Abstract
Drought is one of the main environmental factors affecting plant survival and growth. Atraphaxis bracteata is a common desert plant mainly utilized in afforestation and desertification control. This study analyzed the morphological, physiological and molecular regulatory characteristics of different organs of A. bracteata under drought stress. The results showed that with the intensification of drought stress, the height, root length and leaf area of A. bracteata seedlings decreased, while the content of osmotic substances and antioxidant enzyme activity increased. Furthermore, a total of 63 907 non-redundant transcript sequences, of which 55 574 transcripts were annotated, 248 178 FLNC sequences, 107 112 high-quality consistent sequences and 291 314 CCSs were obtained from Iso-Seq. Meanwhile, a total of 2 039 AS events, 22 919 SSR, 40 404 CDS and 5 902 lncRNA were also obtained. The RNA-Seq analysis results revealed that a total of 2 821, 3 907 and 5 532 DETs were identified from roots, stems and leaves, respectively, and which had significantly enrichment in “circadian rhythm-plant” and “starch and sucrose metabolism” pathway. These results would be great significance for further research on the stress resistance of A. bracteata and these DETs function.
Keywords: Atraphaxis bracteata, Full-length transcripts, Differentially expressed transcript, Drought stress, Circadian rhythm-plant
Subject terms: Genetics, Molecular biology, Physiology, Plant sciences
Introduction
Drought is one of the main climatic factors that seriously affects most of plants survival and growth1,2, which is mainly achieved by influencing photosynthesis, hormone balance and other many key physiological and biochemical processes3. When plants were under drought stress, they usually resisted external stress by regulating changed in photosynthesis, relative water content, protective enzyme systems, osmotic substances, etc. However, with the development of molecular biology, it had been found that plants adapt to drought response through a series of morphological, structural and biochemical changed, which were closely related to the relevant gene expression in organisms4–7. Desert plants have developed strong resistance on the drought, cold, high temperature, sand burial, salt alkali and other characteristics during their long-term continuous adaptation to extreme climate conditions. They have inevitably formed their unique stress resistance molecular mechanisms and are excellent resources for screening stress resistance genes.
Atraphaxis bracteata (A. Los.) is a desert xerophytic broad-leaved shrub of Polygonaceae and Atraphaxis, belongs to the main desert afforestation shrub in desert areas, which is not only suitable for growth in drought soil, but also has a certain tolerance to water and humidity8,9. In addition, it could grow on the mobile sand dunes in both desert and semi desert areas, with high survival rate, large annual growth, developed root systems and a preference for sand burial, so it was also an excellent fuelwood forest resource in desert areas10. In recent years, scholars have also conducted some researches on A. bracteata and found that environmental factors had a significant impact on the sap flow of A. bracteata, of which solar radiation was the most important environmental factor. During the growth season, the sap flow rate of the tree trunk was relatively high and the daytime sap flow was significantly affected by meteorological conditions11–13. Although the transpiration of A. bracteata was lower than that of Elaeagnus angustifolia and Corethrodendron scoparium, it was higher than the typical afforestation tree species Caragana korshinskii and Haloxylon ammodendron in desert areas14. A. bracteata had strong adaptability to high temperature, drought, strong light and sandy environments, just like its antioxidant capacity remained at a high level throughout the entire drought stress period based on its strong antioxidant defense system15–17, and its leaf structure also exhibited typical characteristics of desert plants and strong drought resistance18. In addition, the wild A. bracteata community had few biotypes, simple composition and obvious characteristics of desert grassland vegetation, and which also had strong natural regeneration ability and superiority in different habitats. Therefore, the use of A. bracteata for desert vegetation restoration and reconstruction would have great significance19,20.
With the tremendous growth of science and technology, the transcripts research methods have become diverse and diverse for solving the two major aspects of gene structure and gene function in transcripts21. RNA sequencing (RNA-Seq) technology had the advantages of short sequencing time, low cost, high accuracy and high throughput based on Illumina/Solex sequencing compared to traditional sequencing methods22. It could also obtain low abundance expressed genes and be used for species research with unknown genome sequences. Therefore, it was widely used in many fields23,24. In terms of plant stress resistance, the RNA-seq technology was used to comprehensively and dynamically detect the characteristics of gene expression changes in plants body under stress, excavate some related functional genes, analyze the regulatory mechanism of stress response, and lay on molecular genetic basis for studying plant stress resistance mechanisms and cultivating stress resistant varieties25–29.
It can be said that transcriptome has become one of the essential tools for scholars to study life processes. However, because of the intrinsic limitations of RNA-Seq technology, it is challenging for us to comprehend the intricate meaning of life activity because it is unable to reliably obtain or assemble complete transcripts of organisms or recognize transcripts expressed by isomers, homologous genes, superfamily genes and alleles. Full-length cDNA could be obtained by reverse transcription without the need for interrupting RNA fragments in Isoform-Sequencing (Iso-Seq). Measurable information was obtained from the platform’s super long read (median 10 kb), which provided information about a single full transcript sequence and subsequent analysis did not need assembly30,31. Currently, Iso-Seq was being applied extensively in numerous fields32–37, including studies on drought-resistant plants33,38–41.
Using Iso-Seq in conjunction with RNA-Seq, we examined the molecular regulatory mechanism of the desert plant A. bracteata in response to drought stress. A total of 63 907 non-redundant full length non-chimeric (FLNC) sequences and 291 314 circular consensus sequences (CCSs) with an average read length of 2 062 were acquired. A total of 2 821, 3 907 and 5 532 differential expressed transcripts (DETs) were identified from CKR vs. HR24h, CKS vs. HS24h and CKL vs. HL24h, respectively. As a result, which would offer fresh perspectives on the molecular biological aspects of A. bracteata's drought resilience as well as a theoretical framework for future research and application.
Materials and methods
Plant materials
The experimental materials of A. bracteata were obtained from Gansu Desert Control Research Institute, Lanzhou, P.R. China in April 2022 (Delu Li Researcher officially identified the plant materials, lidlu2008@163.com), and which were cultivated at the comprehensive experimental station of Wuwei Academy of Forestry, Wuwei, P.R. China in May 2022. A specimen was deposited at Wuwei Academy of Forestry under the voucher number WWSLKY-HMS-202204–01. The experimental materials used for A. bracteata were licensed by the Gansu Desert Control Research Institute and Wuwei Academy of Forestry. The A. bracteata seeds with healthy and plump were selected and soaked in clean water for 4 h before planting, and then planted them in the nutrient bowl (10 × 12 cm) which containing some substrate and cultivated in the solar greenhouse (the average temperature was 24 ~ 26 °C, the average humidity was 55 ~ 60%). After the seedlings grew to 40 days, started drought treatment. The sorbic alcohol with osmotic pressure of -0.2 MPa, -0.4 MPa, -0.6 MPa were used to simulate drought treatment, and clear water as the control, and which named T1, T2, T3 and CK, respectively. During treatment, selected the nutrient bowl with basically the same seedling growth and placed them in a tray containing the equal treatment solution.
Once the nutrient bowl had reached saturation with the treatment solution, commenced timing. After 24 h, collected a mixed sample consisting of roots, stems and leaves from the CK, T1, T2 and T3 groups for Iso-Seq analysis. Concurrently, separately collected samples of roots, stems and leaves from the T3 and CK groups for RNA-Seq analysis. Thereafter, maintained the treatment by replenishing the water every three days using the identical treatment protocol. After 15 days, collected the roots, stems and leaves from each sample and stored them at -80°C for the determination of their physiological and biochemical indicators. Additionally, the roots, stems and leaves of T3 and CK used for RNA-Seq analysis were named HR24h, HS24h, HL24h, CKR, CKS and CKL, respectively.
Measurement of morphological and physiological indicators
A tape and a vernier caliper were used to measure the height, root length and leaf area of each treated plant. The content of malondialdehyde (MDA), proline (Pro), soluble protein (SP), as well as the activities of peroxidase (POD), catalase (CAT) and superoxide dismutase (SOD) in the sample were determined according to the methods in “principles and techniques of plant physiology and biochemistry experiments”.
RNA preparation
The total RNA was extracted from the experimental materials according to the specifications of manufacturer of the RNA isolation Kit (Tiangen Biotech, Beijing, China) and each sample had 3 biological replicates. To guarantee the quality of transcriptome sequencing samples, 1% agarose gel electrophoresis and LabChip GX were used to assess the quality and integrity of their total RNA.
Construction and sequencing of PacBio Iso-Seq libraries
Biomarker Technologies Co. Ltd. (Beijing, China) was commissioned for this study to sequence A. bracteata’s Iso-Seq and RNA-Seq. The qualified RNA sample were used to construct the Iso-Seq library constructions, the main process was as follows: (1) The NEBNext Single Cell/Low Input cDNA Synthesis & Amplification Module was utilized to synthesize full-length cDNA of mRNA; (2) PCR amplification of full-length cDNA and performing damage repair/end repair on it; (3) Connection of the single molecule real-time sequencing (SMRT) dumbbell. Following cDNA library quality testing, the PacBioSeque II was employed for Iso-Seq according to the intended offline data.
The RNA-Seq library was constructed using the following steps: Eukaryotic mRNA was enriched with Oligo (dT) magnetic beads. The fragmentation buffer was applied to randomly break mRNA. Using the mRNA as a template and random hexamers, the first cDNA strand was produced by random hexamers. The dNTPs, buffer, RNase H and DNA polymerase I were added to create the second strand of cDNA. The AMPure XP beads were utilized to purify the cDNA, which was subsequently corrected and A-tail added before being coupled to sequencing connections. At last, the cDNA library was generated through PCR enrichment.
Transcriptome raw data analysis
The raw subreads was analyzed according to the Iso-Seq3 pipeline. When conducting full-length transcript analysis, all original sequences were first transformed into CCSs based on the adapters in the sequence, and which were also polished to obtain the quality information of the CCSs. After that, the CCSs were divided to FLNC and non-full-length (NFL) based on the presence or absence of 3’primers, 5'primers and Poly-A tails. Finally, clustering similar sequences from FLNC sequences into a cluster using the Iso-seq3 and extracting high-quality (HQ) isoforms and removing redundancy using cd-hit (identity > 99%). The clean data were extracted from the raw data. Meanwhile, their GC-content, sequence duplication level, Q20 and Q30 were detected, only HQ clean data were selected for subsequent analysis.
Transcripts function analysis
The obtained non-redundant transcript sequences were compared with non-redundant protein databasse (NR)42, swissprot protein database (SWISSPROT) (Consortium)43, gene ontology (GO)44, clusters of Orthologous Groups (COG)45, eukaryotic ortholog groups (KOG)46, Pfam (Protein family)47 and kyoto encyclopedia of genes and genomes (KEGG) (Kanehisa et al.)48 databases by DIAMOND software. InterProScan utilized the InterPro integrated database to analyze the GO methodology results of non-redundant transcript sequences. After calculating amino acid (aa) sequences of non-redundant transcripts, the HMMER software was used to analyze the transcripts annotation information. In addition, GO, COG and KEGG enrichment were taken to further analyze the DETs function.
Structure analysis
CDS detection
TransDecoder was used to detect the candidate coding regions transcript sequences and their corresponding aa sequences. The methods applied were as follows: (1) It was necessary to find an open reading frame (ORF) that satisfies the minimum length in the transcript sequence. (2) When the logarithmic likelihood score of the first reading frame was the greatest compared to the other five reading frames. (3) If the candidate ORF was completely included within the framework of other candidate ORFs, the longest ORF was reported. Otherwise, a single transcript would receive reports of multiple ORFs. (4) As an optional option, the predicted peptides have alignment scores higher than the score threshold in the Pfam domain library.
SSR and AS detection
The simple sequence repeats (SSRs) were identified by MISA software, which could identify seven forms of SSRs by evaluating transcript sequences, including mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide, hexanucleotide and compound SSR. In this study, we selected over 500 bp transcripts for further SSR analysis.
Prediction of AS candidate events based on FLNC sequences of Iso-Seq. The BLAST software was used to compare all sequences in pairs, if comparison results met the undermentioned criteria, it was selected as a candidate AS event. (1) Two sequences were over 1000 bp long and included two high-scoring segment pairs (HSPs). (2) The AS gap exceeded 100 bp and located 100 bp from the 3’/5' end at a minimum. (3) Permit 5 bp overlap.
Quantification of transcript expression levels and differential expression analysis
In present research, the Iso-Seq data were taken as the reference transcriptome. The expression levels of the transcripts were directly tallied and quantified by using the Kallisto software to compare the readings of the fq file to the third-generation transcripts. Fragments per kilobase of transcript per million fragments mapped (FPKM) was used to estimate the expression level of transcript. The differential expression of two comparison groups was investigated using DESeq249. The fold change (FC) ≥ 2 and false discovery rate (FDR) < 0.01 were taken to select the DETs.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed using a quantitative reagent 2X SG Fast qPCR Master Mix (B639271, BBI, Roche) in a LightCycler480 II fluorescence quantitative PCR instrument (Roche, Rotkreuz, Switzerland). Reaction system: 2X SG Fast qPCR Master Mix 5 µL, 10 µM Primers F and R each 0.2 µL, ddH20 3.6 µL, Template (cDNA) 1 µL. The qRT-PCR values were calculated using the comparative cycle threshold method 2−(∆∆ Ct) method, with 3 technical replicates for each sample. The specific gene and internal reference gene (PB-MIX_transcript_58967) primers were shown in Table S1.
Results
The effect of drought stress on the phenotype of A. bracteata
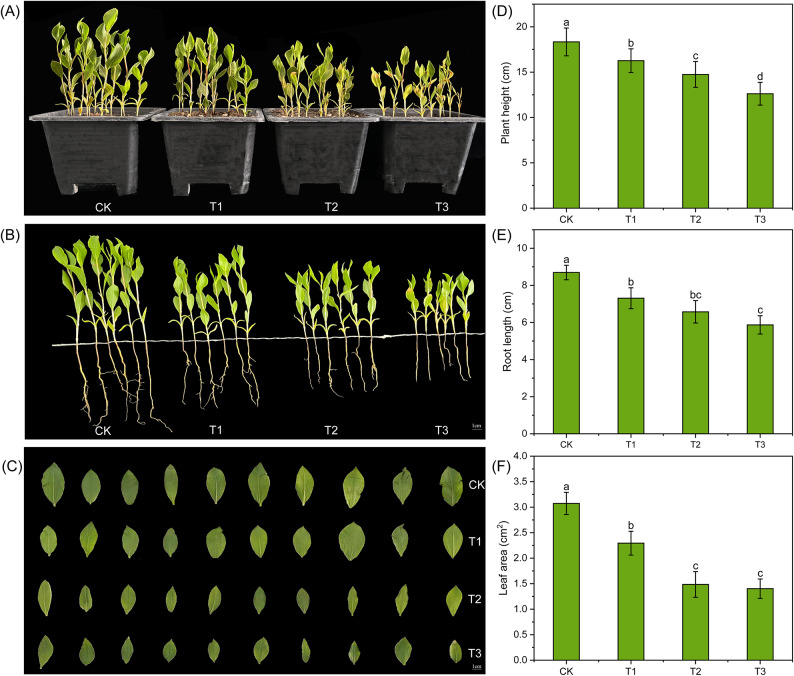
After 15 days of simulated drought treatment with sorbitol, significant phenotypic changes occurred between the treatments and the control, mainly manifested as slower plant growth and smaller leaf area. Analysis of plant height showed that after drought stress, the height of each treatment was lower than CK, and there were significant differences between them, indicating that different degrees of drought stress treatment with sorbitol had a certain inhibitory effect on the growth of A. bracteata plants, and the degree of inhibition increased with the increased of stress level (Fig. 1A, B, D). By analyzing the leaf area of each treatment, it was found that as the degree of drought stress increased, the leaf area gradually decreased, and there were significant differences between each treatment and CK. The treatment T1 had significant differences with T2 and T3, while there was no significant difference between T2 and T3 (Fig. 1C, F). In addition to changes in the phenotype of above ground organs, significant changes were also observed in their underground roots. Root length was significantly inhibited between treatments compared to CK, but except for T1 and T3 had significantly differences, there were no significant differences among other drought treatments (Fig. 1B, E).
Fig. 1.
The phenotype of A. bracteata under drought stress. (A) , (B) and (D) The effect of drought stress on the growth of A. bracteata. (B) and (E) The effect of drought stress on the roots of A. bracteata. (C) and (F) The effect of drought stress on the leaves of A. bracteata.
The effect of drought on physiological indicators of A. bracteata
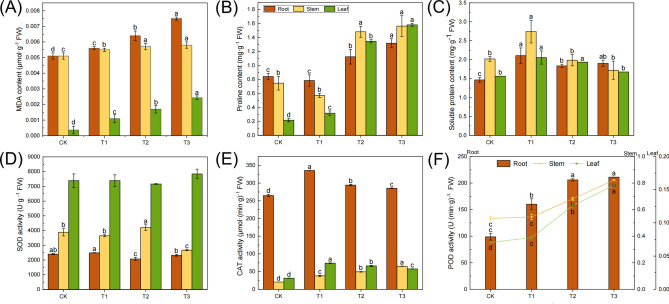
The Fig. 2 illustrated the variation characteristics of the main physiological indicators in different organs of A. bracteata under drought treatment. The MDA content ranged from 0.0003 to 0.0075 µmol·g-1, with the highest levels in roots, followed by stems and then leaves, and which increased with the degree of drought stress (Fig. 2A). As the degree of drought stress increased, the content of Pro gradually increased, while the SP showed a trend of first increasing and then decreasing (Fig. 2B, C). The Fig. 2D–F showed that the POD and CAT activities in the roots were significantly higher than those in the stems and leaves, and the SOD activities were the highest in leaves. Meanwhile, there was no significant trend in SOD activity, while CAT first increased and then decreased (except for those with an increasing trend in the stems), and POD gradually increased trend. These results indicated that different organs of A. bracteata had their own response mechanisms to drought stress, and they could adapt to drought stress by regulating the content of osmotic substances and the activity of antioxidant enzymes.
Fig. 2.
The physiological indicators of A. bracteata under drought stress. (A) The MDA content. (B) The Pro content. (C) The SP content. (D) The activity of SOD. (E) The activity of CAT. (F) The activity of POD.
Iso-Seq data analysis
A total of 31.86 Gb data were obtained from A. bracteata full-length transcriptome, and 291 314 CCSs with an average reading length of 2 062 and a reading base of 600 805 240. In addition, a total of 248 178 FLNC sequences with the percent of 85.19% were obtained after the Iso-Seq polishing. Clustering of FLNC sequences resulted in 107 138 consensus isoforms with an average length of 1 815 and 107 112 high-quality (HQ) consistent sequences with the percent of 99.98%. In addition, a total of 63 907 non-redundant transcript sequences were acquired for further subsequent analysis (Table 1). Contributed raw sequence data to the national center for biotechnology information (NCBI) sequence read archive (SRA), accession number PRJNA 1 028 835.
Table 1.
The Iso-Seq data information of A. bracteata.
| Categories | Datasets |
|---|---|
| Total bases (Gb) | 31.86 |
| cDNA size | 1-6 K |
| CCS number | 291 314 |
| Read bases of CCS | 600 805 420 |
| Mean read length of CCS | 2 062 |
| Mean Number of Passes | 37 |
| Number of FLNC | 248 178 |
| Full-length non-chimeric percentage (FLNC%) | 85.19 |
| Number of consensus isoforms | 107 138 |
| Average consensus isoforms read length | 1 851 |
| Number of high- quality isoforms | 107 112 |
| Number of low-quality isoforms | 26 |
| Percent of high-quality isoforms (%) | 99.98% |
| Number of Non-redundant transcript sequence | 63 907 |
Transcript function annotation
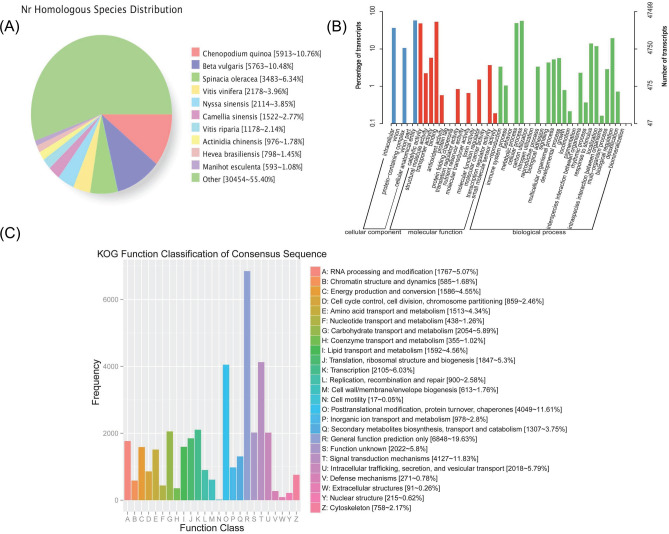
All 63 907 non-redundant transcript were contrasted to NR, COG, GO, eggNOG, KOG, Pfam, Translation of EMBL (TrEMBL), Swissprot and KEGG databases using DIAMOND software, a total of 55 574 transcripts were annotated, with an annotation rate of 86.96% (Table 2). However, there were still 8 333 transcripts that have not been annotated, which suggested that these unannotated transcripts may be specific transcripts of A. bracteata and have high development and utilization value. In addition, we searched for homologous species through the NR database, comparison results displayed that seven most abundant species were Chenopodium quinoa (5 913 ~ 10.76%), Beta vulgaris (5 763 ~ 10.48%), Spinacia oleracea (3 483 ~ 6.34%), Vitis vinifera (2 178 ~ 3.96%), Nyssa sinensis (2 114 ~ 3.85%), Camellia sinensis (1 522 ~ 2.77%) and Vitis riparia (1 178 ~ 2.14%), respectively. However, the proportion of transcript sequences that could annotate homology was only 44.6%, and another 55.4% of transcript sequences still have not annotated homologous species. This indicated that excavating the genes related to the characteristics of A. bracteata play an important role in further searching for its homologous species (Fig. 3A).
Table 2.
The number of annotated transcripts in different databases.
| Annotated databases | Isoform Number | 300 < = length < 1000 | length > = 1000 |
|---|---|---|---|
| COG | 17 581 | 2 096 | 15 479 |
| GO | 47 499 | 7 579 | 39 821 |
| KEGG | 40 398 | 5 886 | 34 446 |
| KOG | 34 880 | 5 062 | 29 767 |
| Pfam | 42 074 | 5 726 | 36 330 |
| Swissprot | 41 871 | 6 378 | 35 412 |
| TrEMBL | 54 880 | 9 207 | 45 555 |
| eggNOG | 48 679 | 7 848 | 40 761 |
| NR | 54 972 | 9 218 | 45 637 |
| All | 55 574 | 9 417 | 46 014 |
Fig. 3.
The functional annotation results of the full-length transcripts of A. bracteata. (A) Statistical diagram of species classification annotated with NR of the full-length transcripts. Different colors represented different species. (B) Statistical diagram of GO annotation classification of the full-length transcripts. The horizontal axis represented GO classification, the left side of the vertical axis represented the percentage of transcripts, and the right side represented the number of transcripts. (C) Statistical diagram of KOG annotation classification of the full-length transcripts. The horizontal axis represented the classification content of KOG, and the vertical axis represented the number of transcripts.
We mainly analyzed the functional annotation results of GO and KOG. The GO function annotation results showed that these transcripts were mainly divided into molecular function (MF), cellular component (CC) and biological process (BP), and most of which belonged to BP. There were three major enrich terms related to cellular anatomical entity, intracellular and protein-containing complex in the CC, whereas biological regulation, metabolic processes and cellular processes were notable enriched in the BP, and binding, transporter activity and catalytic activity were mainly enriched in the MF (Fig. 3B). In addition, biological-process (GO: 0,008,150), cellular process (GO: 0,009,987), metabolic process (GO: 0,008,152), cellular metabolic process (GO: 0,044,237), organic substance metabolic process (GO: 0,071,704), biological regulation (GO: 0,065,007) and primary metabolic process (GO: 0,044,238) were significantly enriched in the category of BP, which unigene number were all over 4 000 (Table S2). The KOG enrichment results showed that these transcripts were classified to 25 functional clusters, with “general function prediction only” (6 848, 19.63%), “signal transduction mechanisms” (4 127, 11.83%), and “posttranslational modification, protein turnover, chaperones” (4 049, 11.61%) were for the top three, accounting for a cumulative proportion of 43.07% (Fig. 3C).
Structure analysis
Prediction analysis of CDS and SSR
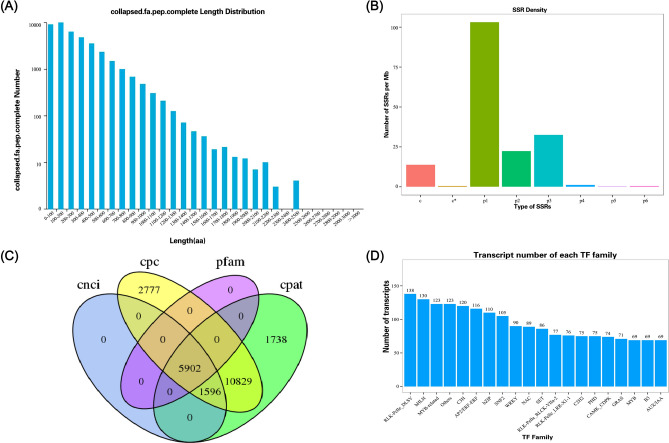
In all, 58 427 ORFs were obtained in this study, including 40 404 complete ORFs which had start and stop codons. In addition, most of the predicting complete ORF region encoded protein sequences were less than 700 aa in length with 37 379 isoforms (92.51%), 2 147 isoforms (5.31%) ranged between 700–1 000 aa, and only 878 (2.17%) isoforms were longer than 1000 aa (Fig. 4A, Table S3).
Fig. 4.
The structure analysis of the full-length transcripts of A. bracteata. (A) The length distribution diagram of prediction CDS encoded protein. (B) Density distribution statistics of different SSR types. (C) The venn diagram of lncRNA predicted by CNCI, CPAT, CPC and Pfam. (D) The prediction classification chart of transcription factors.
SSR analysis was performed out applying the MISA software on a subset of transcripts containing upwards of 500 bp, which revealed a total of 20 868 SSRs. The results showed that seven types SSR were identified including mononucleotide repeats (density: 103.12), dinucleotide repeats (density: 22.27), trinucleotide repeats (density: 32.61), tetranucleotide repeats (density: 1.22), pentanucleotide repeats (density: 0.4), hexanucleotide repeats (density:0.56) and compound SSR (density:13.82) (Fig. 4B).
AS events, LncRNA and TF identification.
After redundancy elimination, the full-length transcriptome was used to predict AS events. A total of 2 093 AS events were recognized (Table S4). Unfortunately, the kinds of AS events in A. bracteata could not be identified because it did not have the reference genome.
To predict long non-coding RNA (lncRNA) in transcripts, we employed the most comprehensive coding potential analytic techniques, primarily the coding potential calculator (CPC), coding-non-coding index (CNCI), pfam protein domain and coding potential assessment tool (CPAT) analyses. The result showed that there were 7 498, 21 104, 5 902 and 20 065 isoforms in the CNCI, CPC, Pfam and CPAT, respectively. Combining the analysis results of four methods, a total of 5 902 lncRNAs were predicted, including 4 788 novel lncRNAs (81.13%) and 1 114 new lncRNAs (18.87%) (Fig. 4C, Table S5).
Transcription factors (TFs) were identified and categorized using the animalTFDB 3.0 and a total of 4 802 TFs were obtained. Meanwhile, the top 10 TF families were RLK-Pelle-DLSV, bHLH, others, MYB-related, C3H, AP2/ERF-ERF, bZIP, SNF2, WRKY and NAC. Among them, “RLK-Pelle-DLSV” family had the most members (138, 2.87%), followed by the “bHLH” family (130, 2.71%), and the third was “others” and “MYB-related” family with 123 members (Fig. 4D, Table S6).
The transcript expression analysis of A. bracteata under drought
Comparison of Iso-seq and RNA-seq
In present study, the spliced transcripts alignment to reference (STAR) was used to compare clean reads with transcripts to obtain positional information on the transcripts. The comparative results demonstrated that the percentage of reads mapped to transcripts loci in clean reads were all greater than 80%, and unique mapped reads accounting for the highest proportion, followed by reads mapped to multiple loci, and reads mapped to too many loci accounting for the lowest proportion (Table 3). These results indicated that the RNA-Seq results were reliable and could be used for subsequent analysis.
Table 3.
The data comparison results between RNA-Seq and Iso-Seq.
| Sample ID | Total reads | % of reads mapped to uniquely loci | % of reads mapped to multiple loci | % of reads mapped to too many loci | Mapped statistics |
|---|---|---|---|---|---|
| CKR-1 | 22 303 062 | 48.71% | 34.76% | 0.51% | 83.98% |
| CKR-2 | 21 292 208 | 50.20% | 35.13% | 0.55% | 85.88% |
| CKR-3 | 20 610 836 | 49.15% | 34.72% | 0.59% | 84.46% |
| CKS-1 | 20 824 428 | 49.53% | 35.08% | 0.94% | 85.55% |
| CKS-2 | 20 525 921 | 46.37% | 35.62% | 2.76% | 84.75% |
| CKS-3 | 20 030 625 | 48.59% | 35.12% | 0.99% | 84.70% |
| CKL-1 | 20 344 166 | 46.85% | 35.52% | 2.24% | 84.61% |
| CKL-2 | 20 374 733 | 48.08% | 36.00% | 1.36% | 85.44% |
| CKL-3 | 21 967 515 | 48.58% | 36.07% | 1.56% | 86.21% |
| HR24h-1 | 21 201 728 | 49.14% | 34.95% | 0.59% | 84.68% |
| HR24h-2 | 23 496 024 | 49.34% | 35.43% | 0.51% | 85.28% |
| HR24h-3 | 21 503 534 | 48.83% | 34.49% | 0.51% | 83.83% |
| HS24h-1 | 21 097 545 | 47.65% | 36.39% | 0.92% | 84.96% |
| HS24h-2 | 23 769 730 | 48.41% | 36.18% | 0.48% | 85.07% |
| HS24h-3 | 20 954 926 | 48.42% | 35.72% | 0.49% | 84.63% |
| HL24h-1 | 21 387 246 | 47.46% | 38.13% | 0.75% | 86.34% |
| HL24h-2 | 22 557 998 | 46.05% | 36.72% | 1.97% | 84.74% |
| HL24h-3 | 20 306 887 | 47.25% | 35.70% | 0.55% | 83.50% |
Transcript and expression quantification analysis of RNA-Seq
In order to clarify the transcripts expression pattern, the RNA-Seq was used to analyze the expression transcripts of different parts of A. bracteata under drought. A total of 384 549 112 clean reads were obtained from 18 samples which included three biological replicates in each group (Table 4). The Q20 of 18 samples was > 97.87%, the Q30 was > 93.9%, and the GC content ranged from 46.32% to 48.34% (Table 2), which showed that the RNA-Seq data was trustworthy and appropriate for additional study. Contributed raw sequence data to the NCBI SRA, accession number PRJNA 1 030 753.
Table 4.
The evaluation statistical table of RNA-Seq data.
| Sample ID | Read number | Base number | GC (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|
| CKR-1 | 22 303 062 | 6 674 759 940 | 46.44 | 98.09 | 94.56 |
| CKR-2 | 21 292 208 | 6 376 208 880 | 46.91 | 97.97 | 94.12 |
| CKR-3 | 20 610 836 | 6 170 104 132 | 46.32 | 97.87 | 93.90 |
| CKS-1 | 20 824 428 | 6 233 818 162 | 46.79 | 98.27 | 94.91 |
| CKS-2 | 20 525 921 | 6 143 561 570 | 47.14 | 98.13 | 94.58 |
| CKS-3 | 20 030 625 | 5 996 556 924 | 46.86 | 98.14 | 94.59 |
| CKL-1 | 20 344 166 | 6 088 844 094 | 48.17 | 98.01 | 94.23 |
| CKL-2 | 20 374 733 | 6 099 525 456 | 47.71 | 98.27 | 94.91 |
| CKL-3 | 21 967 515 | 6 576 086 594 | 48.34 | 97.87 | 93.97 |
| HR24h-1 | 21 201 728 | 6 346 531 032 | 46.59 | 98.12 | 94.58 |
| HR24h-2 | 23 496 024 | 7 032 866 094 | 46.48 | 98.09 | 94.47 |
| HR24h-3 | 21 503 534 | 6 437 821 450 | 46.57 | 98.07 | 94.42 |
| HS24h-1 | 21 097 545 | 6 315 409 508 | 46.92 | 98.15 | 94.62 |
| HS24h-2 | 23 769 730 | 7 115 330 912 | 46.77 | 98.00 | 94.23 |
| HS24h-3 | 20 954 926 | 6 271 772 248 | 46.95 | 98.03 | 94.26 |
| HL24h-1 | 21 387 246 | 6 401 988 224 | 47.51 | 98.19 | 94.71 |
| HL24h-2 | 22 557 998 | 6 753 289 962 | 47.74 | 98.15 | 94.59 |
| HL24h-3 | 20 306 887 | 6 078 726 308 | 47.47 | 98.02 | 94.36 |
DETs analysis
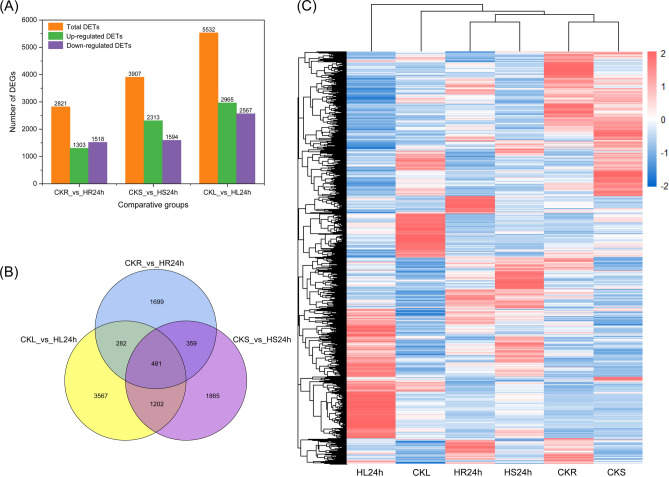
In this study, the screening conditions were set as FDR < 0.01 and FC ≥ 2 to obtain DETs, and the statistical results of DETs in each comparison group were shown in Fig. 5A. According to the findings, there were 2 821 DETs in the CKR vs. HR24h, including 1 303 up-regulated and 1 518 down-regulated transcripts. A total of 3 907 DETs were identified from CKS vs. HS24h, including 2 313 up-regulated and 1 594 down-regulated transcript. Meanwhile, 5 532 DETs were identified from CKL vs. HL24h, of which there were 2 965 up-regulated and 2 567 down-regulated transcripts. A total of 481 identical DETs were identified from roots, stems and leaves of A. bracteata under drought treatment. In addition, there were also have some unique DETs in different organs, such as a maximum number of unique DETs in leaves with 3 567, followed in stems with 1 865, and a minimum in roots with 1 699 (Fig. 5B). By further analyzing the cluster heat map of these DETs, we found that they were mainly clustered into nine categories and had significant differences in six samples (Fig. 5C). This indicated that the transcripts expression level in different organs of A. bracteata had significantly differences, and its leaves were more sensitive to drought stress, followed by the stems and finally the roots.
Fig. 5.
The number analysis of DETs in different organs of A. bracteata under CK and drought treatment. (A) Statistical histogram of the number of DETs in different organs. (B) The venn diagram of DETs in different organs. (C) The cluster heat map of DETs in six samples.
The DETs function analysis
To further analysis the functions of DETs in different organs of A. bracteata under drought stress, the cluster of COG classification statistics, the GO and KEGG enrichment analysis were used. The “carbohydrate transport and metabolism”, “posttranslational modification, protein turnover, chaperones”, “signal transduction mechanisms”, “general function prediction only” and “amino acid transport and metabolism” were the top five functional clusters of three comparative groups in the COG. However, the frequency of each functional clusters in three comparative groups were significantly differences (Fig. S1). This indicated that different organs of A. bracteata may adapt to drought stress by regulating the frequency of DETs in various functional clusters.
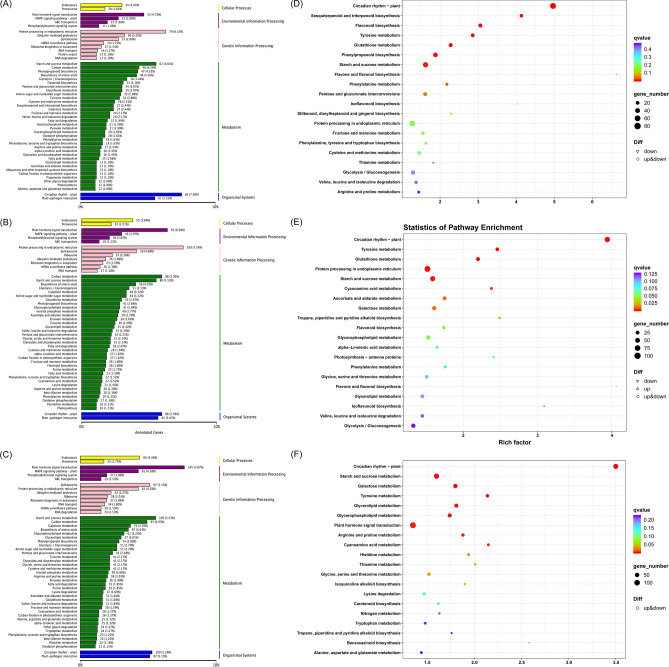
For GO enrichment, a total of 2 201 (998 up-regulated, 1 203 down-regulated), 2 986 (1 681 up-regulated, 1 305 down-regulated), and 4 115 (2 041 up-regulated, 2 074 down-regulated) DETs were enriched in CKR vs. HR24h、CKS vs. HS24h and CKL vs. HL24h, respectively. Concurrently, they were significantly enriched to 47, 52, and 51 GO items in CKR vs. HR24h、CKS vs. HS24h and CKL vs. HL24h, respectively. The top five items in the BP were followed by “cellular process”, “metabolic process”, “response to stimulus”, “single-organism process”, and “biological regulation” (Fig. S2). At the same time, KEGG annotation and pathway enrichment were used to analyse the potential biological functions of these DETs. The KEGG annotation results showed that these DETs were mainly divided into 5 categories, namely “cellular processes”, “environmental information processing”, “genetic information processing”, “metabolism” and “organismal systems”, and most of which involved in “metabolism”. Further analysis of the “metabolism” showed that the top three pathways of these DETs participation were followed by “starch and sucrose metabolism” (62, 5.61%), “carbon metabolism” (48, 4.34%) and “phenylpropanoid biosynthesis” (47, 4.25%) in the CKR vs. HR24h, “carbon metabolism” (86, 5.95%), “starch and sucrose metabolism” (80, 5.53%), and “biosynthesis of amino acids” (58, 4.01%) in the CKS vs. HS24h, “starch and sucrose metabolism” (105, 5.55%), “carbon metabolism” (93, 4.92%) and “galactose metabolism” (70, 3.7%) in the CKL vs. HL24h, respectively (Fig. 6 A-C). To further analysed whether there was a significant difference in DETs on a certain pathway, we conducted KEGG pathway enrichment analysis. The enrichment results showed that “circadian rhythm-plant” (ko04712), “starch and sucrose metabolism” (ko00500) and “phenylpropanoid biosynthesis” (ko00940) in the CKR vs. HR24h, “protein processing in endoplasmic reticulum” (ko04141), “circadian rhythm-plant” and “starch and sucrose metabolism” in the CKS vs. HS24h, and “plant hormone signal transduction”, “starch and sucrose metabolism” and “circadian rhythm-plant” in the CKL vs. HL24h were the top three significant enrichment pathway, respectively. Among which, “starch and sucrose metabolism” and “circadian rhythm-plant” were all significantly enriched in three different parts of A. bracteata (Fig. 6D-F).
Fig. 6.
The function analysis of differential expressed transcripts (DETs) in different organs of A. bracteata under CK and drought treatment. (A)—(C): The KEGG annotation classification diagram of DETs. (A) CKR vs. HR24h. (B) CKS vs. HS24h. (C) CKL vs. HL24h. (D)—(F): The KEGG pathway enrichment diagram of DETs. (D) CKR vs. HR24h. (E) CKS vs. HS24h. (F) CKL vs. HL24h.
Analysis of DETs involved in circadian rhythm-plant
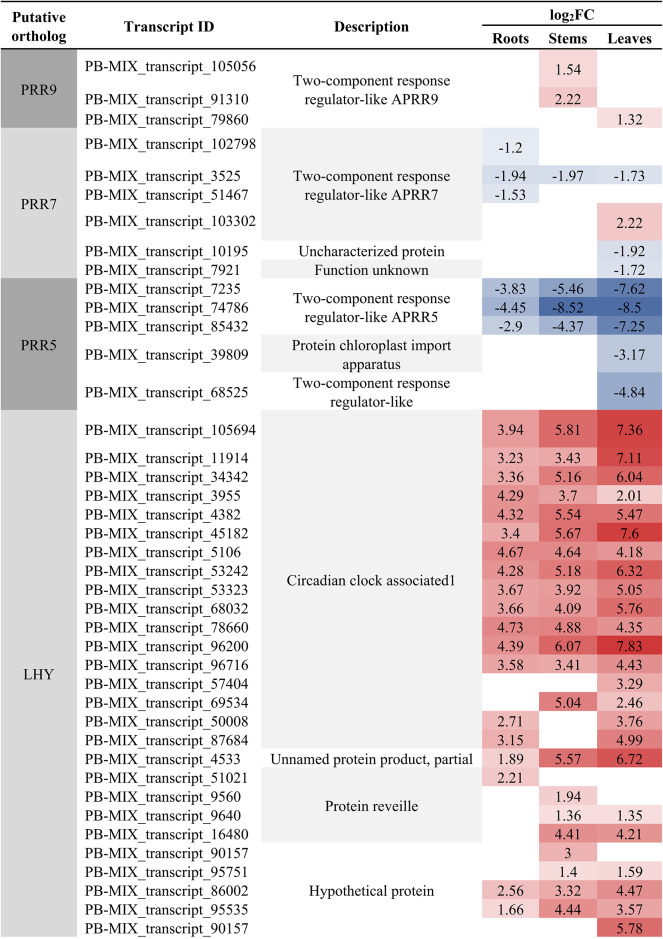
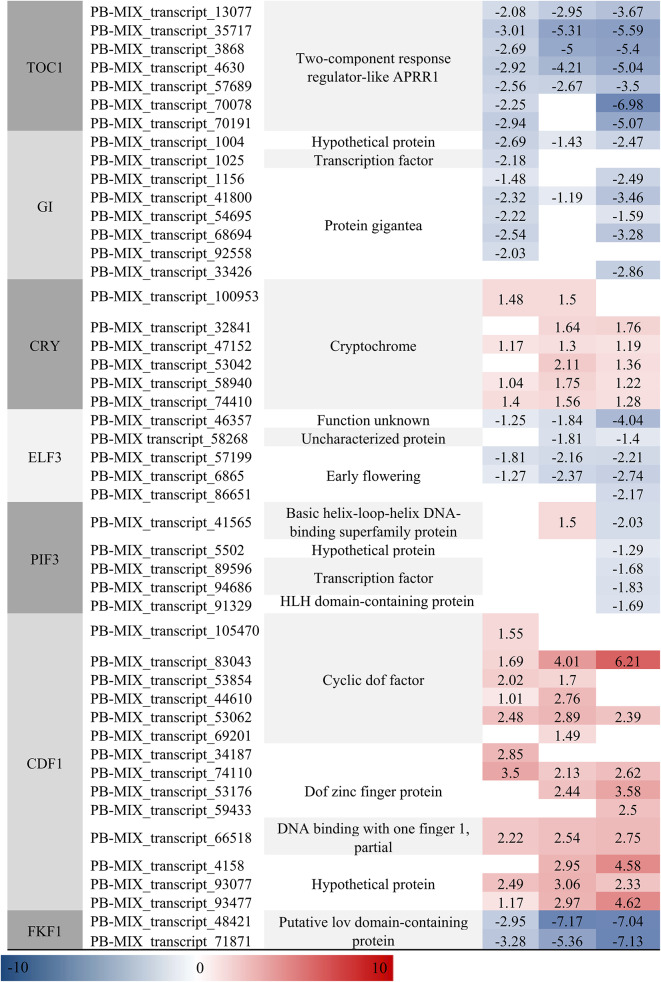
There were 83, 86, and 100 DETs in CKR vs. HR24h, CKS vs. HS24h, and CKL vs. HL24h of the circadian rhythm-plant pathway, respectively (Table S7). However, CRY, LHY and CDF1 were significantly up-regulated in three comparison groups, while PHYA was only up-regulated in the CKR vs. HR24h, PHYB was up-regulated in the CKR vs. HR24h and CKS vs. HS24h, and PRR9 was up-regulated in the CKS vs. HS24h and CKL vs. HL24h, respectively. Other genes in the circadian rhythm-plant pathway which DETs involved in were down-regulated, such as PRR7, PRR5, TOC1, GI, ELF3, PIF3 and FKF1. These results indicated that the variation of circadian rhythm-plant pathway caused by DETs in the different organs of A. bracteata under drought stress was diverse, and most of them were negatively regulated (Table 5). Further analyzed the functions of these DETs, we found that most of up-regulated DETs which related to CDF1 and CHY belonged to “C2C2-Dof” and “MYB-related” family, respectively, and they had a large number of DETs and a high expression level. Most of down-regulated DETs which related to TFs belonged to “other” and “Pseudo ARR-B” family (Table S6). This indicated that “MYB-related”, “C2C2-Dof”, “other” and “Pseudo ARR-B” of TFs members may play an important role in circadian rhythm-plant pathway of A. bracteata under drought stress.
Table 5.
The partial DETs related to circadian rhythm-plant pathway.
Analysis of DETs involved in arginine and proline metabolism
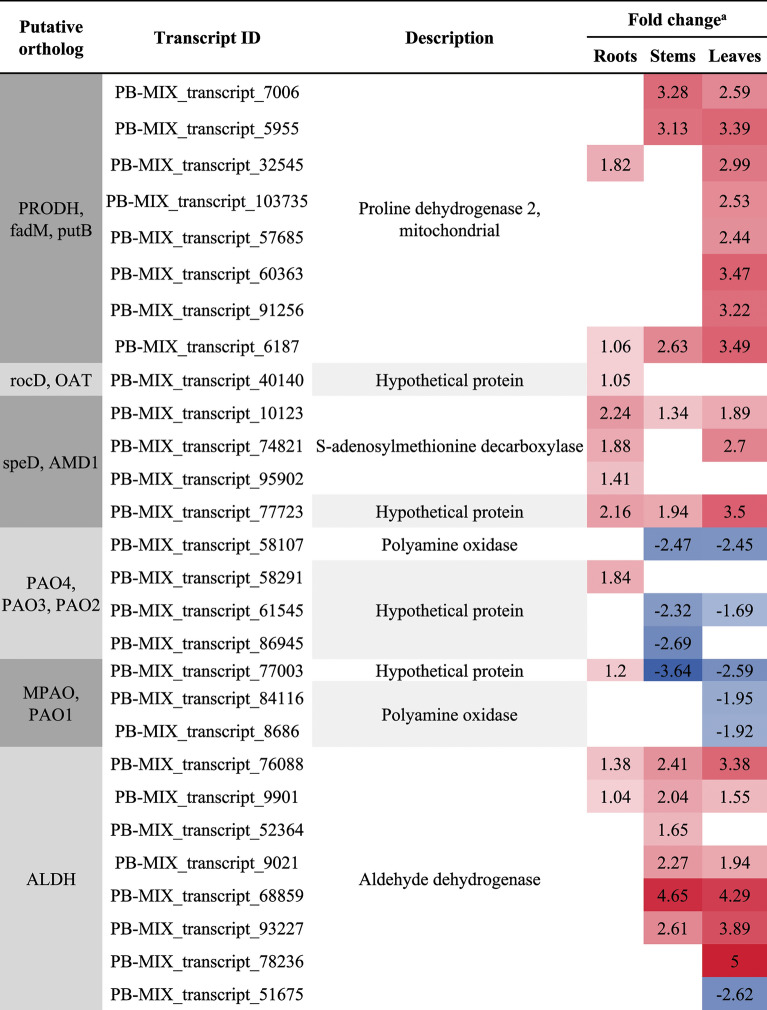
In the arginine and proline metabolism, a total of 17, 20, and 38 DETs were identified in CKR vs. HR24h, CKS vs. HS24h, and CKL vs. HL24h, respectively. Most of these DETs were up-regulated, and the number of DETs was highest in leaves, followed by stems, and lowest in roots. In addition, some DETs involved in the regulation of proline synthesis genes “PRODH, fadM, putB, rocD, OAT, ALDH18A1, P5CS, P4HA” were all up-regulated, and their numbers was also the highest in leaves, and less in roots and stems (Table 6).
Table 6.
The DETs related to arginine and proline metabolism.
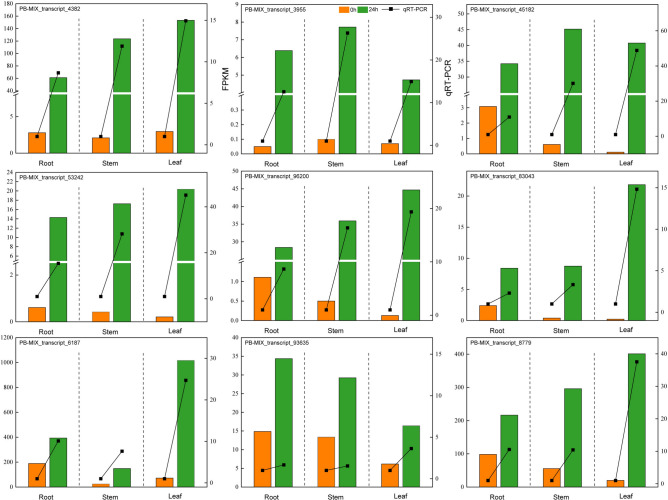
qRT-PCR validation of transcriptome
A total of 9 DETs were carefully chosen from key metabolic and signaling pathways, including “circadian rhythm in plants”, “arginine and proline metabolism”, “plant hormone signal transduction” as well as “starch and sucrose metabolism”. These DETs were selected for qRT-PCR analysis to validate the accuracy of the RNA-Seq data. As depicted in Fig. 7, the qRT-PCR results for the majority of these DETs across various organs of A. bracteata aligned well with the FPKM values from the RNA-Seq analysis. This concordance suggested that the RNA-Seq data obtained in this study was both accurate and reproducible.
Fig.7.
The qRT-PCR validation of RNA-Seq data. Note: The left vertical axis represents RNA-seq-FPKM, the right vertical axis represents qRT-PCR, and the horizontal axis represents different samples.
Discussion
The development and application of drought resistant tree species is a main way to enrich biodiversity in desert areas. A. bracteata as a typical shrub in desert areas, studying its drought resistance mechanism can provide scientific basis for its further development and application. In present study, we analyzed the full-length transcriptome and DETs in its roots, stems and leaves of A. bracteata under drought stress based on third-generation sequencing and combined with second-generation sequencing technology, morphological and physiological indicators. A total of 291 314 CCSs with mean read length of 2 062, and 248 178 FLNC, and 63 907 non-redundant FLNC were obtained (Table 1), meanwhile non-redundant FLNC sequences had an annotation rate of 86.96%. In addition, there were 2 821, 3 907 and 5 532 DETs identified from CKR vs. HR24h, CKS vs. HS24h and CKL vs. HL24h, respectively. In addition, as the degree of drought stress increased, the phenotype, osmotic substances, and antioxidant enzyme activity of different organs of A. bracteata undergo significant changes. It can be inferred that under drought stress, A. bracteata may alter the accumulation of related stress resistant substances and phenotypic changes by activating the expression of some drought resistant genes, and ultimately regulate its drought resistance.
RNA-Seq had become the most mature method for analyzing gene expression differences, RNA variable splicing phenomena, and miRNA regulatory effects in different periods and organs. However, RNA-Seq also had its own inevitable drawbacks, and which was difficult to accurately splice full-length transcripts and had more difficult to recognize long-chain non coding RNAs, fusion transcripts and homologous gene transcripts50. The Iso-Seq could directly reverse transcribe to obtain full-length cDNA, and which included a single complete transcript sequence information based on its ultra long reading, and subsequent analysis did not require assembly31. Because molecular biology research on A. bracteata has just started and there was no report on its genome, SMRT sequencing could provide a foundation for in-depth research in various aspects. In present study, using SMRT sequencing of A. bracteata, an entire of 31.86 Gb of clean data were acquired, and the numbers of CCS、FLNCs and non-redundant FLNC were also quite considerable compared to insects41,51, crops36,52 and trees53–55. That is to say, this study’s comprehensive transcriptome data might offer great support for the early molecular research of A. bracteata.
AS can play an important role on mechanism for gene expression regulation and can enhance the diversity of proteomes while having a smaller genome, adapting to the complexity of gene regulation requirements in higher multicellular organisms, it was also crucial in reacting to abiotic stress56,57. For example, the Oryza Sativa could improve drought resistance based on directly generate OsDREB2B2 by AS and spliced I1, E2, and I2 under drought stress58. Sanchez Martin et al. researched on the disease resistance of Triticum aestivum, and found that there were two ASs enhance its disease resistance by forming endoplasmic reticulum related complex59. In this study, we identified 2 093 AS events, but unfortunately, it could not classify them because of lacking of genomic data on A. bracteata.
TFs were a type of DNA binding proteins whose DNA binding domain (DBD) could specifically bind to specific sequences in the gene promoter region, thereby inhibiting or activating gene expression60. Furthermore, TFs were frequently involved in plant drought resistance, which focused on regulating the expression of genes related to drought response, mostly through activation or inhibition to regulate plant drought resistance. Among them, important transcription factors related to drought resistance include MYC/MYB, DREB, WRKY, bZIP, NAC and so on61. In present study, a total of 4 802 TFs were obtained from 64 families and the top 10 TF families were RLK-Pelle-DLSV, bHLH, others, MYB-related, C3H, AP2/ERF-ERF, bZIP, SNF2, WRKY and NAC, respectively, and most of them were related to plant stress resistance. NAC was unique to plants and played a key role in abiotic stress62,63. WRKY was a type of zinc finger transcription factor that participated in various abiotic stress responses64,65. As a regulatory protein, MYB was crucial for plants, and it widely participated in the response of plants to hormones and environmental factors66. In current study, when analyzed the function of DETs in different parts of A. bracteata under drought stress, the results showed that the “circadian rhythms-plant” pathway were significantly enriched in roots, stems and leaves. So, we further analyzed the circadian rhythm pathway which revealed that most of DETs mainly belonged to the MYB-related family. Therefore, we speculated that the TF of MYB-related played an important regulatory role in A. bracteata under drought stress. That is to say, TFs are quite necessary in studying the drought resistance of A. bracteata, and the TFs identified in present study could help A. bracteata to deal with drought stress through their complex regulatory.
The circadian rhythm is a biological rhythm that undergoes 24-h periodic changes when organisms are affected by various external environments67,68. When plants subjected to abiotic stress, their circadian rhythm also would have changed so as to enable them better to cope with environmental changes. The biological clock could transfer the valuable energy from responding to various stresses to plant growth by controlling various metabolic and physiological activities, which was the core of accurately synchronizing the circadian rhythm with the external environment to improve plant tolerance to abiotic stress67. When plants were subjected to drought stress, most drought response genes exhibited rhythmic expression patterns69, therefore drought was also regulated by circadian rhythms. For example, PRR7 may be closely related to drought, and most ABA and drought related genes may be negatively regulated by PRR7, while plants overexpressing PRR7 exhibited significant high sensitivity to drought stress68,70,71. In this study, DETs were significantly enrichment in the circadian rhythm-plant pathway, and there were 83 DETs, 86 DETs and 100 DETs in the CKR vs. HR24h, CKS vs. HS24h and CKL vs. HL24h, respectively. The majority of DETs participated in the circadian rhythm-plant pathway, including PRR7, were down-regulated in the roots, stems and leaves of A. bracteata following drought treatment, which were consistent with previous research results. In addition, scholars also found that PRR7 was mainly participated in the response of oxidative stress and the regulation of stomata conductance68,72, which would better help us to research the drought resistance ability of A. bracteata.
To further analyze the mechanism of changes in physiological indicators of A. bracteata under drought stress, we analyzed the metabolic pathways of arginine and proline, and found that under severe drought stress, the number of DETs related to proline synthesis regulation genes (“PRODH, fadM, putB, rocD, OAT, ALDH18A1, P5CS, P4HA”) in different organs of A. bracteata changed, with the highest number in leaves and fewer in roots and stems, all of which were up-regulated. This is consistent with the accumulation trend of proline content measured by physiological indicators (Fig. 2B), indicating that under drought stress, the expression regulation of drought resistant genes and physiological responses in different organs synergistically regulate the drought tolerance strategy of A. bracteata. In addition, PRODH is a key enzyme gene in the degradation process of proline, and P5CS is a key enzyme gene in the synthesis of proline. A large number of studies have shown that under stress conditions, the expression of P5CS gene increased, while the expression of degradation enzyme gene PRODH decreased, leading to an increase in the synthesis of proline in plants and a significant accumulation, thus improving stress resistance73. In this study, both P5CS and PRODH genes were all up-regulated, which showed A. bracteata has its own drought resistance regulatory characteristics.
There were many metabolic pathways related to plant stress, such as “phenylpropane biosynthesis”, “flavonoid biosynthesis”, “photosynthesis”, “plant hormone signal transduction”, ect. Our research group had previously studied the “phenylpropane biosynthesis”, “plant hormone signal transduction” and “flavonoid biosynthesis” pathway which were significantly enriched in the leaves of different ages of A. bracteata under natural drought74. In present study, the metabolic pathways of “phenylpropane biosynthesis” and “flavonoid biosynthesis” were also significantly enriched in roots of A. bracteata under drought stress, but relatively less in stems and leaves. The “plant hormone signal transduction” pathway was significantly enriched in leaves of A. bracteata, but relatively less in stems and roots. Therefore, this study focused on analyzing the pathways significantly enriched in all organs of A. bracteata, and has not yet analyzed those pathways specifically enriched in various organs. In the future, it is necessary to further study the specific pathways significantly enriched in different organs of A. bracteata under drought stress, which will provide a basis for further exploration of functional genes.
Conclusion
Under severe drought stress, the phenotype and physiological indicators of A. bracteata have undergone significant changes, and its DETs also have significant differences in different organs. In addition, the full-length transcriptome of A. bracteata under drought was sequenced and analyzed, which obtained a large of transcript data. Through analyzed the transcript raw data, main function and structure, we found that A. bracteata have large research space in terms of homologous species and new gene mining. Meanwhile, using the full-length transcriptome of A. bracteata as reference genome was help for studying the function of DETs under drought stress from RNA-seq. The selected DETs from different organs of A. bracteata were significantly enriched in the “starch and sucrose metabolism” and “circadian rhythm-plant” pathway, which maybe the key pathways in the gene regulation of A. bracteata under drought. Therefore, we could further explore some key genes for drought resistance of A. bracteata through these two pathways in the future research. In summary, this study could provide a reliable molecular basis and new ideas for future research on drought tolerance of A. bracteata.
Supplementary Information
Author contributions
Conceptualization, Y.L.(Yi Li); methodology, C.H. and W.L.; software, M.J. and C.L.; validation, W.L., M.J. and L.S.; formal analysis, L.S. and F.H.; investigation, C.H. and Y.L.(Yuan Liu); resources, C.H. and C.L.; data curation, M.W. and Y.L.(Yuan Liu); writing—original draft preparation, C.H., W.L., M.J. and Y.W.; writing—review and editing, C.H., M.J., Y.L.(Yi Li), Y.W.and P.C.; project administration, Q.Z.; funding acquisition, C.H., Y.L.(Yi Li); L.S. and P.C. All authors have read and agreed to the published version of the manuscript.
Funding
Natural Science Foundation of Gansu Province,24JRRH007,21JR7RH893,Gansu Province Key Research and Development Programme,24YFFH002,22YF7FA117,23YFFA0065,Plan Project of Gansu Forestry Science and Technology,2022kj013,Regular Science and Technology Assistance Projects of the Ministry of Science and Technology to Developing Countries,KY202002011
Data availability
The data of this study has been submitted to NCBI (https://www.ncbi.nlm.nih.gov/) and are publicly available at the number PRJNA 1 030 753 and PRJNA 1 028 835.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yi Li, Email: liyi@gsau.edu.cn.
Qinde Zhang, Email: 573857774@qq.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-80831-2.
References
- 1.He, C. Y., Zhang, G. Y., Zhang, J. G., Duan, A. G. & Luo, H. M. Physiological, biochemical, and proteome profiling reveals key pathways underlying the drought stress responses of Hippophae rhamnoides. Proteomics.16(20), 2688–2697. 10.1002/pmic.201600160 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Li, R. et al. Selection of the reference genes for quantitative gene expression by RT-qPCR in the desert plant Stipagrostis pennata. Sci. Rep.11(1), 21711. 10.1038/s41598-021-00833-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei, Y., Yin, C. & Li, C. Differences in some morphological, physiological, and biochemical responses to drought stress in two contrasting populations of Populus przewalskii. Physiol. Plant.127(2), 182–191. 10.1111/j.1399-3054.2006.00638.x (2010). [Google Scholar]
- 4.Hõrak, H. Learning from the experts: drought resistance in desert plants. New Phytol.216(1), 5–7. 10.1111/nph.14753 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Hu, Q. et al. Genome evolution of the psammophyte pugionium for desert adaptation and further speciation. Proc. Natl. Acad. Sci. U.S.A. 118(42), e2025711118. 10.1073/pnas.2025711118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang, J. et al. Mongolian Almond (Prunus mongolica Maxim): The morpho-physiological, biochemical and transcriptomic response to drought stress. PLoS One.10(4), e0124442. 10.1371/journal.pone.0124442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, H., Zhu, J., Gong, Z. & Zhu, J. K. Abiotic stress responses in plants. Nat. Rev. Genet.23(2), 104–119. 10.1038/s41576-021-00413-0 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Kong, L. An excellent sand fixing plant-Atraphaxis bracteata. Forest Sci. and Technol.10, 39. 10.13456/j.cnki.lykt.2001.10.020 (2001). [Google Scholar]
- 9.Xu, C. An excellent tree species for sand fixation and afforestation - Atraphaxis bracteata. J. Gansu Forest. Sci. Technol.4, 41–43 (1986). [Google Scholar]
- 10.Li, Y. Discussion on the possibility of promoting the application of excellent fuelwood Atraphaxis bracteata in sandy areas. J. Gansu Forest. Sci. Technol.2, 4–8 (1991). [Google Scholar]
- 11.Guo, Y. et al. Stem sap flow dynamics of Atraphaxis bracteata in Mu Us Sandy Area. Bull. Soil Water Conserv.30(5), 22–26. 10.1396/j.cnki.stbctb.2010.05.035 (2010). [Google Scholar]
- 12.Li, W. et al. Characteristics of water consumption of Atraphaxis bracteata A. Los. in the Mu Us Sandy Land of North Central China. J. Resour. Ecol.14(4), 880–892. 10.5814/j.issn.1674-764x.2023.04.020 (2023). [Google Scholar]
- 13.Wu, R. et al. Characteristics of water consumption of Atraphaxis bracteata through transpiration and its environmental impacting factors. Bull. Soil Water Conserv.32(3), 68–72. 10.13961/j.cnki.stbctb.2012.03.056 (2012). [Google Scholar]
- 14.Zhao, M. et al. Study on the transpiration characteristics of 10 species of plants under the different depths of groundwater level. Arid Zone Res.20(4), 286–291. 10.13866/j.azr.2003.04.010 (2003). [Google Scholar]
- 15.Dang Q. Study on antioxidant defense system of six shrubs under drought stress. Northwest A&F University. (2008).
- 16.Luo, Q. H., Ning, H. S., He, M., Ji, X. M. & Lei, C. Y. Ecophysiological responses of five sandy shrubs to drought stress. Sci. Silvae Sinicae.53(11), 29–42. 10.1170/j.1001-7488.20171104 (2017). [Google Scholar]
- 17.Ning H., Ji X., Sun H., Luo Q. Photosynthetic characteristics and related influencing factors of four plants in arid sand area. J. Northwest AF Uni. (Nat. Sci. Ed. ). 42 (11), 113-120. 10.1320/j.cnki.jnwafu.2014.11.096(2014)
- 18.Han, G., Li, S., Xu, P. & Han, E. Analyisis of drought resistance on anatomical structure of leave of six species of shrubs. J. Northwest Forest. Uni.21(4), 43–46. 10.3969/j.issn.1001-7461.2006.04.011 (2006). [Google Scholar]
- 19.Li, D. L. et al. Community structure and species diversity of Atraphaxis bracteata in Minqin Liangucheng Nature Reserve. J. Northwest Forest. Uni.31(5), 85–89. 10.3969/j.issn.1001-7461.2016.05.14 (2016). [Google Scholar]
- 20.Liu, S. et al. Structure and dynamics of Atraphaxis bracteata population in Minqin Liangucheng Nature Reserve. Gansu. Acta Ecol. Sinica.37(8), 1–7. 10.5846/stxb201409281913 (2017). [Google Scholar]
- 21.Li, X., Zhang, W. & Zhai, X. Study on transcriptome in plant stress resistance. J. Henan Forest. Sci. Technol.39(2), 7–10 (2019). [Google Scholar]
- 22.Morozova, O. & Marra, M. A. Applications of next-generation sequencing technologies in functional genomics. Genomics.92(5), 255–264. 10.1016/j.ygeno.2008.07.001 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Gao, H. et al. Application of RNA-seq technology in research on herb, shrub and tree stress resistance. Acta Prataculturae Sinica.28(12), 184–196. 10.11686/cyxb2019352 (2019). [Google Scholar]
- 24.Yan, S., Yang, R., Leng, S., Wang, Q. & Zhou, R. High-fluxed DNA sequencing technology and its application in agricultural science research. Chi. Agric. Sci. Bull.28(30), 171–176. 10.3969/j.issn.1000-6850.2012.30.032 (2012). [Google Scholar]
- 25.Guo, Q., Li, X., Niu, L., Jameson, P. E. & Zhou, W. Transcription-associated metabolomic adjustments in maize occur during combined drought and cold stress. Plant Physiol.186(1), 677–695. 10.1093/plphys/kiab050 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, M., Qi, X., Ni, M. & Lam, H. Silicon era of Carbon-based life: application of genomics and bioinformatics in crop stress research. Int. J. Mol. Sci.14(6), 11444–11483. 10.3390/ijms140611444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen, J. Z. et al. Transcriptomic and metabolomic profiling of Camellia sinensis L. cv. “Suchazao” exposed to temperature stresses reveals modification in protein synthesis and photosynthetic and anthocyanin biosynthetic pathways. Tree Physiol.39(9), 1583–1599. 10.1093/treephys/tpz059 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Wang, J. et al. Integration of transcriptomics and metabolomics for Pepper (Capsicum annuum L.) in response to heat stress. Int. J. Mol. Sci.20(20), 5042. 10.3390/ijms20205042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, B. H. et al. Transcriptomics integrated with metabolomics reveals the effect of regulated deficit irrigation on anthocyanin biosynthesis in Cabernet Sauvignon grape berries. Food Chem.314, 126170. 10.1016/j.foodchem.2020.126170 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Gordon, S. P. et al. Widespread polycistronic transcripts in fungi revealed by Single-molecule mRNA sequencing. PLoS One.10(7), e0132628. 10.1371/journal.pone.0132628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas, S., Underwood, J. G., Tseng, E. & Holloway, A. K. Long-read sequencing of chicken transcripts and identification of new transcript isoforms. PLoS One.9(4), e94650. 10.1371/journal.pone.0094650 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du, W. et al. Full-Length transcriptome characterization and functional analysis of pathogenesis-related proteins in Lilium Oriental hybrid ‘Sorbonne’ infected with botrytis elliptica. Int. J. Mol. Sci.24(1), 425. 10.3390/ijms24010425 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang, Z. et al. Full-length transcriptome of in Medicago sativa L. roots in response to drought stress. Front. Genet.13, 1086356. 10.3389/fgene.2022.1086356 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu, Q. et al. Physiological and full-length transcriptome analyses reveal the dwarfing regulation in Trifoliate Orange (Poncirus trifoliata L.). Plants.12 (2), 271. 10.3390/plants12020271 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He, X. et al. Full-length transcriptome characterization and comparative analysis of Chosenia arbutifolia. Forests.13(4), 543. 10.3390/f13040543 (2022). [Google Scholar]
- 36.Li, X. et al. Full-length transcriptome analysis of maize root tips reveals the molecular mechanism of cold stress during the seedling stage. BMC Plant Biol.22(1), 398. 10.1186/s12870-022-03787-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, D. et al. Full-length transcriptome analysis of Phytolacca americana and its congener P. icosandra and gene expression normalization in three Phytolaccaceae species. BMC Plant Biol.20(1), 396. 10.1186/s12870-020-02608-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, D. X. et al. Glycinebetaine mitigated the photoinhibition of photosystem II at high temperature in transgenic tomato plants. Photosynth. Res.147(3), 301–315. 10.1007/s11120-020-00810-2 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Sun, M. et al. Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol.20(1), 323. 10.1186/s12870-020-02530-0 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, W. et al. Transcriptome analysis and molecular mechanism of linseed (Linum usitatissimum L.) drought tolerance under repeated drought using single-molecule long-read sequencing. BMC Genomics.22(1), 109. 10.1186/s12864-021-07416-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, L. et al. Genome size estimation and full-length transcriptome of Sphingonotus tsinlingensis: genetic background of a drought-adapted grasshopper. Front. Genet. 12, 678625.10.3389/fgene.2021.678625 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng, Y. et al. Integrated NR database in protein annotation system and its localization. Comput. Eng.32(5), 71–74. 10.3969/j.issn.1000-3428.2006.05.026 (2006). [Google Scholar]
- 43.Consortium T.U. UniProt: the universal protein knowledgebase. Nucleic Acids Res.45(D1), D158–D169. 10.1093/nar/gkw1099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet.25(1), 25–29. 10.1038/75556 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucl. Acids Res.28(1), 33–36. 10.1093/nar/28.1.33 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koonin, E. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome biol.5(2), R7. 10.1186/gb-2004-5-2-r7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finn, R. D. et al. Pfam: the protein families database. Nucl. Acids Res.42(D1), D222–D230. 10.1093/nar/gkt1223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics.26(1), 139–140. 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nie, T. Explore the anthocyanin synthesis mechanism of colored potatoes based on full-length transcriptome sequencing and extensive targeted metabolome. Northwest A&F University. (2022). [Google Scholar]
- 51.Zheng, S. Y., Pan, L. X., Cheng, F. P., Jin, M. J. & Wang, Z. L. A global survey of the full-length transcriptome of Apis mellifera by single-molecule long-read sequencing. Int. J. Mol. Sci. 24 (6), 5827.10.3390/ijms24065827 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Min, X., Lin, X., Ndayambaza, B., Wang, Y. & Liu, W. Coordinated mechanisms of leaves and roots in response to drought stress underlying full-length transcriptome profiling in Vicia sativa L. BMC Plant. Biol. 20 (1), 165. 10.1186/s12870-020-02358-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiao, D. et al. Comprehensive identification of the full-length transcripts and alternative splicing related to the secondary metabolism pathways in the tea plant (Camellia sinensis). Sci. Rep.9(1), 2709. 10.1038/s41598-019-39286-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun, X. & Li, H. Full-length transcriptome combined with RNA sequence analysis of Fraxinus chinensis. Genes Genom.45(5), 553–567. 10.1007/s13258-023-01374-w (2023). [DOI] [PubMed] [Google Scholar]
- 55.Zhang, L. et al. Full-length transcriptome and transcriptome sequencing unveil potential mechanisms of Brassinosteroid-induced flowering delay in tree peony. Acta. Hortic. 8(12), 1136. 10.3390/horticulturae8121136 (2022). [Google Scholar]
- 56.Barbazuk, W., Fu, Y. & McGinnis, K. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res.18(9), 1381–1392. 10.1101/gr.053678.106 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez Gallo, M. C. & Uhrig, R. G. Phosphorylation mediated regulation of RNA splicing in plants. Front. plant sci.14, 1249057. 10.3389/fpls.2023.1249057 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsukura, S. et al. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics.283(2), 185–196. 10.1007/s00438-009-0506-y (2010). [DOI] [PubMed] [Google Scholar]
- 59.Sánchez-Martín, J. et al. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat. Plants.7(3), 327–341. 10.1038/s41477-021-00869-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y., Yan, Y. & Wang, Z. The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza. J. Agric. Food Chem.58(23), 12168–12175. 10.1021/jf103203e (2010). [DOI] [PubMed] [Google Scholar]
- 61.Liu, X. & Li, Y. Transcription factors related to plant stress-tolerance. Chi. Agric. Sci. Bull.22(4), 61–65. 10.3969/j.issn.1000-6850.2006.04.016 (2006). [Google Scholar]
- 62.Liu, X. S. et al. Ectopic expression of NAC transcription factor HaNAC3 from Haloxylon ammodendron increased abiotic stress resistance in tobacco. Planta.256, 105. 10.1007/s00425-022-04021-y (2022). [DOI] [PubMed] [Google Scholar]
- 63.Zong, X. et al. Expression analysis of HaNAC2 and identification of stress resistance function in Haloxylon ammodendron. Acta Agriculturae Boreali-occidentalis Sinica.28(8), 1317–1325. 10.7606/j.issn.1004-1389.2019.08.014 (2019). [Google Scholar]
- 64.Javed, T. & Gao, S. WRKY transcription factors in plant defense. Trends in genetics: Trends Genet.39(10), 787–801. 10.1016/j.tig.2023.07.001 (2023). [DOI] [PubMed] [Google Scholar]
- 65.Sun, S., Liu, X., Zhang, T., Yang, H. & Yu, B. GsWRKY23 functional characterisation of the transcription factor gene from in overexpressed Soybean composite plants and under salt stress. Plants (Basel, Switzerland). 12 (17), 3030. 10.3390/plants12173030 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren, C. et al. Overexpression of a Grape MYB transcription factor gene VhMYB2 increases salinity and drought tolerance in Arabidopsis thaliana. Int. J. Mol. Sci.24(13), 10743. 10.3390/ijms241310743 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao, X. et al. The plant circadian rhythm and its response to abiotic stresses. Plant Physiol. J.53(10), 1833–1841. 10.13592/j.cnki.ppj.2017.0142 (2017). [Google Scholar]
- 68.Seo, P. J. & Mas, P. STRESSing the role of the plant circadian clock. Trends Plant Sci.20(4), 230–237. 10.1016/j.tplants.2015.01.001 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Mizuno, T. & Yamashino, T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol.49(3), 481–487. 10.1093/pcp/pcn008 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Nakamichi, N. et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol.50(3), 447–462. 10.1093/pcp/pcp004 (2009). [DOI] [PubMed] [Google Scholar]
- 71.Xu, X., Yuan, L. & Xie, Q. The circadian clock ticks in plant stress responses. Stress Biol. 2(1), 15.10.1007/s44154-022-00040-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu, T., Carlsson, J., Takeuchi, T., Newton, L. & Farré, E. M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J.76(1), 101–114. 10.1111/tpj.12276 (2013). [DOI] [PubMed] [Google Scholar]
- 73.Peng, Z., Lu, Q. & Verma, D. Reciprocal regulation of Δ1-pyrroline-5-carboxylate synthetase and proline dehydrogenase genes controls proline levels during and after osmotic stress in plants. Mol. Gen. Genet.253(3), 334–341. 10.1007/pl00008600 (1996). [DOI] [PubMed] [Google Scholar]
- 74.He, C. et al. The metabolomics and transcriptomics of Atraphaxis bracteata reveal age-related molecular alterations under naturally high temperature and drought conditions. Physiol. Plantarum.176(1), e14129. 10.1111/ppl.14129 (2024). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of this study has been submitted to NCBI (https://www.ncbi.nlm.nih.gov/) and are publicly available at the number PRJNA 1 030 753 and PRJNA 1 028 835.