Abstract
Background
RING finger protein 112 (RNF112) exerts a key role in human tumors. However, its biological function in colorectal cancer (CRC) has not been discussed. We aimed to explore the function and molecular mechanism of RNF112 in CRC.
Results
In this study, RNF112 expression was notably decreased in CRC tissues and cells. Clinical analysis revealed a significant association between low RNF112 expression and tumor size, N classification and TNM stage. In vitro experiments demonstrated that overexpression of RNF112 repressed cell viability, promoted cell cycle arrest and apoptosis, while knocking down RNF112 had the opposite function. The tumor formation results in nude mice supported that RNF112 overexpression exerted anti-tumor effects by inhibiting cell growth and promoting cell apoptosis. Mechanistically, Krüppel-like factor 4 (KLF4) acted as an upstream regulator of RNF112 by mediating its transcription. Furthermore, we explored the downstream mechanism of RNF112 and discovered that it promoted ubiquitination and degradation of oncoprotein N-alpha-acetyltransferase 40 (NAA40) through ubiquitin ligase activity. In addition, overexpression of NAA40 eliminated the effect of RNF112 overexpression on CRC tumorigenesis.
Conclusions
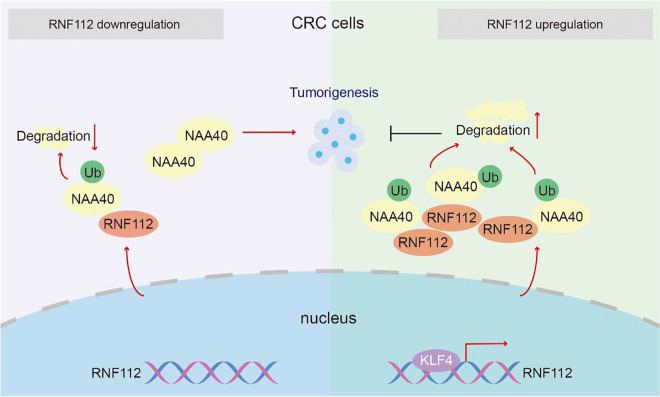
In summary, our findings confirm that RNF112, whose transcription is regulated by KLF4, inhibits CRC growth through promoting ubiquitin-dependent degradation of NAA40. We have unraveled the mechanism of KLF4-RNF112-NAA40 axis in CRC, which shed light on the therapeutic strategies for this disease.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10565-024-09977-z.
Keywords: Colorectal cancer, RNF112, KLF4, NAA40, Ubiquitination
Introduction
Colorectal cancer (CRC) is one of the most common cancers (Li et al. 2022). Recently, the incidence of CRC patients has continued to increase(Biller and Schrag 2021). Many CRC patients will develop metastasis at diagnosis or follow-up (Vayrynen et al. 2020). Although chemotherapy is generally recommended, only a few targeted therapies are suitable for cases with specific mutational signatures (Xu et al. 2021). Therefore, the development of novel molecular targets against CRC is imminent.
RING finger protein 112 (RNF112) has been identified as an E3 ubiquitin ligase (Pao et al. 2011). RNF112 exerted a vital role in neuronal differentiation(Tsou et al. 2017; Wang et al. 2015). RNF112 showed a protective effect on intracerebral hemorrhage through suppressing the TLR-4/NF-κB pathway (Zhang and Zhang 2018). Notably, RNF112 has been demonstrated to participate in cancer progression. Knockdown of RNF112 reduced expression of the negative cell cycle regulators p35 and p27, leading to cell cycle reprogramming in embryonal carcinoma(Pao et al. 2011). RNF112 blocked the malignant behavior of glioma cells via the p53-mediated cell cycle signaling pathway(Lee et al. 2017). RNF112 impeded gastric cancer process by promoting ubiquitination of FOXM1(Zhang et al. 2023). RNF112 may be a promising prognostic biomarker for CRC(Yang et al. 2024). Strikingly, data from mRNA sequencing and GEO databases presented that RNF112 expression was remarkably downregulated in CRC tissues. However, the biological role of RNF112 in CRC has never been discussed.
Krüppel-like factor 4 (KLF4), a member of the evolutionarily conserved zinc finger transcription factor family, regulates many physiological processes (He et al. 2023). Accumulating evidence suggested that KLF4 was a potential tumor suppressor in CRC. For instance, KLF4 inhibited CRC cell proliferation through transcriptional activation of NDRG2 (Ma et al. 2017). KLF4 also enhanced the sensitivity of HCT-15 cells to cisplatin (Yadav et al. 2019). Notably, data from mRNA sequencing and GEO databases revealed that KLF4 expression was obviously lower in CRC tissues than in controls. In addition, JASPAR database predicted the possible binding sites of KLF4 in the RNF112 promoter. However, whether RNF112 is transcriptionally modulated by KLF4 in CRC remains to be confirmed.
N-alpha-acetyltransferase 40 (NAA40) belongs to NAT family (Hole et al. 2011). Increasing studies indicated that NAA40 played a carcinogenic role in CRC. NAA40-mediated metabolic recombination promoted CRC cell resistance to anti-metabolic drug chemotherapy (Demetriadou et al. 2022). NAA40 facilitated CRC progression by controlling PRMT5 expression (Demetriadou et al. 2019). Depletion of NAA40 induced cell apoptosis in CRC (Pavlou and Kirmizis 2016). Of note, IP-LC/MS and Label-Free assays suggested that NAA40 may be a downstream target protein of RNF112, but whether NAA40 affects the function of RNF112 in CRC needs further confirmation.
Here, we want to explore the capabilities of RNF112 in CRC and its potential mechanisms.
Materials and methods
mRNA sequencing
Clinical study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University and conducted based on the Declaration of Helsinki. All subjects provided written informed consent. mRNA sequencing was conducted using 22 CRC tissue samples and 18 adjacent tissue samples by Wuhan Yingzi Gene Technology Co., Ltd. Sample information was shown in supplementary Table 1. Data analysis of mRNA sequencing was as follows: the original sequencing data was obtained through data quality control. Clean data was compared to the reference genome of the corresponding species. Based on the comparison results, the library quality was evaluated, and the sample library data qualified for quality control was analyzed.
Bioinformatics analysis
GSE200427 chip (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE200427) containing 2 normal samples and 2 CRC samples and GSE196006 chip (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196006) containing 21 normal samples and 21 CRC samples were download from NCBI. Differentially expressed genes (DEGs, |log2FC|> 1 and p < 0.01) were identified. GO and KEGG enrichment analysis was then carried out. In addition, list of genes related to E3 ligase was retrieved from GeneCards (https://www.genecards.org/) database using “E3 ubiquitin ligases” as a keyword. Ubiquitin ligase coding and related genes in gene class were obtained from UALCAN database (https://ualcan.path.uab.edu/cgi-bin/Kinase-summary2.pl).
Clinical samples detection
Twenty-nine pairs of fresh primary CRC and adjacent tissues, as well as 92 paraffin-embedded CRC tissues were collected. RNF112 levels were evaluated by immunohistochemistry using a scoring method (Zheng et al. 2022).
Immunohistochemistry
Sections were dewaxed and rehydrated. Afterwards, endogenous peroxidase was blocked with 3% H2O2 for 15 min. RNF112 (1:100, PA5-118,985, Thermo Fisher, USA) or Ki67 (1:100, AF0198, Affinity, Changzhou, China) antibodies were incubated overnight at 4°C, followed by secondary antibody (1:200, D110058, Sangon, Shanghai, China) at 37°C for 0.5 h. DAB was used to develop, and staining was acquired with a microscope.
Cell culture
GP2D and SW1116 cells were obtained from iCell (Shanghai, China). DLD1 and SW620 and NCM460 cells were obtained from Cellverse (Shanghai, China). DLD1, GP2D and NCM460 cells were cultured in 1640 (Solarbio, Beijing, China). SW1116 and SW620 cells were cultured in L-15 (Procell, Wuhan, China). Cells were placed at 37°C and 5% CO2.
Knockdown and overexpression
siRNAs targeting RNF112 were synthesized from JinTuoSi (Wuhan, China). shRNAs targeting RNF112 were cloned to pRNAH1.1 vector. In addition, KLF4 CDS, RNF112 CDS or NAA40 CDS were cloned to pcDNA3.1 vector. Cells were transfected using Liposome 3000 (Invitrogen, USA).
siRNAs targeting RNF112 were shown:
RNF112 siRNA-1, CCUGAGUGCCGGAAGAUAU;
RNF112 siRNA-2, CCUUCCUCCUCAACCAUUU;
RNF112 siRNA-3, GGUGAUGGGCAAGCAUUAU;
RNF112 siRNA-4, AGAGAUUGUCUGGCAGAUA;
RNF112 siRNA-5, CACCCAGAAAGAUGCCAUU.
shRNAs targeting RNF112 were shown:
RNF112 shRNA-1, GGGAAGATATGCAAGCAGAATTCAAGAGATTCTGCTTGC ATATCTTCCTTTTT;
RNF112 shRNA-2, GGGAAGTCCTTCCTCCTCAATTCAAGAGATTGAGGAGGA AGGACTTCCTTTTT.
Cell viability analysis
Cells (5 × 103/ well) were seeded in 96-well plates. Next, after 48 h and 72 h, CCK8 reagent (KeyGEN, Nanjing, China) was added. Finally, OD 450 was tested on a microplate reader (Biotek, USA).
Flow cytometry detection
For cell cycle, cells were treated with 500 µl PI/RNase A (KeyGEN) and incubated for 30 min. For apoptosis, cells were treated with 5 µl AnnexinV-FITC and 5 µl PI and incubated for 10 min. Finally, data was measured by a NovoCyte flow cytometer.
Caspase 3 and Caspase 9 activity
Caspase-3 (C1116, Beyotime, Shanghai, China) and Caspase-9 activity (C1158, Beyotime) was tested by the respective kits.
Xenotransplantation
Animal studies were approved by Medical Ethics Committee of Shengjing Hospital of China Medical University and in lines with the National Research Council’s Guide for the Care and Use of Laboratory Animals. After 1 week of adaptive feeding, BALB/c nude mice (4-week-old) were divided into 5 groups (vector, RNF112 OE, NC, RNF112 shRNA-1 and RNF112 shRNA-2). SW620 or DLD1 cells (2 × 106) were injected subcutaneously. Tumor size was measured every 5 days. After 4 weeks, mice were killed and tumor tissues were collected.
TUNEL staining
After deparaffinized and hydrated, sections were treated with 0.1% Triton X-100 (50 µl, Beyotime) for 8 min and 50 µl TUNEL solution for 1 h. Afterwards, sections were stained with DAPI for 5 min, and staining was acquired under a microscope.
Prediction of binding of KLF4 to RNF112 promoter
RNF112 promoter was retrieved through the UCSC Genome Browser Gateway database (https://genome-asia.ucsc.edu/cgi-bin/hgGateway?hgsid=730609074_3yH3.
QyIyhAexYB3spswXoIohivRN), and the promoter sequence was pasted into the JASPAR database (https://jaspar.elixir.no/). Binding sites of KLF4 on the RNF112 promoter were predicted.
Luciferase reporter assay
Luciferase reporter vector containing the promoter sequence of RNF112 was constructed and transferred into SW620 cells with KLF4 overexpression plasmid. pRL-TK was a control plasmid. Finally, luciferase activity was measured 48 h later.
Chromatin-immunoprecipitation (Ch-IP) assay
Cells were incubated with 1% formaldehyde for 1 h and 10X Glycine Solution. Cells precipitates were then resuspended in SDS lysis. After ultrasonic treatment, 1.8 ml Ch-IP dilution buffer was added to 0.2 ml supernatant. 20 µl supernatant was used as input, and the remain supernatant was added to 70 µl Protein A + G Agarose/Salmon Sperm DNA. After centrifugation, supernatant was added with 1 µg KLF4 antibody (11,880–1-AP, Proteintech) or IgG. Then the mixture was treated with 60 µl Protein A + G Agarose/Salmon Sperm DNA. Afterwards, DNA–protein complex was added with 5 M NaCl, and the purified DNA was used for PCR assay. PCR primers were as follows:
Ch-IP-1
F: 5’-CCTGCCTTGACAACCTTT-3
R: 5’-GATGGGACAATCAGTCTTCAC-3’
Ch-IP-2
F: 5’-GGTAATGGTGGCTCCTC-3’
R: 5’-CCTTCTCATCCCTCCTG-3’
Co-immunoprecipitation (Co-IP)
Cells were lysed and proteins were isolated. The antibody was immobilized, and IP was performed. In brief, IP lysate (200 µl) was added to the resin that solidified the antibody. After elution, the obtained samples were used for western blot.
Ubiquitination assay
For ubiquitination analysis, cells were treated with 10 µM MG132 for 8 h. Western blot was used to detect ubiquitination levels.
IP-LC/MS and Label-Free assays
RNF112 overexpression vector (with Flag tag) and its control vector (with Flag tag) were constructed and transfected into SW620 cells, respectively. After 48 h, IP-LC/MS analysis was performed on the vector-Flag and RNF112-Flag from anti-Flag by Novogene (Beijing, China). According to the files detected, the corresponding database was searched for protein identification. At the same time, mass tolerance distribution of polypeptide, protein and parent ion was analyzed to evaluate the quality of mass spectrometry data.
RNF112 overexpression vector or its control vector were transfected into SW620 cells, respectively. After 48 h, cells were employed for Label-Free. Label-Free was conducted by Qinglian Baiao Technology Co., Ltd. (Beijing, China) in accordance with a standard experimental procedure. Differentially expressed proteins (|log2FC|> 1 and p < 0.05) were identified. Proteins were then subjected to GO and KEGG enrichment analysis.
Immunofluorescence double staining
After blocking with 1% BSA, sections were incubated with Flag (1: 100, 66,008–4-Ig, Proteintech) and NAA40 (1: 100, 16,698–1-AP, Proteintech) antibodies at 4°C overnight, followed by respective secondary antibodies (1: 200, #4408, CST, USA or 1: 200, #4413, CST) for 1 h. Finally, after treating with DAPI, sections were pictured with a microscope.
Real-time PCR
Total RNA was extracted with TRIpure. Next, RNA was transcribed into cDNA by All-in-One First-Strand SuperMix (Magen, Guangzhou, China). Afterwards, real-time PCR was conducted with the SYBR Green kit (Solarbio). Relative mRNA levels were calculated with a 2−ΔΔCt method. Primers were shown as follows:
RNF112, F: 5’-GGACAGACGCCTACTCACG-3’;
RNF112, R: 5’-CTGCCTCACATACTCCTCGA-3’;
KLF4, F: 5’-CCAGAGGAGCCCAAGCCAAAG-3’;
KLF4, R: 5’-TCCACAGCCGTCCCAGTCA-3’;
β-actin F: 5’-GGCACCCAGCACAATGAA-3’;
β-actin R: 5’-TAGAAGCATTTGCGGTGG-3’.
Western blot
Total protein was extracted and quantified by BCA assay kit (Beyotime). 20 µg protein was loaded into each well of a 10% SDS-PAGE gel and transferred to PVDF membranes (Abcam, UK). Next, the blots were incubated with RNF112 (1:1000, PA5-118,985, Thermo Fisher), Cyclin E1 (1:1000, 11,554–1-AP, Proteintech), CyclinD1 (1:5000, 26,939–1-AP, Proteintech) and NAA40 (1:500, 16,698–1-AP, Proteintech) antibodies overnight at 4 °C. Thereafter, the blots were incubated with HRP-labeled goat anti-rabbit IgG (1:5000, A0208, Beyotime) or goat anti-mouse IgG (1:5000, A0216, Beyotime) at 37 °C for 45 min. Finally, the blots were treated with ECL reagent, and data were then analyzed by Gel-Pro-Analyzer software.
Statistical analysis
In this study, data are expressed as mean ± SD. Data between two groups were compared by student’s t-test. Data of multiple groups were compared by one-way or two-way ANOVA. Correlation between RNF112 expression and clinicopathological features was analyzed by Chi-square test. Additionally, correlation between RNF112 mRNA and KLF4 mRNA was evaluated by Pearson. p < 0.05 was considered statistically significant.
Results
RNF112 may be involved in CRC tumorigenesis
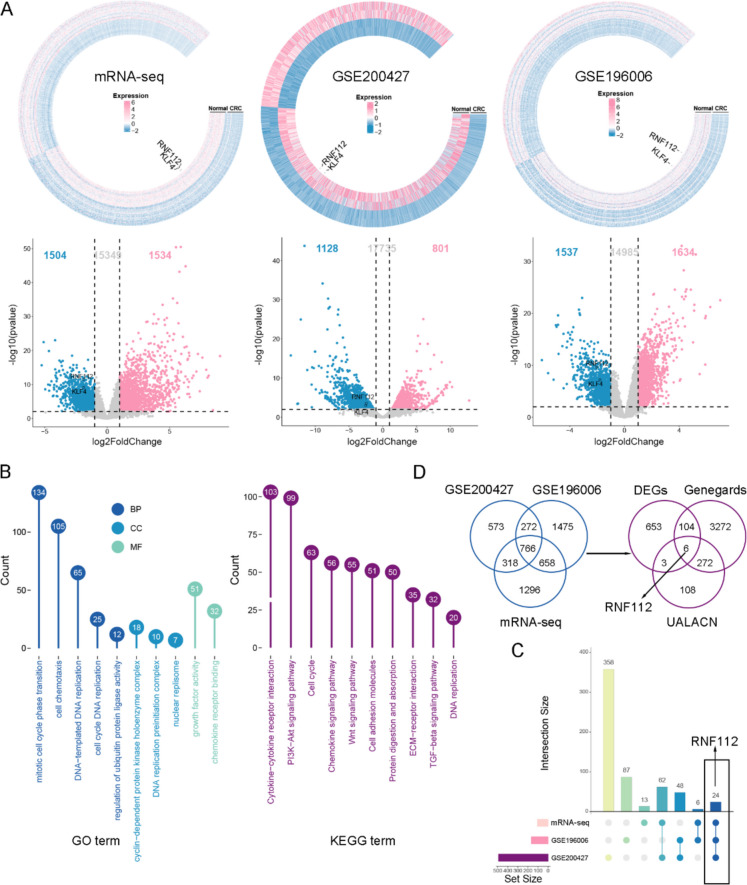
To screen potential molecular targets of CRC, we performed mRNA sequencing using CRC tissue samples and adjacent tissue samples. In addition, we downloaded GSE200427 and GSE196006 chips from NCBI and then carried out bioinformatic analysis of the DEGs in these three databases. Firstly, the ring heat map and volcano map showed the expression of genes in the three datasets. There were 1,534 upregulated genes and 1,504 downregulated genes in mRNA sequencing results (Fig. 1A). In GSE200427 dataset, there were 801 upregulated genes and 1128 downregulated genes (Fig. 1A). In addition, in GSE196006 dataset, there were 1,634 upregulated genes and 1,537 downregulated genes (Fig. 1A). GO and KEGG analysis were conducted. BP results presented that DEGs were enriched in mitotic cell cycle phase transition, DNA-templated DNA replication and regulation of ubiquitin protein ligase activity (Fig. 1B). CC data indicated that DEGs were enriched in cyclin-dependent protein kinase holoenzyme complex and DNA replication preinitiation complex (Fig. 1B). MF results suggested that DEGs were enriched in growth factor activity and chemokine receptor binding (Fig. 1B). Moreover, KEGG enrichment analysis showed that DEGs participated in cytokine-cytokine receptor interaction, PI3K-Akt signaling pathway and cell cycle (Fig. 1B). Upset chart was used to show the number of crossed genes in the three datasets. We found that 24 DEGs had intersections in the three databases (Fig. 1C). We further screened the target factors based on the results of three databases. Firstly, we intersected the DEGs of the three datasets to obtain 766 common DEGs. Next, the 766 common DEGs were intersected with genes related to E3 Ubiquitin ligase in GeneCards and ubiquitin ligase coding and related genes in UALACN database to obtain 6 shared DEGs, including RNF112, UHRF1, ENC1, CCNF, RNF183 and NEDD4L (Fig. 1D). By investigating the function of these 6 DEGs, we found that only the function of RNF112 in CRC was unknown, so RNF112 was selected as a target for subsequent analysis.
Fig. 1.
RNF112 may be involved in CRC tumorigenesis. The data from mRNA sequencing, GSE200427 and GSE196006 profiles were collected, and total DEGs were used for bioinformatics analysis. A The ring heat map and volcano map showed the expression of genes in the three datasets. B GO and KEGG analysis were performed on the total DEGs. C The upset plot showed the intersection of the correlation analysis between three datasets. D Venn diagram showed the common DEGs in the three datasets. The common DEGs of the three datasets were then intersected with genes related to E3 Ubiquitin ligase in GeneCards and ubiquitin ligase coding and related genes in UALACN database. Venn diagram showed the shared DEGs
RNF112 expression was obviously decreased in CRC tissues and cells
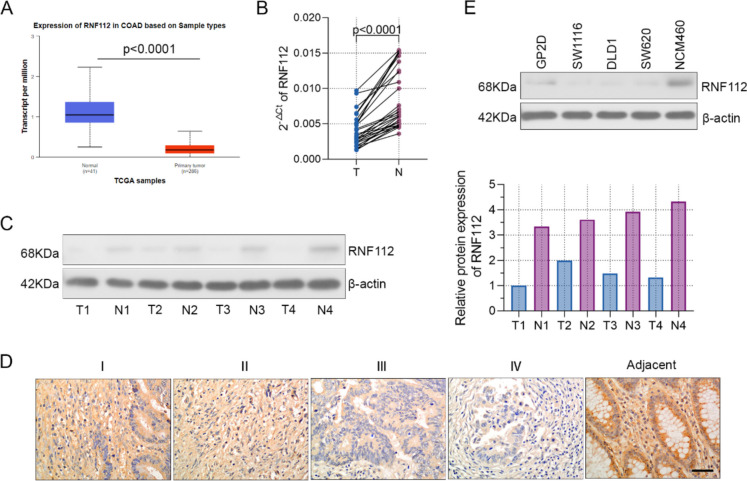
Firstly, we set out to explore the expression of RNF112 in CRC tissues. Data from transcriptome sequencing and GSE200427 and GSE196006 chips showed that RNF112 was obviously decreased in CRC tissues (Fig. 1A). Results from the UALCAN database also indicated that RNF112 levels were markedly reduced in colon cancer tissues in comparison with the controls (Fig. 2A). In addition, we collected 29 pairs of adjacent and tumor tissues from CRC patients. Figure 2B-C displayed that RNF112 expression was overtly decreased in tumor tissues. Immunohistochemistry assay presented that RNF112 levels were increased in adjacent tissues, but gradually decreased in TNM stage (Fig. 2D). To explore the clinicopathological significance of RNF112 in CRC, the correlation between RNF112 expression and the pathologic materials was explored. The analysis suggested that RNF112 low expression was correlated with tumor size, N classification and TNM stage (Table 1).
Fig. 2.
RNF112 expression was significantly decreased in CRC tissues and cells. A RNF112 expression in CRC tissues from the UALCAN database. B-C Real-time PCR (n = 29) and western blot (n = 4) were used to test the expression of RNF112 in CRC and adjacent tissues. D RNF112 expression in different TNM stages (n = 92) and adjacent tissues (n = 32) was determined by immunohistochemistry. (Scale bar = 50 µm). E Western blot analysis of RNF112 expression in CRC cell lines (GP2D, SW1116, DLD1 and SW620) and a normal cell line NCM460 (n = 3). p < 0.0001
Table 1.
Correlations between RNF112 protein expression and clinicopathological characteristics in CRC
| RNF112 expression | |||||
|---|---|---|---|---|---|
| variable | category | high | low | p | |
| Age | ≤ 60 | 15 | 11 | 0.785 | |
| > 60 | 36 | 30 | |||
| Sex | Female | 19 | 20 | 0.266 | |
| Male | 32 | 21 | |||
| Tumor size | ≤ 5 cm | 41 | 24 | 0.022 | |
| > 5 cm | 10 | 17 | |||
| T classification | T1-T2 | 7 | 2 | 0.156 | |
| T3-T4 | 44 | 39 | |||
| N classification | N0 | 34 | 18 | 0.029 | |
| N1-N2 | 17 | 23 | |||
| M classification | M0 | 45 | 36 | 0.950 | |
| M1 | 6 | 5 | |||
| TNM stage | I + II | 34 | 18 | 0.029 | |
| III + IV | 17 | 23 | |||
Furthermore, the abundance of RNF112 was remarkably decreased in the CRC cells including GP2D, SW1116, DLD1 and SW620, compared to NCM460 (Fig. 2E). In addition, we found that RNF112 was moderately expressed in SW620 and DLD1 cells. Therefore, siRNAs targeting RNF112 and its control NC, as well as RNF112 overexpression plasmid and its vector plasmid were transfected into SW620 and DLD1 cells, respective. After transfection for 48 h, the knockdown and overexpression efficiency of RNF112 was verified. These findings indicated that RNF112 was successfully knocked down or overexpressed in CRC cells (Figure S1A-B).
Overexpression of RNF112 suppressed the proliferation of CRC cells
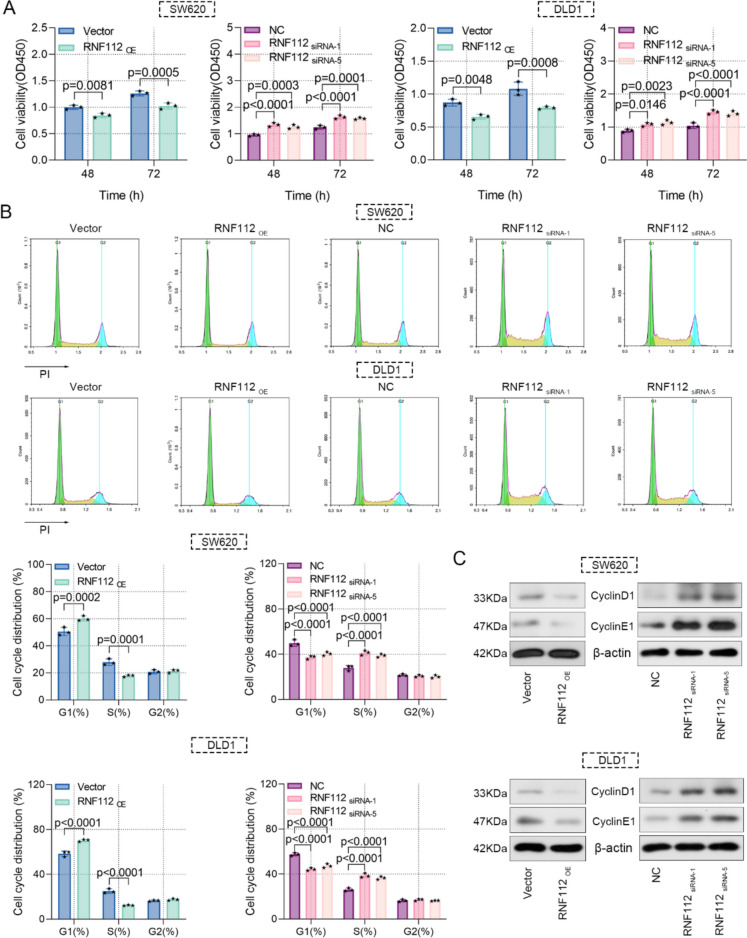
We elucidated the precise role of RNF112 in CRC tumor biology. Functional experiments were carried out after RNF112 overexpression or knockdown. Phenotypically, overexpression of RNF112 repressed cell viability and triggered cell cycle arrest in G1 phase in CRC cells, however, RNF112 knockdown exhibited the opposite effect (Fig. 3A-B). Notably, RNF112 overexpression also inhibited the expression of cyclin E1 and cyclinD1, while RNF112 knockdown elevated their protein levels (Fig. 3C). Taken together, we elucidated that RNF112 suppressed CRC cell viability and cell cycle progression.
Fig. 3.
Overexpression of RNF112 suppressed the proliferation of CRC cells. A Cell viability was detected by CCK8 assay. B Cell cycle was measured by flow cytometry. C Western blot analysis of cyclin E1 and cyclinD1 in RNF112 overexpression or RNF112 knockdown cells. (n = 3). p < 0.05. p < 0.01. p < 0.001. p < 0.0001
Overexpression of RNF112 promoted the apoptosis of CRC cells
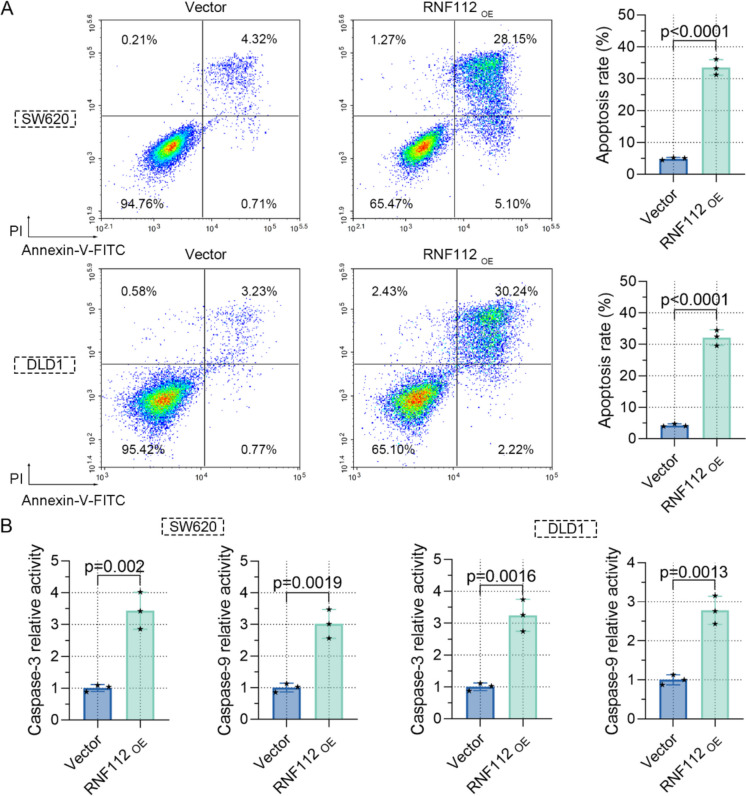
Furthermore, we ascertained the function of RNF112 on apoptosis. Firstly, our results indicated that overexpression of RNF112 distinctly enhanced apoptosis (Fig. 4A). We also assessed the impact of RNF112 on the levels of proapoptotic markers and discovered that overexpression of RNF112 upregulated caspase 3 and caspase 9 activity (Fig. 4B). Therefore, these data uncovered that RNF112 overexpression promoted CRC cell apoptosis.
Fig. 4.
Overexpression of RNF112 promoted the apoptosis of CRC cells. A Cell apoptosis was detected by flow cytometry. B The activity of caspase 3 and caspase 9 in RNF112 overexpression cells was determined by the kit. (n = 3). p < 0.01. p < 0.0001
Overexpression of RNF112 repressed the growth of CRC cells in vivo
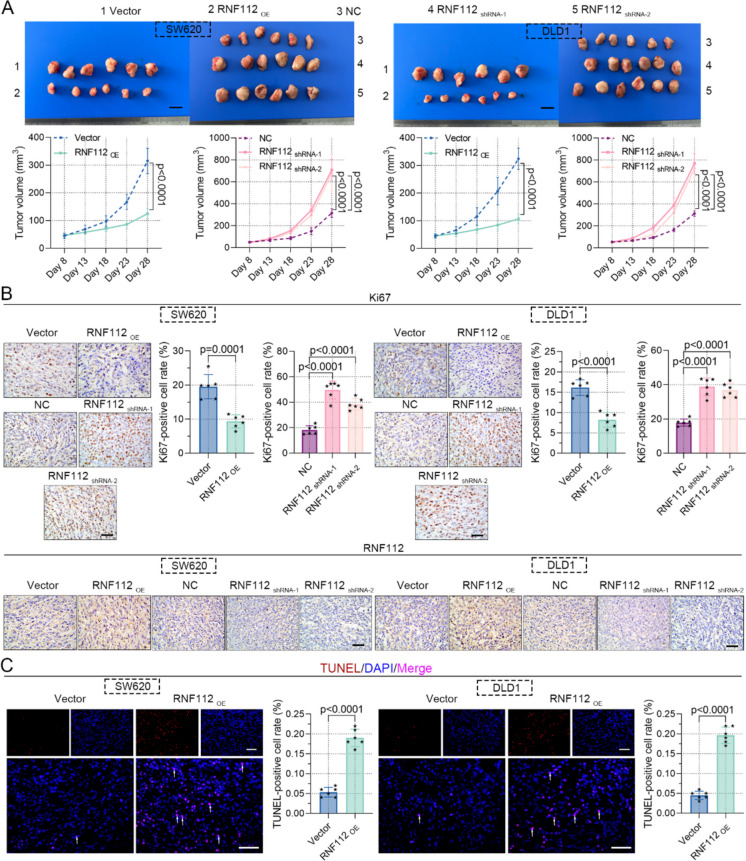
We also determined whether RNF112 is involved in the tumorigenesis of CRC in vivo. RNF112 overexpression significantly retarded xenograft tumor growth, as expected, RNF112 knockdown exhibited the opposite effect (Fig. 5A). In addition, we investigated the role of RNF112 in the expression of tumor growth marker Ki67 (Menon et al. 2019). Immunohistochemistry results illustrated that RNF112 overexpression decreased Ki67 expression, while RNF112 knockdown increased its expression (Fig. 5B). The knockdown and overexpression efficiency of RNF112 was also tested by immunohistochemistry, indicating that RNF112 was successfully overexpressed or knocked down (Fig. 5B). TUNEL staining was used to assess the impact of RNF112 on apoptosis, and the results demonstrated that RNF112 overexpression increased TUNEL-positive cells, supporting its pro-apoptotic effect (Fig. 5C). Collectively, our findings confirmed that overexpression of RNF112 repressed the growth of CRC cells in vivo.
Fig. 5.
Overexpression of RNF112 repressed the growth of CRC cells in vivo. A Representative images of xenografts in nude mice. (Scale bar = 1 cm). Tumor volume was also measured. B Immunohistochemistry analysis of Ki67 and RNF112 in xenografts. (Scale bar = 50 µm). C TUNEL staining was used to test cell apoptosis. (Scale bar = 50 µm). Arrows represented TUNEL-positive cells. (n = 6). p < 0.001. p < 0.0001
KLF4 promoted the transcriptional regulation of RNF112
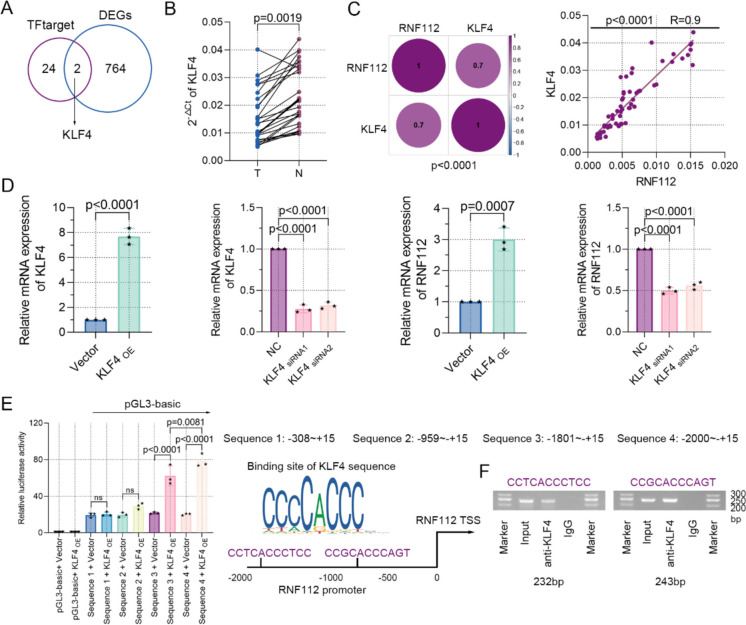
Here, we set out to identify the transcriptional mechanism of RNF112 in CRC. Firstly, the transcription factors predicted by TFtarget database to bind to the RNF112 promoter region were intersected with 766 common DEGs to obtained 2 DEGs, including KLF4 and TCF21 (Fig. 6A). Next, real-time PCR assay indicated that KLF4 levels were notably downregulated in CRC tissues compared to adjacent tissues (Fig. 6B). The correlation of KLF4 mRNA and RNF112 mRNA in mRNA sequencing and CRC clinical samples was then explored. The analysis indicated that their expression was positively correlated (Fig. 6C). Furthermore, KLF4 overexpression significantly upregulated KLF4 and RNF112 mRNA levels in SW620 cells (Fig. 6D). JASPAR database analysis identified the binding sites for KLF4 in the RNF112 promoter, suggesting that KLF4 may regulate the transcription of RNF112. To test this possibility, we conducted a luciferase reporter assay to interrogate the regulation mode of KLF4 on RNF112. As reflected, exogenous KLF4 stimulated a significant increase in luciferase activity at −1801 ~—+ 15bp and −2000 ~—+ 15bp of the promoter region of RNF112 in CRC cells (Fig. 6E). Ch-IP experiments suggested that two motifs of KLF4 bound to the promoter region of RNF112 (Fig. 6F). Therefore, our data revealed that KLF4 was a transcription factor involved in regulating RNF112 expression.
Fig. 6.
KLF4 mediated the transcriptional regulation of RNF112. A The transcription factors predicted by TFtarget (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) to bind to the RNF112 promoter region were intersected with 766 common DEGs. Venn diagram showed the common DEGs. B Real-time PCR was used to detect the expression of KLF4 in CRC and adjacent tissues. C The correlation between the expression of KLF4 mRNA and RNF112 mRNA in mRNA sequencing and CRC clinical samples was analyzed. D The expression of KLF4 and RNF112 in SW620 cells was determined by real-time PCR and western blot. E Luciferase activity in SW620 cells was tested. Potential binding sites of KLF4 on the RNF112 promoter region were also shown. F Binding of KLF4 on the RNF112 promoter region in SW620 cells was determined by Ch-IP. (n = 3). p < 0.01. p < 0.001. p < 0.0001
NAA40 was a downstream protein of RNF112
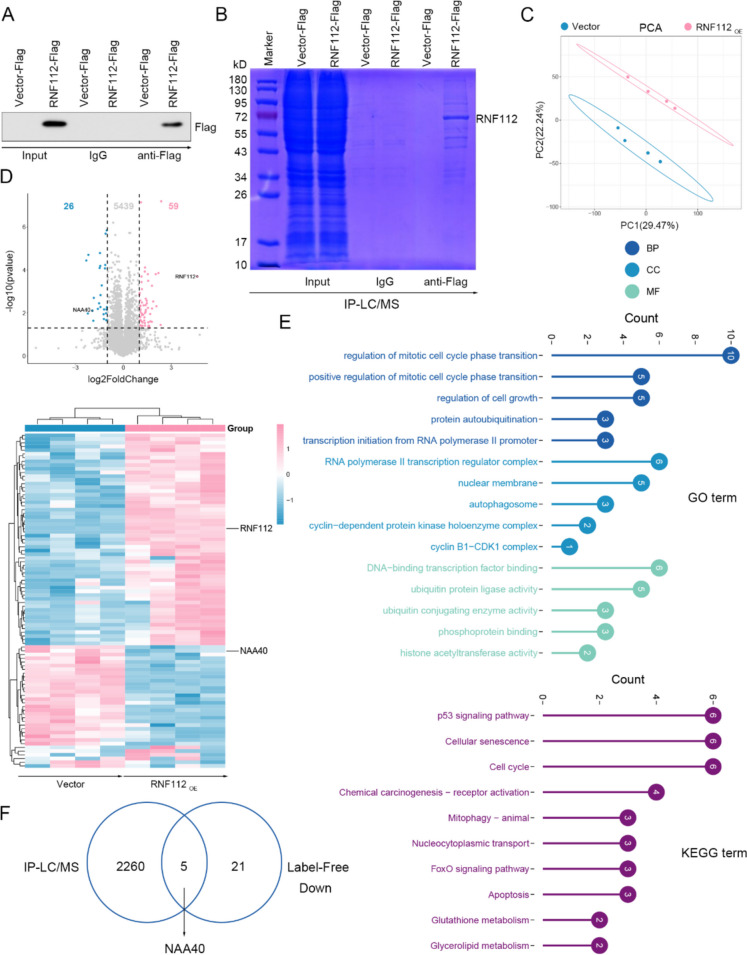
To probe the mechanism downstream of RNF112, IP-LC/MS and Label-Free were conducted. Firstly, 48 h after transfection, the levels of RNF112-Flag in Co-IP precipitation were determined by immunoblotting (Fig. 7A). Co-IP precipitates were then subjected to gel electrophoresis and stained with Coomassie Brilliant Blue (Fig. 7B). IP-LC/MS and Label-Free assays were performed. Label-Free PCA analysis showed that vector samples and RNF112 overexpression samples were clustered, respectively (Fig. 7C). We found that RNF112 overexpression resulted in significant downregulation of 26 proteins and upregulation of 59 proteins (Fig. 7D). GO enrichment results showed that these proteins were related to regulation of mitotic cell cycle phase transition, RNA polymerase II transcription regulator complex and DNA-binding transcription factor binding (Fig. 7E). KEGG data implied that these proteins participated in cellular senescence, p53 signaling pathway and cell cycle (Fig. 7F). Finally, 2265 binding proteins of RNF112 in the IP-LC/MS results were cross-analyzed with the downregulated proteins in the Label-Free results, and 5 common proteins were obtained, including CRABP2, LGALS1, NAA40, NMD3 and MMAB (Fig. 7G). By inquiring the function of these 5 proteins, it was found that only NAA40 played a tumor-promoting role in CRC. But the function of the remaining 4 proteins in CRC is unknown. Thus, we speculated that NAA40 may be a downstream protein of RNF112.
Fig. 7.
NAA40 was a downstream protein of RNF112. A The RNF112 overexpression vector (with Flag tag) and its control vector (with Flag tag) were constructed and transfected into SW620 cells, respectively. After 48 h of transfection, Flag levels in Co-IP precipitate were detected by immunoblotting. B Co-IP precipitates were subjected to gel electrophoresis and stained with Coomassie Brilliant Blue. C The RNF112 overexpression vector and its control vector were transfected into SW620 cells, respectively. After 48 h of transfection, cells were used for Label-Free assay. PCA was performed on the samples (n = 4). D The heat map and volcano map showed the expression of differentially expressed proteins in Label-Free. E GO and KEGG analysis were performed on these proteins. F The intersection of RNF112 binding proteins obtained by IP-LC/MS and downregulated proteins obtained by Label-Free was analyzed. Venn diagram showed 5 common proteins
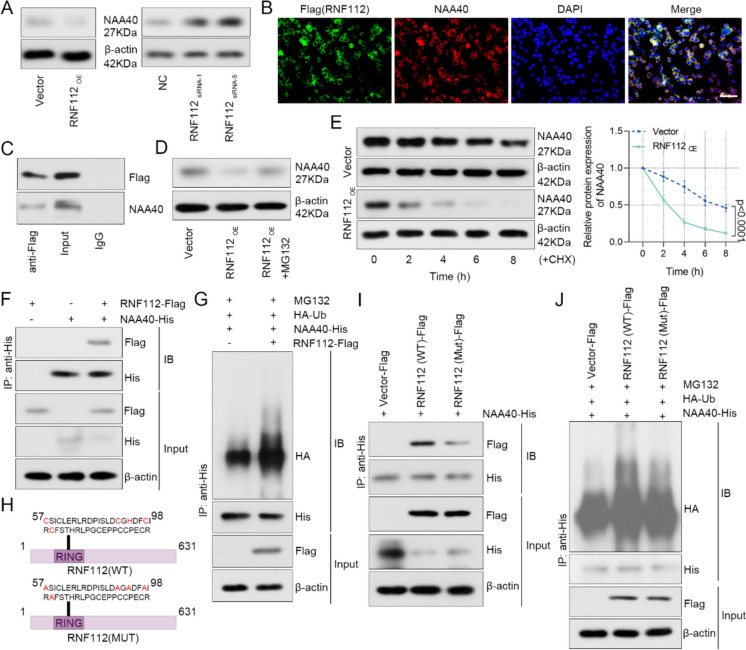
RNF112 interacted with NAA40 and enhanced its ubiquitination degradation
We further validated the regulatory mechanism of RNF112 and NAA40. Firstly, RNF112 overexpression suppressed the protein levels of NAA40, while RNF112 knockdown promoted its protein levels (Fig. 8A). As indicated by immunofluorescence, RNF112 and NAA40 were mainly colocalized in cytoplasm, and rarely colocalized in nucleus (Fig. 8B). Co-IP assay indicated that RNF112-Flag combined with NAA40 in SW620 cells (Fig. 8C). After SW620 cells were treated with MG132 for 8 h, the levels of NAA40 were detected. We noted that RNF112 inhibited the expression of NAA40 via proteasome pathway (Fig. 8D). As revealed in Fig. 8E, overexpression of RNF112 promoted the degradation of NAA40. In addition, HEK293T cells were transfected with NAA40 and RNF112 alone or co-transfected to detect the exogenous combination of RNF112 and NAA40. Co-IP results showed a combination of the two proteins (Fig. 8F). HEK293T cells were also transfected with NAA40, RNF112 and Ub. Co-IP was used to detect the ubiquitination levels. The data indicated that RNF112 promoted the ubiquitination of NAA40 (Fig. 8G).
Fig. 8.
RNF112 interacted with NAA40 and enhanced its ubiquitination degradation. A SW620 cells were transfected with RNF112 overexpression plasmid or siRNA targeting RNF112 sequences. The expression of NAA40 was detected by western blot 48 h after transfection. B The co-localization of Flag (RNF112) and NAA40 in SW620 cells was detected by immunofluorescence double staining. (Scale bar = 50 µm). C Co-IP was used to detect the binding of Flag (RNF112) and NAA40. D The expression of NAA40 was detected by western blot after SW620 cells were treated with 10 µM MG132 for 8 h. E After 100 µg/ml CHX treatment for 0, 2, 4, 6 and 8 h, the expression of NAA40 was tested by western blot, and the degradation rate of NAA40 protein was calculated. F HEK293T cells were transfected with NAA40 vector (with His tag) and RNF112 vector (with Flag tag) alone or co-transfected. After 48 h of transfection, co-IP was used to detect the binding of RNF112 and NAA40. G HEK293T cells were transfected with NAA40 vector (with His tag), RNF112 vector (with Flag tag) and Ub vector (with HA tag). After transfection for 48 h and treatment with 10 µM MG132 for 8 h, co-IP was used to test the ubiquitination levels in the cells. H Schematic diagram of RNF112-Mut lacking E3 ubiquitin ligase activity. I HEK293T cells were transfected with NAA40 vector (with His tag) and RNF112-WT vector (with Flag tag) or RNF112-Mut vector (with Flag tag). After 48 h of transfection, co-IP was used to detect the binding of RNF112 and NAA40. J HEK293T cells were transfected with NAA40 vector (with His tag), RNF112-WT (with Flag tag) or RNF112-Mut (with Flag tag) and Ub vector (with HA tag). After transfection for 48 h and treatment with 10 µM MG132 for 8 h, co-IP was used to test the ubiquitination levels in the cells. (n = 3). p < 0.0001
Since RNF112 is a RING-type E3 ubiquitin ligase, we deleted its E3 ubiquitin ligase activity by mutating the RING domain and named it RNF112-MUT (Fig. 8H). Co-IP results showed a slight reduction in binding of RNF112-Mut and NAA40 compared to RNF112-WT and NAA40 (F8g. 8I). In addition, HEK293T cells were transfected with NAA40, RNF112-WT or RNF112-Mut and Ub. It was found that RNF112-Mut reduced NAA40 ubiquitination levels compared to RNF112-WT (Fig. 8J). Totally, these observations confirmed that RNF112 promoted ubiquitin-dependent degradation of NAA40.
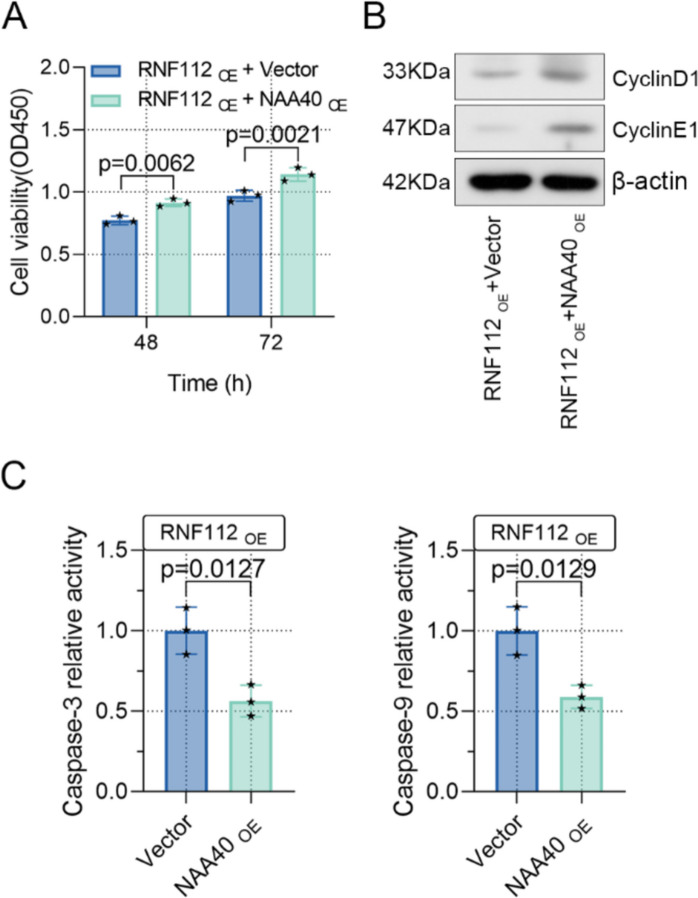
NAA40 overexpression diminished the impact of RNF112 overexpression on CRC tumorigenesis
To further verify whether NAA40 affects the function of RNF112 on CRC tumorigenesis, SW620 cells were transfected with RNF112 and NAA40 overexpression plasmids. As indicated by CCK8 assay, NAA40 overexpression increased cell viability that was inhibited by RNF112 overexpression (Fig. 9A). Furthermore, NAA40 overexpression upregulated the levels of cyclin E1 and cyclinD1, as well as downregulated caspase 3 and caspase 9 activity, further diminishing the role of RNF112 overexpression in CRC (Fig. 9B-C). Overall, our data certified that RNF112 suppressed CRC growth by inhibiting NAA40 levels (Fig. 10).
Fig. 9.
NAA40 overexpression diminished the impact of RNF112 overexpression on CRC tumorigenesis. (A) Cell viability was detected by CCK8 assay. (B) Western blot analysis of cyclin E1 and cyclinD1 in RNF112 overexpression and NAA40 overexpression cells. (C) The activity of caspase 3 and caspase 9 was determined by the kit. (n=3). p<0.05. p<0.01
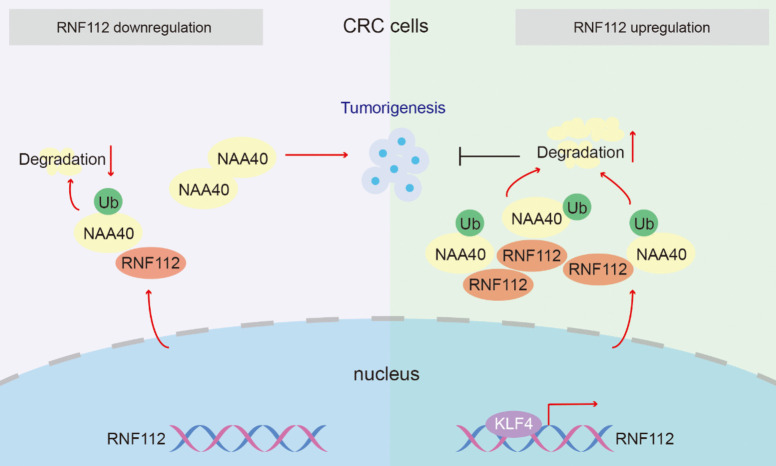
Fig. 10.
Molecular mechanism of antitumor effect of RNF112 in CRC cells
Discussion
As demonstrated, RNF112 reprogramed glioma cells to a more differentiated phenotype and inhibited glioma progression through a p53-mediated cell cycle signaling pathway(Lee et al. 2017). In addition, RNF112 suppressed gastric cancer process by triggering ubiquitination of FOXM1(Zhang et al. 2023). Notably, bioinformatics analysis implied that RNF112 may be implicated in CRC. Our data further indicated that RNF112 expression was overtly decreased in CRC tissues, which was consistent with the results of transcriptome sequencing. Next, we found that the low RNF112 expression was significantly correlated with tumor size, N classification and TNM stage. However, our study did not track the relationship between RNF112 levels and patient survival using clinical data. We believed that exploring the relationship between RNF112 expression and patient survival will deepen the clinical relevance of RNF112 in CRC prognosis, and this investigation will be conducted in future studies. Afterwards, gain or loss of function assays were conducted. Overexpression of RNF112 inhibited cell viability and cell cycle process and induced apoptosis in vitro, as well as reduced the tumorigenesis of CRC cells in vivo. As demonstrated, RNF112 knockdown showed a cancer-promoting effect. Together, our findings verified that RNF112 had an antitumor role in CRC.
In order to explore transcriptional regulatory mechanism of RNF112 in CRC, we analyzed its upstream transcription factors. By cross-analyzing the transcription factors bound to the RNF112 promoter region predicted by TFtarget and 766 DEGs, two genes that may regulate the transcription of RNF112 were obtained, including KLF4 and TCF21. The reason we chose KLF4 as the upstream of RNF112 is that there are more reliable reports that KLF4 plays a cancer-suppressing role in CRC (Xiu et al. 2017; Zhao et al. 2004). Downregulation of KLF4 contributed to metastasis and the epithelial-to-mesenchymal transition of CRC cells (Shao et al. 2019). KLF4 also inhibited the proliferation of CRC cells dependent on NDRG2 signaling(Ma et al. 2017). In addition, KLF4 sensitized colon cancer cell to cisplatin cytotoxicity by regulating HMGB1 and hTERT12 (Yadav et al. 2019). But the role of TCF21 in CRC is less clear. Therefore, we selected KLF4 as the transcription factor for RNF112 for follow-up studies. Notably, results of transcriptome sequencing and GEO databases illustrated that KLF4 expression was largely downregulated in CRC tissues. Our further assays confirmed that KLF4 regulated RNF112 expression. Of note, JASPAR prediction analysis presented that KLF4 had potential binding sites in the RNF112 promoter. Subsequently, our data demonstrated that KLF4 bound to RNF112 promoter and promoted its transcription. Totally, our results proved that KLF4, as an upstream of RNF112, elevated its expression.
A previous study indicated that RNF112 mediated ubiquitination of FOXM1 and altered its stability (Zhang et al. 2023). To elucidate the downstream mechanism of RNF112, IP-LC/MS and Label-Free assays were conducted. Cross-analysis of the 2265 binding proteins of RNF112 in the IP-LC/MS results with the downregulated proteins in the Label-Free results yielded 5 common proteins, including CRABP2, LGALS1, NAA40, NMD3 and MMAB. Through the exploration of the functions of these 5 proteins, only NAA40 was found to play a pro-tumor role in CRC. For instance, loss of NAA40 resulted in altered expression of key oncogenes and tumor suppressor genes that inhibit the growth of CRC cells (Demetriadou et al. 2019). Depletion of NAA40 induced cell apoptosis in CRC (Pavlou and Kirmizis 2016). Therefore, NAA40 with clear function was selected as the downstream factor for study. If other factors with unknown function are selected, it is debatable whether RNF112 works through these factors. Our findings further confirmed that RNF112 interacted with NAA40 and induced its ubiquitination degradation depending on the ubiquitin ligase activity in CRC cells. Recovery assays thus demonstrated that NAA40 partially rescued the function of RNF112, not completely abolished the role of RNF112, but it also reflected that RNF112 did play a role in CRC through NAA40. These data implied that RNF112 showed an anticancer effect by decreasing NAA40 expression, however, the deeper mechanisms of the two proteins still need to be further explored in the future.
Conclusions
Altgether, our findings imply that RNF112, whose transcription is regulated by KLF4, inhibits CRC growth by promoting ubiquitination and degradation of NAA40. Therefore, our study highlights the importance of the KLF4-RNF112-NAA40 signaling axis in CRC tumor biology, which may hold great promise for either diagnosis or therapy.
Supplementary Information
Below is the link to the supplementary material.
(DOCX 16.3 KB)
(DOCX 292 KB)
(PDF 514 KB)
Acknowledgements
We thank the Medical Ethics Committee of Shengjing Hospital of China Medical University for assistance with clinical sample collection.
Author contributions
Chunfei Li and Wenzheng Guan performed the experiments and drafted the manuscript. Yong Feng designed the study and revised the manuscript. Donghua Geng analyzed the data.
Funding
Not applicable.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Ethical approval
Clinical study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University and conducted in accordance with the Declaration of Helsinki.
Animal studies were approved by Medical Ethics Committee of Shengjing Hospital of China Medical University, and all procedures were conducted according to the National Research Council’s Guide for the Care and Use of Laboratory Animals.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chunfei Li and Wenzheng Guan contributed equally.
References
- Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325:669–85. 10.1001/jama.2021.0106. [DOI] [PubMed] [Google Scholar]
- Demetriadou C, Pavlou D, Mpekris F, Achilleos C, Stylianopoulos T, Zaravinos A, Papageorgis P, Kirmizis A. NAA40 contributes to colorectal cancer growth by controlling PRMT5 expression. Cell Death Dis. 2019;10:236. 10.1038/s41419-019-1487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriadou C, Raoukka A, Charidemou E, Mylonas C, Michael C, Parekh S, Koufaris C, Skourides P, Papageorgis P, Tessarz P, Kirmizis A. Histone N-terminal acetyltransferase NAA40 links one-carbon metabolism to chemoresistance. Oncogene. 2022;41:571–85. 10.1038/s41388-021-02113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, He J, Xie K. KLF4 transcription factor in tumorigenesis. Cell Death Discov. 2023;9:118. 10.1038/s41420-023-01416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hole K, Van Damme P, Dalva M, Aksnes H, Glomnes N, Varhaug JE, Lillehaug JR, Gevaert K, Arnesen T. The human N-alpha-acetyltransferase 40 (hNaa40p/hNatD) is conserved from yeast and N-terminally acetylates histones H2A and H4. PLoS ONE. 2011;6:e24713. 10.1371/journal.pone.0024713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Chen CL, Lee YC, Kao TJ, Chen KY, Fang CY, Chang WC, Chiang YH, Huang CC. Znf179 induces differentiation and growth arrest of human primary glioblastoma multiforme in a p53-dependent cell cycle pathway. Sci Rep. 2017;7:4787. 10.1038/s41598-017-05305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Song J, Guo Z, Gong Y, Zhang T, Huang J, Cheng R, Yu X, Li Y, Chen L, Ma X, Sun Y, Wang Y, Xue L. EZH2 inhibitors suppress colorectal cancer by regulating macrophage polarization in the tumor microenvironment. Front Immunol. 2022;13:857808. 10.3389/fimmu.2022.857808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Wu L, Liu X, Xu Y, Shi W, Liang Y, Yao L, Zheng J, Zhang J. KLF4 inhibits colorectal cancer cell proliferation dependent on NDRG2 signaling. Oncol Rep. 2017;38:975–84. 10.3892/or.2017.5736. [DOI] [PubMed] [Google Scholar]
- Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta. 2019;491:39–45. 10.1016/j.cca.2019.01.011. [DOI] [PubMed] [Google Scholar]
- Pao PC, Huang NK, Liu YW, Yeh SH, Lin ST, Hsieh CP, Huang AM, Huang HS, Tseng JT, Chang WC, Lee YC. A novel RING finger protein, Znf179, modulates cell cycle exit and neuronal differentiation of P19 embryonal carcinoma cells. Cell Death Differ. 2011;18:1791–804. 10.1038/cdd.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlou D, Kirmizis A. Depletion of histone N-terminal-acetyltransferase Naa40 induces p53-independent apoptosis in colorectal cancer cells via the mitochondrial pathway. Apoptosis. 2016;21:298–311. 10.1007/s10495-015-1207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Dong D, Shao F. Long non-coding RNA TUG1-mediated down-regulation of KLF4 contributes to metastasis and the epithelial-to-mesenchymal transition of colorectal cancer by miR-153-1. Cancer Manag Res. 2019;11:8699–710. 10.2147/CMAR.S208508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou JH, Yang YC, Pao PC, Lin HC, Huang NK, Lin ST, Hsu KS, Yeh CM, Lee KH, Kuo CJ, Yang DM, Lin JH, Chang WC, Lee YC. Important roles of ring finger protein 112 in embryonic vascular development and brain functions. Mol Neurobiol. 2017;54:2286–300. 10.1007/s12035-016-9812-7. [DOI] [PubMed] [Google Scholar]
- Vayrynen V, Wirta EV, Seppala T, Sihvo E, Mecklin JP, Vasala K, Kellokumpu I. Incidence and management of patients with colorectal cancer and synchronous and metachronous colorectal metastases: a population-based study. BJS Open. 2020;4:685–92. 10.1002/bjs5.50299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM, Lee YC, Ko CY, Lai MD, Lin DY, Pao PC, Chi JY, Hsiao YW, Liu TL, Wang JM. Increase of zinc finger protein 179 in response to CCAAT/enhancer binding protein delta conferring an antiapoptotic effect in astrocytes of Alzheimer’s disease. Mol Neurobiol. 2015;51:370–82. 10.1007/s12035-014-8714-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu DH, Chen Y, Liu L, Yang HS, Liu GF. Tumor-suppressive role of Kruppel-like factor 4 (KLF-4) in colorectal cancer. Genet Mol Res. 2017;16. 10.4238/gmr16019272 [DOI] [PubMed]
- Xu H, Liu L, Li W, Zou D, Yu J, Wang L, Wong CC. Transcription factors in colorectal cancer: molecular mechanism and therapeutic implications. Oncogene. 2021;40:1555–69. 10.1038/s41388-020-01587-3. [DOI] [PubMed] [Google Scholar]
- Yadav SS, Kumar M, Varshney A, Yadava PK. KLF4 sensitizes the colon cancer cell HCT-15 to cisplatin by altering the expression of HMGB1 and hTERT. Life Sci. 2019;220:169–76. 10.1016/j.lfs.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Yang L, Fang C, Zhang R, Zhou S. Prognostic value of oxidative stress-related genes in colorectal cancer and its correlation with tumor immunity. BMC Genomics. 2024;25:8. 10.1186/s12864-023-09879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhang C. Rnf112 deletion protects brain against intracerebral hemorrhage (ICH) in mice by inhibiting TLR-4/NF-kappaB pathway. Biochem Biophys Res Commun. 2018;507:43–50. 10.1016/j.bbrc.2018.10.141. [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang J, Hu W, He L, Tang Q, Li J, Jie M, Li X, Liu C, Ouyang Q, Yang S, Hu C. RNF112-mediated FOXM1 ubiquitination suppresses the proliferation and invasion of gastric cancer. JCI Insight. 2023;8. 10.1172/jci.insight.166698 [DOI] [PMC free article] [PubMed]
- Zhao W, Hisamuddin IM, Nandan MO, Babbin BA, Lamb NE, Yang VW. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene. 2004;23:395–402. 10.1038/sj.onc.1207067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhao Y, Jiang J, Zou B, Dong L. Transmembrane protein 100 inhibits the progression of colorectal cancer by promoting the ubiquitin/proteasome degradation of HIF-1alpha. Front Oncol. 2022;12:899385. 10.3389/fonc.2022.899385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 16.3 KB)
(DOCX 292 KB)
(PDF 514 KB)
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.