Summary
The development of antibody drugs through animal immunization typically requires the humanization of host antibodies to address concerns about immunogenicity in humans. However, employing an animal model capable of producing human antibodies presents the opportunity to develop antibody drugs without the need for humanization. Despite the ratio of human immunoglobulin (Ig) κ to Igλ usage being approximately 60%:40%, the majority of approved antibody therapeutics are kappa antibodies, and the development of lambda antibodies as therapeutic agents has lagged behind. Therefore, in this study, we developed mice carrying the IGH and IGL loci (IGHL), which can produce human lambda antibodies, using mouse artificial chromosome (MAC) vectors. We demonstrated that IGHL mice consistently retain the human lambda antibody locus integrated on the MAC across generations and can be induced to produce specific antibodies upon antigen stimulation. These findings provide a promising platform for advancing lambda antibody drugs, which have historically been neglected.
Subject areas: Immune response, Genetic engineering, Biological sciences research methodologies
Graphical abstract

Highlights
-
•
IGHL mice carry the full-length IGL (2.2Mb) and IGH (1.8Mb) genomic loci
-
•
IGHL mice are capable of producing fully human lambda antibodies
-
•
Antigen administration induced antigen-specific antibody production in IGHL mice
-
•
IGHL mice generate a diverse repertoire of fully human lambda antibodies
Immune response; Genetic engineering; Biological sciences research methodologies
Introduction
Antibodies are a major category of biotherapeutics. Currently, over 200 therapeutic monoclonal antibodies have been approved or are in regulatory trials.1 Their popularity stems from the ability of antibodies to mimic the adaptive immune response, with high affinity and specificity for a wide variety of target pathogens and antigens.2 These characteristics enable antibodies to serve as potent therapeutic agents for various diseases.
Most antibody drugs have historically been derived from rodents, particularly mice. Mice are physiologically similar to humans and prone to immune responses to a wide variety of antigens. However, direct administration of rodent-derived antibodies to humans is fraught with difficulties because of their potential immunogenicity, which can lead to adverse immune responses upon repeated exposure. To address this limitation, efforts have been made to minimize immunogenic reactions by humanizing antibody sequences.3,4 Humanization involves strategically substituting or modifying non-human components in the antibody structure while preserving antigen-binding specificity and affinity. Humanization reduces the likelihood of inducing an immune response upon administration to a human recipient. This approach has been pivotal in the development of safer, more effective antibody therapeutics, which has increased clinical utility and patient safety in a wide variety of therapeutic applications.4
Recent advances in single B cell technology have revolutionized the process of antibody discovery, enabling direct isolation and characterization of human-derived antibodies.5,6 This breakthrough has had a particularly important impact on the fight against infectious diseases, such as coronavirus disease 2019, by facilitating the rapid development of human-donor-derived antibody therapeutics. However, despite the efficacy of human-derived antibodies against common diseases, rodent-based antibody acquisition remains advantageous when the target disease affects a limited patient population or has a high mortality rate, as for rare diseases or certain types of cancer.7,8 Furthermore, in diseases characterized by poorly immunogenic epitopes, such as self-proteins in humans, antibodies raised in rodents may exhibit better antigen recognition.9 Thus, although antibodies obtained directly from humans are a valuable resource in combating a wide range of infectious diseases, rodent-based antibody production is essential to acquire antibodies that bind to niche epitopes and ultimately contribute to advances in precision medicine.
Transgenic mouse models have been developed to mitigate the risks associated with the humanization of rodent antibodies—such as insolubility, aggregation, proteolysis, and potential loss of binding affinity—by enabling the production of human antibodies. Transgenic technology has facilitated the generation of mice capable of producing human antibodies. One approach involves the knockout of endogenous mouse antibody genes, coupled with the introduction of partial human antibody sequences into the mouse genome.10 Another method entails the targeted knocking out of the variable (V) regions responsible for antigen recognition in mice, followed by their replacement with corresponding human sequences.11,12 Recently, mice have been generated using genetic recombination techniques to replace the V regions of immunoglobulins (Ig) with human-derived V regions.13 However, these methods are limited by the difficulty of retaining the full diversity of Ig genes, which can affect the breadth and specificity of the antibodies produced.
Recently, we generated trans-chromosomic–human-antibody-producing (TC-mAb) mice, which are mice that carry the entire human antibody heavy chain locus (1.8 Mb) and kappa light chain locus (1.7 Mb) via mouse artificial chromosome (MAC) vectors (IGHK-NAC).14,15 TC-mAb mice have demonstrated utility in generating human kappa antibodies by retaining full-length immunoglobulin (Ig) genes, resulting in antibody repertoire diversity comparable to that observed in humans. TC-mAb mice can produce human kappa antibodies but not human lambda antibodies. Hence, there is a concern that exclusively relying on antibody production by TC-mAb mice may lead to a loss of diversity in lambda antibody-dependent specificity.
Despite a good understanding of light chain selection during B cell development, significant gaps persist in our knowledge regarding the widely varying bias of light chains across species and their functional significance beyond the phenomenological level. Ig light chains exist as two types, kappa and lambda, and during B cell differentiation, one of these light chains is selected. The usage patterns of light chains vary among species. For instance, in mice, over 90% of produced Igs contain kappa light chains. Conversely, in dogs, cats, horses, and cattle, lambda light chains are used in over 90% of antibodies.16 In humans, approximately 60% of antibodies feature kappa light chains, whereas lambda light chains are utilized in the remaining 40%.17 Light chains contribute less to antibody diversity and antigen specificity than heavy chains because they have fewer V region segments and lack a diversity (D) gene. However, recent reports have indicated biases in light chain usage depending on antigen type.18,19,20,21 Further research is needed to elucidate the immunological factors that contribute to the observed differences in light chain usage across species, but these findings suggest that kappa and lambda light chains may exhibit differential antigen specificity.
In this study, we generated “IGHL mice”, which produce human lambda antibodies, by introducing IGHL-NAC, an MAC vector containing the full-length human Ig (hIg) heavy chain locus (1.8 Mb) and lambda light chain locus (2.2 Mb), in which the endogenous antibody gene has been disrupted. We found that human Ig genes that loaded onto IGHL-NAC effectively function as a B cell receptor (BCR) in mice and promote the production of human lambda antibodies with diversity similar to that observed in humans. Moreover, IGHL mice were capable of producing antigen-specific human lambda antibodies after antigen injection. Our findings suggest that IGHL-NAC is a useful tool for the generation of trans-chromosomal mice that produce human lambda antibodies. Furthermore, IGHL mice are expected to provide a promising platform for the development of human lambda antibody therapeutics.
Results
Generation of mice carrying the full-length human heavy and lambda light chain loci
The MAC was engineered by excising the gene region of the native mouse chromosome.22,23 Leveraging the Cre/loxP and Flp/FRT systems, similar to those used in the generation of the TC-mAb mice, the IGL locus spanning 2.2 Mb on hChr.22 and the IGH locus spanning 1.8 Mb on hChr.14 were translocated onto the MAC in Chinese hamster ovary (CHO) cells, resulting in the creation of IGHL-NAC (Figures 1A and S1–S4). Alongside IGH and IGL, IGHL-NAC harbors the enhanced green fluorescent protein (EGFP) gene under the control of the CAG promoter and the neomycin resistance gene driven by the PGK promoter.15,22
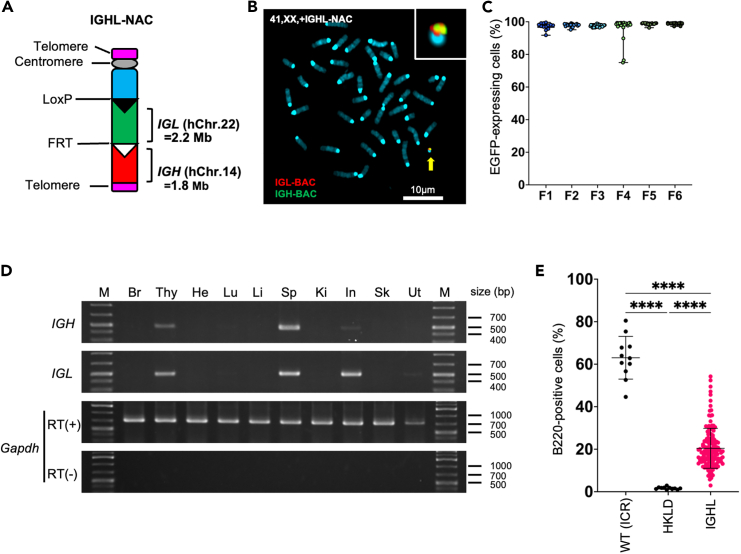
Figure 1.
Generation of mice carrying the full-length human heavy and lambda light chain loci
(A) Representation of the IGHL-NAC structure. The human immunoglobulin heavy chain locus (IGH) derived from human chromosome 14 (hChr.14) and immunoglobulin lambda light chain locus (IGL) derived from human chromosome 22 (hChr.22) are represented on a mouse artificial chromosome (MAC).
(B) Representative image of metaphase fluorescence in situ hybridization analysis with IGL-BAC (CH17-95F2) (red) and IGH-BAC (CH17-212P11) (green) to detect IGHL-NAC in peripheral blood mononuclear cells from an IGHL mouse. The arrow indicates IGHL-NAC, and the inset shows an enlarged image thereof. Scale bar, 10 μm.
(C) Percentage of enhanced green fluorescent protein (EGFP)-expressing cells in peripheral blood mononuclear cells from IGHL mice of different generations. Data are presented as means. Error bars indicate the standard deviation of the data.
(D) Reverse transcription polymerase chain reaction analysis of total RNA from various tissues from a 10-week-old IGHL mouse. Li, liver; In, intestine; Ki, kidney; Sp, spleen; Lu, lung; He, heart; Sk, skeletal muscle; Thy, thymus; Br, brain; and Ut, uterus. M, size marker.
(E) Percentage of B220-positive cells in the lymphocyte fraction of the peripheral blood from 4-week-old wild-type (WT; n = 11, ICR), HKLD (n = 11, Igh−/−, Igk−/−, and Igl1low/low), and IGHL mice (n = 139). Data are presented as means. Error bars indicate the standard deviation of the data. Gating strategies are presented in Figure S7.
To generate IGHL-NAC–carrying mice, IGHL-NAC was introduced into mouse embryonic stem cells (mESCs) from CHO K1 cells by MMCT (Figures S1 and S5A–S5C). The mESCs were derived from endogenous Ig-knockout (HKLD) mice.24
IGHL-NAC-carrying mESCs were transferred into C57BL/6 mouse embryos to generate chimeric mice. Those exhibiting a high degree of coat-color chimerism were subsequently crossed with HKLD mice to produce IGHL-NAC-carrying mice, referred to as IGHL mice. The maintenance of IGHL mice was ensured by continuous mating with HKLD mice. The germline transmission efficiency of IGHL mice was comparable to that observed in the previous study.25 IGHL mice possess only one IGHL-NAC and are maintained by crossing with HKLD mice, which do not have the IGHL-NAC. Therefore, the theoretical probability of IGHL mice being born is 50% due to meiosis. The actual birth frequency of IGHL mice was 69 out of 183 (37.70%) from IGHL males crossed with HKLD females and 96 out of 229 (41.92%) from HKLD males crossed with IGHL females, which is similar to the germline transmission rate observed for MAC1.25 The independent, stable retention of IGHL-NAC in IGHL mice was confirmed through fluorescence in situ hybridization (FISH) chromosome analysis of mouse lymphocytes and by monitoring the expression of the EGFP gene carried under the CAG promoter on IGHL-NAC through multiple generations (Figures 1B, 1C, S5D, S6, and S7). These findings corroborate the successful introduction and stable retention of a single IGHL-NAC, independent of mouse endogenous chromosomes. Furthermore, mRNA expression of the IGH and IGL genes were observed in the spleen, thymus, and intestine, where B cells are abundant, suggested cell-type-specific transcription of hIg genes (Figure 1D).
The BCR plays a pivotal role in the development of B cells. In HKLD mice, characterized by the absence of mouse antibody genes, significantly fewer B220-positive B cells were observed than in wild-type animals. Interestingly, a significant recovery in the number of B220-positive cells was observed in IGHL mice carrying IGHL-NAC, indicating that the hIg genes on IGHL-NAC acted as surrogates for the original mouse antibody genes (Figures 1E and S7). This observation suggests that IGHL-NAC effectively supports B cell development in the absence of endogenous mouse Ig genes.
The recovery of B220-positive B cell production was limited in IGHL mice compared with wild-type mice. This is a common phenomenon with TC-mAb mice.14
IGHL mice produce human lambda light chain antibodies
Subsequently, we investigated the production of human lambda antibodies in the IGHL mice. Analysis of human antibody expression on the surface of B220-positive splenocytes was conducted with anti-hIgM and hIgλ antibodies (Figures 2A and S8; Table S1). The findings validated the production of human lambda antibodies by B220-positive cells, suggesting that the hIg genes on IGHL-NAC were effectively expressed and that the human lambda antibodies served as functional BCRs. The hIgM-positive but hIgλ-negative B cell population is thought to express a complex formed by mouse Igλ from HKLD mice and hIgM from IGHL-NAC. The same B cell population has been reported in TC-mAb mice.14 The expression levels of BCRs were found to be lower in IGHL mice than in wild-type mice (Figures 2B and 2C).
Figure 2.
IGHL mice produce human lambda antibodies
(A) Human B cell receptor (BCR; human immunoglobulin [Ig] μ+/human Ig λ+) expression in B220-positive cells from IGHL mice. Flow cytometric analysis of stained splenocytes from 10-week-old IGHL mice. Gating strategies are presented in Figure S8A.
(B) Mouse BCR (mouse Ig μ+/mouse Ig κ+) expression in B220-positive cells of ICR mice. Flow cytometric analysis of stained splenocytes from 10-week-old ICR mice. Gating strategies are presented in Figure S8B.
(C) Percentages of BCR expression by IGHL mice and ICR mice (n = 3). Data are presented as means. Error bars indicate the standard deviation of three independent mice.
(D) The serum concentrations of different classes of human Igs in 10-week-old IGHL mice. The concentrations of human Ig μ, γ, λ, α, and ε in IGHL mice (hIg μ, γ, λ: n = 13; hIg α and ε: n = 8) were measured.
(E) The serum concentrations of human IgG subclasses. The concentrations of Ig γ1, 2, 3, and 4 were measured in 10-week-old IGHL mice (n = 6). All boxplots in this figure are indicated in terms of minima, maxima, center, bounds of box and whiskers (1.5 interquartile range value), and percentile in the style of Tukey.
We next analyzed the isotypes of the antibodies secreted into the peripheral blood using sandwich enzyme-linked immunosorbent assays (ELISAs) on serum collected from 10-week-old IGHL mice. The results revealed the secretion of not only IgM and IgG but also IgA and IgE antibodies (Figure 2D; Table S2). Moreover, we verified the expression of IgG1, IgG2, IgG3, and IgG4 (Figure 2E). These findings suggest that the regulatory mechanism governing class switching, observed in wild-type mice, functions effectively for the hIg loci in IGHL mice.
Production of antigen-specific antibodies
SARS-CoV-2, the causative agent of coronavirus disease 2019, has undergone multiple mutations since its emergence in 2019, facilitating its continued evasion of immune recognition.26,27,28 Consequently, it remains a critical focus of antibody development efforts. In this study, we evaluated whether IGHL mice can drive an immune response and produce antigen-specific antibodies using receptor-binding domain (RBD) as a model antigen, which is a crucial component of the interaction with human cells during SARS-CoV-2 infection.
IGHL mice were intraperitoneally administered recombinant RBD mixed with adjuvant every 2 weeks for a total of eight doses (Figure 3A). Peripheral blood samples were collected via tail vein puncture 3 or 4 days after each administration to obtain plasma. Additionally, final blood samples were collected through cardiac puncture at the time of euthanasia following the last immunization. By performing ELISAs on the collected sera, we analyzed the production of anti-RBD IgG antibodies. We observed the antigen-administration-dependent production of RBD antigen-specific hIgG in IGHL mice (Figure 3B). Furthermore, the immunized mice had a significantly higher serum IgG concentration and a trend toward more splenocytes than the unimmunized group (Figures 3C and 3D; Table S3).
Figure 3.
Production of antigen-specific antibodies
(A) Immunization schedule.
(B) Antibody titers against the receptor-binding domain were measured by enzyme-linked immunosorbent assay. An anti-human IgG-Fc–horseradish peroxidase conjugate was used as a secondary antibody.
(C) The serum concentration of hIg γ before (6-week-old) and after immunization (20-week-old) of IGHL mice (left) and unimmunized IGHL mice (right). The symbols representing samples collected from the same mouse 6 weeks old and 20 weeks old are connected by a line.
(D) The number of lymphocytes. The numbers of harvested cells were counted using Trypan blue. Data are presented as means. Error bars indicate the standard deviation of three independent mice.
Moreover, we evaluated the immune response by detecting the generation of germinal center (GC) B cells and antigen-specific B cells in the spleen following the final immunization, as well as differentiation into plasma cells (PCs) (Table S1). Significantly more GC B cells were found in the spleens of immunized mice compared with those of unimmunized mice (Figures 4A and S9). Compared to the unimmunized group, the immunized mice showed a tendency toward having more antigen-specific B cells (p = 0.0567) (Figures 4B and S9). There was no significant increase of PCs in the spleen of the immunized group than those of the unimmunized controls (Figures 4C and S10). However, a significant increase in GC B cells and the increase in antigen-specific antibodies suggest the production of antigen-specific PCs (Figures 4A, 3B, and 3C). Taken together, these findings suggest continuity of the humoral immune response in IGHL mice, wherein B cells bearing antigen-specific BCRs undergo class switching to IgG and differentiate into PCs to produce antibodies, thereby enabling IGHL mice to mount an effective humoral immune response against antigens.
Figure 4.
Immune responses in IGHL mice
Flow cytometric analysis of B cell subsets, including germinal center B cells (A), receptor-binding domain (RBD)-specific hIgG-expressing B cells (B), and plasma cells (C), in the spleens of unimmunized (left) and immunized (right) IGHL mice. Data are presented as means. Error bars in this figure indicate the standard deviation of three independent mice. Gating strategies are presented in Figures S9 and S10.
Diversity of antibodies produced by IGHL mice
The antibodies produced by B cells derive their diversity from the random recombination of various V region gene segments during V(D)J recombination. For heavy chains, this process involves the recombination of V, D, and J gene segments, whereas for light chains, it entails the recombination of V and J gene segments. Thus, it is advantageous to utilize a greater variety of gene segments within the antibody V region gene locus to obtain antibodies with the desired paratope.
To analyze the diversity of the human antibody repertoire in IGHL mice, NGS was performed on RNA samples extracted from pooled splenocytes from five unimmunized IGHL mice, one IGHL mouse immunized with the RBD, and, as a comparative control, RNA samples from healthy adult donor human peripheral blood mononuclear cells (PBMCs) (Table S4).
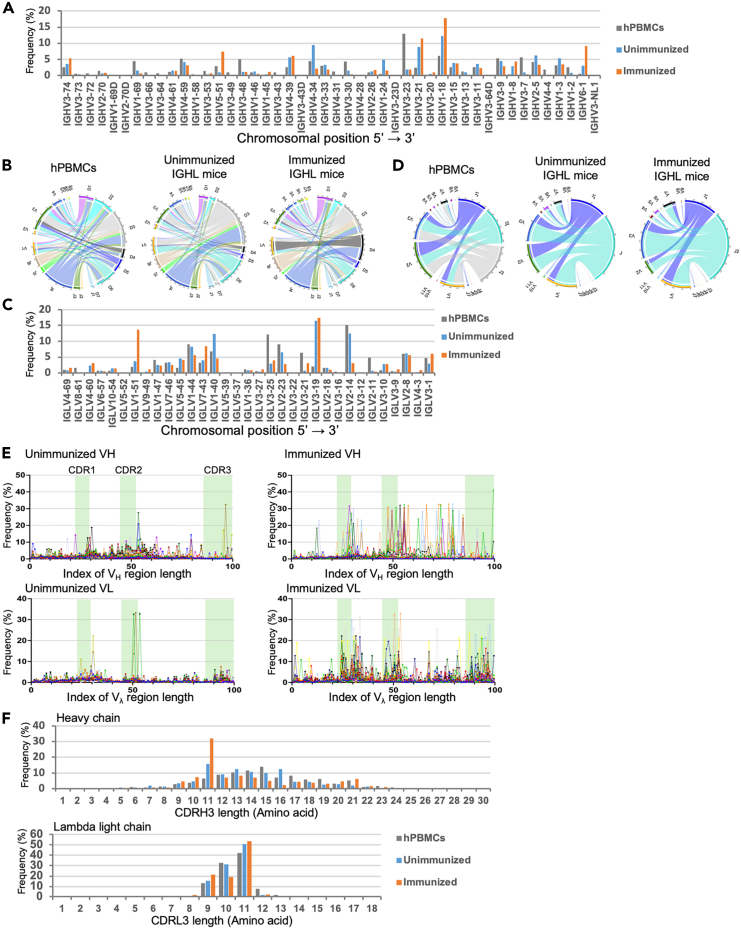
We investigated the combination of the frequency of usage of functional V gene segments as identified by IMGT (https://www.imgt.org/) and the reconstruction of variable regions through V(D)J rearrangement in both heavy and lambda light chains. The results showed that IGHL mice use a diverse repertoire of V, D, and J gene segments for the heavy chain (Figures 5A, S11A, and S11B; Table S5). In IGHL mice, IGHV1–18 was the most frequent segment in both unimmunized and immunized IGHL mice. This frequency was similar to that of IGHV3–23, the most frequently used segment in humans. These findings indicate that the bias in the available V gene segments in IGHL mice is as minimal as that in humans, consistent with previous reports on TC-mAb mice.14 The Shannon–Weaver index, which provides robust estimates of overall immune repertoire diversity and is calculated using Equation 1, indicated that V(D)J recombination in IGHL mice exhibited a similar level of diversity to that observed in humans (Table S6). However, the diversity of the lambda light chain J segments was lower. This discrepancy arises because the IGLJ3∗01 sequence is identical to the IGLJ201 sequence, resulting in IGLJ3 being counted together with IGLJ2. In contrast, human PBMC samples contain the IGLJ3∗02 sequence, which is counted separately as IGLJ3. The IGHL mice generated a variety of V(D)J rearrangement patterns akin to those in humans (Figures 5B and S12). In unimmunized IGHL mice, diverse combinations of V(D)J rearrangements were observed, similar to those in human PBMCs. In immunized IGHL mice, the frequencies of use of specific V-D combinations were higher than those in the unimmunized group. This increase may be due to clonal expansion of antigen-specific B cells in response to immunization. Furthermore, a diverse array of V genes was utilized in the lambda light chains, with varied patterns of VJ rearrangement (Figures 5C, 5D, S11C, and S12; Table S7). These results demonstrate that both the heavy and lambda light chains of hIg genes can undergo similar rearrangement in mice and humans, allowing for the formation of diverse antigen-binding sites. In addition, the use of each segment, regardless of whether it is proximal or distal to the Ig locus, indicates that all of the gene regions introduced into the IGHL mice are functional.
Figure 5.
Diversity of antibodies produced by IGHL mice
(A) Human VH gene utilization in human peripheral blood mononuclear cells (hPBMCs) from healthy donors and in IGHL mice with (female, 20-week-old) and without (female, 18-week-old, n = 5) immunization. The frequencies of V gene segment use (percentages) in healthy human donors (gray) and IGHL mice with (orange) and without immunization (blue) are represented.
(B) Circos plots comparing VDJ gene association. The gene segments are grouped as subfamilies and shown together with the first digit of their allele name. Links indicate the relative frequencies of specific VDJ combinations, and wider links indicate higher frequencies of recombination.
(C) Human VL gene utilization in hPBMCs (gray) and immunized IGHL (orange) and unimmunized IGHL (blue) mice. The same RNA samples as for the heavy chain analysis were used.
(D) Circos plots comparing VJ gene association. The same RNA samples as for the heavy chain analysis were used.
(E) The frequency of somatic hypermutation at every position of the V region was calculated as described in the STAR Methods section and is represented for VH and VL chains in immunized IGHL mice and unimmunized IGHL mice.
(F) CDR3 length comparison of productive rearrangements.
We also investigated whether somatic hypermutation (SHM) was induced in IGHL mice by antigen administration, since SHM is responsible for the generation of B cells producing high-affinity antibodies against an antigen.29 We targeted V(D)J clonotypes with more than 500 reads detected, analyzed mutation rates compared with germline sequences, and aligned the frequencies of SHM in the heavy and light chains of unimmunized and RBD-immunized IGHL mice along their adjusted relative positions (Figure 5E). The results indicated a higher frequency of mutations was introduced in both heavy and light chains of the immunized group, mainly at the complementarity-determining region (CDR) and framework region (FR)3. This suggests that antigen administration drives GC formation, resulting in affinity maturation to select B cells with increased binding strength to and specificity for the antigen. The CDR3 length, P and N nucleotide addition in the CDR3, and amino acid usage in the CDR3 of IGHL mice mimicked those of humans for both the heavy and lambda chains (Figures 5F, S13A, and S13B). These results indicate that it is possible to recapitulate the human antibody reorganization process in IGHL mice, retaining the complete human IGH and IGL loci, and achieve the formation of human-like combinatorial and junctional diversity.
Maps of mutations based on amino acid sequences and the expansion of clonotypes for each clone lineage were drawn using a phylogenetic tree (circular dendrogram). Based on a copy number with ≥500 reads in RBD-immunized IGHL mice, 164 VH clone lineages and 56 VL clone lineages were analyzed. The circular dendrograms of the 10 most frequent clone lineages for VH and VL sequences are shown in Figure S14. The average fold expansion of the top 10 clone lineages was 4.8 for VH sequences and 4.1 for VL sequences (Table S8). These results indicate that antibody affinity maturation, accomplished through SHM and clonal expansion, is induced by immunization. Therefore, upon immunization of IGHL mice, clonal selection and expansion of B cells occur, followed by SHM, leading to the antigen-driven diversification of VH and VL sequences and contributing to the generation of B cells with higher-affinity antibodies.
Discussion
In this study, we generated mice capable of producing human lambda antibodies. Furthermore, we addressed the challenge of limited antibody diversity due to constraints on the loadable Ig region associated with conventional transgenic technology by employing chromosomal engineering techniques. By utilizing an MAC vector, we overcame this limitation and successfully created IGHL mice.
There are evident differences in the binding characteristics of kappa and lambda light chains, with reports indicating that the CDR3 of lambda light chains is generally longer and more hydrophobic.19,21,30 Additionally, the mutation pattern of lambda light chains has been reported to differ from that of kappa light chains.31 These disparities suggest that each light chain may play distinct biological roles. Light chains have lower V gene segment diversity than heavy chains. Although some arguments suggest that the choice between kappa and lambda light chains does not substantively affect the binding activity of antibodies,32 the usage of light chains in antibodies produced against viruses, toxins, and vaccines is biased.19,20 In particular, in patients with human immunodeficiency virus (HIV) and anti-viral vaccine recipients, a significant proportion of antibodies targeting HIV membrane proteins (e.g., gp120) utilize lambda light chains.19 Therefore, the choice of light chain is a critical issue, and lambda antibodies, like kappa antibodies, hold potential value as pharmaceuticals.
However, the proportion of lambda light chain antibodies among currently approved antibody therapeutics is only about 10%. This is notably lower than the 40% usage of lambda chains in human blood. This bias could stem from the development of humanized antibodies from mouse kappa antibodies, which predominate in mice, and the lack of human lambda light chains introduced in many animal models used for human antibody production. Moreover, the CDR3 of the lambda light chain is on average longer, more hydrophobic, and more acidic than that of the kappa light chain. Therefore, the kappa isotype may be intentionally selected based on superior stability or other biophysical properties.18,33
In this study, the introduced IGHL-NAC expressed human lambda antibodies in the mice, which functioned as BCRs. Moreover, the production of various antibody isotypes demonstrated the capability of intrinsic murine Ig gene expression regulatory mechanisms to control the expression of hIg genes. Compared to wild-type mice, IGHL mice have fewer B220-positive B cells. This might be due to the technical challenge of having only one copy of the immunoglobulin locus introduced. Additionally, the lower BCR expression in B220-positive B cells could be attributed to species differences, as the Igα and Igβ proteins involved in BCR expression have not been humanized. However, the successful production of human lambda antibodies by introducing the full length of the human immunoglobulin locus align with findings from previous studies, such as those on the TC-mAb mouse model developed by Satofuka et al., and further support the feasibility of regulating the expression of hIg genes in mice.14 TC-mAb (IGHK) mice and IGHL mice had the same heavy chain locus, and both mice had a similar usage pattern of heavy chain V, D, and J segments. This reproducibility indicates the stability of MACs as gene delivery vectors.
Our observations revealed not only antibody production but also behaviors characteristic of humoral immunity, such as increased numbers of B cells, GC formation, and differentiation into PCs, following antigen administration. Additionally, we confirmed the production of antigen-specific antibodies, suggesting the feasibility of generating lambda light chain antibodies against the target antigen. Furthermore, Ig transcript repertoires in the spleen were analyzed, revealing the utilization of diverse V gene segments for both heavy and lambda light chains and evidence of SHM. These findings suggest the potential for antibody acquisition against various antigens.
Consequently, we anticipate that IGHL mice will be valuable tools in the development of lambda light chain antibody therapeutics, which are currently underrepresented in the pharmaceutical landscape.
Limitation of the study
Immunoglobulin loci normally exist as two copies each of IGH, IGK, and IGL. During B cell differentiation, rearrangements of immunoglobulin light chain genes occur sequentially, starting with the rearrangement of the κ chain followed by the λ chain. The IGHL mice generated in this study have only one copy each of the human immunoglobulin loci IGH and IGL, which may result in partially incomplete B cell development. The generation of TC-mAb and IGHL mice demonstrates that human immunoglobulins function effectively within a mouse model. Therefore, future efforts to generate mice with two copies each of the IGH, IGK, and IGL loci will better emulate human B cell development.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yasuhiro Kazuki (kazuki@tottori-u.ac.jp).
Materials availability
Mouse models generated in this study, namely, IGHL mice, are available from the lead contact’s laboratory upon request.
Data and code availability
-
•
Data are available from the lead contact and corresponding authors. The repertoire analysis data generated in this study have been deposited in the DDBJ database under accession code PRJDB18270. SAMD00795425 (TCK152MG_S41_S43) and SAMD00795428 (TCK152L_S46) are the data of an RBD-immunized IGHL mouse individual. SAMD00795424 (TCK111MG_S56_S62) and SAMD00795427 (TCK111L_S52) are the data of unimmunized IGHL mice (pooled, 5 individuals). SAMD00795432 (TCK041L_S19) is the data of a human PBMC pool sample.
-
•
No original code was reported in this article.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
We thank Y. Sato (DNA Chip Research Inc.) for assistance with generating Circos plots, VH and VL frequency analyses, and circular dendrograms. We thank Y. Sumida, E. Kaneda, A. Ashiba, M. Morimura, K. Yoshida, M. Fukino, F. Adachi, T. Kurosaki, Y. Ohkame, and S. Takata at Tottori University for assistance with generating and maintaining IGHL mice. We thank Mr H. Sugihara of the Technical Department, Tottori University, for his technical support. We also thank Dr H. Kugoh, Dr M. Oshimura, Dr T. Ohbayashi, Dr Y. Nakayama, Dr T. Ohira, Dr S. Hamamichi, Dr H. Satofuka, and Dr Y. Hiramuki at Tottori University and Dr K. Tomizuka and Dr N. Uno at Tokyo University of Pharmacy and Life Sciences. We thank Amanda Holland, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This study was supported in part by the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP18am0301009 (Y.K.), JP21am0101124 (Y.K.), JP24ama121046 (Y.K.), JP24gm1610006 (Y.K.), JP24gm0010010 (Y.K.), JP23a.m.0401002 (Y.K.), and JP24gm1810008 (Y.K.); the Joint Research of the Exploratory Research Center on Life and Living Systems (ExCELLS) (ExCELLS program No. 21-101) (Y.K.); and JST CREST Grant Number JPMJCR18S4, Japan (Y.K.). This research was partly performed at the Tottori Bio Frontier managed by Tottori prefecture.
Author contributions
K.S. wrote the manuscript. Y.K. and S.A. planned the study. S.A., K.K., and A.O. performed cell culture and mouse experiments. K.K. performed cytogenetic analyses. K.S., S.B., Y.M., T.M., and S.A. performed flow cytometric analyses and ELISAs. Y.K. contributed to analysis and discussion of the data. K.S., T.M., S.A., S.B., and Y.K. analyzed the results. Y.K. and S.A. supervised the project.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-IgG(Fc), Human, Goat-Polyclonal Antibody | Bethyl Laboratories | Cat# A80-304A; RRID: AB_10634115 |
| Anti-IgG(Fc), Human, Goat-Poly, HRP | Bethyl Laboratories | Cat# A80-304P; RRID: AB_10631811 |
| Anti-IgM(Fc), Human, Goat-Poly | Bethyl Laboratories | Cat# A80-100A; RRID: AB_67079 |
| Anti-IgM(Fc), Human, Goat-Poly, HRP | Bethyl Laboratories | Cat# A80-100P; RRID: AB_67082 |
| Anti-Igλ Light Chain, Human, Goat-Poly | Bethyl Laboratories | Cat# A80-116A; RRID: AB_67093 |
| Anti-Igλ, Human, Goat-Poly, HRP | Bethyl Laboratories | Cat# A80-116P; RRID: AB_67591 |
| Anti-IgA(α), Human, Goat-Poly | Bethyl Laboratories | Cat# A80-102A; RRID: AB_67044 |
| Anti-IgA, Human, Goat-Poly, HRP | Bethyl Laboratories | Cat# A80-102P; RRID: AB_67047 |
| Anti-IgE(ε), Human, Goat-Poly | Bethyl Laboratories | Cat# A80-108A; RRID: AB_67053 |
| Anti-IgE, Human, Goat-Poly, HRP | Bethyl Laboratories | Cat# A80-108P; RRID: AB_67056 |
| PE anti-mouse/human CD45R/B220 Antibody | Biolegend | Cat# 103207; RRID: AB_312992 |
| PE/Cyanine7 anti-mouse/human CD45R/B220 | Biolegend | Cat# 103221; RRID: AB_313004 |
| BD Horizon™ BUV661 Rat Anti-Mouse CD19 | BD Biosciences | Cat# 612971; RRID: AB_2870243 |
| BD Horizon™ PE-CF594 Mouse Anti-Human IgM | BD Biosciences | Cat# 562539; RRID: AB_2737641 |
| PE/Cyanine7 anti-human IgG Fc Antibody | Biolegend | Cat# 410721; RRID: AB_2750226 |
| PE/Cyanine7 anti-human Ig light chain λ Antibody | Biolegend | Cat# 316622; RRID: AB_2687262 |
| Pacific Blue™ anti-mouse/human GL7 Antigen (T and B cell Activation Marker) Antibody | Biolegend | Cat# 144613; RRID: AB_2563291 |
| BD OptiBuild™ BV650 Rat Anti-Mouse CD38 | BD Biosciences | Cat# 740489; RRID: AB_2740212 |
| PE/Cyanine7 anti-mouse CD138 (Syndecan-1) Antibody | Biolegend | Cat# 142513; RRID: AB_2562197 |
| PE anti-mouse CD267 (TACI) Antibody | Biolegend | Cat# 133403; RRID: AB_2203542 |
| Bacterial and virus strains | ||
| XL10-Gold Ultracompetent Cells | Agilent | Cat# 200315 |
| Chemicals, peptides, and recombinant proteins | ||
| Antibiotic G-418 Sulfate | Promega | Cat#V7983 |
| Blasticidin S | FUJIFILM Wako Pure Chemical Corporation | Cat# 029-18701 |
| Hygromycin B | FUJIFILM Wako Pure Chemical Corporation | Cat# 089-06151 |
| HAT Media Supplement (50×) Hybri-Max™ | Sigma-Aldrich | Cat# H0262-10VL |
| Lipofectamine™ 2000 Transfection Reagent | ThermoFisher Scientific | Cat# 11668027 |
| Sigma Adjuvant System | Sigma-Aldrich | Cat# S6322 |
| Freund's Complete Adjuvant | BD | Cat# DF0638-60-7 |
| Ni-NTA Agarose | QIAGEN | Cat# 30210 |
| Critical commercial assays | ||
| Human IgG Subclass ELISA Kit | ThermoFisher Scientific | Cat# 99-1000 |
| Expi293™ Expression System Kit | ThermoFisher Scientific | Cat# A14635 |
| Deposited data | ||
| Repertoire analysis of mice producing fully human antibody with lambda light chain | This paper | DDBJ: PRJDB18270 |
| Experimental models: Cell lines | ||
| Chinese hamster ovary (CHO) (Hprt-/-) cells | JCRB Cell Bank | JCRB0218; RRID:CVCL_8545 |
| CHO-K1 | RIKEN BRC | RCB0285; RRID:CVCL_0214 |
| DT40 | RIKEN BRC | RCB1464; RRID:CVCL_0249 |
| Endogenous Ig knockout (HKLD) mouse embryonic stem cells | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse:ICR:Jcl:ICR | CLEA | RRID: IMSR_JCL:JCL:mOT-0001 |
| Mouse:C57BL/6N | CLEA | RRID:MGI:3055581 |
| Mouse:HKLD:STOCK Ighmtm1YakazIgkctm1YakazIglc1rs32383745-A | Satofuka et al.14 | N/A |
| Mouse:IGHL:STOCK Tc(MAC;IGL;IGH)1Yakaz IghmIghmtm1YakazIgkctm1YakazIglc1rs32383745-A | This paper | N/A |
| Oligonucleotides | ||
| hChr.22: arm for loxP insertion HindIII553La L 5'-TGTAGCTGACTTTAGCCACCCACAAGTAC-3' AscI553La R 5'-TCGAGGCGCGCCCTCAAACTC CTGGGTGTAAATGATCCTCCTGC-3' (5,743 bp) |
This paper | N/A |
| hChr.22: arm for loxP insertion KpnI553Ra L 5'-TGAG GGTACCGTGCAGTAAAGTATGATTGAGC-3' SalI553Ra R 5'-TCGAGTCGACCTTGCTGATTA TACCTCATCTCCTTCCCTC-3' (3,880 bp) |
This paper | N/A |
| hChr.22: arm for FRT insertion BamHISL350La L 5'-TCGAGGATCCGGCCTCCCAAAGGATTAT AGACGTGAGCCACTGTAscISL350La R-3' AscISL350La R 5'-TCGAGGCGCGCCGGC ACCTCTCCTATTTTCTTCACAGCACTT-3' (3,595 bp) |
This paper | N/A |
| hChr.22: arm for FRT insertion AscISL350Ra L 5'-TCGAG GCGCGCCAGCATGGTGGCCCGCACGTATAGTCGC AGCTA-3' NotISL350Ra R 5'-TCGAGCGGCCGCAAA GAAGGGGCCCGCCTCTGCCTCTAAATCC TGAC-3' (3,872 bp) |
This paper | N/A |
| hChr.22: loxP targeting check 22CeT La L 5'-CCTG CCTTCTTGTTTCAGCTCTCAACTG-3' 22CeT La R 5'-GACGTGCTACTTCCATTTGTCACGTCCT-3' (4,794 bp) |
This paper | N/A |
| hChr.22: loxP targeting check 22CeT Ra L 5'-ATCCCCATGTGTATCACTGGCAAAC TGT-3' 22CeT Ra R 5'-ACACTTTAGTCC CTGTCCCCTCAACGAG-3' (4,791 bp) |
This paper | N/A |
| hChr.22: FRT targeting check 22TeT Ra L 5'-AGCAGAGCTCGTTTAGTGAACCGT CAGA-3' 22TeT Ra R 5'-CTGTCCTATC CTTGCAGCTGTCTTCCAG-3' (4,911 bp) |
This paper | N/A |
| hChr.22 region check SERPIND1 L 5'-ACCT AGAGGGTCTCACCTCC-3' SERPIND1 R 5'-CCCTGGACATCAAGAATG-3' (223 bp) |
This paper | N/A |
| hChr.22 region check 553P-F 5'-AGATCTC TTGAGCCCAGCAGTTTGA-3' 553P-R 5'-TGAAGTTAGCCGGGGATACAG ACG-3' (454 bp) |
This paper | N/A |
| hChr.22 region check PPM1F L 5'-AACGG CAGCCAAACCAAAGA-3' PPM1F R 5'-ACC AGGACTGGCTGGGCATA-3' (207 bp) |
This paper | N/A |
| hChr.22 region check IGLVI-70 L 5'-AGTC TGCGCTGACCCAGGAA-3' IGLVI-70 R 5'-TTGAGCCAGAGAAGCGGTCA-3' (195 bp) |
This paper | N/A |
| hChr.22 region check hVpreB1-F 5'-TGTCCTG GGCTCCTGTCCTGCTCAT-3' hVpreB1-Rm 5'-GGCGGCGACTCCACCCTCTT-3' (589 bp) |
This paper | N/A |
| hChr.22 region check IgL-F 5'-GGAGACCAC CAAACCCTCCAAA-3' IgL-Rm 5'-GAGAGTTG GAGAAGGGGTGACT-3' (482 bp) |
This paper | N/A |
| hChr.22 region check GNAZ L 5'-TCCACTTGG GGGTCTGCATT-3' GNAZ R 5'-TGGTGCTGA GCAGCTGTGTG-3' (202 bp) |
This paper | N/A |
| hChr.22 region check 344-F 5'-ATCATCTGCT CGCTCTCTCC-3' 344-R 5'-CACATCTGTAGT GGCTGTGG-3' (450 bp) |
This paper | N/A |
| hChr.22 region check hλ5-F 5'-AGCCCCAAG AACCCAGCCGATGTGA-3' hλ5-R 5'-GGCAG AGGGAGTGTGGGGTGTTGTG-3' (529 bp) |
This paper | N/A |
| hChr.22 region check hVpreB3-F 5'-CACTGC CTGCCCGCTGCTGGTA-3' hVpreB3-R 5'-GG GCGGGGAAGTGGGGGAGAG-3' (511 bp) |
This paper | N/A |
| hChr.22 region check 350P-F 5'-ACCAGCGC GTCATCATCAAG-3' 350P-R 5'-ATCGCCAG CCTCACCATTTC-3' (461 bp) |
This paper | N/A |
| hChr.22 region check LIF L 5'-TGGGACTTA GGTGGGCCAGA-3' LIF R 5'-GCCTCCCCA AGAGCCTGAAT-3' (199 bp) |
This paper | N/A |
| IGL expression Vλ3LEA1 5'-CCCCCAAGCTT GCCTGGACCCCTCTCTGG-3' |
This paper | N/A |
| IGL expression Cλ1 5'-GGGAATTCGGGTAG AAGTCACTGATCAG-3' |
This paper | N/A |
| IGL expression Vλ3JLEAD 5'-ATCGGCAAAG CTTGGACCCCTCTCTGGCTCAC-3' |
This paper | N/A |
| IGL expression Vλ3BACK4 5'-CCCCCAAGCT TCTCGGCGTCCTTGCTTAC-3' |
This paper | N/A |
| IGL expression Cλ7 5'-GGGAATTCGGG TAGAAGTCACTTACGAG-3' |
This paper | N/A |
| cDNA synthesis BSL-18E 5'-AAAGCGGC CGCATGCTTTTTTTTTTTT TTTTTTVN-3' |
Kitaura et al.34 | N/A |
| Adaptor P10EA 5'-GGGAATTCGG-3' | Kitaura et al.34 | N/A |
| Adaptor P20EA 5'-TAATACGACT CCGAATTCCC-3' |
Kitaura et al.34 | N/A |
| IgG constant region CG1 5'-CACCT TGGTGTTGCTGGGCTT-3' |
Kitaura et al.34 | N/A |
| Second PCR CB2 5'-AGGCAGTATCT GGAGTCATTGAG-3' |
Kitaura et al.34 | N/A |
| Second PCR CG2 5'-TCCTGAGGACT GTAGGACAGC-3' |
Kitaura et al.34 | N/A |
| Second PCR P22EA-ST1 5'-GTCTCGTGG GCTCGGAGATGTGTATAAGAGACAGC TAATACGACTCCGAATTCCC -3' |
Kitaura et al.34 | N/A |
| Second PCR CB-ST1-R 5'-TCGTCGGCAG CGTCAGATGTGTATAAGAGACAGGCT CAAACACAGCGACCTC-3' |
Kitaura et al.34 | N/A |
| Second PCR P22EA-ST1 5'-GTCTCGTGGG CTCGGAGATGTGTATAAGAGACAGCTAA TACGACTCCGAATTCCC -3' |
Kitaura et al.34 | N/A |
| Second PCR CG-ST1-R 5'-TCGTCGGCAGCG TCAGATGTGTATAAGAGACAGTGAGTTCC ACGACACCGTCAC-3' |
Kitaura et al.34 | N/A |
| Recombinant DNA | ||
| IGH-BAC | BACPAC resource center | CH17-212P11 |
| IGL-BAC | BACPAC resource center | CH17-95F2 |
| RBD expression vector | This paper | N/A |
| 5′ HPRT-loxP-PGKHyg unit | Satofuka et al.14 | N/A |
| 5′ HPRT-FRT-CMVBsd unit | Satofuka et al.14 | N/A |
| pBS185 CMV-Cre | Addgene | RRID:Addgene_11916 |
| pCAG-FLPo vector | Satofuka et al.14 | N/A |
| Software and algorithms | ||
| Metafer4 slide scanning platform software V3.10.5 | Meta Systems | https://metasystems-international.com/en/products/metafer/ |
| Cytexpert version 2.3 | Beckman Coulter | https://www.beckman.jp/flow-cytometry/software |
| Isis Fluorescence Imaging System software V5.4.12 | Meta Systems | https://metasystems-international.com/ |
| R software version 3.4.2 | The R Project for Statistical Computing | https://www.r-project.org/ |
| IGBLAST | NIH | https://www.ncbi.nlm.nih.gov/igblast/ |
| IMGT | IMGT | https://www.imgt.org/ |
| CLASTALW | GenomeNet | https://www.genome.jp/tools-bin/clustalw |
| Principal component analysis was performed using the prcomp-function in R (version 3.4.2) | RDocumentation | https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/prcomp |
| R software using circos open-source software | circos | https://circos.ca/ |
| Biotek Gen5 version 3.03 | Agilent | https://www.chem-agilent.com/contents.php?id=1006877 |
| Graphpad Prism 10 | Graphpad | https://www.graphpad.com/updates/prism-1000-release-notes |
| Kaluza software version2.1. | Beckman Coulter | https://www.beckman.jp/flow-cytometry/software/kaluza |
Experimental model and study participant details
Mice
Experimental and control animals were cohoused in a controlled, ambient-temperature environment with a 12-h light/dark cycle in the specific pathogen-free (SPF) animal facility. All animal experiments were approved by the Animal Care and Use Committee of Tottori University (Permit Numbers: 14-Y-23, 15-Y-31, 16-Y-20, 17-Y-28, 20-Y-13, 20-Y-31, 21-Y-26, and 22-Y-36). All animal experiments were performed with 4, 6 or 10-week-old female mice.
IGHL mouse generation
To generate chimeric mice, HKLD mESCs carrying IGHL-NAC were injected into eight-cell-stage embryos derived from C57BL/6N mice (CLEA, Tokyo, Japan) and then transferred into pseudopregnant ICR females (CLEA, Tokyo, Japan). Chimeric mice with a high percentage of coal colour, derived from the injected HKLD mESCs, were crossed with HKLD mice (Igh-/-, Igk-/-, and Igl1low/low) to obtain a mouse strain carrying IGHL-NAC. IGHL mice were maintained by crossing IGHL and HKLD mice.
Materials
CHO (Hprt-/-) cells (JCRB0218) were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan) and CHO K1 (RCB0285) and chicken DT40 (RCB1464) cells were obtained from the Riken BioResource Research Center (Ibraraki, Japan). Primers were purchased from Sigma-Aldrich (St Louis, MO, USA). Escherichia coli strain XL10-Gold was purchased from Agilent Technologies (Santa Clara, CA, USA). The total RNA from human PBMCs (Takara Bio USA, San Jose, CA, USA) used in this study was derived from normal human peripheral leucocytes pooled from 426 male/female Asian individuals aged 18–54 years old.14
Method details
Cell culture
Chicken DT40 cells, CHO (Hprt-/-), and CHO K1 cells were cultured as described in a previous study.14 CHO cells carrying the MAC or IGHL-NAC were cultured in selection medium with 800 μg/ml G418. HKLD mESCs derived from HKLD mice and mouse embryonic fibroblasts as feeder cells were also cultured as in previous studies.14,24
Construction of targeting vectors
Targeting vector to introduce loxP-5′ HPRT into human chromosome 22
To prepare the homology arm, DNA from DT40 cells with human chromosome 22 (hChr.22) was used as a template for polymerase chain reaction (PCR). The left and right arms were amplified using the following primers: HindIII553La L and AscI553La R for the left arm and KpnI and SalI553Ra R for the right arm. The arms (left: 4.4 kb, right: 3.9 kb) were inserted into the vector containing the 5′ HPRT-loxP-PGKHyg unit by HindIII/AscI and KpnI/SalI digestion.
Targeting vector to introduce FRT-5′ HPRT into hChr.22
The left and right arms were amplified using the following primers: BamHISL350La L and AscISL350La R for the left arm and AscISL350Ra L and NotISL350Ra R for the right arm. The arms (left: 3.6 kb, right 3.9 kb) were inserted into the vector containing the 5′ HPRT-FRT-CMVBsd unit by BamHI/AscI and MluI/NotI digestion. The primer sequences are described in key resources table.
Modification of hChr.22 in DT40 cells
Homologous recombination-proficient chicken DT40 cells (1×107) in 0.5 ml RPMI were electroporated at 550 V and 25 μF with 25 μg of linearized targeting vector using a Gene Pulser (Bio-Rad, Hercules, CA, USA). Drug-resistant DT40 clones were obtained in selection medium with 1.5 mg/ml hygromycin or 15 μg/ml blasticidin S. Each homologous recombination on hChr.22 in DT40 hybrids was confirmed by genomic PCR using the primers listed in key resources table.
Microcell-mediated chromosome transfer
Microcell-mediated chromosome transfer (MMCT) was performed as described previously.35 hChr.22loxPFRT was transferred from DT40 cells into CHO cells containing the MAC via MMCT. Microcell hybrids were obtained in selection medium with 800 μg/ml G418, 8 μg/ml blasticidin S, and 10 μM ouabain. The MAC carrying the IGL locus (IGL-NAC) was transferred into CHO K1 cells and hybrid clones were selected in medium with 8 μg/ml blasticidin S and 800 μg/ml G418. IGL-NAC was transferred from CHO K1 cells to CHO (hChr.14-FRT) cells14 and hybrid clones were selected in medium with 800 μg/ml G418 and 8 μg/ml blasticidin S. IGHL-NAC was transferred into CHO K1 cells and HKLD mESCs and selected with 800 μg/ml and 250 μg/ml G418, respectively.
DNA transfection
The pBS185 vector for Cre expression (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) or pCAG-FLPo vector for FLPo expression was transfected into CHO hybrids carrying the modified hChr.22 and MAC vector, or hChr.14 and IGL-NAC, using Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s protocol. After 24 h of culture in basic growth medium, the cells were cultured in medium with 6 μg/ml blasticidin S and 1×HAT (Sigma-Aldrich) for selection.
Genomic PCR
Genomic DNA was purified from each cell line and IGHL mouse tissue samples using a genomic extraction kit (Gentra Systems, Minneapolis, MN, USA). Genomic PCR was conducted using the primers described in key resources table.14
IGHL-NAC construction
We constructed the MAC carrying the human IGH and IGL loci and verified by genomic PCR using the primers described in key resources table, and fluorescence in situ hybridization (FISH) at each step, following the same strategy as in a previous study(Figure S1).14 Modification of hChr.22 was conducted in DT40 cells for recombination-mediated translocation. First, a loxP site was inserted proximal to the IGL locus on hChr.22q by homologous recombination. The targeting vector was introduced into DT40 cells containing an intact hChr.22 with a neomycin resistance gene by electroporation, and drug-resistant clones were obtained in selection medium with 1.0 mg/ml hygromycin. FISH analyses confirmed that a single copy of hChr.22 with the loxP unit was independently maintained in DT40 cells (DT40 hChr.22loxP) (Figures S2A–S2D). Next, the FRT site was inserted distal to the IGL locus on hChr.22q. The targeting vector for FRT insertion was introduced into DT40 cells containing hChr.22loxP, and drug-resistant clones were selected with 15 μg/ml blasticidin S. FISH analyses confirmed that a single copy of hChr.22 with an FRT unit was independently maintained in DT40 cells (DT40 hChr.22loxPFRT) (Figures S2B and S2E). The modified hChr.22 was transferred from DT40 cells to CHO Hprt-/- cells carrying the MAC via MMCT. Microcell hybrids were obtained in selection medium with 800 μg/ml G418, 8 μg/ml blasticidin S, and 10 μM ouabain. The independent and stable maintenance of the MAC and modified hChr.22 in host CHO cells was confirmed by FISH analysis (Figure S3B).
Then, Cre/loxP recombination-mediated translocation of a region of hChr.22 distal from the loxP site, which included the IGL locus, to the MAC was conducted (Figure S3A), and drug-resistant clones were selected in medium with 6 μg/ml blasticidin S and 1×HAT. Each expected recombination junction was detected by genomic PCR, and FISH analysis confirmed the structure of the MAC carrying the IGL locus (IGL-NAC) and its by-product (Figure S3B).
IGL-NAC was transferred into CHO (hChr.14-FRT) cells through CHO K1 cells (Figures S4B and S4C).14 Flp/FRT recombination-mediated translocation between modified hChr.14 and IGL-NAC was performed in CHO Hprt-/- cells to clone the IGH locus into IGL-NAC; HPRT gene reconstruction on the desired product enabled the selection of CHO cells carrying IGL-NAC with the IGH locus (IGHL-NAC) and its by-product (Figure S4A). Drug-resistant clones were obtained in selection medium with 6 μg/ml blasticidin S and 1×HAT. FISH confirmed the independent and stable maintenance of IGHL-NAC and a by-product in host CHO cells (Figures S4D and S4E). The IGHL-NAC was transferred to CHO K1 cells to obtain donor CHO K1 cells carrying a single desired chromosome, IGHL-NAC, for further MMCT. Microcell hybrid clones were obtained in selection medium with 800 μg/ml G418 and were monitored for enhanced green fluorescent protein (EGFP) expression. FISH analyses confirmed the independent and stable maintenance of a single copy of IGHL-NAC in host CHO K1 cells (Figures S5A and S5B).
Reverse transcription PCR
Reverse transcription PCR was performed as described previously.14 The primer sequences are listed in key resources table.14
Antigen
In this study, the receptor-binding domain (RBD), a substructure of the S protein of the SARS-CoV-2 virus, was used as an immunological antigen and protein probe. Specifically, genes containing the Kozak sequence, the signal sequence of the MERS coronavirus spike protein, the RBD region of the SARS-CoV-2 S protein (amino acids 311 to 529), the myc tag sequence, and the 6×His sequence were inserted into the pcDNA3.1 vector. The genes were synthesized by GeneArt (Thermo Fisher Scientific, Waltham, MA, USA), taking into account optimal codon usage in humans. The synthesized RBD expression vector was expressed with the Expi293F Expression System (Thermo Fisher Scientific) protocol. Purification was then performed using Ni-NTA-agarose (Qiagen, Hilden, Germany) binding to His tags.
Immunization
The protein antigen (RBD, 1 mg/ml) was prepared in phosphate-buffered saline (PBS). Freund’s Complete Adjuvant (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and 100 μg of RBD were mixed 1:1 (v/v) to form an emulsion and administered to mice for the primary injection. For booster injection, 50 μg of RBD was mixed with Sigma Adjuvant System (Sigma-Aldrich) at 1:1 (v/v).
Injections were performed in 6-week-old female mice using a 1-ml glass syringe and a 27-gauge needle. Primary and booster injections were given intraperitoneally every 2 weeks for a total of eight times. The injection volumes were determined according to the relevant JP Home Office animal license for the procedure. Final boosters were delivered without adjuvant.
Enzyme-linked immunosorbent assay
Sandwich enzyme-linked immunosorbent assays (ELISAs) were used to measure the quantities of human immunoglobulins (hIgs), including hIgM, hIgG, hIgλ, hIgA, and hIgE. Key resources table provides the list of antibodies that were utilized. Using a mouse monoclonal anti-hIgM antibody (Bethyl Laboratories, Montgomery, TX, USA) fixed on 96-well plates (Nunc MaxiSorp, Thermo Fisher Scientific), the concentration of hIgM was measured. The antibody was then detected using peroxidase-conjugated mouse anti-hIgM (Bethyl Laboratories). Similarly, the capture and detection antibodies given in key resources table were used to evaluate human IgG, human Igλ, human IgA, and human IgE. The antibody conjugates, standards, and samples were all diluted in sample/conjugate buffer (0.05% Tween 20, 1% bovine serum albumin, 0.14 M NaCl, and 50 mM Tris). Utilising 3,3',5,5'-tetramethylbenzidine (TMB; Nacalai Tesque, Kyoto, Japan) as the substrate, a spectrophotometer (BioTek Instruments, Winooski, VT, USA) was used to measure the absorbance at 450 nm. Following the manufacturer's instructions, we used an IgG Subclass Human ELISA Kit (Invitrogen, Thermo Fisher Scientific) to determine the IgG subclasses.
Serum samples taken 3 or 4 days after antigen boost were analyzed by ELISA. We coated 96-well immunoassay plates (Nunc Maxisorp) with 100 μl/well of antigen (1 μg/ml in PBS) for 1 hr at room temperature. The plates were blocked with PBS containing 5% Difco™ skim milk (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) for 30 minutes at room temperature after washing three times with tris-buffered saline-Tween (TBS-T) (0.05% v/v). Following another round of plate washing, 100 μl serially diluted serum samples in TBS-T were added to the wells and allowed to incubate at room temperature for one hour. After incubation, the plates were again washed as above and incubated with 100 μl of anti-hIgG-Fc–horseradish peroxidase conjugate, added at 1:150,000 in TBS-T, for 30 min at room temperature. The plates were washed again as above and developed using 100 μl TMB; the reactions were stopped using 100 μl H2SO4 (1 M). The absorbance was read at 450 nm.
Flow cytometry analysis
To evaluate the phenotype of IGHL mice, bone marrow, spleens, and PBMCs were isolated from 10-week-old adult female mice (3 days after last immunization, where relevant), and single-cell suspensions were prepared. Samples were stained with antibodies (key resources table) and analyzed using a CytoFLEX S (Beckman Coulter, Brea, CA, USA). All staining reactions were performed in 100 μl of staining buffer (5% foetal bovine serum in PBS containing 500 μM EDTA) containing Mouse Seroblock FcR (Bio-Rad Laboratories) with 1×106 cells for detection of antigen-specific B cells or 4×106 cells for detection of GC B cells; the reactions were incubated at 4°C for 30 min. 4′,6-diamidino-2-phenylindole (D523, Dojindo, Kumamoto, Japan) was added to a final concentration of 0.5 μg/ml immediately before analysis.
Deep-sequencing analysis of antibody-coding transcripts
Next-generation sequencing (NGS) analysis was performed using unbiased T-cell receptor/BCR repertoire analysis technology, as used in previous studies and developed by Repertoire Genesis (Osaka, Japan).14,34 The primer sequences are listed in key resources table.
Analysis of the hIg repertoire
The same methods as in previous studies were applied to the analysis of the human antibody repertoire.14 Index sequences were used to classify every paired-end read. The international ImMunoGeneTics information system (IMGT) database (http://www.imgt.org) was used to assign sequences. DNA Chip Research (Tokyo, Japan) originally developed a repertoire analysis software application that was used automatically for data processing, assignment, and merging.
Within the combined sequence population, annotated sequence readings were recognized as unique reads and had BCR genes identified. Using RG software, the counts of identical annotated readings in each sample were automatically calculated and sorted numerically. CDR 3 nucleotide sequences, which include glycine (Gly119) and extend from a conserved cysteine at position 104 (Cys104) to a conserved phenylalanine at position 118 (Phe118), were translated into putative amino acid sequences in accordance with IMGT nomenclature.
Circos analysis was performed as in previous studies.14 Circos open-source software was acquired from www.circos.ca. The data were reformatted using R to meet Circos file requirements. Library sizes were normalized with Circos ideogram (circumference segments) scaling and sizing.
SHM in human VH and VL regions were detected through the comparison of B-cell clone lineages and clonotypes. A clone lineage was defined as a group of B cells descended from the same V(D)J (where J is the joining gene) rearrangement event. A clonotype was defined as a unique antibody sequence comprising the joined CDR1, CDR2, and CDR3 regions. NGS reads matching the CDR1-2-3 sequence were clustered into individual clonotypes. Mutations were identified by comparing each nucleotide position (approximately 315 nucleotides) against the germline sequence, and mutation rates were calculated as percentages. The mutation rate across all positions within the same clone lineage was aggregated as the clone lineage mutation rate. Moreover, annotated reads displayed variable lengths of antibody V regions. Therefore, an average V region length per clone lineage was established and standardized to 100.
To estimate BCR diversity in the deep-sequencing data as the diversity index, we calculated the Shannon–Weaver index (H′) using the following Equation 1.
| (Equation 1) |
In which N is the total number of sequence reads, ni is the number of annotated reads, and S is the species number of annotated reads.36 A greater H′ value reflects greater sample diversity.
Phylogenetic trees (circular dendrograms) were constructed by aligning heavy and light chain CDR3 amino acid sequences using a multiple sequence alignment programme and the neighbour-joining method.37 Subsequently, two phylograms were generated (one for unimmunized IGHL mice and another for RBD-immunized IGHL mice) based on their respective amino acid sequences. Additionally, copies comprising the same heavy chain CDR3 sequences were quantified and superimposed onto the circular dendrogram leaves. A maximum of 50 reads per sequence was represented, with the circle size on the leaves increasing proportionally as the read count increased.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 10 with the two-tailed unpaired Student’s t test. In Figure 3C, a two-tailed paired Student’s t test was performed. Differences with P-values <0.05 were considered significant (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001, and ns: not significant).
Published: October 28, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.111258.
Contributor Information
Satoshi Abe, Email: sabe@tottori-u.ac.jp.
Yasuhiro Kazuki, Email: kazuki@tottori-u.ac.jp.
Supplemental information
References
- 1.The Antibody Society Therapeutic monoclonal antibodies approved or in regulatory review. www.antibodysociety.org/antibody-therapeutics-product-dataTheAntibodySociety
- 2.ElBakri A., Nelson P.N., Abu Odeh R.O. The state of antibody therapy. Hum. Immunol. 2010;71:1243–1250. doi: 10.1016/j.humimm.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Presta L. In: Annual Reports in Medicinal Chemistry. Bristol J.A., editor. Academic Press; 1994. Chapter 32. Humanized Monoclonal Antibodies; pp. 317–324. [DOI] [Google Scholar]
- 4.Safdari Y., Farajnia S., Asgharzadeh M., Khalili M. Antibody humanization methods – a review and update. Biotechnol. Genet. Eng. Rev. 2013;29:175–186. doi: 10.1080/02648725.2013.801235. [DOI] [PubMed] [Google Scholar]
- 5.Pedrioli A., Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42:1143–1158. doi: 10.1016/j.it.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Yu P., Ran J., Yang R., Zhu H., Lu S., Wu Y., Zhao T., Xiong T. Rapid isolation of pan-neutralizing antibodies against Omicron variants from convalescent individuals infected with SARS-CoV-2. Front. Immunol. 2024;15 doi: 10.3389/fimmu.2024.1374913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüggemann M., Osborn M.J., Ma B., Hayre J., Avis S., Lundstrom B., Buelow R. Human Antibody Production in Transgenic Animals. Arch. Immunol. Ther. Exp. 2015;63:101–108. doi: 10.1007/s00005-014-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonberg N., Taylor L.D., Harding F.A., Trounstine M., Higgins K.M., Schramm S.R., Kuo C.-C., Mashayekh R., Wymore K., McCabe J.G., et al. Antigen-specific human antibodies from mice comprising four distinct genetic modifications. Nature. 1994;368:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 9.Kato Y., Kaneko M.K. A Cancer-specific Monoclonal Antibody Recognizes the Aberrantly Glycosylated Podoplanin. Sci. Rep. 2014;4:5924. doi: 10.1038/srep05924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendez M.J., Green L.L., Corvalan J.R., Jia X.-C., Maynard-Currie C.E., Yang X.D., Gallo M.L., Louie D.M., Lee D.V., Erickson K.L., et al. Functional transplant of megabase human immunoglobulin loci recapitulates human antibody response in mice. Nat. Genet. 1997;15:146–156. doi: 10.1038/ng0297-146. [DOI] [PubMed] [Google Scholar]
- 11.Macdonald L.E., Karow M., Stevens S., Auerbach W., Poueymirou W.T., Yasenchak J., Frendewey D., Valenzuela D.M., Giallourakis C.C., Alt F.W., et al. Precise and in situ genetic humanization of 6 Mb of mouse immunoglobulin genes. Proc. Natl. Acad. Sci. USA. 2014;111:5147–5152. doi: 10.1073/pnas.1323896111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy A.J., Macdonald L.E., Stevens S., Karow M., Dore A.T., Pobursky K., Huang T.T., Poueymirou W.T., Esau L., Meola M., et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. USA. 2014;111:5153–5158. doi: 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee E.-C., Liang Q., Ali H., Bayliss L., Beasley A., Bloomfield-Gerdes T., Bonoli L., Brown R., Campbell J., Carpenter A., et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 14.Satofuka H., Abe S., Moriwaki T., Okada A., Kazuki K., Tanaka H., Yamazaki K., Hichiwa G., Morimoto K., Takayama H., et al. Efficient human-like antibody repertoire and hybridoma production in trans-chromosomic mice carrying megabase-sized human immunoglobulin loci. Nat. Commun. 2022;13:1841. doi: 10.1038/s41467-022-29421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriwaki T., Abe S., Oshimura M., Kazuki Y. Transchromosomic technology for genomically humanized animals. Exp. Cell Res. 2020;390 doi: 10.1016/j.yexcr.2020.111914. [DOI] [PubMed] [Google Scholar]
- 16.Arun S.S., Breuer W., Hermanns W. Immunohistochemical Examination of Light-chain Expression (Λ/k Ratio) in Canine, Feline, Equine, Bovine and Porcine Plasma Cells. Zentralblatt für Veterinarmed. A. 1996;43:573–576. doi: 10.1111/j.1439-0442.1996.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 17.Bräuninger A., Goossens T., Rajewsky K., Küppers R. Regulation of immunoglobulin light chain gene rearrangements during early B cell development in the human. Eur. J. Immunol. 2001;31:3631–3637. doi: 10.1002/1521-4141(200112)31:12<3631::AID-IMMU3631>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Raybould M.I.J., Turnbull O.M., Suter A., Guloglu B., Deane C.M. Contextualising the developability risk of antibodies with lambda light chains using enhanced therapeutic antibody profiling. Commun. Biol. 2024;7:62. doi: 10.1038/s42003-023-05744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sajadi M.M., Farshidpour M., Brown E.P., Ouyang X., Seaman M.S., Pazgier M., Ackerman M.E., Robinson H., Tomaras G., Parsons M.S., et al. λ Light Chain Bias Associated With Enhanced Binding and Function of Anti-HIV Env Glycoprotein Antibodies. J. Infect. Dis. 2016;213:156–164. doi: 10.1093/infdis/jiv448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K., Shah H., Muther J.J., Duke A.L., Haley K., James J.A. Antigen nature and complexity influence human antibody light chain usage and specificity. Vaccine. 2016;34:2813–2820. doi: 10.1016/j.vaccine.2016.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend C.L., Laffy J.M.J., Wu Y.-C.B., Silva O’Hare J., Martin V., Kipling D., Fraternali F., Dunn-Walters D.K. Significant Differences in Physicochemical Properties of Human Immunoglobulin Kappa and Lambda CDR3 Regions. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abe S., Honma K., Okada A., Kazuki K., Tanaka H., Endo T., Morimoto K., Moriwaki T., Hamamichi S., Nakayama Y., et al. Construction of stable mouse artificial chromosome from native mouse chromosome 10 for generation of transchromosomic mice. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-99535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takiguchi M., Kazuki Y., Hiramatsu K., Abe S., Iida Y., Takehara S., Nishida T., Ohbayashi T., Wakayama T., Oshimura M. A Novel and Stable Mouse Artificial Chromosome Vector. ACS Synth. Biol. 2014;3:903–914. doi: 10.1021/sb3000723. [DOI] [PubMed] [Google Scholar]
- 24.Tomizuka K., Shinohara T., Yoshida H., Uejima H., Ohguma A., Tanaka S., Sato K., Oshimura M., Ishida I. Double trans-chromosomic mice: Maintenance of two individual human chromosome fragments containing Ig heavy and κ loci and expression of fully human antibodies. Proc. Natl. Acad. Sci. USA. 2000;97:722–727. doi: 10.1073/pnas.97.2.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kazuki K., Takehara S., Uno N., Imaoka N., Abe S., Takiguchi M., Hiramatsu K., Oshimura M., Kazuki Y. Highly stable maintenance of a mouse artificial chromosome in human cells and mice. Biochem. Biophys. Res. Commun. 2013;442:44–50. doi: 10.1016/j.bbrc.2013.10.171. [DOI] [PubMed] [Google Scholar]
- 26.Mahilkar S., Agrawal S., Chaudhary S., Parikh S., Sonkar S.C., Verma D.K., Chitalia V., Mehta D., Koner B.C., Vijay N., et al. SARS-CoV-2 variants: Impact on biological and clinical outcome. Front. Med. 2022;9 doi: 10.3389/fmed.2022.995960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obermeyer F., Jankowiak M., Barkas N., Schaffner S.F., Pyle J.D., Yurkovetskiy L., Bosso M., Park D.J., Babadi M., MacInnis B.L., et al. Analysis of 6.4 million SARS-CoV-2 genomes identifies mutations associated with fitness. Science. 2022;376:1327–1332. doi: 10.1126/science.abm1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragonnet-Cronin M., Nutalai R., Huo J., Dijokaite-Guraliuc A., Das R., Tuekprakhon A., Supasa P., Liu C., Selvaraj M., Groves N., et al. Generation of SARS-CoV-2 escape mutations by monoclonal antibody therapy. Nat. Commun. 2023;14:3334. doi: 10.1038/s41467-023-37826-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papavasiliou F.N., Schatz D.G. Somatic Hypermutation of Immunoglobulin Genes: Merging Mechanisms for Genetic Diversity. Cell. 2002;109:S35–S44. doi: 10.1016/S0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 30.van der Kant R., Bauer J., Karow-Zwick A.R., Kube S., Garidel P., Blech M., Rousseau F., Schymkowitz J. Adaption of human antibody λ and κ light chain architectures to CDR repertoires. Protein Eng. Des. Sel. 2019;32:109–127. doi: 10.1093/protein/gzz012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hershberg U., Shlomchik M.J. Differences in potential for amino acid change after mutation reveals distinct strategies for κ and λ light-chain variation. Proc. Natl. Acad. Sci. USA. 2006;103:15963–15968. doi: 10.1073/pnas.0607581103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeKosky B.J., Lungu O.I., Park D., Johnson E.L., Charab W., Chrysostomou C., Kuroda D., Ellington A.D., Ippolito G.C., Gray J.J., Georgiou G. Large-scale sequence and structural comparisons of human naive and antigen-experienced antibody repertoires. Proc. Natl. Acad. Sci. USA. 2016;113:E2636–E2645. doi: 10.1073/pnas.1525510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goulet D.R., Atkins W.M. Considerations for the Design of Antibody-Based Therapeutics. J. Pharm. Sci. 2020;109:74–103. doi: 10.1016/j.xphs.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitaura K., Yamashita H., Ayabe H., Shini T., Matsutani T., Suzuki R. Different Somatic Hypermutation Levels among Antibody Subclasses Disclosed by a New Next-Generation Sequencing-Based Antibody Repertoire Analysis. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuroiwa Y., Tomizuka K., Shinohara T., Kazuki Y., Yoshida H., Ohguma A., Yamamoto T., Tanaka S., Oshimura M., Ishida I. Manipulation of human minichromosomes to carry greater than megabase-sized chromosome inserts. Nat. Biotechnol. 2000;18:1086–1090. doi: 10.1038/80287. [DOI] [PubMed] [Google Scholar]
- 36.Kim B.-R., Shin J., Guevarra R., Lee J.H., Kim D.W., Seol K.-H., Lee J.-H., Kim H.B., Isaacson R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017;27:2089–2093. doi: 10.4014/jmb.1709.09027. [DOI] [PubMed] [Google Scholar]
- 37.Yermanos A.D., Dounas A.K., Stadler T., Oxenius A., Reddy S.T. Tracing Antibody Repertoire Evolution by Systems Phylogeny. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data are available from the lead contact and corresponding authors. The repertoire analysis data generated in this study have been deposited in the DDBJ database under accession code PRJDB18270. SAMD00795425 (TCK152MG_S41_S43) and SAMD00795428 (TCK152L_S46) are the data of an RBD-immunized IGHL mouse individual. SAMD00795424 (TCK111MG_S56_S62) and SAMD00795427 (TCK111L_S52) are the data of unimmunized IGHL mice (pooled, 5 individuals). SAMD00795432 (TCK041L_S19) is the data of a human PBMC pool sample.
-
•
No original code was reported in this article.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.