Abstract
Background
Targeting glucose uptake by glucose transporter (GLUT) inhibitors is a therapeutic opportunity, but efforts on GLUT inhibitors have not been successful in the clinic and the underlying mechanism remains unclear. We aim to identify the key metabolic changes responsible for cancer cell survival from glucose limitation and elucidate its mechanism.
Methods
The level of phosphorylated YAP was analyzed with Western blotting and Phos-tag immunoblotting. Glucose limitation-induced metabolic changes were analyzed using targeted metabolomics (600MRM). The anti-cancer role of metabolite was examined using colony formation assay and APCmin/+ mice. Co-immunoprecipitation, LS-MS, qRT-PCR, and immunofluorescence were performed to explore the underlying mechanisms.
Results
We found that D-Ribose-5-phosphate (D5P), a product of the pentose phosphate pathway connecting glucose metabolism and nucleotide metabolism, functions as a metabolic checkpoint to activate YAP under glucose limitation to promote cancer cell survival. Mechanistically, in glucose-deprived cancer cells, D5P is decreased, which facilitates the interaction between MYH9 and LATS1, resulting in MYH9-mediated LATS1 aggregation, degradation, and further YAP activation. Interestingly, activated YAP further promotes purine nucleoside phosphorylase (PNP)-mediated breakdown of purine nucleoside to restore D5P in a feedback manner. Importantly, D5P synergistically enhances the tumor-suppressive effect of GLUT inhibitors and inhibits cancer progression in mice.
Conclusions
Our study identifies D5P as a metabolic checkpoint linking glucose limitation stress and YAP activation, indicating that D5P may be a potential anti-cancer metabolite by enhancing glucose limitation sensitivity.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13045-024-01655-1.
Keywords: Metabolic stress, Glucose deprivation, D-ribose-5-phosphate, YAP, LATS1, Purine nucleoside phosphorylase
Background
Metabolic reprogramming during malignant transformation allows cancer cells to undergo extraordinarily vigorous anabolism to synthesize macromolecules that favor survival and replication [1–3]. Such changes are accompanied by high demands of the requisite nutrients, of which glucose is particularly important. Thus, targeting glucose metabolism is a therapeutic opportunity for cancer. However, the efforts on glucose metabolism targeted inhibitors have not been successful to produce effective treatment in clinic [4, 5]. Metabolic heterogeneity should be the key reason, since cancer cells with different metabolic preferences may respond totally differently to the same metabolic stress [6–10]. For example, upon glucose deprivation (GD), some cancer cell lines, such as HCT116, UMRC6, LN18, and U87, exhibit rapid cell death, whereas others (e.g., DLD1, 786-O and LN229) could survive for longer time [8, 11]. Therefore, it is an essential prerequisite to understand the heterogeneity of intrinsic signaling networks in distinct cancer cells for improving the efficacy of glucose-targeted therapy.
Yes-associated protein (YAP) is a crucial transcriptional coactivator that functions in many physiological processes, and its activity is tightly regulated by spatial translocation and posttranslational modifications. In fully confluent cells, YAP is phosphorylated by large tumor suppressors (LATSs) at Serine 127 (S127), which promotes its binding to 14-3-3 protein, resulting in cytoplasmic localization and functional inactivation [12]. Un-phosphorylated YAP is translocated to the nucleus and enhances the transcription of target genes (CTGF, CYR61, NUAK2, THBS1, etc.) to promote proliferation and cell survival [13, 14]. Importantly, YAP is also an essential mediator of the cellular response to nutritional stress [15–21]. It was previously reported that YAP was inactivated by GD and glycolysis inhibition in HEK293, MCF10A and MEF cells [18, 21, 22]. However, the signaling pathways responding to GD, especially mediated through non-glycolysis pathway, such as pentose phosphate pathway (PPP), remain largely unknown.
PPP is an essential branch of glucose metabolism to generate D-Ribose-5-phosphate (D5P), an important metabolite for de novo nucleotide biosynthesis [23]. Thus, D5P acts as a link between glucose metabolism and de novo nucleotide biosynthesis. In addition to the PPP, D5P can also be converted from ribose-1-phosphate (R1P), a product of purine nucleotide degradation, under the catalysis of phosphoglucomutase 2 (PGM2) [24, 25]. Therefore, D5P, as a linker among PPP, de novo nucleotide biosynthesis, and purine nucleotide degradation, may serve as a checkpoint for the balance of these pathways. While, how it responds to glucose limitation and its roles in linking glucose metabolism and nucleotide metabolism need to be further disclosed.
Here, we elucidated the crosstalk between glucose metabolism and nucleotide metabolism under GD through a MYH9/LATS1-YAP-PNP-D5P negative feedback loop. GD-induced D5P downregulation promoted MYH9-mediated LATS1 degradation and consequent YAP activation, which potentiated purine nucleoside degradation by enhancing PNP activity to recover the D5P level. Significantly, D5P-mediated YAP inactivation inhibited the progression of APC mutation-driven colorectal cancer and also sensitized cancer cells to inhibitors of glucose transporters (GLUT). Considering that there are no GLUT inhibitors approved in clinical practice so far, D5P may be a synergist for GLUT inhibitors to improve its clinical efficacy. Importantly, D5P itself showed a significant anti-cancer effect, indicating a potential application in cancer treatment.
Methods
Cell culture and reagents
All cancer cell lines were obtained from the American Type Culture Collection. All cell lines were free from mycoplasma contamination (tested by PCR). Cells were cultured in DMEM with 10% fetal bovine serum (Corning) in a 5% CO2 air atmosphere. DMEM without glucose was purchased from Life Technologies (11966-025, Thermo Fisher Scientific, Waltham, Massachusetts, USA). Nucleosides, nucleotides, guanine were purchased from Merck Sigma‒Aldrich. PNP assay kit (ab204706) was purchased from Abcam. GLUT inhibitor KL-11743 was purchased form MCE. D5P, FDP and 6PG were obtained from Shanghai YuanYe Bio-Technology. The cell death detection kit was purchased from Beyotime (Shanghai, China).
Treatments
For glucose deprivation, cells seeded in 12-well plates or 6 cm dishes were cultured to 90% confluence. After washing with PBS, conditioned medium prewarmed to 37 °C was gently added to each sample, and the cells were cultured for the indicated times. For the recovery experiment, adenosine, guanosine, D5P, guanine or other nucleosides/nucleotides were supplemented in glucose-free medium before being added to the samples, and then the cells were cultured for the indicated times.
Mice, histology and immunofluorescence
All mice were maintained in specific-pathogen-free (SPF) facility and were performed in compliance with the guide for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of the Institutional Animal Care and Use Committee of Zhengzhou University. APCmin/+ mice (Strain NO. T001457) were purchased from GemPharmatech (Nanjing, China). Mice were sacrificed at the indicated time points, and colons were collected immediately after sacrifice, and prepared for experiments. For Histology and Immunofluorescence experiments, tissues were fixed overnight in 4% paraformaldehyde, prepared using the Swiss roll technique, embedded in paraffin, and cut into 7-μm sections.
Organoid experiment
In brief, the tissue from colorectal cancer patient was washed by 1 × PBS and digested by collagenase I. Then a 100 µL mixture of Matrigel (BD Biosciences) and complete growth medium (2:1) was added with a pellet of 500–1000 crypts. After polymerization, 100 µL of complete growth medium [Advanced DMEM/F12 (Invitrogen) containing growth factors: 50 ng/mL EGF, 500 ng/mL R-spondin1, and 100 ng/mL Noggin (PeproTech)] was added and refreshed every 2–3 days. On the fifth day, 6 mM D5P and 3 μM KL-11743 was added. After a 60 h culture, the organoids were photographed to record the volume and measured by MD50 software from Mshot.
Stable cell line generation
The short hairpin RNA (shRNA) used in this paper was constructed with the pLKO.1 system, and the lentiviral plasmid overexpressing YAP-5SA was constructed with the pLVX-IRES-Puro system. Cell lines with specific gene silencing or overexpression were generated as previously described. In brief, HEK293T cells were transfected with either the pLKO.1 or pLV plasmid, together with Δ8.9 and the VSVg third-generation lentiviral packaging system using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s instructions. After transfection, lentivirus particles in the medium were collected every twenty-four hours, and after three collections, the medium was filtered, and the target cell lines were infected. At 48 h after infection, 2 μg/ml puromycin was added to obtain stable cell lines with successful transduction. The sequences of shRNAs used in this study: shYAP-1: CAGGTGATACTATCAACCAAA; shYAP-2: GACCAATAGCTCAGATCCTTT; shPNP: GCTCTCAGTACCTGGAAACAA; shPGM2: CGACTAATAGCAGAAGGTAAT; shMYH9: GACAGCAATCTGTACCGCATT; shMYH10: GCTCGGATGAAGCAGCTTAAA.
Western blotting
Proteins were separated by FuturePAGE™ surePAGE™ and SDS‒PAGE and transferred to PVDF or nitrocellulose membranes with a wet transfer system. Membranes were blocked with TBST containing 5% milk or 5% BSA before overnight incubation with primary antibodies. After incubation with secondary horseradish peroxidase-coupled antibodies, signals were detected using ECL-based chemiluminescence. The primary antibodies and concentrations used for western blotting were as follows: YAP (1:1000, CST, 14074), P-YAP (1:1000, CST, 13008), LATS1 (1:1000, CST, 3477), MYH9 (1:2000, CST, 3403), P-JNK (1:1000, CST, 9255), PNP (1:100, Santa Cruz, sc-365551), LC3A/B (1:1000, CST, 4108), P21(1:1000, CST, 2947), PARP (1:1000, CST, 9542), c-PARP (1:1000, CST, 5625), G3BP1 (1:1000, CST, 61559), GAPDH (1:10000, Proteintech, 66004), actin (1:10000, Proteintech, 66009), and tubulin (1:10000, Proteintech, 66031). Phos-tag acrylamide was purchased from Wako. Phos-tag western blotting was performed in gels containing 8% acrylamide, 50 μM MnCl2 and 25 μM Phos-tag and was followed by immunoblotting.
Protein isolation
Most proteins were collected with 1 × loading buffer. For supernatant and pellet separation, collected cells were lysed with 160 μl of Pierce™ IP Lysis Buffer (#87787 Thermo Fisher Scientific) supplemented with cOmplete protease inhibitor (Roche) and PhosSTOP phosphatase inhibitor (Roche). Then, 80 μl of cell lysate was transferred to a new tube with 80 μl of 3 × loading buffer, and incubated on ice for 15 min. After incubation, the tubes were centrifuged at 13,000 rpm for 15 min, 80 μl of supernatant was transferred to a new tube with 40 μl of 3 × loading buffer, and the insoluble pellet was resuspended in 120 μl of 1 × loading buffer.
Immunoprecipitation (IP) and Mass Spectrometry (MS)
For IP assays, cells were lysed and washed with Pierce™ IP Lysis Buffer (#87787, Thermo Fisher Scientific) supplemented with cOmplete protease inhibitor (Roche) and PhosSTOP phosphatase inhibitor (Roche). Cell lysates were incubated overnight at 4 °C with the indicated primary antibody and Pierce™ Protein A/G Magnetic Beads (#88802, Thermo Fisher Scientific). Beads were washed 4 times with IP lysis buffer and boiled in SDS-loading buffer for 10 min at 95 °C. Samples were run on a SurePAGE™ gel (M00657, Genscript.) and stained with Coomassie brilliant blue staining solution to identify the differential bands. Then, the bands were excised and sent out for MS identification (Biotree, Shanghai, China).
RNA isolation and RT‒qPCR
RNA was isolated using TRIzol followed by RQ1 RNase-free DNase Set treatment (Promega) according to the manufacturer’s instructions. First-strand cDNA was synthesized using Superscript II (Takara), and 1 μg of total RNA was used in each cDNA synthesis reaction. TB Green Master Mix reagents (Takara) and primer mixtures were used for real-time PCR. Student's t test was used for statistical analysis, and a P value less than 0.05 was considered significant. Primers: CTGF: CAGCATGGACGTTCGTCTG and AACCACGGTTTGGTCCTTGG; CYR61: CTCGCCTTAGTCGTCACCC and CGCCGAAGTTGCATTCCAG; THBS1: GCCATCCGCACTAACTACATT and TCCGTTGTGATAGCATAGGGG; NUAK2: GATGCACATACGGAGGGAGAT and GCTGGCATACTCCATGACGAT; PGM2: GAGGCAGTGAAACGACTAATAGC and CTGTCCCAAACTCCATTCGGG; MYH9: CCTCAAGGAGCGTTACTACTCA and CTGTAGGCGGTGTCTGTGAT; MYH10: TGGTTTTGAGGCAGCTAGTATCA and AGTCCTGAATAGTAGCGATCCTT.
RNA-Seq
Samples were subjected to Illumina NovaSeq 6000 performed by Mingma Technologies. mRNA was extracted from total RNA using magnetic beads with oligo-dT and then fragmented randomly. First- and second-strand cDNA were then synthesized using reverse transcriptase. The reverse transcription product was subjected to terminal repair, followed by the addition of an A base at the 3' end. Subsequently, the fragments were ligated with an Illumina universal adapter. After PCR amplification, the ligation products were purified to remove the incompletely ligated fragment and the self-ligated adapter. Library sizes were determined using an Agilent 2100 Bioanalyzer, and library concentrations were ascertained using Qubit 2.0. DNA sequencing was performed according to Illumina's standard protocol. edgeR used the overdispersed Poisson model and empirical Bayes model to estimate the dispersion of all gene expression levels. In this project, raw read counts of gene expression were applied to estimate the differentiation between paired samples or groups by using the edgeR package in R. Genes with CPM and TMM values greater than 1 in at least one sample were selected. The criteria for significantly differentially expressed genes were P-adj < 0.05 and |FC|> = 2. The Ensembl, GO, and KEGG databases were used to annotate differentially expressed genes to illustrate the biological processes related to these genes. Enrichment analysis was based on hypergeometric distribution statistics to determine whether a group of genes was significantly enriched in a certain function or KEGG pathway. In this project, a set of significantly upregulated or downregulated genes was used as the input to examine the significance of gene enrichment in a GO function or KEGG pathway by using the GOstat package in R. The criteria was P value < 0.01.
Cell death analysis
After the indicated treatment, cells were digested and collected. After centrifugation, the medium was removed, and the cells were resuspended in the corresponding staining buffer. After incubation, the samples were analyzed by Attune NxT.
PNP activity assay
Cells seeded in 12-well plates were treated as indicated, and then harvested to detect PNP activity using PNP activity assay kit (ab204706, abcam). Briefly, to prepare cell extract, add 150–300 μL of cold 1 × PNP Assay Buffer containing protease inhibitor cocktail to 1–5×106 fresh cells and pipette several times to disrupt the cells. Transfer cell homogenate including cell debris to a cold microfuge tube and agitate on a rotary shaker at 4 °C for at least 15 min. Centrifuge cell homogenate at 10,000 g, 4 °C for 15 min. Transfer the clarified supernatant to a fresh pre-chilled tube and store on ice. Use lysates immediately to examine PNP activity. The fluorescent product was measured at ex = 535 nm/em = 587 nm in a kinetic mode. And calculate the activity of PNP using the formula provided in the manual.
Immunofluorescence
Cells grown on coverslips were fixed with 4% paraformaldehyde (DF0135, Leagene, Beijing, China) for 15 min at RT, washed for three times with 1 × PBS, blocked in blocking buffer (5% goat serum, 0.3% Triton X-100 in 1 × PBS) for at least one hour at RT, and incubated with primary antibodies diluted in blocking buffer for 2 h at RT or 4 °C overnight. After three washes in 1 × PBS, the samples were treated with secondary antibodies tagged with Alexa Fluor 647 (4414S, CST) for one hour at RT in the dark. Cells were washed twice in 1 × PBS and then stained with DAPI (D9542, Sigma-Aldrich). Glass slides were mounted in ProLong™ Diamond Antifade Mountant (P36965, Invitrogen). Images were acquired using a Leica THUNDER microscope and processed by LAS X. Actin was stained by CellMask™ Green Actin Tracking Stain.
Untargeted metabolomics analysis
Metabolite extraction: Cells were grown in individual 60 mm plates with 6–10 biological replicates per sample group, and each sample had an equivalent amount of protein (~ 10 mg) or number of cells (1 × 107 or 1 × 106). Samples were collected in an EP tube, and 1000 μL of extract solution (methanol: acetonitrile: water = 2:2:1) was added. Then, the samples were homogenized at 60 Hz for 3 min at − 20 °C. Then, the samples were incubated for 1 h at − 20 °C and centrifuged at 12,000 rpm (RCF = 13,800 × g, R = 8.6 cm) for 15 min at 4 °C. The resulting supernatant was transferred to a fresh glass vial for analysis. LC‒MS/MS Analysis: LC‒MS/MS analyzes were performed using an UHPLC system (ExionLC AD, AB SCIEX, USA) with an ACQUITY UPLC BEH Amide column (2.1 mm × 100 mm, 1.7 μm, Waters) coupled to a quadruple time-of-flight mass spectrometer (TripleTOF 5600+, AB SCIEX, USA). The column temperature was maintained at 25 °C. Mobile phase A was 25 mM ammonium hydroxide (NH4OH) + 25 mM ammonium acetate (NH4OAc) in water, and mobile phase B was ACN for both the positive (ESI+) and negative (ESI−) modes. The elution flow rate was 0.3 mL/min, and the gradient was as follows: 0–1 min: 95% B, 1–14 min: 95% B to 65% B, 14–16 min: 65% B to 40% B, 16–18 min: 40% B, 18–18.1 min: 40% B to 95% B, and 18.1–23 min: 95% B. The injection volume was 2 μL. All samples were injected randomly during data acquisition, which occurred in information-dependent acquisition (IDA) mode. The source parameters were set as follows: ion source gas 1 (GAS1), 50 psi; ion source gas 2 (GAS2), 50 psi; curtain gas (CUR), 35 psi; temperature (TEM), 500 °C; declustering potential (DP), 80 V or − 80 V in positive or negative modes, respectively; and ion spray voltage floating (ISVF), 5500 or − 4500 V in positive or negative modes, respectively. The TOF MS scan parameters were set as follows: mass range, 60–1200 Da; accumulation time, 200 ms; and dynamic background subtraction, on. The product ion scan parameters were set as follows: mass range, 25–1200 Da; accumulation time, 50 ms; collision energy, 35 or − 35 V in positive or negative modes, respectively; collision energy spread, 15; resolution, UNIT; charge state, 1 to 1; intensity, 100 cps; exclude isotopes within 4 Da; mass tolerance, 10 ppm; maximum number of candidate ions to monitor per cycle, 10; and exclude former target ions for 4 s after two occurrences. Data preprocessing and annotation for Fig. 2A, B and F: The raw data were processed with XCMSPlus v3.6.3 for peak detection, extraction, alignment, integration, and annotation. The centWave algorithm was used for peak picking with a resolution of 15 ppm, minimum and maximum chromatographic peak widths from 5 to 20 s, and a signal-to-noise ratio of 6. The Obiwarp algorithm was applied for retention time correction.
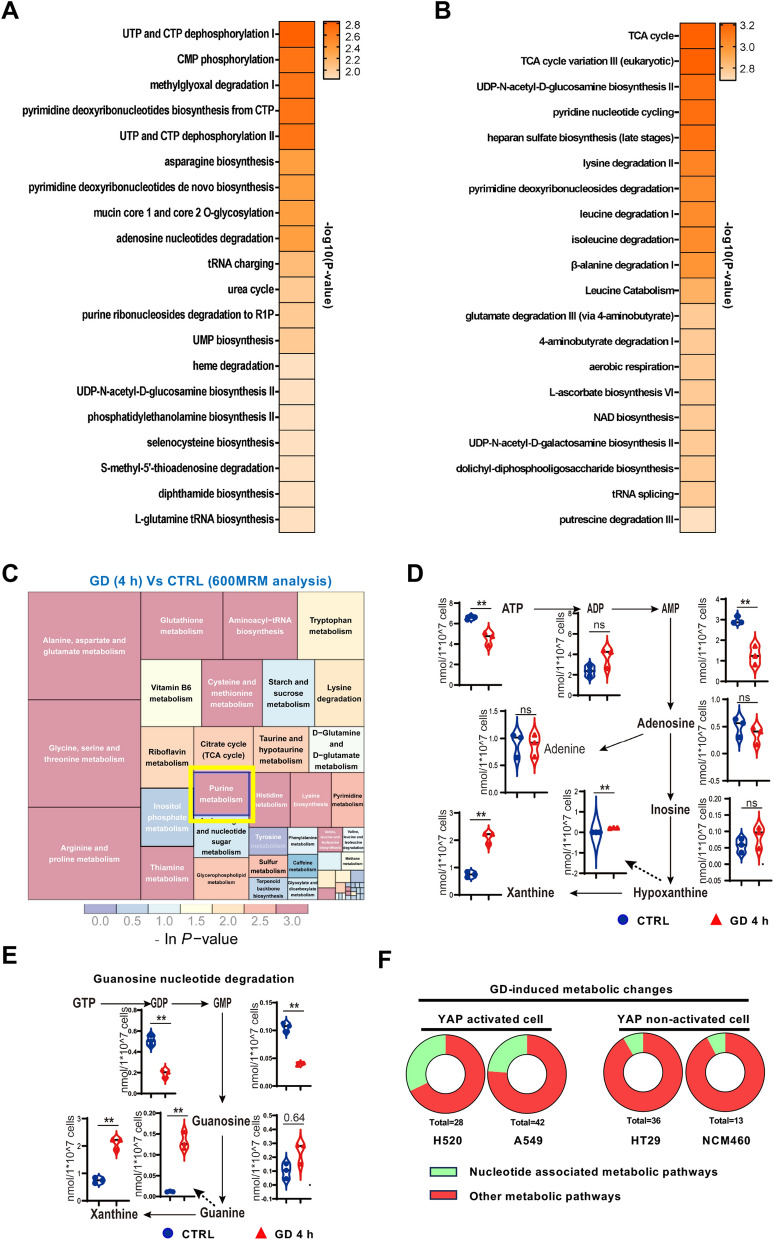
Fig. 2.
GD induces purine nucleotide degradation. A Metabolite set enrichment of the polar metabolites from HCT116 cells with GD versus normal culture as determined by untargeted metabolomics analysis (XCMS). B Metabolite set enrichment of the polar metabolites from DLD1 cells with GD versus normal culture as determined by untargeted metabolomics analysis (XCMS). C Metabolic pathway enrichment analysis of changed metabolites from HCT116 cells under GD determined by targeted metabolomics analysis (quantitative 600MRM analysis). Purine metabolism was marked with a yellow box. D Levels of metabolites involved in adenine nucleotide degradation from HCT116 cells under normal conditions or GD (0 mM, 4 h). E Levels of metabolites involved in guanine nucleotide degradation from HCT116 cells under normal conditions or GD (0 mM, 4 h). F Pie chart of nucleotide associated metabolic pathways among GD-induced changed metabolic pathways in YAP activated or YAP non-activated cells. In (D), (E), data are the mean ± S.D.; P values were calculated using a two-tailed unpaired Student’s t-test. *, P < 0.05; **, P < 0.01; ns, no significance
Targeted metabolomics analysis (600MRM)
We performed 600MRM analysis (Biotree, Shanghai, China) in Figs. 2C and 5A. After the addition of 200 μL of H2O, the samples were vortexed for 30 s. The samples were precooled in dry ice, underwent three freeze‒thaw cycles in liquid nitrogen, and vortexed for 30 s. After the addition of 800 μL of acetonitrile-methanol (1:1, v/v precooled at − 40 °C), the samples were vortexed for 30 s and sonicated for 15 min in an ice water bath. This was followed by incubation at − 40 °C for 2 h and centrifugation at 12,000 rpm (RCF = 13,800 × g, R = 8.6 cm) and 4 °C for 15 min. An 800 μL aliquot of the supernatant of each sample was transferred to a new Eppendorf tube and dried with a centrifugal concentrator. Then, 160 μL of 60% acetonitrile was added to the Eppendorf tube to reconstitute the dried sample, and the Eppendorf tube was vortexed until the powder was dissolved, followed by centrifugation at 12,000 rpm and 4 °C for 15 min. Finally, 100 μL of the supernatant of each sample was transferred to a glass vial for LC‒MS/MS analysis. A mixture of standard metabolites was prepared as a QC sample. Standard Solution Preparation: Different volumes of each standard stock solution were transferred to a flask to create a mixed working standard solution. A series of calibration standard solutions were then prepared by stepwise dilution of this mixed standard solution (containing an isotopically labelled internal standard mixture that had concentrations identical to those in the samples). The criteria for significantly altered metabolites were P < 0.05 and VIP (Variable Importance in the Projection) > 1. The mass spectrum data acquisition and quantitative analysis of metabolites were completed by SCIEX Analyst Work Station Software (1.7.3) and BI0TREEBioBud (2.0.3). The enrichment analysis was performed using R (KEGGgraph) Software (1.46.0, 2.4-2).
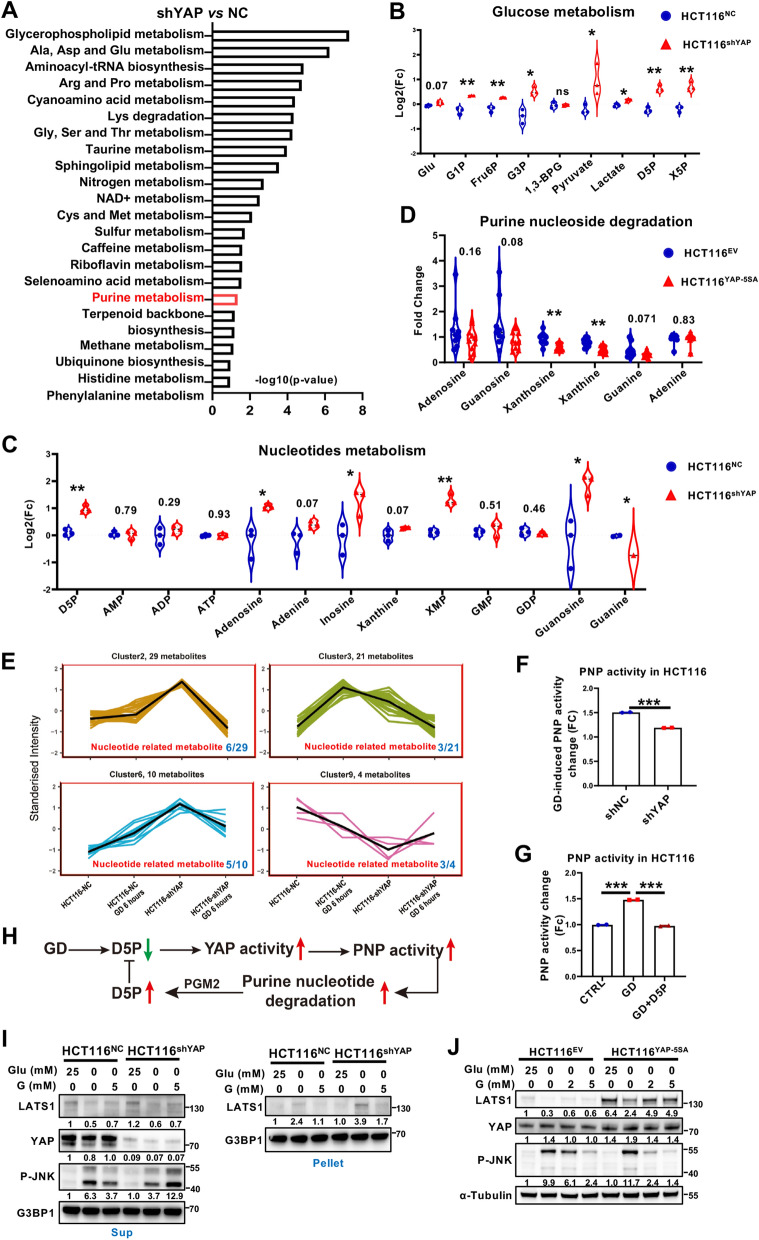
Fig. 5.
Dysregulation of YAP activity reprograms purine nucleotide metabolism and cellular response to GD. A Set enrichment of polar metabolites from HCT116-shYAP and HCT116-NC cells as determined by 600MRM analysis. B Relative levels of metabolites involved in glucose metabolism in HCT116-NC and HCT116-shYAP cells. C Relative levels of metabolites involved in purine (adenine and guanine) nucleotide metabolism in HCT116-NC and HCT116-shYAP cells. D Relative levels of metabolites involved in purine (adenine and guanine) nucleotide metabolism in HCT116-EV- and HCT116-YAP-5SA-overexpressing cells. E Cluster analysis of polar metabolites of HCT116-NC and HCT116-shYAP cells under normal culture or GD conditions (0 mM, 6 h). F Analysis of PNP enzyme activity in HCT116-NC and HCT116-shYAP cells treated under GD. G Analysis of PNP enzyme activity in HCT116 cells treated under GD with or without D5P supplementation. H Schematic diagram depicting that GD-induced D5P downregulation triggers YAP activation, promoting PNP activity to recover D5P level. I Immunoblotting analysis of LATS1, YAP and phosphorylated JNK in the supernatants (Sup) or LATS1 in the pellets of HCT116-NC and HCT116-shYAP cells under the indicated treatment. G3BP1 was used as a loading control. J Immunoblotting analysis of LATS1, YAP and phosphorylated JNK in the supernatants of HCT116-EV- and YAP-5SA-overexpressing cells with the indicated treatment. Tubulin was used as a loading control. The experiments in (I), (J) were repeated twice independently. In (B–D) and (F–G), data are the mean ± S.D.; P values were calculated using a two-tailed unpaired Student’s t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance
Statistical analysis
All experiments in vitro were done independently with at least three biological replicates. The data presentation form and statistical analyses are given in the corresponding figure legends. Data analyses were carried out with GraphPad Prism (v9.0, La Jolla, USA). P values < 0.05 were considered as statistically significant.
Results
Differential responses of YAP activation to glucose deprivation in different cancer cells
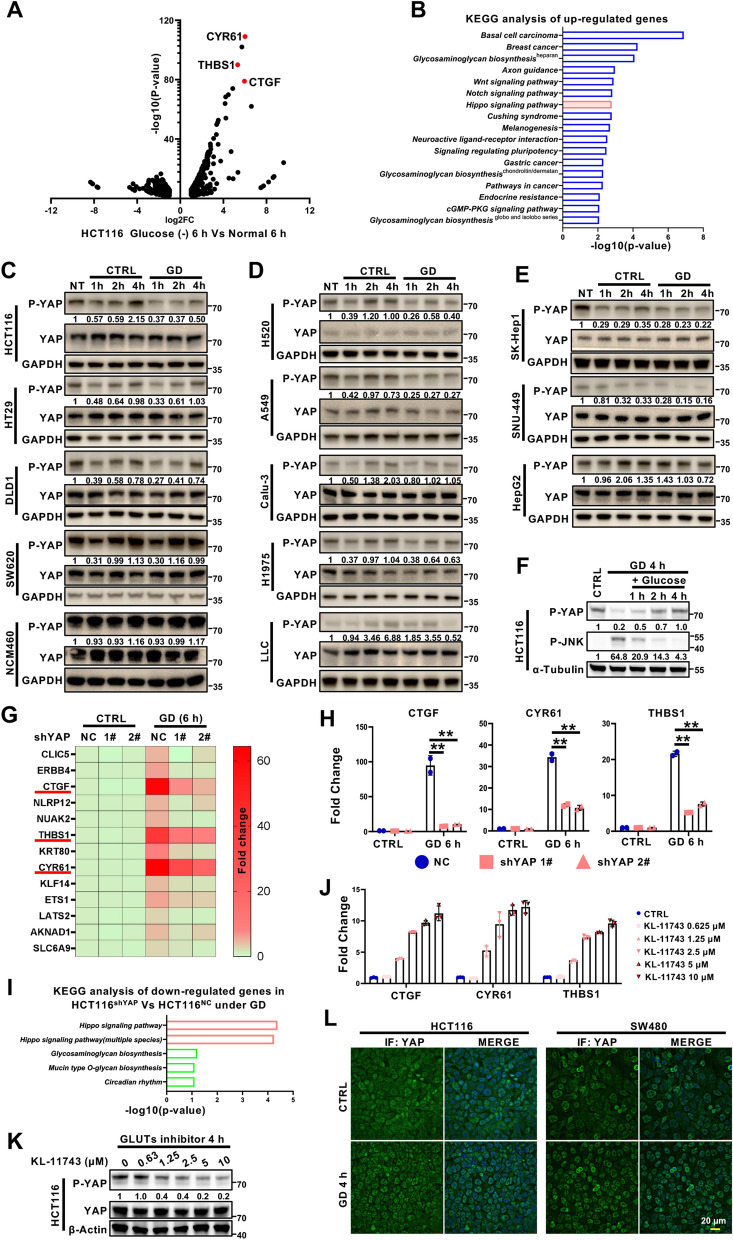
To explore how cells respond to glucose deprivation (GD), colorectal cancer (CRC) HCT116 cells with high confluency were treated with or without GD for 6 h and subjected to RNA sequencing (RNA-seq) analysis. Interestingly, CTGF, THBS1 and CYR61, which are the well-known YAP target genes, were among the top genes induced by GD (Fig. 1A). Moreover, KEGG analysis showed the Hippo signaling pathway, the well-known upstream signaling of YAP activation, was also enriched from these differentially expressed genes (Fig. 1B), suggesting that YAP was activated by GD. This phenotype was not consistent with previous studies reporting that YAP activity was inhibited by GD in some cell lines like HEK293, MCF10A and MEF cells [16, 18, 22], which may be due to the metabolic preference, malignancy or differential responses from different cell types to GD treatment. To further validate the effect of GD on YAP activation, YAP phosphorylation at Serine 127 was examined by immunoblotting in other cell lines, including colorectal cancer cells, lung cancer cells, and hepatocellular carcinoma cells, as well as one immortalized intestinal epithelial cell line. As shown, GD-induced YAP activation was observed in most tested cell lines, but not in some cell lines like HT29, DLD1, and SW620 cells (Fig. 1C–E; Figure S1A and S1B). Meanwhile, consistent with previous reports [16, 18, 22], in NCM460, an immortalized intestinal epithelial cell, GD treatment slightly inactivated YAP by increasing YAP phosphorylation (Fig. 1C). These results suggest that different cells might adopt various strategies to overcome glucose deficiency, and YAP is activated by GD in most tested cancer cells. Additionally, when glucose was re-added to GD-treated HCT116 cells, the inactivated form of YAP with S127 phosphorylation was significantly recovered and GD-induced JNK phosphorylation was diminished [26, 27] (Fig. 1F). In addition to GD, a low concentration (≤ 1.2 mM) of glucose also activated YAP in HCT116 cells (Figures S1C and S1D). Also, GD-induced YAP activation was observed not only in cells with high confluency but also with low confluency (Figures S1E and S1F). Moreover, the RNA-seq data from the HCT116-NC and HCT116-shYAP cell lines showed that GD-induced upregulation of YAP target genes (CTGF, CYR61 and THBS1) and activation of the Hippo signaling pathway were dependent on YAP (Fig. 1G–I). Consistent with GD, GLUT inhibitor KL-11743 treatment, which mimicked glucose limitation, also activated YAP in a dose-dependent manner (Fig. 1J–K). YAP activation is often associated with the increase of its nuclear localization. Consistently, YAP staining demonstrated that GD promoted the nuclear localization of YAP (Fig. 1L). As YAP is a well-known pro-survival signaling [28], we also asked whether YAP activation would increase the probability of cell survival under glucose limitation. Cell apoptosis analysis showed that silencing YAP significantly promoted GD-induced apoptosis (Figure S1G), suggesting that GD-induced YAP activation protected cancer cells from cell death. These results demonstrate that cancer cells response to GD differently, and the relationship between YAP and glucose requires further investigation.
Fig. 1.
Differential responses of YAP activation to glucose deprivation in different cancer cells. A Volcano plot of the genes differentially expressed under glucose deprivation (GD). The target genes of YAP (CTGF, CYR61 and THBS1) were shown in red. B KEGG pathway enrichment analysis of genes upregulated by GD (https://david.ncifcrf.gov/). C–E Immunoblotting analysis of YAP phosphorylation (serine 127) in GD-treated colorectal cancer cells (C), lung cancer cells (D), and hepatocellular carcinoma cells (E). GAPDH was used as a loading control. F Immunoblotting of YAP phosphorylation (serine 127) and JNK phosphorylation (Threonine183/Tyrosine185) in HCT116 cells treated under GD for 4 h, and then treated with glucose for the indicated times. Tubulin was used as a loading control. G RNA-seq analysis of HCT116-shNC and HCT116-shYAP cells treated with or without GD (0 mM, 6 h). H RT‒qPCR analysis of YAP target genes (CTGF, CYR61 and THBS1) in the samples from (G). I KEGG pathway enrichment analysis of genes downregulated by YAP knockdown under GD. J RT‒qPCR analysis of YAP target genes (CTGF, CYR61 and THBS1) in HCT116 cells treated with different doses of GLUT inhibitor KL-11743. K Immunoblotting analysis of YAP phosphorylation (serine 127) in HCT116 cells treated with different doses of GLUT inhibitor KL-11743 for 4 h. Actin was used as a loading control. L Immunofluorescence staining of YAP localization in HCT116 cells or SW480 cells treated with or without GD (0 mM, 4 h). DAPI served as the nuclear stain. The experiments in (C), (D), (E), (F) and (K) were repeated twice independently. In (H) and (J), data are the mean ± S.D.; P values were calculated using a two-tailed unpaired Student’s t-test. **, P < 0.01
GD induces purine nucleotide degradation
GD-induced metabolic changes have been recognized as essential regulators of signal transduction [29]. To investigate whether GD-induced YAP activation was ascribed to the metabolic alterations, we conducted untargeted metabolomics analysis in HCT116 (YAP activated) and DLD1 (YAP non-activated) cells. Interestingly, the metabolic pathways most significantly altered in HCT116 cells were nucleotide metabolism-associated pathways, whereas those in DLD1 cells were the canonical glucose metabolism-associated pathways (Fig. 2A, B; Figure S2A). As untargeted metabolomics analysis only provided relatively quantitative results, we further performed targeted metabolomics analysis of GD-treated HCT116 cells to confirm this result. Quantitative analysis (600MRM analysis) showed that purine metabolism was the most enriched nucleotide metabolism-associated pathway in HCT116 cells (Fig. 2C). Specifically, purine nucleotides were significantly degraded, shown as the reduction of purine nucleotides (AMP, GDP and GMP) and the increase of purine nucleotide degradation products (hypoxanthine, Guanine and xanthine) (Fig. 2D, E). Furthermore, consistent with HCT116, the GD-induced nucleotide metabolic alterations were also observed in other YAP-activated cells (H520 and A549), but not in YAP non-activated cells (HT29 and NCM460), which showed similar metabolic alterations to DLD1 (Fig. 2F and Table S1). These results indicate that the differences in metabolic response may result in the differences in cellular intrinsic signaling responding to GD, and GD mainly induces purine nucleotide degradation in YAP-activated cells.
GD-induced D5P reduction is responsible for GD-induced YAP activation
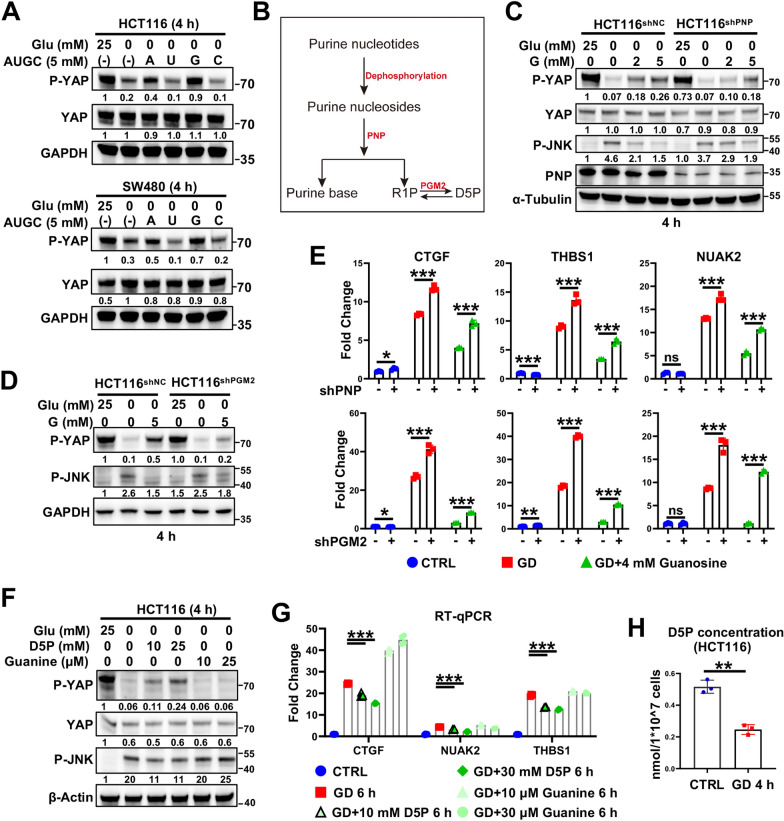
Next, we examined the effect of nucleotide metabolism on GD-induced YAP activation. As nucleotides cannot be directly absorbed by cells, we tested the effects of nucleosides on GD-induced YAP activation. Surprisingly, the activation of YAP and also the cell death induced by GD were significantly reduced by the supplementation of purine nucleosides (A and G, especially G), but not pyrimidine nucleosides (U and C) (Fig. 3A; Figures S2B–S2E), suggesting that purine nucleotides or their downstream metabolites suppressed GD-induced YAP activation. In cells, guanosine cannot be re-phosphorylated to generate GMP, instead, is degraded by purine nucleoside phosphorylase (PNP) to produce ribose-1-phosphate (R1P) and one purine base, which are recycled back into the salvage pathway [30, 31] (Fig. 3B). We then asked whether the effect observed above is ascribed to guanosine itself or its downstream metabolites. Interestingly, blocking purine nucleoside degradation by silencing PNP attenuated the effect of guanosine (Fig. 3C, and 3E upper panel), indicating that the guanosine breakdown product, but not guanosine itself, rescued GD-induced YAP activation. R1P, one of the guanosine breakdown products, can be further converted into D-ribose-5-phosphate (D5P) by Phosphopentomutase 2 (PGM2) [32]. More importantly, PGM2 silencing attenuated the rescuing effect of guanosine by blocking the transformation of R1P to D5P (Fig. 3D, and 3E lower panel; Figure S3A). Additionally, supplementation with D5P, but not guanine (purine base from guanosine degradation), blocked GD-induced or KL11743-induced YAP activation in CRC cells or CRC organoids (Fig. 3F, G; Figure S3B–D). As a well-known GD-responsive signaling, the change of p-JNK level was similar to YAP activation (Fig. 3C, D, F). Consistently, the D5P level was largely decreased in glucose-deprived cells (Fig. 3H; Figure S3E). All these findings indicate the central role of D5P in GD-induced YAP activation. D5P can also be converted from 6PG (6-phosphogluconate) through the PPP. Consistent with the role of D5P, supplementation with 6-PG also diminished GD-induced YAP activation (Figure S3F and S3G), suggesting that GD-induced D5P reduction is responsible for YAP activation.
Fig. 3.
GD-induced D5P downregulation is responsible for GD-induced YAP activation. A Immunoblotting analysis of YAP phosphorylation (serine 127) and YAP expression in glucose-deprived HCT116 (upper panel) and SW480 cells (lower panel) with or without supplementation with adenosine (A), uridine (U), guanosine (G), or cytidine (C). GAPDH was used as a loading control. B Schematic diagram of the purine nucleotide degradation pathway. D5P, D-ribose-5-phosphate; R1P, ribose-1-phosphate. C Immunoblotting of YAP phosphorylation (serine 127) and JNK phosphorylation in HCT116 NC and shPNP cells under normal conditions or GD (0 mM, 4 h) treated with or without G supplementation. D Immunoblotting of YAP phosphorylation (serine 127) and JNK phosphorylation (Threonine 183/Tyrosine 185) in HCT116 NC and shPGM2 cells under normal conditions or GD (0 mM, 6 h) treated with or without G supplementation. E RT‒qPCR analysis of YAP target genes (CTGF, THBS1 and NUAK2) in HCT116 NC and shPNP cells (upper panel) or shPGM2 cells (lower panel) under normal conditions or GD (0 mM, 6 h) treated with or without G supplementation. F Immunoblotting of YAP phosphorylation (serine 127) and JNK phosphorylation in HCT116 cells under normal conditions or GD (0 mM, 4 h) treated with or without D5P or guanine supplementation. G RT‒qPCR analysis of YAP target genes (CTGF, NUAK2 and THBS1) in HCT116 cells under normal conditions or GD (0 mM, 6 h) treated with or without D5P/guanine supplementation. H GD reduced D5P levels in HCT116 cells. The experiments in (A), (C), (D), (F) were repeated twice independently. In (E), (G), (H), data are the mean ± S.D.; P values were calculated using a two-tailed unpaired Student’s t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001
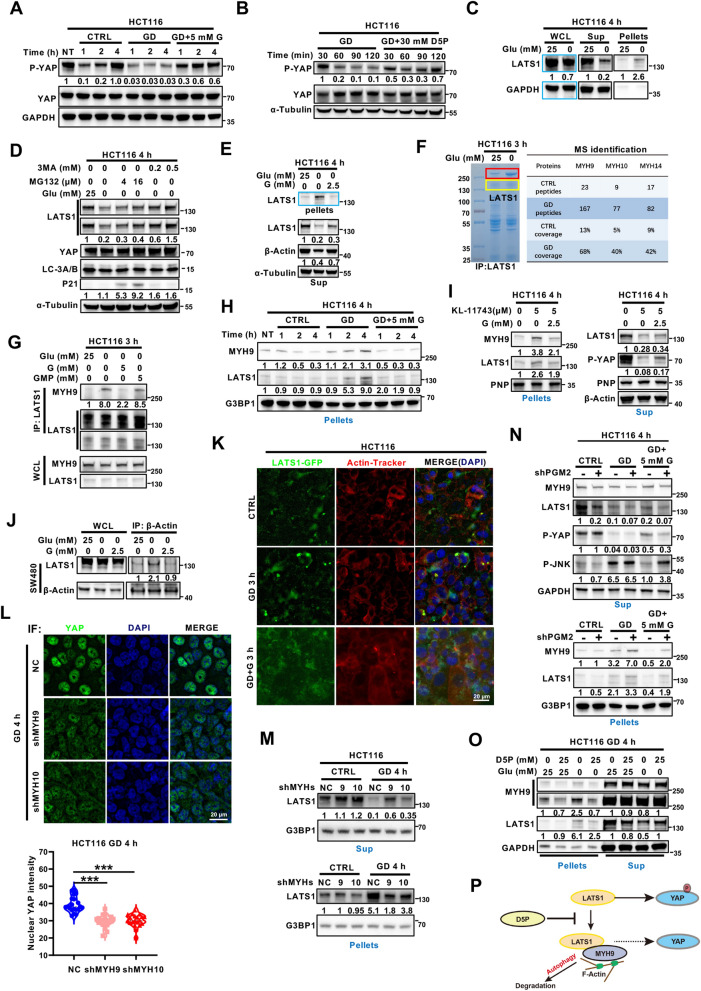
GD-induced D5P downregulation increases MYH9/LATS1 aggregation and LATS1 degradation to activate YAP
To further elucidate the mechanisms of GD-induced YAP activation, we conducted time-course experiments and found that YAP was dephosphorylated within 1 h after medium changing and re-phosphorylated gradually in the control group, guanosine, and D5P supplementation groups, but not in the GD-treated group (Fig. 4A, B; Figure S4A). These findings suggested that the glucose-free condition might restrict the kinase responsible for YAP phosphorylation. As LATS1 is the canonical upstream kinase of YAP [33], we further asked whether LATS1 was involved in GD-induced YAP activation. As expected, LATS1 was significantly downregulated in the whole cell lysate (WCL) of GD-treated HCT116 cells, and this downregulation could be abolished by the autophagy inhibitor 3MA and P62 knockdown, but not the proteasome inhibitor MG132 (Fig. 4C, D; Figure S4B), indicating that LATS1 might be degraded through the autophagy-lysosome pathway upon GD treatment. Autophagy is the main degradation pathway responsible for eliminating abnormal protein aggregates [34]. To examine whether LATS1 had been incorporated into aggregates, we analyzed the LATS1 protein levels in the soluble (supernatant) and insoluble (pellet) fractions of the cell lysate. Interestingly, the GD-induced reduction of LATS1 in the supernatant was more remarkable than that in the WCL, whereas the accumulation of LATS1 was detected in the pellets (Fig. 4C; Figure S4C). Consistent with the role of guanosine in inhibiting the YAP activation, supplementation with guanosine also diminished GD-induced LATS1 accumulation in the pellets (Fig. 4E; Figure S4C). These data suggest that GD promotes the accumulation of LATS1 in aggregates, which may result in autophagy-lysosome mediated LATS1 degradation and further YAP activation.
Fig. 4.
GD-induced D5P downregulation increases MYH9/LATS1 aggregation and LATS1 degradation to activate YAP. A, B Immunoblotting analysis of phosphorylated YAP (serine 127) and YAP in HCT116 cells cultured with normal medium, glucose-free medium or glucose-free medium supplemented with G (A) or D5P (B) for the indicated times. GAPDH or α-Tubulin was used as the loading control. C Immunoblotting analysis of LATS1 in the supernatants (Sup) and pellets of the HCT116 cell lysate treated with or without GD (0 mM, 4 h). GAPDH was used as a loading control. D Immunoblotting analysis of LATS1, YAP, LC3A/B and P21 expression in HCT116 cells under normal conditions or GD (0 mM, 4 h) with or without MG132 or 3MA. α-Tubulin was used as a loading control. E Guanosine supplementation diminished GD-induced LATS1 insolubilization. HCT116 cells were cultured with normal medium, glucose-free medium or glucose-free medium supplemented with G for 4 h. Tubulin was used as a loading control. F Coomassie blue staining and MS analysis of GD-induced LATS1-interacting proteins in HCT116 cells. The bands marked in the red boxes (left) were sent for MS analysis. G The effects of GD and G/GMP supplementation on the interaction between MYH9 and LATS1 in HCT116 cells were examined using Co-IP assay. H HCT116 cells were cultured with normal medium (CTRL), glucose-free medium or glucose-free medium supplemented with 5 mM G for indicated time before immunoblotting analysis of MYH9 and LATS1. G3BP1 was used as a loading control for the pellets. I Guanosine supplementation diminished KL-11743-induced LATS1 insolubilization. HCT116 cells were cultured under normal conditions, treated with or without KL-11743, or KL-11743 plus G supplementation for 4 h. PNP or actin was used as a loading control. J Immunoblotting analysis of the recruitment of LATS1 to actin in SW480 cells under normal conditions, GD (0 mM) or GD supplemented with G. K Representative fluorescence images of LATS1 aggregation and F-actin in LATS1-GFP overexpressing HCT116 cells under normal conditions, GD (0 mM) or GD supplemented with G for 3 h. DAPI served as the nuclear stain. L Representative immunofluorescence images and quantitative analysis of YAP nuclear localization in HCT116 NC, shMYH9 and shMYH10 cells treated with GD (0 mM) for 4 h. DAPI served as the nuclear stain. M Immunoblotting analysis of LATS1 in the supernatants (Sup) and pellets of HCT116 NC, shMYH9, and shMYH10 cells under normal conditions or GD (4 h). G3BP1 was used as a loading control. N Immunoblotting analysis of MYH9, LATS1, phosphorylated YAP (serine 127) and phosphorylated JNK in the supernatants (Sup) and pellets of HCT116 NC and shPGM2 cells treated under normal conditions, GD (4 h) or GD supplemented with G (4 h). GAPDH was used as a loading control. O Immunoblotting analysis of LATS1 and YAP in the supernatants (Sup) and pellets of HCT116 cells treated under normal conditions, GD (0 mM, 4 h) or GD supplemented with D5P. GAPDH was used as a loading control. P Schematic diagram depicting that D5P disrupts the interaction between LATS1 and MYH9/F-actin and recovers the inhibition of LATS1 on YAP. The experiments in (A–E), (G–J), and (M–O) were repeated twice independently. In (L), data are the mean ± S.D.; P values were calculated using a two-tailed unpaired Student’s t-test. ***, P < 0.001
To explore the mechanism underlying GD-triggered LATS1 accumulation in the insoluble fraction, co-immunoprecipitation assays were performed using anti-LATS1 antibody. The band at ~ 250 kDa was notably increased in the immunoprecipitates from GD-treated cells (Fig. 4F, left penal) and could also be observed in its insoluble fraction (Figure S4D). Mass spectral analysis identified that MYH9, MYH10 and MYH14 proteins were the major components of these bands and were significantly increased in GD-treated samples (Fig. 4F, right penal). MYH9 is an actin cytoskeleton protein and involved in autophagy-lysosome mediated degradation pathway [35]. The GD-induced interaction between MYH9 and LATS1, and the suppressive effect of guanosine were further verified using Co-IP and immunoblotting assays (Fig. 4G). Moreover, coaggregation of MYH9 and LATS1 was detected in glucose-deprived cells, and guanosine supplementation attenuated this accumulation (Fig. 4H), which was consistent with the changes of YAP phosphorylation (Fig. 4A). Similar to GD, GLUT inhibitor KL-11743 treatment promoted the accumulation of LATS1 and MYH9 in aggregates while guanosine supplementation attenuated this effect, indicating a consistent mechanism for GLUT inhibitor (Fig. 4I). As MYH9 had been characterized as an actin-binding protein and the involvement of actin in autophagy has also been reported [35], we wondered if LATS1 was anchored on actin filaments. Consistently, the Co-IP assays showed that GD significantly induced the interaction between LATS1 and actin, while guanosine supplementation diminished this interaction (Fig. 4J). Also, LATS1 aggregation and its colocalization with actin were obviously induced upon GD, and rescued by guanosine supplementation (Fig. 4K). Furthermore, MYH9 or MYH10 depletion (Figure S4E) significantly attenuated GD-induced YAP nuclear localization (Fig. 4L), LATS1 accumulation in the pellets (Fig. 4M), and the transcription of YAP target genes (Figure S4F), confirming the effect of MYH9/10 on GD-induced YAP activation. Consistent with the role of D5P, PGM2 silencing resulted in a reduction in the rescue effect of guanosine on LATS1 aggregation (Fig. 4N). Furthermore, the supplementation of D5P in both glucose-free culture medium and the lysate of GD-treated cells significantly mitigated the GD-induced aggregation of LATS1 (Fig. 4O; Figure S4G). To further validate the association between MYH9/10/14 and YAP activation, we analyzed the correlation between MYH9/10/14 expression and YAP signature (including CTGF, CYR61, THBS1, MYC, Gli2, Bcl2, AXL and COX2) in colon adenocarcinoma using the GEPIA database (http://gepia.cancer-pku.cn/index.html), which showed that the expression of MYH9/10, but not MYH14, was positively correlated with YAP downstream genes (Figure S4H). This result further confirmed the regulation of MYH9/10 on YAP activation. Also, we checked the YAP activation under MST1/2 inhibition and cytoskeleton dysregulation, both of these treatments showed little effect on GD-induced YAP activation, further demonstrated the specificity of MYH9/10 for GD-induced YAP activation (Figure S4I, J). Taken together, these results indicate that D5P exhaustion after GD treatment induces the MYH9/actin-mediated incorporation of LATS1 into aggregates, resulting in LATS1 degradation and YAP activation (Fig. 4P).
Dysregulation of YAP activity reprograms purine nucleotide metabolism and cellular response to GD
During the process of cell culture, we observed that YAP activity was negatively correlated with the acidity of the medium (Figures S5A, B). Therefore, it is reasonably hypothesized that YAP involved in regulating cellular metabolism. To analyze the role of YAP in metabolic reprogramming, untargeted metabolomics analysis was performed to identify the metabolic alterations in YAP-silenced and YAP-5SA (a constitutively activated mutant of YAP)-overexpressing HCT116 cells. Interestingly, YAP deficiency enhanced multiple cellular metabolism pathways, including glucose metabolism and purine metabolism (Fig. 5A; Figure S5C). Considering that intermediates of purine metabolism, including guanosine and D5P, regulate GD-induced YAP activation, we further focus on the role of YAP in purine metabolism. Metabolite quantification analysis revealed that YAP deficiency significantly augmented glucose metabolism, particularly D5P production (Fig. 5B). The increases in G3P (Glyceraldehyde-3-phosphate), pyruvate and lactate indicated an enhanced glycolysis in HCT116shYAP cells (Fig. 5B), which accounted for the heightened acidity of the medium. These findings suggest that YAP might regulate the metabolic flux of glucose metabolism. Moreover, purine nucleotide-associated metabolites, including nucleosides (adenosine, guanosine and inosine), were upregulated in HCT116shYAP cells, while the metabolites derived from purine nucleoside degradation (adenine, xanthine and guanine) showed slight accumulations or even decrease (Fig. 5C). Consistently, adenosine and guanosine were downregulated by the continuous activation of YAP in HCT116YAP-5SA cells (Fig. 5D; Figure S5D). In addition, both up- and down-regulation of YAP activity weakly regulated pyrimidine nucleotide-related metabolism in HCT116 cells (Figure S5E, F). These results suggest that YAP activation might be mainly correlated with purine nucleotide metabolism. Further quantitative analysis of the metabolites from HCT116NC and HCT116shYAP cells with or without GD treatment showed that the change trends of three clusters of metabolites induced by GD were completely reversed by YAP silencing, among which 26.5% (17/64) of the metabolites were nucleotide-related metabolites (Fig. 5E; Figure S6A; Table S2). Moreover, we found that the catalytic activity of PNP, the key enzyme catalyzing phosphorolysis of purine nucleosides, was increased by GD treatment in HCT116 cells (Fig. 5G). In contrast, this phenotype was reduced in HCT116shYAP cells (Fig. 5F), indicating that silencing YAP attenuated GD-induced PNP activity. Furthermore, D5P supplementation attenuated GD-induced PNP activity (Fig. 5G), which was consistent with its effect on YAP activation. Together, these findings indicate that GD-activated YAP promotes purine nucleotide degradation by enhancing PNP activity (Fig. 5H).
Theoretically, YAP-mediated purine nucleoside degradation would increase D5P/R1P levels, which further prevent the formation of MYH9/LATS1 aggregates and upregulate LATS1 to inactive YAP via a negative feedback loop. Indeed, our results showed that silencing YAP further increased the level of LATS1 in the insoluble fractions of glucose-deprived cells (Fig. 5I, Lane 2 vs. Lane 5), and attenuated the rescue effect of guanosine (Fig. 5I, Lane 3 vs. Lane 6). Further evidence was obtained from GD-induced JNK activation, a responsive signal that is activated by GD [36]. Under GD treatment, JNK phosphorylation was decreased in YAP-silenced cancer cells, while increased in YAP overexpressed cells (Fig. 5I, Lane 2 vs. Lane 5; Fig. 5J, Lane 2 vs. Lane 6), suggesting a positive correlation between JNK and YAP, which was consistent with previously reports [37]. However, with the supplementation of guanosine, although its rescue effect on JNK phosphorylation was enhanced by consistent YAP activation (Fig. 5J, Lane 3&4 vs. Lane 7&8), YAP silencing reversed this rescuing effect (Fig. 5I, Lane 3 vs. Lane 6). These results suggest that dysregulation of YAP activity reprograms purine nucleotide metabolism and cellular response to GD.
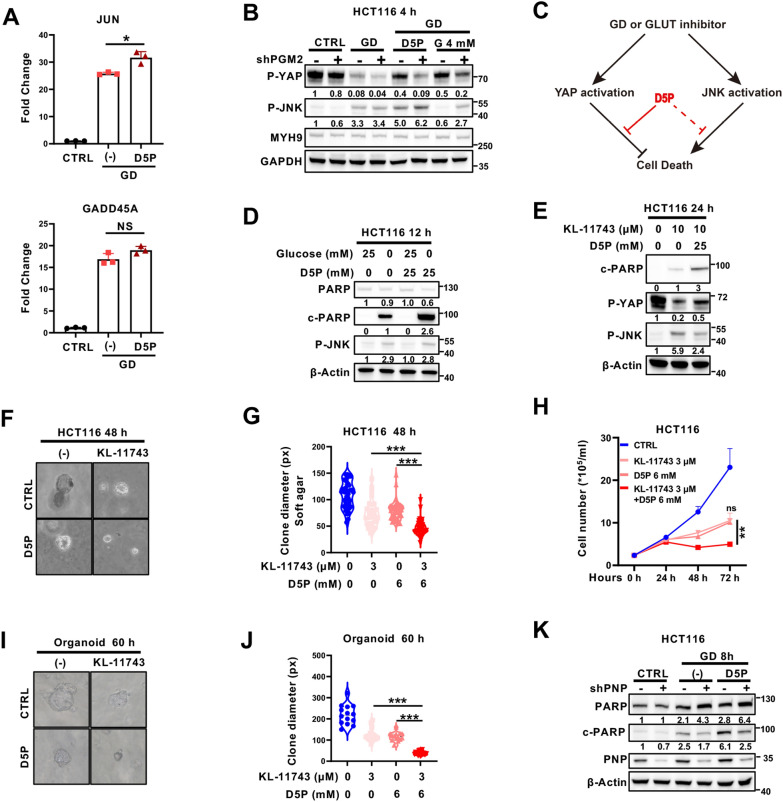
D5P sensitizes cancer cells to GLUT inhibitor by suppressing YAP
GLUT inhibitors have been considered as a promising cancer treatment option [38]. As shown in Figs. 3F, 6A, B and S3C, D5P could attenuate GD-induced YAP activation, while having no impact on GD-induced p-JNK or the expression of JNK downstream genes, including JUN and GADD45A. These findings suggested that D5P may promote GD- or GLUT inhibitor (KL-11743)-induced cell death by specifically suppressing YAP activation (Fig. 6C). In agreement with our hypothesis, supplementing D5P in glucose-free or KL-11743-containing medium significantly promoted cell death, as evidenced by increased cleaved PARP, decreased colony formation and delayed cell growth curves (Fig. 6D–H and Figure S7A). Moreover, CRC organoid culture assay showed that D5P supplementation enhanced the growth inhibitory effect of KL-11743 (Fig. 6I–J). Consistently, GD-induced cleaved PARP was attenuated by PNP deficiency (Fig. 6K, lane 4 vs. lane 3), and this phenotype was partially restored by D5P supplementation (Fig. 6K, lane 6 vs. lane 4). These results indicate that D5P has the potential to enhance the inhibitory effect of GLUT inhibitors by suppressing YAP activation.
Fig. 6.
D5P sensitizes cancer cells to GLUT inhibitor by suppressing YAP. A RT‒qPCR of JNK downstream genes (JUN and GADD45A) in HCT116 cells treated under normal conditions, GD or GD supplemented with D5P. B Immunoblotting analysis of phosphorylated YAP (serine 127) and phosphorylated JNK in HCT116 NC and shPGM2 cells treated under normal conditions, GD (4 h) or GD supplemented with D5P or G (4 h). C Schematic diagram of signaling pathways stimulated by GD or GLUT inhibitor treatment. D Immunoblotting analysis of PARP, cleaved PARP (c-PARP) and phosphorylated JNK in HCT116 cells treated under normal conditions or GD with or without D5P supplementation (12 h). Actin was used as a loading control. E Immunoblotting analysis of cleaved PARP (c-PARP), phosphorylated YAP (serine 127), and phosphorylated JNK in HCT116 cells treated with GLUT inhibitor (KL-11743) supplemented with or without D5P (24 h). Actin was used as a loading control. F, G Representative images (F) and quantitative results (G) of soft agar colony formation assays in HCT116 cells treated with KL-11743 supplemented with or without D5P (48 h). H Cell growth curves of HCT116 cells treated with KL-11743, D5P, or both. I, J Representative images (I) and quantitative results (J) of organoid cultures treated with KL-11743 supplemented with or without D5P (60 h). K Immunoblotting analysis of PARP, cleaved PARP (c-PARP) and PNP in HCT116 NC and shPNP cells treated under normal conditions, GD or GD supplemented with D5P (8 h). Actin was used as a loading control. In (A), (G), (H), data are the mean ± S.D.; P values were calculated using Dunnett’s multiple comparison test or Student’s t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance
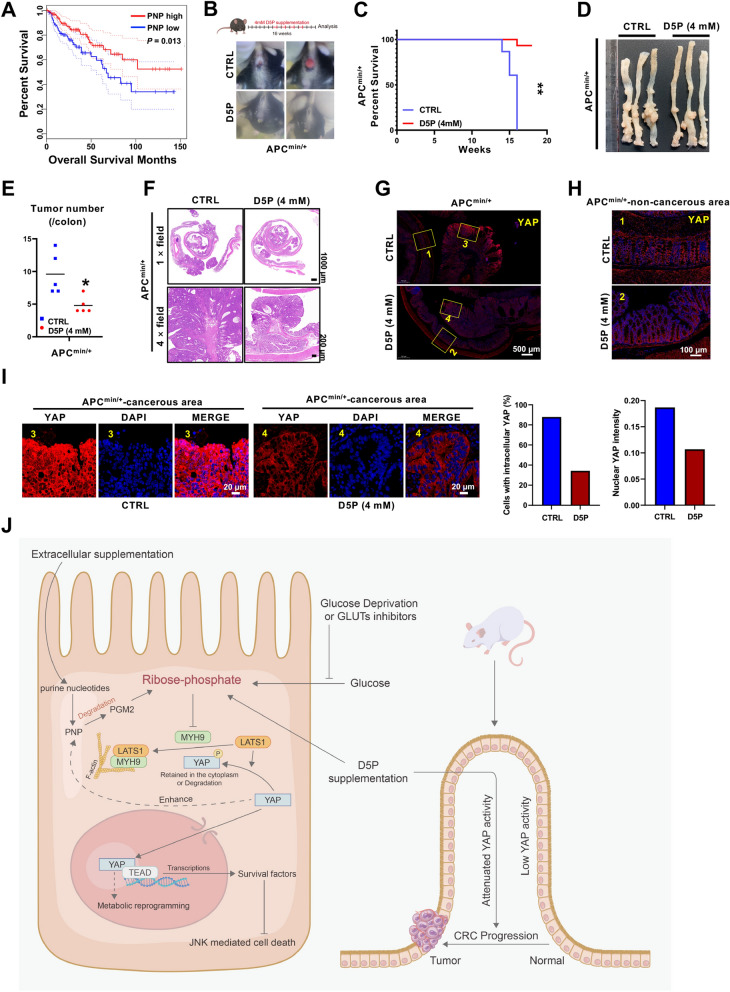
Oral administration of D5P attenuates CRC development in APC mutant mice
Although D5P can synergize with GLUT inhibitor to promote cancer cell death, there are no GLUT inhibitors approved in clinic so far [4, 5]. Therefore, we assessed the therapeutic potential of D5P as a standalone treatment for cancer. First, we investigated the clinical relevance of PNP and PGM2 in colon adenocarcinoma using the GEPIA database (http://gepia.cancer-pku.cn/) [39]. Interestingly, patients with higher PNP or PGM2 expression, especially PNP, had favorable prognosis than those with lower levels (Fig. 7A; Figure S7B), suggesting that higher PNP might inhibit cancer progression through increasing D5P level. Furthermore, we analyzed the relationship between PNP, PGM2 and YAP in another database GSE14333 [40], and found that PNP, but not PGM2, was negatively associated with YAP target genes (Figure S7C, D). To better clarify the function of the PNP-D5P axis in cancer progression, APCmin/+ mice were employed. Surprisingly, the APCmin/+ mice treated with D5P in the drinking water exhibited much less rectal prolapse and longer survival (Fig. 7B, C). Moreover, the numbers of macroscopically visible tumors were significantly decreased from APCmin/+ mice with D5P treatment (Fig. 7D, E). Importantly, haematoxylin & eosin (H&E) staining showed that the tumors in the D5P group exhibited smaller size and lower grade carcinoma (Fig. 7F), indicating an anti-cancer effect of D5P. It is worth noting that the phosphorylation modification of YAP plays an inhibitory role in regulating its protein stability and nuclear localization. Our results showed that the overall level of YAP in tumors in the control group was significantly higher than that in the D5P group (Fig. 7G, H). More importantly, the YAP nuclear localization exhibited a considerably higher intensity in the malignant cells of the control group compared to that of D5P group (Fig. 7I), indicating that anti-cancer effect of D5P might be mediated through inhibiting YAP activation. Taken together, these findings demonstrate that D5P could be a potential anti-cancer selection for patients with high YAP activity.
Fig. 7.
Oral administration of D5P attenuates CRC development in APC mutant mice. A Kaplan–Meier survival analysis of colon adenocarcinoma patients stratified by PNP expression. Data from GEPIA database. B Rectal prolapse observation showing that the onset of CRC in APCmin/+ mice treated with D5P was delayed. C Survival probability analysis of APCmin/+ mice treated with or without D5P (4 mM) in drinking water. D Representative colon images of the APCmin/+ mice treated with or without D5P (4 mM). E The quantification of tumor number presented in (D). Colon tumor number of APCmin/+ mice treated with or without D5P (4 mM). F Representative H&E staining of colon tumor from mice treated with or without D5P (4 mM). G Large field view of YAP staining in colon sections of mice treated with or without D5P (4 mM). The numeric labeled regions were zoom in and showed in (H) and (I). H A representative image of non-cancerous area in (G). YAP staining of the non-cancerous area of colon from mice treated with or without D5P (4 mM). I A representative image of cancerous area in (G) and quantitative results of YAP staining in the cancerous area of colon from mice treated with or without D5P (4 mM). J Schematic diagram showing the anti-cancer function of D5P via the MYH9/LATS1-YAP-PNP-D5P negative feedback loop. *, P < 0.05; **, P < 0.01
Discussion
In this study, we demonstrate that GD or GLUT inhibitor treatment induces YAP activation in multiple cancer cell lines and propose a negative feedback loop of MYH9/LATS1-YAP-PNP-D5P under glucose limitation (Fig. 7J). These findings suggest that D5P, a metabolite derived from both glucose metabolism and purine nucleotide degradation, functions as a metabolic checkpoint for YAP activation. Importantly, our results indicate that D5P synergizes with GLUT inhibitors to promote cancer cell death, and even D5P alone has the potential to attenuate CRC progression in APC mutant mice.
Glucose metabolism has been intensively investigated since the discovery of Otto Warburg effect in the late 1920s. The initial studies mainly focused on the cancer promoting role of glucose and the importance of glycolysis and tricarboxylic acid cycle (TCA) in cancer [41] [42]. Given the importance of glucose metabolism, targeting glucose metabolism has great potential for cancer therapy [43, 44]. Nevertheless, despite nearly 6 decades of research, targeting glucose metabolism for cancer treatment remains challenging. This fact indicates that the role of glucose metabolism in cancer is much more complicated. Recent studies implied that glucose metabolism is highly intertwined with intrinsic signaling45. Interestingly, we found that different cancer cell lines exhibited different responses to GD. GD induced YAP activation in some cells like HCT116, SW480, A549 and SK-Hep1, but not in cells like DLD1, HT29 and SW620. In line with this, glucose withdrawal has been reported to induce supra-physiological levels of phopho-tyrosine signaling in glucose withdrawal-sensitive cells, but not in insensitive cells (Graham et al. 2012). Although GD has been reported to inhibit YAP activation in non-cancer cell lines like HEK293, MCF10A and MEF cells [18, 21, 22], the different responses might potentially arise from the different primary metabolic network coupled with glucose, yet the molecular mechanism governing the divergent responses to GD remains to be elucidated.
Several studies reported that D5P could be converted to intermediates in the glycolytic pathway to fuel central carbon metabolism [46–48]. In agreement with these studies, we analyzed our metabolomic data and found that D5P supplementation also increased the metabolites of glycolytic pathway, further verified the conclusion that D5P fueled central carbon metabolism (Figure S8A). However, our findings showed that D5P enhanced the inhibitory effect of glucose deprivation, seemingly contradictory to the previous studies. We and other groups have demonstrated that GD-induced cell death is largely dependent on the accumulation of ROS [11, 49]. As shown in Figure S8A, theoretically, D5P supplementation could increase glycolytic metabolites, while having no contribution to the production of NADPH, thereby doesn’t influence GD-induced ROS-dependent cell death. Furthermore, D5P inhibits the activation of YAP, a pro-survival signal, to promote cell death under GD. Interestingly, based on the observation of cell lines tested in this study, both HCT116 and SW480 are GD-sensitive and present GD-induced YAP activation, while both DLD1 and RKO are GD-insensitive and GD has no impact on YAP activation in these cells (Figure S8B and S8C), which suggests a correlation between GD-sensitivity and YAP activation. Considering that D5P is undetectable in GD-insensitive DLD1 cells (Figure S8D), we hypothesize this correlation may be ascribed to D5P level. For GD-insensitive cells like DLD1, the absence of D5P leads to the over-activation of YAP under both normal glucose and GD, resulting in GD-resistant capacity. While for GD-sensitive cells, YAP is less activated under normal glucose due to the high level of D5P and tightly controlled by D5P-MYH/LATS1-YAP-PNP feedback loop under GD, which restricts the over-activation of YAP therefore resulting in a GD-sensitive phenotype. Meanwhile, our results showed that suppressing YAP activation by D5P treatment or shYAP significantly promoted GD-induced HCT116 cell death (Fig. 6D and Figure S8B), indicating that GD-induced YAP activation is essential for the survival of GD-sensitive cells.
Ribose-phosphates (D5P and R1P) are the central metabolites that links multiple metabolic pathways, including glucose metabolism associated PPP, de novo nucleotide synthesis, and nucleotide degradation and salvage pathway. However, the role of D5P/R1P in glucose and nucleotide metabolism has not been assessed, and the function of D5P on cancer progression is rarely studied. Here in this study, we revealed that ribose-phosphates served as signaling metabolites to attenuate GD-induced YAP activation and CRC progression in APC mutant mice by inhibiting YAP signaling. Mechanistically, D5P disrupted the interaction between MYH9 and LATS1, and further inhibited MYH9-mediated recruitment and degradation of LATS1. MYH9 is an actin cytoskeleton protein and can be targeted for autophagic degradation through its interaction with NIMA-related kinase 9 (NEK9) [35]. Recently, another study has reported that the interaction between MYH9 and actin was enhanced by GD [11], supporting that MYH9 plays an important role in GD-induced response. Our study offers significant evidence that ribose-phosphates are involved in establishing a connection between cellular signaling and metabolic networks. Furthermore, previous reports have indicated the activation of JNK by GD. Our investigation revealed that the activation of JNK induced by GD was suppressed by the supplementation of glucose, but not D5P. This suggests that the impact of D5P on YAP inactivation is specific. Interestingly, we found that in YAP-unresponding cells such as DLD1 (Figure S8D), the amount of D5P is very low and even undetectable by MS. Therefore, D5P-mediated YAP activation under GD is absent in these cells which might explain why GD cannot induce YAP activation. This result further confirmed the central role of D5P in GD-mediated YAP activation.
PNP is the key enzyme in purine nucleotide degradation and mediates the generation of Ribose-phosphate, and its insufficiency has been reported to associate both with immune deficiency and autoimmunity diseases [30, 50]. Although PNP inhibitors, such as forodesine, have been developed to treat leukemia [51], its role in solid cancer is not clear. This study investigated the impact of the PNP-D5P axis on CRC, revealing that increased PNP expression was associated with a favorable prognosis in patients, and higher PNP expression correlated negatively with YAP activity. Consistently, our findings also demonstrate that D5P, a product of PNP-mediated purine nucleoside degradation, exerts an anti-cancer effect by inhibiting YAP activation in the mouse model of CRC. All these results suggest that PNP may function as a potential clinical biomarker for selecting D5P responsive CRC patients. As PNP expression is negatively correlated with YAP activity, D5P treatment may be beneficial for patients with low PNP expression through inhibiting YAP-dependent cancer progression. Consistent with the regulatory role of PNP on YAP activation, we observed a significant increase of PNP expression in radiation-induced rectum injury (Figure S8E), which required YAP-mediated intestinal stem cell activation for injury repair, indicating that PNP-D5P-YAP axis may play roles in different biological processes.
In summary, we reveal that different cancer cells exhibit distinct metabolic preferences and respond differently to the same nutrient stress. In glucose-sensitive cells, YAP signaling would be activated to assist cancer cells to survive from glucose-limitation stress, whereas D5P supplementation specifically attenuates GD-induced YAP signaling and promote glucose limitation-induced cell death. Our research offers new insights into the regulatory mechanism of cancer cells responding to glucose limitation. Importantly, our findings propose a potential anti-cancer role of D5P, either as a standalone treatment or in combination with glucose transporter inhibitors for cancer therapy.
Supplementary Information
Acknowledgements
We gratefully thank the moral support from Dr. Lei Wang, the former vice president of the Sixth Affiliated Hospital of Sun Yat-sen University, who had been dedicated his lifetime to colorectal cancer and radiation enteritis. Though Dr. Lei Wang has away from us for 5 years, his noteworthy contribution to human health as well as his extraordinary benevolence, dauntlessness and selflessness, as recorded in our previous Lancet Digital Health and Nucleic Acids Research papers, is still engraved on our mind. Dear Dr. Wang, we miss you, very much.
Abbreviations
- GLUT
Glucose transporter
- D5P
D-ribose-5-phosphate
- PPP
Pentose phosphate pathway
- PNP
Purine nucleoside phosphorylase
- GD
Glucose deprivation
- YAP
Yes-associated protein
- LATS
Large tumor suppressors
- R1P
Ribose-1-phosphate
- PGM2
Phosphoglucomutase 2
- G3P
Glyceraldehyde-3-phosphate
- TCA
Tricarboxylic acid cycle
Author contributions
CET, YFL, XJF, XBW, and YLW conceived and designed the research study. CET, YFL and CMJ conducted the experiments and analyzed the data. HWL conducted the non-target metabolic experiments. QL provided critical advice and discussions. CET, YFL, and YLW drafted the manuscript. YLW, PL, XJF, XBW, and MCH supervised the project and wrote the final paper. All authors approved the final version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2022YFA1105300, 2022YFC2503702), the National Natural Science Foundation of China (82102924, 82225040, 82122057, 82373513), the Science and Technology R&D program of Henan province (235200810091), Science and Technology Planning Project of Hebei Academy of Sciences (24308), Science and Technology Projects in Guangzhou (202201010914), Science Fund for Distinguished Young Scholars of Guangdong Province (2021B1515020022), the Young and Middle‑Aged Health Science and Technology Innovation Talent Training Project of Henan Province (JQRC2024006), National Science and Technology Council Taiwan (NSTC 113-2639-B-039-001-ASP and T-Star Center NSTC 113-2634-F-039-001), Ministry of Health and Welfare Taiwan (MOHW113-TDU-B-222-134016), and the Featured Areas Research Center Program by the Ministry of Education (MOE) in Taiwan.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Zhengzhou University. All animal experiments were performed in compliance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and according to the institutional ethical guidelines for animal experiments.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cheng-E Tu, Yong-Feng Liu and Hong-Wei Liu contributed equally to this work.
Contributor Information
Mien-Chie Hung, Email: mhung@cmu.edu.tw.
Peng Li, Email: li-peng@mail.tsinghua.edu.cn.
Xiang-Bo Wan, Email: wanxbo@zzu.edu.cn.
Xin-Juan Fan, Email: fanxjuan@zzu.edu.cn.
Yun-Long Wang, Email: wangylong@zzu.edu.cn.
References
- 1.Ohshima K, Morii E. Metabolic reprogramming of cancer cells during tumor progression and metastasis. Metabolites. 2021;11(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sokolov D, Sullivan LB. A metabolic signaling role for arginine in liver cancer. Life Metab. 2023; 3(1): load046. [DOI] [PMC free article] [PubMed]
- 4.Chen X, Zhao Y, He C, Gao G, Li J, Qiu L, et al. Identification of a novel GLUT1 inhibitor with in vitro and in vivo anti-tumor activity. Int J Biol Macromol. 2022;216:768–78. [DOI] [PubMed] [Google Scholar]
- 5.Katt WP, Lukey MJ, Cerione RA. Starving the devourer: cutting cancer off from its favorite foods. Cell Chem Biol. 2019;26(9):1197–9. [DOI] [PubMed] [Google Scholar]
- 6.Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. 2020;577(7788):115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, et al. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol. 2015;17(12):1556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham NA, Tahmasian M, Kohli B, Komisopoulou E, Zhu M, Vivanco I, et al. Glucose deprivation activates a metabolic and signaling amplification loop leading to cell death. Mol Syst Biol. 2012;8:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J, DeBerardinis RJ. Mechanisms and implications of metabolic heterogeneity in cancer. Cell Metab. 2019;30(3):434–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du X, Hu Y, Huang G, Wei F. The metabolic adaptation in wild vertebrates via omics approaches. Life Metab. 2022;1(3):234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Nie L, Zhang Y, Yan Y, Wang C, Colic M, et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat Cell Biol. 2023;25(3):404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474(7350):179–83. [DOI] [PubMed] [Google Scholar]
- 13.Yuan WC, Pepe-Mooney B, Galli GG, Dill MT, Huang HT, Hao M, et al. NUAK2 is a critical YAP target in liver cancer. Nat Commun. 2018;9(1):4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashiro Y, Thang BQ, Ramirez K, Shin SJ, Kohata T, Ohata S, et al. Matrix mechanotransduction mediated by thrombospondin-1/integrin/YAP in the vascular remodeling. Proc Natl Acad Sci U S A. 2020;117(18):9896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo JH, Guan KL. Interplay between YAP/TAZ and metabolism. Cell Metab. 2018;28(2):196–206. [DOI] [PubMed] [Google Scholar]
- 16.Peng C, Zhu Y, Zhang W, Liao Q, Chen Y, Zhao X, et al. Regulation of the Hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol Cell. 2017;68(3):591-604 e5. [DOI] [PubMed] [Google Scholar]
- 17.Enzo E, Santinon G, Pocaterra A, Aragona M, Bresolin S, Forcato M, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34(10):1349–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mo JS, Meng Z, Kim YC, Park HW, Hansen CG, Kim S, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol. 2015;17(4):500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Qiao Y, Wu Q, Chen Y, Zou S, Liu X, et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun. 2017;8:15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ortillon J, Le Bail JC, Villard E, Leger B, Poirier B, Girardot C, et al. High glucose activates YAP signaling to promote vascular inflammation. Front Physiol. 2021;12:665994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeRan M, Yang J, Shen CH, Peters EC, Fitamant J, Chan P, et al. Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 2014;9(2):495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, Xiao ZD, Li X, Aziz KE, Gan B, Johnson RL, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat Cell Biol. 2015;17(4):490–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camici M, Tozzi MG, Ipata PL. Methods for the determination of intracellular levels of ribose phosphates. J Biochem Biophys Methods. 2006;68(3):145–54. [DOI] [PubMed] [Google Scholar]
- 24.Maliekal P, Sokolova T, Vertommen D, Veiga-da-Cunha M, Van Schaftingen E. Molecular identification of mammalian phosphopentomutase and glucose-1,6-bisphosphate synthase, two members of the alpha-D-phosphohexomutase family. J Biol Chem. 2007;282(44):31844–51. [DOI] [PubMed] [Google Scholar]
- 25.Mascia L, Cappiello M, Cherri S, Ipata PL. In vitro recycling of alpha-D-ribose 1-phosphate for the salvage of purine bases. Biochim Biophys Acta. 2000;1474(1):70–4. [DOI] [PubMed] [Google Scholar]
- 26.Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, et al. Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. Cancer Res. 2007;67(7):3345–55. [DOI] [PubMed] [Google Scholar]
- 27.Kim JW, Kim MJ, Kim KJ, Yun HJ, Chae JS, Hwang SG, et al. Notch interferes with the scaffold function of JNK-interacting protein 1 to inhibit the JNK signaling pathway. Proc Natl Acad Sci U S A. 2005;102(40):14308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavel M, Renna M, Park SJ, Menzies FM, Ricketts T, Fullgrabe J, et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat Commun. 2018;9(1):2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang YP, Lei QY. Metabolite sensing and signaling in cell metabolism. Signal Transduct Target Ther. 2018;3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abt ER, Rashid K, Le TM, Li S, Lee HR, Lok V, et al. Purine nucleoside phosphorylase enables dual metabolic checkpoints that prevent T cell immunodeficiency and TLR7-associated autoimmunity. J Clin Investig. 2022;132(16): e160852. [DOI] [PMC free article] [PubMed]
- 31.Simoni RE, Gomes LN, Scalco FB, Oliveira CP, Aquino Neto FR, de Oliveira ML. Uric acid changes in urine and plasma: an effective tool in screening for purine inborn errors of metabolism and other pathological conditions. J Inherit Metab Dis. 2007;30(3):295–309. [DOI] [PubMed] [Google Scholar]
- 32.Caspi R, Billington R, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Smolen GA, Haber DA. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68(8):2789–94. [DOI] [PubMed] [Google Scholar]
- 34.Bjorkoy G, Lamark T, Johansen T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006;2(2):138–9. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, Chino H, Tsukamoto S, Ode KL, Ueda HR, Mizushima N. NEK9 regulates primary cilia formation by acting as a selective autophagy adaptor for MYH9/myosin IIA. Nat Commun. 2021;12(1):3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song JJ, Lee YJ. Differential activation of the JNK signal pathway by UV irradiation and glucose deprivation. Cell Signal. 2007;19(3):563–72. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Luo JY, Li B, Tian XY, Chen LJ, Huang Y, et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature. 2016;540(7634):579–82. [DOI] [PubMed] [Google Scholar]
- 38.Koppula P, Olszewski K, Zhang Y, Kondiparthi L, Liu X, Lei G, et al. KEAP1 deficiency drives glucose dependency and sensitizes lung cancer cells and tumors to GLUT inhibition. iScience. 2021;24(6):102649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15(24):7642–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–14. [DOI] [PubMed] [Google Scholar]
- 42.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551(7678):115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng B, Geng L, Zeng L, Liu F, Huang Q. AKT2 contributes to increase ovarian cancer cell migration and invasion through the AKT2-PKM2-STAT3/NF-kappaB axis. Cell Signal. 2018;45:122–31. [DOI] [PubMed] [Google Scholar]
- 44.Tornatore L, Thotakura AK, Bennett J, Moretti M, Franzoso G. The nuclear factor kappa B signaling pathway: integrating metabolism with inflammation. Trends Cell Biol. 2012;22(11):557–66. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Wei X, Xiong J, Feng JW, Zhang CS, Lin SC. Hierarchical inhibition of mTORC1 by glucose starvation-triggered AXIN lysosomal translocation and by AMPK. Life Metab. 2023; 2(3): load005. [DOI] [PMC free article] [PubMed]
- 46.Nwosu ZC, Ward MH, Sajjakulnukit P, Poudel P, Ragulan C, Kasperek S, et al. Uridine-derived ribose fuels glucose-restricted pancreatic cancer. Nature. 2023;618(7963):151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skinner OS, Blanco-Fernández J, Goodman RP, Kawakami A, Shen H, Kemény LV, et al. Salvage of ribose from uridine or RNA supports glycolysis in nutrient-limited conditions. Nat Metab. 2023;5(5):765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang T, Gnanaprakasam JNR, Chen X, Kang S, Xu X, Sun H, et al. Inosine is an alternative carbon source for CD8(+)-T-cell function under glucose restriction. Nat Metab. 2020;2(7):635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Tu C, Guo X, Wu C, Gu C, Lai Q, et al. Tumor-suppressive function of EZH2 is through inhibiting glutaminase. Cell Death Dis. 2021;12(11):975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grunebaum E, Cohen A, Roifman CM. Recent advances in understanding and managing adenosine deaminase and purine nucleoside phosphorylase deficiencies. Curr Opin Allergy Clin Immunol. 2013;13(6):630–8. [DOI] [PubMed] [Google Scholar]
- 51.Balakrishnan K, Verma D, O’Brien S, Kilpatrick JM, Chen Y, Tyler BF, et al. Phase 2 and pharmacodynamic study of oral forodesine in patients with advanced, fludarabine-treated chronic lymphocytic leukemia. Blood. 2010;116(6):886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.