Abstract
Objective:
The three steps of brain image processing – preprocessing, segmentation, and classification are becoming increasingly important in patient care. The aim of this article is to present a proposed method in the mentioned three-steps, with emphasis on the preprocessing step, which includes noise removal and contrast enhancement.
Methods:
The fast and adaptive bidimensional empirical mode decomposition and the anisotropic diffusion equation as well as the modified combination of top-hat and bottom-hat transforms are used for noise reduction and contrast enhancement. Fast C-means clustering with enhanced image is used to detect tumors and the tumor cluster corresponds to the maximum centroid. Finally, Ensemble learning is used for classification.
Result:
The Figshare brain tumor dataset contains magnetic resonance images used for data selection. The optimal parameters for both noise reduction and contrast enhancement are investigated using a tumor contaminated with Gaussian noise. The results are evaluated against state-of-the-art results and qualitative performance metrics to demonstrate the dominance of the proposed approach. The fast C-means algorithm is applied to detect tumors using twelve enhanced images. The detected tumors were compared to the ground truth and showed an accuracy and specificity of 99% each, and a sensitivity and precision of 90% each. Six statistical features are retrieved from 150 enhanced images using wavelet packet coefficients at level 4 of the Daubechies 4 wavelet function. These features are used to develop the classifier model using ensemble learning to create a model with training and testing accuracy of 96.7% and 76.7%, respectively. When this model is applied to classify twelve detected tumor images, the accuracy is 75%; there are three misclassified images, all of which belong to the pituitary disease group.
Conclusion:
Based on the research, it appears that the proposed approach could lead to the development of computer-aided diagnosis (CADx) software that physicians can use as a reference for the treatment of rain tumor.
Key Words: Bidimensional empirical mode decomposition (BEMD), Bidimensional intrinsic mode function (BIMF)
Introduction
The development of science and technology has conducted increasing use of medical imaging for non-invasive diagnosis and monitoring of diseases, especially in protected organs such as the brain. Brain tumors, characterized by uncontrolled cell growth in the brain or spinal cord, have a significant impact on patients’ lives. In 2020, approximately 308,102 people worldwide were diagnosed with primary brain or spinal cord tumors [1]. Early detection of brain tumors is crucial for successful treatment, and brain MRI is the preferred diagnostic tool. However, MRI diagnosis by a Radiologist is labor-intensive and time-consuming. Therefore, image processing techniques such as preprocessing, segmentation and classification now play an important role in the detection and classification of tumors, and their results can help Physicians in diagnosis. Image processing is performed on MRI images, which are often noisy and have poor contrast. These include: Preprocessing: This includes contrast enhancement and noise reduction; Segmentation: This involves dividing the image into different sections, typically using thresholding or clustering, and tumor detection is one aspect of segmentation; Classification: Detected tumors can be classified as benign, malignant or normal using machine learning or deep learning techniques. Some articles on noise reduction and contrast enhancement are summarized.
There are various approaches to noise reduction that are used in both spatial and transform domains. These approaches include threshold methods and filters such as Wiener, Gaussian or median filters [2]. In recent decades, Bidimensional Empirical Mode Decomposition (BEMD), which decomposes an image into bidimentional intrinsic mode functions (BIMFs) and residue (Res), has been widely used in many fields [3, 4]. Kommuri use BEMD to decompose the noisy image into BIMF and residue; diffusion filters were used to remove Gaussian noise from all BIMFs and residue [5]. Liu and Chen used l2-norm distances between the probability density functions (PDFs) of BIMFs and the original data to determine the number of noisy BIMFs focusing on low-level BIMFs; a soft interval thresholding method was applied to noisy BIMFs to reduce noise [6]. What is notable about this article is the determination of the number of noisy BIMFs. Lu Y. and Lu R. used BEMD to eliminate vignetting and noise [7]. BEMD was used to decompose the image into BIMFs and residue. First, BIMFs containing noise and residue representing the vignetting effect were removed to obtain the denoised and free vignetting image. A two-stage noise reduction method was proposed by Feng-Ping An [8].The data was first extrapolated using the Support Vector Machine (SVM) model. Next, BEMD was used twice, first decomposing the extrapolated data into BIMF1 and residue1 and then decomposing BIMF1 again into BIMF12 and residue12. The sum of residue1 and residue12 as input to repeat the previous process until a certain analysis number is reached.
Contrast enhancement increases brightness contrasts between objects and backgrounds to improve object detection. Histogram-based and Retinex-based are the two main classes. These methods affect human vision but are less useful for image segmentation Another approach is to use morphological operations such as top-hat transform (TH) for increasingly bright features and bottom-hat transform (TB) for increasingly dark features. According to Anitha and Chandrasekar, contrast enhancement by adding bright features and removing dark features was used, called “dual morphological contrast enhancement” (DMCE) [9]. Somasekara used modified DMCE to improve the contrast of the X-ray image by adjusting the formula of DMCE using the properties of histogram and luminance contrast [10]. Kushol used optimal structure element (SE) to improve DCME and performed it to improve the contrast of video-based images [11]. Widyantara used histogram equalization and DMCE to improve the contrast of video-based images [12]. The Contrast Improvement Ratio (CIR) plot was used by Hassanpour to determine that the optimal SE coincides with the maximum of the CIR [13]. Tekam used the opening by reconstruction to detect the background and combined it with Weber’s law to achieve image contrast improvement for poor quality images [14].
This article is a continuation of the previous articles and fills some gaps by focusing on three phases: image enhancement, tumor identification, and classification. Image enhancement includes image denoising using Bidimensional Empirical Mode Decomposition (BEMD) and Anisotropic Diffusion Equation (PDE), as well as contrast enhancement using morphological operations. C-means clustering is used for tumor detection, and statistical features obtained from wavelet packet coefficients are used to build a classification model through ensemble learning.
The remainder of the article consists of materials and methods including data and proposed method, experimental results, discussions and finally conclusions.
Materials and Methods
Data
Data source
Jun Cheng’s 2015 Figshare brain tumor dataset [15]. It includes 3064 T1-weighted MRI images of the brains of 233 patients, including three tumor types: meningioma (708), glioma (1426), pituitary (930) as well as information about patient ID, disease type and ground truth determined by Radiologists. These skull images were manually removed using the Matlab environment to avoid calculations that could affect tumor detection. In these MRIs, Tumor-3 corrupted by additive Gaussian noise is used to study image enhancement, 12 images are used for tumor detection, and 150 images with 50 images per disease type are used to extract features that create a classification model. In addition, the 004 image of the Brats-2015 dataset is used to compare the image enhancement results of the proposed approach with those of Rao and other filters [16, 17].
Equipment and Software
The proposed algorithm is executed on a laptop computer with an i.5 processor and 8GB RAM in the Matlab environment. Software: Fast and Adaptive BEMD code for BEMD (FABEMD), Anisotropic Diffusion (Perona & Malik) (APDF) and The Fast and Robust Fuzy C-mean clustering [18, 19, 20].
Proposed method
The proposed approach consists of three phases: studying image enhancement including noise reduction and contrast enhancement, segmentation and classification. These phases are presented below.
Image enhancement
Image enhancement includes noise reduction using BEMD and an anisotropic diffusion filter (PDE), as well as contrast enhancement using morphological transforms.
Bidimensional Empirical Mode Decompositon (BEMD) algorithm
Nunes et al. developed the Bidimensional Empirical Mode Decompositon (BEMD) algorithm, which uses the shifting process to decompose stationary or non-stationary data into oscillating components, called bidimensional intrinsic mode functions (BIMFs) and a monotonic component called residue (Res) [3]. BEMD decomposes an image and can be represented by the following equation:
(1)
where, I(m,n): the image, BIMF(m,n): the bidimensional intrinsic mode function, N: number of BIMF, and Res(m,n): the residue.
The BIMFs contain frequencies from high to low, corresponding to the decomposition level from low to high, with BIMF1 having the highest frequency and BIMFN having the lowest frequency, and Res (residue) representing the background.
BEMD decomposes noisy image
The white noise image (I_noise) can be the sum of the original image (OI) and noise, given by the equation:
(2)
Using Equation (1), the noisy image is decomposed by BEMD and can be expressed:
(3)
The image is corrupted by additive Gaussian noise, which has a small amplitude and a very high frequency. Therefore, the noise is contained in BIMF1, the highest frequency component, and sneaks into subsequent BIMFs. As a result, BIMFs can be divided into two groups: noisy BIMFs with lower levels and free noise BIMFs with higher levels:
(4)
Therefore, equation (3) is rewritten:
(5)
The last two terms on the right-hand side of Equation (5) are merged into a single term Res(x,y) since they do not include any noise:
(6)
Determine the number of noisy BIMFs
In order for BEMD to reduce noise, the filter is applied to noisy BIMFs, so the number of noisy BIMFs (n1 in equation (6)) is required first. Lu Y. and Lu R. used the similarity measurement distance graph between the probability density function (PDF) of the original image and the PDF of each BIMF versus to the ordinal number of BIMFs to determine the number of noisy BIMFs [7]. The number of noisy BIMFs is the abscissa of the first minimum corresponding to the ordinal number BIMF. The Hausdorff distance is used [21]:
(7)
where, a and b are points in pdfA and pdfB respectively and d(a,b) is the distance between them, for simplicity the Euclidean distance.
Noise reduction by combining BEMD and PDF
When BEMD is used for noise reduction, noisy data must be decomposed into both BIMF and Residue (Res) so that each BIMFi has a corresponding Resi. To create a denoised image, a filter is applied to each noisy BIMFi and the sum of the denoised BIMFi is then added to the residue of the last noisy BIMF in the sum. PDE is used to filter noise because it eliminates noise while preserving edges. The PDE is given:
(8)
where, is the gradient; div is the divergence and c(.) is the diffusion given by Perona and Malik [22]:
(9)
where, k is a constant selected by the noise level and edge strength; it determines how much diffusion occurs.
The gradients in the 8-direction and the iterative numerical technique are used to solve Equation (8). The image denoising is given:
(10)
Contrast enhancement using morphological operations
In mathematical morphology, operations process the shapes and structures of objects in the image based on the structuring element (SE), a small matrix with the correct shape of the processed image and pixel values of 0 or 1. Therefore, a morphological operation is considered as a filter in an image. The binary/gray image is denoised using the opening operator (denoted as o ), and any small holes in the objects are removed using the closing operator (denoted as ■ ). The top-hat transform, which defines the difference between the image and the opening operator, is used to highlight bright objects on a dark background (the size of which is smaller than the SE size):
(11)
On the other hand, bottom-hat transform, which defines the difference between the image and the closing operator, is used to highlight dark objects on a bright background:
(12)
These transforms are combined with the original image to obtain a contrast enhancement image according to the equation (dual morphological contrast enhancement) [12]:
(13)
The proposed approach involves a modified formula (13) by replacing the original image (OI) with a high-pass image generated by removing the background from the OI. The background image is created by convolving the original image with a weighted average kernel, with brightness in the center and fading toward the edges [23]. The result of the modified equation is superior to that of Equation (13).
Image enhancement
The denoised image and contrast enhancement are performed simultaneously. To achieve image enhancement, these two images are fused together using wavelet-based image fusion [24]. This method was chosen because it performs a selection of dominant features. The quality of image enhancement is evaluated using performance metrics such as Mean Square Error (MSE), Peak Signal-to-Noise Ratio (PSNR) and Effective Measure of Enhancement (EME) [25]:
(14)
(15)
(16)
where, I (i,j): reference image, IE(i,j): image enhancement, IE(i,j) is divided into m1 × n1 blocks and IEmax and IEmin are maximum and minimum values in each block.
Tumor detection
The tumor is detected by C-means clustering. This is a soft clustering technique in machine learning to divide data into multiple clusters (groups) based on the distance between each cluster center and the data point. Using C-means, a data point can belong to one or more clusters. By applying C-means to MRI brain enhancement, the tumor is detected based on one of the extracted clusters, which is the cluster that matches the segmented image.
The C-means algorithm is based on minimizing the objective function:
(17)
where, m is the Fuzziness parameter, any real number greater than 1, N is the number of data, C is the number of clusters, xi is the ith data, cj is the cluster center j and ||*|| is the similarity distance between any data points and the center point.
The objective function is minimized by iteratively updating the membership degrees (ui,j) and cluster centroids (cj) until the ui,j of two consecutive iterations are almost the same [26]. C-means image segmentation has high efficiency when the right number of clusters is selected to obtain a cluster with the tumor in it. Because the background and objects are displayed in the histogram, each histogram peak can be viewed as a representation of a segment in the image. Therefore, the number of clusters is selected equal to the number of peaks in the smooth histogram. After clustering, the cluster corresponding to the largest centroi is selected for tumor detection; tumor is one of the objects in this cluster.
Both tumor detection and classification are evaluated using performance metrics such as [27] :
(18)
(19)
(20)
(21)
where, TP: true positive, TN: true negative, FP: false positive and FN: false negative.
Tumor classification
Classification is used to identify the type of disease in the image or to determine whether the disease is benign or malignant. The proposed approach uses ensemble learning for classification and therefore requires feature extraction first [28]. In our approach, features used for classification are extracted from the enhanced image.
Feature extraction
An essential first step for effective classification is feature extraction, which reveals the hidden image properties of the image. The two main types are low-level (global) features and high-level (local) features [29]. We use low-level features, which include some statistical features (mentioned in the Experimental Results section) derived from the Wavelet Packet Decomposition (WPD) coefficients [30].
Classification
In machine learning, small data size and not enough data samples can be one of the causes of overfitting the machine learning model. From a modeling perspective, using ensemble learners instead of individual learners is one way to solve this problem [31]. Ensemble learning combines the predictions of multiple classifiers, called base classifiers or weak classifiers, such as: neural networks, SVM, Naïve Bayes and decision trees. The two most common types are bagging and boosting [32].
Results
Experimental results
Study of image enhancement
Figure 1(a) shows Tumor-3 from the Fishare database as in the original image. It is corrupted by additive Gaussian noise with a mean of 0 and a variance of 25, as shown in Figure 1(b). This noisy image is used to study image enhancement.
Figure 1.
(a) Skull-stripped Tumor-3, (b) Tumor-3 corrupted by additive Gaussian noise (mean = 0, variance = 25), PSNR = 22.1939
Determine the number of noisy BIMFs
The noisy image is divided into five BIMFs using FABEMD code [17]. Figure 2 shows the graph of the similarity measure of the Hausdorff distance (Equation (7)) between each PDF of BIMFs and the original image plotted against the ordinal BIMFi. The minimum of this diagram appears at BIMF4, i.e. the four noisy BIMFs used for noise reduction.
Figure 2.

PDF Similarity Distance versus Number of BIMFs
Noise reduction using BEMD and anisotropic diffusion filter
Figure 3 shows the noisy BIMFi and the corresponding Residuei (i = 1, 2, 3, 4), these noisy BIMFi are filterd by the PDE filter to reduce the noise [18]. Figure 4 shows the denoised images obtained from the sequential sum of the denoised BIMFi and the residue matching the last BIMFi in the sum. The sum of four denoised BIMFi and the Residue4 gives the good denoised image as shown in Figure 4(d), In addition to visual measurement, measurement indices such as MSE, PSNR and EME are also used and listed in Table 1.
Figure 3.
BEMD Tumor-3. a): BIMF1, b): BIMF2, c): BIMF3, d): BIMF4, e): Res1, f): Res2, g): Res3, h): Res4
Figure 4.
The Image Denoising Using Anisotropic Diffusion Filter (PDE); a): PDE(BIMF1) + Res1, b): PDE(BIMF1) + PDE(BIMF2) +Res2, c): PDE(BIMF1) + PDE(BIMF2) + PDE(BIMF3) + Res3, d): PDE(BIMF1) + PDE(BIMF2) + PDE(BIMF3) + PDE(BIMF4) + Res4
Table 1.
Quality Metrics for Denoised Image Using BEMD and Anisotropic Diffusion Equation
| Method (PDE: Anisotropic diffusion equation) | PSNR | MSE | EME | EMEE |
|---|---|---|---|---|
| PDE(BIMF1) + Res1 | 25.0493 | 203.3048 | 9.5026 | 0.5688 |
| PDE(BIMF1) + PDE(BIMF2) + Res2 | 26.9374 | 131.6257 | 19.0419 | 1.1283 |
| PDE(BIMF1) + PDE(BIMF2) + PDE(BIMF3) + Res3 | 27.9646 | 103.9005 | 18.4217 | 1.1561 |
| PDE(BIMF1) + PDE(BIMF2) + PDE(BIMF3) + PDE(BIMF4) + Res4 | 28.2436 | 97.4371 | 11.002 | 0.7737 |
Contrast enhancement
This section examines how the optimal SE is determined. SE shapes such as disk, square, diamond and sphere are used with sizes of 5, 10, 15 and 20 pixels for each shape. Each SE is applied in the modified Equation (13) to calculate the contrast enhancement of noisy Tumor-3. Table 2 shows PSNRs of contrast-enhanced images with different SE. A high PSNR indicates that the square SE provides an image with better contrast enhancement than other SEs, Figure 5 shows the contrast enhancement images with different square SE size. Among these, the 5-pixel square SE contrast enhancement image is the best and is shown in Figure 5(a), which shows good contrast between the tumor and the other parts of the brain.
Table 2.
PSNR of Contrast Enhancement Using Different Structuring Elements.
| No | Shape / Size | 5 | 10 | 15 | 20 |
|---|---|---|---|---|---|
| 1 | disk | 27.7242 | 26.6416 | 26.0005 | 25.5468 |
| 2 | square | 28.2586 | 27.4221 | 26.8821 | 26.4609 |
| 3 | diamond | 27.7127 | 26.839 | 26.2976 | 25.7776 |
| 4 | sphere | 27.5532 | 26.6099 | 26.0247 | 25.5606 |
Figure 5.
Contrast Enhancement Using Square Structuring Element with Different Size: a): d = 5, b): d = 10, c): d = 15, d): d = 30.
Figure 6.
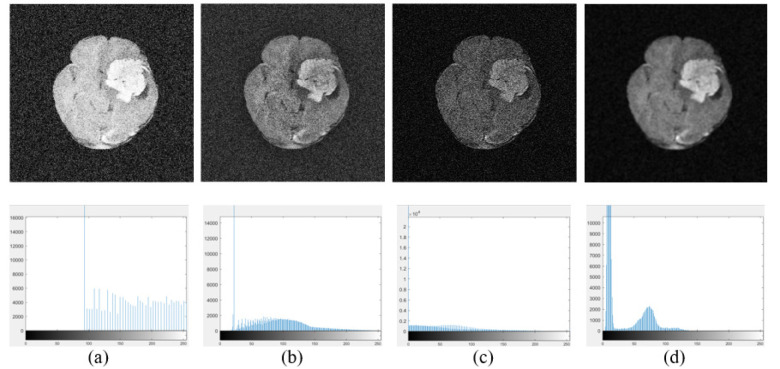
Contrast Enhancement and Histogram, a): HE, b): CLAHE, c): Conventional Morphological contrast enhancement, and d): Modified Morphological contrast enhancement.
Furthermore, contrast enhancement of noisy Tumor-3 is achieved by Histogram Equalization (HE), shown in Supplementary Figure 6(a); Control Limited Adaptive Histogram Equalization (CLAHE), shown in Supplementary Figure 6(b); conventional Equation (9) with a 5-pixel square SE, shown in Supplementary Figure 6(c) and a proposed approach shown in Supplementary Figure 6(d). There is a histogram for each image below and the PSNR indices are listed in Table 3. These images and values in Table 3 are used to compare the results of these methods.
Table 3.
PSNR of the Proposed Contrast Enhancement with Different Methods.
| No | Shape / Size | PSNR |
|---|---|---|
| 1 | Equalization | 5.933 |
| 2 | Adapt-Equalization | 11.965 |
| 3 | Conventional Morphological | 15.5316 |
| 4 | Proposed Morphological | 28.2586 |
Image enhancement
Due to the independent processing of the contrast-enhanced and denoised images. They are fused using DWT-based image fusion to achieve image enhancement as this fusionallows the selection of fusion approaches (“max”, “min”, and “mean”). The “max”-“max,” “mean”-“mean,” and “min”-“min” approaches yield PSNRs of 27.6, 28.6, and 28.5, respectively (noisy image with a PSNR of 22.2). Consequently, the optimal image enhancement is provided by the “mean”-“mean” approach. The images during the fusion process are shown in Supplementary Figure 7(a–c), while Supplementary Figure 7(d) shows the original image. The PDFs of the original, noise, and enhancement images are shown in Supplementary Figure 7(e).
Comparison of the proposed method results with others
The 004 image shown in Supplementary Figure 8(a) is used to compare the quality of image enhancement produced by our method with others. It is corrupted by additive Gaussian noise with a mean of 0 and a variance of 0.05, as shown in Supplementary Figure 8(b). The noisy image is the input for calculating image enhancement using bilateral filter, Wiener filter, Rao-Srinivas deep convolutional network and our approach. PSNR and structural similarity index (SSIM) are listed in Table 4 [17]. This shows that the image enhancement of the proposed approach dominates to compare other results with high PSNR and SSIM close to 1. Supplementary Figure 8(c) shows the result of Rao and Srinivas and Supplementary Figure 8(d) shows the result of the proposed approach. The visual appearance shows that the resulting image from Rao and Srinivas (2019) is similar to the noisy image, while the resulting image from the proposed approach is similar to the original image.
Table 4.
The Quality Metrics for Image Enhancement of the Proposed Approach and Others.
| Metric | Noisy image | Bilateral | Wiener 2 | Rao and | Proposed |
|---|---|---|---|---|---|
| (variance = 0.05) | filter | filter | Srinivas | method | |
| PSNR | 14.74 | 15.4137 | 20.0932 | 20.6564 | 20.9219 |
| SSIM | 0.03 | 15.4137 | 10.0715 | 0.1412 | 0.1655 |
Tumor detection
Fast and Robust Fuzzy C-Means Clustering is used for tumor detection from improved images due to its speed, noise immunity and detail preservation [19]. Automatically determine the initial number of clusters using smooth histogram peak selection. Supplementary Figure 9 shows some images in the tumor detection process; therein, Supplementary Figure 9(a) shows the enhanced Tumor-3, Supplementary Figure 9(b-d) shows all the clusters created by the C-means, and Supplementary Figure 9(f) shows the centroid values corresponding to the clusters. Supplementary Figure 9(c) shows cluster 2, which corresponds to the maximum centroid; therefore, the tumor contained in this cluster and the raw tumors from it are detected, as shown in Supplementary Figure 9(e). Morphological procedures are performed on the raw tumor to fill holes and smooth edges to create the fine tumor. Supplementary Figure 10 shows tumor detection results from twelve brain MRIs, including original tumors, enhanced images, raw tumors, fine tumors, ground truth, and detected tumor with ground truth boundaries.
Based on the areas of the detected tumors and their ground truth, TP, TN, FP and FN are calculated and the performance indices are calculated from these. The results show an accuracy of 99% and a precision, sensitivity and specificity of 90% each. Table 5 lists them and shows the effectiveness of the proposed method. It also shows that meningioma tumors are more difficult to detect than other types.
Table 5.
The Performance Metrics for Tumor Detection.
| No | Disease | Tumor | Acc. | Prec. | Sen. | Spec. |
|---|---|---|---|---|---|---|
| 1 | Meningioma | 3 | 0.9947 | 0.9821 | 0.885 | 0.9993 |
| 2 | Meningioma | 10 | 0.9978 | 0.9237 | 0.9237 | 0.9989 |
| 3 | Meningioma | 17 | 0.9948 | 0.7171 | 0.7625 | 0.997 |
| 4 | Meningioma | 225 | 0.9955 | 0.8636 | 0.9765 | 0.996 |
| 5 | Glioma | 2315 | 0.9981 | 0.9848 | 0.9247 | 0.9997 |
| 6 | Glioma | 2407 | 0.9973 | 0.9498 | 0.8819 | 0.9992 |
| 7 | Glioma | 2447 | 0.9943 | 0.8961 | 0.912 | 0.9968 |
| 8 | Glioma | 2730 | 0.9948 | 0.8658 | 0.9422 | 0.9962 |
| 9 | Pituitary | 1020 | 0.9982 | 0.9886 | 0.865 | 0.9999 |
| 10 | Pituitary | 1023 | 0.9976 | 0.8624 | 0.9849 | 0.9978 |
| 11 | Pituitary | 1144 | 0.998 | 0.8677 | 0.8243 | 0.9992 |
| 12 | Pituitary | 1517 | 0.9982 | 0.8815 | 0.9291 | 0.9988 |
| Average | 0.9966 | 0.8986 | 0.9009 | 0.9985 |
Tumor classification
Feature extraction
Using six statistical features, including mean absolute value, standard deviation, skewness, kurtosis, RMS power and the ratio of the mean absolute values of two consecutive subbands. These features are extracted from 16 coefficients of WPD at level 4 using the Daubechies 4 wavelet function (‘db4’). The dataset includes 150 images for three diseases (50 images per disease): meningioma, glioma and pituitary. Each image provides 95 features, resulting in a classification input matrix of size 150x95. The data is divided into training set (80%) and test set (20%).
Classifier
Ensemble learning is used in Malalb’s fitcensemble function with base decision trees. The predictor combination with Bayesian Optimization and Hyperparameter Optimization is set for automatic selection of optimal parameters and appropriate bagging or boosting for each cycle of 30 cycles [33].
Results
The development of the classifier model is shown in Supplementary Figure 11, where Supplementary Figure 11(a) shows the training confusion matrix with an accuracy rate of 96.7%. The test confusion matrix with an accuracy of 76.7% is shown in Supplementary Figure 11(b). The ensemble learning model has high accuracy on training data, but not yet high accuracy on test data, as shown in Supplementary Figure 11(a,b). When this model is used to classify 12 tumor detection images that are independent of training data, the result shown in Supplementary Figure 11(c) shows the stability of the model with an accuracy of 75%, which is similar to the accuracy of test data.
Discussion
Investigate how BEMD and PDE reduce noise at different noisy BIMFs. The result shows that noise reduction works best when applied to all noisy BIMF, which is consistent with the findings of Liu and Chen and is further supported by low MSE, high PSNR and visual observation [6]. However, the denoised image does not have strong contrast because the EME value does not compare well with the denoising for a portion of the noisy BIMFs. The morphological process of contrast enhancement shows that flat SE with small square shape is suitable for contrast enhancement with high performance index and good vision. Compared with other contrast enhancement techniques, the proposed approach provides a bimodal histogram, showing that image contrast enhancement is suitable for tumor detection, traditional morphological contrast enhancement provides a dark image, HE produces the brightest image, and CLAHE produces an image with a nearly unimodal histogram, which is suitable for human observation. The PSNRs also demonstrate the superior performance and great value of the proposed method. As already mentioned, a denoised image has low contrast, so it is necessary to combine a contrast enhancing image. Image enhancement is achieved through wavelet-based fusion using the ‘mean’-‘mean’ method. The enhanced image reconstructs a nearly flawless original image, as evidenced by the perfect overlap of the PDFs of the two images. In addition, since the tumor in the enhanced image is brighter than in the original images, the maximum PDF of the enhanced image is larger than the maximum PDF of the original image, indicating that the enhanced image is suitable for tumor detection.
Using the C-means algorithm, tumors are detected using 12 images with improved quality. These images show three different disease groups, each with its own characteristics. However, the C-means algorithm is used to accurately detect the tumors of these groups. The good quality of the C-means used for segmentation according to the detected tumor corresponds to the ground truth as well as high accuracy, precision, sensitivity and specificity assessment. To achieve these results, the appropriate clusters should be selected based on the number of peaks of the smooth histogram. It is necessary to ensure that a tumor is present in one of the clusters formed by C-means. Furthermore, the largest centroid cluster is identified as the cluster containing the tumor without requiring any optimization method to find the optimal centroid [34]. These two points can be considered as notable points in tumor segmentation using C-means.
A total of 150 brain MRI images that improved quality in three type of diseases (50 images per disease) were used to extract features with six statistical features. Using 16 WPD coefficients of the Daubechies 4 wavelet at level 4. WPD divides the data into two groups: approximate and detail components, called coefficients, using low-pass and high-pass filters at level 1. To produce finer coefficients at subsequent levels, two filters are applied to the approximate and detail coefficients, and each coefficient has the same frequency width. As the level increases, the frequency bands become narrower, and all frequencies in a level cover the frequency of the data. Consequently, features retrieved using WPD coefficients provide more information than features obtained directly from the image because features are extracted in a sequence of segment images, each of which has a narrow frequency bandwidth. This makes them suitable for creating a classification model. However, when this data set is used to build a classification model for ensemble learning using Matlab function, it allows automatic selection of the optimal bagging or boosting method for each cycle. The training set (80% data set) gives good results, whereas the test set (20% data set) gives quite good results. One of the possible reasons is that a 512x512 image enhancement was used for feature extraction instead of the ROI image,which shows the small surrounding area of the tumor with more tumor features than noise. Therefore, noise could affect the features and the model may not work properly when applied to test data that is different from training data. Applying this model to 12 images (four images per disease) using the Tumor detection section produced the following results. There were four properly classified images for each of the meningioma and glioma groups. In particular, only one case out of four images in the pituitary group was correctly classified; two cases were misclassified as gliomas and one case was misclassified as meningioma. This suggests that pituitary diseases are more difficult to classify than the other diseases.
In conclusion, brain tumors can be effectively treated by physicians if the tumor is correctly detected and classified early due to the development of science and technology. The aim of this article is to develop a program to improve image quality, tumor detection and classification. The results can serve as a reference for physicians. The survey results show that high-quality image enhancement is achieved through noise reduction using edge-preserving filtering such as PDE on BEMD components and high contrast enhancement is achieved through modified dual morphological contrast enhancement using high-pass image instead of the original image. This image enhancement is suitable for tumor detection and can be directly used to extract features for classification. C-means clustering with the appropriate number of cluster numbers ensured high accuracy in tumor detection. For classification, image enhancement is divided into segment images with a narrow frequency range, such as WPD coefficients, which are extracted features and appropriate ensemble learning is selected to generate an effective model for classification.
This article uses C-means for tumor detection and WPD coefficient based feature extraction for the classification model. The future article will use the EMD and BEMD algorithms to achieve the two goals mentioned with a classification model that uses a large number of instances.
Acknowledgements
General
The authors would like to thank Sir Rao GS for sending the 04 brain image, Sir Mruthun Thirumalaisamy and Sir Tao Lei for publishing the programs to calculate a fast and adaptive BEMD and a fast and robust C-means. These documents were used in the implementation of this article.
Data Availability
The data used in this article was downloaded from Jun Cheng’s Figshare Publicity Brain Dataset 2015. The 004 brain image from the Brats-2015 dataset provided by Rao GS.
Consent for publication
Each author agreed to publication.
Conflict of Interest
The authors declare that there is no conflict of interest.
Author Contribution Statement
All authors contributed equally in this study.
Supplementary materials
References
- 1.Brain tumor network. Brain tumor network and unite us partner to increase access to community resources for patients and families [internet] Available from: https://www.Braintumornetwork.Org/brain-tumor-network-and-unite-us-partner-to-increase-access-to-community-resources-for-patients-and-families.
- 2.Vaishali S, Rao KK, Rao GVS. SPACES-2015. Dept of ECE, K L UNIVERSITY.; 2015. A review on noise reduction methods for brain mri images. [Google Scholar]
- 4.Huang J, Zhao B, Chen Y, Zhao P. Bidimensional empirical mode decomposition (bemd) for extraction of gravity anomalies associated with gold mineralization in the tongshi gold field, western shandong uplifted block, eastern china. Computers & Geosciences. 2010;36:987–95. [Google Scholar]
- 5.Kommuri SVR, Singh H, Kumar A, Bajaj V. Bidimensional empirical mode decomposition-based diffusion filtering for image denoising. Circuits, Systems, and Signal Processing. 2020:39. [Google Scholar]
- 6.Liu D, Chen X. Image denoising based on improved bidimensional empirical mode decomposition thresholding technology. Multimed Tools Appl. 2019:78 . [Google Scholar]
- 7.Lu Y, Lu R. Fast bi-dimensional empirical mode decomposition as an image enhancement technique for fruit defect detection. Comput Electron Agric. 2018;152:314–23. [Google Scholar]
- 8.An F-P, Lin D-C, Zhou X-W, Sun Z. Enhancing image denoising performance of bidimensional empirical mode decomposition by improving the edge effect. International Journal of Antennas and Propagation. 2015;2015:1–12. [Google Scholar]
- 9.Anitha J, Dinesh Peter J, Immanuel Alex Pandian S. A dual stage adaptive thresholding (dusat) for automatic mass detection in mammograms. Comput Methods Programs Biomed. 2017;138:93–104. doi: 10.1016/j.cmpb.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Somasekara J, Ramesh G, Ramuc G, Kumar Reddy Pd-K, Madhavi K, Praveen J. Beneficial image preprocessing by contrast enhancement technique for sem images. Indian journal of engineering and materials sciences (ijems) 2023;29(6):832–6. [Google Scholar]
- 11.Kushol R, Rahman ABMA, Salekin S, Raihan MN. Contrast enhancement of medical x-ray image using morphological operators with optimal structuring element. 2018. [Google Scholar]
- 12.Widyantara IM. Image enhancement using morphological contrast enhancement for video based image analysis. 2016. [Google Scholar]
- 13.Hassanpour H, Samadiani N, Salehi SM. Using morphological transforms to enhance the contrast of medical images. Egypt J Radiol Nucl Med. 2015;46:481–9. [Google Scholar]
- 14.Tekam M. Morphological transformations for enhancement of images with poor contrast and detection of background. IJIREEICE. 2015;3:16–20. [Google Scholar]
- 15.Chen J. Figshare: Brain tumor dataset [internet] [2015 aug 17 [update 2017 april 3; cited 2015 oct 8]]. Available from: https://figshare.Com/articles/dataset/brain_tumor_dataset/1512427/5.
- 16.The sicas medical image repository [internet] [C2012 [update 2017 june 08; cited 2015 oct]]. Available from: https://www.Smir.Ch/brats/start2015#!#download.
- 17.Rao G, B S. De-noising of mri brain tumor image using deep convolutional neural network. SSRN Electronic Journal. 2019 [Google Scholar]
- 18.Thirumalaisamy m. Matlab central file exchange. Fast and adaptive multivariate and multidimensional emd [internet] 2019. Available from: https://www.Mathworks.Com/matlabcentral/fileexchange/71270-fast-and-adaptive-multivariate-and-multidimensional-emd.
- 19.Lopes d. Anisotropic diffusion (perona & malik) Matlab central mathworks. 2007 .
- 20.Lei T, Jia X, Zhang Y, He L, Meng H, Nandi A. Significantly fast and robust fuzzy c-means clustering algorithm based on morphological reconstruction and membership filtering. IEEE Transactions on Fuzzy Systems. 2018:1. [Google Scholar]
- 21.Ma W, Yin S, Jiang C, Zhang Y. Variational mode decomposition denoising combined with the hausdorff distance. Rev Sci Instrum. 2017;88(3):035109. doi: 10.1063/1.4978029. [DOI] [PubMed] [Google Scholar]
- 22.Perona p, malik j. Scale-space and edge detection using anisotropic diffusion. Ieee transactions on pattern analysis and machine intelligence. 1990;12(7):629–39. [Google Scholar]
- 23.Mathworks. Change filter strength radially outward [internet] 2024. Available from: https://www.Mathworks.Com/help/images/increase-filter-strength-radially-outward.Html.
- 24.Balakrishna T, Hannan M, Nimishka G, Subba Reddy GV. Dwt-based image fusion technique in matlab: Identifying the most effective method. E3S Web of Conferences. 2023:391. [Google Scholar]
- 25.Akila K, Subramanian J, Vasuki A. Mammographic image enhancement using indirect contrast enhancement techniques – a comparative study. Procedia Computer Science. 2015;47:255–61. [Google Scholar]
- 26.Hamad Y, Simonov K, Naeem M. Detection of brain tumor in mri images, using a combination of fuzzy c-means and thresholding. International Journal of Advanced Pervasive and Ubiquitous Computing. 2019;11:45–60. [Google Scholar]
- 27.Nadeem MW, Goh HG, Ponnusamy V, Andonovic I, Khan MA, Hussain M. A fusion-based machine learning approach for the prediction of the onset of diabetes. Healthcare (Basel) 2021;9:10. doi: 10.3390/healthcare9101393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subbarao M, Grandhi C, Darisi G, Terlapu S. Brain tumor classification using ensemble classifiers. 2022. [Google Scholar]
- 29.Kang J, Ullah Z, Gwak J. Mri-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors (Basel) 2021;21:6. doi: 10.3390/s21062222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang s, wang j, chen h, li s. Feature extraction and classification of tumor based on wavelet package and support vector machines. Inadvances in knowledge discovery and data mining. 11th pacific-asia conference, pakdd 2007; Proceedings 11 2007 (pp. 871-878). Springer berlin heidelberg. nanjing, china: [Google Scholar]
- 31.Beliasau s. Model overfitting or possible ways to defeat it. [internet] 2023 . Available from: https://bestasoff.Medium.Com/model-overfitting-or-possible-ways-to-defeat-it-9310c4fe9130.
- 32.Salakhutdinov r. Research topics in statistical machine learning: Statistical machine learning (sta 4273h) [internet] 2011 . Available from: https://www.Utstat.Toronto.Edu/~rsalakhu/sta4273/notes/lecture12.Pdf.
- 33.Mathworks. Hyperparameter optimization in classification learner app [internet] 2024. Available from: https://www.Mathworks.Com/help/stats/hyperparameter-optimization-in-classification-learner-app.html.
- 34.Kumar D, Satyanarayana D, Prasad M. Mri brain tumor detection using optimal possibilistic fuzzy c-means clustering algorithm and adaptive k-nearest neighbor classifier. Journal of Ambient Intelligence and Humanized Computing. 2021;12:1–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this article was downloaded from Jun Cheng’s Figshare Publicity Brain Dataset 2015. The 004 brain image from the Brats-2015 dataset provided by Rao GS.