Abstract
Background
X chromosome inactivation (XCI) is a female-specific process in which one X chromosome is silenced to balance X-linked gene expression between the sexes. XCI is initiated in early development by upregulation of the lncRNA Xist on the future inactive X (Xi). A subset of X-linked genes escape silencing and thus have higher expression in females, suggesting female-specific functions. One of these genes is the highly conserved gene Kdm6a, which encodes a histone demethylase that removes methyl groups at H3K27 to facilitate gene expression. KDM6A mutations have been implicated in congenital disorders such as Kabuki Syndrome, as well as in sex differences in development and cancer.
Methods
Kdm6a was knocked out (KO) using CRISPR/Cas9 gene editing in hybrid female mouse embryonic stem (ES) cells derived either from a 129 × Mus castaneus (cast) cross or a BL6 x cast cross. In one of the lines a transcriptional stop signal inserted in Tsix results in completely skewed X silencing upon differentiation. The effects of both homozygous and heterozygous Kdm6a KO on Xist expression during the onset of XCI were measured by RT-PCR and RNA-FISH. Changes in gene expression and in H3K27me3 enrichment were investigated using allele-specific RNA-seq and Cut&Run, respectively. KDM6A binding to the Xist gene was characterized by Cut&Run.
Results
We observed impaired upregulation of Xist and reduced coating of the Xi during early stages of differentiation in Kdm6a KO cells, both homozygous and heterozygous, suggesting a threshold effect of KDM6A. This was associated with aberrant overexpression of genes from the Xi after differentiation, indicating loss of X inactivation potency. Consistent with KDM6A having a direct role in Xist regulation, we found that the histone demethylase binds to the Xist promoter and KO cells show an increase in H3K27me3 at Xist, consistent with reduced expression.
Conclusions
These results reveal a novel female-specific role for the X-linked histone demethylase, KDM6A in the initiation of XCI through histone demethylase-dependent activation of Xist during early differentiation.
Plain language summary
X chromosome inactivation is a female-specific mechanism that evolved to balance sex-linked gene dosage between females (XX) and males (XY) by silencing one X chromosome in females. X inactivation begins with the upregulation of the long noncoding RNA Xist on the future inactive X chromosome. While most genes become silenced on the inactive X chromosome some genes escape inactivation and thus have higher expression in females compared to males, suggesting that escape genes may have female-specific functions. One such gene encodes the histone demethylase KDM6A which function is to turn on gene expression by removing repressive histone modifications. In this study, we investigated the role of KDM6A in the regulation of Xist expression during the onset of X inactivation. We found that KDM6A binds to the Xist gene to remove repressive histone marks and facilitate its expression in early development. Indeed, depletion of KDM6A prevents upregulation of Xist due to abnormal persistence of repressive histone modifications. In turn, this results in aberrant overexpression of genes from the inactive X chromosome. Our findings point to a novel mechanism of Xist regulation during the initiation of X inactivation, which may lead to new avenues of treatment to alleviate congenital disorders such as Kabuki syndrome and sex-biased immune disorders where X-linked gene dosage is perturbed.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13293-024-00683-3.
Keywords: Allele-specific, Epigenetics, X inactivation, Histone methylation, Dosage compensation, Escape
Highlights
Kdm6a knockout prevents upregulation of the lncRNA Xist during differentiation of female stem cells, suggesting that KDM6A plays a threshold-dependent role in facilitating Xist expression during the onset of X chromosome inactivation, a female-specific phenomenon.
X chromosome inactivation efficacy is markedly reduced in cells with Kdm6a knockout, leading to increased gene expression from the Xi.
KDM6A binding at the Xist gene during the onset of X inactivation is necessary for the removal of H3K27 methylation, revealing a direct role in regulating Xist expression via H3K27 demethylation-dependent mechanisms.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13293-024-00683-3.
Introduction
In mammalian females X chromosome inactivation (XCI) serves to balance X-linked gene expression between males (XY) and females (XX) [1, 2]. XCI is a robust silencing mechanism characterized by several chromosome-wide epigenetic changes on the inactive X (Xi), including histone de-acetylation, ubiquitination of H2AK119, methylation of H3K27 and H3K9, DNA methylation at CpG islands, and chromatin condensation. XCI is initiated at early stages of development by upregulation of the long non-coding RNA (lncRNA) Xist from the future Xi. Xist RNA coats the Xi and recruits proteins that implement epigenetic changes for silencing. This process can be modeled in mouse embryonic stem (ES) cells, which demonstrate extensive silencing by day 4–7 of differentiation [3, 4].
A subset of X-linked genes escape XCI and remain expressed from the Xi, albeit at a lower level than from the active X chromosome (Xa), resulting in higher expression in females compared to males [5, 6]. XCI and escape are processes unique to females, suggesting that factors involved in these processes such as Xist and other regulatory elements that influence the structure and epigenetic features of the Xi may be partly controlled by genes with female-biased expression [7]. One of these genes is Kdm6a, a highly conserved gene that escapes XCI in all mammalian species tested [8, 9]. The main function of KDM6A is to facilitate gene expression during differentiation and development by resolution of bivalent domains through demethylation of H3K27me3 [10–13]. Additionally, KDM6A plays a role in allelic gene regulation, where it preferentially targets maternal alleles of autosomal genes to increase expression [14]. Interestingly, KDM6A has a Y-encoded homolog (UTY) that has little or no demethylase activity, suggesting that KDM6A may have female-specific roles that depend on its demethylase function [15–17].
Here, we show that KDM6A plays a role in Xist upregulation during the earliest stages of XCI. We find that Kdm6a KO results in decreased levels of Xist RNA and in reduced formation of an Xist cloud during ES cell differentiation. This causes impaired silencing of genes from the Xi. A direct association between H3K27me3 demethylation by KDM6A and Xist expression is supported by findings that KDM6A binds to the Xist promoter region during ES cell differentiation, and that Kdm6a KO results in an increase of H3K27me3 levels along the Xist gene. Thus, the female-biased expression of Kdm6a via escape from XCI may have evolved as a mechanism that contributes to XCI by regulating Xist.
Results
Generation of Kdm6a KO ES cells
To study the potential role of KDM6A at the onset of XCI, we used CRISPR/Cas9 to knockout (KO) Kdm6a in hybrid female mouse ES cells derived from a 129 x Mus castaneus (cast) cross in which alleles can be distinguished by SNPs (Additional File 1: Figure S1A). In order to examine changes specifically on the Xi we used Tsix-stop cells (donated by J. Gribnau, Erasmus MC) in which a transcriptional stop signal is inserted onto the 129 allele of Tsix, resulting in completely skewed silencing of the 129 X chromosome upon differentiation [18]. Stable Kdm6a KO Tsix-stop ES cell clones with a heterozygous or homozygous deletion of part of exon 2 through part of exon 4 (wt/ΔE or ΔE/ΔE, respectively) were generated (Additional File 1: Figure S1A) [14]. PCR and Sanger sequencing verified homozygous editing of Kdm6a in two KO clones (Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21) and heterozygous editing in one KO clone (Tsix-Kdm6awt/ΔE4). Despite residual Kdm6a expression in the homozygous KO clones, western blot analysis showed no evidence of KDM6A protein [14, 15]. We also isolated control Tsix-stop ES cell clones (Tsix-cln1, Tsix-cln2; hereafter called CRISPR controls), which were subject to the CRISPR/Cas9 treatment but did not exhibit a deletion. In a separate experiment, Kdm6a homozygous editing was done by deleting the promoter in another hybrid female mouse ES cell line called E8 (donated by J. Gribnau, Erasmus MC) to derive clone E8-Kdm6aΔP/ΔP13 (Additional File 1: Figure S1A). The E8 cell line derived from a C57BL/6J (BL6) x cast cross undergoes random XCI with partial skewing due to a strong Xce allele that results in a ~2:1 inactivation of the BL6:cast X chromosome upon differentiation [4, 19]. A summary of the clones used in this study can be found in Additional File 2: Table S1.
Kdm6a KO results in expression changes of genes involved in germ layer development after XCI occurs
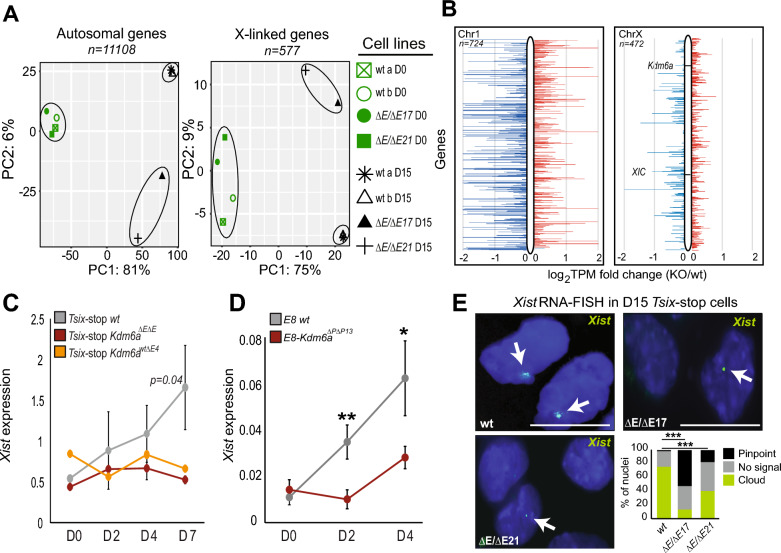
RNA-seq analyses were done to compare gene expression between Tsix-stop wild-type (wt) and Kdm6a KO (Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21) clones before day 0 (thereafter abbreviated D0) and after (D15) differentiation of ES cells into embryoid bodies (EB). For Tsix-stop cells, principal component analysis (PCA) based on either autosomal or X-linked gene expression shows little separation between wt and KO clones at D0, but a clear separation at D15, indicating that the effects of Kdm6a KO are most pronounced after differentiation (Figure 1A). Consistent with PCA clustering, we found only 624 differentially expressed genes (DEGs) between wt and KO cells at D0, while there were 886 DEGs at D15 (≥2 fold change; FDR ≤0.1) (Additional File 1: Figure S1B; Additional File 3: Table S2). These findings are in agreement with KDM6A’s dispensable role in undifferentiated ES cells versus its multiple roles in regulation of genes involved in development and differentiation [11, 14, 20]. Indeed, among the DEGs found at D15 (a time point after XCI has occurred) there is a subset of genes involved in germ layer development, including genes expressed in all three germ layers, which are downregulated in Tsix-stop Kdm6a KO cells at D15, but not at D0 (Additional File 1: Figure S1C; Additional File 4: Table S3). Of the germ layer genes with decreased expression in Tsix-stop KO cells, 19 were expressed (>1 TPM) at D2 and 18 were expressed at D4 of differentiation in E8-Kdm6a wt cells. There was no clear pattern of downregulation of these genes at either time point following Kdm6a KO, strongly suggesting that germ layer gene downregulation occurred after XCI (Additional File 1: Figure S1C; Additional File 4: Table S3). In line with KDM6A’s role in facilitating gene expression by removing the repressive mark H3K27me3, we observed an overall decrease in autosomal gene expression (Figure 1B). In contrast, 217/472 (46%) of X-linked genes assayed showed increased expression (>1.25 TPM fold change) in Tsix-stop Kdm6a KO (average of clones Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21) compared to wt at D15 (Figure 1B).
Fig. 1.
Kdm6a KO results in Xist downregulation. A Principal component analysis (PCA) based on expression of autosomal or X-linked genes in two Tsix-stop wt clones and two Kdm6a KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) before (D0) and after differentiation (D15). Only genes with a minimum of 2.5 CPM in 4 libraries were considered. PCA shows separation of clones based on Kdm6a KO status mainly after differentiation. PCA figures were generated by iDEP.95. B Plots of diploid gene expression from a representative autosome (Chr1, left) with similar gene content as the X, and from the X chromosome (right) in Tsix-stop wt and KO clones (average for Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D15. Kdm6a and the XIC are highlighted. Log2 TPM fold change (KO/wt) is shown. Genes with a decrease of expression in KO versus wt are in blue, and with an increase in red. C qRT-PCR of average Xist expression normalized to Actinβ during ES differentiation in three Tsix-stop wt samples, two Kdm6aΔEΔE KO clones, and one Kdm6aΔwtΔE clone. p-value is derived from comparing wt to Kdm6aΔEΔE biological replicates. Divergence of Xist expression between wt and KO cells begins at D2 and is largest at D7 (p = 0.04). The Kdm6awt/ΔE4 heterozygous KO clone behaves similarly to the homozygous clones. D qRT-PCR of Xist expression in E8 wt and KO clone (E8-Kdm6aΔPΔP13). Expression is normalized to Actinβ. Xist is significantly lower at D2 and D4 in KO cells. P-values derived from two technical replicates of wt and KO cells (**p < 0.01;*p < 0.05). E RNA-FISH with a probe specific for Xist RNA labeled in green in Tsix-stop wt and KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21). Examples of nuclei are shown together with histograms for quantification of signal type (pinpoint, no signal or cloud). Xist clouds are present in 74% of D15 wt cells, but significantly decrease in KO clones (cloud compared to no signal; ***p < 0.00001 for Tsix-Kdm6aΔEΔE17 and < 0.0001 for Tsix-Kdm6aΔEΔE21 fisher’s exact test), while the frequency of pinpoint Xist signals increases in KO clones (pinpoint in wt compared to pinpoint in KO; ***p < 0.00001 for Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21 fisher’s exact test). Nuclei are counterstained with Hoechst 33,342. Scale bar = 10 µm
Xist expression and coating are impaired during early differentiation following Kdm6a KO
One of the earliest events at the onset of XCI is the upregulation of Xist [1, 2]. Therefore, we investigated changes in the dynamics of Xist expression during early mouse ES cell differentiation at D0, D2, D4, D7, and D15 following Kdm6a KO. Compared to Tsix-stop wt and CRISPR-control clones, Kdm6a KO clones (Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21) have significantly reduced levels of Xist expression beginning at D2 of differentiation when Xist upregulation normally starts (Figures 1C and Additional File 5: Figure S2A). Importantly, this finding was confirmed in Kdm6a KO clone (E8-Kdm6aΔP/ΔP13) derived from an independent ES line E8 (Figure 1D). Thus, we infer that KDM6A contributes to the activation of Xist in early differentiation beginning prior to D2 and increasing at D4 and D7 (Figures 1B and Additional File 5: Figure S2B). Analysis in Tsix-stop cells with heterozygous deletion of Kdm6a (Tsix-Kdm6a wt/ΔE4) revealed a similar effect on Xist by D7, suggesting that expression of Kdm6a from both alleles is needed for activation of Xist (Figure 1C). The lower expression of Xist in Kdm6a KO cells does not appear to be due to changes in levels of several of the known Xist repressors (Tsix, Nanog, Pou5f1, Sox2, Rex1) or activators (Yy1, Rlim) as these were not differentially expressed between Tsix-stop wt and KO cells at D0 (Additional File 5: Figure S2C) [21]. Klf4, which is known to repress Xist via activation of Tsix [22], did show an increase in Kdm6a KO cells at D0, but Tsix expression was consistently lower in KO cells at D2, reaching levels similar to those in wt only at D4, suggesting that the increase in Klf4 did not affect Tsix expression at this stage (Additional File 5: Figure S2D and E; Additional File 3: Table S2). Note that despite a small increase in Sox2 expression in Kdm6a KO cells, this gene was not called as a DEG by Deseq2 (Additional File 5: Figure S2C; Additional File 3: Table S2). Previous studies have shown aberrantly increased expression of Xist with Tsix inhibition in undifferentiated ES cells grown in media with serum [23, 24]. However, we verified that the Tsix-stop ES cells used here maintain a low level of Xist expression at D0 (~2 TPM) similar to levels observed in previous studies despite the presence of serum in the media (Additional File 5: Figure S2F) [3, 14].
To determine whether Kdm6a KO affects the formation of an Xist cloud we performed RNA-FISH with a probe that targets Xist in differentiated Tsix-stop Kdm6a KO and wt cells at D15. The percentage of nuclei with Xist clouds sharply decreased after KO, with only 18% and 38% of nuclei with an Xist cloud in clones Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21, respectively, versus 74% of wt nuclei (Figure 1E). Consistent with reduced Xist expression in Kdm6a KO clones, nuclei with a small pinpoint Xist signal rather than a cloud were prevalent in these clones (41% and 23% of nuclei in clones Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21, respectively) compared to 4% in wt nuclei (Figure 1E). Loss of an X chromosome during cell culture is a common occurrence in female mouse ES cells and would confound our analysis of X-linked gene expression. To address this possibility DNA-FISH using probes targeting X chromosome-specific regions was done to verify that minor X chromosome loss occurred in any condition tested (Additional File 6: Figure S3).
Together, these results indicate that KDM6A contributes to upregulation of Xist expression in a threshold dependent manner, and to Xist coating of the Xi during early mouse ES cell differentiation.
Aberrantly increased gene expression from the Xi in differentiated Kdm6a KO cells
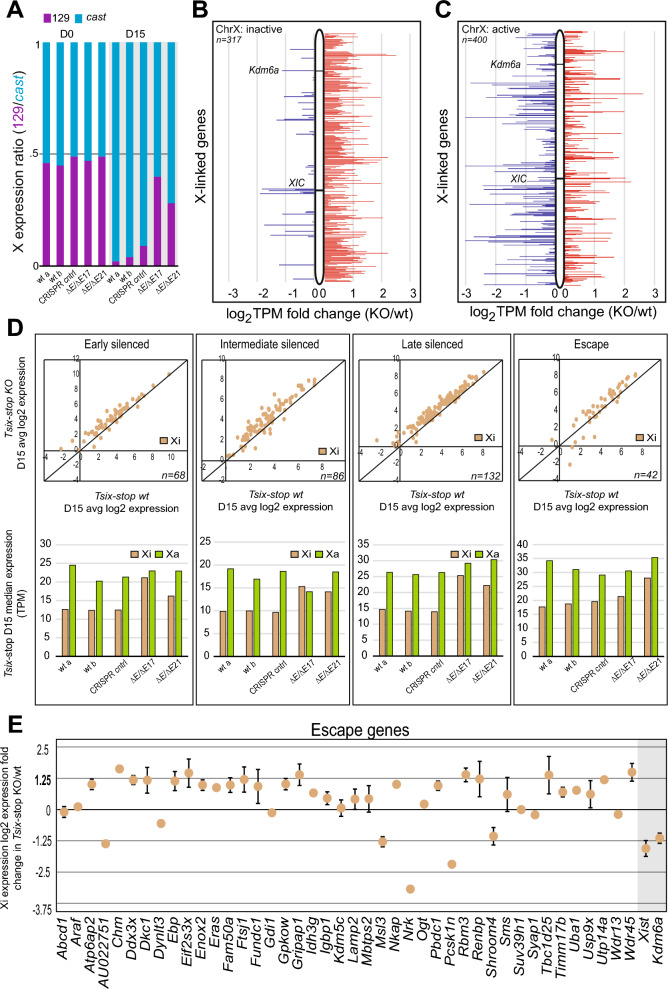
We postulated that reduced Xist upregulation and Xist coating in Kdm6a KO cells may lead to impaired XCI in these cells. Allelic analyses of X-linked gene expression in Tsix-stop cells show that the increase in expression following Kdm6a KO is mainly due to increased expression specifically from the Xi where 76% (242/317) of assayed genes show a ≥ 1.25 TPM fold increase (Figure 2A, B; Additional File 8: Table S4). In contrast, there is no clear directionality for dysregulated genes on the Xa (54% up and 46% downregulated; ≥ 1.25 TPM fold cutoff) (Figure 2C). In addition, we confirmed increased X-linked gene expression in an independent Kdm6a KO clone (E8-Kdm6aΔP/ΔP13) where 42% (208/491) of X-linked genes show a ≥ 1.25 TPM fold increased expression at D4 of differentiation (Additional File 7: Figure S4; Additional File 8: Table S4). In E8 cells, where XCI is largely random, allelic analyses show increased X-linked gene expression following Kdm6a KO coming from the BL6 and cast X (Additional File 7: Figure S4A, B). However, gene expression is slightly more elevated from the BL6 X, suggestive of a specific effect of Kdm6a KO on the Xi, which would tend to be the BL6 X due to partial XCI skewing in E8 cells (Additional File 7: Figure S4C, D). Thus, failure to upregulate Xist leads to increase X-linked gene expression in differentiated cells. Considering autosomal genes, Kdm6a KO resulted in a majority of dysregulated genes with decreased expression (84% and 60% of dysregulated genes in Tsix-stop Kdm6a KO clones and in E8-Kdm6aΔP/ΔP13, respectively), consistent with the role of KDM6A in facilitating gene expression through removal of H3K27me3 (Additional File 3: Table S2).
Fig. 2.
Kdm6a KO leads to increased gene expression from the Xi in differentiated cells. A Allelic X-linked gene expression ratios (129:cast) in Tsix-stop wt, CRISPR control, and KO cells (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D0 and D15 of differentiation. Ratios between expressed X-linked genes (> 1TPM) are shown. There is increased gene expression from the 129 X-chromosome (Xi) following Kdm6a KO differentiation. B, C Plots of expression from the Xi (B) and the Xa (C) (average for Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D15. Kdm6a and the XIC are highlighted. Log2 TPM fold change (KO/wt) is shown. Genes with a decrease of expression in KO versus wt are in blue, and with an increase in red. Gene expression from the Xi is selectively increased following KO. D Top, scatter plots of average log2 expression between Tsix-stop wt and KO clones (average for Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D15 for X-linked genes categorized as those silenced early, silenced at an intermediate time, silenced late, and not silenced (escape) during XCI [3, 25]. KO clones show higher expression of genes in each category compared to wt. n indicates the number of genes in each category. Bottom, histograms of median allelic expression of genes silenced at different times during XCI and of genes that escape XCI is increased in Tsix-stop KO clones compared to wt and CRISPR controls for all categories, while there is little change in expression from the Xa. n indicates the number of genes in each category. E Expression fold changes of individual escape genes from the Xi in Tsix-stop KO clones (average for Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) compared to wt at D15. Most genes show increased expression
Next, we grouped X-linked genes in four defined temporal clusters, early, intermediate, late, and not silenced (or escape), based on their previously reported timing of silencing during differentiation [3, 25]. All gene clusters show a trend of increased expression from the Xi in Tsix-stop Kdm6a KO cells compared to wt or CRISPR controls (Figures 2D, E; Additional File 9: Table S5). Interestingly, the expression of most escape genes is also increased after Kdm6a KO (Figure 2D, E). For all clusters the increase is usually less than ~ 1.25 fold, with few escape genes showing expression changes > 1.25 fold in KO versus wt ES cells. Few escape genes show a decrease in expression and include Xist and Kdm6a, as expected (Figure 2E).
Taken together, these results indicate a reduced potency of XCI in differentiated Kdm6a KO cells, which implicates KDM6A, at least in part, as a regulator of X-linked gene expression dosage via upregulation of Xist during the onset of XCI.
Kdm6a KO results in dysregulated H3K27me3 levels on the X chromosome
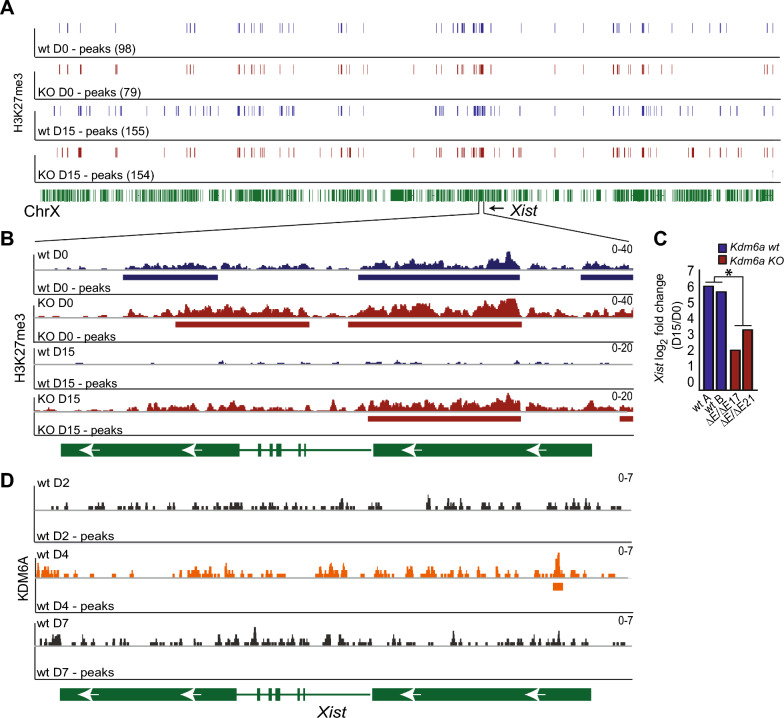
Next, we investigated changes in H3K27me3 profiles using Cut&Run to compare wt Tsix-stop cells to Kdm6a KO clone Tsix-Kdm6aΔE/ΔE17. At D0 and D15, H3K27me3 levels (H3K27me3 peak numbers) appear similar along the whole X chromosome between wt and Kdm6a KO (Figure 3A), but a closer inspection revealed changes at the Xist locus. Initially (D0) H3K27me3 enrichment is only slightly higher across the Xist gene in Kdm6a KO ES cells, consistent with KDM6A’s role in resolving bivalent chromatin (Figure 3B). However, following XCI at D15, there is a marked increase in H3K27me3, which correlates with lower Xist expression that culminates at ~ 2.5 fold upregulation between D0 and D15 in KO cells, compared to ~ 6.0 fold in wt cells (Figure 3B, C). In addition to changes at Xist, H3K27me3 enrichment is also markedly higher at Cdx4, the Tsix/Xist locus, and the Ftx 5’ region in Kdm6a KO versus wt cells at D15 (Additional File 10: Figure S5A). Both Cdx4 and Ftx show little expression change following Kdm6a KO, however, these genes are expressed at very low levels following differentiation (< 1TPM) and thus are not classified as DEGs (Additional File 10: Figures S5B–E). As a control, we analyzed a subset of known KDM6A target genes where we confirmed an increase in H3K27me3 levels associated with decreased expression in KO versus wt cells (Additional File 11: Figure S6A). Genes without significant expression change in KO cells did not show differences in H3K27me3, as expected (Additional File 11: Figure S6B). Consistent with our previous study, the imprinted Dlk1/Meg3 locus showed an increase H3K27me3 levels in Kdm6a KO cells, which is specifically located along the imprinting control regions (ICR), and concordant with decreased gene expression (Additional File 11: Figure S6C; Additional File 3: Table S2) [14].
Fig. 3.
KDM6A binds to the promoter of Xist and Kdm6a KO results in increased H3K27me3. A IGV browser view of H3K27me3 peak profiles across the entire X chromosome in wt Tsix-stop cells (blue) and KO cells (Tsix-Kdm6aΔEΔE17) (red) at D0 and D15. The number of X chromosome peaks for each condition and time point are in parentheses. Green bars underneath represent genes along the X chromosome. The Xist gene is marked. B IGV browser view of H3K72me3 profiles and peaks at the Xist locus in wt Tsix-stop (blue) and KO Tsix-Kdm6aΔEΔE17 cells (red) at D15. There is a slight increase in H3K27me3 levels in KO cells at D0, and a marked increase at D15. C Histogram of log2 fold change of Xist expression in wt (2 isolates: wt A, wt B) and two KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D15 relative to D0. There is a markedly higher level of Xist induction in wt compared to KO clones. D IGV browser view of KDM6A profiles and peaks at D2, D4 and D7 of differentiation at the Xist locus in wt Tsix-stop cells. Merged data from two replicates of each time point are shown (see methods). KDM6A binding at the Xist gene promoter region and exon 1 is increased at D4 and lost at D7 following XCI. The scales of the profiles shown in (A) and (B) and (D) are indicated in the upper right corners
Chromatin analyses using Cut&Run with an antibody for KDM6A in wt Tsix-stop cells at D2, D4, and D7 show that KDM6A binding to the Xist promoter region increases at D4, when Xist expression levels continue to diverge between wt and Kdm6a KO cells, followed by a decrease at D7 (Figures 1C and 3D; Additional File 5: Figure S2A; Additional File 12: Table S6). A total of 582 KDM6A peaks mapped thoughout the entire genome in wt cells correspond to peaks of H3K27me3 enrichment, in contrast, in Kdm6a KO cells there is an increase in the number of H3K27me3 peaks that correspond to KDM6A peaks (870). This was observed both at D4 and D7, with a general decrease of KDM6A at D7 (Additional File 13: Figure S7A, B). Together, these results are consistent with KDM6A’s role in regulating H3K27me3 during cellular differentiation, such as in XCI, and with a genome-wide effect of Kdm6a KO on H3K27me3 levels (Additional File 13: Figure S7A, B). As expected, we observed KDM6A binding at promoters of known target genes with decreased expression in KO cells (Additional File 3: Table S3 and Additional File 13: Figure S7C) [14]. We next investigated the dynamics of KDM6A during early differentiation by qRT-PCR and protein blots, which showed a slight increase of both expression and protein levels during the early stages of differentiation followed by a decrease at D7 after XCI has occurred (Additional File 13: Figure S7D).
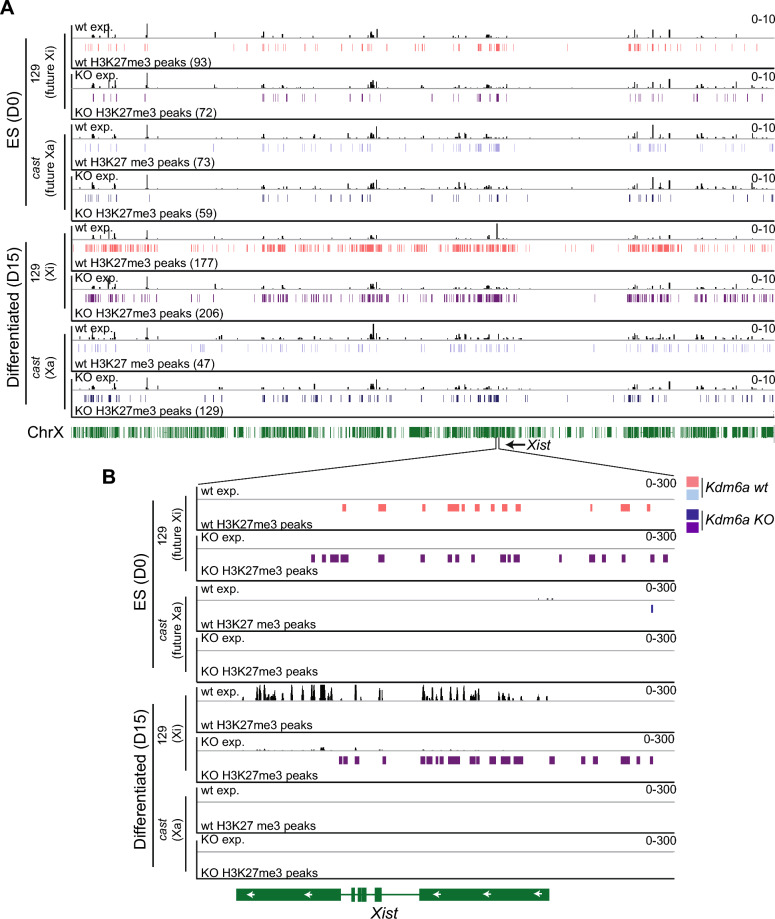
Allelic effects of Kdm6a KO on H3K27me3 enrichment were investigated in Tsix-stop cells in which the Xi is from strain 129 due to skewed XCI [3]. At D0 few differences between alleles and between wt and Kdm6a KO are seen (Figure 4A). However, the future Xi (maternal 129 X) shows enrichment in H3K27me3 along Xist, which is not seen on the future Xa (paternal cast X) (Figure 4A, B). This is evident both in wt and KO cells at D0, and is consistent with previous studies demonstrating maternal-specific H3K27me3 enrichment at Xist [26]. At D15, wt cells show high overall H3K27me3 levels on the 129 Xi compared to the cast Xa, as expected due to the onset of XCI (Figure 4A; Additional File 14: Table S7). In Kdm6a KO cells there is both an increase and a decrease in H3K27me3, the latter being specifically associated with a number of genes that gained expression due to the lack of Xist expression and partial failure of XCI (Fig. 2B; Additional File 15: Figure S8). Overall, the gain in H3K27me3 on the 129 Xi in KO cells was modest. We used the wt sample as a background to call Xi peaks that appear in the KO sample. There were only 50 Xi-specific peaks gained in the KO cells that were absent in the wt, with the majority appearing either in gene bodies or at the Xist gene. As a result, only an additional 0.014% of the Xi was associated with an H3K27me3 peak. Interestingly, only the maternal allele of Xist shows persistence of higher levels of H3K27me3 following Kdm6a KO, which would explain decreased Xist expression and impaired XCI (Figures 4B; Additional File 14: Table S7). The observed genome-wide increase in H3K27me3 probably contributes to the large number of autosomal genes with decreased expression in Kdm6a KO cells versus wt (Figure 1B; Additional File 1: Figure S1B).
Fig. 4.
Kdm6a KO leads to increased H3K27me3 at Xist specifically on the Xi. A IGV browser views of gene expression (exp) and H3K27me3 peaks across the 129 and cast X chromosomes obtained by allelic analysis in Tsix-stop wt and KO cells (Tsix-Kdm6aΔEΔE17) at D0 and D15. The 129 profile is in pink (wt) or purple (KO) and the cast in light blue (wt) or dark blue (KO). XCI is skewed in Tsix-stop cells so that the 129 X will become the Xi and the cast X the Xa. The number of peaks for each condition and time point are in parentheses. There is a marked increase in Xist expression from the Xi in wt cells but not in KO cells at D15. H3K27me3 levels increase following differentiation on the Xi in wt and Kdm6a KO cells. B Same analysis as in (A), but zoomed in on the Xist gene. Allelic RNA-seq tracks of gene expression show very low expression from the 129 Xi allele in KO cells. A correlated Xi-specific increase in H3K27me3 enrichment at the Xist promoter is seen as an increase in peaks. No expression of Xist is observed from the cast allele, which is the Xa. The scales of the profiles are indicated in the upper right corners
We conclude that prior to the onset of XCI, Kdm6a KO has a minimal effect on H3K27me3 levels at Xist. However, following differentiation persistence of high H3K27me3 levels would explain low levels of Xist expression. Although we cannot fully exclude indirect effects of Kdm6a KO on Xist regulation, our data show association of KDM6A with the 5’ end of Xist during differentiation, suggesting that KDM6A may directly facilitate Xist expression during the female specific process of XCI.
Discussion
Our study implicates KDM6A in the upregulation of Xist at the onset of XCI. We find that the female-biased histone demethylase helps Xist upregulation by binding to its promoter region and demethylating H3K27me3 during early mouse embryonic stem cell differentiation. In differentiated cells and adult tissues Kdm6a has higher expression in females compared to males due to escape from XCI, which may have evolved as a mechanism to regulate female-specific processes such as XCI [27]. Here, we show that KDM6A facilitates upregulation of Xist during early stages of XCI in hybrid mouse ES cell models with skewed or near-random XCI. Kdm6a KO leads to increased H3K27me3 levels on the maternal allele of Xist whose expression and ability to coat the Xi decrease following differentiation. In turn, this failure of XCI results in aberrant expression of genes from the Xi. Increased X-linked gene expression due to impaired XCI due to KDM6A deficiency may contribute to phenotypes in a variety of tissues such as those observed in Kabuki and Turner syndromes in human [28, 29].
The ES cell line in which we followed Xist expression and silencing of X-linked genes has skewed XCI of the maternal 129 allele due to a stop mutation at Tsix. It could be argued that this mutation may affect our observations. However, we found that an independent Kdm6a KO line without a Tsix mutation also shows impaired Xist expression and X silencing. Our findings are consistent with previous studies that have shown that maternal imprinting of Xist is maintained by maternal-specific H3K27me3 enrichment at Xist in blastocysts and that demethylation is critical for initiation of Xist expression and XCI [26]. Indeed, injection of Kdm6b, an autosomal gene that encodes another H3K27me3 demethylase, in preimplantation embryos results in maternal allele-specific loss of H3K27me3 and induction of Xist expression [26]. Therefore, removal of the repressive chromatin mark appears necessary to initiate random XCI. However, the question of how the maternal imprint at the Xist locus is maintained in the blastocyst remains. One possibility is that the increased levels of H3K27me3 on the maternal allele in wt Tsix-stop ES cells we observe is due to the influence of Xce. Here, we used hybrid mESCs with different X chromosome controlling element (Xce) [30]. However, the 129 X chromosome (maternal in this model) is more likely to be inactivated compared to the cast X chromosome (paternal is this model) [30]. Therefore, if Xce differences played a major role in biased H3K27me3 levels here, we would expect increased H3K27me3 on the cast allele instead. Another possibility is that KDM6A may play a role in the erasure of the imprint at Xist in mESCs with KO resulting in re-establishment of the imprint, but experiments in mESCs derived from a reciprocal cross would be needed to confirm this. In addition, we have previously demonstrated that KDM6A regulates expression of maternally expressed X-linked Rhox genes in a female-specific manner via removal of H3K27me3, and preferentially regulates maternal alleles of autosomal genes [14, 31]. Transcriptional regulation by KDM6A can occur through histone demethylation-dependent and -independent mechanisms [20]. In contrast, the protein encoded by the Y-linked homolog of Kdm6a, UTY, lacks detectable demethylase activity, suggesting that female-biased expression of Kdm6a may have evolved female-specific functions requiring H3K27me3 demethylation [15, 20]. Consistent with this, homozygous knockout (KO) of Kdm6a is lethal in female mice, while a small number of runted males with hemizygous deletion survive, suggesting that UTY partially compensates for loss of KDM6A through demethylation-independent mechanisms [32].
XCI is necessary for loss of pluripotency and normal differentiation, a mechanism that depends on expression of specific genes [4, 33, 34]. We find that germ layer marker genes have lower expression of Kdm6a in KO cells than in wt, which is consistent with previous analyses demonstrating that KDM6A impairs differentiation. Indeed, KDM6A mutations are associated with increased “stemness” in cancer and Kdm6a KO impairs mesoderm development in vivo largely through histone demethylase-independent mechanisms [20]. However, recent studies of Kdm6a KO models indicate that depletion of the enzyme does not block differentiation, and may even facilitate differentiation to neuroectoderm lineages [35, 36]. Our analysis in an additional cell line E8 with Kdm6a KO revealed no significant trend of downregulation of germ layer genes at time points prior and during XCI. Indeed, we only observed effects on germ layer marker genes at D15 after XCI has occurred, and our KDM6A chromatin binding analyses suggest a direct effect of KDM6A on Xist expression during early XCI rather than an indirect effect via a block on differentiation. In support, allelic single cell RNA-seq analyses in differentiating hybrid mouse ES cells show that XCI is complete prior to lineage specification, suggesting that the effects of Kdm6a KO on Xist expression we observe occur before lineage determination [4]. Thus, our results indicate that impaired XCI is due to failure of Xist upregulation in Kdm6a KO cells leading to abnormally high dosage of X-linked genes, which may then cause a block in differentiation at later time points [37]. However, increased expression of Klf4 and decreased expression of lineage-specific genes following Kdm6a KO suggests a potential delay in differentiation, therefore we cannot fully decouple the potential effects of Kdm6a KO on differentiation from the effects on regulation of Xist expression.
In undifferentiated ES cells the Xist promoter is maintained in a bivalent state, which is probably targeted for resolution by KDM6A via removal of H3K27me3 to facilitate expression during initiation of XCI [13, 38–40]. Consistent with this hypothesis, H3K27me3 levels show an increase in KO cells. A number of repressive epigenetic events occur following coating of the Xi by Xist including deposition of H3K27me3 by PRC2 [2]. One interesting possibility is that KDM6A, in addition to resolving bivalent chromatin for activation, also serves to help protect the Xist locus from being silenced during the XCI process by actively removing H3K27me3 during the process of XCI. However, additional studies are needed to fully address that possibility. KDM5C, which is also encoded by an escape gene, has recently been implicated in facilitating Xist expression by converting H3K4me3 to H3K4me1 at the Xist 5’ end [7]. Interestingly, KDM5C appears to be enriched at a similar location on the Xist gene as KDM6A, suggesting potential cooperation between the two proteins, each encoded by an escapee, that may help form a permissive chromatin environment for Xist expression. This previous study also points to a threshold of higher expression of escapees in females necessary for XCI. Consistent with this, our heterozygous depletion of Kdm6a leads to reduced Xist upregulation similar to our homozygous KO, albeit at a later time point. KDM6A may also regulate the expression of other regulatory elements located in the X inactivation center (XIC), such as Ftx and Jpx, two escape genes with roles in Xist regulation [41]. Transcription of the lncRNA Ftx promotes Xist activation during initiation of XCI [42]. While Ftx has very low expression in Tsix-stop ES cells (< 1TPM), we did observe an increase of H3K27me3 levels at the Ftx promoter and a slight decrease in Ftx expression following Kdm6a KO, which could contribute to a decrease in Xist. Tsix is another important regulator of Xist. However, the consistently lower expression of Tsix we observed during early differentiation of Kdm6a KO cells compared to wt, indicates that the decrease in Xist expression is independent of Tsix-induced repression [43].
The polycomb complexes PRC1/2 that mediate methylation of H3K27 have been implicated in the dynamics of XCI and maternally imprinted genes [26, 44, 45]. Indeed, the absence of polycomb group proteins due to lack of Xist-mediated recruitment leads to a relaxation of transcriptional silencing [46]. Consistent with these results, we observe a significant upregulation of gene expression from the Xi following differentiation in Kdm6a KO cells. A trend of increased expression from the Xi was seen for all temporally defined clusters of silenced genes, as well as genes in the escape category. Genes that escape XCI rarely achieve a level of Xi expression equal to that from the Xa due the repressive chromatin environment of the Xi [28]. Thus, the effects of Kdm6a KO on genes that escape may be due to an overall relaxation of the Xi heterochromatin. Our Xi-specific analysis of H3K27me3 indicates that KDM6A’s main action in XCI is on upregulation of Xist itself, and not through regulation of individual X-linked genes. Indeed, increased gene expression from the Xi was not accompanied by a clear reduction of H3K27me3 peaks on the Xi in differentiated Kdm6a KO cells compared to wt. It is possible that depletion of Xist, together with the absence in histone demethylation on the Xi due to KDM6A KO results in relatively balanced Xi H3K27me3 levels relative to wt.
Here, we illustrate a novel female-specific role for the histone demethylase KDM6A in the threshold dependent regulation of Xist expression for the initiation of XCI through its histone demethylation-dependent function. A recent study also points to a role for KDM6A in regulating XIST expression in human disease [47]. Therefore, extending our studies to human models will help determine the role of KDM6A in regulating X-linked gene expression in cases of sex chromosome aneuploidy as well as Kabuki syndrome and in cancer, where it is frequently mutated.
Perspectives and significance
This study identifies KDM6A as a female-biased novel member of the Xist regulome by demonstrating that KDM6A helps increase Xist expression by removing a repressive histone mark H3K27me3 at the Xist promoter region. The female-biased expression of Kdm6a may have evolved as a mechanism to enhance female-specific processes such as X inactivation. Interestingly, Kdm5c, another gene that also escapes X inactivation and encodes another demethylase critical for enhancer and promoter activity, has also been implicated in facilitating Xist expression. Thus, KDM6A and KDM5C may cooperate in enhancing Xist expression at a critical time during early development to ensure silencing of the X chromosome in females. KDM6A appears to have both histone demethylase-dependent and -independent activities, which are not well understood. Future studies should include additional epigenetic analyses of histones and of other chromatin components, together with analyses of the 3D conformation of chromatin, which may help ascertain the complex network of gene regulation by KDM6A. Our findings may have implications for the understanding of sex differences in disease, including sex-biased immune disorders where X inactivation is often incomplete. Finally, extending our studies to human stem cell models will help understand the role of KDM6A in disease.
Conclusions
Here, we demonstrate a novel role for the female-biased H3K27me3 demethylase KDM6A in the regulation of the female-specific process of X chromosome inactivation. We show that depletion of KDM6A prevents Xist upregulation during early differentiation, resulting in reduced efficacy of X chromosome silencing. Thus, escape from X inactivation may have evolved in part to perform essential roles in female-specific processes such as X inactivation.
Methods
Cell culture and differentiation
The female F1 hybrid mouse ES line Tsix-stop was derived from a cross between 129 and cast followed by the insertion of a transcriptional stop in the Tsix gene on the 129 X chromosome [18]. Differentiation of Tsix-stop ES cells results in skewed XCI where the Xi is always the 129 (maternal) X chromosome. In contrast, differentiation of the female F1 hybrid mouse ES line E8, which is derived from a cross between C57BL/6J (B6) and cast, results in largely random XCI. ES cells (Tsix-stop, E8) were maintained in the presence of 1000 U/ml leukemia inhibitory factor (LIF) (Millipore) on a monolayer of chemically inactivated mouse embryonic fibroblasts (MEFs), in high glucose DMEM media supplemented with 15% fetal bovine serum (FBS), 1% non-essential amino acids, 10 mg/ml APS, 0.1mM 2mercaptoethanol and 25 mM l-glutamine, in a humidified incubator at 37 ºC and 5% CO2. Just prior to use, ES cells were enriched by incubation on 0.1% gelatin-coated dishes for 1h to allow MEFs to attach, followed by transfer to fresh gelatin-coated plates for overnight culture. Following expansion, ES cells were split once (1:10) to further reduce potential MEF contamination. For embryoid body (EB) formation, 4x106 wt control and Kdm6a KO cells were cultured on non-adherent bacterial culture dishes without LIF for 7 days. Cells were collected at D0, D2, D4, and D7 of differentiation. Cells were then plated onto gelatin-coated cell culture plates and maintained for another 8 days prior to collection at D15.
Kdm6a editing using CRISPR/Cas9
We chose a dual sgRNA approach because of the capacity to create large deletions and reduced chance of off-target effects [48]. Editing of Kdm6a in Tsix-stop ES cells was done by constructing CRISPR plasmids containing sgRNAs that target exons 2 and 4 to delete ~ 45kb, as done previously [14, 49]. ES cells were co-transfected with the CRISPR/Cas9 constructs and a plasmid carrying puromycin resistance using UltraCruz® transfection reagent at a 3:1 CRISPR to pPGKpuro ratio in media with no antibiotics. Two days later, cells were selected in ES media containing 1 μg/ml puromycin for 48–72 h, followed by recovery in media with antibiotic. Cells were then cloned into 96-well plates using serial dilutions. Clones were expanded and screened for deletions using PCR and Sanger sequencing to confirm the predicted ~ 45 kb deletion, which was found in three clones including a heterozygous deletion clone (Tsix-Kdm6awt/ΔE4) and two homozygous deletion clones (Tsix-Kdm6aΔE/ΔE17 and Tsix-Kdm6aΔE/ΔE21) (Additional File 1: Figure S1A; Additional File 2: Table S1). Additionally, we confirmed that neither Tsix-cln1 nor Tsix-cln2 CRISPR controls were edited (Additional File 1: Figure S1A). Editing of Kdm6a in E8 ES cells was done by targeting the Kdm6a promoter region using sgRNAs designed using the CHOPCHOP v2 online tool [50]. PCR amplification and Sanger sequencing confirmed the predicted ~5 kb homozygous promoter deletion in one clone (E8-Kdm6aΔP/ΔP13) (Additional File 1: Figure S1A; Additional File 2: Table S1). Plasmid preparations were made using either the Qiagen Mini-prep or Maxi-prep kits according to the manufacturer protocol. Plasmid DNA and RNA lysates were submitted to Eurofin Genomics for Sanger sequencing (http://www.operon.com).
DNA, RNA and protein isolation and analyses
RNA and DNA lysates were prepared using Qiagen RNeasy mini kit and the Qiagen DNeasy blood and tissue kit, respectively. cDNA synthesis was done with the GoScript reverse transcription kit (Promega). Relative transcript levels were determined using SYBR Green PCR master mix on an ABI 7900HT machine. qRT-PCR assays were conducted in triplicates and normalized to Actinβ prior to analysis using the comparative CT method. For protein analyses, western blots were performed using a monoclonal antibodies directed against KDM6A (Cell Signaling #33510) at 1:1000 and GAPDH (GeneTex #GTX627408) at 1:10000 dilution in 5% BSA according to standard methods. 30ug of total protein was added to each lane and visualized with Clarity ECL (Bio-Rad #1705060). Band densities were calculated using Imagej (https://imagej.net/ij/).
DNA-FISH and RNA-FISH
To verify the presence of two X chromosomes in differentiated wt and CRISPR-treated cells, DNA-FISH was done on nuclei fixed in methanol:acetic acid (3:1 volume) using BAC probes (RP23-299L1 for minisatellite repeats or RP24-322N20 for Firre) [51]. Probes were labeled with SpectrumGreen (Vysis #02N32-050) using a nick-translation reagent kit according to the manufacturer’s protocol (Abbott Molecular Inc.). Probes in hybridization buffer were denatured at 70 °C for 5 min and hybridized overnight at 37 °C to slides that were also denatured at 70 °C for 5 min prior to hybridization. On the next day, slides were washed in SSC containing NP40 at 73 °C, followed by washes at room temperature. Counterstaining was done with Hoechst 33342 (2 µg/ml) in phosphate buffered saline (PBS) and slides were mounted in anti-fade solution.
For Xist RNA-FISH cells were plated on chamber slides at D7 of differentiation and allowed to differentiate until D15 when cells were processed directly on the chamber slides as described [52]. Briefly, cells were washed in PBS and permeabilized in CSK buffer with 0.05% Triton. Following fixation in 4% paraformaldehyde, cells were washed in 70% ethanol and dehydrated for 3 min each in 70%, 85%, and 100% ethanol. Dehydrated slides were incubated with a denatured probe for Xist RNA overnight in 50% formamide/50% 2xSSC at 42 °C in a humidified chamber [52]. On the following day, slides were washed three times in 50% formamide/50% 2xSSC at 42 °C for 10 min followed by one wash in 2xSSC at 42 °C for 5 min. Counterstaining was done with 33342 Hoechst in 2xSSC. A minimum of 130 nuclei were scored for each condition to quantify the number of nuclei with either an Xist cloud, a pinpoint Xist signal, or no Xist signal.
RNA-seq and allelic gene expression analysis
RNA-seq indexed libraries were prepared using Illumina TruSeq RNA sample preparation kit V2. For Tsix-stop cells, libraries were prepared from two biological replicates from wt controls, one CRISPR control (cln1) and Tsix-Kdm6aEΔ/EΔ17 and Tsix-Kdm6aEΔ/EΔ21 KO clones at D0 and D15. For E8 cells, libraries were prepared from one wt control and one E8-Kdm6aΔP/ΔP13 clone at D0, D2 and D4. Sequencing on a NextSeq sequencer yielded 75bp single-end reads. Diploid gene expression was estimated using Tophat/v2.0.14 [53] with default parameters and gene-level expression was normalized using TPM (transcripts per kilobase of exon length per million mapped reads). Differentially expressed genes were determined using DESeq2 analysis [54] with a false discovery rate (FDR) threshold of 0.1 and a fold-change cutoff of 2. PCA plots and heatmaps were generated with iDEP.95 using raw read counts as the input. Only genes with a minimum expression value of 2.5 CPM in at least two libraries were included (http://bioinformatics.sdstate.edu/idep95/).
To estimate allele-specific gene expression in mouse hybrid cells, the pseudo-diploid genome of 129xcast was generated using GATK-fastagenerator subcommand [55], taking the mm10 reference genome fasta file and the corresponding VCF file as the input. The VCF file was downloaded from the Sanger mouse genomes project [56], with indels and low-quality SNPs filtered out. RNA-seq reads were mapped using bowtie2 [57] to the diploid pseudo-129 and pseudo-cast genomes and transcriptomes. Only those reads that mapped uniquely and with a high-quality mapping score (MAPQ ≥ 30) were kept for further analyses. After filtering, reads were segregated into three categories: (1) 129-SNP reads containing only 129-specific SNPs; (2) cast-SNP reads containing only cast-specific SNPs; (3) allele-uncertain reads, that is, reads that do not contain valid SNPs. Allelic mapping metrics are shown in Additional File 16: Table S8.
Epigenetic analyses by CUT&RUN
Cut&Run for KDM6A was done in wt cells using an antibody against KDM6A (Cell signaling #33510) according to the manufacturer’s protocol (EpiCypher) except with mild fixation (0.01% formaldehyde for 1 min). Cut&Run for H3K27me3 was performed on clone Tsix-Kdm6aΔE/ΔE17 and wt cells using an antibody against H3K27me3 (Millipore) following a published protocol [58]. Libraries were constructed using TruSeq DNA sample preparation kit (Illumina) following the manufacturer protocol with minor changes including two 1.1x bead purifications during the final selection step.
Libraries were sequenced on a NextSeq sequencer to generate 75bp paired-end reads. In the following text, we refer to paired-end reads simply as “reads”. Demultiplexed fastq files were mapped to NCBi v38/mm10 mouse genome using bowtie2 [57] with the following parameters:
––end–to–end ––very–sensitive ––no–mixed ––no–discordant ––phred33 –I 10 –X 700.
Only paired reads that were mapped to the same chromosome with a fragment length of less than 1 kb were kept for further analyses. Read alignment BAM files were converted to bigWig format for IGV visualization using Bedtools [59]. Read counts were normalized using RPKM; and IgG control was subtracted from the knockout sample using the bamcompare subcommand in deeptools [60]. BAM files of KDM6A Cut&Run replicates for D2, D4, and D7 were merged using SAMtools to facilitate peak calling. The read counts in the BigWig files were normalized based on RPKM. Peak calling for KDM6A and H3K27me3 samples was performed using SEACR [61] on bedGraph files, applying the “norm” option.
For allelic-analysis, the pseudo-diploid genome of 129 × cast was generated using GATK-fastagenerator subcommand [55], taking the mm10 reference genome fasta file and the corresponding VCF file as the input. The VCF file was downloaded from the Sanger mouse genomes project [56], with indels and low-quality SNPs filtered out. Reads were mapped to the diploid pseudo-129 and pseudo-cast genomes using bowtie2 [57] with the following parameters:
––end–to–end ––very–sensitive ––no-mixed ––no–discordant –phred33 –I 10 –X 700.
Reads were further processed through a read tagging step, where read alignments of both pseudo-129 and pseudo-cast genomes were compared to identify the allele origins of the reads. Reads overlapping SNPs were assigned to G1 (129 genome), G2 (cast), or CF (conflicting) based on the sequenced base information at SNP sites, while reads do not overlap with any SNPs were assigned to UA (Unassigned). The G1 and G2 tagged reads were separated into two BAM files, which were later used to generate allele-specific bigWig files for IGV visualization [62]. An allelic mapping summary can be found in Additional File 17: Table S9.
Analysis of overlapping KDM6A and H3K27me3 peaks
We called peaks for all samples using SEACR as described, filtering out H3K27me3 peaks with a signal lower than 10 for the D15 samples. Due to lower coverage, peaks with a signal lower than 5 for the KDM6A sample were excluded. Transcription start sites (TSS) were defined as regions extending 5 kb upstream and downstream from the transcript start site. TSS regions containing peaks were extracted for each sample. These sample-specific TSS regions were used to identify both sample-specific and shared peaks across two or three samples for Venn diagram plotting. Only the number of genes rather than transcripts was considered.
Supplementary Information
Additional file 1: Supplemental figure S1. CRISPR-Cas9 strategy and characteristics of Kdm6a KO cells. (A) Top: Schematic shows the location of the exonic deletion (Kdm6aΔE) that removed exons 2–4 of Kdm6a in female Tsix-stop ES cells and the promoter targeted deletion (Kdm6aΔP) made in female E8 ES cells. Exons are shown as vertical bars. The location of the PCR and RT-PCR primers (color-coded arrows) used to confirm each deletion is indicated. Below left: Images of gels after electrophoresis of PCR products using different sets of primers to confirm Kdm6a homozygous and heterozygous KO (- no deletion; + deletion positive). Actinβ was run as a control. Below right: Partial Kdm6a sequence obtained by Sanger sequencing as verification of deletions in Tsix-stop and E8 homozygous Kdm6a KO clones. Arrows point to the location of non-homologous end-joining and colored arrows correspond to those on the schematic. (B) Heat maps of gene expression differences between Tsix-stop wt (2 replicates, wt a and wt b) and KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D0 and D15. Consistent with PCA clustering, more DEGs were identified at D15 than D0. Heat maps were generated using iDEP.95. (C) Scatter plots of expression of genes involved in germ layer differentiation obtained by RNA-seq. Tsix-stop wt and KO cells after differentiation (D15) show higher expression of germ layer-associated genes in wt versus KO cells (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21). Log2 average TPM values are from two wt replicates and two KO clones. E8-Kdm6a KO cells show no trend of significant difference in expressed germ layer genes at D2 or D4 of differentiation. P-values are calculated using 1-way ANOVA test.
Additional file 5:Supplemental figure S2. Confirmation of Xist expression changes in Kdm6a KO cells. (A) qRT-PCR of Xist expression normalized to Actinβ during ES differentiation in three Tsix-stop wt replicates, the two Kdm6aΔEΔE KO clones, and one Kdm6aΔwtΔEclone. Shown is expression over time for each individual sample. (B) Histogram of qRT-PCR for Xist expression during differentiation in Tsix-stop wt and CRISPR controls (Additional File 1: Table S1). Expression is normalized to Actinβ. Expression at D4 is relative to D0 for each cell line. As expected, Xist is upregulated upon differentiation in wt (wta and wtb replicates) and CRISPR-control clones (cln1 and cln2). (C) Histogram of log2of average TPM values for pluripotency genes and genes known to play a role in Xist repression. Average TPM values are from two Tsix-stop wt and two Kdm6a KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21). Only Klf4 is called as differentially expressed between wt and KO by DESeq2 (***p<0.002). (D, E) Histograms of qRT-PCR of Tsix expression in (D) Tsix-stop wt and Kdm6aΔEΔE KO clones, and (E) E8 wt and KO clone E8-Kdm6aΔP/ΔP13 at D2 and D4. Expression is normalized to Actinβ and relative to wt. (F) Histogram of average TPM values for Xist in Tsix-stop wt, a CRISPR-control clone (cln1), and two KO clones (Tsix-Kdm6aΔEΔE17, Tsix-Kdm6aΔEΔE21) at D0, confirming that Xist expression is very low in ES cells cultured with serum.
Additional file 6 Supplemental figure S3. Two X chromosomes are present in wt and KO Tsix-stop cells. (A, B) DNA-FISH using probes specific for the X-linked gene Firre or an X-linked mini-satellite repeat region (Dxz4) labelled in green in Tsix-stop wt (A) and CRISPR-control (cln1) (B). Examples of nuclei with two green signals representing the two X chromosomes are shown along with histograms of the number of signals in nuclei scored. n indicates the number of nuclei scored. Nuclei are counterstained with Hoechst 33342. (C, D) Same analysis in as in (A, B), but in the Tsix-stop Kdm6a KO cell clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21).
Additional file 7 Supplemental Figure S4. Confirmation of X-linked gene expression changes in E8-Kdm6aΔPΔP13 KO cells. (A) Allelic X-linked gene expression ratios (BL6:cast) in E8 wt and KO cells (E8-Kdm6aΔPΔP13) at D0, D2, and D4 of differentiation. Ratios between expressed X-linked genes (>1TPM) are shown. X expression ratios near 0.5 reflect the largely random nature of XCI in these cells. (B) Plots of expression changes from the BL6 and cast X chromosomes in E8 wt and KO cells (E8-Kdm6aΔPΔP13) at D4. The XIC is highlighted. Log2 TPM fold change (KO/wt) along the X chromosomes is shown. Genes with a decrease of expression in KO versus wt are in blue, and with an increase in red. (D) Venn diagram of the number of X-linked genes with upregulation following Kdm6a KO. The majority of genes are upregulated from the BL6 and cast X chromosomes. (D) Plots of expression changes (based on log2 TPM) for genes with increased expression from each X chromosome (BL6 purple and cast blue) between E8 wt and E8-Kdm6aΔPΔP13 KO cells at D4. The X-axis includes 254 X-linked genes. There is a slightly greater increase in expression from the BL6 X chromosome (purple).
Additional file 10 Supplemental figure S5. H3K27me3 profiles along the XIC in wt and Kdm6a KO cells. (A) IGV browser view of profiles and peaks of H3K27me3 enrichment at the XIC (chrX: 103126305-103891745) show increased H3K27me3 at Cdx4, Xist, and Ftx in differentiated Kdm6a KO cells. Tsix-stop wt (blue) and KO clone Tsix-Kdm6aΔEΔE17 (red). (B, C) Histograms of expression (TPM) of Cdx4 (B) and Ftx (C) in D0 Tsix-stop wt (blue) and Kdm6a KO clones (red). Both wt and KO ES cells show low expression of Cdx4 and Ftx (~1.5TPM or less). (D, E) Same analysis as in (B) and (C) but for D15. Both wt and KO ES cells show low expression of Cdx4 and Ftx (~1TPM or less).
Additional file 11 Supplemental figure S6. Epigenetic and expression changes at known KDM6A target genes. (A) IGV browser views of profiles of H3K27me3 enrichment and peaks at known KDM6A target genes in differentiated Tsix-stop wt cells (blue) and KO clones (Tsix-Kdm6aΔEΔE17) (red). Peaks are represented by colored bars below the signal profiles. Below are RNA-seq expression profiles (black) in wt and KO cells. Histograms show TPM expression values for T and Pitx2 in two differentiated wt clones and KO clones. For Wnt3 and Gata4, both changes in expression at D0-7 obtained by qRT-PCR during differentiation and histograms of TPM values following differentiation are shown (*p<0.05). KO represents an average for the Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21 clones. Values are normalized to Actinβ. (B) Same analysis as in (A), but for Nsdhl and Araf, two genes that do not show significant downregulation following Kdm6a KO. (C) Same analysis as in (A), but at the Dlk1/Meg3 imprinted locus at D15 where H3K27me3 markedly increases at the promoter of Meg3 in KO cells, including at the imprinting control region ICR (2) and the intergenic ICR (1). The scales of the profiles shown in (A), (B) and (C) are indicated in the upper right corners.
Additional file 13 Supplemental figure S7. Characterization of KDM6A binding, expression and protein levels during differentiation of Tsix-stop cells. (A) Venn diagram comparing number of peaks of KDM6A in wt Tsix-stop cells at D4 of differentiation to H3K27me3 peaks in wt and Kdm6a KO cells at D15 of differentiation. (B) Same analysis as in (A) but using KDM6A binding at D7 for comparisons. (C) KDM6A enrichment profiles at Foxr2 and Igsf9, two genes known to be regulated by KDM6A. D4 signal and peaks in wt Tsix-stop cells are shown. Scales are shown in the upper right corner. (D) Top, protein blots of KDM6A in wt Tsix-stop (top) during differentiation. Adjusted density was calculated by normalizing levels to the GAPDH loading control using ImageJ (middle). Bottom, histograms of qRT-PCR of Kdm6a expression in two wt Tsix-stop replicates during differentiation. Expression was normalized using Actinβ and expression relative to D0 is shown.
Additional file 15: Supplemental figure S8. Allelic H3K27me3 enrichment at X-linked genes with Xi-specific expression increases in Kdm6a KO Tsix-stop cells. IGV browser views of Xi H3K27me3 signal and peaks at a subset of genes with increased expression from the Xi in Kdm6a KO cells. Wt cells are in pink and KO cells are in purple. Some genes with Xi-specific increases in expression following differentiation (D15) show lack of H3K27me3 near their TSS, due to failure of XCI and recruitment of PRC complexes.
Acknowledgements
We thank Di Kim Nguyen (University of Washington) for consultation on this study and for critical reading of the manuscript and Charlie Lee (University of Washington) for his help with next-generation sequencing. Hybrid mouse ES cells used in this study were graciously donated by Joost Gribnau (Erasmus MC, University Medical Center).
Author contributions
Conceptualization, J.B. and C.M.D.; Investigation, J.B., J.L., C. K., H.F., C.G.; Formal analysis, H.F., J.Z., L.M., R.M., W.M.; Data Curation, W.M.; Writing–Original Draft, J.B. and C.M.D.; Writing–Review and Editing, J.B., J.L., W.M., G.N.F., and C.M.D.; Visualization, J.B., J.Z., W.M., X.D., and C.M.D.; Supervision, J.B., J.E.Y., and C.M.D.; Funding Acquisition, J.B. and C.M.D.
Funding
This work was supported by NIH grants GM131745 (CMD), HG011586 (CMD), MH105768 (JB), AG073918 (CMD), and NSF DBI175317 (WM). The funders had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All sequencing data that support findings in this study have been deposited in the NCBI GEO database under the accession numbers GSE279278 and GSE279279. All other data and the scripts used for the analyses that support the findings of this study are available from the corresponding authors upon reasonable request. Oligonucleotide sequences used in this study are available in Additional File 2: Table S1.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publications
All authors have given their consent for publication of the manuscript in Biology of Sex Differences.
Competing interests
The authors declare no competing interests
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Josephine Lin and Jinli Zhang are contributed equally to this work.
Contributor Information
Wenxiu Ma, Email: wenxiu.ma@ucr.edu.
Christine M. Disteche, Email: cdistech@uw.edu
Joel B. Berletch, Email: joelb07@uw.edu
References
- 1.Loda A, Collombet S, Heard E. Gene regulation in time and space during X-chromosome inactivation. Nat Rev Mol Cell Biol. 2022;23(4):231–49. 10.1038/s41580-021-00438-7. [DOI] [PubMed] [Google Scholar]
- 2.Fang H, Disteche CM, Berletch JB. X inactivation and escape: epigenetic and structural features. Front Cell Dev Biol. 2019;7:219. 10.3389/fcell.2019.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks H, Kerstens HH, Barakat TS, Splinter E, Dirks RA, van Mierlo G, et al. Dynamics of gene silencing during X inactivation using allele-specific RNA-seq. Genome Biol. 2015;16:1–149. 10.1186/s13059-015-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pacini G, Dunkel I, Mages N, Mutzel V, Timmermann B, Marsico A, et al. Integrated analysis of Xist upregulation and X-chromosome inactivation with single-cell and single-allele resolution. Nat Commun. 2021;12(1):3638. 10.1038/s41467-021-23643-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tukiainen T, Villani AC, Yen A, Rivas MA, Marshall JL, Satija R, et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550(7675):244–8. 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Disteche CM, Berletch JB. X-chromosome inactivation and escape. J Genet. 2015;94(4):591–9. 10.1007/s12041-015-0574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samanta MK, Gayen S, Harris C, Maclary E, Murata-Nakamura Y, Malcore RM, et al. Activation of Xist by an evolutionarily conserved function of KDM5C demethylase. Nat Commun. 2022;13(1):2602. 10.1038/s41467-022-30352-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenfield A, Carrel L, Pennisi D, Philippe C, Quaderi N, Siggers P, et al. The UTX gene escapes X inactivation in mice and humans. Hum Mol Genet. 1998;7(4):737–42. 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- 9.Wainer Katsir K, Linial M. Human genes escaping X-inactivation revealed by single cell expression data. BMC Genomics. 2019;20(1):201. 10.1186/s12864-019-5507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–4. 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 11.Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, et al. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci USA. 2012;109(38):15324–9. 10.1073/pnas.1204166109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shpargel KB, Starmer J, Wang C, Ge K, Magnuson T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc Natl Acad Sci USA. 2017;114(43):E9046–55. 10.1073/pnas.1705011114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang QY, Zhang SF, Dai SK, Liu C, Wang YY, Du HZ, et al. UTX Regulates human neural differentiation and dendritic morphology by resolving bivalent promoters. Stem Cell Reports. 2020;15(2):439–53. 10.1016/j.stemcr.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Fang H, Pease N, Filippova GN, Disteche CM, Berletch JB. Sex-biased and parental allele-specific gene regulation by KDM6A. Biol Sex Differ. 2022;13(1):40. 10.1186/s13293-022-00452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 2012;8(9):e1002964. 10.1371/journal.pgen.1002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508(7497):494–9. 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508(7497):488–93. 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 18.Luikenhuis S, Wutz A, Jaenisch R. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol Cell Biol. 2001;21(24):8512–20. 10.1128/MCB.21.24.8512-8520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattanach BM, Rasberry C. Identification of the Mus castaneus Xce allele. Mouse Genome. 1994; 114.
- 20.Tran N, Broun A, Ge K. Lysine demethylase KDM6A in differentiation, development, and cancer. Mol Cell Biol. 2020;40:20. 10.1128/MCB.00341-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. 2011;130(2):223–36. 10.1007/s00439-011-1008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarro P, Oldfield A, Legoupi J, Festuccia N, Dubois A, Attia M, et al. Molecular coupling of Tsix regulation and pluripotency. Nature. 2010;468(7322):457–60. 10.1038/nature09496. [DOI] [PubMed] [Google Scholar]
- 23.Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99(1):47–57. 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- 24.Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O’Neill LP, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5(12):e326. 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol. 2017;24(3):226–33. 10.1038/nsmb.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue A, Jiang L, Lu F, Zhang Y. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev. 2017;31(19):1927–32. 10.1101/gad.304113.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Disteche CM. Dosage compensation of the sex chromosomes and autosomes. Semin Cell Dev Biol. 2016;56:9–18. 10.1016/j.semcdb.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarro-Cobos MJ, Balaton BP, Brown CJ. Genes that escape from X-chromosome inactivation: potential contributors to Klinefelter syndrome. Am J Med Genet C Semin Med Genet. 2020;184(2):226–38. 10.1002/ajmg.c.31800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peeters SB, Korecki AJ, Baldry SEL, Yang C, Tosefsky K, Balaton BP, et al. How do genes that escape from X-chromosome inactivation contribute to Turner syndrome? Am J Med Genet C Semin Med Genet. 2019;181(1):28–35. 10.1002/ajmg.c.31672. [DOI] [PubMed] [Google Scholar]
- 30.Sun KY, Oreper D, Schoenrock SA, McMullan R, Giusti-Rodriguez P, Zhabotynsky V, et al. Bayesian modeling of skewed X inactivation in genetically diverse mice identifies a novel Xce allele associated with copy number changes. Genetics. 2021;218:1. 10.1093/genetics/iyab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9(5):e1003489. 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpargel KB, Starmer J, Yee D, Pohlers M, Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10(8):e1004507. 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navarro P, Chambers I, Karwacki-Neisius V, Chureau C, Morey C, Rougeulle C, et al. Molecular coupling of Xist regulation and pluripotency. Science. 2008;321(5896):1693–5. 10.1126/science.1160952. [DOI] [PubMed] [Google Scholar]
- 34.Minkovsky A, Barakat TS, Sellami N, Chin MH, Gunhanlar N, Gribnau J, et al. The pluripotency factor-bound intron 1 of Xist is dispensable for X chromosome inactivation and reactivation in vitro and in vivo. Cell Rep. 2013;3(3):905–18. 10.1016/j.celrep.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lang A, Whongsiri P, Yilmaz M, Lautwein T, Petzsch P, Greife A, et al. Knockdown of UTX/KDM6A enriches precursor cell populations in urothelial cell cultures and cell lines. Cancers. 2020;12:4. 10.3390/cancers12041023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng Y, Zhang T, Zheng R, Ding S, Yang J, Liu R, et al. Depletion of demethylase KDM6 enhances early neuroectoderm commitment of human PSCs. Front Cell Dev Biol. 2021;9: 702462. 10.3389/fcell.2021.702462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, et al. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14(2):203–16. 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–8. 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 39.Dhar SS, Lee SH, Chen K, Zhu G, Oh W, Allton K, et al. An essential role for UTX in resolution and activation of bivalent promoters. Nucleic Acids Res. 2016;44(8):3659–74. 10.1093/nar/gkv1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cerase A, Young AN, Ruiz NB, Buness A, Sant GM, Arnold M, et al. Chd8 regulates X chromosome inactivation in mouse through fine-tuning control of Xist expression. Commun Biol. 2021;4(1):485. 10.1038/s42003-021-01945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galupa R, Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu Rev Genet. 2018;52:535–66. 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 42.Furlan G, Gutierrez Hernandez N, Huret C, Galupa R, van Bemmel JG, Romito A, et al. The Ftx noncoding locus controls X chromosome inactivation independently of its RNA products. Mol Cell. 2018;70(3):462-72 e8. 10.1016/j.molcel.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 43.Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci USA. 2001;98(18):10232–7. 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature. 2017;547(7664):419–24. 10.1038/nature23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue A, Chen Z, Yin Q, Zhang Y. Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev. 2018;32(23–24):1525–36. 10.1101/gad.318675.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bousard A, Raposo AC, Zylicz JJ, Picard C, Pires VB, Qi Y, et al. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019;20(10):e48019. 10.15252/embr.201948019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo RY, Song S, Wang JQ, Guo JY, Liu J, Jia Z, et al. Downregulation of lncRNA XIST may promote Th17 differentiation through KDM6A-TSAd pathway in neuromyelitis optica spectrum disorders. Mult Scler Relat Disord. 2023;76: 104801. 10.1016/j.msard.2023.104801. [DOI] [PubMed] [Google Scholar]
- 48.Byrne SM, Ortiz L, Mali P, Aach J, Church GM. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015;43(3):e21. 10.1093/nar/gku1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44(W1):W272–6. 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang H, Bonora G, Lewandowski JP, Thakur J, Filippova GN, Henikoff S, et al. Trans- and cis-acting effects of Firre on epigenetic features of the inactive X chromosome. Nat Commun. 2020;11(1):6053. 10.1038/s41467-020-19879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a Mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ayadi A, Birling MC, Bottomley J, Bussell J, Fuchs H, Fray M, et al. Mouse large-scale phenotyping initiatives: overview of the European mouse disease clinic (EUMODIC) and of the Wellcome trust sanger institute mouse genetics project. Mamm Genome. 2012;23(9–10):600–10. 10.1007/s00335-012-9418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13(5):1006–19. 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 59.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–5. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meers MP, Tenenbaum D, Henikoff S. Peak calling by Sparse Enrichment Analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;1:42. 10.1186/s13072-019-0287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–92. 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental figure S1. CRISPR-Cas9 strategy and characteristics of Kdm6a KO cells. (A) Top: Schematic shows the location of the exonic deletion (Kdm6aΔE) that removed exons 2–4 of Kdm6a in female Tsix-stop ES cells and the promoter targeted deletion (Kdm6aΔP) made in female E8 ES cells. Exons are shown as vertical bars. The location of the PCR and RT-PCR primers (color-coded arrows) used to confirm each deletion is indicated. Below left: Images of gels after electrophoresis of PCR products using different sets of primers to confirm Kdm6a homozygous and heterozygous KO (- no deletion; + deletion positive). Actinβ was run as a control. Below right: Partial Kdm6a sequence obtained by Sanger sequencing as verification of deletions in Tsix-stop and E8 homozygous Kdm6a KO clones. Arrows point to the location of non-homologous end-joining and colored arrows correspond to those on the schematic. (B) Heat maps of gene expression differences between Tsix-stop wt (2 replicates, wt a and wt b) and KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21) at D0 and D15. Consistent with PCA clustering, more DEGs were identified at D15 than D0. Heat maps were generated using iDEP.95. (C) Scatter plots of expression of genes involved in germ layer differentiation obtained by RNA-seq. Tsix-stop wt and KO cells after differentiation (D15) show higher expression of germ layer-associated genes in wt versus KO cells (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21). Log2 average TPM values are from two wt replicates and two KO clones. E8-Kdm6a KO cells show no trend of significant difference in expressed germ layer genes at D2 or D4 of differentiation. P-values are calculated using 1-way ANOVA test.
Additional file 5:Supplemental figure S2. Confirmation of Xist expression changes in Kdm6a KO cells. (A) qRT-PCR of Xist expression normalized to Actinβ during ES differentiation in three Tsix-stop wt replicates, the two Kdm6aΔEΔE KO clones, and one Kdm6aΔwtΔEclone. Shown is expression over time for each individual sample. (B) Histogram of qRT-PCR for Xist expression during differentiation in Tsix-stop wt and CRISPR controls (Additional File 1: Table S1). Expression is normalized to Actinβ. Expression at D4 is relative to D0 for each cell line. As expected, Xist is upregulated upon differentiation in wt (wta and wtb replicates) and CRISPR-control clones (cln1 and cln2). (C) Histogram of log2of average TPM values for pluripotency genes and genes known to play a role in Xist repression. Average TPM values are from two Tsix-stop wt and two Kdm6a KO clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21). Only Klf4 is called as differentially expressed between wt and KO by DESeq2 (***p<0.002). (D, E) Histograms of qRT-PCR of Tsix expression in (D) Tsix-stop wt and Kdm6aΔEΔE KO clones, and (E) E8 wt and KO clone E8-Kdm6aΔP/ΔP13 at D2 and D4. Expression is normalized to Actinβ and relative to wt. (F) Histogram of average TPM values for Xist in Tsix-stop wt, a CRISPR-control clone (cln1), and two KO clones (Tsix-Kdm6aΔEΔE17, Tsix-Kdm6aΔEΔE21) at D0, confirming that Xist expression is very low in ES cells cultured with serum.
Additional file 6 Supplemental figure S3. Two X chromosomes are present in wt and KO Tsix-stop cells. (A, B) DNA-FISH using probes specific for the X-linked gene Firre or an X-linked mini-satellite repeat region (Dxz4) labelled in green in Tsix-stop wt (A) and CRISPR-control (cln1) (B). Examples of nuclei with two green signals representing the two X chromosomes are shown along with histograms of the number of signals in nuclei scored. n indicates the number of nuclei scored. Nuclei are counterstained with Hoechst 33342. (C, D) Same analysis in as in (A, B), but in the Tsix-stop Kdm6a KO cell clones (Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21).
Additional file 7 Supplemental Figure S4. Confirmation of X-linked gene expression changes in E8-Kdm6aΔPΔP13 KO cells. (A) Allelic X-linked gene expression ratios (BL6:cast) in E8 wt and KO cells (E8-Kdm6aΔPΔP13) at D0, D2, and D4 of differentiation. Ratios between expressed X-linked genes (>1TPM) are shown. X expression ratios near 0.5 reflect the largely random nature of XCI in these cells. (B) Plots of expression changes from the BL6 and cast X chromosomes in E8 wt and KO cells (E8-Kdm6aΔPΔP13) at D4. The XIC is highlighted. Log2 TPM fold change (KO/wt) along the X chromosomes is shown. Genes with a decrease of expression in KO versus wt are in blue, and with an increase in red. (D) Venn diagram of the number of X-linked genes with upregulation following Kdm6a KO. The majority of genes are upregulated from the BL6 and cast X chromosomes. (D) Plots of expression changes (based on log2 TPM) for genes with increased expression from each X chromosome (BL6 purple and cast blue) between E8 wt and E8-Kdm6aΔPΔP13 KO cells at D4. The X-axis includes 254 X-linked genes. There is a slightly greater increase in expression from the BL6 X chromosome (purple).
Additional file 10 Supplemental figure S5. H3K27me3 profiles along the XIC in wt and Kdm6a KO cells. (A) IGV browser view of profiles and peaks of H3K27me3 enrichment at the XIC (chrX: 103126305-103891745) show increased H3K27me3 at Cdx4, Xist, and Ftx in differentiated Kdm6a KO cells. Tsix-stop wt (blue) and KO clone Tsix-Kdm6aΔEΔE17 (red). (B, C) Histograms of expression (TPM) of Cdx4 (B) and Ftx (C) in D0 Tsix-stop wt (blue) and Kdm6a KO clones (red). Both wt and KO ES cells show low expression of Cdx4 and Ftx (~1.5TPM or less). (D, E) Same analysis as in (B) and (C) but for D15. Both wt and KO ES cells show low expression of Cdx4 and Ftx (~1TPM or less).
Additional file 11 Supplemental figure S6. Epigenetic and expression changes at known KDM6A target genes. (A) IGV browser views of profiles of H3K27me3 enrichment and peaks at known KDM6A target genes in differentiated Tsix-stop wt cells (blue) and KO clones (Tsix-Kdm6aΔEΔE17) (red). Peaks are represented by colored bars below the signal profiles. Below are RNA-seq expression profiles (black) in wt and KO cells. Histograms show TPM expression values for T and Pitx2 in two differentiated wt clones and KO clones. For Wnt3 and Gata4, both changes in expression at D0-7 obtained by qRT-PCR during differentiation and histograms of TPM values following differentiation are shown (*p<0.05). KO represents an average for the Tsix-Kdm6aΔEΔE17 and Tsix-Kdm6aΔEΔE21 clones. Values are normalized to Actinβ. (B) Same analysis as in (A), but for Nsdhl and Araf, two genes that do not show significant downregulation following Kdm6a KO. (C) Same analysis as in (A), but at the Dlk1/Meg3 imprinted locus at D15 where H3K27me3 markedly increases at the promoter of Meg3 in KO cells, including at the imprinting control region ICR (2) and the intergenic ICR (1). The scales of the profiles shown in (A), (B) and (C) are indicated in the upper right corners.
Additional file 13 Supplemental figure S7. Characterization of KDM6A binding, expression and protein levels during differentiation of Tsix-stop cells. (A) Venn diagram comparing number of peaks of KDM6A in wt Tsix-stop cells at D4 of differentiation to H3K27me3 peaks in wt and Kdm6a KO cells at D15 of differentiation. (B) Same analysis as in (A) but using KDM6A binding at D7 for comparisons. (C) KDM6A enrichment profiles at Foxr2 and Igsf9, two genes known to be regulated by KDM6A. D4 signal and peaks in wt Tsix-stop cells are shown. Scales are shown in the upper right corner. (D) Top, protein blots of KDM6A in wt Tsix-stop (top) during differentiation. Adjusted density was calculated by normalizing levels to the GAPDH loading control using ImageJ (middle). Bottom, histograms of qRT-PCR of Kdm6a expression in two wt Tsix-stop replicates during differentiation. Expression was normalized using Actinβ and expression relative to D0 is shown.
Additional file 15: Supplemental figure S8. Allelic H3K27me3 enrichment at X-linked genes with Xi-specific expression increases in Kdm6a KO Tsix-stop cells. IGV browser views of Xi H3K27me3 signal and peaks at a subset of genes with increased expression from the Xi in Kdm6a KO cells. Wt cells are in pink and KO cells are in purple. Some genes with Xi-specific increases in expression following differentiation (D15) show lack of H3K27me3 near their TSS, due to failure of XCI and recruitment of PRC complexes.
Data Availability Statement
All sequencing data that support findings in this study have been deposited in the NCBI GEO database under the accession numbers GSE279278 and GSE279279. All other data and the scripts used for the analyses that support the findings of this study are available from the corresponding authors upon reasonable request. Oligonucleotide sequences used in this study are available in Additional File 2: Table S1.