Abstract
Background
The emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae (CRKP) is a significant public health concern, as colistin has been the last resort for treating such infections. This study aimed to investigate the prevalence and molecular characteristics of colistin-resistant CRKP isolates in Central South China.
Methods
CRKP isolates from twelve hospitals in Central South China were screened for colistin resistance using broth microdilution. The epidemiological characteristics, virulome, resistome, plasmid replicons and two-component systems associated with colistin resistance of colistin-resistant isolates were explored by whole-genome sequencing. The mgrB gene and the relative expression of the pmrC and pmrK genes were analyzed by polymerase chain reaction (PCR) and real-time quantitative PCR, respectively. The bacterial virulence was evaluated through a Galleria mellonella larvae infection model.
Results
Of the 429 nonduplicate CRKP isolates, 26 (6.1%) were colistin-resistant and they included eight clonal clusters. Six distinct sequence type (ST)-capsule loci (KL) types were identified: ST11-KL64, ST11-KL47, ST963-KL16, ST307-KL102, ST751-KL64 and ST5254-KL47. 88.5% (23/26) of them were found to carry at least one carbapenemase gene, including blaKPC−2 (65.4%, 17/26) and blaNDM−1 (7.7%, 2/26), as well as coharbouring blaKPC−2 and blaNDM−1 (15.4%, 4/26). Diverse mutations of colistin resistance-related genes were observed, with mgrB inactivation by insertions and the T157P deleterious mutation in pmrB being detected in 57.7% and 42.3% of the colistin-resistant isolates, respectively. In addition, a novel deleterious mutation, R248P, in the crrB gene was found in two ST11 isolates. 88.5% of the 26 isolates presented an increase in pmrK transcription, and 69.2% of them had an overexpression of the pmrC gene. All the 16 ST11-KL64 isolates and one ST751-KL64 isolate (65.4%, 17/26) carried at least two hypervirulence biomarkers and showed high virulence in vivo.
Conclusions
This study highlights the presence of different colistin resistance mechanisms in isolates belonging to the same clone and identified multiple clonal transmission clusters in colistin resistant isolates, including the globally high-risk ST11 and ST307 clones, of which a significant proportion exhibited high virulence. Consequently, it is crucial to enforce measures to prevent the ongoing spread of colistin resistance.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12941-024-00769-1.
Keywords: Colistin resistance, CRKP, Whole-genome sequencing, MgrB, Hypervirulence, Central South China
Background
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is a persistent threat to human health worldwide due to its multidrug resistance and ability to cause various infections with high morbidity and mortality rates [1]. The worldwide spread of CRKP is mainly linked to plasmid-mediated dissemination of carbapenemase-encoding genes, particularly, K. pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), and oxacillinase-48 (OXA-48) [2]. Currently, available data indicate that the popular sequence types (STs) among CRKP are ST258, ST11, ST307, ST15 and ST147 [3], with ST11 being the most common type in Asia and South America [4, 5]. Moreover, hypervirulent CRKP (hv-CRKP) has emerged through the continuous evolution of plasmids with hypervirulence or carbapenem resistance and can lead to more serious infections and higher mortality rates [6].
Colistin is considered one of the last-resort treatment options for CRKP and hv-CRKP [7]. It was approved for the treatment of gram-negative bacterial (GNB) infections in the 1960s and was largely prohibited in human medicine in the 1970s owing to its adverse effects, primarily nephrotoxicity and neurotoxicity [8]. However, in recent years, colistin has been reintroduced into the clinical setting to eradicate multiresistant GNB. Unfortunately, with the increasing use of colistin in the livestock industry and hospitals, the worldwide emergence and dissemination of colistin-resistant CRKP (ColR-CRKP), even hypervirulent ColR-CRKP poses a great challenge to global public health [9, 10]. A high mortality rate of 69.3% in bloodstream infections caused by ColR-CRKP was observed among Indian patients [11]. The colistin resistance rate among CRKP isolates ranges from 13 to 61% in Germany, the US, South Africa and India [12–15]. In addition, the prevalence rate of colistin resistance in CRKP from mainland China was 11.8% in 2023, according to data from the China Antimicrobial Surveillance Network (CHINET, https://www.chinets.com/).
Colistin resistance in K. pneumoniae is mainly chromosomally mediated and facilitated by various distinct mechanisms, particularly lipopolysaccharide (LPS) modification [16–18]. The chromosomal mechanism includes mutational alterations in the two-component regulatory systems (TCSs), including PmrA/B, PhoP/Q and CrrA/B, and MgrB, which is a negative regulator of the PhoP/Q system [16, 17]. Recently, mobile colistin resistance (mcr) genes (mcr-1 to mcr-10) have spread globally, further aggravating the situation of colistin resistance [19]. Owing to the complex and diverse nature of colistin resistance mechanisms, efforts to expand our understanding of these mechanisms and identify potential targets for future antimicrobial therapies are highly essential.
The current study aimed to determine the prevalence and molecular characteristics of colistin-resistant isolates among CRKP isolates collected from twelve tertiary hospitals in Central South China.
Materials and methods
Bacterial isolates and identification
We evaluated 429 nonduplicate CRKP clinical isolates retrieved from 12 hospitals in Central South China from January to December 2021 (Fig. S1). Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics GmbH, Bremen, Germany) was used to identify the isolates at the species level.
Antimicrobial susceptibility testing
Susceptibility tests of CRKP isolates to carbapenems (imipenem, meropenem and ertapenem ) were performed using Kirby-Bauer disk diffusion method with disks from Oxoid (Hampshire, UK) according to the 2022 Clinical and Laboratories Standard Institute (CLSI) recommendations [20]. CRKP isolates were screened for colistin resistance by the standard broth microdilution (BMD) method using cation-adjusted Mueller Hinton broth and colistin (Sigma-Aldrich, St. Louis, MO) following the protocols of CLSI M100-S32, which suggest that the resistance minimum inhibitory concentration (MIC) breakpoint of colistin is ≥ 4 mg/L for Enterobacterales [20].
The VITEK 2 compact system (BioMérieux, Marcy-l’Étoile, France) was used to evaluate the antimicrobial susceptibility of the ColR-CRKP isolates, with the exception of tigecycline and ceftazidime/avibactam. The antimicrobial agents tested in the automated analyzer included extended-spectrum cephalosporins (ceftriaxone and cefepime), enzyme-inhibitor complexes (piperacillin/tazobactam), monobactam (aztreonam), carbapenems (imipenem and ertapenem), aminoglycosides (amikacin, gentamicin and tobramycin), fluoroquinolones (ciprofloxacin and levofloxacin) and folate pathway inhibitors (trimethoprim/sulfamethoxazole). Bacterial susceptibility to tigecycline and ceftazidime/avibactam was assessed by the BMD method according to the CLSI M100-S32 guidelines [20]. The susceptibility results were interpreted following the CLSI breakpoints, with the exception of tigecycline, which is based on the Food and Drug Administration (FDA) breakpoints for Enterobacteriaceae available at http://www.fda.gov.E. coli ATCC 25,922 served as the quality control isolate. The ColR-CRKP isolates are categorized as multidrug resistant(MDR), extensively drug resistant“XDR”and pandrug resistant“PDR” [21].
Clinical Data Collection
The demographic information, admission ward and clinical specimens of the total patients infected with CRKP isolates were obtained from the electronic medical record system. More clinical details were collected for each patient infected with ColR-CRKP, such as type of infection, length of stay before the isolation of ColR-CRKP, hospitalization days, underlying disease and prior exposure to antimicrobials and the outcome.
Whole-genome sequencing (WGS)
Genomic DNA was extracted from fresh cultures of all the ColR-CRKP isolates using TIANamp Bacteria DNA Kit (catalog no. DP302; Tiangen Biotech, Beijing, China) following the manufacturer’s protocol. A Nanodrop-1000 spectrophotometer and 0.8% agarose gel were used to measure the concentration and quality of the DNA samples. DNA library preparation was performed according to the manufacturer’s protocol (VAHTS Universal DNA Library Prep Kit for Illumina V3, catalog no. ND607; Vazyme, Nanjing China). The libraries were subsequently sequenced on the BGI DNBSEQ-T7 platform, and the raw reads in FASTQ format were generated for downstream analysis.
Bioinformatic analysis
The version and website information of the software and databases used for bioinformatic analysis was available in Table S1. The raw data underwent filtration using fastp software, followed by quality control of the clean data with FastQC. Then, the data were assembled and corrected utilizing Unicycler to obtain the final genome sequence. The quality of the sequenced genome was evaluated by Samtools for sequencing depth analysis and the Checkm for genome completeness. Finally, genome annotation was performed using prodial.
Multilocus sequence typing (MLST) was identified using the Bacterial Isolate Genome Sequence Database (BIGSdb). Capsule locis (KLs) were analyzed using Kleborate with Kaptive. Virulence genes were detected using the virulence factor database (VFDB) and the virulence-associated integrative conjugative element of K. pneumoniae (ICEKp) was identified by Kleborate. Antimicrobial resistance determinants were predicted by Comprehensive Antibiotic Resistance Database (CARD) and plasmid replicons were identified by PlasmidFinder.
A phylogenetic tree of the ColR-CRKP isolates in this study was constructed by BacWGSTdb based on single nucleotide polymorphism (SNP) strategy and visualized using MEGA v7.0.26 [22]. The pairwise SNP distances were calculated using SNP-dist. The putative transmission events between ColR-CRKP isolates were identified on the bases of pairwise SNPs with a threshold of 21 SNPs [23].
Molecular characterization of genes related to colistin resistance
The amino acid mutations in colistin resistance-related sequences of TCSs (PmrA/B, PhoP/Q and CrrA/B) were defined by alignment with K. pneumoniae MGH 78,578 (NC_009648.1) with Snippy. The phenotypic effects of these mutations on protein biological functions were predicted with protein variation effect analyzer (PROVEAN). If the PROVEAN score is equal to or below the threshold of -2.5, the protein variant indicates a “deleterious” effect, whereas a score above − 2.5 indicates “neutral”.
As reported earlier, the regions within and around the mgrB gene, an important regulator of the PhoP/Q system, cannot be fully sequenced by WGS, polymerase chain reaction (PCR) using specific primers was performed to investigate the sequence alterations of mgrB gene [24, 25]. The PCR products were Sanger sequenced on an ABI 3730XL DNA analyzer (Life Technologies, USA). The nucleotide sequences were compared to those of the colistin-susceptible strain K. pneumoniae MGH 78,578. The ISfinder database platform was used to identify the insertion sequence (IS) elements.
To determine whether the upregulation of pmrCAB and pmrHFIJKLM operons mediates colistin resistance, the relative expression levels of the pmrC gene encoding for phosphoethanolamine (pEtN) transferase and the pmrK gene encoding for 4-amino-4-deoxy-l-arabinose (L-Ara4N) transferase were assessed by real-time quantitative PCR (RT-qPCR). Total RNA was extracted from bacterial cultures in the mid-log phase using RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, Beijing, China). The cDNA was synthesized using the HiScript® II Q RT SuperMix for qPCR (+ gDNA wiper) Kit (Vazyme, China). RT-qPCR was carried out with ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) on an ABI Q5 real-time PCR system (Applied Biosystems, Thermo Fisher Scientific) under the following conditions: 1 cycle at 95 °C for 30 s, 40 cycles at 95 °C for 10 s and 60 °C for 30 s. For each run, melt curve analysis was performed to ensure that a single product was synthesized. Melting curves were generated at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. Relative expression levels of the target genes were calculated using the 2−ΔΔCT formula. The internal controls were based on the ribosomal gene rpsL. The specific oligonucleotide primers of pmrC, pmrK and rpsL genes were synthesized based on previously published sequences [26].
Virulence phenotypic analysis
The infection model of G. mellonella larvae was used to evaluate the in vivo virulence of the ColR-CRKP isolates as described previously [27]. For each isolate, 10 larvae were randomly selected and injected into their second left gastropod with 10 µl of bacterial suspension at a concentration of 1 × 108 CFU/mL. Hypervirulent K. pneumoniae NTHU-K2044, classic K. pneumoniae ATCC700603 and sterile phosphate-buffered saline (PBS) were used as positive, negative and blank controls, respectively. The larvae were then incubated in a dark environment at 37 °C in sterile petri dishes, and the three-day survival rate was determined every 12 h. The tests were repeated three times. Hypervirulent K. pneumoniae (hvKp) was defined based on both virulence factors (rmpA, rmpA2, iucA, iroB or peg-344) and the G. mellonella infection model [28, 29].
Statistical analysis
The relative expression levels of the pmrC and pmrK genes were presented as the means ± standard deviations from three independent experiments. The survival distributions of the G. mellonella infection models were estimated by the Kaplan-Meier method, and the survival curve differences were assessed by the log-rank (Mantel-Cox) test using GraphPad Prism 9.0.
Results
Sources of CRKP isolates
The 429 CRKP isolates were recovered from 321 male and 108 female patients aged 0–95 years (mean 38.7 years). The predominant department where the patients were located was central intensive care unit (ICU) (21.9%), followed by respiratory ICU (8.2%), neurosurgical ward (7.5%) and neurosurgical ICU(7.2%). The isolates were isolated from a wide variety of clinical specimens, mainly from lower respiratory tract (56.2%), urine tract (15.2%) and blood (14.7%). The general information of the patients infected with CRKP was shown in Table S2.
Colistin MIC distribution and antimicrobial susceptibility
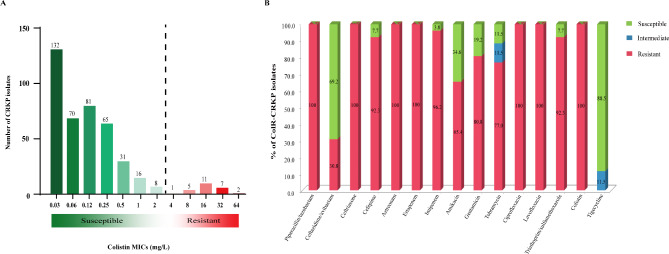
Colistin susceptibility testing revealed that 403 isolates (93.9%) were susceptible to colistin (≤ 2 mg/L), whereas 26 isolates (6.1%) showed colistin resistance, with the MIC ranging from 4 to 64 mg/L (median: 16 mg/L) (Fig. 1A). Isolates from Changsha had a colistin resistance rate of 7.0% (21/301), containing more than three quarters of the ColR-CRKP isolates in this study (21/26, 80.8%). Two colistin-resistant isolates were detected in Chenzhou, one was detected in Hengyang, Yongzhou and Zhuzhou, respectively (Fig. S1).
Fig. 1.
Antibiotic susceptibility results. A Distribution of colistin MIC results for the 429 CRKP isolates in this study; B Results of the antimicrobial sensitivity test for the ColR-CRKP isolates
As shown in Fig. 1B, All 26 ColR-CRKP isolates were resistant to piperacillin-tazobactam, ceftriaxone, aztreonam, ertapenem, ciprofloxacin and levofloxacin. Moreover, 96.1% (n = 25), 92.3% (n = 24), 92.3% (n = 24), 80.8% (n = 21), 77.0% (n = 20) and 65.4% (n = 17) were resistant to imipenem, cefepime, trimethoprim/sulfamethoxazole, gentamicin, tobramycin and amikacin, respectively. While they exhibited striking resistance to 12/14 of the tested antimicrobial agents, ceftazidime/avibactam and tigecycline were effective against 69.2% (n = 18) and 88.5% (n = 23) of the ColR-CRKP isolates, respectively. 73.1% (n = 19) of the ColR-CRKP isolates showed the XDR phenotype, and a smaller proportion (26.9%, n = 7) of them presented the MDR phenotype.
Clinical details of patients with ColR-CRKP infections
Twenty-six ColR-CRKP isolates were collected from diverse clinical specimens, including sputum (n = 9), urine (n = 7), blood (n = 3), bronchial secretions (n = 3), cerebrospinal fluid (n = 2), pleural fluid (n = 1) and bronchoalveolar lavage fluid (n = 1). Sixteen male and ten female hospitalized patients aged 16–86 years (mean 51.3 years) were infected with ColR-CRKP in our study. The clinical details of the infected patients were shown in Table 1. Seventeen patients (65.4%) were admitted to different intensive care unit (ICU) departments, including the central ICU (n = 9), neurosurgical ICU (n = 4), neurology ICU (n = 2), respiratory ICU (n = 1) and emergency ICU (n = 1). The most common infection type was lung infection (14/26, 53.8%), followed by the urinary tract infection (7/26, 27.0%). Fifteen patients (57.7%) had brain diseases. The mean duration of hospital stay before ColR-CRKP isolation was 31.5 days. The patients were previously treated with an average of three kinds of antibiotics before ColR-CRKP isolation, and 14 patients (53.8%) had prior exposure to polymyxin B. Ten patients (38.5%) experienced fatal outcomes with the failure of antibiotic treatment.
Table 1.
Clinical characteristics of 26 patients infected with ColR-CRKP isolates
| Clinical Characteristics | Results |
|---|---|
| Mean age (Min-Max) | 51.3 (16–86) |
| Gender | n (%) |
| Male | 16 (61.5) |
| Female | 10 (38.5) |
| Admission Unit | n (%) |
| Intensive care unit (ICU) | 9 (34.6) |
| Neurosurgical ICU | 5 (19.2) |
| Haematology ward | 3 (11.5) |
| Neurology ICU | 2 (7.7) |
| Rehabilitation medicine | 2 (7.7) |
| Respiratory ICU | 1 (3.8) |
| Emergency ICU | 1 (3.8) |
| Others | 3 (11.5) |
| Hospitalization days median (Min-Max) | 51.3 (13–231) |
| Underlying disease | n (%) |
| Brain disease | 15 (57.7) |
| Severe pneumonia | 4 (15.4) |
| Hematologic malignancy | 3 (11.5) |
| Liver disease | 2 (7.7) |
| Kidney disease | 2 (7.7) |
| Healthcare-associated infections | n (%) |
| Lower respiratory tract infection | 10 (38.5) |
| Urinary tract infection | 7 (26.9) |
| Ventilator-associated pneumonia | 4 (15.4) |
| Blood stream infection | 3 (11.5) |
| Central nervous system infection | 2 (7.7) |
| Prior exposure to antimicrobials | n (%) |
| Carbapenems | 20 (77.0) |
| beta-lactam/beta-lactamase inhibitor | 17 (65.4) |
| Polymyxin B | 14 (53.8) |
| Fluoroquinolone | 7 (26.9) |
| Cephalosporins | 7 (26.9) |
| Tigecycline | 6 (23.1) |
| Aminoglycosides | 4 (15.4) |
| Ceftazidime-avibactam | 1 (3.8) |
| Outcomes | n (%) |
| Improved | 16 (61.5) |
| Gave up | 7 (26.9) |
| Death | 3 (11.5) |
Epidemiological characteristics of ColR-CRKP isolates
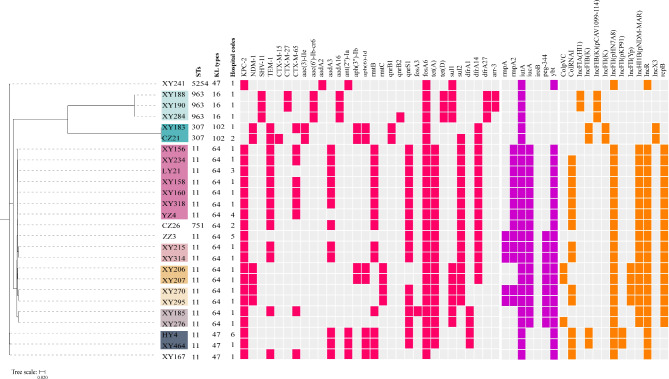
WGS short-read sequencing was performed for the 26 ColR-CRKP isolates. According to in silico MLST analysis using the genome sequence data, five distinct sequence types (STs) featured the studied isolates: ST11 (19/26), ST963 (3/26), ST307 (2/26), ST751 (1/26) and ST5254 (1/26) (Fig. 2, Table S3). Among them, ST963 has not been reported before, whereas ST751 and ST5254 are rare. ST751 and ST5254 were single-locus variants of ST11 in rpoB gene and phoE gene, respectively. These data suggest that ST751 and ST5254 may be derived from ST11. Four different KL types were identified: KL64 (17/26), KL47 (4/26), KL16 (3/26) and KL102 (2/26). Among the nineteen ST11 isolates, sixteen (84.2%, 16/19) harbored KL64, while the other three (15.8%, 3/19) harbored KL47. Interestingly, KL64 and KL47 were not exclusively present in ST11, with strain CZ26 belonging to ST751-KL64 and strain XY241 belonging to ST5254-KL47. The ST963 and ST307 isolates were KL16 and KL102, respectively.
Fig. 2.
Phylogenetic relationships and profiles of antimicrobial resistance genes, virulence factors and plasmid replicons of the 26 ColR-CRKP isolates in this study. The phylogenetic tree was constructed based on single nucleotide polymorphism (SNP) strategy with ST11 K. pneumoniae HS11286 (GenBank accession number CP003200) as a reference. The isolates labeled with the same colorful background belonged to the same cluster, with a SNP threshold of 21 between isolates. The presence of antimicrobial genes, virulence factors and plasmid replicons in each isolate is indicated by filled rose red, purple and orange squares, respectively. STs, KL types and hospital codes are also shown in this figure
Phylogenetic relatedness analysis revealed eight putative transmission events in this study, on the bases of a cut-off of 21 SNPs between isolates (Fig. 2, Table S4). Six clones among the nineteen ST11 isolates were disseminated within hospital wards or between hospitals, with the largest clone comprising seven isolates from three different hospitals. The two ST307 isolates (XY183 and CZ21) from different hospitals were highly clonal, differing by 2 SNPs. The three ST963 isolates (XY188, XY190 and XY284) from different wards of the same hospital were also highly clonal, with SNP differences ranging from 2 to 7.
Antimicrobial resistance determinants of ColR-CRKP isolates
Multiple acquired antibiotic resistance genes (ARGs) were identified among the 26 ColR-CRKP genomes in this study using the CARD database (Fig. 2), which were consistent with the XDR and MDR phenotypes of the isolates. All the ColR-CRKP isolates were carbapenemase producers except for the three ST963 isolates, which included blaKPC-2 (65.4%, 17/26), blaNDM-1 (7.7%, 2/26), and coharbouring blaKPC-2 and blaNDM-1 (15.4%, 4/26). The extended-spectrum β-lactamase (ESBL) genes blaCTX-M-15, blaCTX-M-27 and blaCTX-M-65 were found in 46.2% (12/26) of the studied isolates, with blaCTX-M-65 (34.6%, 9/26) being the most commonly detected gene. Other β-lactamase genes, blaTEM-1 and blaSHV-11, were also detected in 53.9% (14/26) and 11.5% (3/26) of the isolates, respectively. Ten genes contributed to aminoglycoside resistance in our study: aac(3)-IIe, aac(6’)-Ib-cr6, aadA2, aadA3, aadA16, ant(2’’)-Ia, aph(3’’)-Ib, aph(6)-Id, rmtB and rmtC. Among these genes, rmtB was the most common (n = 14), followed by aadA3 (n = 12). Moreover, genes responsible for fluoroquinolone resistance (qnrB1, qnrB2 and qnrS1) were detected in 20 isolates, with qnrS1 being the most abundant gene (n = 17). Different sul and dfrA variants coexisted in 76.9% of the isolates conferring resistance to sulfamethoxazole and trimethoprim, respectively. The sul2 gene exhibited higher detection among the isolates, reaching a prevalence of 61.5%, compared to sul1 being detected in 38.5% of the isolates. Additionally, other ARGs were identified for various classes of antimicrobials such as tet (A) and tet (D), which mediat resistance to tetracycline; fosA3 and fosA6, which encode fosfomycin resistance; and arr-3, which encods rifamycin resistance. However, mcr genes (mcr-1 to mcr-10) were absent in our study.
Molecular mechanism of chromosome-mediated colistin resistance
As the mcr genes were not detected among the ColR-CRKP isolates, we further studied potential chromosomal mechanisms that caused resistance to colistin. Sequence analysis of pmrA, pmrB, phoP, phoQ, crrA and crrB genes of the isolates revealed missense nucleotide mutations at four different positions of pmrB gene, three positions of crrB gene, two positions of pmrA gene and one position of phoP gene (Table 2, Table S5). Of these mutations, two in pmrB (R256G and T157P), one in pmrA (G53S) and one in crrB (R248P) were predicted as deleterious mutations by the PROVEAN tool (Table S5). The most common deleterious mutation was R256G amino acid substitution of pmrB in 92.3% (24/26) of the isolates, followed by T157P substitution of pmrB in 42.3% (11/26) of the isolates. Interestingly, the mutation R248P (G743C) of crrB in two ST11 isolates (XY206 and LY21) was not reported earlier. The sequences of phoQ and crrA genes of all the 26 ColR-CRKP isolates showed no differences with the wild-type sequences from colistin-susceptible reference strain (K. pneumoniae MGH78578).
Table 2.
Colistin MICs, chromosomal mutations in genes related to colistin resistance and relative expression levels of the pmrC and pmrK genes
| Strains | COL MIC (mg/L) |
Chromosomal mutations related to colistin resistance# | Relative expression level* | |||||
|---|---|---|---|---|---|---|---|---|
| mgrB | pmrA | pmrB | phoP | crrB | pmrC | pmrK | ||
| XY156 | 16 | IS903B | WT | R256G T246A | WT | WT | 1.28 ± 0.08 | 18.46 ± 1.43 |
| XY158 | 32 | WT | WT | R256G T246A T157P | WT | WT | 5.63 ± 0.58 | 2.60 ± 0.21 |
| XY160 | 16 | WT | G53S | R256G T246A T157P | WT | WT | 7.56 ± 0.78 | 1.93 ± 0.19 |
| XY167 | 16 | WT | WT | R256G T246A T157P | WT | WT | 13.47 ± 0.88 | 2.35 ± 0.16 |
| XY183 | 4 | WT | A41T | T246A L213M T157P | E82K | C68S | 14.67 ± 0.84 | 1.25 ± 0.20 |
| XY185 | 16 | WT | WT | R256G T246A T157P | WT | WT | 31.19 ± 2.00 | -a |
| XY188 | 16 | IS903B | WT | R256G T246A | WT | WT | 1.58 ± 0.22 | 11.80 ± 1.01 |
| XY190 | 16 | IS903B | WT | R256G T246A | WT | I27V C68S | 1.25 ± 0.07 | 15.08 ± 1.53 |
| XY206 | 16 | ISKpn74 | WT | R256G T246A | WT | R248P ▲ | -a | 4.87 ± 0.17 |
| XY207 | 16 | ISKpn74 | WT | R256G T246A | WT | WT | -a | 5.38 ± 0.40 |
| XY215 | 32 | ISKpn26 | G53S | R256G T246A | WT | WT | 1.68 ± 0.09 | 6.72 ± 0.48 |
| XY234 | 16 | ISKpn26 | WT | R256G T246A | WT | WT | -a | 10.86 ± 0.88 |
| XY241 | 8 | ISKpn26 | WT | R256G T246A | WT | WT | -a | 5.44 ± 0.38 |
| XY270 | 16 | WT | WT | R256G T246A T157P | WT | WT | 11.58 ± 0.85 | 1.82 ± 0.08 |
| XY276 | 32 | ISKpn26 | WT | R256G T246A | WT | WT | 1.89 ± 0.18 | 7.21 ± 0.52 |
| XY284 | 8 | ISKpn74 | G53S | R256G T246A | WT | I27V C68S | 1.30 ± 0.06 | 4.06 ± 0.30 |
| XY295 | 8 | WT | WT | R256G T246A T157P | WT | WT | 8.18 ± 0.51 | -a |
| XY314 | 64 | ISKpn26 | WT | R256G T246A | WT | WT | 1.10 ± 0.05 | 7.62 ± 0.79 |
| XY318 | 32 | WT | WT | R256G T246A T157P | WT | WT | 10.81 ± 1.00 | -a |
| XY464 | 64 | ISKpn14 | WT | R256G T246A | WT | WT | -a | 5.42 ± 0.25 |
| CZ21 | 8 | WT | A41T | T246A L213M T157P | E82K | C68S | 3.48 ± 0.21 | 2.58 ± 0.13 |
| CZ26 | 8 | WT | WT | R256G T246A T157P | WT | WT | 14.33 ± 1.09 | 1.55 ± 0.16 |
| HY4 | 64 | ISKpn14 | WT | R256G T246A | WT | WT | -a | 6.20 ± 0.28 |
| LY21 | 8 | WT | WT | R256G T246A T157P | WT | R248P ▲ | 8.73 ± 0.50 | 1.92 ± 0.29 |
| YZ4 | 32 | ISKpn14 | WT | R256G T246A | WT | WT | -a | 7.00 ± 0.16 |
| ZZ3 | 64 | ISKpn26 | WT | R256G T246A | WT | WT | -a | 4.91 ± 0.35 |
Abbreviations: COL: colistin; MIC, minimal inhibitory concentration; mg/L, milligram per liter; WT: wild-type
# Colistin-susceptible K. pneumoniae MGH 78,578 (NC_009648.1) was used as reference for mutational analysis. The insertion mutations of mgrB and missense mutations of two-component regulatory system genes were listed in this table. The mutations predicted as deleterious by PROVEAN were in bold and the novel mutation was marked with ▲
* Relative fold change of pmrC and pmrK genes in the ColR-CRKP isolates were detected by RT-qPCR compared with colistin-susceptible strain K. pneumoniae ATCC700603. Transcription levels were normalized to levels of rpsL gene. Values were represented by the mean ± standard deviation of three replicates using 2−ΔΔCt method
a Unaltered expression levels compared to K. pneumoniae ATCC700603
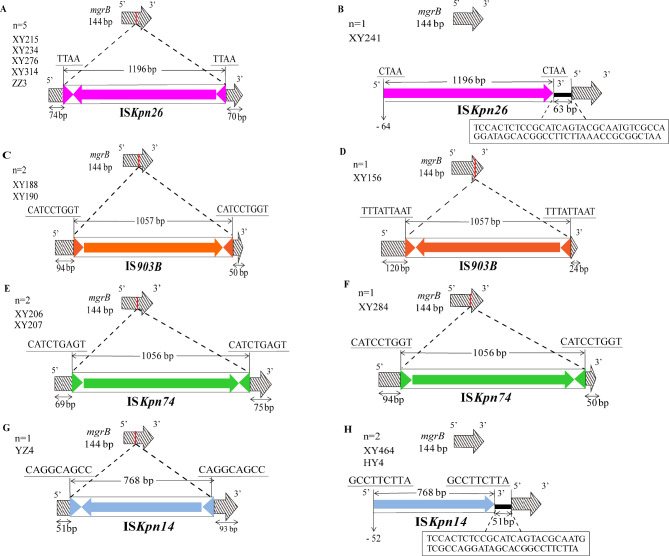
In addition, genetic alterations in the mgrB gene were also investigated by PCR-based Sanger sequencing. All 26 ColR-CRKP isolates amplified the mgrB gene, and 57.7% (15/26) of them had amplicons larger than its normal size, demonstrating the presence of ISs at the coding region of the mgrB gene. Four types of IS elements were found, either within the promoter or open reading frame (ORF) of the mgrB gene, including ISkpn26 (n = 6), IS903B (n = 3), ISkpn74 (n = 3) and ISkpn14 (n = 3). The orientations, insertion sites, repeat regions and IS element lengths within mgrB were illustrated in Fig. 3. Diverse locations of IS elements were observed in the mgrB gene in our study. Among the six isolates with ISKpn26 (a member of the IS5 family) insertional inactivation, five harbored insertions at nucleotide position + 74, and one harbored the insertion at position − 64 upstream of the start codon (Fig. 3A and B). Among the three isolates with IS903B (a member of the IS5 family), two presented insertions at position + 94 and one at + 120 (Fig. 3C and D). Disruption of mgrB at nucleotide positions + 69 and + 94 by a member of the IS5 family, ISKpn74, was detected in two isolates and one isolate, respectively (Fig. 3E and F). Among the three isolates with ISKpn14 (a member of the IS1 family), one had an insertion at nucleotide position + 51, and two had an insertion at position − 52.
Fig. 3.
Schematic representation of insertion sequences (ISs) observed within the mgrB gene. Four ISs were identified: ISKpn26, IS903B, ISKpn74 and ISKpn14. The Red lines on mgrB represent the insertion sites. The directions of the insertion events are shown as colorful arrows, with the IS lengths marked above each IS. Underlined nucleotide sequences were direct repeats (DRs). Triangles flanking each IS were left and right inverted repeats (IRLs and IRRs). (1) For the six isolates harboring the ISKpn26 insertion: five had insertions at position + 74 (A); one had an insertion at -64 bp, located in the promoter region upstream of the start codon (B); (2) among the three isolates with the IS903B insertion: two displayed insertions at + 94 (C); one revealed an insertion at + 120 (D); (3) for the three isolates bearing the ISKpn74 insertion: two demonstrated insertions at + 69 bp (E); one presented an insertion at + 94 bp (F); (4) among the three isolates with the ISKpn14 insertion: one showed an insertion at + 51 bp (G); two had insertions at -52 bp, situated in the promoter region upstream of the start codon (H)
RT-qPCR was used to analyze the relative transcription levels of the pmrC and pmrK genes encoding proteins involved in LPS decoration with pEtN and L-Ara4N, respectively. Compared with K. pneumoniae ATCC700603, 69.2% (18/26) of the ColR-CRKP isolates presented an increase in the pmrC gene ranging from 1.10-fold to 31.19-fold, whereas 88.5% (23/26) of them presented an increase in pmrK transcription between 1.25- and 18.46-fold (Table 2).
Virulence phenotypes and genetic characteristics of ColR-CRKP isolates
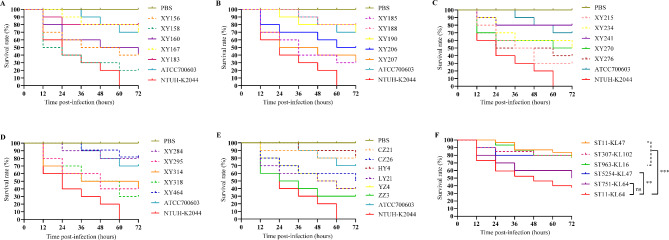
The in vivo virulence of the ColR-CRKP isolates was evaluated in a G. mellonella larvae infection model. The number of larvae deaths every 12 h after bacterial inoculation (106 CFU) for 72 h was recorded, and survival analysis was performed by Kaplan-Meier method (Fig. 4A and E). All the ST11-KL64 isolates (n = 16) and the ST751-KL64 isolate (CZ26) showed high virulence, with a 72-h larvae survival rate ≤ 50%. The other isolates in our study exhibited low virulence, with a 72-h larvae survival rate ≥ 80%. The ColR-CRKP isolates were classified into six groups according to their ST-KL type. The survival rate of the larvae infected with the ST11-KL64 isolates was significantly lower than those infected with the ST11-KL47, ST963-KL16, ST307-KL102 (p < 0.001) and ST5254-KL47 isolates (p < 0.01) (Fig. 4F). However, there was no statistically significant difference in the survival rate of the larvae infected with the ST11-KL64 isolates compared with those infected with the ST751-KL64 isolate (Fig. 4F).
Fig. 4.
Kaplan-Meier survival curves of the G. mellonella larvae infection model. A-E Survival curves of larvae infected with ColR-CRKP isolates in our study (1 × 106 CFU). Hypervirulent K. pneumoniae NTHU-K2044, classic K. pneumoniae ATCC700603 and PBS were used as positive, negative and blank controls, respectively. F The larvae survival distribution of the six groups of ColR-CRKP isolates according to ST-KL type. Survival curve differences were assessed by the log-rank (Mantel-Cox) test. ∗∗∗P value < 0.001; ∗∗P value < 0.01; and ns (not significant), P value > 0.05
The virulence genetic characteristics of the ColR-CRKP isolates were studied to infer the likelihood of causing serious clinical infections and explain the virulence phenotype in the G. mellonella larvae infection model. A large number of virulence factors coding for adherence, iron uptake and regulation were identified in the ColR-CRKP isolates. All the ColR-CRKP isolates carried the fimABCDEFGHIK and mrkABCDFHIJ genes, encoding type I and type I fimbriae, which are associated with the adherence and biofilm formation, respectively, as well as the entABCDEFS and fepABCDGfes genes, encoding the siderophore enterobactin, which is responsible for scavenging iron in vivo. The ColR-CRKP belonging to ST11, ST751 and ST5254 harbored the siderophore yersiniabactin-encoding ybt genes (ybtAEPQSTUX), typed as ybt9 located on the ICEKp3-like mobile genetic element by Kleborate. Two siderophore salmochelin-encoding genes (iroN and iroE), were harbored by all the ColR-CRKP isolates, while iroBCD genes were not detected in any of them. The siderophore aerobactin-encoding iuc genes (iucABCD), especially iucA, a hypervirulent biomarker gene, were found in all the ST11-KL64 and ST751-KL64 isolates (65.4%, 17/26), which carried at least two hypervirulent biomarkers and exhibited high virulence in the larvae infection model (Fig. 2). Other hypervirulent biomarker genes except iroB gene were also detected in our study, with distributions in the ColR-CRKP isolates as follows: rmpA2 (50.0%, 13/26), peg344 (34.6%, 9/26) and rmpA (19.2%, 5/26) (Fig. 2).
Plasmid replicon analysis
WGS-based analysis showed 13 different plasmid replicons in the ColR-CRKP isolates. They possessed one to six plasmid replicons, with an average of four incompatibility (Inc) groups per isolate (Fig. 2). The IncFII group was found in all the carbapenemase-producing isolates, including the IncFII(pHN7A8) backbone in blaKPC−2-positive isolates, IncFII(Yp) type in the four isolates co-harbouring blaKPC−2 and blaNDM−1, and IncFII(K) type in the two blaNDM−1-positive isolates. The IncHI1B(pNDM-MAR)/repB plasmid was found only in isolates carrying hypervirulence genes. Notably, IncFII(pHN7A8) and IncHI1B(pNDM-MAR)/repB plasmids were identified in all the isolates co-carrying blaKPC−2 and hypervirulent biomarkers.
Discussion
In recent years, ColR-CRKP isolates have emerged as a serious threat to public health worldwide [7]. The prevalence and underlying mechanisms of colistin resistance in CRKP isolates should be investigated continuously. Here, we described the colistin MIC distributions of 429 clinical CRKP isolates collected from hospitalized patients at twelve tertiary hospitals in Central South China and studied the prevalence, antibiotic resistance, molecular characteristics and virulence phenotypes of colistin-resistant isolates among CRKP. The colistin resistance rate among the CRKP isolates in our study was 6.1%, which was slightly higher than that reported by the CHINET in 2021 (5.3%). There has been an increasing trend in the prevalence rate of colistin resistance in CRKP from mainland China, reaching 8.2% in 2022 and 11.8% in 2023, according to CHINET surveillance. Furthermore, the prevalence of colistin resistance has recently increased worldwide, attracting global public health concern. During the period between 2014 and 2019, the detection rate of colistin resistance in clinical Enterobacterales increased from 2.4 to 3.4% in Europe, from 2.7 to 4.3% in Latin America, and from 3.3 to 6.7% in Asia [9]. The continuous increase in resistance rates may be the result of the massive use of colistin in hospital settings, veterinary medicine and global trade.
Among the ColR-CRKP isolates in our study, resistance rates to almost all the antimicrobial agents were alarmingly high (65.4-100%), except for tigecycline and ceftazidime/avibactam, which displayed higher activity against 88.5% and 69.2% of the isolates, respectively, and were proposed as therapeutic options for combating ColR-CRKP infections. Notably, when ceftazidime/avibactam is used to treat infections caused by isolates coproducing KPC and NDM carbapenemases, NDM carbapenemases may confer a selective advantage and could potentially replace KPC, thereby further restricting available therapeutic options [30–32]. For the management of patients infected with CRKP coharboring KPC and NDM, the recommended treatment strategies are consistent with the guidelines established by the Infectious Diseases Society of America (IDSA) for metallo-β-lactamases (MBL)-producing CRE. These guidelines advocate for either a combination therapy utilizing ceftazidime-avibactam and aztreonam or monotherapy with cefiderocol [30, 31]. Consistent with previous studies [33, 34], ColR-CRKP infections were mostly prevalent in ICU wards with 65.4% of patients admitted to the ICU in this study. This could be attributed to the excessive use of antibiotics to initiate empiric anti-infective treatment immediately in ICU wards due to delayed culture and antibiotic susceptibility results. Therefore, laboratory-based accurate and constant surveillance is essential so that clinicians can provide better empirical treatment on the basis of the up-to-date local epidemiological resistance data.
Genomic analysis of the twenty-six ColR-CRKP isolates revealed that they had diverse genetic backgrounds, including multiple STs and KL types, thus highlighting the fear of genomic plasticity in the circulating isolates from Central South China. ST11, the dominant epidemic clone among CRKP isolates in South America and Asia, especially in China [5, 35], was found in 73.1% of our ColR-CRKP populations, which cocarried blaKPC−2 and blaNDM−1 or blaKPC−2, the predominant carbapenemase in China [36]. Among the other STs identified in this study, ST307, an endemic high-risk clone in Italy, Colombia, the United States (Texas) and South Africa [37], was observed in two blaNDM−1-positive isolates (XY183 and CZ21). More surprisingly, three carbapenemase-negative isolates (XY188, XY190 and XY284) belonged to ST963, which has not been previously reported. In addition, KL64 accounted for 84.2% of the isolates in this study, which is in line with previous findings that KL64 has become the dominate capsule type among clinical CRKP isolates in eastern and central regions of China [38], and has been reported globally [39–41]. The phylogenetic analysis further revealed that the ST11 isolates formed different clonal transmission clusters, suggesting that many ST11 ColR-CRKP clones were circulating in Central South China. Furthermore, the three ST963 and two ST307 isolates were clonally disseminated in the same hospital and between two hospitals, respectively. We speculated that the medical flow pattern between the provincial capital city of Changsha and surrounding cities facilitates cross-regional clonal transmission. Our findings indicate that the ColR-CRKP isolates belonging to globally high-risk ST11 and ST307 clones could be emerging threats to global public health. Hence, it is crucial to track different clones of ColR-CRKP isolates by genotyping to limit their further transmission and even prevent outbreaks.
The molecular mechanism of colistin resistance in CRKP is highly diverse because of various mutational mechanisms [42, 43]. Different chromosomal mutations, particularly disruptions in mgrB caused by IS elements, were identified in this study. MgrB is a negative feedback regulator of the PhoP/Q system, and its inactivation is the most common underlying mechanism for colistin resistance in K. pneumoniae, as found in our study [17, 44]. MgrB inactivation caused by IS-mediated insertion was present in 57.7% of the 26 tested isolates, and no other types of mgrB alterations were detected in the present study. Our results indicate that insertional inactivation is the dominant alteration type in mgrB, which is in agreement with the findings of previous studies [17, 45]. Three types of IS5 family elements (ISKpn26, IS903B and ISKpn74) were detected in twelve isolates, with ISKpn26 (n = 5) being the main insertion type. As a member of the IS1 family, ISKpn14 was detected in three isolates and was reported to be the most common type of insertion sequence in other literatures [40, 46]. Geographical variation likely accounts for the different prevalent insertion sequences in mgrB. For TCS-related genes, deleterious mutations in pmrA (G53S), pmrB (T157P, R256G), and crrB (R248P) predicted by the PROVEAN tool were detected in our collected isolates. Notably, to our knowledge, the novel point mutation R248P in the crrB gene, potentially involved in colistin resistance, was discovered in two isolates (XY206 and LY21). However, mutations in pmrA (G53S) and pmrB (R256G) have also been reported in colistin-susceptible K. pneumoniae in other studies [47, 48], so their roles in colistin resistance remain controversial. The T157P substitution in PmrB was identified in 42.3% of the isolates and has been confirmed to be responsible for the colistin resistance phenotype [49]. Moreover, it is crucial to highlight the occurrence of different mechanisms to colistin resistance among isolates from the same clones in our study. For example, the three ST963 isolates showed colistin-resistant phenotype with different IS interruptions in mgrB gene. The seven ST11 isolates belonging to the same clone also showed various resistance mechanisms. This finding may suggest independent acquisition of disseminated clones. Finally, our data on the expression levels of the pmrC and pmrK genes also indicate that colistin resistance in CRKP isolates is associated with increased expression of the pmrCAB and pmrHFIJKLM operons, which leads to the modification of lipid A in the structure of LPS, as reported in a prior study [50]. Therefore, targeting these mutations might be a promising strategy for overcoming resistance to colistin.
Virulence analysis highlighted the presence of numerous virulence factors, mainly capsular polysaccharide, fimbrial adhesion and siderophore systems, which are essential pathogenicity determinants of causing invasive infectious diseases, in the ColR-CRKP isolates [51]. Among the siderophore systems, yersiniabactin, encoded by ybt, is the most common high-virulence factor in K. pneumoniae, where ybt is located within the ICEKp [52]. ICEKp is an integrative conjugative element that mobilizes the ybt locus and is made up of various sublineages [52]. In this study, ybt-9 located on ICEKp3 elements was present in the ST11, ST751 and ST5254 ColR-CRKP isolates, and was carried mainly by ST11 K. pneumoniae, as previously reported [53]. Notably, the siderophore aerobactin encoding iucA, which was harbored by all the ST11-KL64 and ST751-KL64 isolates demonstrating high virulence in a larvae infection model in this study, has been identified as a promising marker of hypervirulence in K. pneumoniae [54]. Our data were the first to show that there was a greater prevalence (65.4%) of hypervirulent strains among ColR-CRKP isolates, than the proportion of hypervirulent strains (45.7%) among CRKP isolates in 2022 in China [38]. Previous studies have shown that the mgrB mutation and the presence of CrrAB regulatory system can enhance K. pneumoniae virulence in a G. mellonella infection model [55, 56]. Notably, the emergence and clonal spread of hypervirulent ColR-CRKP could speed up the lack of effective treatments, posing a serious challenge to antimicrobial therapy. Our results highlight the importance of carrying out proactive monitoring strategies and effective infection control measures to prevent potential hazards caused by hypervirulent ColR-CRKP in nosocomial settings.
Previous researches have demonstrated that IncFII plasmids functioned as the primary vectors for the blaKPC gene, while IncHIB/repB predominantly represented virulence-associated plasmids [57, 58]. In our study, the IncFII(pHN7A8) and IncHI1B(pNDM-MAR)/repB plasmids were co-detected only in the isolates co-harbouring blaKPC-2 and hypervirulence genes. This finding suggested that the two types of plasmids formed a highly successful partnership and it was most likely that hv-CRKP primarily evolved from CRKP through the acquisition of another plasmid harboring hypervirulence genes. According to previous studies, the IncFII conjugative plasmids carrying blaKPC gene appear to function as a pivotal driver in the mobilization of the non-conjugative IncHI1B virulence plasmid and the co-occurrence of blaKPC plasmids and virulence plasmids enhances the formation of hv-CPKP [59, 60]. Previous studies confirmed that the conserved origin of transfer (oriT) region of virulence plasmids and the widespread of conjugative helper plasmids (such as blaKPC-harboring plasmids) represent potential factors for the mobilization of virulence plasmids, which contribute to the emergence and dissemination of both highly resistant and hypervirulent phenotype in blaKPC-positive isolates which mainly belong to ST11 [59, 60]. A heightened focus should be directed towards the rapid dissemination of virulence plasmids and the pervasive prevalence of hv-CRKP.
This study has several limitations. Firstly, the present study collected a relatively small number of isolates and could be useful as a preliminary data source for carrying out a broader regional surveillance. Secondly, colistin resistance may be associated with the mutations observed in this study, but the underlying mechanism is not fully understood. Further studies are needed to validate the precise functions of these mutations in colistin resistance. Thirdly, Long-read sequencing will be conducted on convergent isolates to identify antimicrobial resistance and hypervirulence plasmids for further investigating the genetic context of resistance and virulence genes.
Conclusions
This multicenter study revealed a colistin resistance prevalence of 6.1% among CRKP isolates obtained from Central South China in 2021. The predominant mechanism underlying colistin resistance is inactivation of mgrB via insertion sequences, and a novel deleterious mutation, R248P, in CrrB was identified. Furthermore, it is important to highlight the presence of different mechanisms of colistin resistance in isolates belonging to the same clones. Clonal transmissions of the ColR-CRKP isolates occurred in globally high-risk ST11 and ST307 clones, and there was a high prevalence of hypervirulent strains among ColR-CRKP isolates. These findings emphasize the urgent need for global public health initiatives to prioritize continuous genomic surveillance to spot high-risk clones early, particularly those with hypervirulence, to prevent further transmission.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all of the microbiology staff from the 12 hospitals for their contributions in identifying the CRKP isolates and collecting the clinical data.
Abbreviations
- CRKP
Carbapenem-resistant Klebsiella pneumoniae
- KPC
K. pneumoniae carbapenemase
- NDM
New Delhi metallo-β-lactamase
- OXA-48
Oxacillinase-48
- hv-CRKP
Hypervirulent CRKP
- GNB
Gram-negative bacterial
- ColR-CRKP
Colistin-resistant CRKP
- CHINET
China Antimicrobial Surveillance Network
- LPS
Lipopolysaccharides
- TCSs
Two-component regulatory systems
- Mcr
Mobile colistin resistance
- MALDI-TOF
Matrix-Assisted Laser Desorption Ionization-Time of Flight
- CLSI
Clinical and Laboratories Standard Institute
- BMD
Broth microdilution
- MIC
Minimum inhibitory concentration
- FDA
Food and Drug Administration
- MDR
Multidrug-resistant
- XDR
Extensively drug-resistant
- PDR
Pandrug-resistant
- WGS
Whole-genome sequencing
- MLST
Multilocus sequence typing
- KL
Capsule loci
- VFDB
Virulence factor database
- ICEKp
Integrative conjugative element of K. pneumoniae
- CARD
Comprehensive Antibiotic Resistance Database
- SNP
Single nucleotide polymorphism
- PROVEAN
Protein variation effect analyzer
- PCR
Polymerase chain reaction
- IS
Insertion sequence
- pEtN
Phosphoethanolamine
- L-Ara4N
4-amino-4-deoxy-l-arabinose
- RT-qPCR
Real-time quantitative PCR
- PBS
Phosphate buffered saline
- hvKp
Hypervirulent K. pneumoniae
- ICU
Intensive care unit
- STs
Sequence types
- ARGs
Antibiotic resistance genes
- ESBL
Extended spectrum β-lactamase
- ORF
Open reading frame
- Inc
Incompatibility
- IDSA
Infectious Diseases Society of America
- MBL
Metallo-β-lactamases
- OriT
Origin of transfer
Author contributions
W.L. and Z.J. contributed to the experimental conception and design. Z.J., Y.L. and P.L. collected the isolates and performed the experiments. J.W. collected clinical information. Z.J., Y.L. and Z.W. conducted the bioinformatics analysis and data analysis. Z.J. drafted the first version of this manuscript. W.L. and Q.Y. revised the manuscript. All the authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (grant no. 2018YFC2000203).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The studied isolates were routinely collected for clinical diagnosis and the interests and privacy of the patients involved were not affected. Consequently, there was no need for written individual informed consent or approval from an ethics committee, and Chinese legal standards were rigorously upheld.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Global mortality associated. With 33 bacterial pathogens in 2019: a systematic analysis for the global burden of Disease Study 2019. Lancet. 2022;400(10369):2221–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopotsa K, Osei Sekyere J, Mbelle NM. Plasmid evolution in carbapenemase-producing Enterobacteriaceae: a review. Ann N Y Acad Sci. 2019;1457(1):61–91. [DOI] [PubMed] [Google Scholar]
- 3.Heng H, Yang X, Ye L, Tang Y, Guo Z, Li J, Chan EW, Zhang R, Chen S. Global genomic profiling of Klebsiella pneumoniae: a spatio-temporal population structure analysis. Int J Antimicrob Agents. 2024;63(2):107055. [DOI] [PubMed] [Google Scholar]
- 4.Wang J, Feng Y, Zong Z. Worldwide transmission of ST11-KL64 carbapenem-resistant Klebsiella pneumoniae: an analysis of publicly available genomes. mSphere. 2023;8(4):e0017323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. [DOI] [PubMed] [Google Scholar]
- 7.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, et al. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis. 2015;15(2):225–34. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. [DOI] [PubMed] [Google Scholar]
- 9.Binsker U, Käsbohrer A, Hammerl JA. Global colistin use: a review of the emergence of resistant enterobacterales and the impact on their genetic basis. FEMS Microbiol Rev 2022, 46(1). [DOI] [PMC free article] [PubMed]
- 10.Huang YH, Chou SH, Liang SW, Ni CE, Lin YT, Huang YW, Yang TC. Emergence of an XDR and carbapenemase-producing hypervirulent Klebsiella pneumoniae strain in Taiwan. J Antimicrob Chemother. 2018;73(8):2039–46. [DOI] [PubMed] [Google Scholar]
- 11.Kaur A, Gandra S, Gupta P, Mehta Y, Laxminarayan R, Sengupta S. Clinical outcome of dual colistin- and carbapenem-resistant Klebsiella pneumoniae bloodstream infections: a single-center retrospective study of 75 cases in India. Am J Infect Control. 2017;45(11):1289–91. [DOI] [PubMed] [Google Scholar]
- 12.Koppe U, von Laer A, Kroll LE, Noll I, Feig M, Schneider M, Claus H, Eckmanns T, Abu Sin M. Carbapenem non-susceptibility of Klebsiella pneumoniae isolates in hospitals from 2011 to 2016, data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob Resist Infect Control. 2018;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satlin MJ, Chen L, Patel G, Gomez-Simmonds A, Weston G, Kim AC, Seo SK, Rosenthal ME, Sperber SJ, Jenkins SG et al. Multicenter Clinical and Molecular Epidemiological Analysis of Bacteremia due to Carbapenem-Resistant Enterobacteriaceae (CRE) in the CRE Epicenter of the United States. Antimicrob Agents Chemother 2017, 61(4). [DOI] [PMC free article] [PubMed]
- 14.Kopotsa K, Mbelle NM, Osei Sekyere J. Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb Genom 2020, 6(12). [DOI] [PMC free article] [PubMed]
- 15.Shankar C, Jacob JJ, Sugumar SG, Natarajan L, Rodrigues C, Mathur P, Mukherjee DN, Sharma A, Chitnis DS, Bharagava A, et al. Distinctive Mobile genetic elements observed in the Clonal expansion of Carbapenem-resistant Klebsiella pneumoniae in India. Microb Drug Resist. 2021;27(8):1096–104. [DOI] [PubMed] [Google Scholar]
- 16.Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and Resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58(10):5696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Y, Hao J, Xiao W, Ye C, Xiao X, Jian C, Tang M, Li G, Liu J, Zeng Z. Role of efflux pumps, their inhibitors, and regulators in colistin resistance. Front Microbiol. 2023;14:1207441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JH, Liu YY, Shen YB, Yang J, Walsh TR, Wang Y, Shen J. Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol. 2024;32(4):365–78. [DOI] [PubMed] [Google Scholar]
- 20.CLSI. Performance standards for antimicrobial susceptibility testing, 31st ed. CLSI Supplement M100. Wayne: Clinical and Laboratory Standards Institute; 2021. [DOI] [PMC free article] [PubMed]
- 21.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Zou S, Chen H, Yu Y, Ruan Z. BacWGSTdb 2.0: a one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021;49(D1):D644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadjirin NF, van Tonder AJ, Blane B, Lees JA, Kumar N, Delappe N, Brennan W, McGrath E, Parkhill J, Cormican M et al. Dissemination of carbapenemase-producing enterobacterales in Ireland from 2012 to 2017: a retrospective genomic surveillance study. Microb Genom 2023, 9(3). [DOI] [PMC free article] [PubMed]
- 24.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents. 2014;44(6):500–7. [DOI] [PubMed] [Google Scholar]
- 25.Berglund B, Hoang NTB, Tärnberg M, Le NK, Svartström O, Khu DTK, Nilsson M, Le HT, Welander J, Olson L, et al. Insertion sequence transpositions and point mutations in mgrB causing colistin resistance in a clinical strain of carbapenem-resistant Klebsiella pneumoniae from Vietnam. Int J Antimicrob Agents. 2018;51(5):789–93. [DOI] [PubMed] [Google Scholar]
- 26.Kong Y, Li C, Chen H, Zheng W, Sun Q, Xie X, Zhang J, Ruan Z. In vivo emergence of Colistin Resistance in Carbapenem-resistant Klebsiella pneumoniae mediated by premature termination of the mgrB Gene Regulator. Front Microbiol. 2021;12:656610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis. 2014;14:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. Identification of biomarkers for differentiation of Hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 2018, 56(9). [DOI] [PMC free article] [PubMed]
- 29.Li G, Shi J, Zhao Y, Xie Y, Tang Y, Jiang X, Lu Y. Identification of hypervirulent Klebsiella pneumoniae isolates using the string test in combination with Galleria mellonella infectivity. Eur J Clin Microbiol Infect Dis. 2020;39(9):1673–9. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Xiao X, Wang X, Xia M, Liu S. In Vitro Antimicrobial susceptibility differences between Carbapenem-resistant KPC-2-Producing and NDM-1-Producing Klebsiella pneumoniae in a Teaching Hospital in Northeast China. Microb Drug Resist. 2020;26(2):94–9. [DOI] [PubMed] [Google Scholar]
- 31.Tamma PD, Aitken SL, Bonomo RA, Mathers AJ, van Duin D, Clancy CJ. Infectious Diseases Society of America 2023 Guidance on the treatment of Antimicrobial resistant gram-negative infections. Clin Infect Dis 2023. [DOI] [PubMed]
- 32.Rodríguez-Medina N, Rodríguez-Santiago J, Alvarado-Delgado A, Sagal-Prado A, Silva-Sánchez J, De la Cruz MA, Ares MA, Sánchez-Arias M, Morfín-Otero R, Hernández-Castro R, et al. Comprehensive study reveals phenotypic heterogeneity in Klebsiella pneumoniae species complex isolates. Sci Rep. 2024;14(1):5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.da Silva KE, Baker S, Croda J, Nguyen TNT, Boinett CJ, Barbosa LS, Tetila A, Simionatto S. Risk factors for polymyxin-resistant carbapenemase-producing Enterobacteriaceae in critically ill patients: an epidemiological and clinical study. Int J Antimicrob Agents. 2020;55(3):105882. [DOI] [PubMed] [Google Scholar]
- 34.Sodhi K, Mittal V, Arya M, Kumar M, Phillips A, Kajla B. Pattern of colistin resistance in Klebsiella isolates in an Intensive Care Unit of a tertiary care hospital in India. J Infect Public Health. 2020;13(7):1018–21. [DOI] [PubMed] [Google Scholar]
- 35.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–12. [DOI] [PubMed] [Google Scholar]
- 36.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peirano G, Chen L, Kreiswirth BN, Pitout JDD. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother 2020, 64(10). [DOI] [PMC free article] [PubMed]
- 38.Hu F, Pan Y, Li H, Han R, Liu X, Ma R, Wu Y, Lun H, Qin X, Li J, et al. Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-centre study. Nat Microbiol. 2024;9(3):814–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallal Ferreira Raro O, Nordmann P, Dominguez Pino M, Findlay J, Poirel L. Emergence of carbapenemase-producing hypervirulent Klebsiella pneumoniae in Switzerland. Antimicrob Agents Chemother. 2023;67(3):e0142422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attalla ET, Khalil AM, Zakaria AS, Baker DJ, Mohamed NM. Genomic characterization of colistin-resistant Klebsiella pneumoniae isolated from intensive care unit patients in Egypt. Ann Clin Microbiol Antimicrob. 2023;22(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sundaresan AK, Vincent K, Mohan GBM, Ramakrishnan J. Association of sequence types, antimicrobial resistance and virulence genes in Indian isolates of Klebsiella pneumoniae: a comparative genomics study. J Glob Antimicrob Resist. 2022;30:431–41. [DOI] [PubMed] [Google Scholar]
- 42.Otter JA, Doumith M, Davies F, Mookerjee S, Dyakova E, Gilchrist M, Brannigan ET, Bamford K, Galletly T, Donaldson H, et al. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci Rep. 2017;7(1):12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu H, Shi Q, Zhang P, Quan J, Han X, Zhao D, Zhang H, Wang Q, Jiang Y, Yu Y. Prevalence and molecular characteristics of colistin-resistant isolates among clinically isolated carbapenem-resistant Klebsiella pneumoniae in China. Int J Antimicrob Agents. 2023;62(2):106873. [DOI] [PubMed] [Google Scholar]
- 44.Huang PH, Cheng YH, Chen WY, Juan CH, Chou SH, Wang JT, Chuang C, Wang FD, Lin YT. Risk factors and mechanisms of in vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents. 2021;57(6):106342. [DOI] [PubMed] [Google Scholar]
- 45.Yang TY, Wang SF, Lin JE, Griffith BTS, Lian SH, Hong ZD, Lin L, Lu PL, Tseng SP. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents. 2020;55(3):105894. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Liu X, Lei Z, Li C, Zhang F, Wu Y, Yang X, Zhao J, Zhang Y, Hu Y, et al. Genetic diversity of polymyxin-resistance mechanisms in clinical isolates of Carbapenem-resistant Klebsiella pneumoniae: a Multicenter Study in China. Microbiol Spectr. 2023;11(2):e0523122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother. 2015;59(5):2909–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elias R, Spadar A, Phelan J, Melo-Cristino J, Lito L, Pinto M, Gonçalves L, Campino S, Clark TG, Duarte A, et al. A phylogenomic approach for the analysis of colistin resistance-associated genes in Klebsiella pneumoniae, its mutational diversity and implications for phenotypic resistance. Int J Antimicrob Agents. 2022;59(6):106581. [DOI] [PubMed] [Google Scholar]
- 49.Jayol A, Poirel L, Brink A, Villegas MV, Yilmaz M, Nordmann P. Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob Agents Chemother. 2014;58(8):4762–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azam M, Gaind R, Yadav G, Sharma A, Upmanyu K, Jain M, Singh R. Colistin Resistance among multiple sequence types of Klebsiella pneumoniae is Associated with Diverse Resistance mechanisms: a Report from India. Front Microbiol. 2021;12:609840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. 2020;18(6):344–59. [DOI] [PubMed] [Google Scholar]
- 52.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 2018, 4(9). [DOI] [PMC free article] [PubMed]
- 53.Zhu C, Li C, Lai CKC, Ng R, Chau KY, Wong KT, Lo NWS, Barua N, Yang Y, Liyanapathirana V, et al. Longitudinal genomic characterization of carbapenemase-producing Enterobacteriaceae (CPE) reveals changing pattern of CPE isolated in Hong Kong Hospitals. Int J Antimicrob Agents. 2021;58(5):106430. [DOI] [PubMed] [Google Scholar]
- 54.Shankar C, Basu S, Lal B, Shanmugam S, Vasudevan K, Mathur P, Ramaiah S, Anbarasu A, Veeraraghavan B. Aerobactin seems to be a promising marker compared with unstable RmpA2 for the identification of Hypervirulent Carbapenem-resistant Klebsiella pneumoniae: in Silico and in Vitro evidence. Front Cell Infect Microbiol. 2021;11:709681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kidd TJ, Mills G, Sá-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9(4):430–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SJ, Shin JH, Kim H, Ko KS. Roles of crrAB two-component regulatory system in Klebsiella pneumoniae: growth yield, survival in initial colistin treatment stage, and virulence. Int J Antimicrob Agents. 2024;63(1):107011. [DOI] [PubMed] [Google Scholar]
- 57.Fu P, Tang Y, Li G, Yu L, Wang Y, Jiang X. Pandemic spread of bla((KPC-2)) among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents. 2019;54(2):117–24. [DOI] [PubMed] [Google Scholar]
- 58.Hu D, Li Y, Ren P, Tian D, Chen W, Fu P, Wang W, Li X, Jiang X. Molecular Epidemiology of Hypervirulent Carbapenemase-producing Klebsiella pneumoniae. Front Cell Infect Microbiol. 2021;11:661218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian D, Liu X, Chen W, Zhou Y, Hu D, Wang W, Wu J, Mu Q, Jiang X. Prevalence of hypervirulent and carbapenem-resistant Klebsiella pneumoniae under divergent evolutionary patterns. Emerg Microbes Infect. 2022;11(1):1936–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Y, Zhang J, Wang M, Liu M, Liu G, Qu H, Liu J, Deng Z, Sun J, Ou HY, et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae. Genome Med. 2021;13(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.