Abstract
Background
Trimethylaminuria (TMAU) is a rare recessive genetic disorder with limited global prevalence. To date, there have been no official reports of TMAU cases documented in Saudi Arabia.
Purpose
In this study, we developed a liquid chromatography–mass spectrometry (LC-MS) method for the analysis of trimethylamine (TMA) and Trimethylamine N-Oxide (TMAO) in urine and plasma samples for the first reported case of TMAU in Saudi Arabia.
Patients and Methods
A 41-year-old Saudi man was diagnosed with TMAU in National Guard Hospital. Blood and urine samples were collected to confirm the diagnosis of TMAU. In this study, we have studied LC-MS, cell culture, flow cytometry, adhesion assay and Sanger sequencing analysis. Additionally, in this study, we have selected 5 healthy controls.
Results
The results have revealed elevated TMA levels were present in both urine and plasma samples, while TMAO levels were significantly lower compared to control group. Further, we utilized plasma sample from the TMAU patient as novel model to investigate the potential effect of low TMAO on monocyte and endothelial cell function in vitro. DNA sequencing analysis identified a c.622G >T (p.Glu208*) which creates a premature stop codon in FMO3 gene.
Conclusion
Our findings revealed differential responses in monocytes and endothelial cells stimulated with plasma from the TMAU patient compared to plasma from non-TMAU patients. These distinct responses may be key modulators of endothelial function and contributes to vascular damage.
Keywords: Trimethylaminuria, TMAU, LC-MS, cell culture, flow cytometry, adhesion assay and Sanger sequencing analysis
Introduction
Trimethylaminuria (TMAU) is a rare recessive genetic disorder associated with mutations in the FMO3 gene that cause loss of function in the FMO3 protein. This enzyme is responsible for conversion of trimethylamine (TMA) to trimethylamine N‐oxide (TMAO) in the liver. TMA containing nutrient precursors, choline, betaine, and carnitine present in different types of foods, such as egg, fish, and meat, are metabolized by gut microbiota to TMAO.1 In healthy individuals, more than 90% of TMA is converted (by FMO3) to nonodorous TMAO and excreted in the urine.2 Defective FMO3 enzyme can interrupt the conversion of TMA to TMAO, leading to accumulation of TMA in the body. This eventually leads to a strong fishy odor excreted from breath, sweat, and urine, which gives this disorder another name as fish odor syndrome.3
TMAU affects an estimated range of one to five cases per million people worldwide.4 However, underdiagnoses and lack of awareness suggest the true prevalence is likely higher. No physical symptoms associated with TMAU have been reported and patients often appear healthy. However, individuals diagnosed with TMAU experience major psychological and social problems including depression, embarrassment, social isolation, low self-esteem, and attempted suicide due to the fishy odor they suffer from.5–8
Relying solely on a rotten-fish body odor for diagnosing TMAU can be inaccurate. The odor can be episodic, and not all clinicians/physicians have the same sensitivity to the smell.4 TMAU is diagnosed and classified by a urine test that measures both TMA and TMAO concentrations. Gene sequencing of the FMO3 gene is used to confirm a diagnosis of primary TMAU. Based on both analyses, TMAU can be categorized into two types; primary and secondary (acquired). In Primary TMAU, patients have defective FMO3 enzyme, which can be affected by single or multiple mutations in the FMO3 gene. On the other hand, with secondary TMAU patients had elevated TMA levels in the urine due to multiple factors including an excessive dietary intake of TMA precursors, viral, and gut flora and hepatic disease that will lead to decreased activity of FMO3 enzyme.9 Several mutations in the FMO3 gene (OMIM: 136132 and LOVD database) have been reported including missense and nonsense types.10–15
The biochemical analysis of TMAU is achieved by measuring the levels of TMA and TMAO in urine and then calculates the TMA: TMAO ratio. Furthermore, following the biochemical analysis, the “FMO3 metabolic capacity” can be calculated using the following equation: TMAO/(TMA + TMAO), which can be used to classify TMAU into mild, moderate, or severe.15 Individuals with TMAU exhibit elevated levels of TMA associated with low levels of TMAO. From the broader perspective, several studies suggest a link between elevated plasma TMAO level and cardiovascular diseases.16 High TMAO level has been linked to endothelial cell dysfunction and monocyte activation.17 However, the impact of low TMAO levels, as observed in TMAU patients, on endothelial function and inflammation has remained unclear.18,19 The role of TMAO in cardiovascular disease remains controversial and two recent cohort studies have found no link.
Several analytical methods have been developed to determine TMA and TAMO in urine samples including proton nuclear magnetic resonance spectroscopy (H-NMR), matrix-assisted laser desorption/ionization with time-of-flight MS (MALDI-TOF-MS), fast atom bombardment mass spectrometry (FAB-MS), and capillary gas chromatography-mass spectrometry (GC-MS).20–23 Some of these methods lack sensitivity, specificity, or require time-intensive sample preparation.24 In contrast, stable isotope dilution liquid chromatography selected reaction monitoring (SID-LC-SRM) showed accurate and precise method to detect and quantify TMA and TMAO in biological samples.25,26
This study investigated the first documented case of an adult patient in Saudi Arabia presenting to our genetic clinic with Trimethylaminuria and experiencing unpleasant body odor. A comprehensive diagnostic approach was employed, incorporating clinical evaluation, genetic analysis, and LC-ESI-MS/MS for the quantification of TMA, TMAO, and their precursors. Further, this study investigated the impact of TMAO level on endothelial dysfunction in Trimethylaminuria patient using models of monocytes and macrophage cells.
Methods
Chemicals and Reagents
Trimethylamine Hydrochloride, Trimethylamine N-Oxide Dihydrate, choline chloride, and L-Carnitine were purchased from TCI Company. Betaine monohydrate and Trimethyl-d9-amine hydrochloride were purchased from Sigma Aldrich. Trimethylamine N-Oxide (d9), choline chloride (Trimethyl-d9), Betaine (d11) were purchased from Cambridge Isotope Laboratories. The Ultra PFP Propyl Column was purchased from Restek.
Patient and Study Approval
A 41-year-old Saudi male presented to our genetics clinic with a main complaint history of unpleasant body odor was included in our study. A control group of five healthy individuals, matched for age and ethnicity, was also recruited. The study was approved by the institutional review board of King Abdullah international medical research center (RC20/171 and RC-19-120-R). Written informed consent was obtained from participants. Also, this study compiles with the Declaration of Helsinki.
Preparation of Stock Solutions, Calibrators and Quality Control Samples
Stock solution (1 mg/mL) for standards and QCs were prepared in water using accurately weighed amounts of TMA, TMAO, Choline, Carnitine, and Betaine and stored at −20°C. For calibration standards, the stock solution was serially diluted in water to obtain ten different concentrations of (125, 62.5, 31.25, 15.6, 7.8, 3.9, 1.95, 0.97, 0.48, and 0.24, µM) for TMA, TMAO, and Choline and ten different concentrations of (15.6, 7.8, 3.9, 1.95, 0.97, 0.48, 0.24, 0.12, 0.06 and 0.03 µM) for Carnitine and Betaine. The quality control (QC) samples were prepared in the same way at three levels: lower – LQC with 1.95µM for TMA, TMAO, and Choline, and 0.24µM for Carnitine and Betaine. Medium – MQC with 7.8 µM for TMA, TMAO, and Choline, and 0.97µM for Carnitine and Betaine. High – HQC with 62.5 µM for TMA, TMAO, and Choline, and 7.8µM for Carnitine and Betaine. During calibration, a blank sample spiked only with an internal standard was examined using the standard procedure.
Internal Standards Information
Stock solutions of TMA-d9, TMAO-d9, Betaine-d11, and Choline-d9 were accurately weighed and dissolved at a concentration of 1 mg/mL in water. The stock solution was further diluted into by Methanol as the internal standard solution.
Sample Preparation
Urine samples were collected from five healthy controls and one patient. Aliquots of 1.5 mL were stored at −80°C. Samples were thawed at room temperature. Next, urine samples were diluted into 1:100 in water. Ten calibration standards were prepared from serially diluted stock solution. 20ul of the samples were spiked with 150ul of IS working solution. Subsequently, sample centrifugation at 12000 rpm for 5 min at 4°C was done, after that, the supernatant was transferred into autosampler vial and injected.
LC-MS/MS
The analysis was performed using the QTRAP® 5500 (SCIEX, Canada) coupled with Agilent 1260 Infinity HPLC system (Agilent, Germany). 0.5 µL of the sample was injected into an Agilent Pursuit 3 PFP column (30 × 3.0mm), and the flow rate was set at 150 µL/min. Mobile phase A consist of 5mM ammonium formate in water with pH = 3.0. Mobile phase B contains 90% acetonitrile in water and 10% of 5mM ammonium formate in water with pH = 5.8. The scan mode was operated with multiple reaction monitoring (MRM) with positive ion mode. The TurboIonSpray source parameters were as follows: CUR = 20; CAD = low; IS = 5500 V; source temperature = 500°C; GS1 and GS2 = 40 and 20, respectively. DP = 38.00, EP = 10.00, and CXP = 10.00.
Single Gene Sequencing
Genomic DNA was obtained from 2mL of peripheral blood collected in an EDTA vacutainer using a Qiagen extraction kit. Next, a NanoDrop spectrophotometer was used to measure the DNA quality. Genotyping for (c.622C>T) variant is present in the FMO3 gene was carried out with 50µL reactions including the Qiagen master mix, oligonucleotide sets designed for FMO3 gene and DNA templates. The PCR products were run on 2% agarose gel electrophoresis to sustain the bands prior to performing the Sanger sequencing analysis for FMO3 gene.
Cell Culture and Plasma Incubation
Human umbilical vein endothelial cells (HUVECs) were purchased from ATCC (CRL-1730 USA) and were maintained in completed Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human leukemia monocytic cell line ATCC (TIB-202 USA) was cultured in RPMI 1640 supplemented with 2-mercaptoethanol to a final concentration of 0.05 mm, 10% fetal bovine serum (Gibco), and 1% penicillin/streptomycin antibiotic (Gibco). All cells were cultured in a humidified 5% CO2 atmosphere at 37°C. For the differentiation of monocyte to monocyte-like-macrophages, THP-1 cells were stimulated for 3 days with 100 nM phorbol-12-myristate 13-acetate (PMA) (MilliporeSigma, Burlington, Massachusetts, United States). After that, the cells were washed three times with PBS and incubated with fresh media. Cells were incubated for 24 h, and then the culture medium was removed and replaced with fresh complete medium supplemented with 10% healthy control or patient plasma. Cells were incubated for another 24h before harvesting.
Adhesion Assay
THP-1 cells were stained with 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE) for 30 min, then incubated with endothelial cells for 2 hours. Cells were washed three times with PBS and used for imaging by EVOS® FL Auto Imaging System (Thermo Fisher Scientific).
Flow Cytometry
Cells were harvested, then washed with 1X PBS and incubated for 30 minutes at 4°C with PE-conjugated antibodies (ICAM-1, CD11C and TNFα). For the unconjugated primary antibody (V-CAM1) a secondary antibody and FITC Streptavidin were added. After that cells were washed twice with PBS and resuspended with 300 ul FACS buffer, then analyzed using LSRFortessa BD FACS machine (BD Biosciences) and data were analyzed using FACSDiva software. For reactive oxygen species (ROS) detection, CellROX™ Deep Red Reagent (1 µL) (Invitrogen) was diluted in RPMI Medium (Gibco) (4mL), then cells were incubated for 20 min in 100 µL of this mixture at 37°C. Flow cytometry was used to evaluate the cells after they were washed in PBS. Annexin-V binding assay was used to determine apoptosis.
3D Protein Model Analysis
Multiple Alignment Sequences for p.Glu208Ter Variant
The protein sequence of p.Glu208* variant and normal sequences were translated into in-silico analysis using the Ensembl tool used online. The p.Glu208Glu and p.Glu208* amino acid sequences will be further converted into FASTA sequences and will be used to analyze the Sanger sequencing analysis for c.622G>T (p.Glu208*) variant present in the FMO3 gene.
Prediction Analysis of 3D Protein Structure in p.Glu208Ter
In this study, we will use a web server known as I-TASSER, which will be used to predict the 3D protein structure and function of the FMO3 protein. With the help of I-TASSER, we will predict the protein structure, homology analysis of protein type proximity based on 3D structure, biological functional analysis and protein homology analysis type proximity based on biological functional analysis. PyMol software will be used to visualize the predicted structures.
Secondary Structure
The secondary structure of the protein was studied in detail for normal and mutated sequences present in p.Glu208Glu and p.Glu208* region. The secondary structure consists of α-helices, β-turn extended strand, β-sheet and coils. Using the PDM sum tool, both the secondary motif map and topology diagram were calculated.27
Results
A 41-year-old Saudi male who was referred to our clinic 2 years ago and noticed to have offensive fishy body odor since adolescent. No other clinical manifestation that he has a part of this, and type 2 diabetes mellitus diagnosed one year ago, and he is on metformin and controlled. No history of hypertension, tachycardia, anemia or neutropenia or recurrent infections. He is married and has 2 healthy daughters. No other similar illness in his family. The offensive fishy odor has an impact on his psychosocial life as it makes him live in coastal cities, he is currently live in Vietnam.
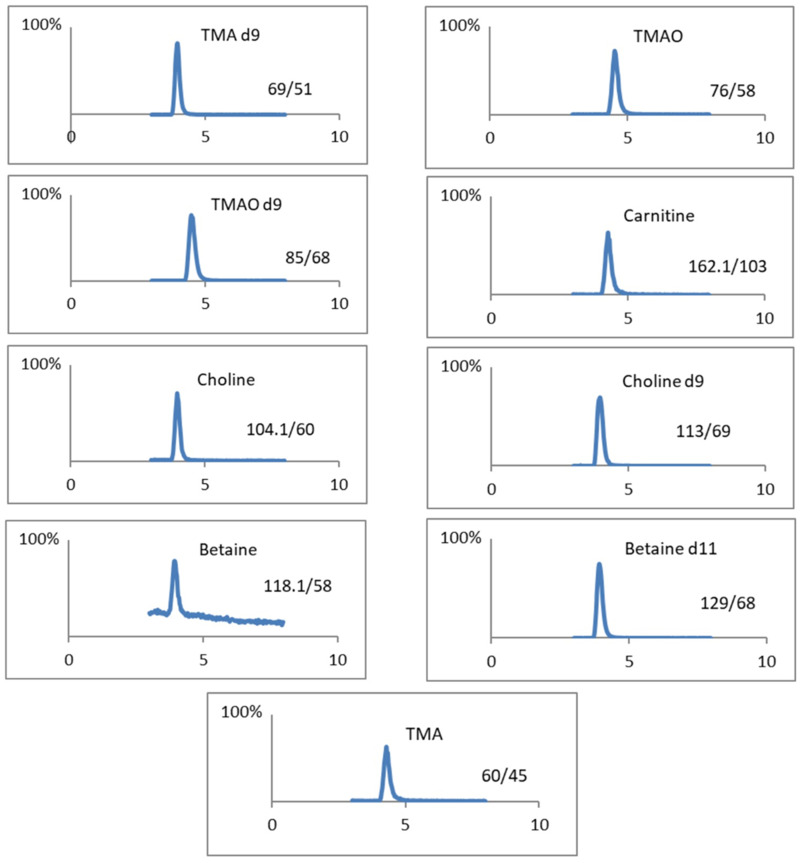
Given the absence of prior reported cases of Trimethylaminuria in Saudi Arabian medical clinics, the initial priority was to establish a biomedical assay for the measurement of TMA and TMAO. This assay will serve as a rapid diagnostic tool for identifying future cases in Saudi Arabia and can be used to confirm the results of FMO3 gene sequencing. TMA precursors including choline, carnitine, and betaine were added to the study to evaluate their potential dietary influence on TMA levels in urine samples for the participants (patient). The present study describes an adaptation of the previously published LC-SRM method employing pentafluorophenyl (PFP) stationary phase for the analysis of TMA and TMAO without pre-derivatization.28 The mobile phase composition was modified. Ammonium formate buffer (pH 3.0) was employed for mobile phase A, while in the mobile phase B, the combination of acetonitrile and ammonium formate buffer (pH 5.8) was used. Chromatographic separation of TMA and TMAO was satisfactory, as illustrated in Figure 1.
Figure 1.
Extracted Ion Chromatogram (XIC). LC-MS chromatogram for all compounds (TMA, TMAO, Betaine, Choline, and Carnitine) and their internal standards.
Quantification was achieved using deuterium isotope internal standards to ensure analytical accuracy and precision. Since a method for determining TMA and TMAO using a pentafluorophenyl (PFP) stationary phase has already been established and validated, this study assessed only linearity and precision. All calibration curves demonstrated excellent linearity, with correlation coefficients exceeding 0.996 for all compounds. To assess reproducibility, three quality controls (QC) samples were incorporated at low, medium, and high concentration during analytical runs. The coefficient of variation (CV) for the high and medium concentration levels was below 5%, while the CV for the low concentration level remained under 9%. The concentrations of TMA, TMAO, Carnitine, betaine, and choline in urine samples have been normalized to the creatinine levels. The patient with the highest TMA concentration exhibited a level of 164.8 µmol/mmol exceeding all other participants in the study. The patient’s TMAO concentration was 7.49 µmol/mmol, and lower than that of all of the control individuals. The patient showed a markedly low TMAO/TMA ratio of 0.05, significantly lower than the healthy control group. Urine levels for TMA precursors including choline, carnitine, and betaine were not significantly different compared to healthy controls. Table 1 summarizes the quantitative analysis of all measured compounds in the urine samples collected from the patient and healthy controls. Further, the plasma levels of TMA and TMAO were measured in our patient, with results indicating 114.1 and 0.43 μmol/L, respectively.
Table 1.
TMA, TMAO, and Their Precursor’s Concentrations in Urine Samples. All Results Normalized to Creatinine Level
| Sample Name | Carnitine µmol/mmol | Betaine µmol/mmol | Choline µmol/mmol | TMA µmol/mmol |
TMAO µmol/mmol | TMAO/TMA ratio | TMAO/(TMA+TMAO)*100 |
|---|---|---|---|---|---|---|---|
| Patient 1 | 11.04 | 10.10 | 21.86 | 164.81 | 7.49 | 0.05 | 4.35 |
| Ctrl 1 | 16.11 | 12.64 | 34.22 | 13.35 | 47.88 | 3.59 | 78.20 |
| Ctrl 2 | 17.88 | 12.03 | 52.09 | 38.90 | 67.72 | 1.74 | 63.51 |
| Ctrl 3 | 4.12 | 9.63 | 31.98 | 8.94 | 39.84 | 4.46 | 81.67 |
| Ctrl 4 | 23.06 | 6.84 | 32.66 | 5.80 | 14.86 | 2.56 | 71.92 |
| Ctrl 5 | 2.43 | 5.87 | 27.02 | 7.07 | 24.38 | 3.45 | 77.51 |
To verify the diagnosis based on the urinary TMA and TMAO level, genetic analysis was subsequently performed. Genetic analysis of the patient identified a homozygous non-sense mutation in the FMO3 gene; c.622G > T p. (Glu208*). This mutation was reported as a pathogenic variant in the ClinVar, LOVD and OMIM databases.
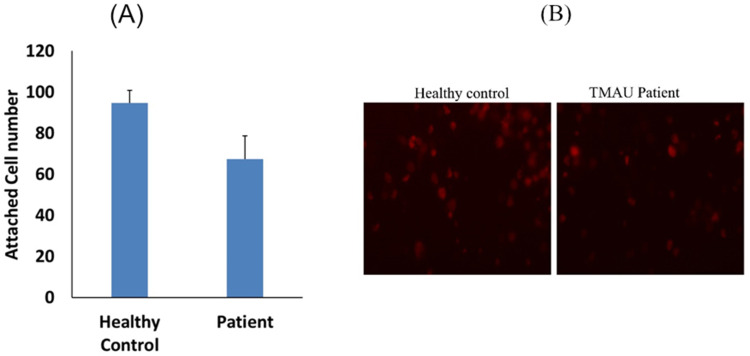
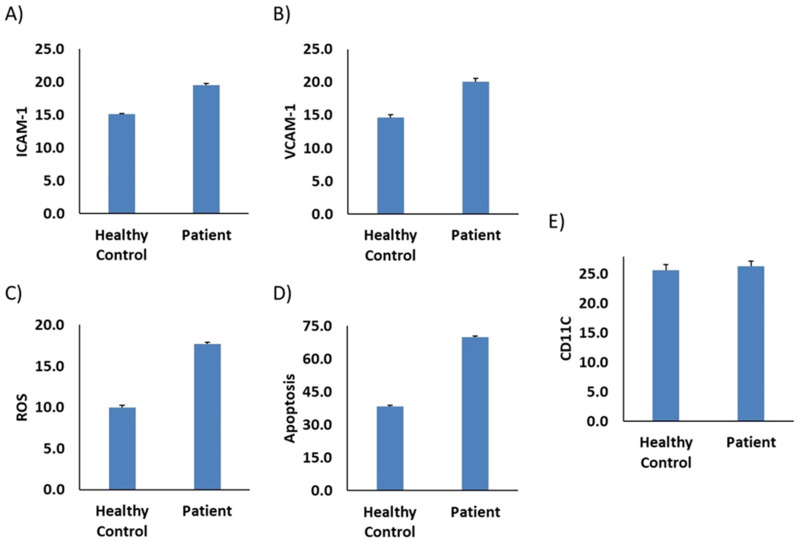
The observation of sustained reduced levels of TMAO in our patient, who is on a low-protein diet, and is suffering from a rare genetic defect in FMO3, prompts us to further investigate the impact of his plasma on endothelial function and inflammatory processes. We used plasma from our patient as model for TMAO low level and we evaluate the potential interaction with monocytes and endothelial function. We find that monocytes stimulated by plasma of the patient are less adherent to endothelial cells compared to healthy control (Figure 2A and B), while the adhesion molecules ICAM-1 and VCAM-1, were increased (Figure 3A and B). Furthermore, an elevated level of ROS marker was observed in our patient compared to control sample (Figure 3C). We further evaluated the apoptosis process where we observe an increase in apoptotic cells after media supplementation with plasma from the patient (Figure 3D) compared to control. No difference was observed in CD11C marker between patient and control samples (Figure 3E).
Figure 2.
Plasma from TMA/patient decreased monocyte adhesion to endothelial cells. Thp-1 monocytes were incubated with plasma from (TMAU patient) compared to healthy donors (healthy control). (A) Number of adherent cells and (B) representative image for healthy control and patient.
Figure 3.
Plasma from TMA/patient stimulates monocyte adhesion markers, oxidative stress and apoptosis. Thp-1 monocytes were incubated with plasma from (TMA/patient) compared to healthy donors (healthy control). (A) ICAM-1, (B) VCAM-1 and (E) CD11C were measured by flow cytometry, bars are presented as percentage of positive cells (C) ROS detection was measured by flow cytometry using CellROX™ Deep Red Reagent and (D) Apoptosis measurement was detected using Annexin V assay. The data are presented as mean ± SEM, n=3-4.
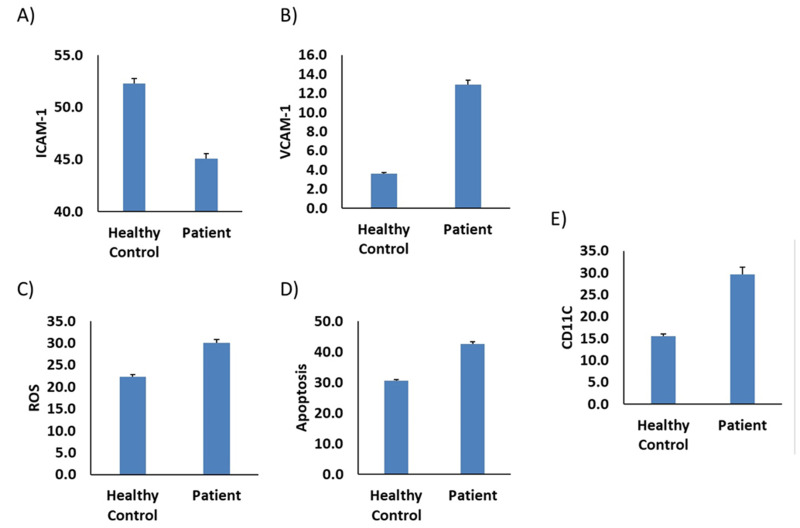
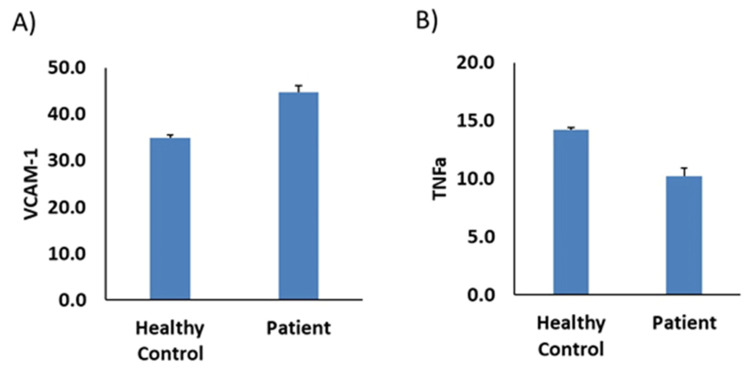
Further, we investigate the potential interaction with macrophages and endothelial function using the same markers. The result was the same (Figure 4B–D) except ICAM-1, the only marker that was completely reversed (Figure 4A). Furthermore, the CD11C marker was increased in the patient sample. We further evaluate the expression of VCAM-1 and TNF-α on endothelial cells. Results revealed an increase level of VCAM-1 and decrease level TNF-α in patient sample compared to health control as shown in Figure 5A and B, respectively.
Figure 4.
Plasma from TMA/patient stimulates monocyte-like-macrophage adhesion markers, oxidative stress and apoptosis. Thp-1-like macrophages cells were incubated with plasma from (TMA/patient) compared to healthy donors (healthy control). (A) ICAM-1, (B) VCAM-1 and (E) CD11C were measured by flow cytometry, bars are presented as percentage of positive cells (C) ROS detection was measured by flow cytometry using CellROX™ Deep Red Reagent and (D) Apoptosis measurement was detected using Annexin V assay. The data are presented as mean ± SEM, n=3-4.
Figure 5.
Plasma from TMA/patient stimulates VCAM-1 and TNF-α expression in endothelial cells. HUVEC cells were incubated with plasma from (TMA/patient) compared to healthy donors (healthy control). (A) Percentage of VCAM-1 positive cells and (B) percentage of TNF-α positive cells, measured by flow cytometry. The data are presented as mean ± SEM, n=3-4.
The sequence identification of both normal and mutated proteins was confirmed by 82.86% and 91.35%. Multiple sequence alignments of the FMO3 gene protein present in p.Glu208Ter were performed to identify the conserved residues. In our study, we could not show mutated protein due to the deletion of termination codon in the amino acid sequence present after the 208th position in the FMO3 gene, and it was revealed from GRCh38.p13 in the Ensembl genome browser along with the transcript ID: ENSG00000007933.13.
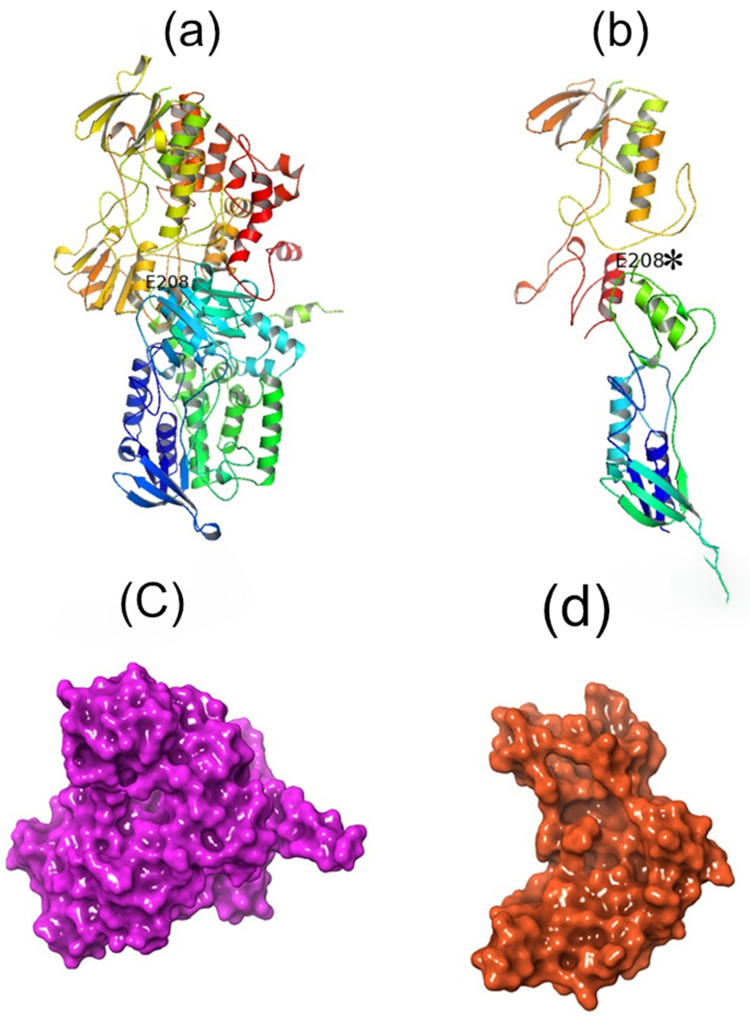
With the support of I-TASSER tool, we have created a couple of protein 3D models for both the normal and mutated protein present in 208th position in the FMO3 gene. Figure 6 shows the different forms of 3D protein models shown between the normal and mutated protein.
Figure 6.
Predicted protein structures present for Glu208Glu and Glu208Ter proteins appear in FMO3 gene. (a) contains normal protein and (b) mutated protein modelling (c) displays as surface view of normal protein structure and (d) mutated protein structure in the form of surface view.
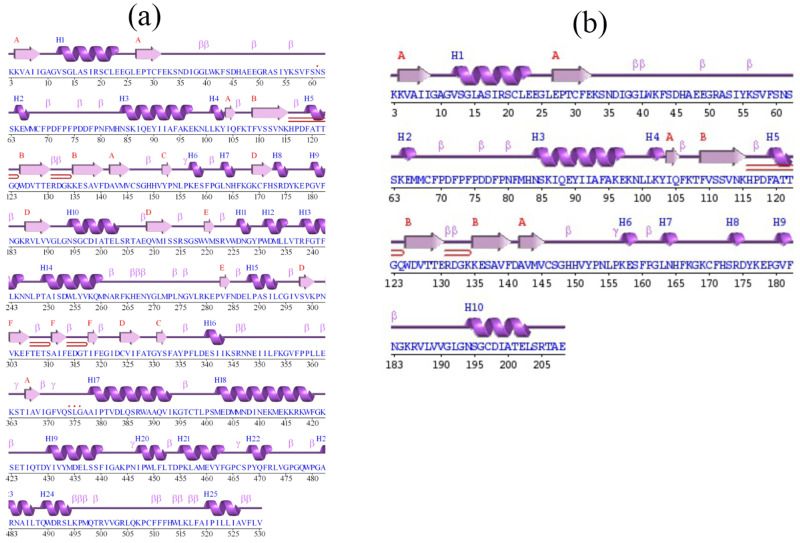
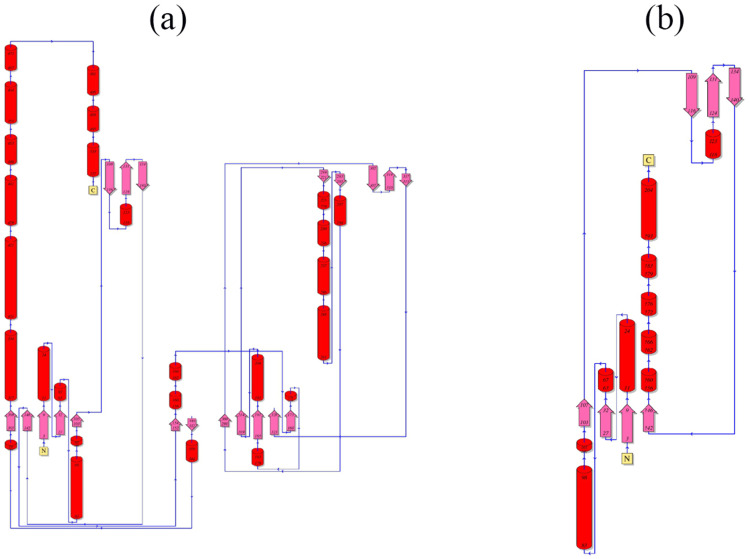
The knowledge towards the secondary structural analysis is to comprehend confirmation modification in 3D structures of protein. In this study, we have performed the secondary structural element for both the normal and mutated protein for the position 208 in the FMO3 gene ie, Glu-208-Glu and Glu-208-Ter. The predicted model for secondary structures for both the normal and mutated proteins was generated using PDMsum tool, and it is shown in Figure 7. The Figure 7a and b show the normal and mutated proteins and it shows the Helices strands ie, H1-H25 and H1-H10. The α-helices were labelled with H and β-strands indicates the presence of upper letters and additionally, β, γ. Moreover, hairpin turns were also labelled. In the normal protein of the secondary structure, 25 helices ie, H1-H25 had appeared in multiple helix–helix interactions, while various β-sheet motifs are composed of 34 β-strands. The normal protein sequence of this mutation does not come under any of the helix, and it was seen in the Figure 7; precisely 7a. The topology of this mutation was documented in the Figure 8a. When it comes to the mutated protein sequence, 10 helices had appeared as H1-H10 among the helix–helix interactions. The mutated protein sequence also does not come under any of the helixes, and it was seen in the Figure 7b. The topology of this mutation was present in the Figure 8b. Further, in Figure 8, it was showed the arrangement and connectivity of the helices and strands in a protein. Both the protein chain consists of C and N terminal domains, and it was folded into a mixed α/β topology. The helices were represented as cylinders and arrows define the β-strands.
Figure 7.
Predicted secondary structure for normal and mutated protein sequence present at 208th position in FMO3 gene. (a) consists of normal protein structure ie, Glu208Glu (b) consists of termination codon present at 208th position.
Figure 8.
Exposition of Topology diagram for both normal and terminated proteins present in 208th position in FMO3 gene. (a) consists of normal protein structure and (b) consists of termination codon.
Discussion
TMAU is a rare genetic disorder with limited reported cases in the global population. Based on the availability of the documented literature, no studies were present on TMAU cases in Saudi Arabia. Among, a 41-year-old man with extensive history of strong fishy body odor was reported to genetic division, department of pediatrics, King Abdullah Specialist Children Hospital, in Riyadh. He complained of disturbing social and work life that made him depressed and anxious. Five healthy individuals’ males and age matched served as controls alongside the patient in this study.
This study presents the first documented case of TMAU in the Saudi population. The unpleasant odor had a significant negative impact on his mental health, prompting his relocation to Vietnam, a coastal nation, where he sought to minimize the comments on his odor every day. The patient’s distinctive fish odor provoked us to investigate whether similar cases of TMAU with the same genetic mutation have been previously reported. It has been found that one single case was reported previously for an Asian patient with the same mutation.29 The TMA and TMAO urine levels in the Asian patient were 48.7 and 70.0 μmol/mmol creatinine, respectively. Despite having the same mutation, the significantly lower TMAO/TMA ratio in our patient (0.05) compared to the Asian patient (1.4) may be associated with more severe body odor observed in our patient.
Plasma levels of TMAO are also independently associated with cardiovascular diseases as reported in multiple cohorts.18,19,30–34 Lowering TMAO levels for prevention of atherosclerosis and cardiac events has become of significant interest.35,36 Production of TMAO depends on diet, gut microbes, and FMO3 considered the three potential therapeutic targets for such interventions. Our gut microbiota metabolizes some dietary nutrients, particularly carnitine, choline and phosphatidylcholine into TMA. In the liver and in the presence of FMO3, a class of hepatic FMO enzymes, TMA is oxidized into TMAO. The observation of consistently reduced plasma levels of TMAO in our patient (TMAO = 0.43μmol/L compared to the reference interval of 1.28–19.67μmol/L defined as 2.5th–97.5th percentile,37 who is on a low-protein diet and is suffering from a rare genetic defect in FMO3, prompts us to further investigate the impact of his plasma on endothelial function and inflammatory processes.
Endothelial dysfunction of Vasculature is an early key marker for the development of vascular damages and inflammation leading to cardiovascular complications.38 It was demonstrated that TMAO modulates oxidative stress and inflammation as well as expression of adhesion molecules, and that high level of TMAO stimulates endothelial cell dysfunction and induces monocyte activation.39,40 However, there is no clear study reported on the effect of low circulating level of TMAO in endothelial function or inflammation. We employed our patient plasma, which had a low level of TMAO, as a model to investigate potential interactions with monocytes and endothelial function. Adhesion of monocytes to endothelial cells is a crucial process during the pathophysiology of inflammation and cardiovascular diseases. Our findings indicate that monocytes stimulated by the patient’s plasma less adhere to endothelial cells than those stimulated by healthy control plasma, despite the upregulation of adhesion molecules ICAM-1 and VCAM-1. This effect may be explained by the excessive production of ROS, which leads to endothelial dysfunction and abnormal monocyte adhesion. Furthermore, upon adding the patient plasma to the media, we observed a rise in the number of apoptotic cells, suggesting an enhanced apoptotic process.
The plasticity of monocyte differentiation throughout the inflammatory phase alters their biological activity, we differentiate thp-1 cells into thp-1-like macrophages and evaluate the same markers used on monocytes. While all other markers displayed similar trends, ICAM-1 exhibited a complete reversal. Decrease of ICAM-1 in monocyte during differentiation may also explain at least in part the loss of adhesion property and the potential activation and change of function after stimulation with plasma from patient, supported by the increase of CD11C. This marker plays a major role in modulating migration and adhesion of monocyte and regulation of immune responses throughout the inflammatory process. We further evaluate the expression of VCAM-1 and TNF-α on endothelial cells. VCAM-1 was increased in cells cultured with media supplemented by plasma from a patient compared to control. However, TNF-α, which played a role in monocyte recruitment, adhesion and migration, was decreased. These results support the hypothesis that the decrease of monocyte adhesion is due mostly to activation of these cells rather than dysfunction of adhesion molecules in endothelial cells. Overall, decrease of TMAO levels in our patient may be the cause of some hidden abnormalities in the circulatory system, specifically the interaction between monocytes and endothelial cells. However, these effects did not reach the pathologic level that affects the clinical phenotype. Furthermore, additional investigation with a large number of subjects is required to confirm these preliminary results, which have evident limitations in terms of the quantity of samples used.
The supplementary Figure-1 and supplementary Table-1 consists of previous studies carried out between different variants present in the FMO3 gene on TMAU. In these global wide studies, we have documented 147 variants studied in the FMO3 gene in different populations (Supplementary File) and majority from the Japanese population.41–53 Among 147 variants, 3.4% belong to the Intronic region, 7.5% be appropriate to UTR region and 89.1% were present in eight exonic regions ie, between exons 2–9. Almost 22.9% of FMO3 gene variants are present in exon-9. The protein sequence Glu308Gly was commonly documented in the global population.13,14,42,43,46,47,49,54–57 Next, P135L and G158L variants were well documented in the global wide populations. Additionally, V257M, A205C, C197Ter, Arg500Ter, Arg388Ter, Gln470Ter, Gln475Asp, I441T, Asn114Ser, M166I, Gly180Val, Thr201Lys, Ser310Leu (S.Table-1) were mainly documented. In our study, we have documented the p.Glu208Ter variant and it was previously reported in the UK population58 among a nine-year-old boy. When it comes to our study, we have found this variant in adult men with 41 years of age.
The FMO3 gene variants in TMAU disorder were focused deeply and primarily studied in the Japanese population by Shimizu et al studies and reported many variants including novel mutation.41–53 Based on the preceding aspect, we assume that FMO3 genetic variants have frequently appeared in the Japanese population, which could be attributable to the predominance of common ethnicity in TMAU disorder. We also assume that as a dietary factor, seafood can contribute to TMAU disease in this population because seafood consists of elevated TMA levels. In addition, it is worth noting that a small number of mutations identified in certain patients can have a significant impact on a large number of patients.
Previous studies have confirmed the elevated levels of TMAO are strongly associated with obesity59 and diabetes.60 The patient had a BMI of 29.4kg/m2 and was the child of parents who were consanguineous. He was married to a woman who was non-consanguineous to him. In our study, we found that the parents of the patient had a consanguineous history, which is a common factor in this autosomal recessive inherited disorder. This is the first documented diagnosed case in his family. One of the limitations of this study could be the detailed analysis of the molecular tests for the family members.
This sequence change creates a premature translational stop signal (p.Glu208*) in the FMO3 gene. It is expected to result in an absent or disrupted protein product. Loss-of-function variants in FMO3 are known to be pathogenic (PMID: 20301282). This variant is present in population databases (rs559643079, gnomAD 0.06%). This premature translational stop signal has been observed in individual with TMAU (PMID: 27118741). ClinVar contains an entry for this variant (Variation ID: 1322927). For these reasons, this variant has been classified as Pathogenic.
Conclusion
This study concludes that the termination codon was detected at the 208th protein position in the FMO3 gene. Moreover, we have developed the LC-MS method for quantifying urine TMA and TMAO concentration levels. This method will facilitate disease diagnosis and support other clinical centers within the Kingdom with analyzing samples from suspected patients. Nevertheless, our results require further examination to be confirmed because we are limited in the quantity of samples.
Acknowledgment
The authors would like to express their gratitude to King Abdullah International Medical Research Center, Riyadh for funding the research project (RC20/171/R).
Disclosure
All the authors declare as there is no conflict of interest towards this publication.
References
- 1.Ruocco V, Florio M, Filioli FG, Guerrera V, Prota G. An unusual case of trimethylaminuria. Br J Dermatol. 1989;120(3):459–461. doi: 10.1111/j.1365-2133.1989.tb04175.x [DOI] [PubMed] [Google Scholar]
- 2.Fraser-Andrews EA, Manning NJ, Ashton GHS, Eldridge P, McGrath J, Menagé Hd P. Fish odour syndrome with features of both primary and secondary trimethylaminuria. Clin Exp Dermatol. 2003;28(2):203–205. doi: 10.1046/j.1365-2230.2003.01230.x [DOI] [PubMed] [Google Scholar]
- 3.Mitchell SC, Smith RL. Trimethylaminuria: the fish malodor syndrome. Drug Metab Dispos. 2001;29(4):517–521. [PubMed] [Google Scholar]
- 4.Wise PM, Eades J, Tjoa S, Fennessey PV, Preti G. Individuals reporting idiopathic malodor production: demographics and incidence of trimethylaminuria. Am J Med. 2011;124(11):1058–1063. doi: 10.1016/j.amjmed.2011.05.030 [DOI] [PubMed] [Google Scholar]
- 5.Mountain H, Brisbane JM, Hooper AJ, Burnett JR, Goldblatt J. Trimethylaminuria (fish malodour syndrome): a “benign” genetic condition with major psychosocial sequelae. Med J Aust. 2008;189(8):468. doi: 10.5694/j.1326-5377.2008.tb02126.x [DOI] [PubMed] [Google Scholar]
- 6.Todd WA. Psychosocial problems as the major complication of an adolescent with trimethylaminuria. J Pediatr. 1979;94(6):936–937. doi: 10.1016/S0022-3476(79)80224-3 [DOI] [PubMed] [Google Scholar]
- 7.Shephard EA, Treacy EP, Phillips IR. Clinical utility gene card for: trimethylaminuria - update 2014. Eur J Hum Genet. 2015;23(9):1269. doi: 10.1038/ejhg.2014.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolphin CT, Janmohamed A, Smith RL, Shephard EA, Phillips L. Missense mutation in flavin-containing mono-oxygenase 3 gene, FMO3, underlies fish-odour syndrome. Nature Genet. 1997;17(4):491–494. doi: 10.1038/ng1297-491 [DOI] [PubMed] [Google Scholar]
- 9.Mackay RJ, McEntyre CJ, Henderson C, Lever M, George PM. Trimethylaminuria: causes and diagnosis of a socially distressing condition. Clin Biochem Rev. 2011;32(1):33–43. [PMC free article] [PubMed] [Google Scholar]
- 10.Akerman BR, Lemass H, Chow LML, et al. Trimethylaminuria is caused by mutations of the FMO3 gene in a North American cohort. Mol Gene Metabol. 1999;68(1):24–31. doi: 10.1006/mgme.1999.2885 [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Cho SM, Chae J-H. A compound heterozygous mutation in the FMO3 gene: the first pediatric case causes fish odor syndrome in Korea. Korean J Pediatr. 2017;60(3):94–97. doi: 10.3345/kjp.2017.60.3.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira F, Esteves S, Almeida LS, et al. Trimethylaminuria (fish odor syndrome): genotype characterization among Portuguese patients. Gene. 2013;527(1):366–370. doi: 10.1016/j.gene.2013.05.025 [DOI] [PubMed] [Google Scholar]
- 13.D’Angelo R, Esposito T, Calabrò M, et al. FMO3 allelic variants in Sicilian and Sardinian populations: trimethylaminuria and absence of fish-like body odor. Gene. 2013;515(2):410–415. doi: 10.1016/j.gene.2012.12.047 [DOI] [PubMed] [Google Scholar]
- 14.Motika MS, Zhang J, Zheng X, Riedler K, Cashman JR. Novel variants of the human flavin-containing monooxygenase 3 (FMO3) gene associated with trimethylaminuria. Mol Gene Metabol. 2009;97(2):128–135. doi: 10.1016/j.ymgme.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyle S, O’Byrne JJ, Nesbitt M, et al. The genetic and biochemical basis of trimethylaminuria in an Irish cohort. JIMD Rep. 2019;47(1):35–40. doi: 10.1002/jmd2.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ringel C, Dittrich J, Gaudl A, et al. Association of plasma trimethylamine N-oxide levels with atherosclerotic cardiovascular disease and factors of the metabolic syndrome. Atherosclerosis. 2021;335:62–67. doi: 10.1016/j.atherosclerosis.2021.09.026 [DOI] [PubMed] [Google Scholar]
- 17.Singh GB, Zhang Y, Boini KM, Koka S. High mobility group box 1 mediates TMAO-induced endothelial dysfunction. Int J Mol Sci. 2019;20(14):3570. doi: 10.3390/ijms20143570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andraos S, Jones B, Lange K, et al. Trimethylamine N-oxide (TMAO) is not associated with cardiometabolic phenotypes and inflammatory markers in children and adults. Curr Dev Nutr. 2021;5(1):nzaa179. doi: 10.1093/cdn/nzaa179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhuiya J, Notsu Y, Kobayashi H, et al. Neither trimethylamine-N-oxide nor trimethyllysine is associated with atherosclerosis: a cross-sectional study in older Japanese adults. Nutrients. 2023;15(3):759. doi: 10.3390/nu15030759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam C-W, Law C-Y, To KK-W, et al. NMR-based metabolomic urinalysis: a rapid screening test for urinary tract infection. Clin Chim Acta. 2014;436:217–223. doi: 10.1016/j.cca.2014.05.014 [DOI] [PubMed] [Google Scholar]
- 21.Hsu W-Y, Lo W-Y, Lai -C-C, et al. Rapid screening assay of trimethylaminuria in urine with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2007;21(12):1915–1919. doi: 10.1002/rcm.3043 [DOI] [PubMed] [Google Scholar]
- 22.Mamer OA, Choinière L, Treacy EP. Measurement of trimethylamine and trimethylamine N-oxide independently in urine by fast atom bombardment mass spectrometry. Anal Biochem. 1999;276(2):144–149. doi: 10.1006/abio.1999.4351 [DOI] [PubMed] [Google Scholar]
- 23.daCosta K-A, Vrbanac JJ, Zeisel SH. The measurement of dimethylamine, trimethylamine, and trimethylamine N-oxide using capillary gas chromatography-mass spectrometry. Anal Biochem. 1990;187(2):234–239. doi: 10.1016/0003-2697(90)90449-J [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Zeisel SH, Zhang S. Rapid LC-MRM-MS assay for simultaneous quantification of choline, betaine, trimethylamine, trimethylamine N-oxide, and creatinine in human plasma and urine. ELECTROPHORESIS. 2015;36(18):2207–2214. doi: 10.1002/elps.201500055 [DOI] [PubMed] [Google Scholar]
- 25.Veeravalli S, Karu K, Phillips IR, Shephard EA. A highly sensitive liquid chromatography electrospray ionization mass spectrometry method for quantification of TMA, TMAO and creatinine in mouse urine. MethodsX. 2017;4:310–319. doi: 10.1016/j.mex.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Li S, Kellermann G. A simple dilute and shoot approach incorporated with pentafluorophenyl (PFP) column based LC-MS/MS assay for the simultaneous determination of trimethylamine N-oxide and trimethylamine in spot urine samples with high throughput. J Chromatogr B. 2017;1067:61–70. doi: 10.1016/j.jchromb.2017.09.049 [DOI] [PubMed] [Google Scholar]
- 27.Hasan R, Rony MNH, Ahmed R. In silico characterization and structural modeling of bacterial metalloprotease of family M4. J Genet Eng Biotechnol. 2021;19(1):25. doi: 10.1186/s43141-020-00105-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XS, Li S, Kellermann G. A simple dilute and shoot approach incorporated with pentafluorophenyl (PFP) column based LC-MS/MS assay for the simultaneous determination of trimethylamine N-oxide and trimethylamine in spot urine samples with high throughput. J Chromatogr B Analyt Technol Biomed Life Sci. 2017;1067:61–70. doi: 10.1016/j.jchromb.2017.09.049 [DOI] [PubMed] [Google Scholar]
- 29.Sabir N, Jones EA, Padmakumar B. Trimethylaminuria. BMJ Case Rep. 2016;2016:bcr2015213742. doi: 10.1136/bcr-2015-213742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Tang WH, Buffa JA, et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur Heart J Apr. 2014;35(14):904–910. doi: 10.1093/eurheartj/ehu002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang WH, Wang Z, Fan Y, et al. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J Am Coll Cardiol. 2014;64(18):1908–1914. doi: 10.1016/j.jacc.2014.02.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cantero MA, Guedes MRA, Fernandes R, Lollo PCB. Trimethylamine N-oxide reduction is related to probiotic strain specificity: a systematic review. Nutr Res. 2022;104:29–35. doi: 10.1016/j.nutres.2022.04.001 [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gessner A, Di Giuseppe R, Koch M, Fromm MF, Lieb W, Maas R. Trimethylamine-N-oxide (TMAO) determined by LC-MS/MS: distribution and correlates in the population-based PopGen cohort. Clin Chem Lab Med. 2020;58(5):733–740. doi: 10.1515/cclm-2019-1146 [DOI] [PubMed] [Google Scholar]
- 38.Barhoumi T, Fraulob-Aquino JC, Mian MOR, et al. Matrix metalloproteinase-2 knockout prevents angiotensin II-induced vascular injury. Cardiovasc Res. 2017;113(14):1753–1762. doi: 10.1093/cvr/cvx115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanmugham M, Devasia AG, Chin YL, et al. Time-dependent specific molecular signatures of inflammation and remodelling are associated with trimethylamine-N-oxide (TMAO)-induced endothelial cell dysfunction. Sci Rep. 2023;13(1):20303. doi: 10.1038/s41598-023-46820-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Querio G, Antoniotti S, Geddo F, Levi R, Gallo MP. Modulation of endothelial function by TMAO, a gut microbiota-derived metabolite. Int J Mol Sci. 2023;24(6):5806. doi: 10.3390/ijms24065806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu M, Makiguchi M, Hishinuma E, Saito S, Hiratsuka M, Yamazaki H. Rare but impaired flavin-containing monooxygenase 3 (FMO3) variants reported in a recently updated Japanese mega-databank of genome resources. Drug Metab Pharmacokinet. 2024;55:100539. doi: 10.1016/j.dmpk.2023.100539 [DOI] [PubMed] [Google Scholar]
- 42.Shimizu M. Individual differences of drug-metabolizing enzymes as determinants for the metabolic fate of chemicals--a study of trimethylamine and flavin-containing monooxygenase 3. Yakugaku Zasshi. 2009;129(11):1351–1356. doi: 10.1248/yakushi.129.1351 [DOI] [PubMed] [Google Scholar]
- 43.Shimizu M, Allerston CK, Shephard EA, Yamazaki H, Phillips IR. Relationships between flavin‐containing mono‐oxygenase 3 (FMO3) genotype and trimethylaminuria phenotype in a J apanese population. Br J Clin Pharmacol. 2014;77(5):839–851. doi: 10.1111/bcp.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu M, Denton T, Kozono M, Cashman JR, Leeder JS, Yamazaki H. Developmental variations in metabolic capacity of flavin‐containing mono‐oxygenase 3 in childhood. Br J Clin Pharmacol. 2011;71(4):585–591. doi: 10.1111/j.1365-2125.2010.03876.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimizu M, Fujita H, Aoyama T, Yamazaki H. Three novel single nucleotide polymorphisms of the FMO3 gene in a Japanese population. Drug Metab Pharmacokinet. 2006;21(3):245–247. doi: 10.2133/dmpk.21.245 [DOI] [PubMed] [Google Scholar]
- 46.Shimizu M, Hirose N, Kato M, et al. Further survey of genetic variants of flavin-containing monooxygenase 3 (FMO3) in Japanese subjects found in an updated database of genome resources and identified by phenotyping for trimethylaminuria. Drug Metab Pharmacokinet. 2022;46:100465. doi: 10.1016/j.dmpk.2022.100465 [DOI] [PubMed] [Google Scholar]
- 47.Shimizu M, Kobayashi Y, Hayashi S, Aoki Y, Yamazaki H. Variants in the flavin-containing monooxygenase 3 (FMO3) gene responsible for trimethylaminuria in a Japanese population. Mol Gene Metabol. 2012;107(3):330–334. doi: 10.1016/j.ymgme.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 48.Shimizu M, Koibuchi N, Mizugaki A, et al. Genetic variants of flavin-containing monooxygenase 3 (FMO3) in Japanese subjects identified by phenotyping for trimethylaminuria and found in a database of genome resources. Drug Metab Pharmacokinet. 2021;38:100387. doi: 10.1016/j.dmpk.2021.100387 [DOI] [PubMed] [Google Scholar]
- 49.Shimizu M, Makiguchi M, Yokota Y, et al. Simple confirmation methods for rare but impaired variants of human flavin-containing monooxygenase 3 (FMO3) found in an updated genome resource databank. Drug Metab Pharmacokinet. 2023;53:100528. doi: 10.1016/j.dmpk.2023.100528 [DOI] [PubMed] [Google Scholar]
- 50.Shimizu M, Origuchi Y, Ikuma M, Mitsuhashi N, Yamazaki H. Analysis of six novel flavin-containing monooxygenase 3 (FMO3) gene variants found in a Japanese population suffering from trimethylaminuria. Mol Gene Metabol Rep. 2015;5:89–93. doi: 10.1016/j.ymgmr.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu M, Tomioka S, Murayama N, Yamazaki H. Missense and nonsense mutations of the flavin-containing monooxygenase 3 gene in a Japanese cohort. Drug Metab Pharmacokinet. 2007;22(1):61–64. doi: 10.2133/dmpk.22.61 [DOI] [PubMed] [Google Scholar]
- 52.Shimizu M, Yamamoto A, Makiguchi M, et al. A family study of compound variants of flavin-containing monooxygenase 3 (FMO3) in Japanese subjects found by urinary phenotyping for trimethylaminuria. Drug Metab Pharmacokinet. 2023;50:100490. doi: 10.1016/j.dmpk.2023.100490 [DOI] [PubMed] [Google Scholar]
- 53.Shimizu M, Yoda H, Igarashi N, Makino M, Tokuyama E, Yamazaki H. Novel variants and haplotypes of human flavin-containing monooxygenase 3 gene associated with Japanese subjects suffering from trimethylaminuria. Xenobiotica. 2019;49(10):1244–1250. doi: 10.1080/00498254.2018.1539279 [DOI] [PubMed] [Google Scholar]
- 54.Akerman B, Forrest S, Chow L, Youil R, Knight M, Treacy E. Two novel mutations of the FMO3 gene in a proband with trimethylaminuria. Human Mutation. 1999;13(5):376–379. doi: [DOI] [PubMed] [Google Scholar]
- 55.Alibrandi S, Nicita F, Donato L, et al. Adaptive modelling of mutated FMO3 enzyme could unveil unexplored scenarios linking variant haplotypes to TMAU phenotypes. Molecules. 2021;26(22):7045. doi: 10.3390/molecules26227045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo Y, Hwang L-D, Li J, et al. Genetic analysis of impaired trimethylamine metabolism using whole exome sequencing. BMC Med Genet. 2017;18(1):1–9. doi: 10.1186/s12863-016-0468-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Treacy E, Akerman B, Chow L, et al. Mutations of the flavin-containing monooxygenase gene (FMO3) cause trimethylaminuria, a defect in detoxication. Human Molecular Genetics. 1998;7(5):839–845. doi: 10.1093/hmg/7.5.839 [DOI] [PubMed] [Google Scholar]
- 58.Sabir N, Jones EA, Padmakumar B. Trimethylaminuria. Case Reports. 2016;2016:bcr2015213742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dehghan P, Farhangi MA, Nikniaz L, Nikniaz Z, Asghari‐Jafarabadi M. Gut microbiota‐derived metabolite trimethylamine N‐oxide (TMAO) potentially increases the risk of obesity in adults: an exploratory systematic review and dose‐response meta‐analysis. Obesity Rev. 2020;21(5):e12993. doi: 10.1111/obr.12993 [DOI] [PubMed] [Google Scholar]
- 60.Dambrova M, Latkovskis G, Kuka J, et al. Diabetes is associated with higher trimethylamine N-oxide plasma levels. Exp Clin Endocrinol Diabetes. 2016;124(04):251–256. doi: 10.1055/s-0035-1569330 [DOI] [PubMed] [Google Scholar]