Abstract
Dementia refers to an umbrella phenotype of many different underlying pathologies with Alzheimer’s disease (AD) being the most common type. Neuropathological examination remains the gold standard for accurate AD diagnosis, however, most that we know about AD genetics is based on Genome-Wide Association Studies (GWAS) of clinically defined AD. Such studies have identified multiple AD susceptibility variants with a significant portion of the heritability unexplained and highlighting the phenotypic and genetic heterogeneity of the clinically defined entity. Furthermore, despite women’s increased susceptibility to dementia, there is a lack of sex-specific genetic studies and understanding of sex-specific background for the disorder. Here, we aim to tackle the heterogeneity of AD by specifically concentrating on neuropathological features and pursuing sex-specific analysis. We bring together 14 different genomic and neuropathology datasets (6960 individuals) and we integrate our GWAS findings with transcriptomic and phenotypic data aiming to also identify biomarkers for AD progression. We uncover novel genetic associations to AD neuropathology, including BIN1 and OPCML. Our sex-specific analysis points to a role for BIN1 specifically in women as well as novel AD loci including QRFPR and SGCZ. Post-GWAS analyses illuminate the functional and biological mechanisms underlying AD and reveal sex-specific differences. Finally, through PheWAS and Mendelian Randomization analysis, we identify causal links with AD neuropathology pointing to disrupted lipid metabolism, as well as impaired peripheral immune response and liver dysfunction as part of a vicious cycle that fuels neurodegeneration.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40478-024-01909-6.
Keywords: Alzheimer;s disease, Neuropathology, Genomewide association study, Sex-specific analysis
Introduction
Dementia, characterized as a persistent acquired disorder of mental processes involving memory problems, personality shifts, and impaired reasoning, ranks among the most prevalent age-related illnesses worldwide and is associated with great public health burden and costs [1]. While Alzheimer's disease (AD) is the most widespread form, other types of dementia, like vascular dementia, Lewy body dementia, and frontotemporal dementia also exist, sharing common clinical features. Thus, accurate clinical diagnosis of the specific type of dementia is challenging because multiple pathologies can give rise to similar clinical syndromes [2, 3]. AD diagnosis based on cognitive function assessments carries an approximate 24% misdiagnosis rate while neuropathological findings provide a more accurate approach [4]. This challenge also hampers genetic studies which are largely based on clinically defined AD and leave a large portion of AD heritability still unexplained [5]. Furthermore, despite higher dementia risk in women, genomic studies that investigate sex-specific genetic background are lacking. These challenges and lack of related studies leave a critical gap in our efforts to understand and tackle the clinical and sex-specific heterogeneity of dementia with a goal to drive accurate diagnosis and effective patient management.
Genome-wide Association Studies (GWAS) based on clinical AD diagnosis have led to the identification of more than 70 AD susceptibility common variants and rare genetic factors [6]. However, such studies are hampered by inclusion of pre-clinical patients in the control group and the use of a pathologically heterogeneous disease phenotype [7] which may partly explain the large portion of heritability that remains unaccounted for. Furthermore, the extent to which identified variants are risk factors for AD pathology, coexisting pathologies, or other neurobiological indices is unclear [8, 9] limiting the potential use of GWAS findings to guide drug design for AD and inform clinical trials. In fact, Farfel et al. [10], showed that many recently discovered genomic variants for AD dementia are not associated with the pathology of AD. Through neuropathological examinations, such as post-mortem brain analyses, it is possible to uncover distinctive brain abnormality patterns associated with AD, marked by the presence of neuritic plaques and tau neurofibrillary tangles. Demonstrating the power of this approach and despite using a much smaller sample than traditional GWAS, Beecham et al. [11] were able to confirm association to APOE to common AD pathologies. There is thus a need to further extend GWAS based on neuropathologically-confirmed AD.
Another factor to take into consideration in AD genomic studies is the importance of studying sex-specific differences given the observed prevalence and progression of the disease in men versus women. Women are more likely to develop AD than men, and they also exhibit more tau protein tangles in their brains, leading to faster cognitive decline compared to men [12]. Dumitrescu et al. [13] performed a sex-stratified GWAS on AD neuropathology measurements in a sample of 2,701 males and 3,275 females, the majority of whom were diagnosed with AD at autopsy. They found that, outside of the APOE region, one locus on chromosome 7 (rs34331204) showed a sex-specific association with binary NP score among males but not females, implicating a novel locus that confers male-specific protection from tau pathology. Such studies highlight the value of assessing genetic associations in a sex-specific manner.
Although post-mortem studies ensure an accurate AD diagnosis and can help tackle the heterogeneity of AD, biomarkers are needed in order to move towards risk prediction or early diagnosis and early intervention that would prevent or delay symptoms. Recent work has shown that dementia-associated pathological changes may start 20–30 years before clinical onset [14–16] and newer AD drugs are being tested on pre-symptomatic participants aiming to halt or slow down cognitive decline before substantial damage has been done to the brain. There is thus an urgent need to predict early pre-symptomatic individuals as well as aim to differentiate towards specific neuropathology that leads to cognitive decline.
Here, we tackled the phenotypic and sex-specific heterogeneity presenting a large-scale integrative analysis investigating the genomic background of neuropathologically-confirmed AD (ncAD) as well as continuous measures of Braak stage and NP score. Through neuropathology-based GWAS, sex-specific analysis and multi-omics approaches we aimed to help disentangle the heterogeneous nature of the dementia clinical phenotype to help unravel the complex pathways that lead to neurodegeneration and set the foundation for targeted therapies. Our work uncovered genes that underlie AD neuropathology and revealed insights into a vicious cycle that fuels neurodegeneration through impaired lipid metabolism, immune response, and liver dysfunction.
Methods
Datasets
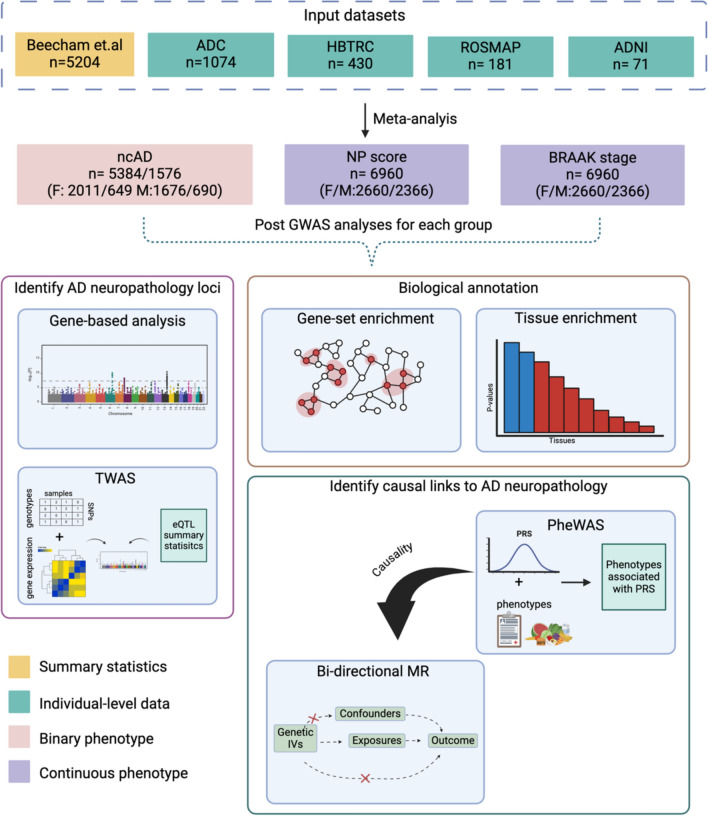
Figure 1 illustrates the complete workflow of our analysis, from data integration to data analysis. We analyze a sample of 6,960 individuals integrating 14 large-scale genetic, clinical, and neuropathology datasets from multiple sources (Supplementary Table S1 for full details). We included summary statistics datasets as described in Beecham et al. [11] along with additional individual-level datasets, to increase the sample size compared to the prior Alzheimer's Disease Genetics Consortium (ADGC) study (full details provided in Supplementary Table S1). We expanded datasets by 1,756 samples with neuropathology assessments, including collaborative studies within ADGC, namely: (1) Alzheimer’s Disease Center (ADC) (with an increased sample size 1074), and (2) Religious Orders Study and Memory and Aging Project (ROSMAP) (with an increased sample size by 181 samples). Furthermore, we included data from The Harvard Brain Tissue Resource Center (HBTRC) study (N = 430) and the Alzheimer's Disease Neuroimaging Initiative (N = 71). The assessment of neuropathological changes on the included samples is described with more details in Supplementary Methods.
Fig. 1.
illustrates the comprehensive workflow of our study, outlining each key step from data integration to final analyses. We analyzed a total of 6,960 individuals by integrating data from 14 large-scale genetic, clinical, and neuropathology datasets, expanding upon previous Alzheimer’s Disease Genetics Consortium (ADGC) studies (Supplementary Table S1). These datasets included neuropathology assessments from the Alzheimer’s Disease Center (ADC), Religious Orders Study and Memory and Aging Project (ROSMAP), The Harvard Brain Tissue Resource Center (HBTRC), and the Alzheimer’s Disease Neuroimaging Initiative (ADNI), contributing 1756 additional samples. We conducted genome-wide association studies (GWAS) on three phenotypes: neuropathology-confirmed Alzheimer’s disease (ncAD), Braak stage, and NP score, applying stringent quality control procedures for both genotypes and variants. We performed sex-specific GWAS meta-analyses on 2660 females and 2366 males and conducted post-GWAS analyses using gene-based and gene-set approaches with FUMA and MAGMA, examining tissue specificity and gene set enrichment. Furthermore, transcriptome-wide association studies (TWAS) were performed across 13 GTEx v8 brain tissues, using the Joint-Tissue Imputation (JTI) method to identify gene regulation patterns. In addition, we conducted a PheWAS-based analysis using polygenic risk scores (PRS) to explore associations with 2248 UK Biobank phenotypes and performed Mendelian Randomization (MR) analysis to assess causal relationships between neuropathology traits and blood biomarkers from UK Biobank. Replication of significant findings was carried out using independent datasets. *ncAD: neuropathology-confirmed AD
Genomewide association studies
Genotyping and quality control process
The genotyping platforms that were used to assay samples in each cohort can be found in Supplementary Table S1. Standard quality control per dataset was performed as described previously [17]. Briefly, samples with call rate < 98%, heterozygosity rate > 0.2, genomic sex discrepancy with reported sex, and formation of pairs with relatedness (pi-hat) > 0.4, were excluded from the downstream analyses. Variant-level quality control was performed to exclude markers with call rate < 95%, and Hardy–Weinberg equilibrium p-value < 10−6. To identify samples with European ancestry, Principal Component Analysis was performed with EIGENSTRAT [18] using 1000 Genomes as reference. Imputation on each dataset was performed via IMPUTE2 with 1000 Genomes as reference panel [19] using data phased by SHAPEIT [20].
Genetic association, meta-analysis
In each dataset association tests were performed for three phenotypes: ncAD case/control (binary), Braak stage (ordinal), and the NP score (ordinal) through PLINK [21] using the appropriate regression model (logistic for the binary phenotype and linear for the ordinal) and including the first three Principal Components (PCs) based on inspection of the data, age at death and sex as covariates. Only variants with minor allele frequency > 0.01 and info score > 0.7 (imputation quality metric) were included in the analyses. Following all quality controls in our study of AD neuropathological traits, a final meta-analysis was conducted using a total of 6960 samples (Supplementary Table S1). A fixed-effects meta-analysis was conducted using METAL, using the analytical strategy suggested by METAL authors, due to the unequal case–control ratios and study sample [22]. Variants with heterogeneity (Cochran’s Q test p < 0.05) and those present in less than half of the subjects were excluded.
Sex-specific GWAS
We performed sex-specific GWAS meta-analysis of 2660 females and 2,366 males integrating seven datasets that also had sex information available for part of their samples (Supplementary Table S2). We used the approach that was described earlier here (Methods). The sex specific GWAS for the three phenotypes was performed through PLINK including the first three PCs and age at death as covariates. For top hits, we also used GWAMA to examine heterogeneity in allelic effects between males and females, equivalent to testing genotype-sex interactions under an additive model [23].
Post GWAS analyses
Gene-based and gene-set GWAS analyses
Gene-based analyses were conducted within FUMA [24] using MAGMA [25], with the 1000 Genomes dataset as a reference, and including gene density and gene size as covariates. The significance level was calculated after Bonferroni correction accounting for the tested genes. Tissue specificity analysis was performed using MAGMA with default parameters, incorporating gene expression data from GTEx v8 RNA-seq for each tissue [26], with significance set at P < 1.67 × 10–3 (after Bonferroni correction for 30 tissues tested) and P < 9.43 × 10–4 (after Bonferroni correction for 54 tissues tested). We further used MAGMA to perform gene set analyses interrogating the “GO terms” from Msigdb v7.0. Pbon < 0.05 was set as the significance threshold for gene-set analysis accounting for multiple-tests.
Transcriptome-wide association study (TWAS)
TWAS was performed using the Joint-Tissue Imputation (JTI) method [27] and with a goal of identifying genes regulated by disease-associated variants on 13 GTEx v8 brain tissues. We combined TWAS p-values from multiple tissues using the Aggregated Cauchy Association Test (ACAT) method [28] and performed the Bonferroni method to control for multiple tests.
Identification of in biomarkers for AD
Phenome-wide association analysis (PheWAS)
In order to explore additional phenotypes associated with AD genetic risk, we performed a PRS-PheWAS analysis on UK Biobank (Supplementary Methods). As base for the PRS calculations we used the GWAS summary statistics of the three neuropathology-based phenotypes (ncAD, Braak stage, NP stage) that we performed on the full dataset and we repeated the analyses for each sex separately (Nfemales = 178,604; Nmales = 152,237). For the PheWAS analysis we used the PHESANT tool [29] to test for PRS association on 2248 UK Biobank phenotypes (Supplementary Table S3), adjusting for the appropriate covariates (Supplementary Methods) and used FDR correction to determine the significant associations.
PheWAS-based on mendelian randomization (MR) analysis of blood biomarkers
We used a standard approach for two-sample MR analysis to examine the potential causal relationship between AD neuropathology traits and PheWAS significant blood assays traits in UK biobank. As exposure variables we considered SNPs with p < 10−5 in our AD neuropathological features GWAS (ncAD, Braak stage, NP stage) and the GWAS of blood assays from the UK Biobank as the outcome. Using multiple MR methods to triangulate findings provides the strongest support for causal inference. In our study, the IVW method served as the primary analysis, with the other methods (Weighted median, MR-EGGER) used for sensitivity assessments. FDR correction was applied to account for multiple testing. To validate the significant associations identified in our analyses, we used independent GWAS datasets of blood assays [30–33] as outcomes, excluding UK Biobank participants, and repeated the MR analyses. For full details, refer to the Supplementary Methods, and see Supplementary Table 4 for information on the datasets.
Results
Genome-wide association studies of AD neuropathology
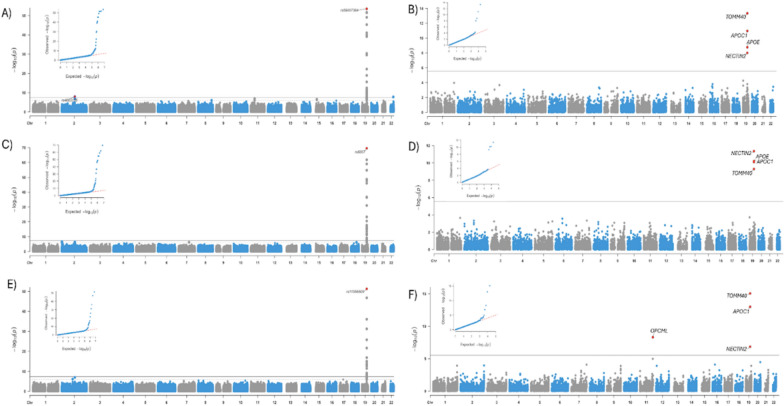
First, we integrated 14 large-scale genetic, clinical, and neuropathology datasets from multiple sources and conducted GWAS meta-analyses for three neuropathology-based phenotypes: ncAD (neuropathology-confirmed AD) on a total of 5384 cases and 1576 controls, Braak stage, and NP stage on 6960 individuals (see Methods as well as Supplementary Materials, Supplementary Table S1). We identified two genomewide significant loci associated to AD neuropathology (see Table 1, Supplementary Fig. 2 for regional plots and Supplementary Fig. 3 for forest plots). The top and only locus shared by the three GWAS that we performed was 19q13.32 near the APOE region (spanning TOMM40, APOE, NECTIN2 and APOC1) (Table 1, Fig. 2). The second genome-wide significant locus shared by case–control ncAD GWAS was on chromosome 2q14 on the BIN1 gene (Table 1, Fig. 2, Supplementary Figure S2 for regional plot).
Table 1.
Genome-wide association study of neuropathology-based AD traits
| Locus | CHR:POS | Top SNP ID | AD traits | A1 | P | Effect | Nearest Gene Position (distance) | MAF |
|---|---|---|---|---|---|---|---|---|
| 2q14.3 | chr2:127,133,851 | rs4663105 | ncAD | C | 7.94 × 10–9 | − 5.77 | BIN1b/intergenic (26,495) | 0.4 |
| chr19:44,893,408 | rs59007384 | ncAD | T | 3.9 × 10−31 | 15.52 | APOE regiona/intronic | 0.30 | |
| 19q13.32 | chr19:44,888,997 | rs6857 | Braak stage | T | 2.7 × 10−70 | 17.73 | APOE regiona/intronic | 0.25 |
| chr19:44,892,887 | rs11556505 | NP score | T | 5.2 × 10−52 | 15.18 | APOE regiona/intronic | 0.22 |
The total sample for the ncAD case–control GWAS was 5384 cases and 1576 controls on 6,394,125 SNPs. The total sample for Braak stage and NP score GWASs was 6960 individuals on 6,542,713 and 6,475,755 SNPs respectively. CHR chromosome; SNP, single-nucleotide polymorphism; A1, effect allele; MAF: minor allele frequency; Effect: Z score effect of A1 allele; Significant genome-wide association P < 5 × 10−8 a: Genes have been identified as genome-wide significant in both previous clinical AD GWAS and neuropathological AD GWAS. b: Genes have been identified as genome-wide significant in previous clinical AD GWAS
ncAD: neuropathology-confirmed AD
Fig. 2.
Manhattan and QQ plots of SNP-based and Gene-Based genome-wide association results of neuropathological features of AD (n = 6960). Dotted red lines represent the threshold for genome-wide significance (P < 5 × 10−8) and Bonferroni correction for the gene-based analyses. A Neuropathologically-confirmed AD case–control GWAS. B Gene-based analysis for neuropathologically confirmed AD case–control sample. C Braak stage GWAS. D Gene-based analysis for Braak stage. E NP score GWAS. F Gene-based analysis for NP score
In the gene-based analyses, the TOMM40, NECTIN2, and APOC1 genes were significantly associated with all three studied neuropathology-based AD-related phenotypes (Fig. 2B, D, F). The APOE gene was also significant in the ncAD and Braak stage meta-analysis (Fig. 2B, D), while one novel gene, OPCML, outside of the 19q13.32 region was significant in the NP score GWAS (Fig. 2F).
Sex-specific genome-wide association studies of AD neuropathology
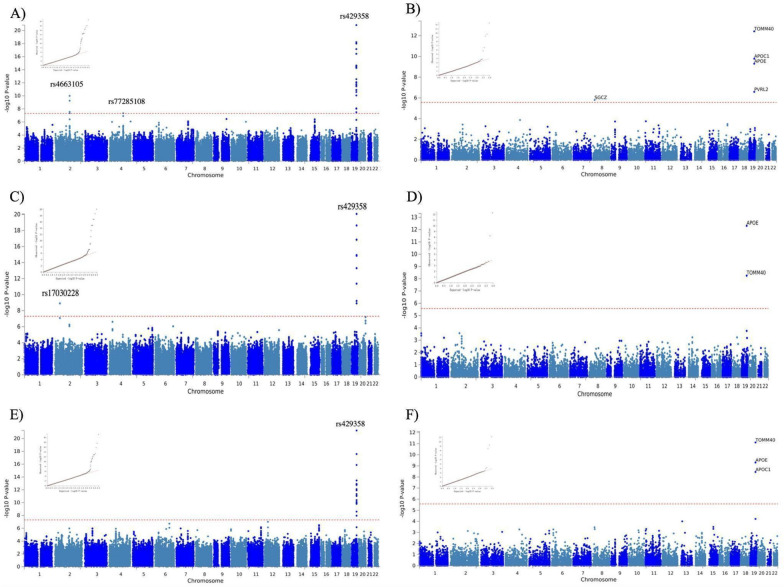
Next, aiming to investigate sex-specific genetic associations with AD neuropathology traits, we performed sex-stratified GWAS after merging seven datasets with available sex information (Supplementary Table S2 and Fig. S6 forest plots). This resulted in a total of 2366 males and 2660 females. As expected, for both sexes, the top locus in all three AD neuropathology phenotypes that we studied (ncAD, Braak stage, NP score) was 19q13.32 with the top SNPs located in the APOE region (Table 2, Fig. 3, Supplementary Figure S1). No additional significant loci were identified in the male specific GWAS for any of the three phenotypes (Table 2). However, SNPs at two additional loci exceeded the genomewide significance level in the female-specific GWAS for Braak stage and ncAD: 2q14 close to the BIN1 gene and 4q27 close to QRFPR were significantly associated with ncAD in females (top SNPs: (2q14) rs4663105, p-value = 1.01 × 10–10; (4q27) rs77285108, p-value = 1.23 × 10–9) (Fig. 3C, E, Supplementary Figure S5A-B for regional plots). Furthermore, rs17030228 close to the LOC102723854 was significant in the female-specific Braak stage GWAS (top SNP:rs17030228, p-value = 8.5 × 10–8) (Table 2, Fig. 3C and Supplementary Figure S5C for regional plot). The sex heterogeneity test revealed that the effects of rs4663105, rs17030228 and rs77285108 were significantly different between males and females (all sex heterogeneity p-value < 0.05 and Supplementary Table S5). In sex-specific gene-based analysis, several genes in the APOE region (APOC1 TOMM40, PVRL2) were significant for all three AD neuropathology traits for both sexes, while the SGCZ gene was significant in female specific ncAD only (Fig. 3B, D, F, Supplementary Figure S1B, S1D, S1F).
Table 2.
Sex-specific genome-wide association study of neuropathology-based AD traits
| Locus | CHR(POS) | SNP ID | AD traits | A1 | Female only p-value | Female only Effect size | Male only p-value | Male only Effect size | Nearest Gene Position (dis) | MAF |
|---|---|---|---|---|---|---|---|---|---|---|
| 2q14 | chr2:127,133,851 | rs4663105 | ncAD | C | 1.01 × 10–10 | − 6.47 | 7.5 × 10–4 | − 3.37 | BIN1b/intergenic (26,495) | 0.4 |
| 2q | chr2:42,953,187 | rs17030228 | Braak stage | A | 1.23 × 10–9 | − 6.08 | 0.49 | − 0.17 | AC016735.1 lncRNAsc/intergenic (48,167) | 0.012 |
| 4q27 | chr4:121,361,613 | rs77285108 | ncAD | G | 4.67 × 10–8 | 5.46 | 0.52 | − 0.65 | QRFPRc/intronic | 0.15 |
| 19q13.32 | chr19:44,908,684 | rs429358 | ncAD | C | 1.50 × 10–21 | − 9.54 | 8.65 × 10–32 | − 11.73 | APOE regiona/exonic | 0.16 |
| chr19:44,908,684 | rs429358 | Braak stage | C | 8.50 × 10–21 | − 9.35 | 1.96 × 10–34 | − 12.24 | APOE regiona/exonic | 0.16 | |
| chr19:44,908,684 | rs429358 | NP score | C | 5.65 × 10–22 | − 9.64 | 1.08 × 10–27 | − 10.91 | APOE regiona/exonic | 0.16 |
The total sample for the ncAD female case–control GWAS was 2660 (2011 case + 649 control) and male was 2366 (1676 case + 690 control) on 7,045,108 and 7,075,025 SNPs respectively. The total sample for female Braak stage GWASs and female NP score GWASs was 2660 on 7,083,498 and 61,890,055 SNPs respectively. The total sample for male Braak stage GWASs and male NP score GWASs was 2366 individuals on 7,153,692 and 6,475,237. CHR chromosome; SNP, single-nucleotide polymorphism; A1, effect allele; MAF: minor allele frequency; Effect: Z score effect of A1 allele; Significant genome-wide association P < 5 × 10−8.a: Genes have been identified as genome-wide significant in both previous clinical AD GWAS and neuropathological AD GWAS. b: Genes have been identified as genome-wide significant in previous clinical AD GWAS. c: Genes have not been identified as genome-wide significant in previous studies
ncAD neuropathology-confirmed AD
Fig. 3.
Manhattan and QQ plots of SNP-based and Gene-Based female-specific GWAS results of neuropathological features of AD (n = 2660). Dotted red lines represent the threshold for genomewide significance (P < 5 × 10−8) and Bonferroni correction for the gene-based analyses. A ncAD female-specific GWAS. B Gene-based analysis for ncAD female-specific case–control sample. C Braak stage female-specific GWAS. D Gene-based analysis for Braak stage in females. E NP score female-specific GWAS. F Gene-based analysis for NP score in females
Post-GWAS analysis for AD neuropathology
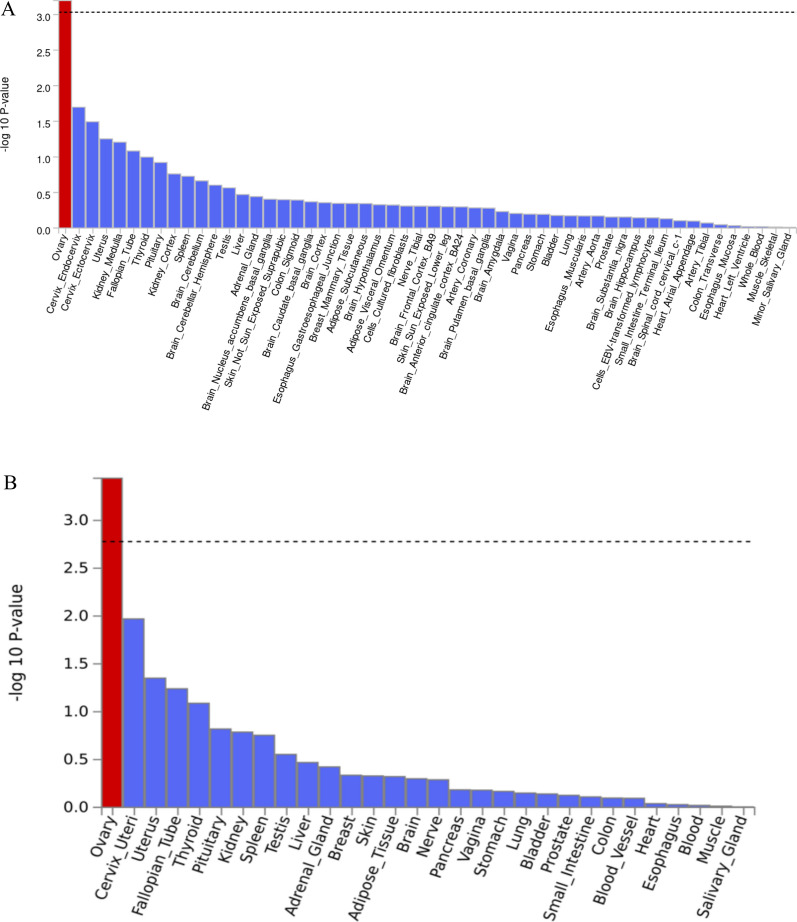
To identify tissue specificity of our AD neuropathology GWAS findings, we performed tissue enrichment analysis by MAGMA (Supplementary Figure S4 and S7). Notably, a significant association was observed between NP score in females and genes expressed in ovary tissue across both the 54- and 30-tissue enrichment analyses (Fig. 4). In our gene-set analysis, we found several significant enrichments: In males, the NP score showed enrichment for “positive regulation of mitochondrial calcium ion concentration” (Pbon = 3.6 × 10–2), while for male ncAD, "histone pre mRNA 3' end processing complex" (Pbon = 3.5 × 10–2) was significant. In females, the NP score revealed "regulation of luteinizing hormone secretion" (Pbon = 1.2 × 10–5) and "positive regulation of gonadotropin secretion" (Pbon = 1.7 × 10–3). Additionally, in females, "positive regulation of receptor catabolic process" (Pbon = 2.6 × 10–2) and "fibroblast migration" (Pbon = 2.2 × 10–2) were enriched for Braak stage and case–control analysis, respectively. (Supplementary Table S6).
Fig. 4.
Post-GWAS analysis results for Tissue enrichment analysis. Tissue enrichment analysis for NP score GWAS results in females. The analysis was performed in MAGMA using GTEx v8 RNA-seq data 54 and 30 general tissue types. With red are shown the significant results after multiple testing corrections. A MAGMA tissue expression analysis using gene expression per tissue based on GTEx RNA-seq data for 54 specific tissue types. B MAGMA tissue expression analysis using gene expression per tissue based on GTEx RNA-seq data for 30 specific tissue types
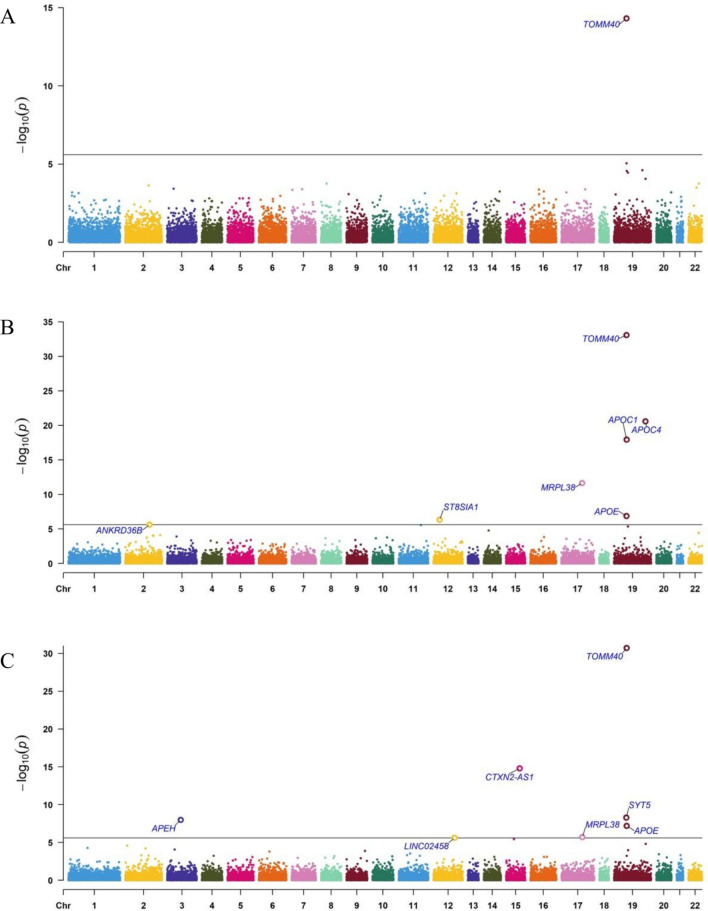
To identify candidate genes whose genetically regulated expression is associated with neuropathological features of AD, we conducted TWAS (see Table 3 and Fig. 5). This analysis identified 11 protein-coding and two significant long non-coding RNA gene hits whose transcript expression was significantly associated with neuropathological features of AD. Notably, TWAS identified six novel loci including genes (ST8SIA1 p-value = 4.69 × 10–7; ANKRD36B p-value = 3.38 × 10–6; MRPL38 p-value = 2.34 × 10–12 and 1.97 × 10–6, APEH p-value = 1.03 × 10–8; CTXN2-AS1 p-value = 1.60 × 10–15; LINC02458 p-value = 2.41 × 10–6) and one novel gene in the APOE region (SYT5 p-value = 5.29 × 10–9). These genes have not been implicated in previous AD-related GWAS or TWAS and are novel findings of this study.
Table 3.
Transcriptome-wide association study of neuropathology-based AD traits
| GWAS summary | Gene | Region | ACAT P | Leading tissues |
|---|---|---|---|---|
| Braak Stage | ST8SIA1 c | 12p12.1 | 4.69 × 10–7 | Brain_Cerebellar_Hemisphere, Brain_Cerebellum |
| APOC1 a | 19q13.32 | 1.18 × 10–18 | Nucleus_accumbens_basal_ganglia | |
| APOE a | 19q13.32 | 1.37 × 10–7 | Brain_Caudate_basal_ganglia, | |
| TOMM40 a | 19q12.32 | 8.33 × 10–34 | Pituitary | |
| ANKRD36B c | 2q11.2 | 3.38 × 10–6 |
Brain_Cortex Brain_Hippocampus Nucleus_accumbens_basal_ganglia Brain_Spinal_cord_cervical_c-1 Brain_Substantia_nigra |
|
| MRPL38 c | 17q25.1 | 2.34 × 10–12 | Brain_Hypothalamus | |
| APOC4 a | 19q13.31 | 2.67 × 10–21 |
Brain_Caudate_basal_ganglia, Brain_Hypothalamus Nucleus_accumbens_basal_ganglia |
|
| ncAD | TOMM40 a | 19q13.32 | 5.43 × 10–16 | Pituitary |
| NP score | SYT5 c | 19q13.42 | 5.29 × 10–9 | Brain_Cerebellar_Hemisphere |
| APOE a | 19q13.32 | 6.51 × 10–8 | Brain_Caudate_basal_ganglia | |
| TOMM40a | 19q13.32 | 1.88 × 10–31 | Pituitary | |
| APEHc | 3p21.31 | 1.03 × 10–8 | Brain_Cerebellar_Hemisphere | |
| MRPL38c | 17q25.1 | 1.97 × 10–6 | Brain_Hypothalamus | |
| LINC02458 c | 12q21.33 | 2.41 × 10–6 | Brain_Frontal_Cortex_BA9 | |
| CTXN2-AS1 c | 15q21.1 | 1.60 × 10–15 | Pituitary |
Transcriptome-wide association study (TWAS) identifies 11 unique and 7 novel genes significantly associated with neuropathological features of AD in GTEx Brain tissues. GWAS: Genome-wide association study; TWAS: Transcriptome-wide association study; ACAT P: Aggregated Cauchy Association Test based combined TWAS p-value Bonferroni correction thresholds are p-value < 2.48 × 10–6 based on 20,205, 20,198, and 20,207 tests in Braak stage, ncAD case–control, and NP score combined JTI-ACAT TWAS analysis respectively. a: Genes have been identified as significant in previous clinical or neuropathology-based AD GWAS. b: Genes have been identified as significant in previous AD-related TWAS alone, c: Genes have not been identified as significant in previous AD GWAS or TWAS studies. * ncAD: neuropathology-confirmed AD
Fig. 5.
Transcriptome-wide association study (TWAS) for AD neuropathological features. The x-axis of Manhattan plot represents the genomic position of the corresponding gene, and the y-axis of Manhattan plot represents -log10-transformed association combined P-value using ACAT. Each dot represents the association for one specific gene. The line shows combined P value 9.71 × 10–6. A TWAS for neuropathologically confirmed AD case–control sample. B TWAS for Braak stage. C TWAS for NP scor
Investigating potentially causal links with AD neuropathology
PheWAS and PheWAS-based on MR
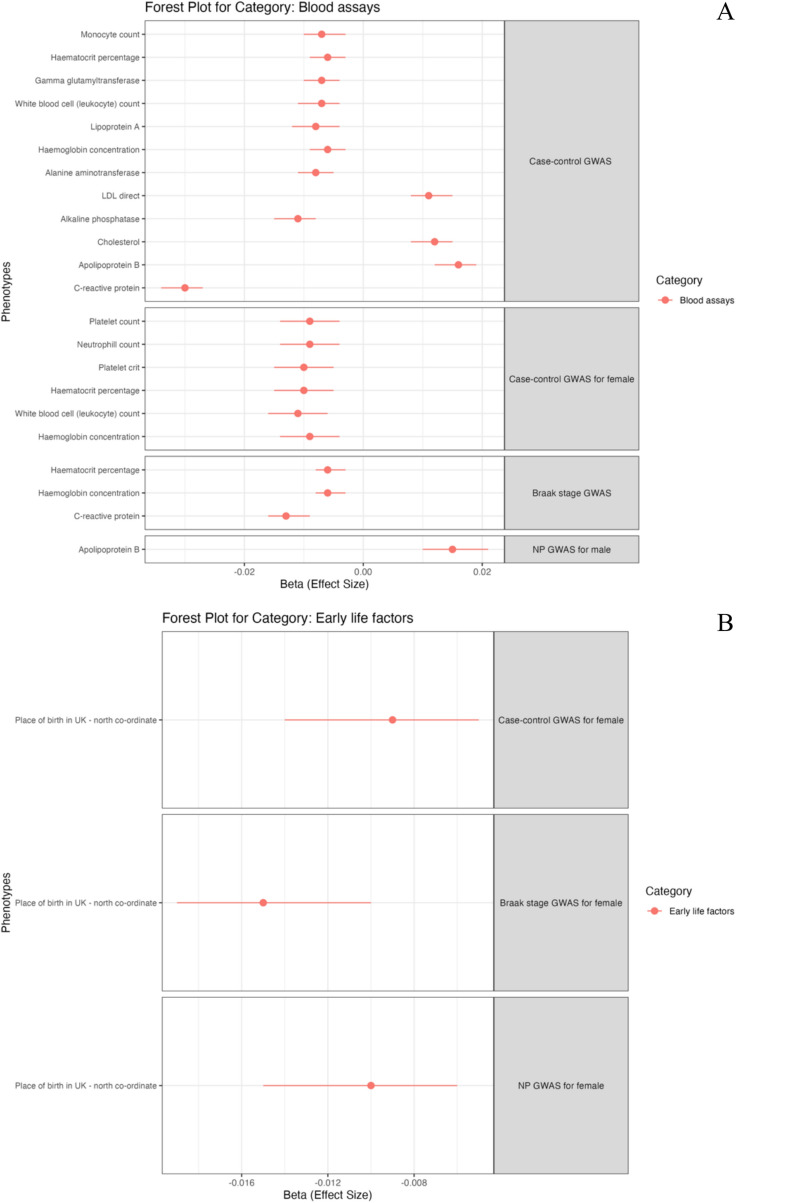
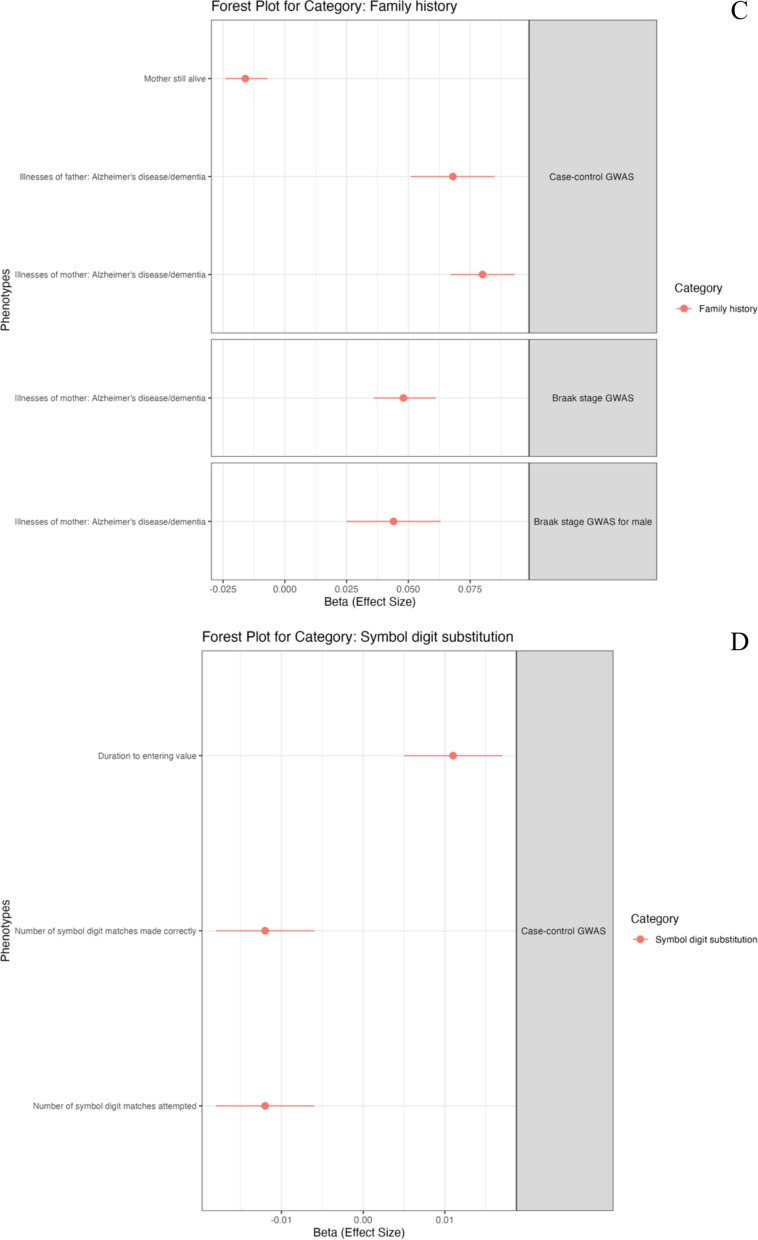
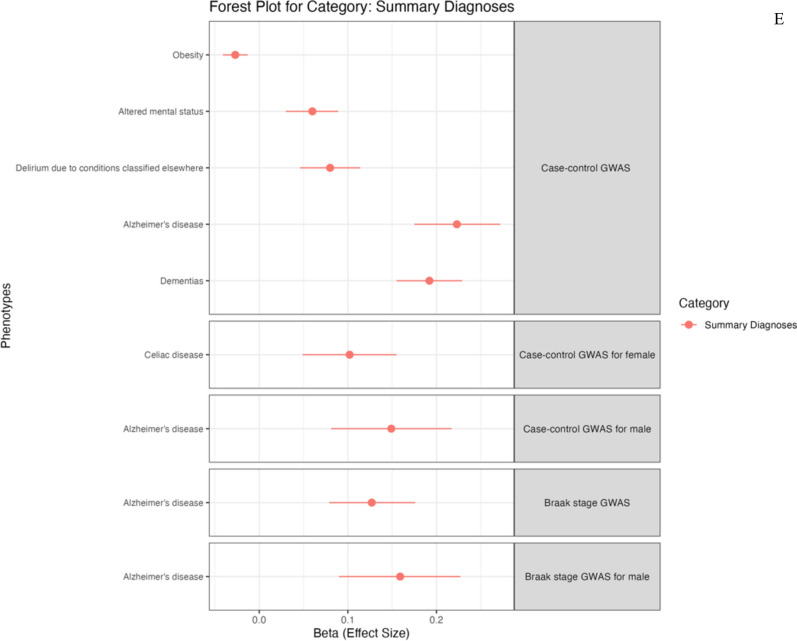
We continued to perform PheWAS to identify associations between genetic variation and phenotypic variation in European populations from the UK Biobank dataset. Our PRS-PheWAS results are presented in Fig. 6 and Supplementary Table 7. The ncAD PRS showed significant associations with 36 traits while we found 10 associations with Braak stage PRS. Interestingly, in the blood biomarkers category ncAD was associated with increased lipid metabolism (apolipoprotein B, LDL direct and cholesterol) and decreased transferase (alkaline phosphatase, alanine aminotransferase and gamma glutamyltransferase). It was also negatively associated with C-reactive protein (CRP) and blood cells counts measurements. Braak stage PRS was also associated with elevated lipid metabolism and decreased CRP. Another interesting result was the inverse relationship between ncAD PRS and obesity. In the sex-specific PheWAS analysis, the majority of associations were linked to blood biomarkers specifically in females. Among disease diagnoses, female ncAD PRS was linked to celiac disease, while male ncAD and Braak stage PRSs were associated with AD (more details in supplementary table 7 and supplementary results).
Fig. 6.
Phenome-wide association analysis (PheWAS) for PRS of neuropathology-based AD GWAS. Forest plot showing phenotypes significantly associated with TS PRS, grouped by categories The x-axis shows the (Beta) effect size for each phenotype estimated by PheWAS. A Forest plot for blood assay. B Forest plot for early life factors C Forest plot for family history. D Forest plot for cognitive function of symbol digit substitution. E Forest plot for ICD10 diagnosis summary
We proceeded to further explore the PheWAS blood assay associations investigating potential causality. To do this, we conducted a bi-directional MR analysis (Table 4). We identified a causal relationship where neuropathology, particularly in ncAD, led to changes in blood assay traits. Specifically, there were positive causal associations between ncAD and lipid metabolism markers, including cholesterol, LDL direct, and apolipoprotein B. In contrast, several negative causal relationships were found, with ncAD linked to decreased levels of CRP, alkaline phosphatase, alanine aminotransferase, and blood cell count measurements. Braak stage also showed a negative causal link with CRP and a positive causal relationship with most lipid metabolism markers. Additionally, we successfully replicated the causal associations of ncAD with cholesterol, CRP, platelet crit, LDL direct, and red cell count in independent blood assay datasets. We also replicated the causal effect of Braak stage In the reverse direction, LDL was found to directly contribute to Braak stage.on lipid metabolism markers although this could not be replicated when using independent GWAS. Sex-specific analysis did not reveal significant results. Full details of methods and results are provided in Supplementary text and Table S8.
Table 4.
Mendelian randomization analysis and replication results
| Exposure | Outcome | MR analysis | Replication | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N SNPs | Beta | SE | P value | FDR | N SNPs | Beta | SE | p value | ||
| Case–control GWAS | Alanine aminotransferase | 20 | − 0.024 | 0.012 | 0.041 | 0.057 | ||||
| Alkaline phosphatase | 20 | − 0.030 | 0.012 | 0.015 | 0.032*a | |||||
| Apolipoprotein A | 20 | − 0.068 | 0.021 | 0.012 | 0.032* | 20 | − 0.003 | 0.022 | 0.895 | |
| Apolipoprotein B | 19 | 0.126 | 0.054 | 0.019 | 0.032* | 20 | 0.064 | 0.091 | 0.483 | |
| Cholesterol | 20 | 0.082 | 0.035 | 0.019 | 0.032* | 20 | 0.096 | 0.035 | 0.006* | |
| C-reactive protein | 20 | − 0.192 | 0.064 | 0.003 | 0.032* | 20 | − 0.181 | 0.077 | 0.018* | |
| Gamma glutamyltransferase | 20 | − 0.008 | 0.008 | 0.341 | 0.38 | |||||
| Haematocrit percentage | 20 | − 0.008 | 0.008 | 0.341 | 0.39 | |||||
| Haemoglobin concentration | 20 | − 0.001 | 0.004 | 0.803 | 0.803 | |||||
| HDL cholesterol | 20 | − 0.028 | 0.014 | 0.043 | 0.050 | |||||
| LDL direct | 20 | 0.095 | 0.041 | 0.020 | 0.032* | 20 | 0.093 | 0.040 | 0.020* | |
| Monocyte count | 20 | − 0.006 | 0.008 | 0.475 | 0.50 | |||||
| Platelet count | 20 | − 0.020 | 0.008 | 0.014 | 0.032* | 20 | − 0.041 | 0.025 | 0.094 | |
| Platelet crit | 20 | − 0.025 | 0.010 | 0.014 | 0.032* | 20 | − 0.054 | 0.018 | 0.003* | |
| Red blood cell (erythrocyte) count | 20 | − 0.014 | 0.005 | 0.007 | 0.032* | 20 | − 0.004 | 0.019 | 0.845 | |
| Red blood cell (erythrocyte) distribution width | 20 | − 0.037 | 0.013 | 0.004 | 0.032* | 20 | − 0.041 | 0.015 | 0.007* | |
| Braak stage GWAS | Apolipoprotein A | 25 | − 0.061 | 0.024 | 0.012 | 0.020* | 26 | − 0.063 | 0.022 | 0.004* |
| Apolipoprotein B | 24 | 0.005 | 0.005 | 0.324 | 0.320 | |||||
| Cholesterol | 24 | 0.007 | 0.006 | 0.256 | 0.320 | |||||
| C-reactive protein | 25 | − 0.176 | 0.058 | 0.002 | 0.008* | 25 | − 0.087 | 0.069 | 0.210 | |
| LDL direct | 26 | 26.000 | 0.126 | 0.003 | 0.008* | 26 | 0.133 | 0.042 | 0.002* | |
The table presents the results of a Mendelian Randomization (MR) analysis and subsequent replication studies investigating the causal relationships between genetic risk factors (exposures) from ncAD and Braak stage GWAS and various blood biomarkers (outcomes) identified from previous PheWAS analysis. Exposure: Either ncAD GWAS or Braak stage GWAS, representing the genetic risk factors being tested. Outcome: The specific blood biomarkers identified through PheWAS analysis, which are the traits being evaluated for causal relationships with the genetic exposures. Replication: Results from replication studies, including the number of SNPs (N SNPs), beta coefficient, standard error (SE), and p-value for select outcomes that were re-tested to confirm consistency in the results. This table focuses on the primary inverse variance-weighted (IVW) analysis, with additional methods and sensitivity tests available in Supplementary Table 8. An asterisk (*) indicates a significant cutoff; The letter "a" indicates that the replicate dataset is not available
Discussion
We performed a large-scale AD-neuropathology-based GWAS, revealing sex-specific AD pathways and novel AD loci that had not been previously found in clinical AD GWAS. The BIN1 gene, which has been previously identified as associated with clinical AD [34] and had not been associated in an original AD neuropathology GWAS [11] is highlighted by our analysis. Importantly, we show, for the first time, that BIN1 is specifically associated with AD neuropathology in females and not in males. BIN1 plays a prominent role in regulating endocytosis and synaptic vesicle trafficking, and it is implicated in the generation of amyloid beta, mediation tau pathology and the propagation of Tau [35–38]. Recent sex-specific clinical GWAS and APOEε4 status GWAS for AD, also identified BIN1 as having a female-specific association [39, 40]. Another investigation, estimating hazard ratios, also showed that BIN1 contributes to a higher risk in females compared to males [41]. Moreover, GTEx RNAseq analysis has previously underscored the sex-heterogeneous effect of BIN1 in brain tissue [42]. These findings suggest that the effects of BIN1 may be sex-dependent, particularly in the context of AD neuropathology.
The different factors that operate towards progression to AD in men and women and the increased risk in women are multifactorial and both sex hormones and sex chromosomes have been implicated [43]. Our sex-specific analysis of AD neuropathology supports an important role for sex hormones. We found high expression of our top GWAS hits in the ovary and our gene-set analysis connected NP score to pathways that are related to the secretion of luteinizing hormone (LH) and gonadotropins in females. LH is a component of the Hypothalamus-Pituitary-Gonads axis and becomes dysregulated during aging, particularly in menopause. Although the potential role of estrogen [44, 45] in AD has received a lot of attention, emerging data [46] suggest an important role for luteinizing hormone in the function of the central nervous system and post-menopausal women have up to tenfold more LH than men [47, 48]. Mounting evidence suggests that such hormone changes during perimenopause contribute to female vulnerability to AD and our work here highlights this mechanism [49, 50].
Besides BIN1, which we discussed earlier here, our sex-specific analysis identified novel AD genes with female-specific association to AD neuropathology. These include genes QRPFR, SGCZ, and the long non-coding RNA (lncRNA) AC016735.1. QRPFR (GPR103) is highly expressed in the brain and acts as a receptor for the orexigenic neuropeptide, influencing the regulation of feeding behavior and circadian rhythms [51, 52]. Interestingly, intra-hippocampal administration of orexin has been shown to mitigate learning and memory impairment, highlighting its potential therapeutic role in AD [53]. Furthermore, disrupted circadian rhythms have been previously linked to AD development, further supporting a potential role of QRPFR in AD pathology [54, 55]. Additionally, QRPFR exhibits a neuroprotective effect, and its expression is reduced in AD due to amyloid-beta and tau pathology. SGCZ, another novel female-specific AD-neuropathology gene that we identified, has been shown to play a role in forming the sarcoglycan complex and exhibits gender-biased expression levels in the brain, as observed in animal models [56]. Mutations in sarcoglycanopathy can lead to protein misfolding and aggregation, which could potentially be connected to the development of AD [57]. A single-cell analysis study revealed elevated expression of SGCZ in a subset of oligodendrocytes when comparing individuals with AD to those without the condition [58]. We also found AD neuropathology gene (OPCML: Opioid-Binding Protein/Cell Adhesion Molecule) that had not been previously identified in neuropathology GWAS or the largest clinical AD GWAS. OPCML belongs to the immunoglobulin protein superfamily and contributes to synaptogenesis in the brain [59]. It has been implicated in AD based on previous GWAS studies in the Dutch population [60].
Through TWAS we also identified seven additional novel genes as regulated by ncAD GWAS variants. (ST8SIA1, ANKRD36B, MRPL38, APEH and CTXN2-AS1, SYT5, and LINC02458). Intriguingly, ST8SIA1 encodes GD3 synthase which is involved in regulating amyloid-beta plaque load [61]. APEH has been linked to endogenous beta-amyloid levels and is also associated with decreased red blood cell counts in AD patients [62, 63]. Additionally, the two lncRNA genes that are implicated by our analysis, LINC02458 and CTXN2-AS1 have been found to influence processes like amyloid beta aggregation, tau hyperphosphorylation, and the interaction of key enzymes in AD by acting as a decoy or scaffold [64]. Further biological experimentation is warranted to provide additional support for the implication of these genes in AD pathology.
Intriguingly, our MR analysis provided evidence that AD-related neuropathology leads to disruption of lipid metabolism, and increased levels of cholesterol, LDL, and apolipoprotein B. Previously, high cholesterol, and LDL have been implicated as risk factors for AD [65] and here we show evidence also for a reverse relationship, pointing to a vicious cycle that fuels neurodegeneration. We also showed that AD neuropathology can cause a decrease in CRP levels, as well as lower levels of liver enzymes, pointing to liver dysfunction, and reduced peripheral immune response that could further aggravate neurodegeneration. Lower CRP levels, an inflammatory biomarker, have been associated with a higher risk of AD in a large population study [66]. Although sex-specific MR did not reveal significant results of causality, in PheWAS we observed significant associations between blood assays, sociodemographic traits, and neuropathological feature PRS, with notable sex differences. In females, we identified negative associations between ncAD PRS and white cell, platelet, and red cell counts, aligning with previous studies that reported decreased peripheral blood cells in AD [67, 68].
Conclusions
In summary, we used a multi-omics approach centered on GWAS to explore the genetic basis of AD neuropathology, with a focus on sex-specific mechanisms and investigating potential causal links for AD. Our GWAS identified strong associations with AD neuropathology, including novel loci such as BIN1, which was highlighted as having a female-specific association. Further analyses through tissue-specificity, gene-set, TWAS, and MR provided additional insights into the role of these genetic variants, implicating key biological pathways. Notably, our findings suggest that sex-specific factors, such as hormone regulation, may play a critical role in the progression of AD, particularly in females. MR analysis further integrated GWAS findings with clinical data from PheWAS, identifying an intriguing bi-directional link to lipid metabolism. The combined approach of GWAS, TWAS, MR, and post-GWAS analyses helped us provide novel insights into the progression of AD neuropathology. Further research is warranted to further validate and understand the suggested interplay between lipid dysregulation, liver function, and inflammation in AD which may offer new therapeutic opportunities to break the cycle and slow down disease progression.
Supplementary Information
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Abbreviations
- ncAD
Neuropathologically-confirmed AD
- AD
Alzheimer’s disease AD
- GWAS
Genome-wide association studies
- ADGC
Alzheimer’s disease genetics consortium
- ADC
Alzheimer’s disease center
- ROSMAP
Religious orders study and memory and aging project
- HBTRC
Harvard Brain Tissue Resource Center
- PCs
Principal components
- TWAS
Transcriptome-wide association study
- JTI
Joint-tissue imputation
- ACAT
Aggregated Cauchy association test
- PheWAS
Phenome-wide association analysis
- MR
Mendelian randomization
- CRP
C-reactive protein
- LH
Luteinizing hormone
- lncRNA
Long non-coding RNA
- OPCML
Opioid-Binding Protein/Cell Adhesion Molecule
Author contributions
Yin conducted the full analysis, and Yin and Apostolia wrote the main manuscript. Sudhanshu and Guanxin prepared Fig. 3, while Alicia and Bryce provided feedback. The research was conducted under the supervision of Professors Paschou, Drines, and Chris. All authors reviewed the manuscript.
Funding
This work was funded by the Purdue Institute for Drug Discovery and NSF grants 1715202 and 2006929 awarded to Dr. Peristera Paschou.
Data Availability
The ADGC cohorts’ data is available at niagads (https://www.niagads.org/), while the ADNI dataset can be accessed from https://adni.loni.usc.edu/. Other data from Religious Orders Study and Memory and Aging Project, Mayo Clinic Alzheimer's Disease Research Center, and Harvard Brain Tissue Resource Center are accessible on https://www.synapse.org.
Declarations
Ethics approval and consent to participate
NA. We used public datasets to perform our analysis. The ADGC cohorts’ data is available at niagads (https://www.niagads.org/), while the ADNI dataset can be accessed from https://adni.loni.usc.edu/. Other data from Religious Orders Study and Memory and Aging Project, Mayo Clinic Alzheimer's Disease Research Center, and Harvard Brain Tissue Resource Center are accessible on https://www.synapse.org.
Consent for publication
All authors have provided their consent for the publication of this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arvanitakis Z, Shah RC, Bennett DA (2019) Diagnosis and management of dementia: review. JAMA J Am Med Assoc. 10.1001/jama.2019.4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider JA, Arvanitakis Z, Bang W, Bennett DA (2007) Mixed brain pathologies account for most dementia cases in community-dwelling older persons. Neurology. 10.1212/01.wnl.0000271090.28148.24 [DOI] [PubMed] [Google Scholar]
- 3.Jellinger KA, Attems J (2015) Challenges of multimorbidity of the aging brain: a critical update. J Neural Transm. 10.1007/s00702-014-1288-x [DOI] [PubMed] [Google Scholar]
- 4.Fischer CE, Qian W, Schweizer TA, Ismail Z, Smith EE, Millikin CP et al (2017) Determining the impact of psychosis on rates of false-positive and false-negative diagnosis in Alzheimer’s disease. Alzheimer’s Dement Transl Res Clin Interv. 10.1016/j.trci.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medway C, Morgan K (2014) Review: the genetics of Alzheimer’s disease; putting flesh on the bones. Neuropathol Appl Neurobiol 40:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N et al (2022) New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat Genet 54(4):412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnen JA, Santa Cruz K, Hemmy LS, Woltjer R, Leverenz JB, Montine KS et al (2011) Ecology of the aging human brain. Arch Neurol 68:1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett DA, Wilson RS, Boyle PA, Buchman AS, Schneider JA (2012) Relation of neuropathology to cognition in persons without cognitive impairment. Ann Neurol 72:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett DA, Schneider JA, Wilson RS, Bienias JL, Arnold SE (2004) Neurofibrillary tangles mediate the association of amyloid load with clinical alzheimer disease and level of cognitive function. Arch Neurol 61:378–384 [DOI] [PubMed] [Google Scholar]
- 10.Farfel JM, Yu L, Buchman AS, Schneider JA, De Jager PL, Bennett DA (2016) Relation of genomic variants for Alzheimer disease dementia to common neuropathologies. Neurology. 10.1212/WNL.0000000000002909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ et al (2014) Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet 10:e1004606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.2023 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 2023 [DOI] [PubMed]
- 13.Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, Bush WS et al (2019) Sex differences in the genetic predictors of Alzheimer’s pathology. Brain. 10.1093/brain/awz206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan CH, Fan CC, Mormino EC, Sugrue LP, Broce IJ, Hess CP et al (2018) Polygenic hazard score: an enrichment marker for Alzheimer’s associated amyloid and tau deposition. Acta Neuropathol 135:85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen WJ, Ossenkoppele R, Knol DL, Tijms BM, Scheltens P, Verhey FRJ et al (2015) Prevalence of cerebral amyloid pathology in persons without dementia: a meta-analysis. JAMA J Am Med Assoc 313:1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM et al (2011) Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging- Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement 7:280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naj AC, Leonenko G, Jian X, Grenier-boley B, Dalmasso C, Bellenguez C, et al. (2021) Genome-wide meta-analysis of late-onset Alzheimer’s disease using rare variant imputation in 65,602 subjects identifies novel rare variant locus NCK2 : the International Genomics of Alzheimer’s Project (IGAP). medRxiv.
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 19.Howie BN, Donnelly P, Marchini J (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5:e1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaneau O, Coulonges C, Zagury JF (2008) Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinform. 10.1186/1471-2105-9-540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR (2010) METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mägi R, Morris AP (2010) GWAMA: software for genome-wide association meta-analysis. Bioinformatics 11:288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, Van Bochoven A, Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 9:215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leeuw CA, Mooij JM, Heskes T, Posthuma D (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 11(4):e1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguet F, Barbeira AN, Bonazzola R, Brown A, Castel SE, Jo B et al (2020) The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369:1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, Gamazon ER (2020) A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat Genet. 10.1038/s41588-020-0706-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Chen S, Li Z, Morrison AC, Boerwinkle E, Lin X (2019) ACAT: a fast and powerful p value combination method for rare-variant analysis in sequencing studies. Am J Hum Genet 5:e1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millard LAC, Davies NM, Gaunt TR, Smith GD, Tilling K (2018) Software application profile: PHESANT: A tool for performing automated phenome scans in UK Biobank. Int J Epidemiol 1:123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J et al (2018) Genomic atlas of the human plasma proteome. Nature. 10.1038/s41586-018-0175-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack S, Coassin S, Rueedi R, Yousri NA, Seppälä I, Gieger C et al (2017) A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J Lipid Res. 10.1194/jlr.M076232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis JK, Sealock JM, Straub P, Lee YH, Hucks D, Actkins KE et al (2021) Clinical laboratory test-wide association scan of polygenic scores identifies biomarkers of complex disease. Genome Med 21:168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL et al (2016) The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167(5):1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 10.1016/j.jalz.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV et al (2013) Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry 18(11):1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyagawa T, Ebinuma I, Morohashi Y, Hori Y, Chang MY, Hattori H et al (2016) BIN1 regulates BACE1 intracellular trafficking and amyloid-β production. Hum Mol Genet. 10.1093/hmg/ddw14 [DOI] [PubMed] [Google Scholar]
- 37.Crotti A, Sait HR, McAvoy KM, Estrada K, Ergun A, Szak S et al (2019) BIN1 favors the spreading of Tau via extracellular vesicles. Sci Rep. 10.1038/s41598-019-45676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rossi P, Nomura T, Andrew RJ, Masse NY, Sampathkumar V, Musial TF et al (2020) Neuronal BIN1 regulates presynaptic neurotransmitter release and memory consolidation. Cell Rep. 10.2139/ssrn.3443599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eissman JM, Archer DB, Mukherjee S, Lee ML, Choi SE, Scollard P et al (2023) Sex-specific genetic architecture of late-life memory performance. Alzheimer’s Dement. 10.1002/alz.13507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Borgne J, Amouyel P, Andreassen O, Frikke-Schmidt R, Hiltunen M, Ingelsson M et al (2024) Association of MGMT and BIN1 genes with Alzheimer’s disease risk across sex and APOE ε4 status. Alzheimer’s Dement. 10.1002/alz.13550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan CC, Banks SJ, Thompson WK, Chen CH, McEvoy LK, Tan CH et al (2020) Sex-dependent autosomal effects on clinical progression of Alzheimer’s disease. Brain. 10.1093/brain/awaa164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le GY, Eger SJ, Belloy ME, Kennedy G, He Z, Napolioni V et al (2021) Sex-heterogenous effect on Alzheimer’s disease risk at the BIN1 locus. Alzheimers Dement 17:e053616 [Google Scholar]
- 43.Lopez-Lee C, Torres ERS, Carling G, Gan L (2024) Mechanisms of sex differences in Alzheimer’s disease. Neuron. 10.1016/j.neuron.2024.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uddin MS, Rahman MM, Jakaria M, Rahman MS, Hossain MS, Islam A et al (2020) Estrogen signaling in alzheimer’s disease: molecular insights and therapeutic targets for Alzheimer’s dementia. Mol Neurobiol 57:2654 [DOI] [PubMed] [Google Scholar]
- 45.Manly JJ, Merchant CA, Jacobs DM, Small SA, Bell K, Ferin M et al (2000) Endogenous estrogen levels and Alzheimer’s disease among postmenopausal women. Neurology. 10.1212/WNL.54.4.833 [DOI] [PubMed] [Google Scholar]
- 46.Bhatta S, Blair JA, Casadesus G (2018) Luteinizing hormone involvement in aging female cognition: not all is estrogen loss. Front Endocrinol (Lausanne). 10.3389/fendo.2018.00544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burger HG, Hale GE, Robertson DM, Dennerstein L (2007) A review of hormonal changes during the menopausal transition: Focus on findings from the Melbourne Women’s Midlife Health Project. Hum Reprod Update. 10.1093/humupd/dmm020 [DOI] [PubMed] [Google Scholar]
- 48.Wiacek M, Hagner W, Zubrzycki IZ (2011) Measures of menopause driven differences in levels of blood lipids, follicle-stimulating hormone, and luteinizing hormone in women aged 35 to 60 years. Menopause. 10.1097/gme.0b013e3181e7060b [DOI] [PubMed] [Google Scholar]
- 49.Zhao L, Mao Z, Brinton RD (2009) A select combination of clinically relevant phytoestrogens enhances estrogen receptor β-binding selectivity and neuroprotective activities in vitro and in vivo. Endocrinology. 10.1210/en.2008-0715 [DOI] [PubMed] [Google Scholar]
- 50.Burger HG, Dudley EC, Robertson DM, Dennerstein L (2002) Hormonal changes in the menopause transition. Recent Prog Horm Res. 10.1210/rp.57.1.257 [DOI] [PubMed] [Google Scholar]
- 51.Takayasu S, Sakurai T, Iwasaki S, Teranishi H, Yamanaka A, Williams SC et al (2006) A neuropeptide ligand of the G protein-coupled receptor GPR103 regulates feeding, behavioral arousal, and blood pressure in mice. Proc Natl Acad Sci USA. 10.1073/pnas.0602371103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Z, Jiang W, Zhang EE (2016) Orexin signaling regulates both the hippocampal clock and the circadian oscillation of Alzheimer’s disease-risk genes. Sci Rep. 10.1038/srep36035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen XY, Du YF, Chen L (2019) Neuropeptides exert neuroprotective effects in alzheimer’s disease. Front Mol Neurosci 11:493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Milán-Tomás Á, Shapiro CM (2018) Circadian rhythms disturbances in Alzheimer disease. Alzheimer Dis Assoc Disord 32(2):162 [DOI] [PubMed] [Google Scholar]
- 55.Musiek ES, Xiong DD, Holtzman DM (2015) Sleep, circadian rhythms, and the pathogenesis of Alzheimer Disease. Exp Mol Med. 10.1038/emm.2014.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou H, Chan C, Yuki KE, Sokolowski D, Roy A, Qu R et al (2022) Postnatal developmental trajectory of sex-biased gene expression in the mouse pituitary gland. Biol Sex Differ 6:13289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandonà D, Betto R (2009) Sarcoglycanopathies: molecular pathogenesis and therapeutic prospects. Expert Rev Mol Med 11:e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sadick JS, O’Dea MR, Hasel P, Dykstra T, Faustin A, Liddelow SA (2022) Astrocytes and oligodendrocytes undergo subtype-specific transcriptional changes in Alzheimer’s disease. Neuron. 10.1016/j.neuron.2022.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashimoto T, Maekawa S, Miyata S (2009) IgLON cell adhesion molecules regulate synaptogenesis in hippocampal neurons. Cell Biochem Funct. 10.1002/cbf.1600 [DOI] [PubMed] [Google Scholar]
- 60.Liu F, Arias-Vásquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P et al (2007) A genomewide screen for late-onset Alzheimer disease in a genetically isolated Dutch population. Am J Hum Genet. 10.1086/518720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernardo A, Harrison FE, McCord M, Zhao J, Bruchey A, Davies SS et al (2009) Elimination of GD3 synthase improves memory and reduces amyloid-β plaque load in transgenic mice. Neurobiol Aging. 10.1016/j.neurobiolaging.2007.12.022 [DOI] [PubMed] [Google Scholar]
- 62.Yamin R, Zhao C, O’Connor PB, McKee AC, Abraham CR (2009) Acyl peptide hydrolase degrades monomeric and oligomeric amyloid-beta peptide. Mol Neurodegener. 10.1186/1750-1326-4-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmieri G, Cocca E, Gogliettino M, Valentino R, Ruvo M, Cristofano G et al (2017) Low erythrocyte levels of proteasome and acyl-peptide hydrolase (APEH) activities in Alzheimer’s disease: a sign of defective proteostasis? J Alzheimer’s Dis 60(3):1097–1106. 10.3233/JAD-170389 [DOI] [PubMed] [Google Scholar]
- 64.Asadi MR, Hassani M, Kiani S, Sabaie H, Moslehian MS, Kazemi M et al (2021) The perspective of dysregulated LncRNAs in Alzheimer’s Disease: a systematic scoping review. Front Aging Neurosci 13:709568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chew H, Solomon VA, Fonteh AN (2020) Involvement of lipids in Alzheimer’s disease pathology and potential therapies. Front Physiol 11:598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hegazy SH, Thomassen JQ, Rasmussen IJ, Nordestgaard BG, Tybjærg-Hansen A, Frikke-Schmidt R (2022) C-reactive protein levels and risk of dementia—observational and genetic studies of 111,242 individuals from the general population. Alzheimer’s Dement. 10.1002/alz.12568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiang YX, Deng YT, Zhang YR, Wang HF, Zhang W, Dong Q et al (2023) Associations of blood cell indices and anemia with risk of incident dementia: A prospective cohort study of 313,448 participants. Alzheimer’s Dement. 10.1002/alz.13088 [DOI] [PubMed] [Google Scholar]
- 68.Wang R, Jin D, Li Y, Liang QC (2013) Decreased mean platelet volume and platelet distribution width are associated with mild cognitive impairment and Alzheimer’s disease. J Psychiatr Res. 10.1016/j.jpsychires.2013.01.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ADGC cohorts’ data is available at niagads (https://www.niagads.org/), while the ADNI dataset can be accessed from https://adni.loni.usc.edu/. Other data from Religious Orders Study and Memory and Aging Project, Mayo Clinic Alzheimer's Disease Research Center, and Harvard Brain Tissue Resource Center are accessible on https://www.synapse.org.