Abstract
The presence of high-risk human papillomavirus (HR-HPV) contributes to the development of cervical lesions and cervical cancer. Recent studies suggest that an imbalance in the cervicovaginal microbiota might be a factor in the persistence of HR-HPV infections. In this study, we collected 156 cervicovaginal fluid (CVF) of women with HR-HPV infection, which were divided into three groups (negative for intraepithelial lesions = 78, low/high-grade squamous intraepithelial lesions = 52/26). We performed metabolomics and 16 S rRNA sequencing to identify changes in metabolites and cervicovaginal microbiota among patients with HR-HPV infection and varying grades of cervical lesions. We detected 164 metabolites and 389 flora types in the three groups. Ten CVF metabolites—N-methylalanine, phenylacetaldehyde, succinic acid, 2-3-dihydroxypyridine, DL-p-hydroxylphenyllactic acid, gluconic acid lactone, guanine, glucose-6-phosphate, erythrose, and sucrose showed significant associations with disease severity and distinct separation patterns in HR-HPV-infected patients with LSIL and HSIL, with an area under the curve of 0.928. The most abundant microbial communities in the CVF were Gardnerella. Gardnerella was found to be associated with increased levels of succinic acid, thereby highlighting distinct metabolic profiles. These findings suggest that the development of cervical lesions resulting from persistent HR-HPV infection is associated with significant alterations in systemic metabolism and shifts in the cervicovaginal microbiota, providing valuable insights into the metabolic and microbiota changes linked to disease severity.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84796-0.
Keywords: Metabolomics, 16S rRNA sequencing, Human papillomavirus, Cervicovaginal fluid
Subject terms: Microbial communities, Metabolomics, Viral infection, Infection
Introduction
High-risk human papillomavirus (HR-HPV) infection is identified in nearly 90% of cervical squamous intraepithelial lesions (SIL) and 99% of cervical cancer tissue samples, as demonstrated by a 15-year population-based cohort study conducted in China1. Recent research indicates that disruptions in the vaginal microbiota are associated with bacterial vaginosis and various infectious disorders. The persistence of HR-HPV infection is influenced by complex interactions between host immune responses, viral factors, and the delicate balance of the vaginal microbiome2. Several studies suggest that disturbances in the cervicovaginal microbiota can lead to persistent HR-HPV infections and may contribute to the development of SILs3,4. Cervical SILs are prevalent conditions that pose a significant threat to women’s health and are closely associated with invasive cervical cancer5,6. This malignancy cauesd by SILs is characterized by a rising incidence rate and an emerging trend toward earlier onset, which adversely affects women’s health and overall quality of life. However, the pathogenesis of SILs remains unclear and warrants further investigation.
Metabolomics, with its capability to detect thousands of small molecules within an ecosystem, has significantly advanced the exploration of metabolites. Paired microbiome-metabolome studies have provided valuable mechanistic insights into host-microbiome interactions across a variety of pathologies7. Specifically, the vaginal metabolome of women with HR-HPV infection (14 out of 39 cases) exhibits distinct profiles compared to that of HPV-negative counterparts, with notable differences in biogenic amines, glutathione, glycogen, and lipid metabolism-related metabolites8. The HR-HPV-positive group demonstrates a reduction in the number of metabolites compared to HPV-negative individuals, while the cervical cancer group shows an increase. This non-linear trend suggests that HR-HPV infection and SILs may deplete metabolites in certain amino acid and nucleotide pathways, whereas cervical cancer is associated with metabolite enrichment from diverse lipid pathways9. Furthermore, HR-HPV infection can activate various metabolic pathways, directly influencing enzymes in the glycolytic pathway, enhancing glucose uptake and glycolysis, and promoting the pentose phosphate pathway, and lactate dehydrogenase A synthesis10.
Academic reports indicate that HR-HPV infection influences mucosal metabolism and host immune status. Conversely, vaginal microbiota and host immunity can affect HR-HPV infection11–13. Individuals infected with HR-HPV harbor a highly diverse cervicovaginal microbiome14. Chronic HR-HPV infection has been associated with imbalances in vaginal microecology and damage to the cervicovaginal mucosal barrier15. Prolonged inflammation within the cervix and vagina may contribute to cervical epithelial degeneration and necrosis, thereby compromising the integrity of the cervical epithelial barrier and triggering an immune response. This increased vulnerability can facilitate the propagation of other pathogenic organisms, perpetuating HR-HPV infection and potentially leading to the development of squamous intraepithelial lesions (SILs)16. Emerging research reveals that cervical microbial metabolites can enter systemic circulation, thereby influencing the overall health of patients at high risk for SILs17. Consequently, metabolites derived from or altered by the microbiome have garnered considerable attention, highlighting their potential to impact the host both locally and systemically16–19.
Taken together, current studies reveal that a disturbed vaginal microbiota not only impairs immune responses but also facilitates the maintenance of HR-HPV infection, potentially leading to the progression of SILs. Alterations in metabolites attributed to the vaginal flora may indicate the risks associated with cervical cancer among patients infected with HR-HPV20. Cervical lavage fluid, which originates from secretions within the cervicovaginal cavity, can be collected through a procedure that is more straightforward and less invasive compared to the collection of cervical exfoliated cells. Consequently, it may serve as a viable tool for monitoring biomarkers to evaluate the risk of cervical cancer in women with HR-HPV infection. In the present study, we collected cervicovaginal lavage fluid samples from women infected with HR-HPV who exhibited various degrees of cervical lesions. We characterized the corresponding changes in microbial flora and associated metabolites in search of potential biomarkers capable of predicting the risk of maintaining HR-HPV infection and the progression of SILs. The findings presented here offer valuable insights into diagnostic and monitoring strategies designed to prevent the advancement of SILs in women with HR-HPV infection.
Results
Baseline characteristics
The clinical baseline data were presented in Table 1, which included age in years, gravidity times, and parity times, all presented as the mean ± standard deviation. The table revealed no significant differences in age (P = 0.293), gravidity times (P = 0.452), and parity times (P = 0.266) among the three groups of women.Clinical information regarding leucorrhea for each group is presented in Table 1.
Table 1.
Basic information of the participants.
| NILM (N = 78) |
LSIL (N = 52) |
HSIL (N = 26) |
P-Value | |
|---|---|---|---|---|
| Age(years) | 35.60 ± 7.843 | 37.50 ± 9.648 | 37.91 ± 3.22 | 0.293 |
| Gravidity(times) | 2.40 ± 1.761 | 2.04 ± 1.414 | 2.27 ± 1.343 | 0.452 |
| Parity(times) | 1.12 ± 0.581 | 0.94 ± 0.639 | 1.12 ± 0.711 | 0.266 |
| Leucorrhea cleanliness(n%) | ||||
| I-II° | 47(60.3%) | 25(48.1%) | 8(30.8%) | 0.028 |
| III° | 26(33.3%) | 16(30.8%) | 9(34.6%) | |
| IV° | 5(6.4%) | 11(21.2%) | 9(34.6%) | |
| H2O2 positive(n%) | 69(88.5%) | 50(89.3%) | 25(96.2%) | 0.200 |
| Sialidase positive(n%) | 19(24.4%) | 14(8.7%) | 9(34.6%) | 0.028 |
| Leukocyte esterase positive(n%) | 18(23.1%) | 10(17.9%) | 7(26.9%) | 0.735 |
| single infections | 44 | 34 | 15 | - |
| dual infections | 22 | 14 | 7 | - |
| Multiple infections | 12 | 4 | 4 | - |
Leucorrhea cleanliness values of grades III and IV indicate poor cleanliness. The vaginal microbiota of HR-HPV infected patients increases with the severity of cervical lesions, and the number of cases with poor vaginal discharge cleanliness and sialidase positive increases. The positivity of H2O2, sialidase, and leukocyte esterase was associated with bacterial vaginitis. There is a statistically significant difference in the Leucorrhea cleanliness values, with a P-value of 0.028 according to the ANOVA test. The difference in the positivity rate of sialidase is statistically significant, with a P-value of 0.028. There was no statistically significant difference in the positive rates of H2O2(P = 0.200) and leukocyte esterase(P = 0.735).
Among the patients with HR-HPV infection under study, 93 (59.6%) exhibited single-infections. The most common type was HPV 16, affecting 28 cases (17.9%), followed by HPV 52 with 12 cases (7.7%), HPV 18 with 10 cases (6.4%), and HPV 58 with 10 cases (6.4%). There were 43 cases (27.6%) with dual infections and 20 cases (12.8%) with multiple infections.
Analysis of CVF sample metabolites
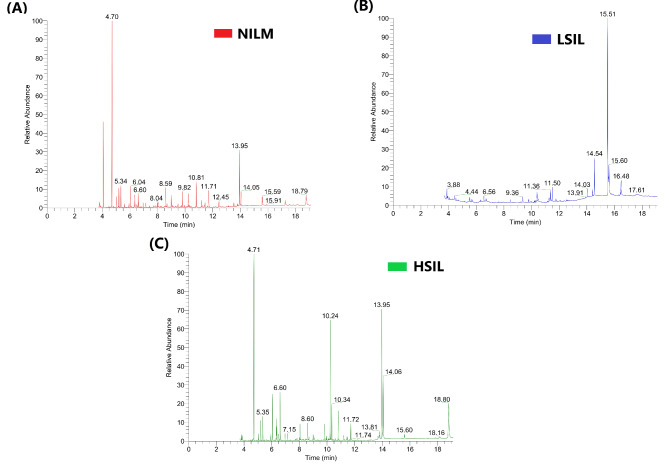
A total of 164 metabolites were identified in the CVF samples. The total ion chromatograms (TICs) of the CVF samples, which were obtained using GC-MS, were shown in Fig. 1. The relative standard deviation (RSD) of the internal standard was less than 12%, while the RSD of the quality control (QC) sample was less than 17.2%. The quality accuracy was controlled within 10 ppm, and the variation in retention time was less than 0.1 min.
Fig. 1.
Total Ion Chromatograms (TICs) of the CVF samples using gas chromatography-mass spectrometry (GC-MS) for the (A) NILM groups, (B) LSIL groups, and (C)HSIL group.
OPLS-DA models Fig. 2 (A and C) clearly separated the CVF samples into two groups: NILM and SIL (LSIL + HSIL) groups, as well as the LSIL and HSIL groups. The goodness of fit and predictive capabilities of the OPLS-DA model were estimated using R2Y, where the first value represented the fit and the second value represented the predictive capability. A permutation p-value less than 0.001 confirmed the model’s reliability and indicated the absence of overfitting, as shown in Fig. 2 (B and D). These results suggest that the metabolic patterns in HSIL patients were significantly different from those in NILM and LSIL patients.
Fig. 2.
OPLS-DA score plots for the NILM and SIL groups (A) and the LSIL and HSIL groups (C). Panels (B) and (D) present the results of the corresponding 1000 permutations (p < 0.001), indicating the reliability of the permutation tests. For the NILM and SIL comparison, panel (B) shows R2Y = 0.558 and Q2 = 0.296, while panel (D) for the LSIL and HSIL comparison shows R2Y = 0.977 and Q2 = 0.673. Here, R2Y indicates the proportion of variance explained by the current model, while Q2 represents the proportion of data variance that the model can predict.
Differential metabolites and pathway analysis in cervical lesions
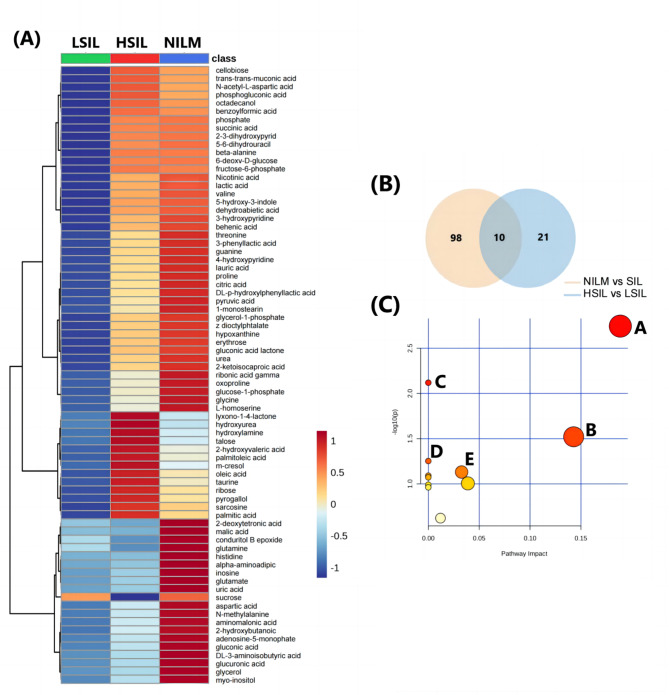
A total of 108 differential metabolites were identified between the NILM group and the SIL group (LSIL + HSIL), with the highest-ranking metabolites including O-phosphothreonine, N-methylglutamic acid, tryptophan, melezitose, glutamine, gamma-aminobutyric acid, alpha-aminoadipic acid, sedoheptulose, N-methylalanine, and spermidine. Detailed information was provided in Table S1.
A total of 31 differential metabolites were identified between the LSIL group and the HSIL group, with the highest-ranking metabolites including sucrose, erythrose, iminodiacetic acid, glucose-6-phosphate, pyrogallol, 2-3-dihydroxypyridine, N-methylalanine, tyrosine minor and erythronic acid lactone. Data was presented in Table S2.
The heatmap results showed that most differential metabolites with low abundances were found in LSIL compared to NILM and HSIL (Fig. 3A). There are 10 different metabolites among the NILM and SIL groups, and the LSIL and HSIL groups (Fig. 3B). After using MetaboAnalyst to analyze the metabolic pathways of the aforementioned 10 potential markers, pathway topology analysis was performed to determine the relevant pathways with an impact value > 0.1. Further pathway analysis also found 5 metabolic pathways that may be related to them, including: (A) starch and sucrose metabolism, (B) phenylalanine metabolism, (C) neomycin, kanamycin, and gentamicin biosynthesis, (D) butanoate metabolism, and (E) TCA cycle (Fig. 3C).
Fig. 3.
Differential metabolites and metabolic pathways of CVF with different degrees of lesion. (A) Heatmap of the 75 most abundant metabolites found in the three groups of samples. (B) Venn plot of differential metabolites between NILM and SIL compared to LSIL and HSIL. (C) The metabolic pathways through which the cervicovaginal microbiota affects the occurrence of SIL. The pathways that mainly affect SIL are A: starch and surose metabolism; B: phenylalanine metabolism; C: neomycin, kanamycin and gentamicin biosynthesis; D: butanoate metabolism; and E: TCA cycle.
Diagnostic performance of a 10-metabolite panel in cervical lesions
These 10 differential metabolites show significant differences between the LSIL group and the HSIL group (Fig. 4A), including the following metabolites: 2,3-dihydroxypyridine, DL-p-hydroxylphenyllactic acid, erythrose, glucose-6-phosphate, gluconic acid lactone, guanine, N-methylalanine, and phenylacetaldehyde, which were increased in the HSIL group, while succinic acid and sucrose were decreased in the HSIL group. Additionally, they show significant differences between the NILM group and the SIL group (Figure S1).
Fig. 4.
Differential metabolites. (A) Changes in the levels of 10 differential metabolites among the LSIL and HSIL groups (P value details can be found in the supplementary materials). (B) The performance of the 10-metabolite panel for HR-HPV-infected cervical lesions was assessed using the area under the curve (AUC) calculated by the random forest algorithm. The metabolite panel distinguished between the NILM group and the SIL group (LSIL + HSIL), yielding an AUC of 0.811. (C) The metabolite panel distinguished between the LSIL group and the HSIL group, with an AUC of 0.928.
The performance of the 10-metabolite panel for HR-HPV-infected cervical lesions was assessed using the area under the curve (AUC) calculated by the random forest algorithm. The metabolite panel in CVF demonstrated good accuracy in distinguishing between the NILM group and the SIL group (LSIL + HSIL), with an AUC of 0.811 (Fig. 4B), and between the LSIL group and the HSIL group, with an AUC of 0.928 (Fig. 4C).
Analysis of the bacterial community in cervical lesions
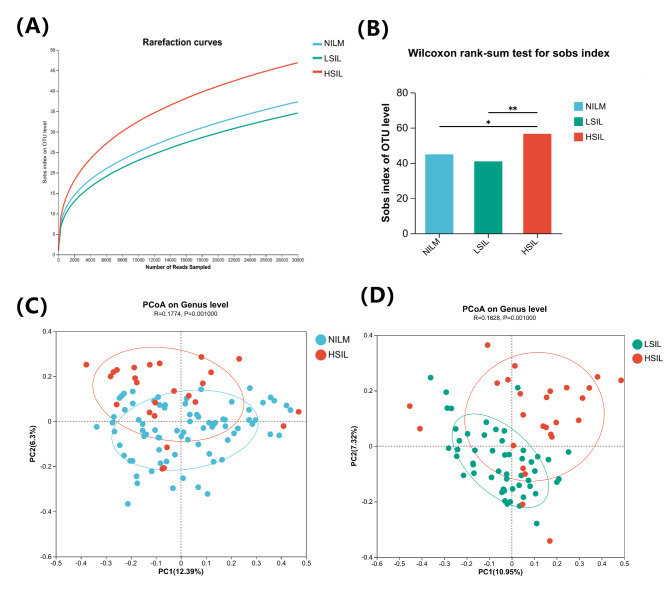
We analyzed 156 CVF samples to assess the bacterial composition in the vagina. Alpha diversity was compared using dilution curves, showing a steeper curve in the HSIL group compared to the others, while the LSIL group had a gentler curve than the NILM group, indicating that vaginal microbiota diversity may initially decrease before significantly increasing (Fig. 5A). Using the Sobs index, we found significant differences in alpha diversity between the HSIL group and the NILM and LSIL groups. Microbial diversity slightly decreased from NILM to LSIL (P = 0.8391) but significantly increased from LSIL to HSIL (P = 0.0042) and from NILM to HSIL (P = 0.0182) (Fig. 5B). Beta diversity analysis via principal coordinate analysis (PCoA) showed distinct separation of the HSIL group from the others based on binary Jaccard distance (ANOSIM test, P = 0.001), with no significant difference between the NILM and LSIL groups (P = 0.7190) (Fig. 5C, D).
Fig. 5.
Alpha diversity and beta diversity of microbial communities in three groups of CVF. (A) Comparison of the Alpha diversity index between NILM, LSIL, and HSIL groups. (B) The Sobs index was used to compare the Alpha diversity of vaginal microbiota in the three groups. (C) Comparison of the Beta diversity index between the NILM and HSIL groups. (D) Comparison of the Beta diversity index between the LSIL and HSIL groups.
At the genus level, we identified 152 microorganisms that were common across the three groups. Lactobacillus was the most abundant genera, followed by Gardnerella. As the severity of cervical lesions progressed, the abundance of Atopobium and Sneathia increased, while Streptococcus decreased (Fig. 6A, C). The NILM group exhibited 149 unique genera, the LSIL group had 86, and the HSIL group had 73 unique genera (Fig. 6B). The Vaginal Microbial Health Index (VMHI) serves as a robust measure for evaluating health status based on species-level classification features of vaginal microbiome samples. Our analysis revealed that the HSIL group had significantly lower VMHI scores compared to the LSIL group, and there was also a notable decrease in VMHI scores in the SIL group compared to the NILM group (Figure S2).
Fig. 6.
The number and composition of microorganisms in the three NILM, LSIL, and HSIL groups. (A) The heatmap of the 50 microbial communities with the highest abundance among the three groups. (B) Venn plots of three unique or shared microbial communities. (C) Bar plot of species composition at the genus level for three groups.
Correlation between vaginal microbiota and metabolites
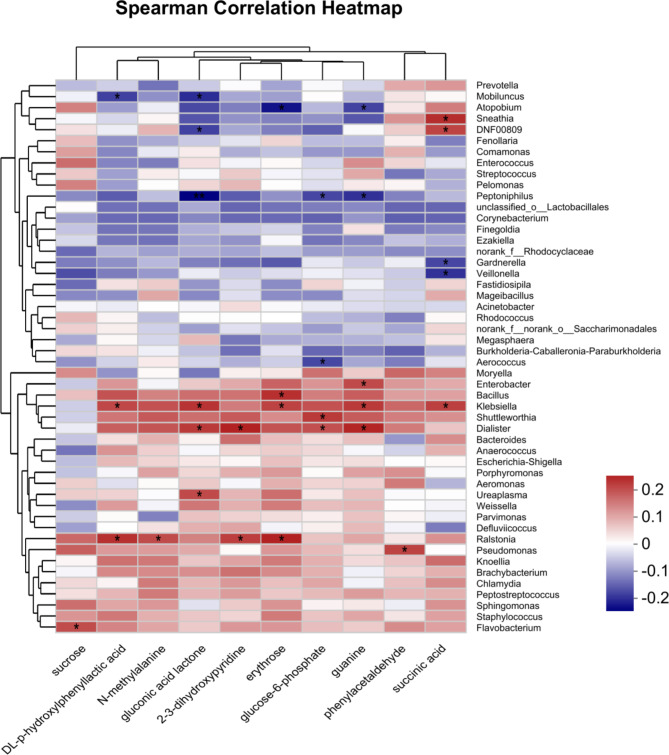
We analyzed the correlation between 10 differentially expressed metabolites and the top 50 bacterial populations based on abundance. Metabolites were screened using variance inflation factor (VIF) analysis, excluding those with a VIF greater than 10 due to weak correlations. The correlation coefficients between the selected metabolites and the top 50 abundant bacterial genera were computed through heatmap analysis, revealing significant associations (Fig. 7). Sucrose positively correlated with Flavobacterium, Succinic acid showed positive correlations with Klebsiella, DNF00809, and Sneathia, but negative correlations with Gardnerella and Veillonella. Glucose-6-phosphate positively correlated with Shuttleworthia and Dialist, while negatively correlating with Aerococcus and Peptoniphilus.
Fig. 7.
Heatmap illustrating the correlation between vaginal microbiota and differentially expressed metabolites in the samples.
Discuss
We performed metabolomics to identify changes in metabolites among patients with HR-HPV infection and varying grades of cervical lesions. We detected 164 metabolites in the three groups. There are ten CVF metabolites—N-methylalanine, phenylacetaldehyde, succinic acid, 2-3-dihydroxypyridine, DL-p-hydroxylphenyllactic acid, gluconic acid lactone, guanine, glucose-6-phosphate, erythrose, and sucrose showed significant associations with disease severity.
Phenylacetaldehyde is an aromatic fatty aldehyde primarily derived from the metabolic processes of fatty acid in animal bodies. It has been shown to inhibit cancer cell proliferation and promote apoptosis in these cells. Furthermore, phenylacetaldehyde reduces the expression of cancer stem cell (CSC) marker genes, including CD44+/CD24- and ALDH1. It also preferentially induces the production of reactive oxygen species (ROS), decreases the phosphorylation of nuclear Stat3, and lowers IL-6 secretion, thereby inhibiting the activation of the Stat3 signaling pathway. Because of these properties, phenylacetaldehyde is considered a potential therapeutic agent for cancer and cancer stem cells14. In our study, we observed that phenylacetaldehyde levels were increased in the HSIL group, suggesting a protective compensatory mechanism in cervical lesions.
Glucose-6-phosphate is a product formed by the phosphorylation of various sugars that are metabolized into glucose after entering cells15. In our study, we observed an upregulation of glucose-6-phosphate and a downregulation of sucrose, indicating that glucose metabolism is enhanced in cervical lesions. This increased metabolism may contribute to viral replication and exacerbate the severity of the lesions16,17.
Succinic acid is a metabolic intermediate of amino acids in the tricarboxylic acid (TCA) cycle within host cells. It can be metabolized by somatic cells, and current research indicates that succinic acid and its derivatives exhibit anticancer activity by inducing cell apoptosis18–20. In our study, we found that succinic acid levels were decreased, which suggests that high-risk HPV infection leads to increased cellular proliferation. This proliferation may exacerbate disease progression and raise the potential for further advancement toward cervical cancer.
We analyzed 156 CVF samples to assess the bacterial composition in the vagina. At the genus level, we identified 152 microorganisms that were common across the three groups. Lactobacillus was the most abundant genera, followed by Gardnerella. This study found a close association between cervicovaginal microbiota and cervical cancer in patients with persistent HR-HPV infection. The cervicovaginal environment is home to various bacteria, with Lactobacillus species frequently recognized as biomarkers of health21. These bacteria play a crucial role in maintaining a balanced micro-ecosystem, which can be disrupted by changes in the host, leading to inflammation and viral infections. HPV infection and clearance rates are correlated with the presence of Langerhans cells and a microbiome dominated by Lactobacillus geeseri22,23. Additionally, Lactobacillus enhances the immune response by stimulating phagocytes and promoting the production of cytokines, including IL-10, IL-12, IFN-γ, and TNF-α24. A 16-week cohort study revealed that HPV-positive women had a vaginal microbiota predominantly composed of Lactobacillus iners and anaerobic bacteria22. Notably, Lactobacillus indolent may hinder HPV clearance, as women with this species experience lower clearance rates compared to those with Lactobacillus crispatus13.
A study has shown that in women lacking Lactobacillus crispatus, the competitive proliferation of three bacterial groups—Lactobacillus iners, Gardnerella vaginalis, and Anaerococcus vaginalis—can lead to the development of squamous intraepithelial lesions (SIL)25. Additionally, some research indicates that Lactobacillus may have a plasmid-like function that allows it to integrate the HPV16 gene, thereby preventing the virus from integrating with host cells26. The loss of Lactobacillus dominance promotes the colonization of anaerobic bacterial species, increasing microbial diversity. This shift often results in changes to immunity and epithelial homeostasis through various mechanisms, which can, in turn, facilitate HPV infection4.
While the vaginal microbiota of most HPV-infected women primarily consists of Lactobacillus and Gardnerella, the alpha diversity of vaginal microbiota tends to decrease initially with increasing cervical lesion grade before significantly rising. The biological diversity of vaginal microecology in patients with high-grade squamous intraepithelial lesions (HSIL) is markedly different from that in patients with low-grade squamous intraepithelial lesions (LSIL). As the grade of cervical lesions increases, the abundance of Atopobium and Sneathia rises, while Streptococcus abundance decreases. These changes in microbial composition may affect the metabolic capacity of the cervical and vaginal microenvironment.
Sucrose positively correlated with Flavobacterium, a Gram-negative bacterium that can produce acid from sugar, potentially causing inflammation when the immune system is weak27. Phenylacetaldehyde was positively correlated with Pseudomonas, where phenylacetaldehyde dehydrogenase converts it to phenylacetic acid in the detoxification pathway28. Succinic acid showed positive correlations with Klebsiella, DNF00809, and Sneathia, but negative correlations with Gardnerella and Veillonella. Gardnerella, a key pathogen in bacterial vaginosis (BV), increases succinic acid concentrations, contributing to inflammation29. Glucose-6-phosphate positively correlated with Shuttleworthia and Dialist, while negatively correlating with Aerococcus and Peptoniphilus. The consumption of key amino acid metabolites indicates disrupted cell metabolism in HPV-positive and SIL patients, suggesting potential HPV-induced cervical-vaginal malnutrition30. This study observed an increase in the expression of sugar metabolism-related enzymes like erythritol and glucose-6-phosphate in SIL. This indicates that glycolysis is already enhanced at the SIL stage, potentially contributing to the development of cervical cancer.
Conclusions
In summary, this study clarifies the pivotal roles of metabolic dysregulation and microbiome alterations in the escalation of HR-HPV infection to cervical lesions. The analysis of cervicovaginal fluids from women with HR-HPV infections uncovered a distinctive metabolic profile, characterized by ten distinct metabolites. These metabolites are significantly associated with lesion severity and can effectively differentiate between LSIL and HSIL. The prevalence of Lactobacillus and Gardnerella in the cervicovaginal microbiota, along with a negative correlation between succinic acid levels and Gardnerella, indicates an intricate interaction between microbial composition and metabolic processes. These findings offer new insights into the metabolic and microbial landscape of HR-HPV infections, emphasizing the need for further research to investigate the clinical implications of these metabolic and microbial changes in cervical disease. There are numerous types of HR-HPV, and the sample size of this study is relatively small, making it impossible to conduct more detailed grouping comparisons based on specific types of HR-HPV infection, which may lead to bias in the research results, and we plan to conduct multi center, large sample case collection and comparison in the future.This study only observed data from one CVF sample and was unable to continuously observe the status of the microbiota and vaginal metabolites in patients with persistent HR-HPV infection. Further studies will consider increasing the number of follow-up visits.
Materials and methods
Subjects of the study
Between May and July 2020, a total of 156 women were recruited from the Gynecology Department of Jiangsu Hospital of Chinese Medicine, who were persistently infected with HR-HPV genotypes. Laboratory results confirmed HR-HPV infection through the detection of specific genotypes, HPV E6/E7 mRNA test, or HPV-DNA typing (HPV testing methods are detailed in the supplementary materials). The high risk HPV types included: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68. Low risk HPV types include: 6, 11, 42, 43, 44, 81, while suspected high-risk types included: 53, 73, and 82. Persistent HR-HPV infection was defined as the detection of two or more consecutive positive tests, with each test occurring at least six months apart. Women with atypical squamous cells of undetermined significance (ASCUS) or more significant cervical abnormalities, detected via thinPrep cytology test (TCT), were referred for colposcopy and cervical biopsy to confirm the presence of lesions and to enable further histopathological analysis for definitive diagnosis.
The inclusion criteria were as follows: non-menopausal women aged 30 years and older with regular menstrual cycles, without a history of vulvovaginal candidiasis, sexually transmitted infections (e.g., chlamydia), or hormonal contraception use, and with comprehensive medical records available. Exclusion criteria included a history of total hysterectomy, autoimmune disorders, immunosuppression, systemic diseases, recent treatment for cervical lesions, pregnancy, recent use of vaginal products, and sexual activity within 48 h prior to sampling. Further details are provided in the supplementary material.
The participants were divided into three cervical biopsy-based groups: women with Negative for Intraepithelial Lesions (NILM, n = 78), those with Low-Grade Squamous Intraepithelial Lesions (LSIL, n = 52), and those with High-Grade Squamous Intraepithelial Lesions (HSIL, n = 26). Ethical approval was obtained from the Ethics Committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (approval number 2021NL-025-03). All experiments were carried out in accordance with relevant guidelines and regulations, and informed consent was obtained from each participant.
Collection of cervicovaginal fluid (CVF)
A sterile cotton swab was employed to collect secretions from the cervix and the superior one-third of the vaginal side wall of patients positioned in the lithotomy position. The swabs were subsequently transferred to a sterile test tube for routine leucorrhea detection, with the process initiated within 15 min of collection. The cervix and the superior one-third of the vaginal wall were rinsed with 5 mL of 0.9% sodium chloride solution. Following this, 3–4 mL of CVF was aspirated, homogenized, and 2 mL of the supernatant CVF was isolated. All CVF samples were immediately preserved at -80 °C for future investigation. The preserved samples were then centrifuged at 4 °C and 18,000 rpm for 10 min, resulting in the separation of the supernatant from the precipitate.
Chemicals and reagents
For metabolomics analysis, 1,2–13 C myristic acid, which contains 1% BSTFA (trimethylchlorosilane), the C8-C40 alkane series, pyridine, and methoxyamine, were acquired from Sigma-Aldrich (USA). For 16 S rRNA sequencing, the following reagents and kits were used: a DNA extraction kit from Omega Bio-Tek (Norcross, GA, USA), biowest agarose from biowest (ESP), FastPfu Polymerase from TransGen (China), the AxyPrep DNA Gel Extraction Kit from Axygen Biosciences (Union City, CA, USA), the NEXTFLEX Rapid DNA-Seq Kit from Bioo Scientific (USA), and the MiSeq Reagent Kit v3 from Illumina (USA).
Experimental instruments and equipment
A Trace 1310 gas chromatography system, coupled with a TSQ 8000 mass spectrometer (GC-MS, Thermo, Waltham, USA), is equipped with an AS 1310 automatic injector. It features a Tg-5ms gas chromatography column (0.25 mm × 30 m, 0.25 μm) suitable for GC-MS applications. Moreover, a Savant SPD1010 vacuum centrifugal concentrator (Thermo Fisher Scientific, USA) was used for sample drying. In addition, the following equipment is required for flora detection: a NanoDrop™ 2000 UV-visible spectrophotometer (Thermo Scientific, Wilmington, USA), an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, USA), a Quantus™ Fluorometer (Promega, USA), and a PE300 platform (Illumina, San Diego, USA).
16 S rRNA sequencing
The centrifugation of CVF samples was conducted at 4 °C and at 18,000 rpm for a period of 10 min. The precipitate was subsequently collected. Microbial community genomic DNA was extracted from the collected precipitate using the E.Z.N.A.® soil DNA kit, following the manufacturer’s instructions. The purity and concentration of the extracted DNA were assessed using a NanoDrop 2000 UV-vis spectrophotometer and confirmed on a 1% agarose gel. The hypervariable region V3-V4 of the bacterial 16 S rRNA gene was amplified using the primer pairs 338 F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), and the PCR amplification was carried out in a thermocycler. The PCR protocol included an initial denaturation step at 95 °C for 3 min, followed by 27 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 45 s, a final extension phase at 72 °C for 10 min, and a termination step at 4 °C. The PCR mixture contained 4 µL of 5× TransStart FastPfu buffer, 2 µL of 2.5 mM dNTPs, 0.8 µL of each primer (5 µM), 0.4 µL of TransStart FastPfu DNA Polymerase, 10 ng of template DNA, and ddH2O to bring the final volume to 20 µL. Triplicate PCR reactions were performed.
The PCR products were purified from a 2% agarose gel using the AxyPrep DNA Gel Extraction Kit, and purified amplicons were pooled in equimolar amounts before being paired-end sequenced on an Illumina MiSeq PE300 platform, utilizing the standard protocols provided by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Operational taxonomic unit (OTU) clustering was performed on the sequencing results of the samples, classifying them into multiple groups based on their similarity, with each group representing one OTU. To obtain the relative species classification information of each OTU, the RDP classifier Bayesian algorithm was used for taxonomic analysis of representative sequences of OTUs at a 97% similarity threshold. The community species composition of each sample was statistically analyzed at each taxonomic level. The raw sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) under accession number SUB9475651.
Metabolomic analysis of cervicovaginal CVF samples
The CVF samples were centrifuged at 4 °C at 18,000 rpm for 10 min to obtain the supernatant, which was then utilized for metabolomic analysis. The supernatant (500 µL) was concentrated to a dry pellet using a vacuum centrifugal concentrator for 2 h. The dried supernatant from the CVF samples was reconstituted with 100 µL of sterile, distilled water. Subsequently, to each sample, 400 µL of an ice-cold methanol solution containing 1,2–13 C myristic acid (12.5 µg/mL) was added. The solutions were vigorously vortexed for 3 min and centrifuged for 10 min at 4 °C and 18,000 rpm. Thereafter, 200 µL of the supernatant was transferred to a centrifugal concentrator to evaporate for another 2 h. Next, 30 µL of methoxyamine pyridine (10 mg/mL) was added to the samples. The samples were vortexed for 5 min and incubated at 30 °C with gentle shaking at 450 rpm for 1.5 h. Following this, 30 µL of N, O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) was introduced into the samples. The samples were vortexed again for 5 min and then allowed to incubate at 37 °C with gentle shaking at 450 rpm for 0.5 h. Finally, the samples were centrifuged for 10 min at 4 °C and 18,000 rpm. The supernatant (50 µL) was collected for metabolite analysis. Concurrently, 3 µL of supernatant from each sample was taken and mixed in equal amounts to create quality control (QC) samples for methodological validation. After analyzing a sequence of 10 samples, a QC sample was inserted to assess the system’s applicability and stability.
Statistical analysis
The clinical characteristics analysis was performed using PASW Statistics 23 (SPSS, Chicago, USA). Data preprocessing was conducted using Xcalibur 2.2. For the processes of raw peak extraction, peak alignment, deconvolution analysis, and identification, MS-DIAL software (Tsugawa et al., 2015) was employed, in conjunction with the Fiehn BinBase DB. Normalization was achieved via Probability Quotient Normalization (PQN). Subsequently, multivariate analyses were conducted including Principal Component Analysis (PCA), orthogonal partial least squares discriminant analysis (OPLS-DA), and pathway enrichment analysis, all executed using MetaboAnalyst 6.0 (https://www.metaboanalyst.ca/faces/home.xhtml). These analytical methods enabled the differentiation of metabolomic profiles within the CVF in HPV-positive female patients versus those who were HPV-positive and concurrently diagnosed with cervical lesions. Further statistical analyses were then performed using GraphPad Prism (version 8.0.2) and R Studio (version 3.4.1) software.
Comparisons of data between two groups were made through the two-sample t-test. For the screening of microbial profiles, Variance Inflation Factor (VIF) analysis was employed. The statistical significance of identified metabolites was determined using non-parametric analysis of variance (ANOVA) tests. A subsequent post-hoc analysis was performed using the Wilcoxon Mann-Whitney test. False discovery rate (FDR < 0.05, corrected by the Benjamini-Hochberg test) and fold change (FC > 1.2 or < 0.833, based on the ratio of medians) were used to screen for significant metabolites.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82074478).
Abbreviations
- HR
HPV High-risk human papillomavirus
- CVF
Cervicovaginal fluid
- NILM
Negative for Intraepithelial Lesions
- LSIL
Low-grade squamous intraepithelial lesions
- HSIL
High-grade squamous intraepithelial lesions
- TICs
Total ion chromatograms
- RSD
Relative standard deviation
- QC
quality control
- GC-MS
Gas chromatography-mass spectrometry
- AUC
Area under the curve
- CSC
Cancer stem cell
- ROS
Reactive oxygen species
- PCoA
Principal coordinate analysis
- VIF
Variance inflation factor
- ATP
Adenosine triphosphate
Author contributions
Su Shen, Shixian Zhao contributed to metabolomic profiling analysis, data collection, and statistical analysis and interpretation. Su Shen, Jin-jun Shan and Qingling Ren contributed substantially to the study’s concept and design, as well as the collection of samples and clinical data. Su Shen, Shixian Zhao contributed to sample collection and statistical analysis of data. Su Shen and Qingling Ren contributed to interpretation and drafting of the manuscript. All authors have read and approved the final manuscript.
Data availability
Figures 5, 6 and 7 and Supplementary Fig. 2 are publicly available in the NCBI database under BioProject PRJNA722355 (www.ncbi.nlm.nih.gov/bioproject/PRJNA722355). Data supporting Table 1 is not publicly available to protect patient privacy. Figures 2, 3 and 4 were generated using the publicly available prognosis tool at www.metaboanalyst.ca/faces/home.xhtml. Figure 4 and Supplementary Fig. 1 are included in the supplementary materials. Raw data supporting Fig. 1 and all other raw data for metabolomics are available from the corresponding author upon reasonable request (email: yfy0047@njucm.edu.cn).
Declarations
Competing interests
The authors declare no competing interests.
Ethical statement
This study was conducted in accordance with the Declaration of Helsinki. The experimental procedures complied with relevant privacy protection laws and guidelines. Ethical approval was obtained from the Ethics Committee of the Affiliated Hospital of Nanjing University of Chinese Medicine (approval number 2021NL-025-03). All experiments were carried out in accordance with the relevant guidelines and regulations, and informed consent was obtained from each participant.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shang-Ying, H. et al. Outcomes in women with biopsy-confirmed cervical intraepithelial neoplasia grade 1 or normal cervix and related cofactors: a 15-year population-based cohort study from China. Gynecol. Oncol.156 (3), 616–623. 10.1016/j.ygyno.2019.12.027 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Brusselaers, N., Shrestha, S., van de Wijgert, J. & Verstraelen, H. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: systematic review and meta-analysis. Am. J. Obstet. Gynecol.221 (1), 9–18e8. 10.1016/j.ajog.2018.12.011 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Srinivasan, S. et al. Metabolic signatures of bacterial vaginosis. mBio. ; 6(2): e00204-15. Published 2015 Apr 14. (2015). 10.1128/mBio.00204-15 [DOI] [PMC free article] [PubMed]
- 4.Torcia, M. G. Interplay among vaginal Microbiome, Immune Response and sexually transmitted viral infections. Int. J. Mol. Sci.20 (2), 266. 10.3390/ijms20020266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y. et al. Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect. Dis.20 (1), 629. 10.1186/s12879-020-05324-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Champer, M. et al. The role of the vaginal microbiome in gynaecological cancer. BJOG125 (3), 309–315. 10.1111/1471-0528.14631 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Mitra, A. et al. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep.5, 16865. 10.1038/srep16865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindschuh, W. F. et al. Preterm birth is associated with xenobiotics and predicted by the vaginal metabolome. Nat. Microbiol.8 (2), 246–259. 10.1038/s41564-022-01293-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borgogna, J. C. et al. The vaginal metabolome and microbiota of cervical HPV-positive and HPV-negative women: a cross-sectional analysis. BJOG127 (2), 182–192. 10.1111/1471-0528.15981 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ilhan, Z. E. et al. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine44, 675–690 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sitarz, K., Czamara, K., Szostek, S. & Kaczor, A. The impact of HPV infection on human glycogen and lipid metabolism - a review. Biochim. Biophys. Acta Rev. Cancer. 1877 (1), 188646. 10.1016/j.bbcan.2021.188646 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Zhu Ruoxi, H. A. O. et al. Relationship between changes in vaginal microbiological metabolites and the risk of high-risk human papillomavirus infection and cervical intraepithelial neoplasia. Chin. J. Practical Gynecol. Obstet.35 (07), 797–802. 10.19538/j.fk2019070119 (2019). [Google Scholar]
- 13.Łaniewski, P. et al. Linking cervicovaginal immune signatures, HPV and microbiota composition in cervical carcinogenesis in non-hispanic and hispanic women. Sci. Rep.8 (1), 7593. 10.1038/s41598-018-25879-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, H. S., Kim, S. L., Kim, J. H., Ko, Y. C. & Lee, D. S. Plant volatile, phenylacetaldehyde targets breast Cancer stem cell by induction of ROS and Regulation of Stat3 Signal. Antioxid. (Basel). 9 (11), 1119. 10.3390/antiox9111119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park, S., Dingemans, J., Gowett, M. & Sauer, K. Glucose-6-Phosphate acts as an Extracellular Signal of SagS to modulate Pseudomonas aeruginosa c-di-GMP levels, attachment, and Biofilm formation. mSphere6 (1), e01231–e01220. 10.1128/mSphere.01231-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu, L. et al. The glycolytic switch in tumors: how many players are involved? J. Cancer. 8 (17), 3430–3440. 10.7150/jca.21125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, X. et al. Metabolomics study reveals the potential evidence of metabolic reprogramming towards the Warburg effect in precancerous lesions. J. Cancer. 12 (5), 1563–1574. 10.7150/jca.54252 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connors, J., Dawe, N. & Van Limbergen, J. The role of Succinate in the regulation of intestinal inflammation. Nutrients11 (1), 25. 10.3390/nu11010025 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iplik, E. S., Catmakas, T. & Cakmakoglu, B. A new target for the treatment of endometrium cancer by succinic acid. Cell Mol Biol (Noisy-le-grand). ; 64(1): 60–63. (2018). 10.14715/cmb/2018.64.1.11 [DOI] [PubMed]
- 20.Ertugrul, G., Iplik, B. & Cakmakoglu, E. S. The apoptotic efficacy of succinic acid on renal cancer cell lines. Med. Oncol.38 (12), 144. 10.1007/s12032-021-01577-9 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Petrova, M. I., Lievens, E., Malik, S., Imholz, N. & Lebeer, S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol.6, 81. 10.3389/fphys.2015.00081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brotman, R. M. et al. Interplay between the temporal dynamics of the vaginal microbiota and human papillomavirus detection. J. Infect. Dis.210 (11), 1723–1733. 10.1093/infdis/jiu330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon, B. et al. Association of HPV infection and clearance with cervicovaginal immunology and the vaginal microbiota. Mucosal Immunol.10 (5), 1310–1319. 10.1038/mi.2016.129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaji, R., Kiyoshima-Shibata, J., Tsujibe, S., Nanno, M. & Shida, K. Short communication: probiotic induction of interleukin-10 and interleukin-12 production by macrophages is modulated by co-stimulation with microbial components. J. Dairy. Sci.101 (4), 2838–2841. 10.3168/jds.2017-13868 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Oh, H. Y. et al. The association of uterine cervical microbiota with an increased risk for cervical intraepithelial neoplasia in Korea. Clin. Microbiol. Infect.21 (7), 674. .e1-674.e6749 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Sanchooli, A. et al. VLP production from recombinant L1/L2 HPV-16 protein expressed in Pichia pastoris. Protein Pept. Lett.25 (8), 783–790. 10.2174/0929866525666180809124633 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Enisoglu-Atalay, V. et al. Chemical and molecular characterization of metabolites from Flavobacterium Sp. PLoS One. 13 (10), e0205817. 10.1371/journal.pone.0205817 (2018). Published 2018 Oct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crabo, A. G. et al. Structure and biochemistry of phenylacetaldehyde dehydrogenase from the Pseudomonas putida S12 styrene catabolic pathway. Arch. Biochem. Biophys.616, 47–58. 10.1016/j.abb.2017.01.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham, L. S., Krass, L., Zariffard, M. R., Spear, G. T. & Mirmonsef, P. Effects of Succinic Acid and other Microbial Fermentation products on HIV expression in macrophages. Biores Open. Access.2 (5), 385–391. 10.1089/biores.2013.0013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilhan, Z. E. et al. Deciphering the complex interplay between microbiota, HPV, inflammation and cancer through cervicovaginal metabolic profiling. EBioMedicine44, 675–690. 10.1016/j.ebiom.2019.04.028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Figures 5, 6 and 7 and Supplementary Fig. 2 are publicly available in the NCBI database under BioProject PRJNA722355 (www.ncbi.nlm.nih.gov/bioproject/PRJNA722355). Data supporting Table 1 is not publicly available to protect patient privacy. Figures 2, 3 and 4 were generated using the publicly available prognosis tool at www.metaboanalyst.ca/faces/home.xhtml. Figure 4 and Supplementary Fig. 1 are included in the supplementary materials. Raw data supporting Fig. 1 and all other raw data for metabolomics are available from the corresponding author upon reasonable request (email: yfy0047@njucm.edu.cn).