63:4862–4871. [PubMed] [Google Scholar]
- 51.Nees M., Geoghegan J.M., Munson P., Prabhu V., Liu Y., Androphy E., et al. Human papillomavirus type 16 E6 and E7 proteins inhibit differentiation-dependent expression of transforming growth factor-β2 in cervical keratinocytes. Cancer Res. 2000;60:4289–4298. [PubMed] [Google Scholar]
- 52.Dall K.L., Scarpini C.G., Roberts I., Winder D.M., Stanley M.A., Muralidhar B., et al. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68:8249–8259. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 53.Rheinwald J.G., Green H. Epidermal growth factor and the multiplication of cultured human epidermal keratinocytes. Nature. 1977;265:421–424. doi: 10.1038/265421a0. [DOI] [PubMed] [Google Scholar]
- 54.Coursey T.L., McBride A.A. Development of keratinocyte cell lines containing extrachromosomal human papillomavirus genomes. Curr Protoc. 2021;1:e235. doi: 10.1002/cpz1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.James C.D., Lewis R.L., Fakunmoju A.L., Witt A.J., Youssef A.H., Wang X., et al. Fibroblast stromal support model for predicting human papillomavirus-associated cancer drug responses. 2024. 2024.04.09.588680. [DOI] [PMC free article] [PubMed]
- 56.Koneva L.A., Zhang Y., Virani S., Hall P.B., McHugh J.B., Chepeha D.B., et al. HPV integration in HNSCC correlates with survival outcomes, immune response signatures, and candidate drivers. Mol Cancer Res MCR. 2018;16:90–102. doi: 10.1158/1541-7786.MCR-17-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBride A.A., Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akagi K., Li J., Broutian T.R., Padilla-Nash H., Xiao W., Jiang B., et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2014;24:185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balaji H., Demers I., Wuerdemann N., Schrijnder J., Kremer B., Klussmann J.P., et al. Causes and consequences of HPV integration in head and neck squamous cell carcinomas: state of the art. Cancers. 2021;13:4089. doi: 10.3390/cancers13164089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamal M., Lameiras S., Deloger M., Morel A., Vacher S., Lecerf C., et al. Human papilloma virus (HPV) integration signature in Cervical Cancer: identification of MACROD2 gene as HPV hot spot integration site. Br. J. Cancer. 2021;124:777–785. doi: 10.1038/s41416-020-01153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeon S., Allen-Hoffmann B.L., Lambert P.F. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 1995;69:2989–2997. doi: 10.1128/JVI.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu L., Majerciak V., Lobanov A., Mirza S., Band V., Liu H., et al. HPV oncogenes expressed from only one of multiple integrated HPV DNA copies drive clonal cell expansion in cervical cancer. mBio. 2024;15 doi: 10.1128/mbio.00729-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan J., Fu Y., Peng W., Li X., Shen Y., Guo E., et al. Multi-omics characterization of silent and productive HPV integration in cervical cancer. Cell Genomics. 2023;3 doi: 10.1016/j.xgen.2022.100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mainguené J., Vacher S., Kamal M., Hamza A., Masliah‐Planchon J., Baulande S., et al. Human papilloma virus integration sites and genomic signatures in head and neck squamous cell carcinoma. Mol. Oncol. 2022;16:3001–3016. doi: 10.1002/1878-0261.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaglia M.M., Munger K. More than just oncogenes: mechanisms of tumorigenesis by human viruses. Curr Opin Virol. 2018;32:48–59. doi: 10.1016/j.coviro.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLaughlin-Drubin M.E., Meyers J., Munger K. Cancer associated human papillomaviruses. Curr Opin Virol. 2012;2:459–466. doi: 10.1016/j.coviro.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mirabello L., Yeager M., Yu K., Clifford G.M., Xiao Y., Zhu B., et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174.e6. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McBride A.A., Münger K. Expert views on HPV infection. Viruses. 2018;10:94. doi: 10.3390/v10020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chapman S., Liu X., Meyers C., Schlegel R., McBride A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu X., Ory V., Chapman S., Yuan H., Albanese C., Kallakury B., et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dakic A., DiVito K., Fang S., Suprynowicz F., Gaur A., Li X., et al. ROCK inhibitor reduces Myc-induced apoptosis and mediates immortalization of human keratinocytes. Oncotarget. 2016;7:66740–66753. doi: 10.18632/oncotarget.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu B., Quintero J., Baker C.C. Keratinocyte growth conditions modulate telomerase expression, senescence, and immortalization by human papillomavirus type 16 E6 and E7 oncogenes. Cancer Res. 2003;63:7815–7824. [PubMed] [Google Scholar]
- 73.Evans M.R., James C.D., Loughran O., Nulton T.J., Wang X., Bristol M.L., et al. An oral keratinocyte life cycle model identifies novel host genome regulation by human papillomavirus 16 relevant to HPV positive head and neck cancer. Oncotarget. 2017;8:81892–81909. doi: 10.18632/oncotarget.18328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans M.R., James C.D., Bristol M.L., Nulton T.J., Wang X., Kaur N., et al. Human papillomavirus 16 E2 regulates keratinocyte gene expression relevant to cancer and the viral life cycle. J. Virol. 2019;93 doi: 10.1128/JVI.01941-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan I.M., DiNardo L.J., Windle B. Integration of human papillomavirus genomes in head and neck cancer: is it time to consider a paradigm shift? Viruses. 2017;9:208. doi: 10.3390/v9080208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fontan C.T., James C.D., Prabhakar A.T., Bristol M.L., Otoa R., Wang X., et al. A critical role for p53 during the HPV16 life cycle. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.00681-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prabhakar A.T., James C.D., Fontan C.T., Otoa R., Wang X., Bristol M.L., et al. Human papillomavirus 16 E2 interaction with TopBP1 is required for E2 and viral genome stability during the viral life cycle. J. Virol. 2023;97 doi: 10.1128/jvi.00063-23. 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prabhakar A.T., James C.D., Das D., Fontan C.T., Otoa R., Wang X., et al. Interaction with TopBP1 is required for human papillomavirus 16 E2 plasmid segregation/retention function during mitosis. J. Virol. 2022;96 doi: 10.1128/jvi.00830-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Song D., Li H., Li H., Dai J. Effect of human papillomavirus infection on the immune system and its role in the course of cervical cancer. Oncol. Lett. 2015;10:600–606. doi: 10.3892/ol.2015.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westrich J.A., Warren C.J., Pyeon D. Evasion of host immune defenses by human papillomavirus. Virus Res. 2017;231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nunes R.A.L., Morale M.G., Silva G.Á.F., Villa L.L., Termini L. Innate immunity and HPV: friends or foes. Clin Sao Paulo Braz. 2018;73:e549s. doi: 10.6061/clinics/2018/e549s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feller L., Khammissa R.A., Wood N.H., Lemmer J. Epithelial maturation and molecular biology of oral HPV. Infect. Agents Cancer. 2009;4:16. doi: 10.1186/1750-9378-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Longworth M.S., Laimins L.A. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol Mol Biol Rev MMBR. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee J.H., Yi S.M.P., Anderson M.E., Berger K.L., Welsh M.J., Klingelhutz A.J., et al. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc. Natl. Acad. Sci. USA. 2004;101:2094–2099. doi: 10.1073/pnas.0308615100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nulton T.J., Olex A.L., Dozmorov M., Morgan I.M., Windle B. Analysis of the Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget. 2017;8:17684–17699. doi: 10.18632/oncotarget.15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bristol M.L., Wang X., Smith N.W., Son M.P., Evans M.R., Morgan I.M. DNA damage reduces the quality, but not the quantity of human papillomavirus 16 E1 and E2 DNA replication. Viruses. 2016;8:175. doi: 10.3390/v8060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.James C.D., Saini S., Sesay F., Ko K., Felthousen-Rusbasan J., Iness A.N., et al. Restoring the DREAM complex inhibits the proliferation of high-risk HPV positive human cells. Cancers. 2021;13:489. doi: 10.3390/cancers13030489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.James C.D., Prabhakar A.T., Otoa R., Evans M.R., Wang X., Bristol M.L., et al. SAMHD1 regulates human papillomavirus 16-induced cell proliferation and viral replication during differentiation of keratinocytes. mSphere. 2019;4 doi: 10.1128/mSphere.00448-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 91.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinforma Oxf Engl. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 92.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Babraham bioinformatics - FastQC A quality control tool for high throughput sequence data n.d. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 96.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinforma Oxf Engl. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma Oxf Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinforma Oxf Engl. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen Y., Lun A.T.L., Smyth G.K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research. 2016;5:1438. doi: 10.12688/f1000research.8987.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Myers J.E., Zwolinska K., Sapp M.J., Scott R.S. An exonuclease V–qPCR assay to analyze the state of the human papillomavirus 16 genome in cell lines and tissues. Curr Protoc Microbiol. 2020;59:e119. doi: 10.1002/cpmc.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X., Smits A.H., van Tilburg G.B., Ovaa H., Huber W., Vermeulen M. Proteome-wide identification of ubiquitin interactions using UbIA-MS. Nat. Protoc. 2018;13:530–550. doi: 10.1038/nprot.2017.147. [DOI] [PubMed] [Google Scholar]
- 106.Huber W., von Heydebreck A., Sültmann H., Poustka A., Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinforma Oxf Engl. 2002;18(Suppl 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 107.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Facompre N.D., Rajagopalan P., Sahu V., Pearson A.T., Montone K.T., James C.D., et al. Identifying predictors of HPV-related head and neck squamous cell carcinoma progression and survival through patient-derived models. Int. J. Cancer. 2020;147:3236–3249. doi: 10.1002/ijc.33125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nulton T.J., Kim N.-K., DiNardo L.J., Morgan I.M., Windle B. Patients with integrated HPV16 in head and neck cancer show poor survival. Oral Oncol. 2018;80:52–55. doi: 10.1016/j.oraloncology.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Williams V.M., Filippova M., Soto U., Duerksen-Hughes P.J. HPV-DNA integration and carcinogenesis: putative roles for inflammation and oxidative stress. Future Virol. 2011;6:45–57. doi: 10.2217/fvl.10.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cheng Y., Huang N., Yin Q., Cheng C., Chen D., Gong C., et al. LncRNA TP53TG1 plays an anti-oncogenic role in cervical cancer by synthetically regulating transcriptome profile in HeLa cells. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.981030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Isaka S., Takei Y., Tokino T., Koyama K., Miyoshi Y., Suzuki M., et al. Isolation and characterization of a novel TP53-inducible gene, TP53TG5, which suppresses growth and shows cell cycle-dependent transition of expression. Genes Chromosomes Cancer. 2000;27:345–352. doi: 10.1002/(SICI)1098-2264(200004)27:4<345::AID-GCC2>3.0.CO;2-352. [DOI] [PubMed] [Google Scholar]
- 113.Ge X., Xu M., Cheng T., Hu N., Sun P., Lu B., et al. TP53I13 promotes metastasis in glioma via macrophages, neutrophils, and fibroblasts and is a potential prognostic biomarker. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.974346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.RB1 RB transcriptional corepressor 1 [Homo sapiens (human)] - Gene - NCBI n.d. https://www.ncbi.nlm.nih.gov/gene/5925 (accessed August 30, 2024).
- 115.RBL1 RB transcriptional corepressor like 1 [Homo sapiens (human)] - Gene - NCBI n.d. https://www.ncbi.nlm.nih.gov/gene/5933 (accessed August 30, 2024).
- 116.RB1CC1 RB1 inducible coiled-coil 1 [Homo sapiens (human)] - Gene - NCBI n.d. https://www.ncbi.nlm.nih.gov/gene/9821 (accessed August 30, 2024).
- 117.Vélez-Cruz R., Johnson D.G. The retinoblastoma (RB) tumor suppressor: pushing back against genome instability on multiple fronts. Int. J. Mol. Sci. 2017;18:1776. doi: 10.3390/ijms18081776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moody C.A. Regulation of the innate immune response during the human papillomavirus life cycle. Viruses. 2022;14:1797. doi: 10.3390/v14081797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hong S., Mehta K.P., Laimins L.A. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J. Virol. 2011;85:9486–9494. doi: 10.1128/JVI.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Valle-Mendiola A., Gutiérrez-Hoya A., Soto-Cruz I. JAK/STAT signaling and cervical cancer: from the cell surface to the nucleus. Genes. 2023;14 doi: 10.3390/genes14061141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morgan E.L., Macdonald A. Manipulation of JAK/STAT signalling by high-risk HPVs: potential therapeutic targets for HPV-associated malignancies. Viruses. 2020;12:977. doi: 10.3390/v12090977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Psyrri A., DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat. Clin. Pract. Oncol. 2008;5:24–31. doi: 10.1038/ncponc0984. [DOI] [PubMed] [Google Scholar]
- 123.Huh W.K., Joura E.A., Giuliano A.R., Iversen O.-E., de Andrade R.P., Ault K.A., et al. Final efficacy, immunogenicity, and safety analyses of a nine-valent human papillomavirus vaccine in women aged 16–26 years: a randomised, double-blind trial. Lancet. 2017;390:2143–2159. doi: 10.1016/S0140-6736(17)31821-4. [DOI] [PubMed] [Google Scholar]
- 124.Kavanagh K., Pollock K.G., Cuschieri K., Palmer T., Cameron R.L., Watt C., et al. Changes in the prevalence of human papillomavirus following a national bivalent human papillomavirus vaccination programme in Scotland: a 7-year cross-sectional study. Lancet Infect. Dis. 2017;17:1293–1302. doi: 10.1016/S1473-3099(17)30468-1. [DOI] [PubMed] [Google Scholar]
- 125.Meeting IWG on the E of CR to H, Cancer IA for R on. Human Papillomaviruses. World Health Organization; 2007. [Google Scholar]
- 126.Vats A., Trejo-Cerro O., Thomas M., Banks L. Human papillomavirus E6 and E7: what remains? Tumour Virus Res. 2021;11 doi: 10.1016/j.tvr.2021.200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xu Y, Sun Y, Chang H, Cai J, Cao C, Zhang B, Zhang Y, Liu Y. The Expression of HPV E6/E7 mRNA In Situ Hybridization in HPV Typing-negative Cervical Cancer. Int J Gynecol Pathol. 2023 Jan 1;42(1):11-20. doi: 10.1097/PGP.0000000000000870. Epub 2022 Apr 12. [DOI] [PMC free article] [PubMed]
- 128.Coursey T.L., Van Doorslaer K., McBride A.A. Regulation of human papillomavirus 18 genome replication, establishment, and persistence by sequences in the viral upstream regulatory region. J. Virol. 2021;95 doi: 10.1128/JVI.00686-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rossi N.M., Dai J., Xie Y., Lou H., Boland J.F., Yeager M., et al. Extrachromosomal amplification of human papillomavirus episomes as a mechanism of cervical. Carcinogenesis. 2021:2021;10.22 doi: 10.1101/2021.10.22.465367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wu T., Yang W., Sun A., Wei Z., Lin Q. The role of CXC chemokines in cancer progression. Cancers. 2022;15:167. doi: 10.3390/cancers15010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hughes C.E., Nibbs R.J.B. A guide to chemokines and their receptors. FEBS J. 2018;285:2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Meng P., Liu C., Li J., Fang P., Yang B., Sun W., et al. CXC chemokine receptor 7 ameliorates renal fibrosis by inhibiting β-catenin signaling and epithelial-to-mesenchymal transition in tubular epithelial cells. Ren. Fail. 2024;46 doi: 10.1080/0886022X.2023.2300727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiu M.-X., Liu Y.-M., Kuang B. The role of DLLs in cancer: a novel therapeutic target. OncoTargets Ther. 2020;13:3881–3901. doi: 10.2147/OTT.S244860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kumar V., Vashishta M., Kong L., Wu X., Lu J.J., Guha C., et al. The role of Notch, hedgehog, and wnt signaling pathways in the resistance of tumors to anticancer therapies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.650772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sales-Dias J., Silva G., Lamy M., Ferreira A., Barbas A. The Notch ligand DLL1 exerts carcinogenic features in human breast cancer cells. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang J., Hua X., Qi N., Han G., Yu J., Yu Y., et al. MiR-27b suppresses epithelial–mesenchymal transition and chemoresistance in lung cancer by targeting Snail1. Life Sci. 2020;254 doi: 10.1016/j.lfs.2019.117238. [DOI] [PubMed] [Google Scholar]
- 137.Chae Y.K., Chang S., Ko T., Anker J., Agte S., Iams W., et al. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci. Rep. 2018;8:2918. doi: 10.1038/s41598-018-21061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim W.K., Kwon Y., Jang M., Park M., Kim J., Cho S., et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-54890-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Benczik M., Galamb Á., Koiss R., Kovács A., Járay B., Székely T., et al. Claudin-1 as a biomarker of cervical cytology and histology. Pathol. Oncol. Res. 2016;22:179–188. doi: 10.1007/s12253-015-9990-z. [DOI] [PubMed] [Google Scholar]
- 140.Cunniffe C., Brankin B., Lambkin H., Ryan F. The role of claudin-1 and claudin-7 in cervical tumorigenesis. Anticancer Res. 2014;34:2851–2857. [PubMed] [Google Scholar]
- 141.Uc P.Y., Miranda J., Raya-Sandino A., Alarcón L., Roldán M.L., Ocadiz-Delgado R., et al. E7 oncoprotein from human papillomavirus 16 alters claudins expression and the sealing of epithelial tight junctions. Int. J. Oncol. 2020;57:905–924. doi: 10.3892/ijo.2020.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhu J., Jiang Q. Twist1-mediated transcriptional activation of Claudin-4 promotes cervical cancer cell migration and invasion. Oncol. Lett. 2023;26:335. doi: 10.3892/ol.2023.13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu Y.-N., Deng M.-S., Liu Y.-F., Yao J., Xiao Z.-Y. Tight junction protein CLDN17 serves as a tumor suppressor to reduce the invasion and migration of oral cancer cells by inhibiting epithelial-mesenchymal transition. Arch. Oral Biol. 2022;133 doi: 10.1016/j.archoralbio.2021.105301. [DOI] [PubMed] [Google Scholar]
- 144.de Freitas Silva B.S., Yamamoto-Silva F.P., Pontes H.A.R., Pinto Júnior D. dos S. E-cadherin downregulation and Twist overexpression since early stages of oral carcinogenesis. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol. 2014;43:125–131. doi: 10.1111/jop.12096. [DOI] [PubMed] [Google Scholar]
- 145.Franco H.L., Casasnovas J., Rodríguez-Medina J.R., Cadilla C.L. Redundant or separate entities?—roles of Twist1 and Twist2 as molecular switches during gene transcription. Nucleic Acids Res. 2011;39:1177–1186. doi: 10.1093/nar/gkq890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tang H., Massi D., Hemmings B.A., Mandalà M., Hu Z., Wicki A., et al. AKT-ions with a TWIST between EMT and MET. Oncotarget. 2016;7:62767–62777. doi: 10.18632/oncotarget.11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Caglar H.O., Biray Avci C. Alterations of cell cycle genes in cancer: unmasking the role of cancer stem cells. Mol. Biol. Rep. 2020;47:3065–3076. doi: 10.1007/s11033-020-05341-6. [DOI] [PubMed] [Google Scholar]
- 148.Jamasbi E., Hamelian M., Hossain M.A., Varmira K. The cell cycle, cancer development and therapy. Mol. Biol. Rep. 2022;49:10875–10883. doi: 10.1007/s11033-022-07788-1. [DOI] [PubMed] [Google Scholar]
- 149.Doorbar J., Egawa N., Griffin H., Kranjec C., Murakami I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015;25(Suppl 1):2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu P.-F., Chen C.-F., Shu C.-W., Chang H.-M., Lee C.-H., Liou H.-H., et al. UBE2C is a potential biomarker for tumorigenesis and prognosis in tongue squamous cell carcinoma. Diagnostics. 2020;10:674. doi: 10.3390/diagnostics10090674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Du R., Huang C., Liu K., Li X., Dong Z. Targeting AURKA in Cancer: molecular mechanisms and opportunities for Cancer therapy. Mol. Cancer. 2021;20:15. doi: 10.1186/s12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jiang A., Zhou Y., Gong W., Pan X., Gan X., Wu Z., et al. CCNA2 as an immunological biomarker encompassing tumor microenvironment and therapeutic response in multiple cancer types. Oxid. Med. Cell. Longev. 2022;2022 doi: 10.1155/2022/5910575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Soboleva A., Arutyunyan I., Jumaniyazova E., Vishnyakova P., Zarubina D., Nimatov E., et al. Gene-expression patterns of tumor and peritumor tissues of smoking and non-smoking HPV-negative patients with head and neck squamous cell carcinoma. Biomedicines. 2024;12:696. doi: 10.3390/biomedicines12030696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Symington L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chatterjee S., Starrett G.J. Microhomology-mediated repair machinery and its relationship with HPV-mediated oncogenesis. J. Med. Virol. 2024;96 doi: 10.1002/jmv.29674. [DOI] [PubMed] [Google Scholar]
- 156.Day R.S., McDade K.K., Chandran U.R., Lisovich A., Conrads T.P., Hood B.L., et al. Identifier mapping performance for integrating transcriptomics and proteomics experimental results. BMC Bioinf. 2011;12:213. doi: 10.1186/1471-2105-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang K.C., Gorski S.M. Protocol for analysis of RNA-sequencing and proteome profiling data for subgroup identification and comparison. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins∗. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. M111.014050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kosti I., Jain N., Aran D., Butte A.J., Sirota M. Cross-tissue analysis of gene and protein expression in normal and cancer tissues. Sci. Rep. 2016;6 doi: 10.1038/srep24799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 161.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357 doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 162.Uhlén M., Björling E., Agaton C., Szigyarto C.A.-K., Amini B., Andersen E., et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics MCP. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 163.Doorbar J. Model systems of human papillomavirus-associated disease. J. Pathol. 2016;238:166–179. doi: 10.1002/path.4656. [DOI] [PubMed] [Google Scholar]
- 164.De Gregorio V., Urciuolo F., Netti P.A., Imparato G. In vitro organotypic systems to model tumor microenvironment in human papillomavirus (HPV)-Related cancers. Cancers. 2020;12:1150. doi: 10.3390/cancers12051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Roberts S., Evans D., Mehanna H., Parish J.L. Modelling human papillomavirus biology in oropharyngeal keratinocytes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019;374 doi: 10.1098/rstb.2018.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bristol M.L., James C.D., Wang X., Fontan C.T., Morgan I.M. Estrogen attenuates the growth of human papillomavirus-positive epithelial cells. mSphere. 2020;5 doi: 10.1128/mSphere.00049-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu X., He T., Tong Z., Liao L., Huang S., Fakhouri W.D., et al. Molecular mechanisms of TWIST1-regulated transcription in EMT and cancer metastasis. EMBO Rep. 2023;24 doi: 10.15252/embr.202356902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989 Dec 1;8(12):3905-10. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed]
- 169.Costello L.C., Franklin R.B. The genetic/metabolic transformation concept of carcinogenesis. Cancer Metastasis Rev. 2012;31:123–130. doi: 10.1007/s10555-011-9334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Alcocer-González J.M., Berumen J., Taméz-Guerra R., Bermúdez-Morales V., Peralta-Zaragoza O., Hernández-Pando R., et al. In vivo expression of immunosuppressive cytokines in human papillomavirus-transformed cervical cancer cells. Viral Immunol. 2006;19:481–491. doi: 10.1089/vim.2006.19.481. [DOI] [PubMed] [Google Scholar]
- 171.Hernández-Monge J., Garay E., Raya-Sandino A., Vargas-Sierra O., Díaz-Chávez J., Popoca-Cuaya M., et al. Papillomavirus E6 oncoprotein up-regulates occludin and ZO-2 expression in ovariectomized mice epidermis. Exp. Cell Res. 2013;319:2588–2603. doi: 10.1016/j.yexcr.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 172.Zhang W.-N., Li W., Wang X.-L., Hu Z., Zhu D., Ding W.-C., et al. CLDN1 expression in cervical cancer cells is related to tumor invasion and metastasis. Oncotarget. 2016;7:87449–87461. doi: 10.18632/oncotarget.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zihni C., Mills C., Matter K., Balda M.S. Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016;17:564–580. doi: 10.1038/nrm.2016.80. [DOI] [PubMed] [Google Scholar]
- 174.Peh W.L., Middleton K., Christensen N., Nicholls P., Egawa K., Sotlar K., et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 2002;76:10401–10416. doi: 10.1128/jvi.76.20.10401-10416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McBride A.A., White E.A. HPV integration can drive the formation of virus-host extrachromosomal DNA in tumors. Cancer Discov. 2023;13:814–816. doi: 10.1158/2159-8290.CD-23-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Warburton A., Della Fera A.N., McBride A.A. Dangerous liaisons: long-term replication with an extrachromosomal HPV genome. Viruses. 2021;13:1846. doi: 10.3390/v13091846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Khurana S, Markowitz TE, Kabat J, McBride AA. Spatial and Functional Organization of Human Papillomavirus Replication Foci in the Productive Stage of Infection. mBio. 2021 Dec 21;12(6):e0268421. doi: 10.1128/mBio.02684-21. Epub 2021 Nov 9. [DOI] [PMC free article] [PubMed]
- 178.Mk J., K S., Aa M. Papillomavirus genomes associate with BRD4 to replicate at fragile sites in the host genome. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Boteva L., Nozawa R.-S., Naughton C., Samejima K., Earnshaw W.C., Gilbert N. Common fragile sites are characterized by faulty condensin loading after replication stress. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Anacker D.C., Moody C.A. Modulation of the DNA damage response during the life cycle of human papillomaviruses. Virus Res. 2017;231:41–49. doi: 10.1016/j.virusres.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bienkowska-Haba M., Luszczek W., Myers J.E., Keiffer T.R., DiGiuseppe S., Polk P., et al. A new cell culture model to genetically dissect the complete human papillomavirus life cycle. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Woodby B., Scott M., Bodily J. In: Pruitt K., editor. vol. 144. Academic Press; 2016. Chapter five - the interaction between human papillomaviruses and the stromal microenvironment; pp. 169–238. (Prog. Mol. Biol. Transl. Sci.). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Following the 2023 NIH data management and sharing policy, all data resulting from the development of projects will be available in scientific communications presented at conferences and in manuscripts that will be published in peer-reviewed scientific journals. Data will be deposited in the Open Science Framework (OSF) platform. OSF can be accessed at https://osf.io. VCU is an OSF institutional member, and OSF is an approved generalist repository for the 2023 NIH data management and sharing policy.
Data will be made available on request.