Abstract
Artificial intelligence (AI) has significantly impacted various fields, including oncology. This comprehensive review examines the current applications and future prospects of AI in lung cancer research and treatment. We critically analyze the latest AI technologies and their applications across multiple domains, including genomics, transcriptomics, proteomics, metabolomics, immunomics, microbiomics, radiomics, and pathomics in lung cancer research. The review elucidates AI’s transformative role in enhancing early detection, personalizing treatment strategies, and accelerating therapeutic innovations. We explore AI’s impact on precision medicine in lung cancer, encompassing early diagnosis, treatment planning, monitoring, and drug discovery. The potential of AI in analyzing complex datasets, including genetic profiles, imaging data, and clinical records, is discussed, highlighting its capacity to provide more accurate diagnoses and tailored treatment plans. Additionally, we examine AI’s potential in predicting patient responses to immunotherapy and forecasting survival rates, particularly in non-small cell lung cancer (NSCLC). The review addresses technical challenges facing AI implementation in lung cancer care, including data quality and quantity issues, model interpretability, and ethical considerations, while discussing potential solutions and emphasizing the importance of rigorous validation. By providing a comprehensive analysis for researchers and clinicians, this review underscores AI’s indispensable role in combating lung cancer and its potential to usher in a new era of medical breakthroughs, ultimately aiming to improve patient outcomes and quality of life.
Keywords: artificial intelligence, lung cancer, precision medicine, early diagnosis, personalized treatment
1. Introduction
Lung cancer is a major challenge in oncology, with a substantial impact on global health metrics. Non-small cell lung cancer (NSCLC) is the predominant form, constituting the majority of cases (1, 2). When diagnosed at an advanced stage, the prognosis is quite poor, with long-term survival rates being relatively low (3, 4). However, if detected at an early stage, the chances of survival improve significantly. For those with locally advanced disease, the survival rates are moderate but still concerning (5). Small cell lung cancer (SCLC) is less common but has an even more serious outlook, with survival rates dropping significantly when the disease has spread extensively. The financial and emotional impact of these cancers on patients and their families is considerable, affecting the quality of life in a meaningful way.
The critical importance of early screening for lung cancer has been increasingly recognized, as it substantially enhances the chances of early detection and treatment. However, even those diagnosed at an early stage are not exempt from the risk of relapse (6). Recurrence often leads to the progression to advanced stages, drastically diminishing the survival prognosis. The pathogenesis and progression of lung cancer are governed by intricate regulatory networks, underscoring the complexity of this disease. Furthermore, the precise mechanisms driving lung cancer initiation and progression, chemotherapeutic resistance, and resistance to targeted therapies such as those against Epidermal Growth Factor Receptor (EGFR) and Anaplastic Lymphoma Kinase (ALK), as well as immune resistance, remain areas necessitating further investigation.
A comprehensive glossary of essential artificial intelligence (AI) terminology used in this manuscript is presented in Table 1 .
Table 1.
Glossary of key terms artificial intelligence.
| Artificial Intelligence (AI) | The ability of computer systems to perform tasks that typically require human intelligence, such as visual perception, speech recognition, decision-making, and language translation. |
| Machine learning (ML) | A subset of AI that focuses on the development of algorithms and statistical models that enable computer systems to improve their performance on a specific task through experience, without being explicitly programmed. |
| Deep Learning (ML) | A subset of machine learning based on artificial neural networks with multiple layers, capable of learning complex patterns in large amounts of data. |
| Natural Language Processing (NLP) | A field of AI that focuses on the interaction between computers and humans using natural language, enabling machines to understand, interpret, and generate human language. |
| Computer vision | A field of AI that enables computers to gain high-level understanding from digital images or videos, aiming to automate tasks that the human visual system can do. |
| Robotics | The branch of technology that deals with the design, construction, operation, and use of robots, often incorporating AI for decision-making and task execution. |
| Deep neural networks (DNNs) | Artificial neural networks with multiple layers between the input and output layers, capable of modeling complex non-linear relationships. |
| Convolutional Neural Networks (CNNs) | A class of deep neural networks most commonly applied to analyze visual imagery, designed to automatically and adaptively learn spatial hierarchies of features. |
| Recurrent Neural Networks (RNNs) | A class of neural networks where connections between nodes form a directed graph along a temporal sequence, allowing it to exhibit temporal dynamic behavior. |
| Support vector machines (SVMs) | Supervised learning models used for classification and regression analysis, effective in high-dimensional spaces. |
| Principal Component Analysis (PCA) | A statistical procedure that uses orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of linearly uncorrelated variables. |
| t-Distributed Stochastic Neighbor Embedding (t-SNE) | A machine learning algorithm for visualization that reduces dimensionality based on similarity of datapoints. |
| Uniform Manifold Approximation and Projection (UMAP) | A dimension reduction technique that can be used for visualization similarly to t-SNE, but also for general non-linear dimension reduction. |
| Principal Component Analysis (PCA) | AI systems that can provide human-understandable explanations for their decisions or predictions. |
| Explainable Artificial Intelligence (XAI) | AI systems that can provide human-understandable explanations for their decisions or predictions. |
| Gradient-weighted Class Activation Mapping (Grad-CAM) | A technique for producing visual explanations for decisions made by convolutional neural networks. |
| Local Interpretable Model-agnostic Explanations (LIME) | A technique that explains the predictions of any classifier in an interpretable and faithful manner. |
| Area Under the Curve of Receiver Operating Characteristic (AUC-ROC) | A performance measurement for classification problems at various thresholds settings, representing the degree of separability between classes. |
| Precision-recall curves | A graphical plot that illustrates the trade-off between precision and recall for different thresholds in a binary classifier system. |
| F1 scores | The harmonic mean of precision and recall, providing a single score that balances both metrics. |
| Calibration plots | Graphical representations of the agreement between predicted probabilities and observed frequencies, used to assess the calibration of probabilistic predictions. |
| Decision curve analysis | A method for evaluating and comparing prediction models that accounts for the clinical consequences of using a model. |
| SHapley Additive exPlanations (SHAP) | A game theoretic approach to explain the output of any machine learning model. It connects optimal credit allocation with local explanations using the classic Shapley values from game theory and their related extensions. |
In recent years, AI has emerged as a transformative force in biomedical research, particularly in the domain of lung cancer. AI enhances the potential for early diagnosis through high-precision imaging and pattern recognition, offering new avenues for predicting and monitoring disease progression (7–9). Additionally, AI-driven approaches are crucial in unraveling the complex molecular and genetic landscapes of lung cancer, thus aiding in the identification of novel therapeutic targets and the development of personalized treatment strategies (10, 11). As lung cancer research continues to evolve, the integration of AI technology promises to revolutionize the field, paving the way for more effective and tailored interventions.
AI, a branch of computer science, focuses on creating systems capable of performing tasks traditionally requiring human intelligence. The overarching goal of AI is to enable machines to emulate and execute functions such as perception, learning, reasoning, planning, and natural language processing. Categorically, AI encompasses various technologies including machine learning (ML), deep learning (DL), natural language processing (NLP), computer vision, and robotics (12–14). ML, a pivotal subset of AI, involves techniques that allow computers to learn from data and enhance their performance over time (15). Within ML, methods such as supervised learning, unsupervised learning, semi-supervised learning, and reinforcement learning are employed to extract patterns and make predictions. DL, a further specialized subset of ML, leverages neural networks with multiple layers (deep neural networks, DNNs) to mimic the human brain’s processing mechanism, significantly advancing applications like lung Computed Tomography (CT) radiomics (16). NLP technology enables machines to recognize, understand, and generate human languages, facilitating tasks such as text processing, language translation, semantic analysis, and the development of chatbots (17). In the domain of computer vision, AI grants computers the ability to interpret and understand visual information, encompassing object detection, image classification, and facial recognition, which are particularly relevant in the pathological analysis of lung cancer (18). Robotics, another facet of AI, involves designing and building robots capable of performing tasks requiring human-like intelligence, including perception, movement, and navigation—for instance, the robotic-assisted lung lobectomy (Robot-L) (19). Presently, the applications of AI in the domain of lung cancer research are extensive, encompassing the comprehensive analysis of genomic data, which includes the fields of genomics, transcriptomics, and epigenomics. Additionally, AI methodologies are applied in the realms of radiomics for imaging analysis, digital pathology for the examination of tissue specimens, and the integration and interpretation of real-world multimodal datasets. A notable example is the recent publication in Nature by Harvard University and Massachusetts Institute of Technology, describing PathChat, a versatile AI framework for visual and linguistic analysis in human pathology (20). Through these varied applications, AI not only enhances diagnostic accuracy and therapeutic planning in lung cancer but also promises to usher in a new era of personalized medicine, ultimately improving patient outcomes.
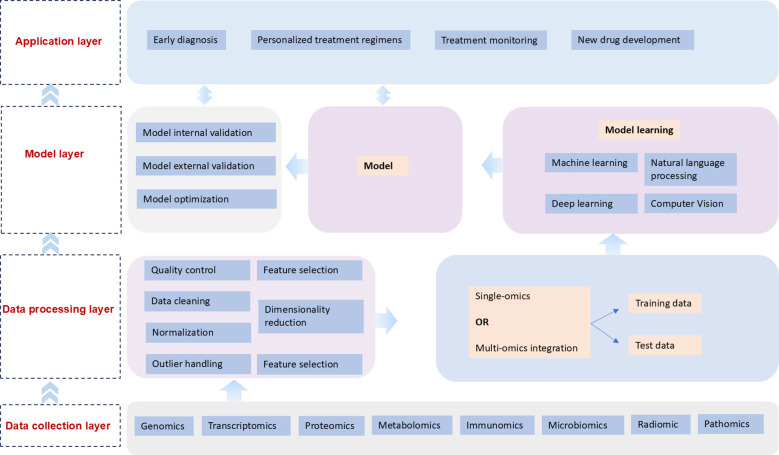
AI has become a pivotal force in lung cancer research, revolutionizing the methodologies by which we understand, diagnose, and treat this pervasive disease. AI techniques are being increasingly applied to various -omics domains, including genomics, transcriptomics, proteomics, metabolomics, immunogenomics, microbiomics, radiomics, and pathology. These applications have significantly enhanced our ability to decipher complex biological data, providing unprecedented insights into the molecular mechanisms underlying lung cancer. Moreover, AI’s integration into precision medicine promises transformative advancements in early diagnosis, therapeutic monitoring, and novel drug development. In this review, we delve into the multifaceted roles AI plays across these diverse fields, exploring current applications and envisioning the promising future of AI-driven innovations in lung cancer research and treatment. Figure 1 illustrates how AI transforms diverse biological data to enhance lung cancer diagnosis, personalize treatment, and accelerate drug development, thereby advancing precision oncology. This comprehensive application underscores AI’s immense potential in addressing the full spectrum of lung cancer management, from early screening to therapeutic monitoring and novel drug discovery. As lung cancer research continues to evolve, the integration of AI technologies promises to revolutionize the field, paving the way for more effective and individualized interventions. In this review, we delve into the multifaceted roles AI plays across these diverse domains, exploring current applications and envisioning the promising future of AI-driven innovations in lung cancer research and treatment. We examine how AI’s capabilities in data analysis, pattern recognition, and predictive modeling are being harnessed to improve early detection rates, optimize treatment strategies, and expedite the development of targeted therapies. Furthermore, we discuss the potential of AI to overcome existing challenges in lung cancer management, such as the complexity of tumor heterogeneity and the need for more precise prognostic tools. By critically assessing the current state of AI applications in lung cancer and projecting future developments, this review aims to provide a comprehensive overview of the transformative impact of AI on the landscape of lung cancer research and clinical practice.
Figure 1.
Artificial intelligence-driven multi-omics framework for lung cancer precision medicine.
2. AI in lung cancer: current applications
The application of AI in lung cancer primarily focuses on several areas: genomics, transcriptomics, proteomics, metabolomics, immunomics, microbiomics, radiomics, and pathomics. A comprehensive comparative analysis of AI applications across these omics fields in lung cancer research is presented in Table 2 .
Table 2.
Comparative analysis of AI applications across omics fields in lung cancer research.
| Omics Field | AI Techniques | Key Applications | Performance/Strengths | Limitations |
|---|---|---|---|---|
| Genomics | Deep Learning, Machine Learning | - Gene mutation analysis | - High accuracy in mutation detection | - Requires large datasets |
| - Genomic instability assessment | - Improved early diagnosis | - Complexity in interpreting genetic variations | ||
| - Epigenetics study | - Non-invasive screening (liquid biopsy) | |||
| - Liquid biopsy enhancement | ||||
| Transcriptomics | Deep Learning, Natural Language Processing | - Gene expression analysis | - Identification of novel biomarkers | - Challenges in handling noise in expression data |
| - RNA sequencing data interpretation | - Insight into gene regulation | - Difficulty in interpreting complex gene interactions | ||
| Proteomics | Machine Learning, Deep Neural Networks | - Protein-protein interaction prediction | - High-throughput analysis | - Limited by proteome complexity |
| - Biomarker discovery | - Identification of drug targets | - Challenges in data integration | ||
| Metabolomics | Machine Learning, Deep Learning | - Metabolic pathway analysis | - Early diagnosis potential | - Metabolite variability challenges |
| - Biomarker identification | - Treatment response monitoring | - Need for standardized data collection | ||
| Immunomics | Deep Learning, Machine Learning | - Immune escape mechanism study | - Personalized immunotherapy approaches | - Complexity of immune system interactions |
| - Neoantigen discovery | - Improved understanding of tumor-immune interactions | - Limited availability of comprehensive immune data | ||
| - Immunotherapy response prediction | ||||
| Microbiomics | Machine Learning | - Microbiome profile analysis | - Novel insights into lung cancer etiology | - Challenges in standardizing microbiome data |
| - Host-microbiome interaction study | - Potential for microbiome-based therapies | - Complexity in interpreting microbial diversity | ||
| Radiomics | Convolutional Neural Networks, Deep Learning | - Image feature extraction | - Non-invasive diagnosis | - Variability in imaging protocols |
| - Tumor classification | - High accuracy in image analysis | - Need for large, diverse image datasets | ||
| - Treatment response prediction | ||||
| Pathomics | Convolutional Neural Networks, Deep Learning | - Automated tissue analysis | - Improved diagnostic accuracy | - Challenges in standardizing tissue preparation |
| - Cancer subtype classification | - Efficient histopathological analysis | - Need for extensive pathologist validation |
2.1. AI in lung cancer genomics
The application of AI in lung cancer genomics encompasses the analysis of gene mutations and variations, the assessment of genomic instability, the study of epigenetics, and the enhancement of non-invasive liquid biopsy techniques. These advancements facilitate early detection, precision treatment, and a comprehensive understanding of the genetic landscape of lung cancer, with the potential to improve patient outcomes and inform therapeutic strategies.
2.1.1. Gene mutations and variations
In the realm of lung cancer genomics, AI has revolutionized the identification and analysis of genetic mutations and variations, playing a pivotal role in advancing precision medicine. The integration of DL models, such as convolutional neural networks (CNNs) and recurrent neural networks (RNNs), facilitates the detailed analysis of large-scale genomic data, enabling the precise identification and classification of specific genetic mutations like EGFR, Kirsten Rat Sarcoma virus (KRAS), and ALK (21–24). Additionally, NLP techniques can autonomously extract mutation-related information from scientific literature and genetic databases, thereby enriching the knowledge base for lung cancer genomics (25–27). ML algorithms further extend the capability of AI by estimating the frequency and distribution of particular gene mutations across diverse populations, thus aiding in the formulation of personalized treatment plans (10, 28).
Moreover, AI excels in the integration of multi-omics data for a comprehensive understanding of the impact of genetic mutations on disease progression and prognosis (29–31). This integration not only enhances molecular characterization but also identifies novel therapeutic targets. In drug discovery, virtual screening powered by AI accelerates the identification of potential drug candidates tailored to specific genetic mutations while predicting their efficacy and side effects (32, 33). Individualized therapeutic approaches are meticulously honed by the AI-driven dissection of each patient’s unique mutational profiles, thereby maximizing the efficacy of treatment outcomes.
Predictive models utilizing ML analyze gene mutation data to forecast disease prognosis, treatment responses, and survival rates, thereby assisting clinicians in decision-making processes (24, 34). Furthermore, the application of DL in pathological image analysis automates the identification of tissue and cellular characteristics correlated with specific gene mutations, increasing diagnostic accuracy and efficiency (35–37). Leading AI-based initiatives, such as those by Foundation Medicine, Tempus, and IBM Watson for Oncology, exemplify the transformative potential of AI in enhancing the precision of lung cancer diagnosis and treatment (38). Additionally, AI-driven large-scale databases like The Cancer Genome Atlas (TCGA) and the Genomic Data Commons (GDC) provide invaluable resources for ongoing research and clinical applications.
2.1.2. Genomic instability
In the domain of AI application on genomic instability in lung cancer, significant advancements have been made across various dimensions, including changes in chromosome number, structural alterations of chromosomes, and gene amplification and deletions (39).
Firstly, alterations in chromosome number, such as triploidy and tetraploidy, are hallmark indicators of genomic instability. AI has been leveraged to study these phenomena through two primary modalities: image analysis and genomic sequencing data analysis. For example, DL techniques using CNNs can now automatically analyze chromosomal smears to identify changes in chromosome number and morphology (40). Concurrently, ML algorithms applied to whole-genome sequencing data can detect aneuploidy states, thereby identifying chromosome number instability (28).
Secondly, AI plays a crucial role in detecting chromosomal structural variations such as deletions, duplications, and inversions. Advanced DL models can analyze high-throughput sequencing data with high precision, accurately pinpointing structural variations on chromosomes. Integration of multi-omics data, including genomics and transcriptomics, further enhances AI’s capability to detect structural changes, utilizing ensemble learning models to combine multiple data sources to improve accuracy and sensitivity (41, 42).
Thirdly, gene amplification and deletions represent another aspect of genomic instability in lung cancer. AI’s involvement in this arena includes the detection of gene amplification through AI analysis of copy number variation (CNV) data. Advanced deep learning models such as CNNs and RNNs have been successfully employed to identify CNVs (43, 44). Similarly, ML algorithms can analyze high-throughput sequencing data to detect gene deletions, and recent developments also include the use of AI models to analyze single-cell sequencing data, revealing cellular heterogeneity and gene loss events with higher resolution.
Notable instances of AI application include Google’s DeepVariant, a deep learning tool for genomic research that accurately detects genomic variants, and CopyNumberNet, a deep learning-based algorithm designed to identify CNVs from whole-genome sequencing data (45). Additionally, databases such as TCGA and Gene Expression Omnibus (GEO) provide extensive genomic and gene expression data, which researchers can analyze using AI techniques to identify genomic instability events in lung cancer.
2.1.3. Epigenetics
The integration of AI into lung cancer epigenetics research has marked a significant advancement in understanding the epigenetic alterations associated with lung cancer, which play crucial roles in tumorigenesis, progression, and treatment responses. Unlike genetic changes, epigenetics focuses on the regulatory mechanisms of gene expression, encompassing DNA methylation, histone modifications, non-coding RNA (ncRNA) regulation, and chromatin remodeling (46–50). The potential of AI in lung cancer epigenetics is becoming increasingly evident, particularly in the analysis of DNA methylation and histone modifications, as well as the regulatory roles of ncRNAs.
In the realm of DNA methylation, AI algorithms, including ML and DL, are employed to analyze extensive methylation datasets, enabling the identification of specific methylation patterns associated with lung cancer (51, 52). These patterns serve as biomarkers for early detection and classification of different lung cancer types, and AI-driven analysis of methylation data is utilized to predict patient prognosis, aiding in the development of personalized treatment plans (53, 54). Histone modifications also benefit from the capabilities of AI; by analyzing acetylation and methylation data, AI can construct detailed histone modification maps to elucidate their distribution and changes in lung cancer (55). Moreover, AI algorithms assist in decoding how these modifications influence gene expression, revealing potential regulatory mechanisms that are critical to the pathogenesis of lung cancer.
The regulatory impacts of ncRNAs, particularly microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are further elucidated through the analytical capabilities of AI. ML models predict miRNA target genes and assess their roles in lung cancer, identifying new therapeutic targets (29, 56, 57). Similarly, AI predicts the expression patterns and potential functions of lncRNAs, uncovering their regulatory effects in lung cancer and aiding in the discovery of novel therapeutic mechanisms (58). Additionally, integrated multi-omics analysis facilitated by AI merges various epigenetic datasets, offering comprehensive insights into the complex regulatory networks underlying lung cancer and allowing for the creation of personalized treatment models based on epigenetic data and clinical information. A notable study utilizing DL models identified multiple DNA methylation sites related to lung cancer prognosis, while integrated analyses revealed coordinated epigenetic regulation of gene expression in lung cancer (30).
These applications underscore AI’s significant potential in decoding the intricate epigenetic landscapes of lung cancer, providing innovative strategies and tools for future research and therapeutic interventions.
2.1.4. Non-invasive liquid biopsy
The application and research of AI in non-invasive liquid biopsy for lung cancer have seen remarkable advancements in recent years, especially in the detection of circulating tumor DNA (ctDNA) from blood samples. Liquid biopsy is an innovative method for diagnosing and monitoring lung cancer by analyzing cancer-related substances in blood or other bodily fluids. Within this domain, AI has significantly enhanced the early detection, monitoring, and personalized treatment of lung cancer. For early diagnosis, AI algorithms coupled with high-sensitivity techniques like digital Polymerase Chain Reaction (dPCR) and next-generation sequencing (NGS) can accurately identify lung cancer-specific mutations and molecular markers from large datasets, distinguishing early-stage lung cancer patients from healthy individuals (59). In terms of monitoring treatment efficacy, ctDNA levels are dynamically measured throughout the treatment process to immediately assess tumor burden and response to therapy (60, 61). AI models can predict treatment outcomes and potential resistance, providing real-time insights (62, 63). Furthermore, AI-driven analysis of ctDNA enables early detection of relapse post-primary treatment by identifying high-risk patients (64, 65). Personalized treatment is another critical area where AI contributes significantly; analyzing the mutation spectrum in ctDNA provides detailed genomic information aiding the selection of the most suitable targeted and immunotherapies (66). Additionally, AI can optimize treatment regimens by integrating ctDNA data with clinical characteristics and treatment history to enhance therapeutic effectiveness (67, 68). Prognostic evaluation is yet another facet where AI demonstrates its utility; it analyzes ctDNA to predict patient survival and disease progression risk, facilitating informed decision-making for treatment strategies and life planning (61, 69). Development of AI-based intelligent diagnostic systems that combine liquid biopsy results with imaging further enhances diagnostic comprehensiveness and accuracy (66). To validate these applications, multicenter clinical trials are essential, ensuring the accuracy and reliability of AI-integrated liquid biopsy approaches.
2.2. AI in lung cancer transcriptomics
Transcriptomics in lung cancer involves investigating the changes in gene expression and transcription levels utilizing high-throughput sequencing technologies such as RNA Sequencing (RNA-Seq). This methodology provides comprehensive insights into the messenger RNA (mRNA) and other transcription products within lung cancer cells and tissues, revealing their gene expression profiles. AI has revolutionized the field of lung cancer genomics, particularly in the study and application of lung cancer transcriptomics. The application of AI in the transcriptomics of lung cancer predominantly focuses on gene expression profiling, analysis of ncRNA, mRNA modification, single-cell transcriptomics, spatiotemporal transcriptomics, and transcription factor regulatory networks.
2.2.1. Gene expression profiling
A gene expression profile specifies the particular pattern of genes that are activated or suppressed in lung cancer cells, compared to normal lung cells. Certain genes may be overexpressed, indicating elevated expression levels, while others may be downregulated, showing decreased expression. These distinct gene expression patterns are crucial in identifying lung cancer types, progression stages, and responses to various treatments.
The role of AI in analyzing lung cancer gene expression profiles is multifaceted and pivotal. Firstly, AI excels in handling and analyzing massive, high-dimensional gene expression datasets, efficiently extracting valuable information from complex data (70). Secondly, through ML algorithms like random forests, support vector machines (SVM), and DL, AI can discern key genetic features associated with lung cancer from vast datasets (71, 72). Thirdly, AI models based on gene expression profiles enhance the accuracy of disease classification and diagnosis by distinguishing between benign and malignant tumors or different lung cancer types (73, 74). Furthermore, AI can predict patient prognosis and treatment efficacy using gene expression data, aiding physicians in formulating personalized treatment plans to improve outcomes (75). Additionally, AI facilitates new drug discovery and targeted therapy development by identifying potential drug targets and biomarkers from gene expression profiles, furthering the advancement of precision medicine (76). Lastly, through pathway analysis, AI can identify biological pathways related to lung cancer, deepening our understanding of cancer mechanisms and guiding effective treatment strategies (77).
In summary, AI in lung cancer gene expression profiling significantly enhances diagnostic and therapeutic precision and supports the progression of personalized medicine.
2.2.2. Analysis of ncRNA
In recent years, significant strides have been made in uncovering the role of ncRNAs in lung cancer, facilitated by advancements in AI. ncRNAs, which include miRNAs, lncRNAs, and circular RNAs (circRNAs), do not translate into proteins but are crucial in regulating gene expression, RNA processing, chromatin structure, and various cellular functions. AI techniques have revolutionized the identification and analysis of these ncRNAs, offering promising avenues for lung cancer research and treatment.
First, AI-assisted miRNA profiling has enabled the identification of novel miRNA biomarkers associated with lung cancer, aiding in gene expression regulation. ML methods applied to miRNA datasets have uncovered potential diagnostic and therapeutic targets (78). Similarly, lncRNAs have been the focus of predictive modeling using sophisticated algorithms such as LDAenDL, which can forecast lung cancer-related lncRNA biomarkers (79). These discoveries are critical for gene regulation and chromatin modification, proposing new therapeutic strategies (80).Furthermore, circRNAs have been studied through ML and AI-based integrative analyses, revealing their potential roles in cancer biomarkers and their importance in gene expression and signaling pathways regulation (81). By leveraging the capabilities of AI in big data analytics and pattern recognition, these studies significantly propel the understanding and application of ncRNAs in lung cancer, offering new horizons for diagnostics and treatment advancements.
2.2.3. AI in mRNA modification for lung cancer
In lung cancer transcriptomics, AI advancements have significantly enhanced the study and application of mRNA modifications, particularly N6-methyladenosine (m6A) (82). As an essential player in gene expression, mRNA undergoes methylation, impacting its stability and translation. ML models like SVM, random forests, and DL networks (CNNs, RNNs) have effectively identified m6A modification sites. These technologies utilize high-throughput data, such as RNA-Seq, to elucidate m6A’s effects on mRNA half-life and degradation. Combining transcriptomic and proteomic data, AI has revealed how m6A influences translation efficiency and mRNA localization. AI analyses have also linked m6A patterns to lung cancer prognosis and treatment responses, facilitating the discovery of biomarkers and personalized therapies. This integration of AI has propelled m6A research, offering promising insights into lung cancer diagnostics and therapeutics.
2.2.4. AI in single-cell transcriptomics
AI has revolutionized single-cell transcriptomics in lung cancer, providing deep insights into cellular heterogeneity, the tumor microenvironment, cancer stem cells, and therapeutic responses and resistance mechanisms. Single-cell transcriptomics, a high-throughput sequencing technology, enables detailed gene expression analysis at the single-cell level, essential for understanding lung cancer pathogenesis and immune evasion. AI algorithms, such as clustering techniques, identify and classify different cell populations within tumors, unveiling intratumoral heterogeneity (83). ML uncovers critical genes associated with specific cell types, delineating unique cellular characteristics (84).
AI reconstructs cellular development trajectories, shedding light on transitions from normal to malignant cells. In the tumor microenvironment, AI analyzes ligand-receptor interactions and integrates single-cell RNA-seq with spatial transcriptomics to map the spatial distributions of cells (85). It also enhances the identification of immune cell populations, deepening the understanding of immune evasion. In cancer stem cell research, AI identifies specific markers and drug resistance mechanisms, unveiling potential therapeutic targets (86). AI predicts therapeutic responses by analyzing pretreatment and posttreatment transcriptomics data, aiding personalized treatment strategies and showcasing the potential of AI potential in precision medicine for lung cancer (86).
2.2.5. AI in spatiotemporal transcriptomics of lung cancer
In the realm of spatial-temporal transcriptomics, AI has become indispensable for dissecting the intricate landscape of lung cancer. This emerging technology scrutinizes cellular transcriptional activity across different spatial and temporal contexts, offering insights into cellular interactions, migration patterns, and the tumor microenvironment. AI-based algorithms, including ML, DL, and image processing, handle the massive datasets generated, facilitating the integration of data across various time points and spatial locations (87, 88). Through AI-enhanced image analysis, such as cell segmentation and microenvironmental parsing, DL methods like CNNs can automatically identify and classify cell types and structures within complex images. Pattern recognition capabilities of AI further enable the identification of dynamic gene expression patterns and interactions among cellular populations, essential for predicting disease progression and therapeutic responses. Dimensionality reduction and visualization techniques, optimized by AI, such as Principal Component Analysis (PCA), t-Distributed Stochastic Neighbor Embedding (t-SNE), and Uniform Manifold Approximation and Projection (UMAP), provide intuitive interpretations of high-dimensional data, revealing the spatial and temporal distribution of different cell types and gene expressions. Moreover, AI can elucidate gene-environment interactions, pinpointing how spatial and temporal variations impact lung cancer development. This approach also aids in predicting drug responses, paving the way for personalized therapies by associating specific gene expression profiles with drug sensitivity or resistance. The integration of AI in spatial-temporal transcriptomics not only enhances the accuracy and efficiency of data analysis but also empowers researchers to unravel the complex biological processes underlying lung cancer, driving the advancement of novel therapeutic strategies.
2.2.6. AI in transcription factor regulatory networks in lung cancer
AI has shown immense potential in enhancing the understanding and treatment of lung cancer by focusing on transcription factor regulatory networks. These networks, which involve complex interactions between transcription factors and their target genes, are critical in lung cancer development and progression. AI excels in managing and integrating vast datasets, such as gene expression and clinical data, revealing hidden patterns and associations. Techniques like genomics and transcriptomics allow AI to analyze high-throughput sequencing data, identifying crucial transcription factors and target genes. Additionally, AI algorithms, including ML and DL, facilitate the construction and inference of intricate regulatory networks (89).ML algorithms play a pivotal role in selecting key regulatory factors from extensive datasets, highlighting potential therapeutic targets. AI also contributes to the discovery of novel biomarkers for early diagnosis and prognosis of lung cancer. In drug discovery and repurposing, AI-driven virtual screening identifies compounds that disrupt critical regulatory networks, offering new therapeutic avenues. Moreover, AI supports personalized medicine by integrating genomic, regulatory network, and clinical data to formulate individualized treatment plans and predict patient outcomes. By modeling and simulating regulatory networks, AI aids in uncovering underlying biological mechanisms, thereby advancing our comprehension of lung cancer. The promising applications of AI in this domain are expected to drive innovative solutions and propel lung cancer research and treatment forward.
2.3. AI in proteomics of lung cancer
Proteomics, the large-scale study of proteins, generates massive datasets that require sophisticated tools for efficient and accurate analysis. AI excels in this aspect, offering rapid data processing and analysis capabilities. ML algorithms are adept at recognizing complex patterns within these datasets, identifying key protein expression profiles and biomarkers associated with lung cancer. This enables researchers to discern important biological insights that traditional methods might overlook.
In the realm of feature selection and classification, AI plays a pivotal role. It can efficiently identify which protein features are most distinctive between lung cancer patients and healthy individuals, thereby enhancing diagnostic precision (90). AI-based classification models, such as decision trees, random forests, and SVM, can predict the presence of lung cancer in samples with high accuracy (91). Moreover, the application of ML algorithms in biomarker discovery has been transformative. These algorithms analyze proteomics data to identify potential biomarkers for early diagnosis, prognosis prediction, and monitoring therapeutic efficacy (92). DL models further augment this process by extracting high-dimensional features from complex data, revealing insights that might be missed by conventional approaches.
AI also contributes to the construction of protein interaction networks, which are crucial for understanding the molecular mechanisms of lung cancer (93). By performing network analyses, AI can identify key nodes and pathways, thereby highlighting critical points of intervention and potential drug targets. Additionally, AI tools can predict protein-protein interactions that are pivotal in lung cancer progression, providing deeper biological understanding and guiding the development of targeted therapies.
Lastly, the automation and optimization of experimental design through AI significantly enhance research efficiency and cost-effectiveness. AI can design and optimize experiments, reducing the number of required trials and associated costs while increasing accuracy. By simulating and modeling experimental outcomes, researchers can expedite their studies, accelerating the overall research process. Overall, the integration of AI in lung cancer proteomics has exponentially expanded the depth and breadth of biomedical research, facilitating the discovery of effective diagnostic and therapeutic methods with unprecedented speed and precision.
2.4. AI in lung cancer metabolomics
AI has been prominently utilized in the field of metabolomics for lung cancer, significantly advancing both research methods and clinical practices. Lung cancer metabolomics, the study and analysis of metabolic changes within patients, involves handling vast and complex datasets generated by high-throughput technologies such as mass spectrometry and nuclear magnetic resonance spectroscopy. These initial datasets often contain significant noise and complex structures, necessitating robust processing techniques. AI plays a critical role in this phase by automating data cleaning and denoising, ensuring high-quality results by removing outliers and reducing noise. Furthermore, AI employs ML and DL algorithms for feature extraction, discerning vital metabolic features pertinent to lung cancer diagnosis and research (94, 95). It integrates data from diverse sources like blood, tissue, and urine samples, creating a comprehensive database. For instance, using autoencoders in ML, researchers can capture nonlinear variations and distinguish significant metabolic features between cancer patients and healthy controls, enhancing data processing efficiency and accuracy (95, 96).
The contribution of AI extends to the discovery of biomarkers critical for early diagnosis and treatment monitoring (96). Through pattern recognition techniques, such as supervised or unsupervised learning algorithms including neural networks, SVM, and random forests, AI identifies metabolites significantly associated with lung cancer, pinpointing potential new diagnostic biomarkers. Additionally, AI bolsters disease classification and personalized diagnosis by utilizing DL models on metabolomics data to distinguish lung cancer subtypes and even precancerous lesions automatically (97). These models, like convolutional neural networks, enable high-accuracy classification supporting the development of personalized treatment strategies based on individual metabolic data. Moreover, AI aids in prognostic prediction, leveraging metabolomics data to forecast disease progression and patient outcomes (98). By integrating AI with survival analysis models such as Cox regression, researchers can identify key prognostic factors, predict survival time of patients and recurrence risk, aiding in tailored therapeutic decision-making. Furthermore, in drug development and repurposing, AI utilizes metabolomics data to identify target enzymes and proteins, predict drug efficacy, and explore existing new potential uses for drugs in lung cancer treatment, thereby expediting drug development processes.
2.5. AI in lung cancer immuno-oncology
“Lung cancer immunomics” is an emerging term that combines immunology and omics research to delve into the immune mechanisms involved in lung cancer and their relationship with genomic characteristics. Our review primarily focuses on this topic, emphasizing the application of AI in studying tumor immune evasion mechanisms, immune-related gene mutations, tumor mutation burden (TMB), neoantigen discovery, immune microenvironment analysis, and the personalization of immunotherapy.
2.5.1. AI in immune evasion of lung cancer
The application of AI in studying the immune escape mechanisms in lung cancer has yielded significant advancements, bringing a more nuanced understanding of the complex tumor-immune interactions. One crucial area of application is the immunophenotyping analysis, where DL and ML techniques are utilized to analyze Fluorescence-activated Cell Sorting (FACS) data (99). This allows for the precise identification and classification of various immune cell subtypes, elucidating their roles in immune escape (100). Another pivotal use of AI lies in the analysis of T-cell receptor (TCR) and B-cell receptor (BCR) repertoires. ML models can decode the complex clonal dynamics associated with immune escape by analyzing the rearrangement sequences of TCRs and BCRs (101). Additionally, single-cell immunogenomics, supported by AI algorithms, has enabled a granular investigation of the functional states and interactions of heterogeneous immune cells within the Tumor Microenvironment (TME) (102). AI techniques also contribute to a comprehensive analysis of immune escape-related genes and pathways, identifying key drivers and mechanisms of immune evasion. In the realm of therapeutic applications, ML models predict patient responses to immunotherapies, such as Programmed Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) inhibitors, thereby aiding in the personalization of treatment strategies (101). Moreover, through the simulation and modeling of the tumor immune microenvironment, AI provides insights into how cancer cells exploit these alterations to evade immune surveillance, identifying novel therapeutic targets (102). Collectively, AI not only enhances our understanding of lung cancer immune escape mechanisms but also plays a crucial role in the development of more effective immunotherapies (101).
2.5.2. AI in immune-related gene mutations of lung cancer
The ability of AI to process vast amounts of genomic data has proven indispensable in identifying mutations pertinent to the immune response, such as those in the PD-L1, Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4), or Human Leukocyte Antigen (HLA) genes (103). By decoding these genetic variations, AI aids in elucidating the interactions between tumors and the immune system, thereby facilitating the design of personalized treatment plans.
Furthermore, AI significantly enhances the prediction of responses to immunotherapy. By scrutinizing immune-related genetic information, AI models can forecast their potential reactions to immunotherapeutic agents like PD-1/PD-L1 inhibitors (99). This predictive capability enables clinicians to tailor immunotherapy regimens more accurately, optimizing therapeutic efficacy and reducing the likelihood of adverse reactions. The integration of AI into the analysis of immune-related gene mutations thus represents a transformative progression in lung cancer treatment, providing deeper insights and fostering more precise interventions (104).
As our understanding of the complex interplay between the immune system and cancer deepens, AI-driven approaches are poised to become even more central in identifying vital genetic mutations and tailoring immunotherapies. The implications for personalized medicine are profound, underscoring the necessity for continued research and development in this promising intersection of AI and lung cancer immunogenomics.
2.5.3. AI in TMB of lung cancer
AI has emerged as a transformative tool in the estimation and prediction of TMB in lung cancer. Utilizing comprehensive whole-genome sequencing data or specific gene panel data, AI, powered by advanced DL and ML algorithms, can meticulously analyze extensive genomic information. This enables the precise estimation of TMB, which is crucial for understanding tumor profile (105). Moreover, TMB is a significant biomarker for determining the efficacy of immunotherapies, such as PD-1/PD-L1 inhibitors. High TMB levels often correlate with better responses to these immuno-oncology agents. By leveraging AI to integrate TMB data with other clinical parameters, healthcare professionals can develop more personalized treatment plans, thereby enhancing therapeutic outcomes. Additionally, AI models that combine TMB with other biomarkers can provide valuable prognostic insights, aiding in the prediction of lung cancer patients’ outcomes. This prognostic capability is vital for shaping long-term treatment strategies and follow-up plans. The continuous advancements in AI technologies herald a promising future for its application in the precise and personalized management of lung cancer, particularly through the lens of TMB analysis.
2.5.4. AI in the discovery of neoantigens in lung cancer immunotherapy
AI models excel in predicting the likelihood of mutant proteins being processed into peptide fragments recognizable as neoantigens by the immune system, with tools such as DeepNovo or PepFormer aiding in this endeavor (106, 107). Additionally, AI algorithms critically evaluate the stability of these peptides within biological environments, ensuring their viability as neoantigens. The binding affinity of mutated peptides to Major Histocompatibility Complex (MHC) molecules is a crucial determinant of neoantigen effectiveness, with machine learning models like NetMHCpan and MHCflurry being instrumental in this prediction (108, 109). AI also models the complex process of neoantigen generation and MHC presentation, filtering out the most promising candidates. This AI-driven approach not only enhances the accuracy of neoantigen identification but also accelerates the development of personalized cancer immunotherapies.
2.5.5. AI in immune microenvironment of lung cancer
AI algorithms have demonstrated remarkable efficacy in single-cell RNA sequencing analysis, enabling the identification and classification of various immune cell types within the TME, such as T cells, B cells, and natural killer (NK) cells (101). ML techniques further facilitate lineage tracing, revealing the developmental and differentiation pathways of immune cells in the TME (102). Additionally, AI aids in functional gene expression analysis, evaluating immune cell states such as activation, inhibition, and exhaustion, and constructing interaction networks between immune cells and tumor or stromal cells, elucidating their functional roles within the TME. AI technologies also excel at characterizing immunosuppressive microenvironments by identifying factors like Tregs and Myeloid-Derived Suppressor Cells (MDSCs), and their impact on immune responses. Dynamic monitoring of the TME is achieved through AI-assisted liquid biopsy technologies, enabling real-time surveillance of ctDNA and immune cell states, thus assessing immunotherapy efficacy. Furthermore, using ML for spatial analysis of TME can predict the efficacy of immunotherapy in SCLC patients (110). The crucial role of AI in lung cancer immunology research promises even greater advancements in the field, heralding innovative and breakthrough diagnostics and therapies.
2.6. AI in lung cancer microbiomics
AI technologies, notably DL and ML algorithms, have become pivotal in several aspects of microbiome research. Through AI-driven approaches, researchers can accurately identify and classify microbial species, as well as assess their relative abundances within the lung microbiome (111). This capability is invaluable, given the complexity and sheer volume of data generated by high-throughput sequencing techniques.
One key area of AI application is the prediction of microbial functions and metabolic pathways. By analyzing microbial genomes using AI models, researchers can infer potential functions and metabolic activities of various microbes, thereby elucidating their roles in the pathogenesis and progression of lung cancer (111). This functional prediction is crucial for understanding how specific microbes may contribute to cancer development, providing insights that could lead to new therapeutic strategies.
Pattern recognition and feature extraction are another vital area where AI proves indispensable. ML algorithms are adept at extracting significant features from complex microbiome data sets (112). These features can then be utilized to explore the associations between certain microbial species or communities and lung cancer. This process aids in the identification of microbiome markers that could be correlated with disease presence, progression, or even patient outcomes.
In addition, AI enhances the ability to perform comprehensive association analyses by integrating clinical characteristics with microbiome data to identify microbial biomarkers related to lung cancer. This integration facilitates the identification of potential microbial indicators that could be used in diagnostic assays or as therapeutic targets. Furthermore, AI algorithms are employed to build and validate predictive models for lung cancer risk assessment, enhancing early screening and diagnostic accuracy by incorporating both microbiome and clinical features (113). Such risk assessment models are crucial for early intervention and improving patient prognosis.
Prognostic models based on microbiome data powered by AI can also predict patient outcomes, including survival time and response to treatment (113). This predictive capability is essential for personalized medicine, as it allows for tailored therapeutic approaches based on unique microbiome profile. Additionally, AI aids in drug discovery and development, where it can be used to screen for potential anti-cancer or health-promoting microbial metabolites. By identifying these promising compounds, AI can contribute to the development of new therapeutics for lung cancer treatment.
In summary, the applications of AI in lung cancer microbiome research have significantly accelerated the research process, providing powerful tools for precise diagnosis, personalized treatment, and the development of new drugs. The integration of AI with microbiome research holds immense potential to advance our understanding of lung cancer and improve patient outcomes.
2.7. AI in radiomics of lung cancer
Radiomics, as a computational technique, focuses on extracting a vast array of features from medical images, which include parameters such as shape, texture, intensity, and wavelet attributes. Detailed analysis of these features provides comprehensive information about lung tumors, aiding in diagnosis, prognostic prediction, and the assessment of therapeutic responses. AI’s application in lung cancer radiomics is extensive, significantly enhancing the automation and accuracy of image analysis.
One of the primary applications of AI in lung cancer radiomics is automated image segmentation. Utilizing DL algorithms, particularly CNNs, AI can precisely segment tumor regions within lung images, thereby minimizing human error and reducing time consumption (16). Architectures such as U-Net and Mask R-CNN are extensively applied in medical image segmentation, demonstrating high efficacy in delineating tumor boundaries from surrounding tissues (114, 115). In terms of feature extraction and selection, ML algorithms play a pivotal role. These algorithms automatically extract high-dimensional imaging features, such as shape, texture, and wavelet characteristics, and subsequently select features with the most significant diagnostic and prognostic value (116). Techniques like Random Forest, SVM, and Principal Component Analysis (PCA) are commonly employed for feature selection and dimensionality reduction, enabling the creation of robust predictive models.AI-driven diagnostic support systems, such as computer-aided diagnosis (CAD), enhance early detection accuracy by identifying abnormalities in CT scans and highlighting regions of interest for clinicians. Notable systems like IBM Watson Health and Google-owned DeepMind have developed advanced solutions that support early lung cancer diagnosis, underscoring the potential of AI in clinical practice.
Prognostic prediction models, powered by DL and other ML techniques, analyze radiomic data to forecast patient outcomes, including survival rates, disease progression risks, and treatment responses (117). For instance, by combining survival analysis models with imaging features, personalized prognostic predictions can be made, aiding in informed clinical decision-making (118).
AI also significantly contributes to personalized treatment planning and evaluation. By analyzing radiomic features, AI algorithms recommend tailored treatment plans, encompassing the optimal combination of immunotherapy, surgery, and radiotherapy (119–121).
To handle the extensive volume of radiomic data, AI leverages cloud computing and distributed computing technologies. Platforms such as Amazon Web Services (AWS) and Google Cloud provide scalable solutions for efficiently processing and analyzing large-scale medical imaging datasets, facilitating advanced research and clinical applications.
Furthermore, AI-enhanced radiomics is instrumental in predicting lymph node metastasis in early-stage NSCLC and improving the accuracy and efficiency of lung nodule detection and diagnosis during screenings. Studies combining AI with Positron Emission Tomography/Computed Tomography (PET/CT) imaging have demonstrated significant advancements in detecting and evaluating NSCLC, particularly in identifying occult lymph node metastasis and predicting responses to immunotherapy based on radiomic features (122).
2.8. Leveraging AI for pathomics in lung cancer
AI represents a transformative technology in the domain of pathological histology of lung cancer, offering substantial potential to revolutionize diagnostic accuracy and efficiency, optimization of treatment plans, and support for personalized medicine. Pathological histology, referring to the microscopic study of lung cancer tissues, is integral to the precise diagnosis, categorization, staging, and formulation of targeted treatment strategies. By leveraging AI in this field, clinicians and pathologists can significantly enhance the accuracy and speed of diagnostic processes. DL technologies, particularly CNNs, are adept at automating the analysis of pathological tissue section images (123). These AI algorithms can identify and quantify the presence, type, density, and distribution of cancer cells, thus aiding pathologists in making rapid and accurate diagnoses.
Furthermore, AI-driven image classification systems classify pathological images into distinct lung cancer types, thereby facilitating more precise diagnostic outcomes (124, 125). In terms of feature extraction and quantification, AI systems can harvest a plethora of data from pathological images, including cell morphology, arrangement patterns, and staining intensity, which are critical for assessing cancer severity and progression (126). The role of AI extends beyond diagnosis by also predicting patient prognosis based on a combination of pathological images and clinical data, such as survival rates and recurrence risks (127). This predictive capability is essential for formulating personalized treatment plans tailored to individual patient profiles.
Additionally, the ability of AI to predict treatment response by analyzing genomic data alongside pathological images enables the selection of the most effective treatment modalities, such as chemotherapy, targeted therapy, or immunotherapy, suited to unique characteristics (128). AI also excels in data integration and information management, combining data from diverse sources, including genomic data, electronic health records (EHRs), and pathological images, to provide comprehensive patient information that supports critical decision-making processes. By automating repetitive workflows, such as digitizing tissue sections, pre-processing images, and performing preliminary analyses, AI enhances the efficiency of pathology laboratories.
Real-time diagnostic assistance during microscopic examinations further exemplifies the capability of AI, offering diagnostic suggestions that help pathologists detect subtle abnormalities (129). The AI systems’ ability to continuously learn and improve from new data ensures sustained accuracy in diagnostic results. Moreover, AI platforms facilitate cross-institutional collaboration and knowledge sharing by enabling the sharing of data and models, fostering innovation and collective expertise in pathological research.
Several studies illustrate the practical application of AI in pathological histology of lung cancer. For instance, researchers at the University of Washington utilized ML algorithms to predict brain metastasis in early-stage non-small cell lung cancer using 118 lung biopsy samples (130). At the University of Texas Southwestern Medical Center, the combination of AI with traditional pathology expedited the formulation of treatment plans (131). Researchers at New York University Langone Health demonstrated that AI tools capable of autonomous learning could analyze lung tissues to predict cancer recurrence, aiding in the prognostication and severity assessment of lung cancer (132).
An article in “Nature” discusses PathChat, a multimodal generative AI assistant specifically developed for human pathologists (20). By utilizing visual language models, PathChat effectively processes and integrates various data modalities. This innovative approach significantly enhances diagnostic accuracy and efficiency, offering robust support to pathologists in their professional analyses. The future applications of AI in lung cancer pathological histology are vast, promising substantial improvements in diagnostic precision and efficiency while advancing personalized medicine and pathological research.
3. The new era of precision oncology: AI-driven revolution in lung cancer care and future prospects
The field of precision medicine in lung cancer is undergoing a paradigm shift, largely driven by advances in AI. Precision medicine refers to the approach of tailoring medical treatment to the individual characteristics of each patient, such as genetic profile, tumor molecular characteristics, and other biomarkers. By aligning treatment options with the unique biological context of each patient, precision medicine aims to enhance therapeutic efficacy while minimizing unnecessary adverse effects. AI holds considerable promise in the future of lung cancer precision medicine, including early diagnosis, personalized treatment regimens, treatment monitoring, and new drug development.
For early diagnosis, the potential of AI potential in image recognition is monumental. By applying AI and image recognition technologies to vast collections of CT scans, AI can automatically identify early-stage lung nodules and minute pathological changes, thereby facilitating early diagnosis and increasing the chances of successful treatment (133). This capability is being seamlessly integrated into clinical workflows, enhancing the efficiency and accuracy of radiologists’ work. For instance, AI-powered computer-aided detection (CAD) systems are now being employed in many hospitals to assist radiologists in detecting and characterizing lung nodules on CT scans. These systems can rapidly analyze hundreds of images, flagging suspicious areas for further review by radiologists. This not only speeds up the diagnostic process but also improves the detection rate of small nodules that might be overlooked by human eyes alone. Moreover, AI algorithms are being developed to differentiate between benign and malignant nodules, potentially reducing unnecessary biopsies and follow-up scans. For example, a deep learning model developed by researchers at Google Health and Northwestern University demonstrated the ability to detect lung cancer from CT scans with a performance on par with or better than radiologists (134). Such tools are gradually being incorporated into clinical practice, serving as a “second reader” to support radiologists’ decision-making processes. AI’s role in early detection extends beyond imaging. Complementarily, AI’s ability to analyze ctDNA from liquid biopsies could detect genetic mutations associated with lung cancer at an early stage (135). This non-invasive approach is particularly promising for screening high-risk populations and monitoring disease recurrence. AI algorithms can analyze complex patterns in ctDNA data, potentially identifying cancer-specific signatures before traditional diagnostic methods can detect the disease (136). In pathology, AI’s automated analysis of histopathological slides can swiftly and accurately identify cancerous regions, thereby improving diagnostic precision and efficiency. These AI systems are being integrated into digital pathology workflows, assisting pathologists in quantifying biomarkers, grading tumors, and identifying specific histological patterns associated with different lung cancer subtypes (126).Importantly, these AI applications are not limited to NSCLC but are also being developed and refined for SCLC detection. Given the aggressive nature of SCLC and the critical importance of early detection, AI tools are being tailored to identify the unique radiological and pathological features of SCLC, potentially leading to earlier diagnosis and improved outcomes for this challenging subtype of lung cancer (137).
In the realm of personalized treatment plans, AI promises to revolutionize gene analysis and targeted therapy. AI systems, leveraging DL and extensive datasets, will be able to process and analyze large volumes of genetic data to pinpoint mutations and biomarkers associated with lung cancer (138, 139). By predicting the response of specific genetic profiles to targeted therapies, AI can help devise highly customized treatment protocols (140). Additionally, AI can integrate multidisciplinary data, encompassing genetic information, imaging data, clinical records, and lifestyle habits, to holistically evaluate patient health (133). This multimodal approach will lead to more precise and effective treatment plans, improving therapeutic outcomes and reducing side effects. Furthermore, adaptive AI systems will dynamically adjust treatment strategies based on real-time data, such as changes in biomarker concentrations or imaging results, thus refining the precision and personalization of lung cancer therapies.
For cases of advanced-stage cancer, AI is increasingly aiding oncologists in creating personalized treatment plans, leveraging predictive analytics to optimize chemotherapy, targeted therapies, and immunotherapies. For instance, AI algorithms are being developed to predict the efficacy of tyrosine kinase inhibitors (TKIs) in patients with specific EGFR mutations, allowing for more precise treatment selection (141). Similarly, AI models are being used to predict response to ALK inhibitors in patients with ALK-positive NSCLC, potentially guiding treatment decisions and improving outcomes (142).Additionally, AI can integrate multidisciplinary data, encompassing genetic information, imaging data, clinical records, and lifestyle habits, to holistically evaluate patient health (143). This multimodal approach leads to more precise and effective treatment plans, improving therapeutic outcomes and reducing side effects. For example, AI models that incorporate radiomics features from CT scans, along with clinical and genetic data, have shown promise in predicting response to immunotherapy in NSCLC patients (144).Furthermore, adaptive AI systems can dynamically adjust treatment strategies based on real-time data, such as changes in biomarker concentrations or imaging results, thus refining the precision and personalization of lung cancer therapies. This is particularly valuable in managing treatment-related toxicities and adapting dosages to maximize efficacy while minimizing side effects. In the rapidly evolving sphere of oncology, particularly within the realm of immunotherapy for lung cancer patients, the integration of AI has introduced significant advancements in therapeutic precision and personalization. This burgeoning field is underscored by recent research that showcases the development of cutting-edge AI and machine learning models, which are meticulously designed to predict patient responses to immunotherapy (145, 146). These models extend their utility by forecasting progression-free survival and overall survival rates, specifically tailored for individuals battling NSCLC (147). Such AI-driven tools leverage standard clinical data to evaluate the efficacy of immune checkpoint inhibitors, thus equipping clinicians with vital insights that inform the selection of optimal treatment regimens. Consequently, these technological innovations are revolutionizing the landscape of personalized medicine, granting healthcare providers the capability to discern which patients are predisposed to benefit most from specific immunotherapy treatments.
In the context of treatment monitoring, AI’s ability to conduct real-time data surveillance is nothing short of transformative. By enabling the continuous monitoring of vital physiological metrics, genetic profiles, and treatment responses through sophisticated smart devices and sensors, AI facilitates timely modifications to treatment plans that align with evolving data patterns, thereby maximizing therapeutic efficacy. For instance, AI algorithms can analyze serial CT scans to assess tumor response to treatment, potentially detecting subtle changes that might indicate the need for treatment modification before clinical symptoms appear (148). Similarly, AI models can monitor changes in circulating tumor DNA levels during treatment, providing early indications of treatment response or resistance (64).Furthermore, predictive models empowered by AI serve a pivotal role in not only forecasting treatment outcomes and patient prognoses but also in crafting individualized predictions regarding disease trajectories (110, 149, 150). These insights are invaluable in helping clinicians devise comprehensive, long-term management strategies. For example, AI models that integrate clinical, pathological, and genomic data have shown promise in predicting long-term survival in NSCLC patients, potentially guiding decisions about treatment intensity and follow-up protocols. The importance of AI is further emphasized by its capacity to predict and signal potential complications through real-time analysis of physiological data and clinical records, which is crucial in preempting adverse medical events and guiding proactive interventions, ultimately mitigating the risks associated with immunotherapy treatments.
In the realm of drug development for lung cancer, significant strides have been made through the integration of AI. AI platforms have revolutionized the pharmaceutical industry by enhancing the speed and reducing the costs of drug discovery processes (151). AI are being used to streamline various stages, from identifying druggable targets to optimizing lead compounds (152–154). AI’s ability to rapidly analyze biological datasets is exemplified by projects like those at Lawrence Livermore National Laboratory and BridgeBio, which have advanced to clinical trials for medications targeting genetic mutations in cancer. The application of AI facilitates virtual drug screening, optimizes therapeutic molecule development, and enhances clinical trial design, thereby shortening trial durations and minimizing costs (155). Additionally, AI is pivotal in personalized medicine, creating tailored treatments based on genetic profiles, thus maximizing efficacy and minimizing side effects compared to conventional approaches.
The future applications of AI in lung cancer precision medicine are vast and multifaceted. AI is playing an increasingly pivotal role in early detection, personalized treatment planning, treatment monitoring, and new drug development. By harnessing the power of genetic, clinical, and imaging data, AI can deliver highly accurate and effective medical solutions, significantly improving patient survival rates and quality of life. As technology advances, the integration of AI into lung cancer precision medicine will continue to expand and deepen, heralding a new era of personalized healthcare. The seamless incorporation of AI tools into clinical workflows is enhancing the ability to detect both NSCLC and SCLC at earlier stages, while also providing oncologists with powerful decision support systems for crafting personalized treatment plans for advanced-stage patients. However, it’s important to note that while AI holds great promise, its implementation in clinical practice should be done cautiously and ethically. Rigorous validation studies, regulatory approvals, and ongoing monitoring of AI systems in real-world settings are crucial to ensure their safety and efficacy. Additionally, efforts should be made to address potential biases in AI algorithms and to ensure equitable access to AI-enhanced healthcare across diverse populations. As we move forward, the synergy between human expertise and AI capabilities will likely define the future of lung cancer care, offering hope for improved outcomes and quality of life for patients worldwide.
4. Technical challenges and solutions
AI holds considerable promise for advancing lung cancer diagnosis, treatment, and prognosis. Nonetheless, realizing this potential requires addressing several key challenges related to data quality and quantity, model interpretability, statistical validation, and ethical and privacy considerations.
Initially, addressing data quality and quantity is crucial. Medical datasets often contain noise, missing values, and errors due to human input, all of which can adversely affect AI model performance. Moreover, the development of large-scale, high-quality annotated datasets is fraught with difficulties, limiting the robustness and generalizability of AI models. Data heterogeneity further complicates this issue, as variations across healthcare institutions or imaging devices often lead to a lack of standardization. Additionally, integrating diverse data types, such as imaging, genomic, and clinical records, calls for sophisticated multimodal analysis.
To confront these challenges, several strategies are recommended. Implementing rigorous data cleaning and standardization protocols can significantly mitigate noise and rectify errors, thereby improving data quality. Enhancing data volume through multi-center collaborations and secure data-sharing platforms is essential, all while maintaining strict data privacy and security. Furthermore, employing data augmentation techniques, including image rotation, translation, and scaling, can bolster training sample sizes. The creation of unified data formats and processing standards is crucial for the standardization and integration of multimodal data, thereby maximizing the utility of available information.
Equally important is the challenge of model interpretability, particularly with DL models like CNNs. While CNNs excel in image diagnostics, their “black box” nature makes understanding their decision-making processes difficult, thus impeding trust in their predictions. To enhance interpretability, several approaches are suggested. Employing Explainable AI (XAI) techniques, such as Gradient-weighted Class Activation Mapping (Grad-CAM), helps produce heat maps that visualize decision-making focal areas. Additionally, incorporating attention mechanisms into CNNs analyzing CT scans can highlight suspicious areas, providing visual cues of the decision process to clinicians. The use of Local Interpretable Model-agnostic Explanations (LIME) can further clarify individual predictions, enhancing clinicians’ comprehension of data feature contributions. A notable addition to these interpretability methods is the SHapley Additive exPlanations (SHAP) approach. SHAP, based on game theory concepts, offers a unified framework for interpreting predictions. It provides both global and local explanations of model behavior, which is particularly valuable in medical imaging where understanding the importance of different image regions in model decisions is crucial. SHAP’s model-agnostic nature and strong theoretical foundation make it especially suitable for complex deep learning models used in lung cancer detection, potentially increasing clinicians’ trust in AI-assisted diagnoses.
In parallel, rigorous statistical validation is needed to ensure the reliability and generalizability of AI models in lung cancer research. Applying stratified k-fold cross-validation ensures that cancer stages or subtypes are consistently represented across folds, yielding robust model performance estimates. External validation on independent datasets is critical for assessing model performance on unobserved data from various institutions, unveiling potential biases or limitations. Comprehensive performance metrics should also be evaluated, including Area Under the Curve of Receiver Operating Characteristic (AUC-ROC), precision-recall curves, and F1 scores, alongside traditional metrics. Calibration plots and decision curve analysis can effectively gauge the clinical utility of these models across different threshold probabilities.
Moreover, ethical and privacy considerations are foundational to the application of AI in healthcare. The use of patient data raises complex ethical issues, necessitating robust safeguards for privacy protection and adherence to ethical guidelines. Transitioning from research to clinical practice further underscores the need for models capable of operating reliably across diverse clinical environments and conditions.
In summary, systematically addressing these challenges through strategic measures can markedly enhance the applicability and effectiveness of AI in lung cancer research and clinical settings. Enhancing model interpretability and ensuring thorough statistical validation are key to building clinician trust and encouraging clinical integration. Developing comprehensive interpretability frameworks and validation protocols will enable healthcare professionals to confidently rely on AI diagnostic results, thereby improving diagnostic accuracy and treatment efficacy for patients.
5. Conclusion
The integration of AI into lung cancer research and treatment has already demonstrated substantial advancements, enhancing diagnostic accuracy, treatment planning, and patient outcome prediction. AI’s application across various -omics fields, radiomics, and pathology has provided unprecedented insights into the molecular mechanisms of lung cancer, facilitating early detection and personalized treatment strategies. Despite challenges such as data quality, interpretability, and ethical considerations, ongoing technological advancements and strategic solutions promise to overcome these hurdles. The future of AI in lung cancer is poised for transformative impact, with potential to revolutionize precision medicine, accelerate drug discovery, and improve patient care. As AI continues to evolve, its role in lung cancer management will deepen, offering innovative solutions and contributing to a more comprehensive understanding of the disease, ultimately leading to better patient outcomes and enhanced quality of life.
Glossary
- AI
Artificial Intelligence
- NSCLC
Non-small cell lung cancer
- SCLC
Small cell lung cancer
- EGFR
Epidermal Growth Factor Receptor
- ALK
Anaplastic Lymphoma Kinase
- ML
Machine learning
- DL
Deep Learning
- NLP
Natural Language Processing
- DNNs
Deep neural networks
- CT
Computed Tomography
- CNNs
Convolutional Neural Networks
- RNNs
Recurrent Neural Networks
- KRAS
Kirsten Rat Sarcoma virus
- TCGA
The Cancer Genome Atlas
- GDC
Genomic Data Commons
- CNV
Copy number variation
- GEO
Gene Expression Omnibus
- ncRNA
non-coding RNA
- miRNAs
microRNAs
- lncRNAs
long non-coding RNAs
- ctDNA
circulating tumor DNA
- dPCR
Digital Polymerase Chain Reaction
- NGS
Next-generation sequencing
- RNA-Seq
RNA Sequencing
- mRNA
messenger RNA
- SVMs
Support vector machines
- circRNAs
circular RNAs
- m6A
N6-methyladenosine
- PCA
Principal Component Analysis
- t-SNE
t-Distributed Stochastic Neighbor Embedding
- UMAP
Uniform Manifold Approximation and Projection
- TMB
Tumor mutation burden
- FACS
Fluorescence-activated Cell Sorting
- TCR
T-cell receptor
- BCR
B-cell receptor
- TME
Tumor Microenvironment
- PD-1
Programmed Death-1
- PD-L1
Programmed Death-Ligand 1
- CTLA-4
Cytotoxic T-Lymphocyte Antigen 4
- HLA
Human Leukocyte Antigen
- MHC
Major Histocompatibility Complex
- NK
Natural killer
- MDSCs
Myeloid-Derived Suppressor Cells
- PCA
Principal Component Analysis
- CAD
computer-aided diagnosis
- TKIs
Tyrosine kinase inhibitors
- AWS
Amazon Web Services
- PET/CT
Positron Emission Tomography/Computed Tomography
- EHRs
Electronic health records
- XAI
Explainable Artificial Intelligence
- HIPAA
Health Insurance Portability and Accountability Act
- GDPR
General Data Protection Regulation
- Grad-CAM
Gradient-weighted Class Activation Mapping
- LIME
Local Interpretable Model-agnostic Explanations
- SHAP
SHapley Additive exPlanations
- AUC-ROC
Area Under the Curve of Receiver Operating Characteristic
Funding Statement
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Author contributions
DH: Writing – original draft, Writing – review & editing. ZL: Writing – original draft, Writing – review & editing. TJ: Writing – original draft, Writing – review & editing. CY: Writing – original draft, Writing – review & editing. NL: Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Sharma R. Mapping of global, regional and national incidence, mortality and mortality-to-incidence ratio of lung cancer in 2020 and 2050. Int J Clin Oncol. (2022) 27:665–75. doi: 10.1007/s10147-021-02108-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Henschke CI, Yip R, Shaham D, Markowitz S, Cervera Deval J, Zulueta JJ, et al. A 20-year follow-up of the international early lung cancer action program (I-ELCAP). Radiology. (2023) 309:e231988. doi: 10.1148/radiol.231988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang CY, Lin YT, Lin LJ, Chang YH, Chen HY, Wang YP, et al. Stage shift improves lung cancer survival: real-world evidence. J Thorac Oncol. (2023) 18:47–56. doi: 10.1016/j.jtho.2022.09.005 [DOI] [PubMed] [Google Scholar]
- 4. van der Drift MA, Karim-Kos HE, Siesling S, Groen HJ, Wouters MW, Coebergh JW, et al. Progress in standard of care therapy and modest survival benefits in the treatment of non-small cell lung cancer patients in the Netherlands in the last 20 years. J Thorac Oncol. (2012) 7:291–8. doi: 10.1097/JTO.0b013e31823a01fb [DOI] [PubMed] [Google Scholar]
- 5. Kubo N, Suefuji H, Nakajima M, Tokumaru S, Okano N, Yoshida D, et al. Five-year survival outcomes after carbon-ion radiotherapy for operable stage I NSCLC: A Japanese national registry study (J-CROS-LUNG). J Thorac Oncol. (2024) 19:491–9. doi: 10.1016/j.jtho.2023.10.016 [DOI] [PubMed] [Google Scholar]
- 6. Tran HT, Heeke S, Sujit S, Vokes N, Zhang J, Aminu M, et al. Circulating tumor DNA and radiological tumor volume identify patients at risk for relapse with resected, early-stage non-small-cell lung cancer. Ann Oncol. (2024) 35:183–9. doi: 10.1016/j.annonc.2023.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacobs C, van Ginneken B. Google’s lung cancer AI: a promising tool that needs further validation. Nat Rev Clin Oncol. (2019) 16:532–3. doi: 10.1038/s41571-019-0248-7 [DOI] [PubMed] [Google Scholar]
- 8. Shin H, Choi BH, Shim O, Kim J, Park Y, Cho SK, et al. Single test-based diagnosis of multiple cancer types using Exosome-SERS-AI for early stage cancers. Nat Commun. (2023) 14:1644. doi: 10.1038/s41467-023-37403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang R, Dai W, Gong J, Huang M, Hu T, Li H, et al. Development of a novel combined nomogram model integrating deep learning-pathomics, radiomics and immunoscore to predict postoperative outcome of colorectal cancer lung metastasis patients. J Hematol Oncol. (2022) 15:11. doi: 10.1186/s13045-022-01225-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirienko M, Sollini M, Corbetta M, Voulaz E, Gozzi N, Interlenghi M, et al. Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur J Nucl Med Mol Imaging. (2021) 48:3643–55. doi: 10.1007/s00259-021-05371-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rong R, Sheng H, Jin KW, Wu F, Luo D, Wen Z, et al. A deep learning approach for histology-based nucleus segmentation and tumor microenvironment characterization. Modern Pathol. (2023) 36:100196. doi: 10.1016/j.modpat.2023.100196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang H, Fu T, Du Y, Gao W, Huang K, Liu Z, et al. Scientific discovery in the age of artificial intelligence. Nature. (2023) 620:47–60. doi: 10.1038/s41586-023-06221-2 [DOI] [PubMed] [Google Scholar]
- 13. Marcus HJ, Ramirez PT, Khan DZ, Layard Horsfall H, Hanrahan JG, Williams SC, et al. The IDEAL framework for surgical robotics: development, comparative evaluation and long-term monitoring. Nat Med. (2024) 30:61–75. doi: 10.1038/s41591-023-02732-7 [DOI] [PubMed] [Google Scholar]
- 14. Chua IS, Gaziel-Yablowitz M, Korach ZT, Kehl KL, Levitan NA, Arriaga YE, et al. Artificial intelligence in oncology: Path to implementation. Cancer Med. (2021) 10:4138–49. doi: 10.1002/cam4.3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. (2022) 23:40–55. doi: 10.1038/s41580-021-00407-0 [DOI] [PubMed] [Google Scholar]
- 16. Zhou J, Hu B, Feng W, Zhang Z, Fu X, Shao H, et al. An ensemble deep learning model for risk stratification of invasive lung adenocarcinoma using thin-slice CT. NPJ Digital Med. (2023) 6:119. doi: 10.1038/s41746-023-00866-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo X, Deng Z, Yang B, Luo MY. Pre-trained language models in medicine: A survey. Artif Intell Med. (2024) 154:102904. doi: 10.1016/j.artmed.2024.102904 [DOI] [PubMed] [Google Scholar]
- 18. Chen Y, Nazhamaiti M, Xu H, Meng Y, Zhou T, Li G, et al. All-analog photoelectronic chip for high-speed vision tasks. Nature. (2023) 623:48–57. doi: 10.1038/s41586-023-06558-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alvarado CE, Worrell SG, Sarode AL, Jiang B, Halloran SJ, Argote-Greene LM, et al. Comparing thoracoscopic and robotic lobectomy using a nationally representative database. Am Surgeon. (2023) 89:5340–8. doi: 10.1177/00031348221148347 [DOI] [PubMed] [Google Scholar]
- 20. Lu MY, Chen B, Williamson DFK, Chen RJ, Zhao M, Chow AK, et al. A multimodal generative AI copilot for human pathology. Nature. (2024) 634:466–73. doi: 10.1038/s41586-024-07618-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hou R, Li X, Xiong J, Shen T, Yu W, Schwartz LH, et al. Predicting tyrosine kinase inhibitor treatment response in stage IV lung adenocarcinoma patients with EGFR mutation using model-based deep transfer learning. Front Oncol. (2021) 11:679764. doi: 10.3389/fonc.2021.679764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moreno S, Bonfante M, Zurek E, Cherezov D, Goldgof D, Hall L, et al. A radiogenomics ensemble to predict EGFR and KRAS mutations in NSCLC. Tomography (Ann Arbor Mich.). (2021) 7:154–68. doi: 10.3390/tomography7020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song Z, Liu T, Shi L, Yu Z, Shen Q, Xu M, et al. The deep learning model combining CT image and clinicopathological information for predicting ALK fusion status and response to ALK-TKI therapy in non-small cell lung cancer patients. Eur J Nucl Med Mol Imaging. (2021) 48:361–71. doi: 10.1007/s00259-020-04986-6 [DOI] [PubMed] [Google Scholar]
- 24. Wang S, Yu H, Gan Y, Wu Z, Li E, Li X, et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. (2022) 4:e309–19. doi: 10.1016/s2589-7500(22)00024-3 [DOI] [PubMed] [Google Scholar]
- 25. Kehl KL, Xu W, Gusev A, Bakouny Z, Choueiri TK, Riaz IB, et al. Artificial intelligence-aided clinical annotation of a large multi-cancer genomic dataset. Nat Commun. (2021) 12:7304. doi: 10.1038/s41467-021-27358-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singhal A, Simmons M, Lu Z. Text mining genotype-phenotype relationships from biomedical literature for database curation and precision medicine. PloS Comput Biol. (2016) 12:e1005017. doi: 10.1371/journal.pcbi.1005017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang N, Zhang H, Liu Z, Dai Z, Wu W, Zhou R, et al. An artificial intelligence network-guided signature for predicting outcome and immunotherapy response in lung adenocarcinoma patients based on 26 machine learning algorithms. Cell Proliferation. (2023) 56:e13409. doi: 10.1111/cpr.13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wood DE, White JR, Georgiadis A, Van Emburgh B, Parpart-Li S, Mitchell J, et al. A machine learning approach for somatic mutation discovery. Sci Trans Med. (2018) 10:eaar7939. doi: 10.1126/scitranslmed.aar7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cui S, Ten Haken RK, El Naqa I. Integrating multiomics information in deep learning architectures for joint actuarial outcome prediction in non-small cell lung cancer patients after radiation therapy. Int J Radiat Oncol Biol Phys. (2021) 110:893–904. doi: 10.1016/j.ijrobp.2021.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sui Q, Chen Z, Hu Z, Huang Y, Liang J, Bi G, et al. Cisplatin resistance-related multi-omics differences and the establishment of machine learning models. J Transl Med. (2022) 20:171. doi: 10.1186/s12967-022-03372-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han T, Bai Y, Liu Y, Dong Y, Liang C, Gao L, et al. Integrated multi-omics analysis and machine learning to refine molecular subtypes, prognosis, and immunotherapy in lung adenocarcinoma. Funct Integr Genomics. (2024) 24:118. doi: 10.1007/s10142-024-01388-x [DOI] [PubMed] [Google Scholar]
- 32. Gogleva A, Polychronopoulos D, Pfeifer M, Poroshin V, Ughetto M, Martin MJ, et al. Knowledge graph-based recommendation framework identifies drivers of resistance in EGFR mutant non-small cell lung cancer. Nat Commun. (2022) 13:1667. doi: 10.1038/s41467-022-29292-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saraon P, Snider J, Kalaidzidis Y, Wybenga-Groot LE, Weiss K, Rai A, et al. A drug discovery platform to identify compounds that inhibit EGFR triple mutants. Nat Chem Biol. (2020) 16:577–86. doi: 10.1038/s41589-020-0484-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rakaee M, Andersen S, Giannikou K, Paulsen EE, Kilvaer TK, Busund LR, et al. Machine learning-based immune phenotypes correlate with STK11/KEAP1 co-mutations and prognosis in resectable NSCLC: a sub-study of the TNM-I trial. Ann Oncol. (2023) 34:578–88. doi: 10.1016/j.annonc.2023.04.005 [DOI] [PubMed] [Google Scholar]
- 35. Coudray N, Ocampo PS, Sakellaropoulos T, Narula N, Snuderl M, Fenyö D, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. (2018) 24:1559–67. doi: 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer. (2020) 1:789–99. doi: 10.1038/s43018-020-0087-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang K, Mo Z, Zhu W, Liao B, Yang Y, Wu FX. Prediction of target-drug therapy by identifying gene mutations in lung cancer with histopathological stained image and deep learning techniques. Front Oncol. (2021) 11:642945. doi: 10.3389/fonc.2021.642945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang S, Wang R, Hu D, Zhang C, Cao P, Huang J. Machine learning reveals diverse cell death patterns in lung adenocarcinoma prognosis and therapy. NPJ Precis Oncol. (2024) 8:49. doi: 10.1038/s41698-024-00538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. (2022) 185:563–75.e511. doi: 10.1016/j.cell.2022.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Rijthoven M, Balkenhol M, Siliņa K, van der Laak J, Ciompi F. HookNet: Multi-resolution convolutional neural networks for semantic segmentation in histopathology whole-slide images. Med Image Anal. (2021) 68:101890. doi: 10.1016/j.media.2020.101890 [DOI] [PubMed] [Google Scholar]
- 41. Jaksik R, Szumała K, Dinh KN, Śmieja J. Multiomics-based feature extraction and selection for the prediction of lung cancer survival. Int J Mol Sci. (2024) 25:3661. doi: 10.3390/ijms25073661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang Y, Xu L, Sun L, Zhang P, Farid SS. Machine learning application in personalised lung cancer recurrence and survivability prediction. Comput Struct Biotechnol J. (2022) 20:1811–20. doi: 10.1016/j.csbj.2022.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dwivedi K, Rajpal A, Rajpal S, Kumar V, Agarwal M, Kumar N. Enlightening the path to NSCLC biomarkers: Utilizing the power of XAI-guided deep learning. Comput Methods Programs Biomed. (2024) 243:107864. doi: 10.1016/j.cmpb.2023.107864 [DOI] [PubMed] [Google Scholar]
- 44. Ren C, Li J, Zhou Y, Zhang S, Wang Q. Typical tumor immune microenvironment status determine prognosis in lung adenocarcinoma. Trans Oncol. (2022) 18:101367. doi: 10.1016/j.tranon.2022.101367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Poplin R, Chang PC, Alexander D, Schwartz S, Colthurst T, Ku A, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. (2018) 36:983–7. doi: 10.1038/nbt.4235 [DOI] [PubMed] [Google Scholar]
- 46. Karlow JA, Pehrsson EC, Xing X, Watson M, Devarakonda S, Govindan R, et al. Non-small cell lung cancer epigenomes exhibit altered DNA methylation in smokers and never-smokers. Genom Proteomics Bioinf. (2023) 21:991–1013. doi: 10.1016/j.gpb.2023.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou J, Cheng T, Li X, Hu J, Li E, Ding M, et al. Epigenetic imprinting alterations as effective diagnostic biomarkers for early-stage lung cancer and small pulmonary nodules. Clin Epigenet. (2021) 13:220. doi: 10.1186/s13148-021-01203-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, et al. Systematic in vivo inactivation of chromatin-regulating enzymes identifies setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. (2017) 77:1719–29. doi: 10.1158/0008-5472.Can-16-2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z, Yang Y, Zhang X. MiR-770 inhibits tumorigenesis and EMT by targeting JMJD6 and regulating WNT/β-catenin pathway in non-small cell lung cancer. Life Sci. (2017) 188:163–71. doi: 10.1016/j.lfs.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 50. LaFave LM, Kartha VK, Ma S, Meli K, Del Priore I, Lareau C, et al. Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell. (2020) 38:212–28.e213. doi: 10.1016/j.ccell.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Qi J, Hong B, Tao R, Sun R, Zhang H, Zhang X, et al. Prediction model for Malignant pulmonary nodules based on cfMeDIP-seq and machine learning. Cancer Sci. (2021) 112:3918–23. doi: 10.1111/cas.15052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jamshidi A, Liu MC, Klein EA, Venn O, Hubbell E, Beausang JF, et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell. (2022) 40:1537–49.e1512. doi: 10.1016/j.ccell.2022.10.022 [DOI] [PubMed] [Google Scholar]
- 53. Heeke S, Gay CM, Estecio MR, Tran H, Morris BB, Zhang B, et al. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell. (2024) 42:225–37.e225. doi: 10.1016/j.ccell.2024.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu X, Yang Q, Wang D, Li Z, Chen N, Kong DX. Predicting lung adenocarcinoma disease progression using methylation-correlated blocks and ensemble machine learning classifiers. PeerJ. (2021) 9:e10884. doi: 10.7717/peerj.10884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaneko S, Mitsuyama T, Shiraishi K, Ikawa N, Shozu K, Dozen A, et al. Genome-wide chromatin analysis of FFPE tissues using a dual-arm robot with clinical potential. Cancers. (2021) 13:2126. doi: 10.3390/cancers13092126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu P, Zhang C, Tang X, Li D, Zhang G, Zi X, et al. Pan-cancer characterization of cell-free immune-related miRNA identified as a robust biomarker for cancer diagnosis. Mol Cancer. (2024) 23:31. doi: 10.1186/s12943-023-01915-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang XW, Qi GX, Liu MX, Yang YF, Wang JH, Yu YL, et al. Deep learning promotes profiling of multiple miRNAs in single extracellular vesicles for cancer diagnosis. ACS Sensors. (2024) 9:1555–64. doi: 10.1021/acssensors.3c02789 [DOI] [PubMed] [Google Scholar]
- 58. Li HY, You ZH, Wang L, Yan X, Li ZW. DF-MDA: An effective diffusion-based computational model for predicting miRNA-disease association. Mol Ther. (2021) 29:1501–11. doi: 10.1016/j.ymthe.2021.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang S, Meng F, Li M, Bao H, Chen X, Zhu M, et al. Multidimensional cell-free DNA fragmentomic assay for detection of early-stage lung cancer. Am J Respir Crit Care Med. (2023) 207:1203–13. doi: 10.1164/rccm.202109-2019OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Giroux Leprieur E, Hélias-Rodzewicz Z, Takam Kamga P, Costantini A, Julie C, Corjon A, et al. Sequential ctDNA whole-exome sequencing in advanced lung adenocarcinoma with initial durable tumor response on immune checkpoint inhibitor and late progression. J Immunother Cancer. (2020) 8:e000527. doi: 10.1136/jitc-2020-000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Abbosh C, Frankell AM, Harrison T, Kisistok J, Garnett A, Johnson L, et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature. (2023) 616:553–62. doi: 10.1038/s41586-023-05776-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Assaf ZJF, Zou W, Fine AD, Socinski MA, Young A, Lipson D, et al. A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat Med. (2023) 29:859–68. doi: 10.1038/s41591-023-02226-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Helzer KT, Sharifi MN, Sperger JM, Shi Y, Annala M, Bootsma ML, et al. Fragmentomic analysis of circulating tumor DNA-targeted cancer panels. Ann Oncol. (2023) 34:813–25. doi: 10.1016/j.annonc.2023.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Widman AJ, Shah M, Frydendahl A, Halmos D, Khamnei CC, Øgaard N, et al. Ultrasensitive plasma-based monitoring of tumor burden using machine-learning-guided signal enrichment. Nat Med. (2024) 30:1655–66. doi: 10.1038/s41591-024-03040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chabon JJ, Hamilton EG, Kurtz DM, Esfahani MS, Moding EJ, Stehr H, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. (2020) 580:245–51. doi: 10.1038/s41586-020-2140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sujit SJ, Aminu M, Karpinets TV, Chen P, Saad MB, Salehjahromi M, et al. Enhancing NSCLC recurrence prediction with PET/CT habitat imaging, ctDNA, and integrative radiogenomics-blood insights. Nat Commun. (2024) 15:3152. doi: 10.1038/s41467-024-47512-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liang N, Li B, Jia Z, Wang C, Wu P, Zheng T, et al. Ultrasensitive detection of circulating tumour DNA via deep methylation sequencing aided by machine learning. Nat Biomed Eng. (2021) 5:586–99. doi: 10.1038/s41551-021-00746-5 [DOI] [PubMed] [Google Scholar]
- 68. Hellmann MD, Nabet BY, Rizvi H, Chaudhuri AA, Wells DK, Dunphy MPS, et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L)1 blockade in NSCLC. Clin Cancer Res. (2020) 26:2849–58. doi: 10.1158/1078-0432.Ccr-19-3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jee J, Lebow ES, Yeh R, Das JP, Namakydoust A, Paik PK, et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med. (2022) 28:2353–63. doi: 10.1038/s41591-022-02047-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Borisov N, Sorokin M, Tkachev V, Garazha A, Buzdin A. Cancer gene expression profiles associated with clinical outcomes to chemotherapy treatments. BMC Med Genomics. (2020) 13:111. doi: 10.1186/s12920-020-00759-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yuan F, Lu L, Zou Q. Analysis of gene expression profiles of lung cancer subtypes with machine learning algorithms. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165822. doi: 10.1016/j.bbadis.2020.165822 [DOI] [PubMed] [Google Scholar]
- 72. Liu W, Dong R, Gao S, Shan X, Li M, Yu Z, et al. Assessing the prognostic capability of immune-related gene scoring systems in lung adenocarcinoma. J Oncol. (2022) 2022:2151396. doi: 10.1155/2022/2151396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu LP, Lu L, Zhao QQ, Kou QJ, Jiang ZZ, Gui R, et al. Identification and validation of the pyroptosis-related molecular subtypes of lung adenocarcinoma by bioinformatics and machine learning. Front Cell Dev Biol. (2021) 9:756340. doi: 10.3389/fcell.2021.756340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Abdelwahab O, Awad N, Elserafy M, Badr E. A feature selection-based framework to identify biomarkers for cancer diagnosis: A focus on lung adenocarcinoma. PloS One. (2022) 17:e0269126. doi: 10.1371/journal.pone.0269126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lai YH, Chen WN, Hsu TC, Lin C, Tsao Y, Wu S. Overall survival prediction of non-small cell lung cancer by integrating microarray and clinical data with deep learning. Sci Rep. (2020) 10:4679. doi: 10.1038/s41598-020-61588-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yang ZY, Liu XY, Shu J, Zhang H, Ren YQ, Xu ZB, et al. Multi-view based integrative analysis of gene expression data for identifying biomarkers. Sci Rep. (2019) 9:13504. doi: 10.1038/s41598-019-49967-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Park S, Lim JM, Chun JN, Lee S, Kim TM, Kim DW, et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in non−small cell lung cancer. Int J Oncol. (2020) 56:559–67. doi: 10.3892/ijo.2019.4953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lou Z, Cheng Z, Li H, Teng Z, Liu Y, Tian Z. Predicting miRNA-disease associations via learning multimodal networks and fusing mixed neighborhood information. Briefings Bioinf. (2022) 23:bbac159. doi: 10.1093/bib/bbac159 [DOI] [PubMed] [Google Scholar]
- 79. Su Z, Lu H, Wu Y, Li Z, Duan L. Predicting potential lncRNA biomarkers for lung cancer and neuroblastoma based on an ensemble of a deep neural network and LightGBM. Front Genet. (2023) 14:1238095. doi: 10.3389/fgene.2023.1238095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yue W, Wang J, Lin B, Fu Y. Identifying lncRNAs and mRNAs related to survival of NSCLC based on bioinformatic analysis and machine learning. Aging. (2024) 16:7799–817. doi: 10.18632/aging.205783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Pedraz-Valdunciel C, Giannoukakos S, Giménez-Capitán A, Fortunato D, Filipska M, Bertran-Alamillo J, et al. Multiplex analysis of circRNAs from plasma extracellular vesicle-enriched samples for the detection of early-stage non-small cell lung cancer. Pharmaceutics. (2022) 14:2034. doi: 10.3390/pharmaceutics14102034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lei M, Luo C, Zhang J, Cao W, Ge J, Zhao M. A m(6)A methyltransferase-mediated immune signature determines prognosis, immune landscape and immunotherapy efficacy in patients with lung adenocarcinoma. Cell Oncol (Dordrecht Netherlands). (2022) 45:931–49. doi: 10.1007/s13402-022-00697-2 [DOI] [PubMed] [Google Scholar]
- 83. Song Q, Su J, Zhang W. scGCN is a graph convolutional networks algorithm for knowledge transfer in single cell omics. Nat Commun. (2021) 12:3826. doi: 10.1038/s41467-021-24172-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cheng Q, Zhao W, Song X, Jin T. Machine-learning and scRNA-Seq-based diagnostic and prognostic models illustrating survival and therapy response of lung adenocarcinoma. Genes Immun. (2024) 25:356–66. doi: 10.1038/s41435-024-00289-0 [DOI] [PubMed] [Google Scholar]
- 85. Charytonowicz D, Brody R, Sebra R. Interpretable and context-free deconvolution of multi-scale whole transcriptomic data with UniCell deconvolve. Nat Commun. (2023) 14:1350. doi: 10.1038/s41467-023-36961-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang P, Feng J, Rui M, Xie J, Zhang L, Zhang Z. Integrating machine learning and single-cell analysis to uncover lung adenocarcinoma progression and prognostic biomarkers. J Cell Mol Med. (2024) 28:e18516. doi: 10.1111/jcmm.18516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kazdal D, Endris V, Allgäuer M, Kriegsmann M, Leichsenring J, Volckmar AL, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol. (2019) 14:1935–47. doi: 10.1016/j.jtho.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 88. Bergenstråhle J, Larsson L, Lundeberg J. Seamless integration of image and molecular analysis for spatial transcriptomics workflows. BMC Genomics. (2020) 21:482. doi: 10.1186/s12864-020-06832-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lei Y, Huang XT, Guo X, Hang Katie Chan K, Gao L. DeepGRNCS: deep learning-based framework for jointly inferring gene regulatory networks across cell subpopulations. Briefings Bioinf. (2024) 25:bbae334. doi: 10.1093/bib/bbae334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Patel AJ, Tan TM, Richter AG, Naidu B, Blackburn JM, Middleton GW. A highly predictive autoantibody-based biomarker panel for prognosis in early-stage NSCLC with potential therapeutic implications. Br J Cancer. (2022) 126:238–46. doi: 10.1038/s41416-021-01572-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu J, Chang X, Qian L, Chen S, Xue Z, Wu J, et al. Proteomics-derived biomarker panel facilitates distinguishing primary lung adenocarcinomas with intestinal or mucinous differentiation from lung metastatic colorectal cancer. Mol Cell Proteomics: MCP. (2024) 23:100766. doi: 10.1016/j.mcpro.2024.100766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu J, Li H, Zhang N, Dong Q, Liang Z. Multiomics analysis of disulfidptosis patterns and integrated machine learning to predict immunotherapy response in lung adenocarcinoma. Curr Med Chem. (2024) 31:4034–55. doi: 10.2174/0109298673313281240425050032 [DOI] [PubMed] [Google Scholar]
- 93. Coker EA, Stewart A, Ozer B, Minchom A, Pickard L, Ruddle R, et al. Individualized prediction of drug response and rational combination therapy in NSCLC using artificial intelligence-enabled studies of acute phosphoproteomic changes. Mol Cancer Ther. (2022) 21:1020–9. doi: 10.1158/1535-7163.Mct-21-0442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hu T, Allam M, Cai S, Henderson W, Yueh B, Garipcan A, et al. Single-cell spatial metabolomics with cell-type specific protein profiling for tissue systems biology. Nat Commun. (2023) 14:8260. doi: 10.1038/s41467-023-43917-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Huang L, Wang L, Hu X, Chen S, Tao Y, Su H, et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat Commun. (2020) 11:3556. doi: 10.1038/s41467-020-17347-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang Q, Luo J, Xu H, Huang L, Zhu X, Li H, et al. Metabolomic investigation of urinary extracellular vesicles for early detection and screening of lung cancer. J Nanobiotechnol. (2023) 21:153. doi: 10.1186/s12951-023-01908-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L, Zhang M, Pan X, Zhao M, Huang L, Hu X, et al. Integrative serum metabolic fingerprints based multi-modal platforms for lung adenocarcinoma early detection and pulmonary nodule classification. Adv Sci (Weinheim Baden-Wurttemberg Germany). (2022) 9:e2203786. doi: 10.1002/advs.202203786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shen J, Sun N, Zens P, Kunzke T, Buck A, Prade VM, et al. Spatial metabolomics for evaluating response to neoadjuvant therapy in non-small cell lung cancer patients. Cancer Commun (London England). (2022) 42:517–35. doi: 10.1002/cac2.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ricciuti B, Lamberti G, Puchala SR, Mahadevan NR, Lin JR, Alessi JV, et al. Genomic and immunophenotypic landscape of acquired resistance to PD-(L)1 blockade in non-small-cell lung cancer. J Clin Oncol. (2024) 42:1311–21. doi: 10.1200/jco.23.00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lamort AS, Kaiser JC, Pepe MAA, Lilis I, Ntaliarda G, Somogyi K, et al. Prognostic phenotypes of early-stage lung adenocarcinoma. Eur Respir J. (2022) 60:2101674. doi: 10.1183/13993003.01674-2021 [DOI] [PubMed] [Google Scholar]
- 101. Wu Y, Kang K, Han C, Wang L, Wang Z, Zhao A. Single-cell profiling comparisons of tumor microenvironment between primary advanced lung adenocarcinomas and brain metastases and machine learning algorithms in predicting immunotherapeutic responses. Biomolecules. (2023) 13:185. doi: 10.3390/biom13010185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhao Z, Yin W, Peng X, Cai Q, He B, Shi S, et al. A machine-learning approach to developing a predictive signature based on transcriptome profiling of ground-glass opacities for accurate classification and exploring the immune microenvironment of early-stage LUAD. Front Immunol. (2022) 13:872387. doi: 10.3389/fimmu.2022.872387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zaarour RF, Sharda M, Azakir B, Hassan Venkatesh G, Abou Khouzam R, Rifath A, et al. Genomic analysis of waterpipe smoke-induced lung tumor autophagy and plasticity. Int J Mol Sci. (2022) 23:6848. doi: 10.3390/ijms23126848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lin L, Zhang W, Chen Y, Ren W, Zhao J, Ouyang W, et al. Immune gene patterns and characterization of the tumor immune microenvironment associated with cancer immunotherapy efficacy. Heliyon. (2023) 9:e14450. doi: 10.1016/j.heliyon.2023.e14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Liu Z, Lin G, Yan Z, Li L, Wu X, Shi J, et al. Predictive mutation signature of immunotherapy benefits in NSCLC based on machine learning algorithms. Front Immunol. (2022) 13:989275. doi: 10.3389/fimmu.2022.989275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tran NH, Qiao R, Xin L, Chen X, Liu C, Zhang X, et al. Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nat Methods. (2019) 16:63–6. doi: 10.1038/s41592-018-0260-3 [DOI] [PubMed] [Google Scholar]
- 107. Cheng H, Rao B, Liu L, Cui L, Xiao G, Su R, et al. PepFormer: end-to-end transformer-based siamese network to predict and enhance peptide detectability based on sequence only. Analytical Chem. (2021) 93:6481–90. doi: 10.1021/acs.analchem.1c00354 [DOI] [PubMed] [Google Scholar]
- 108. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. (2020) 48:W449–w454. doi: 10.1093/nar/gkaa379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. O’Donnell TJ, Rubinsteyn A, Laserson U. MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst. (2020) 11:42–48.e47. doi: 10.1016/j.cels.2020.06.010 [DOI] [PubMed] [Google Scholar]
- 110. Shibaki R, Fujimoto D, Nozawa T, Sano A, Kitamura Y, Fukuoka J, et al. Machine learning analysis of pathological images to predict 1-year progression-free survival of immunotherapy in patients with small-cell lung cancer. J Immunother Cancer. (2024) 12:e007987. doi: 10.1136/jitc-2023-007987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Guan SW, Lin Q, Wu XD, Yu HB. Weighted gene coexpression network analysis and machine learning reveal oncogenome associated microbiome plays an important role in tumor immunity and prognosis in pan-cancer. J Transl Med. (2023) 21:537. doi: 10.1186/s12967-023-04411-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhang Y, Chen X, Wang Y, Li L, Ju Q, Zhang Y, et al. Alterations of lower respiratory tract microbiome and short-chain fatty acids in different segments in lung cancer: a multiomics analysis. Front Cell Infect Microbiol. (2023) 13:1261284. doi: 10.3389/fcimb.2023.1261284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Shi S, Chu Y, Liu H, Yu L, Sun D, Yang J, et al. Predictable regulation of survival by intratumoral microbe-immune crosstalk in patients with lung adenocarcinoma. Microbial Cell (Graz Austria). (2024) 11:29–40. doi: 10.15698/mic2024.02.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ronneberger O, Fischer P, Brox T. Springer International Publishing, 234–41. [Google Scholar]
- 115. He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. IEEE transactions on pattern analysis and machine intelligence. (2020) 42:386–97. doi: 10.1109/tpami.2018.2844175 [DOI] [PubMed] [Google Scholar]
- 116. Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, et al. Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. (2019) 290:783–92. doi: 10.1148/radiol.2018180910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. NIHR Global Health Research Unit on Global Surgery. STARSurg Collaborative . A prognostic model for use before elective surgery to estimate the risk of postoperative pulmonary complications (GSU-Pulmonary Score): a development and validation study in three international cohorts. Lancet Digit Health. (2024) 6:e507–19. doi: 10.1016/s2589-7500(24)00065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhong Y, She Y, Deng J, Chen S, Wang T, Yang M, et al. Deep learning for prediction of N2 metastasis and survival for clinical stage I non-small cell lung cancer. Radiology. (2022) 302:200–11. doi: 10.1148/radiol.2021210902 [DOI] [PubMed] [Google Scholar]
- 119. Saad MB, Hong L, Aminu M, Vokes NI, Chen P, Salehjahromi M, et al. Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study. Lancet Digit Health. (2023) 5:e404–20. doi: 10.1016/s2589-7500(23)00082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lin CY, Guo SM, Lien JJ, Lin WT, Liu YS, Lai CH, et al. Combined model integrating deep learning, radiomics, and clinical data to classify lung nodules at chest CT. La Radiol Med. (2024) 129:56–69. doi: 10.1007/s11547-023-01730-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lou B, Doken S, Zhuang T, Wingerter D, Gidwani M, Mistry N, et al. An image-based deep learning framework for individualizing radiotherapy dose. Lancet Digit Health. (2019) 1:e136–47. doi: 10.1016/s2589-7500(19)30058-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhong Y, Cai C, Chen T, Gui H, Deng J, Yang M, et al. PET/CT based cross-modal deep learning signature to predict occult nodal metastasis in lung cancer. Nat Commun. (2023) 14:7513. doi: 10.1038/s41467-023-42811-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Le Page AL, Ballot E, Truntzer C, Derangère V, Ilie A, Rageot D, et al. Using a convolutional neural network for classification of squamous and non-squamous non-small cell lung cancer based on diagnostic histopathology HES images. Sci Rep. (2021) 11:23912. doi: 10.1038/s41598-021-03206-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cao L, Wang J, Zhang Y, Rong Z, Wang M, Wang L, et al. E2EFP-MIL: End-to-end and high-generalizability weakly supervised deep convolutional network for lung cancer classification from whole slide image. Med Image Anal. (2023) 88:102837. doi: 10.1016/j.media.2023.102837 [DOI] [PubMed] [Google Scholar]
- 125. Rigamonti A, Viatore M, Polidori R, Rahal D, Erreni M, Fumagalli MR, et al. Integrating AI-powered digital pathology and imaging mass cytometry identifies key classifiers of tumor cells, stroma, and immune cells in non-small cell lung cancer. Cancer Res. (2024) 84:1165–77. doi: 10.1158/0008-5472.Can-23-1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pan X, AbdulJabbar K, Coelho-Lima J, Grapa AI, Zhang H, Cheung AHK, et al. The artificial intelligence-based model ANORAK improves histopathological grading of lung adenocarcinoma. Nat Cancer. (2024) 5:347–63. doi: 10.1038/s43018-023-00694-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhang Y, Yang Z, Chen R, Zhu Y, Liu L, Dong J, et al. Histopathology images-based deep learning prediction of prognosis and therapeutic response in small cell lung cancer. NPJ Digital Med. (2024) 7:15. doi: 10.1038/s41746-024-01003-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Vanguri RS, Luo J, Aukerman AT, Egger JV, Fong CJ, Horvat N, et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat Cancer. (2022) 3:1151–64. doi: 10.1038/s43018-022-00416-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Janßen C, Boskamp T, Le'Clerc Arrastia J, Otero Baguer D, Hauberg-Lotte L, Kriegsmann M, et al. Multimodal lung cancer subtyping using deep learning neural networks on whole slide tissue images and MALDI MSI. Cancers. (2022) 14:6181. doi: 10.3390/cancers14246181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhou H, Watson M, Bernadt CT, Lin SS, Lin CY, Ritter JH, et al. AI-guided histopathology predicts brain metastasis in lung cancer patients. J Pathol. (2024) 263:89–98. doi: 10.1002/path.6263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wang S, Rong R, Zhou Q, Yang DM, Zhang X, Zhan X, et al. Deep learning of cell spatial organizations identifies clinically relevant insights in tissue images. Nat Commun. (2023) 14:7872. doi: 10.1038/s41467-023-43172-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Claudio Quiros A, Coudray N, Yeaton A, Yang X, Liu B, Le H, et al. Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides. Nat Commun. (2024) 15:4596. doi: 10.1038/s41467-024-48666-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Guo D, Wang Y, Chen J, Liu X. Integration of multi-omics data for survival prediction of lung adenocarcinoma. Comput Methods Programs Biomed. (2024) 250:108192. doi: 10.1016/j.cmpb.2024.108192 [DOI] [PubMed] [Google Scholar]
- 134. Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. (2019) 25:954–61. doi: 10.1038/s41591-019-0447-x [DOI] [PubMed] [Google Scholar]
- 135. Rolfo C, Russo A. Navigating into a stormy sea: liquid biopsy enters peri-operative management in early-stage non-small cell lung cancer. Ann Oncol. (2024) 35:147–9. doi: 10.1016/j.annonc.2023.12.010 [DOI] [PubMed] [Google Scholar]
- 136. Nguyen VTC, Nguyen TH, Doan NNT, Pham TMQ, Nguyen GTH, Nguyen TD, et al. Multimodal analysis of methylomics and fragmentomics in plasma cell-free DNA for multi-cancer early detection and localization. eLife. (2023) 12:RP89083. doi: 10.7554/eLife.89083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Salimi Y, Hajianfar G, Mansouri Z, Sanaat A, Amini M, Shiri I, et al. Organomics: A concept reflecting the importance of PET/CT healthy organ radiomics in non-small cell lung cancer prognosis prediction using machine learning. Clin Nucl Med. (2024) 49:899–908. doi: 10.1097/rlu.0000000000005400 [DOI] [PubMed] [Google Scholar]
- 138. Annapragada AV, Niknafs N, White JR, Bruhm DC, Cherry C, Medina JE, et al. Genome-wide repeat landscapes in cancer and cell-free DNA. Sci Trans Med. (2024) 16:eadj9283. doi: 10.1126/scitranslmed.adj9283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yao X, Ouyang S, Lian Y, Peng Q, Zhou X, Huang F, et al. PheSeq, a Bayesian deep learning model to enhance and interpret the gene-disease association studies. Genome Med. (2024) 16:56. doi: 10.1186/s13073-024-01330-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang L, Zhang X, Guan M, Zeng J, Yu F, Lai F. Identification of a novel ADCC-related gene signature for predicting the prognosis and therapy response in lung adenocarcinoma. Inflammation Res. (2024) 73:841–66. doi: 10.1007/s00011-024-01871-y [DOI] [PubMed] [Google Scholar]
- 141. Lou N, Cui X, Lin X, Gao R, Xu C, Qiao N, et al. Development and validation of a deep learning-based model to predict response and survival of T790M mutant non-small cell lung cancer patients in early clinical phase trials using electronic medical record and pharmacokinetic data. Trans Lung Cancer Res. (2024) 13:706–20. doi: 10.21037/tlcr-23-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hanley MJ, Yeo KR, Tugnait M, Iwasaki S, Narasimhan N, Zhang P, et al. Evaluation of the drug-drug interaction potential of brigatinib using a physiologically-based pharmacokinetic modeling approach. CPT: Pharmacometrics Syst Pharmacol. (2024) 13:624–37. doi: 10.1002/psp4.13106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Batra U, Nathany S, Nath SK, Jose JT, Sharma T, Preeti P, et al. AI-based pipeline for early screening of lung cancer: integrating radiology, clinical, and genomics data. Lancet Regional Health Southeast Asia. (2024) 24:100352. doi: 10.1016/j.lansea.2024.100352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. de Miguel-Perez D, Ak M, Mamindla P, Russo A, Zenkin S, Ak N, et al. Validation of a multiomic model of plasma extracellular vesicle PD-L1 and radiomics for prediction of response to immunotherapy in NSCLC. J Exp Clin Cancer Res: CR. (2024) 43:81. doi: 10.1186/s13046-024-02997-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Lee J, Kim D, Kong J, Ha D, Kim I, Park M, et al. Cell-cell communication network-based interpretable machine learning predicts cancer patient response to immune checkpoint inhibitors. Sci Adv. (2024) 10:eadj0785. doi: 10.1126/sciadv.adj0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Zeng S, Chen L, Tian J, Liu Z, Liu X, Tang H, et al. Integrative analysis of pan-cancer single-cell data reveals a tumor ecosystem subtype predicting immunotherapy response. NPJ Precis Oncol. (2024) 8:205. doi: 10.1038/s41698-024-00703-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Tohidinezhad F, Bontempi D, Zhang Z, Dingemans AM, Aerts J, Bootsma G, et al. Computed tomography-based radiomics for the differential diagnosis of pneumonitis in stage IV non-small cell lung cancer patients treated with immune checkpoint inhibitors. Eur J Cancer (Oxford England: 1990). (2023) 183:142–51. doi: 10.1016/j.ejca.2023.01.027 [DOI] [PubMed] [Google Scholar]
- 148. Jin W, Tian Y, Xuzhang W, Zhu H, Zou N, Shen L, et al. Non-linear modifications enhance prediction of pathological response to pre-operative PD-1 blockade in lung cancer: A longitudinal hybrid radiological model. Pharmacol Res. (2023) 198:106992. doi: 10.1016/j.phrs.2023.106992 [DOI] [PubMed] [Google Scholar]
- 149. Ye G, Wu G, Qi Y, Li K, Wang M, Zhang C, et al. Non-invasive multimodal CT deep learning biomarker to predict pathological complete response of non-small cell lung cancer following neoadjuvant immunochemotherapy: a multicenter study. J Immunother Cancer. (2024) 12:e009348. doi: 10.1136/jitc-2024-009348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Li C, Zhou Z, Hou L, Hu K, Wu Z, Xie Y, et al. A novel machine learning model for efficacy prediction of immunotherapy-chemotherapy in NSCLC based on CT radiomics. Comput Biol Med. (2024) 178:108638. doi: 10.1016/j.compbiomed.2024.108638 [DOI] [PubMed] [Google Scholar]
- 151. Kargbo RB. AI in pharma: transforming drug discovery and strategic management with MYC-modulating compounds and BET protein inhibitors. ACS Med Chem Lett. (2024) 15:334–6. doi: 10.1021/acsmedchemlett.4c00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Choung OH, Vianello R, Segler M, Stiefl N, Jiménez-Luna J. Extracting medicinal chemistry intuition via preference machine learning. Nat Commun. (2023) 14:6651. doi: 10.1038/s41467-023-42242-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Huang L, Xu T, Yu Y, Zhao P, Chen X, Han J, et al. A dual diffusion model enables 3D molecule generation and lead optimization based on target pockets. Nat Commun. (2024) 15:2657. doi: 10.1038/s41467-024-46569-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. He D, Liu Q, Mi Y, Meng Q, Xu L, Hou C, et al. De novo generation and identification of novel compounds with drug efficacy based on machine learning. Adv Sci (Weinheim Baden-Wurttemberg Germany). (2024) 11:e2307245. doi: 10.1002/advs.202307245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Li BY, Li P, Wei LY, Zou J, Wang YH, You QD, et al. Discovery and development of NLRP3 inhibitors targeting the LRR domain to disrupt NLRP3-NEK7 interaction for the treatment of rheumatoid arthritis. J Med Chem. (2024) 67:9869–95. doi: 10.1021/acs.jmedchem.3c02407 [DOI] [PubMed] [Google Scholar]