Summary
In this issue of Cell Host & Microbe, Huang et al., determine that an oncogenic bacterium contributes to colorectal cancer progression and resistance to receptor tyrosine kinase inhibitors. These findings highlight the need for an integrative approach for cancer treatment that considers the influence of the microbiome.
Within the microbiome, some microbes have the potential to benefit host immunity and metabolism to maintain overall health. However, in the context of disease, the microbiome can enter a state of dysbiosis and some microbes have the capacity to negatively impact the host. In this issue of Cell Host & Microbe, Huang et al. have highlighted a significant microbial adversary in the battle against cancer treatment resistance. They report that Peptostreptococcus stomatis (P. stomatis) is an oncogenic bacterium and a contributing factor for non-responsiveness to receptor tyrosine kinase (RTK) inhibitors in colorectal cancer (CRC) (1). CRC is a major health concern worldwide, involving a combination of genetic, epigenetic, and environmental factors in its pathogenesis. Among these, the gut microbiota has emerged as a crucial player. Previous studies have established a link between gut dysbiosis and CRC, identifying certain bacteria, e.g., Fusobacterium nucleatum, Bacteroides fragilis, pks(+) E. coli, and Salmonella, as promoters of colorectal tumorigenesis (2) (3, 4) (5) and there are efforts to identify global microbial signatures, specific for CRC (6, 7). However, the specific mechanisms by which these bacteria influence cancer treatment resistance are not fully understood.
Huang et al. demonstrate that P. stomatis is enriched in patients with CRC and plays a causal role in accelerating colonic tumorigenesis. In more detail, the authors first confirmed the enrichment of P. stomatis in CRC through metagenome sequencing in multiple cohorts. The bacterium’s role in promoting CRC was demonstrated using adenomatous polyposis coli (Apc) multiple intestinal neoplasia (Min)/+ and Azoxymethane (AOM)/Dextran sodium sulfate (DSS) mouse models, where P. stomatis significantly increased tumor multiplicities and load, compared to controls. Mechanistic studies revealed that P. stomatis promotes cell proliferation and inhibits apoptosis through the activation of the erythroblastic oncogene B (ERBB2)-MAPK pathway. The bacterial surface protein fructose-1,6-bisphosphate aldolase (FBA) from P. stomatis was found to bind specifically to the integrin α6/β4 receptor on CRC cells, triggering a signaling cascade involving ERBB2 and its downstream effectors, such as MEK, ERK, and p90RSK. Disruption of this binding through genetic or pharmacological interventions reduced the tumor-promoting effects of P. stomatis. Moreover, the study highlighted that P. stomatis-driven ERBB2 activation bypasses the effects of EGFR inhibitors, providing an explanation for the observed drug resistance in CRC patients with high levels of this bacterium. This suggests that targeting P. stomatis or its interaction with integrin α6/β4 could enhance the efficacy of RTK inhibitors and improve therapeutic outcomes in CRC.
Traditionally, cancer research has focused predominantly on genetic and environmental factors, often overlooking the potential contributions of the microbiome. The study from Huang et al. (1) focuses on P. stomatis promoting colonic tumorigenesis and receptor tyrosine kinase inhibitor resistance and provides a compelling perspective on the intricate relationship between the microbiome and CRC. This research opens new avenues for understanding the multifaceted mechanisms through which gut bacteria influence cancer progression and treatment resistance. It also indicates that the oral pathogen P. stomatis might be an additional factor in regulating ERBB2-associated resistance to RTK inhibitors in CRC. However, there are still several unanswered questions, offering interesting prospects for future mechanistic research: 1. How does P. stomatis interact with other microbiota? Understanding the interaction of P. stomatis with other members of the gut microbiota and how these interactions influence CRC progression and drug resistance is crucial. This could uncover synergistic or antagonistic relationships that affect disease outcomes. 2. How does the host immune system respond to the presence of P. stomatis? The impact of P. stomatis on the host immune response and CRC progression in the tumor microenvironment is not fully understood. Future studies could focus on the immune-modulatory effects of P. stomatis and its interactions with immune cells in the tumor microenvironment. 3. What are the exact molecular mechanisms by which P. stomatis impairs intestinal barrier function? While the study shows that P. stomatis compromises barrier integrity, the precise molecular mechanisms remain unclear. Future research could explore how P. stomatis interacts with intestinal epithelial cells to impair barrier function. Answers to these questions will further advance the field and help translate these findings from basic research to clinical practice (Fig. 1).
Fig. 1. Fighting invisible foes (e.g., P. stomatis) in the personalized medicine for cancer therapy.
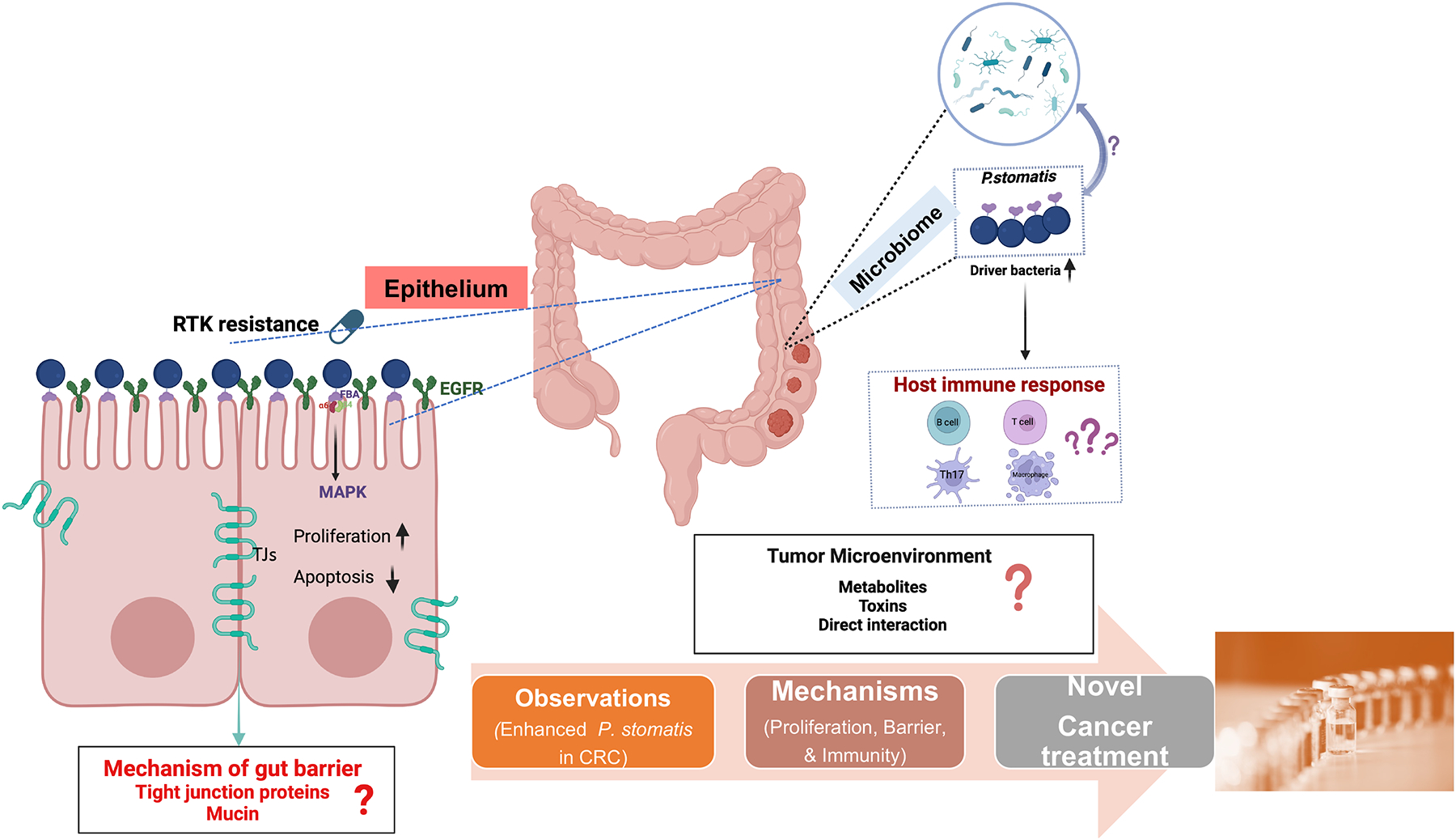
P. stomatis is a key driver in CRC progression and exhibits resistance to RTK inhibitors by activating the ERBB2 signaling pathway through its surface protein FBA. It engages in complex interactions with other gut microbes, significantly influencing CRC progression and drug resistance. The presence of P. stomatis triggers varied responses from the host immune system, although the specific effects on immune cells such as B cells, T cells, Th17 cells, and macrophages within the tumor microenvironment are not yet fully understood. Additionally, P. stomatis has been shown to impair gut barrier function, potentially by affecting tight junction proteins and mucin production, which compromises barrier functions and promotes tumorigenic conditions. Further study of these mechanisms as well as how P. stomatis interacts with other microbes may offer avenues to improve colorectal cancer outcomes and perhaps other cancer types.
Other papers support these observations and provide further evidence of microbes acting in a pro-tumor fashion, highlighted the progress of microbiome research in cancer biology. P. stomatis, along with other bacteria typically originating from the oral microbiome, are present at an increased abundance in CRC patients, highlighting composition changes in the microbiome associated with CRC.(8). Furthermore, P. stomatis was identified along with Parvimonas micra, Fusobacterium nucleatum, and Akkermansia muciniphila as a bacteria biomarker panel of CRC (9). These bacteria might serve as biomarkers for CRC detection and underline the necessity to explore their roles in CRC initiation and progression (9). P. stomatis accelerates colonic tumorigenesis by inducing cell proliferation, inhibiting apoptosis, and impairing intestinal epithelial barrier function (1). Taken together, recent research provides a perspective on the role of microbiome, e.g., P. stomatis, in CRC, emphasizing a bacterium’s multiple roles in promoting tumorigenesis and mediating drug resistance via manipulation of key host signaling pathways.
The translational significance and broader implications of the current findings are profound. Microbial diagnostics could become a crucial component of personalized cancer treatment, whereby the composition of microbiome could inform therapeutic choices. It will open a door to novel therapeutic interventions aimed at modulating the microbiome to counteract cancer progression and resistance. For example, probiotics or bacteriophage therapy targeting P. stomatis could be explored as adjuncts to conventional cancer therapies. RTK inhibitors are used in clinical oncology not only in management of advanced metastatic CRC, but also as the first- or second-line therapy for the treatment of other types of cancer. Thus, this study prompts a re-evaluation of the gut microbiome’s role in cancer beyond CRC, potentially influencing research into other microbiome-associated cancers. Viruses and fungi are also known to be influence tumorigenesis and responses to cancer therapies. This highlights the need for an integrative approach to cancer treatment that considers the influence of microbiome crossing kingdoms, paving the way for personalized medicine. It also encourages the scientific community to explore the in-depth symbiotic relationships between human cells and microbial inhabitants. We believe that microbiome research should not be just observational and we strongly advocate for the mechanistic and functional studies (10). A greater understanding of host–microbial interactions and validation in human cohorts will advance the field for cancer diagnostic, prognostic, or therapeutic purposes.
Acknowledgements/Funds
We would like to acknowledge the DOD CDMRP BC191198, VA Merit Award 1 I01BX004824-01, the NIDDK/National Institutes of Health grant R01 DK134343-01, and R01DK114126 to Jun Sun. The study sponsors play no role in the study design, data collection, analysis, and interpretation of data. The contents do not represent the views of the United States Department of Veterans Affairs or the United States Government.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Pingmei Huang FJ, Alvin Ho-Kwan Cheung, Kaili Fu, Qiming Zhou, Xiao, Ding DC, Yufeng Lin, Luyao Wang, Ying Jiao, Eagle SH Chu, Wei, Kang KFT, Jun Yu, Chi Chun Wong. Peptostreptococcus stomatis promotes colonic tumorigenesis and receptor tyrosine kinase inhibitor resistance by activating ERBB2-MAPK. Cell Host & Microbe. 2024. [DOI] [PubMed] [Google Scholar]
- 2.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23(2):203–14 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Elsland DM, Duijster JW, Zhang J, Stevenin V, Zhang Y, Zha L, et al. Repetitive non-typhoidal Salmonella exposure is an environmental risk factor for colon cancer and tumor growth. Cell Rep Med. 2022;3(12):100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai Z, Coker OO, Nakatsu G, Wu WK, Zhao L, Chen Z, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nature medicine. 2019;25(4):679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loftus M, Hassouneh SA-D, Yooseph S. Bacterial community structure alterations within the colorectal cancer gut microbiome. BMC Microbiology. 2021;21(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osman MA, Neoh H-m, Ab Mutalib N-S, Chin S-F, Mazlan L, Raja Ali RA, et al. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Scientific Reports. 2021;11(1):2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Y, Sun J. Applied Microbiome Statistics: Correlation, Association, Interaction and Composition. Boca Raton, FL, USA: Chapman & Hall/CRC; 2024. [Google Scholar]


