Summary
Bruton’s tyrosine kinase (BTK) inhibitor, ibrutinib, has been shown to synergize in vitro with proteasome inhibitors (PIs) in reducing the viability of cells derived from B cell malignancies, but the mechanism is not known. We report here that an off-target effect of ibrutinib causes synergy because not all BTK inhibitors exhibited the synergistic effect, and those that synergized did so even in cells that do not express BTK. The allosteric BTK inhibitor CGI-1746 showed the strongest synergy. Co-treatment of cells with CGI-1746 increased PI-induced accumulation of ubiquitin conjugates and expression of heat shock proteins and NOXA and decreased a ratio of reduced to oxidized glutathione. CGI-1746, but not other BTK inhibitors, inhibited ATPase activity and all three peptidase activities of the 26S proteasome. The effect demonstrates a conceptually novel mode of proteasome inhibition that may aid the development of more potent PIs.
Subject areas: Biological sciences, Biochemistry, Cell biology, Cancer
Graphical abstract

Highlights
-
•
Some BTK inhibitors exhibit synergistic cytotoxicity with proteasome inhibitors
-
•
Synergy is observed in cell lines that do not express BTK
-
•
Allosteric BTK inhibitor CGI-1746 shows the strongest synergy
-
•
CGI-1746 inhibits peptidase and ATPase activities of the 26S proteasome
Biological sciences; Biochemistry; Cell biology; Cancer
Introduction
The 26S proteasome is essential for maintaining protein homeostasis in every eukaryotic cell, including the quality control of nascent polypeptides.1,2,3,4 Multiple myeloma cells are highly dependent on proteasome activity because these malignant plasma cells synthesize and secrete large amounts of immunoglobulins, complex four-chain molecules that contain multiple disulfide bridges.5,6 Partial inhibition of the proteasome causes selective apoptosis of myeloma cells, and proteasome inhibitors bortezomib, carfilzomib, and ixazomib7,8,9 are approved by the Food and Drug Administration (FDA) for the treatment of multiple myeloma. The initial response rates are high, but many patients develop resistance.
De-differentiation into stem-like plasma cell precursors that do not produce immunoglobulins is one of the mechanisms of myeloma resistance to proteasome inhibitors.10,11 These cells express Bruton’s tyrosine kinase (BTK), an essential kinase in the development and proliferation of immature B cells and many B cell leukemias and lymphomas.12,13,14 Three BTK inhibitors, ibrutinib, acalabrutinib, and zanubrutinib, are approved for the treatment of various B cell neoplasms.15,16 BTK inhibitors have been clinically tested for the treatment of multiple myeloma as a proposed therapy, which in combination with proteasome inhibitors, should overcome resistance to proteasome inhibitors in multiple myeloma.17
Previous experiments by the Driessen laboratory showed that the combination of proteasome inhibitors with a BTK inhibitor, ibrutinib, causes synergistic cell death in cells derived from myeloma and mantle cell lymphoma.18 Furthermore, this pronounced synergy was observed regardless of BTK expression levels. Additionally, the synergy was observed at a hundred times the concentration of ibrutinib needed to completely inhibit BTK activity. These data suggested an off-target effect of the BTK inhibitor, but attempts to identify a target responsible for the synergy were unsuccessful. The initial goal of this study was to obtain additional evidence to support an off-target effect of BTK inhibitors and identify the target.
The 26S proteasome consists of a 20S proteolytic core and one or two 19S regulatory complexes. The 20S core contains three pairs of proteolytic active sites, β5 (PSMB5 or chymotrypsin-like), β2 (PSMB2 or trypsin-like), and β1 (PSMB1 or caspase-like). β5 sites are the most important for protein degradation and the prime targets of FDA-approved proteasome inhibitors.19 Inhibitors of β2 and β1 sites have also been developed20 and have been found to potentiate the anti-neoplastic effects of β5 inhibitors while having little effect on cell proliferation, viability, and protein breakdown when used as single agents.21,22,23 Surprisingly, the synergy with ibrutinib was more pronounced with the specific inhibitor of proteasome β2 sites, LU-102, than with FDA-approved β5 inhibitors, bortezomib and carfilzomib.18 To the best of our knowledge, this is the only example where the biological effect of a β2-specific inhibitor is stronger than that of β5 inhibitors.
The 19S regulatory complex (PA700) contains ubiquitin receptors, deubiquitylating enzymes, and 6 ATPases, which unfold protein substrates and control access to the proteasome.24,25 While several inhibitors of the isopeptidases26,27,28,29 and three ATPase binders30,31,32 have been described, it has not been reported whether these ATPase binders inhibit 19S ATPase activity. Here, we describe how an effort to identify an off-target of ibrutinib led to a surprising discovery that a non-competitive BTK inhibitor CGI-1746, but not other BTK inhibitors, is an allosteric inhibitor of both the 19S ATPase activities and all three proteolytic sites of the proteasome.
Results
The majority of, but not all, BTK inhibitors synergize with proteasome inhibitors in BTK-expressing myeloma cells
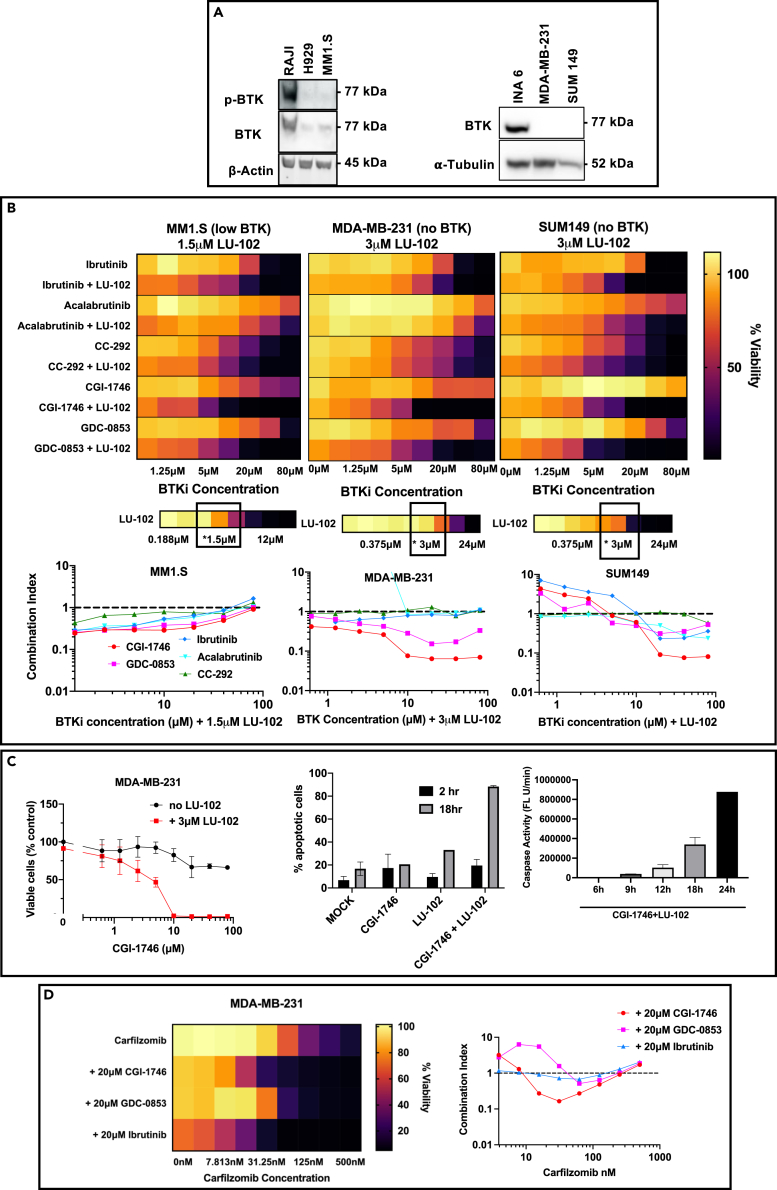
Ibrutinib inhibits many different kinases,33,34,35,36 and if synergy with proteasome is due to the inhibition of one of these kinases, more specific second-generation BTK inhibitors should not synergize. Therefore, we determined whether other BTK inhibitors with different kinome specificity profiles synergize with proteasome inhibitors in various cell lines, starting with BTK-expressing cells. We chose two irreversible inhibitors, acalabrutinib and CC-292 (spebrutinib), which, similar to ibrutinib, covalently bind to the cysteine in the active site, and two reversible allosteric inhibitors, CGI-1746 and GDC-0853 (fenebrutinib). Acalabrutinib is more selective than ibrutinib, CC-292 is less selective,37 while CGI-1746 and GDC-0853 are much more selective.38,39 Specifically, CGI-1746 does not inhibit any other kinase at 1 μM.38 We focused on the β2-specific inhibitor, LU-102, because it showed stronger synergy than bortezomib and carfilzomib. Cells were co-treated with 3 μM LU-102, which caused only 10% loss of viability when used alone, and various concentrations of BTK inhibitors for 48 h. Their viability was then measured by the Alamar Blue mitochondrial dye conversion assay (Figure 1A). Ibrutinib, acalabrutinib, CGI-1746, and GDC-0853 exhibited various degrees of synergy, while CC-292, which was more cytotoxic when used as a single agent, was less synergistic. Cells were then treated with a sub-toxic 10 μM concentration of ibrutinib, GDC-0853, and CGI-1746 and different concentrations of LU-102; the strongest synergy was observed in a range of concentrations from 1 to 6 μM (Figure 1B), indicating that synergy with BTK inhibitors requires inhibition of both β2 sites because inhibition of β2 activity at 1 μM LU-102 reaches 90%.21,40 Synergy was lost at higher concentrations because LU-102 is cytotoxic as a single agent, most likely because it co-inhibits β5 sites.21 We confirmed that combinations reduced cell viability because of apoptosis (Figure 1C). BTK inhibitors exhibited similar synergy with bortezomib and carfilzomib (Figure 1D).18 Similar patterns of synergy were observed in a BTK-expressing lymphoma cell line, Raji (Figure S1D). While the strong synergistic activity of the highly specific inhibitor CGI-1746 can be interpreted as synergy caused by BTK inhibition, complete inhibition of BTK was observed at a concentration at least 100-fold lower than the 10 μM concentration (Figures 1E and S1E) at which maximal synergy was observed (Figures 1A and S1D), strongly suggesting that an off-target effect is responsible for synergy.
Figure 1.
The majority of, but not all, BTK inhibitors synergize with proteasome inhibitors in BTK-expressing myeloma cells
(A) INA-6 BTK-expressing cells were treated with 2-fold serial dilutions of BTK inhibitors (ibrutinib, acalabrutinib, CC-292, CGI-1746, and GDC-0853) alone and in combination with a sub-toxic concentration (3 μM) of β2-site-specific proteasome inhibitor, LU-102, for 48 h, after which viability was determined by Alamar Blue dye conversion assay, and expressed as percentage of mock-treated controls, in a heatmap. Raw data for heatmaps are presented in Figure S1A (n > 3). A single row map demonstrates the results of treatments with 2-fold dilutions of LU-102 in the absence of BTK inhibitors, and a square shows the concentration used in combination experiments. Graphs showing synergy by combination indices—which was determined by the Chou-Talalay method41 using CalcuSyn software (Synergy <1, Antagonism >1, Additive = 1)—are below the heatmaps.
(B) Same as in (A) except that INA-6 cells were co-treated with sub-toxic doses of CGI-1746, GDC-0853, and ibrutinib and indicated concentrations of LU-102. Raw data for heatmaps are presented in Figure S1B (n > 3).
(C) Cells were treated with 3 μM LU-102 and 10 μM CGI-1746 for times indicated. Apoptosis was determined by caspase 3/7 activity in cell extracts using Ac-DEVD-AMC (n = 3). Error bars represent S.D.
(D) INA-6 cells were treated with inhibitors of proteasome β5 sites, bortezomib, and carfilzomib, alone or with sub-toxic doses of CGI-1746, GDC-0853, and ibrutinib. Viability and synergy were determined as in (A). Raw data are presented in Figure S1C (n = 3).
(E) Western blot showing the inhibition of BTK (reduction in autophosphorylated BTK) by indicated concentrations of BTK inhibitors in INA-6 cells treated for 1 h.
BTK inhibitors synergize with LU-102 in cells that do not express BTK
If synergy between ibrutinib and other BTK inhibitors is caused by an off-target effect of BTK inhibitors, the effect will be observed in cells that do not express BTK. To investigate this possibility, we performed the previous experiments in MM1.S multiple myeloma cells, which have very low BTK expression and no phosphorylated (active) BTK expression, as well as in two triple-negative breast cancer cell lines, MDA-MB-231 and SUM149, which have no BTK expression (Figure 2A) but are very sensitive to proteasome inhibitors.40,42,43 The results of these experiments are summarized in Figure 2B. Similar to INA-6 and Raji cells, CC-292 was cytotoxic to all cell lines with IC50 in the 10 μM range but did not show any synergy with LU-102. Ibrutinib appeared to be less synergistic in MM1.S and MDA-MB-231 cells. Acalabrutinib was not synergistic in these cells. Two allosteric inhibitors, GDC-0853 and CGI-1746, maintained synergy. The effect of CGI-1746 on breast cancer cells was the most remarkable because, unlike in INA-6 cells, this compound was not cytotoxic to breast cancer cells yet essentially showed synthetic lethality when combined with LU-102 (Figure 2C), leading to robust apoptosis within 18–24 h of exposure. We have also analyzed LU-102 combinations with three other covalent BTK inhibitors, zanubrutinib, evobrutinib, and ONO-4053 in MDA-MB-231 cells (Figure S2B), and identified synergy which was similar to ibrutinib. Finally, carfilzomib showed synergy with ibrutinib, CGI-1746, and GDC-0853 in MDA-MB-231 cells (Figure 2D). We conclude that the synergy between ibrutinib, CGI-1746, and GDC-0853, and proteasome inhibitors is caused by an off-target effect of BTK inhibitors.
Figure 2.
BTK inhibitors synergize with LU-102 in cells that do not express BTK
(A) The expression of active (phosphorylated) BTK and total BTK was determined by western blots.
(B) Heatmaps summarizing effects of 48 h co-treatments of MM1.S, MDA-MB-231, and SUM149 cells with BTK inhibitors and sub-toxic concentrations of LU-102 indicated on top of each graph. A single-row heatmaps summarize cell response to 2-fold serial dilutions of LU-102 in the absence of BTK inhibitors. Raw data are presented in Figure S2A (n > 3). The bottom graphs show combination indices.
(C) Detailed analysis of effects of CGI-1746/LU-102 combination on MDA-MB-231 cells. Cell viability data on the left graph were used to generate the left heatmap in (A) and is also presented in Figure S2A (n = 3). Apoptosis of cells treated with 10 μM CGI-1746, and 3 μM LU-102 was determined by flow cytometry using caspase 3/7 probe (middle) and by measuring caspase 3/7 in extracts using Ac-DEVD-AMC (right; n = 3). Error bars represent S.E.M.
(D) MDA-MB-231 cells were pulse-treated with carfilzomib for 1 h, after which carfilzomib-containing media was replaced with drug-free media or media containing sub-toxic concentrations of CGI-1746, GDC-0853, and ibrutinib. 48 h after treatment, viability was determined by Alamar Blue (n = 3). Combination indices were determined as on Figure 1A.
The combination of BTK and proteasome inhibitors shows hallmarks of proteasome inhibition in cells
To identify the secondary target of BTK inhibitors that is responsible for the synergy with LU-102, we decided to focus on triple-negative breast cancer cells so that any effects related to direct inhibition of BTK are excluded and to focus on CGI-1746 because it was the most synergistic and less toxic when used as a single agent. We performed next-generation RNA sequencing of MDA-MB-231 cells treated with CGI-1746 at 20 μM, LU-102 at 3 μM, the combination of the two drugs, and a vehicle control. The cells were treated for 4 h and then collected, and RNA was isolated and sequenced in order to identify early changes in the cell that precede the onset of apoptosis. Upon RNA sequencing analysis, two top upregulated mRNAs in cells treated with the drug combination were HSAP1A and HSPA1B, which encode the Hsp70 molecular chaperon (Figure 3A). These genes are usually upregulated by proteasome inhibitors.44,45 This was unsurprising as LU-102 is a proteasome inhibitor. What was surprising is that CGI-1746 alone also significantly upregulated these genes but LU-102 did not. Western blot analysis confirmed Hsp70 upregulation upon treatment with LU-102 and CGI-1746, but not by a combination of LU-102 with other BTK inhibitors (Figure 3B). These data suggest that CGI-1746 acts as a proteasome inhibitor.
Figure 3.
Detailed analysis of MDA-MB-231 cell response to treatment with CGI-1736 and LU-102
(A) Cells were treated with 10 μM CGI-1746 and 3 μM LU-102 (alone and in combination) for 4 h. RNA expression was determined by RNA sequencing. Genes whose expression increased significantly (p < 0.05) by > 1.5-fold over mock-treated samples are presented. No genes in LU-102-treated samples met these criteria. No genes in CGI-1746 and combination-treated cells decreased significantly by > 1.5-fold.
(B) Cells were treated for the indicated length of time with BTK inhibitors (20 μM) alone, or in combination with LU-102 (3 μM), and protein expression was determined by western blots.
(C) Cells were treated for the indicated length of time with CGI-1746 (20 μM) alone, or in combination with LU-102 (3 μM), and protein expression was determined by western blots. Bortezomib (Btz) (25 nM, 4 h) was used as a positive control.
(D) Cells were pulse-treated with carfilzomib (CFZ) for 1 h, after which carfilzomib-containing media was replaced with drug-free media or media containing 20 μM of CGI-1746. Protein expression was determined by western blots.
(E) Cells were treated with CGI-1746 and LU-102 (top image) as in (C) or 100 nM Btz (bottom image) for the indicated length of time. Protein expression was determined by western blots.
(F) Cells were treated with 10 μM of CGI-1746, 3 μM of LU-102, a combination of CGI-1746 and LU-102, 50 nM of bortezomib, or untreated (Mock) for 3 and 8 h. GSH to GSSG ratio was determined using GSH/GSSG-Glo Assay (n = 3). Error bars represent S.E.M.
Accumulation of ubiquitin conjugates is the hallmark of intracellular proteasome inhibition. Indeed, we found that the combination of LU-102 with CGI-1746, GDC-0853, and ibrutinib caused the accumulation of conjugates. In contrast, a combination with acalabrutinib did not cause accumulation (Figure 3B). In fact, a combination of CGI-1746 and LU-102 caused a more robust accumulation of conjugates in MDA-MB-231 cells than bortezomib (Figure 3C). However, they caused little or no accumulation when used as a single agent (Figure 3C). CGI-1746 also enhanced carfilzomib-induced conjugate accumulation (Figure 3D).
We have confirmed (Figure S3A) that BTK inhibitors and proteasome inhibitors did not inhibit deubiquitylating enzymes, which could also account for conjugate accumulation. Inhibition of valosin-containing protein (VCP/p97) has also been shown to cause ubiquitin conjugate accumulation.46,47 Because BTK inhibitors are kinase inhibitors, there is a possibility that these inhibitors interfered with VCP ATPase function.48,49 We performed an ATPase assay and found that CGI-1746 did not inhibit VCP ATPase activity (Figure S3B). These data strongly suggest that CGI-1746 reduces proteasome activity and that this inhibition is responsible for synergistic cytotoxicity.
To further confirm that the CGI-1746 and LU-102 combination exert their effects in the ubiquitin-proteasome pathway, we investigated their effects on NOXA accumulation and generation of reactive oxygen species, both hallmarks of proteasome inhibition.50,51,52,53,54 We found that CGI-1746 and LU-102 individually caused the accumulation of NOXA in MDA-MB-231 cells; the combination treatment caused substantially more accumulation of NOXA, starting at 9 h of treatment, which is only 3 h slower than bortezomib-induced NOXA accumulation (Figure 3E). We also found that, similar to bortezomib, the combination treatment increased the ratio of reduced glutathione (GSSG) to oxidized glutathione (GSH), while the individual treatments of CGI-1746 and LU-102 did not do it (Figure 3F).
Proteasome activity is regulated by phosphorylation on different sites by various kinases.55,56,57,58,59 Since CGI-1746 is a kinase inhibitor, we first tested whether treatment with CGI-1746 changes the pattern of phosphorylation of proteasome subunits. We isolated proteasomes from CGI-1746-treated MDA-MB-231 cells and used a phosphorylation-specific stain to identify phosphorylated subunits. We did not find any changes (Figure S3C). Thus, CGI-1746 does not inhibit proteasome by reducing its phosphorylation.
CGI-1746 inhibits peptidase and ATPase activities of the 26S proteasome
If CGI-1746 acts as a proteasome inhibitor in cells, it should inhibit proteolytic activities of the purified proteasomes. Indeed, CGI-1746 inhibited all three peptidase (Figure 4A) activities of the purified 26S proteasome. Except for weaker inhibition of β1 sites, ibrutinib and GDC-0853 did not inhibit these activities. Moreover, CGI-1746, but not Ibrutinib or GDC-0853, inhibited ATPase activities of 26S proteasomes and 19S particles (Figure 4B). Inhibition of ATPase activities of 19S particles was observed at all concentrations of ATP (Figure 4C, right panel). CGI-1746 significantly reduced Vmax and appeared to reduce Km (Table 1) suggesting either non-competitive or uncompetitive allosteric inhibition. However, concentration dependence of activity appeared to be biphasic or even triphasic, probably reflecting different affinities for ATPase subunits. When data at lower concentrations of ATP (0–250 μM) were fitted to the Michaelis-Menten equation (Figure 4C, right panel), CGI-1746 decreased Km but not Vmax (Table 1), indicating competitive inhibition. The most likely interpretation of these data is that CGI-1746 competitively inhibits the 19S subunit(s) with the highest affinity for ATP,60 and that binding to this subunit also allosterically inhibits the subunits with lower ATP affinity. Although the high background of the ADP-Glo assay that we used to measure ATPase activity prevented us from measuring inhibition at physiological 5 mM concentrations of ATP, the allosteric nature of inhibition at high ATP concentration raises little doubt that CGI-1746 will inhibit hydrolysis at physiologic ATP concentrations.
Figure 4.
CGI-1746 inhibits the 26S proteasome and proteasomal ATPases
(A) Effects of BTK inhibitors on proteolytic sites of the human 26S proteasomes were measured using Suc-LLVY-AMC (β5), Ac-RLR-AMC (β2), and Ac-nLPnLD-AMC (β1) (n = 3).
(B) The hydrolysis of 100 μM ATP by human 26S proteasomes and of 50 μM ATP by bovine 19S particles was measured in the presence or absence of the indicated BTK inhibitors (n = 3).
(C) ATPase activity of bovine 26S proteasome was measured at different concentrations of ATP in the presence or absence of 20 μM CGI-1746 (n = 3) and fit into the Michaelis-Menten equation using different concentration ranges indicated in Table 1.
(D) Peptidase activities of rabbit 26S proteasomes were measured in the presence or absence of CGI-1746 (20 μM for β1 and β5 sites, 40 μM for β2 sites) using the indicated concentrations of site-specific substrates (n = 3) and fit into equations listed in Table 1.
(E) Effects of CGI-1746 on proteolytic activities of human 26S proteasomes were measured as in (A), except that MgCl2 was omitted from the assay buffer (n = 4).
(F) Effects of CGI-1746 on activities of the 20S proteasomes were measured using peptidase substrates of individual active sites (n = 4).
(G) Peptidase activities of the 20S proteasomes were measured in the presence or absence of 20 μM CGI-1746 at different concentrations of site-specific substrates (n = 3) and fit into equations listed in Table 1. Poor solubility of Ac-nLPnLD-amc at >1 mM prevented us from conducting measurements at saturating concentrations.
Error bars on all panels represent S.E.M.
Table 1.
Effect of CGI-1746 on kinetic parameters of ATP hydrolysis by the 19S regulatory particle and on peptidase activities of 26S and 20S proteasomes
| PR | Active site | CGI -1746 (μM) | Curve fit parameters (95% CI) |
|||||
|---|---|---|---|---|---|---|---|---|
| S range (μM) | Equation | Km or K1/2, μM | Vmax, nmole/(min∗mg) | Hill coefficient | R2 | |||
| 19S | Rpt1-6 | 0 | 0–1,000 | MM | 722 (291–2,752) | 186 (117–456) | – | 0.85 |
| – | – | 20 | 0–1,000 | MM | 452 (222–1,053) | 76 (56–118) | – | 0.91 |
| – | – | 0 | 0–250 | MM | 99 (30–126) | 48 (27–270) | – | 0.81 |
| – | – | 20 | 0–250 | MM | 226 (90–1,091) | 48 (30–144) | – | 0.91 |
| 26S | β5 | 0 | 0–400 | Hill | 101 (81–140) | 0.72 (0.64–0.85) | 1.4 (1.1–1.7) | 0.98 |
| – | – | 20 | 0–400 | Hill | 119 (87–223) | 0.27 (0.21–0.42) | 2.8 (1.4–?) | 0.88 |
| – | β1 | 0 | 0–800 | MM | 769 (395–2,190) | 0.46 (0.31–0.87) | – | 0.94 |
| – | – | 20 | 0–800 | Hill | 205 (?–234) | 0.09 (0.08–0.1) | 24.7 (?) | 0.95 |
| – | β2 | 0 | 0–800 | Hill | 190 (81–57,500) | 0.22 (0.16–2.17) | 0.7 (0.4–1.1) | 0.96 |
| – | – | 40 | 0–400 | Hill | 158 (137–179) | 0.062 (0.050–0.070) | 4.6 (3–8.5) | 0.97 |
| 20S | β5 | 0 | 0–400 | SI | 0.37 (?–34) | 0.41 (0.35–?) | – | 0.66 |
| – | – | 20 | 0–400 | MM | 57 (28–120) | 0.20 (0.16–0.26) | – | 0.88 |
| – | β1 | 0 | 0–800 | SI | 516 (205–?) | 2.1 (1.0–∞) | – | 0.98 |
| – | – | 20 | 0–800 | MM | 24,543 (582–∞) | 3.5 (0.16–∞) | – | 0.70 |
| – | β2 | 0 | 0–800 | MM | 205 (115–378) | 1.17 (0.95–1.5) | – | 0.90 |
| – | – | 20 | 0–800 | (MM) | 435 (259–729) | 0.23 (0.18–0.31) | – | 0.94 |
Analysis of data on Figures 4C, 4D, and 4G.
Equations:
Hill: V=Vmax[S]n/(K1/2n+[S]n), n-Hill coefficient.
MM, Michaelis-Menten: V=Vmax[S]/(Km+[S]).
SI-substrate inhibition: V=Vmax[S]/(Km+[S](1+[S]/Ki)).
CGI-1746 reduced Vmax for all three peptidase activities of the 26S particles (Table 1; Figure 4D), clearly indicating allosteric inhibition. It also induced positive cooperativity between all three pairs of active sites, as evidenced by higher Hill coefficients (Table 1). Furthermore, CGI-1746 reduced K1/2 (an equivalent of Km in the Hill equation) for β1 sites, indicating uncompetitive inhibition. Finally, CGI-1746 induced biphasic kinetics in the β2 sites, where activity appeared to plateau between 200 and 400 μM of substrate but then increased again at 800 μM (Figure 4D). When 26S peptidase activities were measured in the absence of magnesium to block ATP hydrolysis, CGI-1746 inhibition was less potent (Figure 4E). These data suggest that the binding of CGI-1746 and inhibition of ATP hydrolysis by 19S particles contribute to the allosteric inhibition of peptidase activities.
If allosteric inhibition due to the binding to 19S ATPases is the only mechanism of inhibition, CGI-1746 should not inhibit 20S proteasomes. However, CGI-1746 inhibited all three peptidase activities of the 20S core (Figure 4F), albeit with IC50s that were higher than for 26S proteasomes in the presence of magnesium, and similar to 26S in the absence of magnesium (Figure 4E). These data indicate that it also binds to the 20S core triggering a different mechanism of peptidase site inhibition. CGI-1746 decreased Km values for all three peptidase activities, and decreased Vmax only for the β2 sites (Figure 4G; Table 1), indicative of competitive inhibition of β1 and β5 sites and mixed inhibition of β2 sites. In addition, CGI-1746 eliminated substrate inhibition of β5 and β1 site (Figure 4G), which was observed in 20S but not 26S, and reported previously for the β5 activity of 20S.61 We conclude that CGI-1746 has binding sites on the 19S and 20S and inhibits peptidase activities by different mechanisms.
Discussion
The major goals of this work were to prove that ibrutinib synergy with proteasome inhibitors, with a focus on our β2-specific inhibitor LU-102, is not caused by the BTK inhibition and to identify a novel ibrutinib target responsible for this synergy. We used five different BTK inhibitors and found that while CGI-1746 and GDC-0853 behaved similarly to bortezomib, two, acalabrutinib and CC-292 (spebrutinib), showed little or no synergy. For inhibitors that were synergistic, we confirmed the previous finding that synergy occurs at a much higher concentration than the concentration that completely inhibits BTK. Finally, synergy was observed in cell lines that do not express BTK. Thus, synergistic cytotoxicity between BTK and proteasome inhibitors is caused by binding to a novel target.
We have switched our target identification efforts from ibrutinib to CGI-1746 because the latter has shown stronger synergy, and because CG-1746’s off-target effect was the most surprising as this is the most specific BTK inhibitor among those tested in this study.38 The compound surprised us again when we found that CGI-1746 is a dual inhibitor of 26S proteasome ATPases and peptidase activities. To the best of our knowledge, this is the first example of such a dual inhibitor and the first ATPase inhibitor that is not a non-hydrolyzable ATP analog. Several ATPase binders are described in the literature but their ability to inhibit their ATPase activity has not been reported.30,31,32 Because of the uniqueness of this observation, we made understanding the mechanisms of ATPase inhibition by CGI-1746 the main goal of this work. We conducted kinetic analysis in the free 19S particles to exclude a potential allosteric effects caused by binding of CGI-1746 to the second site on the 20S allosterically. CGI-1746 acts as a competitive ATPase inhibitor of 19S at lower concentrations of ATP and as a non-competitive inhibitor at higher concentrations. Although CGI-1746 can inhibit peptidase activities of the 20S proteasome in a 19S-independent fashion, and under the conditions when ATP hydrolysis by the 26S proteasome is blocked, inhibition of peptidases is stronger when ATP hydrolysis is permissible. Under these conditions, the inhibition is allosteric and occurs at similar concentrations of CGI-1746 as inhibition of ATPase activities. These data can be explained if binding of CGI-1746 to an active site(s) of the 19S ATPase, which has the highest affinity for ATP, causes competitive inhibition of this ATPase(s) and allosteric inhibition of remaining ATPases and of peptidase activities. CGI-1746 is also capable of inhibiting proteasome peptidases by a different mechanism because it competitively inhibits free 20S, albeit with lower potency. When peptide hydrolysis is stimulated by ATP hydrolysis, this competitive inhibition is simply overridden by an allosteric inhibition due to stronger binding to 19S ATPase(s). The future identification of CGI-1746 binding sites by cryoelectron microscopy should help clarify these models and help in the future design of more potent inhibitors.30,31,32
There are several reasons to believe that the observed inhibition of proteasome ATPase (and perhaps proteolytic activities) is the major contributor to synergy. ATPase inhibition was observed at the same concentration as synergy. Gene expression profiling was consistent with a combination of CGI-1746 and LU-102 acting as proteasome inhibitors. The combination also synergized in causing the accumulation of ubiquitylated proteins in cells. Finally, it induced NOXA and dramatically lowered the ratio of reduced to oxidized glutathione. Except for the degradation of highly basic proteins, β2 inhibitors are much weaker inhibitors of protein degradation,62 and as such, LU-102 may benefit more from additional inhibition of ATPase activities than carfilzomib or bortezomib, thus explaining stronger synergy of the LU-102/CGI-1746 combination.
The micromolar concentration at which synergy is observed is too high for the observed effect to have any therapeutic value, especially for CGI-1746, which had to be used at 100 mg/kg to achieve BTK inhibition in mice.38 While analysis of proteasome inhibition by other kinase inhibitors is underway, the possibility that some of them will be more potent inhibitors of the proteasome cannot be ruled out. In fact, mTOR inhibitor rapamycin allosterically inhibits 20S and interferes with 26S assembly at a lower concentration than CGI-1746.63 This study adds the 26S proteasome to the long list of off-target effects of protein kinase inhibitors64 warranting counter-screening of all new kinase inhibitors for proteasome inhibition.
In summary, we have identified a dual inhibitor of 20S proteolytic core and 19S regulatory particles. While it is not yet clear whether this discovery has therapeutic implications, this inhibitor should at least serve as a novel tool to dissect proteasome mechanisms and as a starting point for the development of specific inhibitors of proteasomal ATPases.
Limitations of the study
Although several experimental observations support our conclusion that inhibition of ATPase activities contributes to synergy between CG-1746 and proteasome inhibitors, they do not rule out alternative mechanisms of synergy. The alternative mechanism should also be responsible for the synergy between proteasome inhibitors and ibrutinib, which we did not fully analyze in this study. Ibrutinib and GDC-0853, which synergize with LU-102 in cell killing, also synergized in causing ubiquitin conjugate accumulation. However, they did not inhibit ATPase activities of 19S and p97/VCP and inhibited only the β1 sites of the 26S proteasome, with activity plateauing at 50%. It remains to be determined whether this inhibition of β1 sites is sufficient to cause synergy or if other mechanisms are involved.
It is also not clear whether partial inhibition of ATPase by CGI-1746 is sufficient to inhibit protein breakdown in cells. The most common model substrate of the 26S proteasome, Ubn-Sic1PY, is modified by K63-linked polyubiquitin chains65 that do not target proteins for intracellular degradation.66 Inhibition of its degradation by CGI-1746 (or lack thereof) would not necessarily mean the same effect on intracellular protein degradation. Other model 26S substrates are unfolded proteins, which do not require ATP hydrolysis for their degradation,67 and we therefore did not test CGI-1746 effects on their degradation.
We also did not explore whether a combination of LU-102 and CGI-1746 preferentially inhibits the degradation of basic proteins,62 as it could be inferred from our past work that degradation of purified basic proteins is more sensitive to the inhibitors of β2 activity. In the future, it would be interesting to study how treatment of cells with interferon-gamma that stimulates β2 activity by replacing the β2 (PSMB7) subunit with a more active PSMB10 (MECL1) subunit68,69 or overexpression of PA28γ that selectively activated β2 activity70 affects LU-102 and CGI-1746 synergy.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alexei Kisselev (afk0006@auburn.edu).
Materials availability
This study did not generate new unique reagents. Antibodies, reagents, and cell lines used were obtained from commercial or other sources as outlined in the key resources table.
Data and code availability
-
•
RNA-seq data have been deposited at GEO (GEO: GSE239482) and is publicly available as of the date of publication. key resources table Original western blot images have been deposited at Mendeley (Data set http://doi:10.17632/7n36xfmfj9.1) and are publicly available as of the date of publication.key resources table
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This paper is dedicated to the memory of Dr. Alfred L. Goldberg who was the post-doctoral mentor to G.N.D.M. and A.F.K. This work was supported by the NIH grants R01CA213223 to A.F.K. and R01GM129088 to G.N.D.M.
Author contributions
A.F.K. conceived the project and secured funding. A.F.K. and O.A.A. designed and performed the experiments and wrote the manuscript. M.B.P. and J.G.S. performed the experiments. A.K.M. analyzed RNA sequencing data. G.N.D.M. provided critical reagents, expertise, and feedback.
Declaration of interests
A.F.K. is a founder and Chief Scientific Officer of InhiProt, LLC.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| K-48 Polyubiquitin | Cell Signaling Technologies | Cat#12805S; RRID:AB_2798031 |
| HSP70 | Cell Signaling Technologies | Cat#4876S; RRID:AB_2119693 |
| α-Tubulin | Cell Signaling Technologies | Cat#3873S; RRID:AB_1904178 |
| GAPDH | Abcam | Cat#AC027; RRID:AB_2769572 |
| β-Actin | Cell Signaling Technologies | Cat#3700S; RRID:AB_2242334 |
| Proteasome α6 | Purified from hybridoma provided by Tanaka laboratory | Clone Mab 2-17 |
| HA Tag Monoclonal Antibody | Thermo Fisher | Cat#32-6700; RRID:AB_2533092 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technologies | Cat#7074S; RRID:AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technologies | Cat#7076S; RRID:AB_330924 |
| Alexa Fluor 647, Goat anti-rabbit IgG (H+L) | Thermo Fisher | Cat#A21245; RRID:AB_2535813 |
| IRDye 800 CW, Goat anti-mouse | LI-COR Biosciences | Cat#926-32210; RRID:AB_621842 |
| Bacterial and virus strains | ||
| Rosetta™(DE3) competent cells | Millipore Sigma | Cat#70954 |
| NEB 5-alpha | New England Biolabs | Cat#C2987H |
| Chemicals, peptides, and recombinant proteins | ||
| LU-102 | Provided by Overkleeft lab | CAS#1421639-62-4 |
| Bortezomib | LC Laboratories | Cat#B-1408 |
| Carfilzomib | LC Laboratories | Cat#C-3022 |
| Ibrutinib | MedChemExpress | Cat#HY-10997 |
| Acalabrutinib | MedChemExpress | Cat#HY-17600 |
| ONO-4059 | MedChemExpress | Cat#HY-15771 |
| CC-292 | MedChemExpress | Cat#HY-18012 |
| CGI-1746 | MedChemExpress | Cat#HY-11999 |
| GDC-0853 | MedChemExpress | Cat#HY-19834 |
| Evobrutinib | MedChemExpress | Cat#HY-101215 |
| Zanubrutinib | MedChemExpress | Cat#HY-101474A |
| Ac-DEVD-AMC (N-Acetyl-Asp-Glu-Val-Asp-7-amido-4-Methylcoumarin) | Bachem | Cat#I-1660 |
| Suc-LLVY-AMC | Bachem | Cat#1-1395 |
| Ac-RLR-AMC | ChinaPeptide | Custom synthesis |
| Ac-nLPnLD-AMC | ChinaPeptide | Custom synthesis |
| NMS-873 | Selleckchem | Cat#S7285 |
| HA-Ub-VS | Enzo Life Sciences | Cat#BML-UW0155-0025 |
| Alamar Blue | Thermo Fisher | Cat#DAL1025 |
| Critical commercial assays | ||
| SuperSignal™ West Femto Maximum Sensitivity Substrate | Thermo Fisher | Cat#34095 |
| CellEvent™ Caspase 3/7 Green Detection Reagent | Thermo Fisher | Cat#C10423 |
| SYTOX™ Red Dead cell stain | Thermo Fisher | Cat#S34859 |
| GSH/GSSG-Glo™ Assay | Promega | V6611 |
| ADP-Glo™ Kinase Assay | Promega | Cat#V6930 |
| Phosphostain | Thermo Fisher | Cat#P33301 |
| GelCode™ Blue Safe Protein Stain | Thermo Fisher | Cat#1860957 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| Deposited data | ||
| Raw and analyzed RNA sequencing data | This paper | GSE239482 |
| Full Western Blots | This paper | https://doi.org/10.17632/7n36xfmfj9.1 |
| Experimental models: Cell lines | ||
| MDA-MB-231 | ATCC | RRID:CVCL_0062 |
| MDA-MB-468 | ATCC | RRID:CVCL_0419 |
| SUM149 | Asterand | RRID:CVCL_3422 |
| INA-6 | DSMZ | RRID:CVCL_5209 |
| MM1.S | Dr. Steven Rosen | RRID:CVCL_8792 |
| Raji | ATCC | RRID:CVCL_0511 |
| NCI-H929 | ATCC | RRID:CVCL_1600 |
| Recombinant DNA | ||
| pDEST15-UBL-hHR23B | Dr. Alfred Goldberg | |
| pET26b-His10-UIM2-hS5a | Dr. Alfred Goldberg | |
| Software and algorithms | ||
| Prism | GraphPad | |
| CalcuSyn | Biosoft | |
Experimental model and study participant details
Cell culture
MDA-MB-231, Raji, NCI-H929, and MDA-MB-468 cells were obtained from ATCC. INA-6 cells were obtained from DSMZ, and SUM149 cells were obtained from Asterand. MM1.S cells were a kind gift from Dr. Steven Rosen.71 All cell lines were validated by short tandem repeat (STR) DNA fingerprinting.40 MDA-MB-231 and SUM149 cells were cultured in DMEM/F12 50:50 media supplemented with 5% fetal bovine serum (FBS). In addition, SUM149 media was supplemented with 1 μg/mL hydrocortisone, 4.8 ug/mL human recombinant insulin, 10 mM HEPES (pH 7.3), and 4 mM L-glutamine. Multiple myeloma and lymphoma cells were cultured in RPMI-1640 media supplemented with 10% FBS, and INA-6 cells media was supplemented with 1 ng/ml IL-6 and 50 μM β-mercaptoethanol. All media used was further supplemented with penicillin, streptomycin, amphotericin-B (0.25 μg/ml), and ciprofloxacin (0.2 μg/ml). Cell viability was assayed with resazurin (Alamar Blue, ThermoFisher) after cells were treated for the times indicated. Combination indices were calculated using CalcuSyn software. Apoptosis was measured by flow-cytometry on BD Accuri C6 Plus flow-cytometer using CellEvent™ Caspase-3/7 Green Detection Reagent and SYTOX cell viability dye (Thermo). Data were analyzed using BD CSampler Plus software. Alternatively, caspase-3,7 activity was measured in extracts using Ac-DEVD-AMC as described by Britton et al.23
Method details
Inhibitors and substrates
Carfilzomib and bortezomib were obtained from LC Laboratories. BTK inhibitors were obtained from MedChemExpress. Suc-LLVY-AMC was obtained from Bachem, Ac-RLR-AMC, and Ac-nLPnLD-AMC were custom synthesized by ChinaPeptides. LU-102 was synthesized as described by Geurink et al.21
Immunoblotting and antibodies used
Cells were lysed in CHAPS buffer (0.5% CHAPS, 10% glycerol, 5 mM MgCl2, and 1 mM EDTA in 50 mM Tris-HCl (pH 7.5)), incubated for 5 min on ice, and centrifuged at 20,000 × g for 15 min. 1X PhosSTOP™ (Roche) was added to the lysis buffer in experiments requiring the study of phosphorylation. Samples were quantified using Bradford assay, and 20 μg of total protein per sample was heated for 15 minutes with LDS buffer (Novex NP0007) at 72°C, fractionated on Bis-Tris gels (Genscript) using MES electrode buffer, and transferred to 0.2μm PVDF membrane (Immobilon, ISEQ00010). Blots were blocked with 2% or 5% non-fat milk. The membrane was incubated with primary antibodies, BTK (Cell Signaling, 8547S), Y223-pBTK (Cell Signaling, 5082S), K48 polyubiquitin (Cell Signaling, 12805S); HSP70 (Cell Signaling, 4876S); NOXA (Cell Signaling, 14766S); α-Tubulin (Cell Signaling, 3873S); β-actin (Cell Signaling, 3700S) and GAPDH (Abcam, AC027), overnight at 4°C and with the secondary antibodies (fluorescent goat anti-mouse (LI-COR, 92668070), and fluorescent anti-rabbit (Thermo Fisher, A21245, HRP-linked secondary antibodies (Cell Signaling; anti-rabbit, 7074S; anti-mouse, 7076S)) for 1h at room temperature, and imaged on Azure c600, directly if fluorescently labeled secondary antibodies were used, or after brief incubation with Pierce SuperSignal West Femto Maximum Sensitivity Substrate of HRP-conjugated secondary antibodies were used.
RNA sequencing
MDA-MB-231 cells were treated for four hours with CGI-1746, LU-102, or a combination of the two. RNA was extracted using the Qiagen RNeasy Mini Kit according to the manufacturer’s protocol, frozen, and shipped on dry ice to LCSciences (Houston, TX) for total RNA sequencing (150 bp PE, 40 million reads per sample). RNAseq data analysis was performed using a command-line-based analysis pipeline. Briefly, raw reads were pre-processed and mapped to the hg38 human genome build using the STAR Aligner tool. Next, mapped read counts were normalized for sequencing depth, and counts per million (CPM) of mapped reads were generated for each transcript. Differential gene expression analysis was performed between the Gene Expression Profiles (GEP) of each treatment group using GSA, as described earlier.72
Protein isolation
Human 26S proteasomes were isolated from mammalian cells via the UBL-affinity purification method.73 HeLa cells were collected by centrifugation and lysed by sonication in affinity purification buffer (APB), 25 mM HEPES-KOH (pH 7.4), 10% (v/v) glycerol, 5 mM MgCl2, 1 mM ATP, and 1 mM DTT, using six ten second bursts (12-micron amplitude) with at least ten-second recovery periods on ice. Lysates were cleared by centrifugation at 4°C, 100,000g for 30 minutes, and the supernatant was then further centrifuged for 3 hours at 4°C, 150,000g. The proteasome-rich pellet was suspended in APB and then incubated with GST-UBL74 and pre-equilibrated Glutathione-Sepharose for two hours at 4°C. The Glutathione-Sepharose resin was washed with APB. Bound proteasomes were eluted by fifteen-minute incubation with His10-UIM74 in APB. His10-UIM was removed from eluted proteasomes by incubation with pre-equilibrated Ni-NTA resin for twenty minutes, followed by resin removal by centrifugation. Rabbit 20S and 26S proteasomes were purified from frozen muscles as described.23,75 19S and VCP were isolated from bovine erythrocytes as described.76,77
Activity assays
Peptidase activities of proteasomes were determined from the slope of the reaction progress curves obtained by continuously following the fluorescence of 7-amino-4-methyl coumarin (AMC), released upon cleavage of Suc-LLVY-AMC (β5 sites), Ac-RLR-AMC (β2 sites), or Ac-nLPnLD-AMC (β1 sites),23 by ∼580 pM of purified proteasomes in the presence or absence of inhibitors. Each substrate was at 100 μM unless stated otherwise. The reaction buffer was 50 mM Tris-HCl (pH 7.5), 1 mM DTT for 20S measurements, supplemented with 5 mM MgCl2 (unless otherwise stated), 100 μM ATP, 2 mM EDTA, and 40 mM KCl for 26S activity.
ATPase assays. 2 μl of purified 19S, 26S and VCP were pre-incubated with BTK inhibitors for two hours in 40 mM Tris (pH 7.5), 20 mM MgCl2, 0.1 mg/ml BSA. 10 μL of ATP in the same buffer were added, and incubation continued for 30 minutes. The final concentrations of enzymes were 14 nM for the 19S regulatory particle, 3 nM for the 26S proteasome, and 80 nM for VCP. The amount of ADP produced was then determined using Promega ADP-Glo™ Kinase Assay according to the manufacturer protocol.
ROS assay
2000 MDA-MB-231 cells/ well were plated on a 96-well plate for 24 hours after which cells were treated with 10 μM of CGI-1746, 3 μM of LU-102, a combination of 10 μM of CGI-1746 and 3 μM of LU-102, 50 nM of bortezomib, and 0.05% of vehicle (DMSO/MOCK) for 3 and 8 hours. The ratio of reduced glutathione to oxidized glutathione was determined using GSH/GSSG-Glo™ Assay (Promega), which was performed according to the manufacturer’s protocol.
Quantification and statistical analysis
Statistical analysis was done using GraphPad Prism. All data points on graphs are averages of n biological replicates. n> 3 indicates 4–7 replicates. Error bars on all graphs indicate S.E.M, except for Figure 1C where they indicate S.D. Combination indexes were calculated using CalcuSyn.
Published: September 24, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110961.
Supplemental information
References
- 1.Gallastegui N., Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends Biochem. Sci. 2010;35:634–642. doi: 10.1016/J.TIBS.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., Martin A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018;87:697–724. doi: 10.1146/ANNUREV-BIOCHEM-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voges D., Zwickl P., Baumeister W. The 26S Proteasome: A Molecular Machine Designed for Controlled Proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/ANNUREV.BIOCHEM.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.Marshall R.S., Vierstra R.D. Dynamic regulation of the 26S proteasome: From synthesis to degradation. Front. Mol. Biosci. 2019;6:40. doi: 10.3389/FMOLB.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cenci S., Oliva L., Cerruti F., Milan E., Bianchi G., Raule M., Mezghrani A., Pasqualetto E., Sitia R., Cascio P. Pivotal Advance: Protein synthesis modulates responsiveness of differentiating and malignant plasma cells to proteasome inhibitors. J. Leukoc. Biol. 2012;92:921–931. doi: 10.1189/JLB.1011497. [DOI] [PubMed] [Google Scholar]
- 6.Cenci S., van Anken E., Sitia R. Proteostenosis and plasma cell pathophysiology. Curr. Opin. Cell Biol. 2011;23:216–222. doi: 10.1016/J.CEB.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Bross P.F., Kane R., Farrell A.T., Abraham S., Benson K., Brower M.E., Bradley S., Gobburu J.V., Goheer A., Lee S.L., et al. Approval Summary for Bortezomib for Injection in the Treatment of Multiple Myeloma. Clin. Cancer Res. 2004;10:3954–3964. doi: 10.1158/1078-0432.CCR-03-0781. [DOI] [PubMed] [Google Scholar]
- 8.Herndon T.M., Deisseroth A., Kaminskas E., Kane R.C., Koti K.M., Rothmann M.D., Habtemariam B., Bullock J., Bray J.D., Hawes J., et al. U.S. Food and Drug Administration Approval: Carfilzomib for the Treatment of Multiple Myeloma. Clin. Cancer Res. 2013;19:4559–4563. doi: 10.1158/1078-0432.CCR-13-0755. [DOI] [PubMed] [Google Scholar]
- 9.Shirley M. Ixazomib: First Global Approval. Drugs. 2016;76:405–411. doi: 10.1007/S40265-016-0548-5. [DOI] [PubMed] [Google Scholar]
- 10.Balleari E., Ghio R., Falcone A., Musto P. Possible Multiple Myeloma Dedifferentiation Following Thalidomide Therapy: A Report of Four Cases. Leuk. Lymphoma. 2004;45:735–738. doi: 10.1080/10428190310001617231. [DOI] [PubMed] [Google Scholar]
- 11.Leung-Hagesteijn C., Erdmann N., Cheung G., Keats J.J., Stewart A.K., Reece D.E., Chung K.C., Tiedemann R.E. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/J.CCR.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G., Liu Y., Zhang B., Meng F., Liu X. Expression of Bruton’s Tyrosine Kinase in Multiple Myeloma. Blood. 2007;110:4751. doi: 10.1182/BLOOD.V110.11.4751.4751. [DOI] [Google Scholar]
- 13.Liu Y., Dong Y., Jiang Q.L., Zhang B., Hu A.M. Bruton’s tyrosine kinase: potential target in human multiple myeloma. Leuk. Lymphoma. 2014;55:177–181. doi: 10.3109/10428194.2013.794458. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Shi J., Gu Z., Salama M.E., Das S., Wendlandt E., Xu H., Huang J., Tao Y., Hao M., et al. Bruton tyrosine kinase is a therapeutic target in stem-like cells from multiple myeloma. Cancer Res. 2015;75:594–604. doi: 10.1158/0008-5472.CAN-14-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Claro R.A., McGinn K.M., Verdun N., Lee S.L., Chiu H.J., Saber H., Brower M.E., Chang C.J.G., Pfuma E., Habtemariam B., et al. FDA Approval: Ibrutinib for Patients with Previously Treated Mantle Cell Lymphoma and Previously Treated Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2015;21:3586–3590. doi: 10.1158/1078-0432.CCR-14-2225. [DOI] [PubMed] [Google Scholar]
- 16.Markham A., Dhillon S. Acalabrutinib: First Global Approval. Drugs. 2018;78:139–145. doi: 10.1007/S40265-017-0852-8. [DOI] [PubMed] [Google Scholar]
- 17.Dasmahapatra G., Patel H., Dent P., Fisher R.I., Friedberg J., Grant S. The Bruton tyrosine kinase (BTK) inhibitor PCI-32765 synergistically increases proteasome inhibitor activity in diffuse large-B cell lymphoma (DLBCL) and mantle cell lymphoma (MCL) cells sensitive or resistant to bortezomib. Br. J. Haematol. 2013;161:43–56. doi: 10.1111/BJH.12206. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 18.Kraus J., Kraus M., Liu N., Besse L., Bader J., Geurink P.P., De Bruin G., Kisselev A.F., Overkleeft H., Driessen C. The novel β2-selective proteasome inhibitor LU-102 decreases phosphorylation of I kappa B and induces highly synergistic cytotoxicity in combination with ibrutinib in multiple myeloma cells. Cancer Chemother. Pharmacol. 2015;76:383–396. doi: 10.1007/s00280-015-2801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kisselev A.F., Van Der Linden W.A., Overkleeft H.S. Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 2012;19:99–115. doi: 10.1016/J.CHEMBIOL.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kisselev A.F. Site-Specific Proteasome Inhibitors. Biomolecules. 2021;12:54. doi: 10.3390/BIOM12010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geurink P.P., Van Der Linden W.A., Mirabella A.C., Gallastegui N., De Bruin G., Blom A.E.M., Voges M.J., Mock E.D., Florea B.I., Van Der Marel G.A., et al. Incorporation of Non-natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites. J. Med. Chem. 2013;56:1262–1275. doi: 10.1021/JM3016987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirabella A.C., Pletnev A.A., Downey S.L., Florea B.I., Shabaneh T.B., Britton M., Verdoes M., Filippov D.V., Overkleeft H.S., Kisselev A.F. Specific Cell-Permeable Inhibitor of Proteasome Trypsin-like Sites Selectively Sensitizes Myeloma Cells to Bortezomib and Carfilzomib. Chem. Biol. 2011;18:608–618. doi: 10.1016/J.CHEMBIOL.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britton M., Lucas M.M., Downey S.L., Screen M., Pletnev A.A., Verdoes M., Tokhunts R.A., Amir O., Goddard A.L., Pelphrey P.M., et al. Selective inhibitor of proteasome’s caspase-like sites sensitizes cells to specific inhibition of chymotrypsin-like sites. Chem. Biol. 2009;16:1278–1289. doi: 10.1016/J.CHEMBIOL.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong Y., Zhang S., Wu Z., Li X., Wang W.L., Zhu Y., Stoilova-McPhie S., Lu Y., Finley D., Mao Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2019;565:49–55. doi: 10.1038/S41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bard J.A.M., Goodall E.A., Greene E.R., Jonsson E., Dong K.C., Martin A. Structure and Function of the 26S Proteasome. Annu. Rev. Biochem. 2018;87:697–724. doi: 10.1146/ANNUREV-BIOCHEM-062917-011931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Zhang Y., Da Silva Sil Dos Santos B., Wang F., Ma Y., Perez C., Yang Y., Peng J., Cohen S.M., Chou T.F., et al. Epidithiodiketopiperazines Inhibit Protein Degradation by Targeting Proteasome Deubiquitinase Rpn11. Cell Chem. Biol. 2018;25:1350–1358.e9. doi: 10.1016/J.CHEMBIOL.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Yakushi T., Parlati F., MacKinnon A.L., Perez C., Ma Y., Carter K.P., Colayco S., Magnuson G., Brown B., et al. Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat. Chem. Biol. 2017;13:486–493. doi: 10.1038/NCHEMBIO.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Arcy P., Brnjic S., Olofsson M.H., Fryknäs M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 2011;17:1636–1640. doi: 10.1038/NM.2536. [DOI] [PubMed] [Google Scholar]
- 29.Lee B.H., Lee M.J., Park S., Oh D.C., Elsasser S., Chen P.C., Gartner C., Dimova N., Hanna J., Gygi S.P., et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/NATURE09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inobe T., Genmei R. Inhibition of the 26S proteasome by peptide mimics of the coiled-coil region of its ATPase subunits. Biochem. Biophys. Res. Commun. 2015;468:143–150. doi: 10.1016/J.BBRC.2015.10.144. [DOI] [PubMed] [Google Scholar]
- 31.Lim H.S., Cai D., Archer C.T., Kodadek T. Periodate-Triggered Cross-Linking Reveals Sug2/Rpt4 as the Molecular Target of a Peptoid Inhibitor of the 19S Proteasome Regulatory Particle. J. Am. Chem. Soc. 2007;129:12936–12937. doi: 10.1021/ja075469+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandu C., Chandramouli N., Glickman J.F., Molina H., Kuo C.L., Kukushkin N., Goldberg A.L., Steller H. Thiostrepton interacts covalently with Rpt subunits of the 19S proteasome and proteasome substrates. J. Cell Mol. Med. 2015;19:2181–2192. doi: 10.1111/JCMM.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Y., Guryanova O.A., Zhou W., Liu C., Huang Z., Fang X., Wang X., Chen C., Wu Q., He Z., et al. Ibrutinib inactivates BMX-STAT3 in glioma stem cells to impair malignant growth and radioresistance. Sci. Transl. Med. 2018;10 doi: 10.1126/SCITRANSLMED.AAH6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim E., Hurtz C., Koehrer S., Wang Z., Balasubramanian S., Chang B.Y., Müschen M., Davis R.E., Burger J.A. Ibrutinib inhibits pre-BCR+ B-cell acute lymphoblastic leukemia progression by targeting BTK and BLK. Blood. 2017;129:1155–1165. doi: 10.1182/BLOOD-2016-06-722900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H., Wang A., Zhang W., Wang B., Chen C., Wang W., Hu C., Ye Z., Zhao Z., Wang L., et al. Ibrutinib selectively and irreversibly targets EGFR (L858R, Del19) mutant but is moderately resistant to EGFR (T790M) mutant NSCLC Cells. Oncotarget. 2015;6:31313–31322. doi: 10.18632/ONCOTARGET.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dubovsky J.A., Beckwith K.A., Natarajan G., Woyach J.A., Jaglowski S., Zhong Y., Hessler J.D., Liu T.M., Chang B.Y., Larkin K.M., et al. Ibrutinib is an irreversible molecular inhibitor of ITK driving a Th1-selective pressure in T lymphocytes. Blood. 2013;122:2539–2549. doi: 10.1182/BLOOD-2013-06-507947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaptein A., de Bruin G., Emmelot-van Hoek M., van de Kar B., de Jong A., Gulrajani M., Demont D., Covey T., Mittag D., Barf T. Potency and Selectivity of BTK Inhibitors in Clinical Development for B-Cell Malignancies. Blood. 2018;132:1871. doi: 10.1182/BLOOD-2018-99-109973. [DOI] [Google Scholar]
- 38.Di Paolo J.A., Huang T., Balazs M., Barbosa J., Barck K.H., Bravo B.J., Carano R.A.D., Darrow J., Davies D.R., Deforge L.E., et al. Specific Btk inhibition suppresses B cell– and myeloid cell–mediated arthritis. Nat. Chem. Biol. 2011;7:41–50. doi: 10.1038/nchembio.481. [DOI] [PubMed] [Google Scholar]
- 39.Crawford J.J., Johnson A.R., Misner D.L., Belmont L.D., Castanedo G., Choy R., Coraggio M., Dong L., Eigenbrot C., Erickson R., et al. Discovery of GDC-0853: A Potent, Selective, and Noncovalent Bruton’s Tyrosine Kinase Inhibitor in Early Clinical Development. J. Med. Chem. 2018;61:2227–2245. doi: 10.1021/ACS.JMEDCHEM.7B01712. [DOI] [PubMed] [Google Scholar]
- 40.Weyburne E.S., Wilkins O.M., Sha Z., Williams D.A., Pletnev A.A., de Bruin G., Overkleeft H.S., Goldberg A.L., Cole M.D., Kisselev A.F. Inhibition of the Proteasome β2 Site Sensitizes Triple-Negative Breast Cancer Cells to β5 Inhibitors and Suppresses Nrf1 Activation. Cell Chem. Biol. 2017;24:218–230. doi: 10.1016/J.CHEMBIOL.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 42.Sato K., Rajendra E., Ohta T. The UPS: A promising target for breast cancer treatment. BMC Biochem. 2008;9:S2. doi: 10.1186/1471-2091-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrocca F., Altschuler G., Tan S.M., Mendillo M.L., Yan H., Jerry D.J., Kung A.L., Hide W., Ince T.A., Lieberman J. A Genome-wide siRNA Screen Identifies Proteasome Addiction as a Vulnerability of Basal-like Triple-Negative Breast Cancer Cells. Cancer Cell. 2013;24:182–196. doi: 10.1016/j.ccr.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitsiades N., Mitsiades C.S., Poulaki V., Chauhan D., Fanourakis G., Gu X., Bailey C., Joseph M., Libermann T.A., Treon S.P., et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA. 2002;99:14374–14379. doi: 10.1073/PNAS.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiita A.P., Ziv E., Wiita P.J., Urisman A., Julien O., Burlingame A.L., Weissman J.S., Wells J.A. Global cellular response to chemotherapy-induced apoptosis. Elife. 2013;2:e01236. doi: 10.7554/ELIFE.01236.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Magnaghi P., D’Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., Gasparri F., Cozzi L., Cucchi U., Orrenius C., et al. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nat. Chem. Biol. 2013;9:548–556. doi: 10.1038/NCHEMBIO.1313. [DOI] [PubMed] [Google Scholar]
- 47.Anderson D.J., Le Moigne R., Djakovic S., Kumar B., Rice J., Wong S., Wang J., Yao B., Valle E., Kiss von Soly S., et al. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/J.CCELL.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skrott Z., Mistrik M., Andersen K.K., Friis S., Majera D., Gursky J., Ozdian T., Bartkova J., Turi Z., Moudry P., et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–199. doi: 10.1038/NATURE25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szczęśniak P.P., Heidelberger J.B., Serve H., Beli P., Wagner S.A. VCP inhibition induces an unfolded protein response and apoptosis in human acute myeloid leukemia cells. PLoS One. 2022;17 doi: 10.1371/JOURNAL.PONE.0266478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipchick B.C., Fink E.E., Nikiforov M.A. Oxidative stress and proteasome inhibitors in multiple myeloma. Pharmacol. Res. 2016;105:210–215. doi: 10.1016/J.PHRS.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park W.H., Kim S.H. MG132, a proteasome inhibitor, induces human pulmonary fibroblast cell death via increasing ROS levels and GSH depletion. Oncol. Rep. 2012;27:1284–1291. doi: 10.3892/OR.2012.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pérez-Galán P., Roué G., Villamor N., Montserrat E., Campo E., Colomer D. The proteasome inhibitor bortezomib induces apoptosis in mantle-cell lymphoma through generation of ROS and Noxa activation independent of p53 status. Blood. 2006;107:257–264. doi: 10.1182/BLOOD-2005-05-2091. [DOI] [PubMed] [Google Scholar]
- 53.Nikiforov M.A., Riblett M., Tang W.H., Gratchouck V., Zhuang D., Fernandez Y., Verhaegen M., Varambally S., Chinnaiyan A.M., Jakubowiak A.J., Soengas M.S. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc. Natl. Acad. Sci. USA. 2007;104:19488–19493. doi: 10.1073/PNAS.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin J.Z., Ziffra J., Stennett L., Bodner B., Bonish B.K., Chaturvedi V., Bennett F., Pollock P.M., Trent J.M., Hendrix M.J.C., et al. Proteasome Inhibitors Trigger NOXA-Mediated Apoptosis in Melanoma and Myeloma Cells. Cancer Res. 2005;65:6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- 55.Kors S., Geijtenbeek K., Reits E., Schipper-Krom S. Regulation of Proteasome Activity by (Post-)transcriptional Mechanisms. Front. Mol. Biosci. 2019;6 doi: 10.3389/FMOLB.2019.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lokireddy S., Kukushkin N.V., Goldberg A.L. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc. Natl. Acad. Sci. USA. 2015;112:E7176–E7185. doi: 10.1073/PNAS.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L., Zhang Y., Shu X., Chen Q., Wei T., Wang H., Wang X., Wu Q., Zhang X., Liu X., et al. Proteasome regulation by reversible tyrosine phosphorylation at the membrane. Oncogene. 2021;40:1942–1956. doi: 10.1038/s41388-021-01674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VerPlank J.J.S., Goldberg A.L. Exploring the regulation of proteasome function by subunit phosphorylation. Methods Mol. Biol. 2018;1844:309–319. doi: 10.1007/978-1-4939-8706-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.VerPlank J.J.S., Goldberg A.L. Regulating Protein Breakdown Through Proteasome Phosphorylation. Biochem. J. 2017;474:3355–3371. doi: 10.1042/BCJ20160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith D.M., Fraga H., Reis C., Kafri G., Goldberg A.L. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144:526–538. doi: 10.1016/j.cell.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stein R.L., Melandri F., Dick L. Kinetic characterization of the chymotryptic activity of the 20S proteasome. Biochemistry. 1996;35:3899–3908. doi: 10.1021/BI952262X. [DOI] [PubMed] [Google Scholar]
- 62.Kisselev A.F., Callard A., Goldberg A.L. Importance of the Different Proteolytic Sites of the Proteasome and the Efficacy of Inhibitors Varies with the Protein Substrate. J. Biol. Chem. 2006;281:8582–8590. doi: 10.1074/JBC.M509043200. [DOI] [PubMed] [Google Scholar]
- 63.Osmulski P.A., Gaczynska M. Rapamycin allosterically inhibits the proteasome. Mol. Pharmacol. 2013;84:104–113. doi: 10.1124/MOL.112.083873. [DOI] [PubMed] [Google Scholar]
- 64.Munoz L. Non-kinase targets of protein kinase inhibitors. Nat. Rev. Drug Discov. 2017;16:424–440. doi: 10.1038/NRD.2016.266. [DOI] [PubMed] [Google Scholar]
- 65.Saeki Y., Isono E., Toh-E A. Preparation of Ubiquitinated Substrates by the PY Motif-Insertion Method for Monitoring 26S Proteasome Activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 66.Nathan J.A., Kim H.T., Ting L., Gygi S.P., Goldberg A.L. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J. 2013;32:552–565. doi: 10.1038/EMBOJ.2012.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Manfredonia A.J., Kraut D.A. The 26S Proteasome Switches between ATP-Dependent and -Independent Mechanisms in Response to Substrate Ubiquitination. Biomolecules. 2022;12:750. doi: 10.3390/BIOM12060750/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basler M., Kirk C.J., Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 2013;25:74–80. doi: 10.1016/J.COI.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Rock K.L., Goldberg A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999;17:739–779. doi: 10.1146/ANNUREV.IMMUNOL.17.1.739. [DOI] [PubMed] [Google Scholar]
- 70.Thomas T.A., Smith D.M. Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. J. Biol. Chem. 2022;298 doi: 10.1016/J.JBC.2022.102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shabaneh T.B., Downey S.L., Goddard A.L., Screen M., Lucas M.M., Eastman A., Kisselev A.F. Molecular Basis of Differential Sensitivity of Myeloma Cells to Clinically Relevant Bolus Treatment with Bortezomib. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar H., Mazumder S., Chakravarti S., Sharma N., Mukherjee U.K., Kumar S., Baughn L.B., Van Ness B.G., Mitra A.K. secDrug: a pipeline to discover novel drug combinations to kill drug-resistant multiple myeloma cells using a greedy set cover algorithm and single-cell multi-omics. Blood Cancer J. 2022;12 doi: 10.1038/s41408-022-00636-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuo C.L., Collins G.A., Goldberg A.L. Methods to Rapidly Prepare Mammalian 26S Proteasomes for Biochemical Analysis. Methods Mol. Biol. 2018;1844:277–288. doi: 10.1007/978-1-4939-8706-1_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Besche H.C., Haas W., Gygi S.P., Goldberg A.L. Isolation of Mammalian 26S Proteasomes and p97/VCP Complexes Using the Ubiquitin-like Domain from HHR23B Reveals Novel Proteasome-Associated Proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/BI802198Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kisselev A.F., Kaganovich D., Goldberg A.L. Binding of Hydrophobic Peptides to Several Non-catalytic Sites Promotes Peptide Hydrolysis by All Active Sites of 20 S Proteasomes: Evidence for Peptide-Induced Channel Opening in the α-Rings. J. Biol. Chem. 2002;277:22260–22270. doi: 10.1074/JBC.M112360200. [DOI] [PubMed] [Google Scholar]
- 76.DeMartino G.N. Purification of PA700, the 19S regulatory complex of the 26S proteasome. Methods Enzymol. 2005;398:295–306. doi: 10.1016/S0076-6879(05)98024-5. [DOI] [PubMed] [Google Scholar]
- 77.Wahlman J., DeMartino G.N., Skach W.R., Bulleid N.J., Brodsky J.L., Johnson A.E. Real-Time Fluorescence Detection of ERAD Substrate Retrotranslocation in a Mammalian In Vitro System. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA-seq data have been deposited at GEO (GEO: GSE239482) and is publicly available as of the date of publication. key resources table Original western blot images have been deposited at Mendeley (Data set http://doi:10.17632/7n36xfmfj9.1) and are publicly available as of the date of publication.key resources table
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.