Abstract
Label noise is a common and important issue that would affect the model’s performance in artificial intelligence. This study assessed the effectiveness and potential risks of automated label cleaning using an open-source framework, Cleanlab, in multi-category datasets of fundus photography and optical coherence tomography, with intentionally introduced label noise ranging from 0 to 70%. After six cycles of automatic cleaning, significant improvements are achieved in label accuracies (3.4–62.9%) and dataset quality scores (DQS, 5.1–74.4%). The majority (86.6 to 97.5%) of label errors were accurately modified, with minimal missed (0.5–2.8%) or misclassified (0.4–10.6%). The classification accuracy of RETFound significantly improved by 0.3–52.9% when trained with the datasets after cleaning. We also developed a DQS-guided cleaning strategy to mitigate over-cleaning. Furthermore, external validation on EyePACS and APTOS-2019 datasets boosted label accuracy by 1.3 and 1.8%, respectively. This approach automates label correction, enhances dataset reliability, and strengthens model performance efficiently and safely.
Subject terms: Retinal diseases, Medical imaging
Introduction
Retinal diseases are some leading causes of irreversible blindness and impose both medical and economic burdens on society1,2. Screening based on retinal images, including color fundus photography (CFP) and optical coherence tomography (OCT), has been shown to play a pivotal role in preventing blindness3,4. Over the past decade, the application of artificial intelligence (AI) techniques, including deep learning, in CFP and OCT has successfully reduced the workforce in image grading and provides a promising avenue for large-scale blindness prevention, which is particularly pronounced in underdeveloped regions with limited medical resources5,6.
The development of supervised AI models requires large, high-quality, labeled datasets7. However, labeling is a subjective process. To overcome this disadvantage, majority-voting consensus and multi-level verification can be used. Both of them demand significant time and effort, especially in medicine, which requires a skilled team with high professionalism8,9. However, despite these efforts, there is still room for errors, omissions, or inaccuracies in labels, called label noise10,11. It is reported that the mainstream public datasets, which were widely and repeatedly used, have a proportion of noisy labels ranging from 0.15 to 38.5%12–16. In EyePACS, a commonly employed public CFP database, labeling errors were reported up to 40%17.
The accuracy of labels directly impacts the model’s performance, and mislabeling within the dataset can mislead data scientists into selecting a suboptimal model for deployment18,19. Models trained on noisy samples from these datasets can pose potential risks in decision-making, hindering the clinical and real-world implementation of AI20. Hence, minimizing label noise is an indispensable component in the development of robust medical artificial intelligence10.
Although manual re-labeling can be used to clean the noise in the label17, it is very labor-intensive and time-consuming. Furthermore, it cannot avoid the problem of graders’ subjectivity. Therefore, it becomes imperative to explore objective, efficient, interpretable, and automated means of label refinement to handle the voluminous data effectively. Confident learning, an emerging branch of weakly supervised learning and noisy learning, can identify label errors by estimating the uncertainty in dataset labels21. An open-source framework known as Cleanlab is developed based on confident learning and presents a potential solution to reduce label noise and annotation workload effectively21. Cleanlab has shown prominent performance in previous studies to find label errors in natural imaging datasets such as MNIST, CIFAR-10, CIFAR-100, Caltech-256, ImageNet, and QuickDraw15. However, there are few reports of Cleanlab in medical images, especially retinal images. Furthermore, Cleanlab can be deployed in a code-free or simple implementation manner, which does not require profound coding expertise and can be driven by clinicians.
In this study, we aimed to comprehensively assess the efficacy and potential risks of multi-iterations Cleanlab in automated rectifying label errors in CFP and OCT datasets with different noise rates, and whether data cleaning improves the performance of the deep learning model in the classification of retinal images. Furthermore, we explored a strategy to optimize the number of cleaning iterations. Finally, we conducted external validation on publicly available datasets to demonstrate the feasibility of our approach.
Results
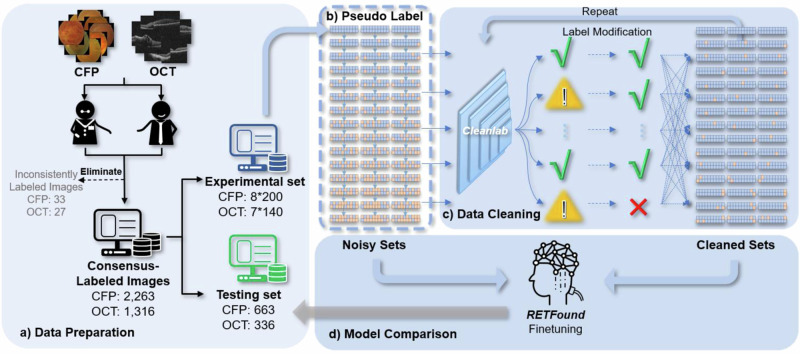
The overall process of the study is depicted in Fig. 1. Initially, we gathered 8 categories of CFP and 7 categories of OCT images, which were subsequently annotated and divided into experimental sets for label cleaning and testing sets for performance evaluation via RETFound finetuning. Within the experimental sets, a proportional pseudo-label strategy was employed to generate multiple noisy subsets for both CFP and OCT. Cleanlab was then iteratively applied to identify and correct mislabeling within the dataset. To assess the impact of dataset noise on model accuracy, the RETFound model was independently fine-tuned on the datasets both before and after label cleaning. The classification accuracy of these models was subsequently compared using the identical hold-out testing set.
Fig. 1. Schematic diagram of the overall label cleaning process.
a Data preparation: A total of 2263 CFP and 1316 OCT images were collected and annotated b pseudo-label strategy: Label noise was deliberately injected by randomly select 5% from one category and evenly distributed to other categories progressively to create 45 subsets. c Data cleaning: Cleanlab was applied to detect and correct label errors repeatedly. d Model comparison: The RETFound foundation model was fine-tuned on datasets before and after label cleaning and tested on a holdout testing set.
Label accuracy and dataset quality score
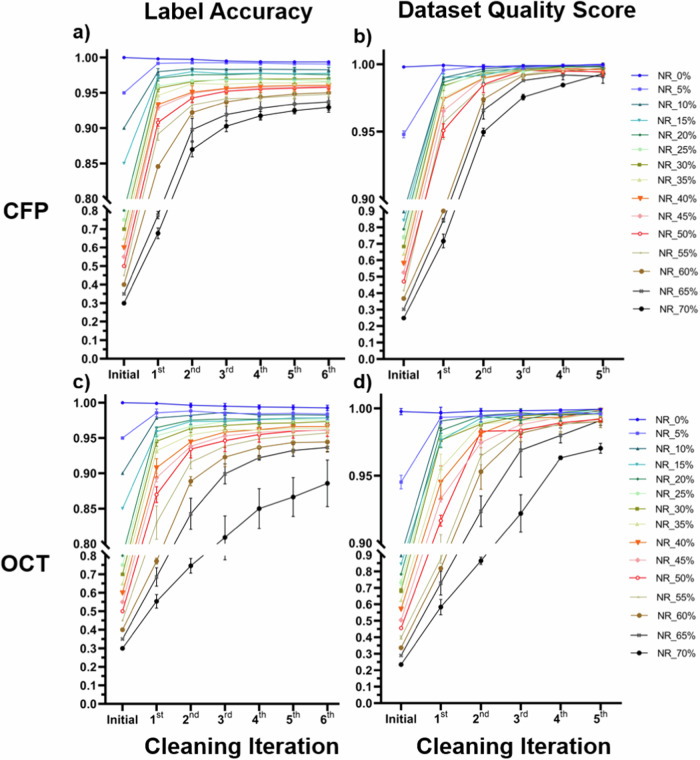
The label accuracies and dataset quality scores (DQSs) before and after each cleaning iteration are shown in Fig. 2. After completing all six iterations of unsupervised label cleaning, most label accuracies and DQSs exhibited remarkable improvements, except in the original noise-free datasets. An example of confusion matrixes for the datasets with a 40% noise rate is shown in Supplementary Fig. 1. The final label accuracy ranged from 92.9 to 99.4% in CFP and from 88.6 to 99.3% in the OCT. The noisier rate in the original dataset, the more improvement of label accuracy; however, the lower final label accuracy in general. The improvement is more in the initial iteration and less in later iterations. However, it is found that in some datasets with low noise label rates, the label accuracy improved at the initial iterations but decreased later. In the original noise-free datasets, there is a slight decrease in label accuracy after automated cleaning.
Fig. 2. Line charts of label accuracy and dataset quality scores after repeated cleaning iterations.
After repeated label cleaning, most of the label accuracies (a, c) and dataset quality scores (b, d) of CFP (a, b) and OCT (c, d) increased significantly and stabilized at high levels, except in the original noise-free datasets.
Dataset quality score guiding cleaning strategy
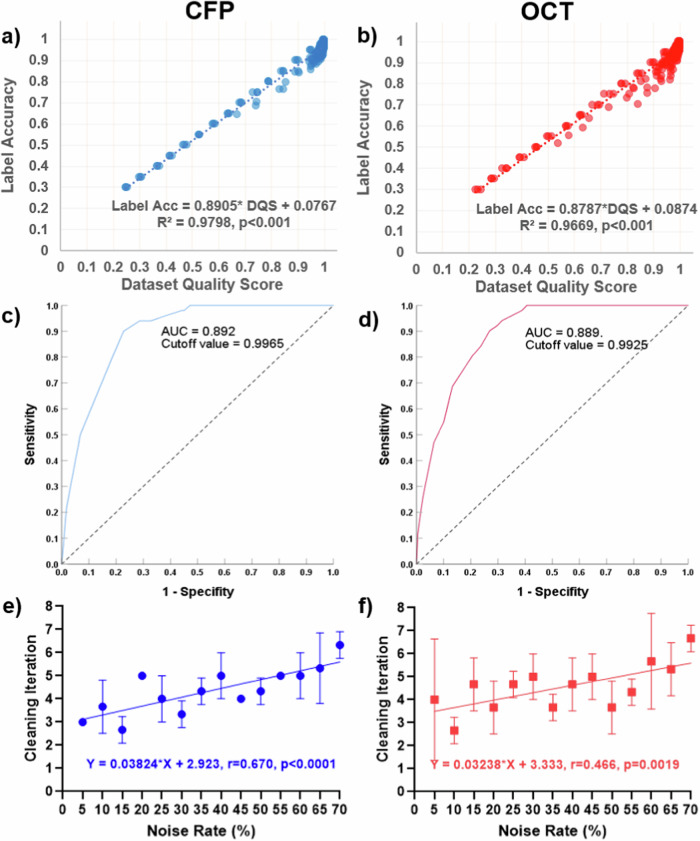
The DQS highly correlated with the label accuracy, with R2 of 0.9798 and 0.9669 for CFP and OCT, respectively (both p < 0.001) (Fig. 3a, b). The receiver operating characteristic curves show that when the accuracy of 0.98 was set as the criteria, the corresponding cutoff values of DQS were 0.9965 for CFP and 0.9925 for OCT (Fig. 3c, d). Then, we used these thresholds to determine whether the dataset should be cleaned. The cleaning iteration was stopped when the DQS decreased or remained unchanged. Using this strategy, the number of iterations needed for various noisy datasets is shown in Fig. 3e, f. Generally, most datasets needed three to seven rounds of cleaning. The noisier the dataset, the more iterations needed (r = 0.670 and 0.466 for CFP and OCT, respectively, both p < 0.01).
Fig. 3. Label cleaning strategy guided by dataset quality scores.
a, b showed the strong linear correlation between dataset quality score (DQS) and label accuracy. c, d showed receiver operating characteristic curves for predicting label accuracy with DQS. The cutoff values of DQS for label accuracy >0.98 were determined to be 0.9965 and 0.9925. e, f The correlation between noise rate and cleaning iterations using DQS-guided strategy.
Cleaning effectiveness and potential risk
Both the 6-iteration strategy and DQS-guided cleaning strategy improved label accuracy (Tables 1, 2). The miss rate of pseudo-labels was low, ranging from 1.4 to 2.1% for CFP and 0.5 to 2.8% for OCT, respectively. The Correct modification rate was high (92.3 to 97.5% for CFP and 86.6 to 95.7% for OCT) after six-iteration or DQS-guided cleaning. However, it should be noted that while some label noise was successfully detected, there were instances where it was mistakenly modified to incorrect categories, accounting for 0.4 to 5.9% for CFP and 2.9 to 10.6% for OCT. Furthermore, some correct labels are falsely labeled and mis-modified (0.7 to 5.6% for CFP and 0.7 to 10.7% for OCT). DQS-guided cleaning reduced the chance of this false labeling in the datasets with a low noise rate (0–15%).
Table 1.
Efficiency and risk of automated label cleaning in color fundus photograph
| NR | Six-Iteration strategy | DQS-guided strategy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC | CMR | EMR | MR | FLR | ACC | CMR | EMR | MR | FLR | |
| 0% | 0.994 ± 0.001 | - | - | - | 0.7% ± 0.7% | 1.000 ± 0.000 | - | - | - | 0.0% ± 0.0% |
| 5% | 0.991 ± 0.002 | 97.5% ± 1.3% | 0.4% ± 0.7% | 2.1% ± 0.7% | 0.9% ± 0.1% | 0.993 ± 0.002 | 96.7% ± 1.4% | 0.4% ± 0.7% | 2.9% ± 0.7% | 0.6% ± 0.1% |
| 10% | 0.983 ± 0.003 | 96.7% ± 2.0% | 1.5% ± 2.0% | 1.9% ± 0.0% | 1.6% ± 0.5% | 0.983 ± 0.002 | 95.2% ± 1.0% | 1.5% ± 1.6% | 3.3% ± 0.7% | 1.4% ± 0.1% |
| 15% | 0.978 ± 0.009 | 95.8% ± 0.8% | 2.4% ± 0.6% | 1.8% ± 0.6% | 1.9% ± 1.0% | 0.977 ± 0.007 | 94.3% ± 2.1% | 2.6% ± 0.9% | 3.1% ± 1.3% | 1.7% ± 0.8% |
| 20% | 0.976 ± 0.002 | 96.3% ± 1.3% | 2.0% ± 0.9% | 1.8% ± 0.7% | 2.1% ± 0.4% | 0.977 ± 0.001 | 96.0% ± 1.1% | 2.1% ± 1.0% | 1.9% ± 0.6% | 2.0% ± 0.3% |
| 25% | 0.966 ± 0.003 | 95.3% ± 1.6% | 2.7% ± 0.5% | 2.0% ± 1.1% | 2.8% ± 0.3% | 0.969 ± 0.001 | 94.9% ± 1.5% | 2.8% ± 0.6% | 2.3% ± 0.9% | 2.4% ± 0.4% |
| 30% | 0.970 ± 0.006 | 96.0% ± 1.4% | 2.6% ± 0.8% | 1.4% ± 0.6% | 2.6% ± 0.2% | 0.970 ± 0.003 | 95.3% ± 1.4% | 2.6% ± 0.5% | 2.1% ± 0.9% | 2.4% ± 0.2% |
| 35% | 0.964 ± 0.005 | 95.2% ± 0.6% | 3.2% ± 0.4% | 1.6% ± 0.2% | 2.8% ± 0.1% | 0.964 ± 0.003 | 94.6% ± 0.3% | 3.2% ± 0.4% | 2.3% ± 0.3% | 2.7% ± 0.2% |
| 40% | 0.961 ± 0.009 | 95.1% ± 0.9% | 3.2% ± 0.5% | 1.7% ± 0.5% | 3.3% ± 0.9% | 0.959 ± 0.009 | 94.7% ± 0.9% | 3.3% ± 0.5% | 1.9% ± 0.5% | 3.3% ± 0.9% |
| 45% | 0.959 ± 0.001 | 95.1% ± 0.3% | 3.1% ± 0.5% | 1.8% ± 0.3% | 3.4% ± 0.1% | 0.957 ± 0.002 | 94.4% ± 0.4% | 3.4% ± 0.5% | 2.2% ± 0.5% | 3.2% ± 0.0% |
| 50% | 0.958 ± 0.003 | 95.1% ± 0.4% | 3.5% ± 0.3% | 1.4% ± 0.3% | 3.5% ± 0.3% | 0.956 ± 0.003 | 94.6% ± 0.7% | 3.8% ± 0.6% | 1.6% ± 0.3% | 3.6% ± 0.2% |
| 55% | 0.948 ± 0.004 | 93.9% ± 0.4% | 4.2% ± 0.4% | 1.9% ± 0.6% | 4.2% ± 0.3% | 0.946 ± 0.005 | 93.5% ± 0.7% | 4.4% ± 0.3% | 2.1% ± 0.7% | 4.1% ± 0.3% |
| 60% | 0.950 ± 0.006 | 94.4% ± 0.7% | 4.0% ± 0.4% | 1.5% ± 0.4% | 4.1% ± 0.8% | 0.930 ± 0.016 | 94.1% ± 1.1% | 4.2% ± 0.6% | 1.8% ± 0.5% | 4.2% ± 0.7% |
| 65% | 0.942 ± 0.010 | 93.3% ± 1.5% | 4.9% ± 1.4% | 1.8% ± 0.2% | 4.2% ± 1.1% | 0.932 ± 0.020 | 92.2% ± 2.5% | 5.4% ± 1.7% | 2.4% ± 0.9% | 4.9% ± 1.3% |
| 70% | 0.929 ± 0.004 | 92.3% ± 0.6% | 5.9% ± 0.2% | 1.8% ± 0.5% | 5.6% ± 0.8% | 0.929 ± 0.005 | 92.3% ± 0.7% | 5.9% ± 0.3% | 1.8% ± 0.5% | 5.7% ± 0.8% |
NR noise rate, ACC label accuracy, CMR correct modification rate, EMR error modification rate, MR miss rate, FLR false labeling rate.
Table 2.
Efficiency and risk of automated label cleaning in optical coherence tomography
| NR | Six-Iteration strategy | DQS-guided strategy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ACC | CMR | EMR | MR | FLR | ACC | CMR | EMR | MR | FLR | |
| 0% | 0.993 ± 0.004 | - | - | - | 0.7% ± 0.7% | 1.000 ± 0.000 | - | - | - | 0.0% ± 0.0% |
| 5% | 0.984 ± 0.008 | 94.6% ± 9.4% | 4.1% ± 7.1% | 1.4% ± 2.4% | 1.4% ± 0.5% | 0.988 ± 0.006 | 94.6% ± 9.4% | 2.7% ± 4.7% | 2.7% ± 4.7% | 1.0% ± 0.5% |
| 10% | 0.982 ± 0.003 | 94.6% ± 2.6% | 3.4% ± 1.6% | 2.0% ± 1.0% | 1.4% ± 0.6% | 0.986 ± 0.005 | 92.5% ± 6.2% | 2.0% ± 1.8% | 5.4% ± 5.0% | 0.8% ± 0.3% |
| 15% | 0.978 ± 0.003 | 95.0% ± 2.1% | 3.9% ± 2.2% | 1.1% ± 1.0% | 1.7% ± 0.1% | 0.979 ± 0.001 | 94.8% ± 1.7% | 3.9% ± 2.2% | 1.4% ± 1.2% | 1.6% ± 0.2% |
| 20% | 0.978 ± 0.002 | 95.1% ± 2.1% | 4.4% ± 2.1% | 0.5% ± 0.0% | 1.5% ± 0.3% | 0.979 ± 0.002 | 94.4% ± 1.3% | 4.3% ± 1.9% | 1.4% ± 0.8% | 1.3% ± 0.5% |
| 25% | 0.975 ± 0.007 | 95.2% ± 1.6% | 3.7% ± 1.8% | 1.1% ± 0.2% | 1.7% ± 0.4% | 0.975 ± 0.005 | 94.8% ± 1.7% | 3.9% ± 2.0% | 1.2% ± 0.4% | 1.5% ± 0.1% |
| 30% | 0.973 ± 0.006 | 95.6% ± 1.4% | 3.5% ± 1.4% | 0.9% ± 0.4% | 1.9% ± 0.3% | 0.972 ± 0.006 | 95.4% ± 1.7% | 3.4% ± 1.2% | 1.2% ± 0.5% | 2.0% ± 0.3% |
| 35% | 0.964 ± 0.008 | 95.7% ± 1.4% | 2.9% ± 1.2% | 1.4% ± 0.9% | 3.2% ± 0.6% | 0.962 ± 0.004 | 94.3% ± 1.1% | 3.8% ± 0.5% | 1.9% ± 1.0% | 2.7% ± 0.1% |
| 40% | 0.966 ± 0.009 | 95.0% ± 1.2% | 4.2% ± 1.1% | 0.9% ± 0.5% | 2.2% ± 0.9% | 0.964 ± 0.005 | 94.6% ± 1.2% | 4.1% ± 0.9% | 1.3% ± 1.0% | 2.4% ± 0.4% |
| 45% | 0.961 ± 0.005 | 95.2% ± 1.5% | 3.9% ± 1.2% | 1.0% ± 0.3% | 3.1% ± 1.1% | 0.960 ± 0.008 | 95.0% ± 1.5% | 3.8% ± 1.1% | 1.2% ± 0.5% | 3.2% ± 1.0% |
| 50% | 0.961 ± 0.009 | 95.2% ± 1.7% | 3.9% ± 1.9% | 1.0% ± 0.7% | 2.9% ± 0.5% | 0.950 ± 0.018 | 92.9% ± 3.4% | 4.2% ± 2.2% | 2.9% ± 1.2% | 2.9% ± 0.8% |
| 55% | 0.957 ± 0.010 | 94.6% ± 1.0% | 3.9% ± 1.0% | 1.5% ± 1.0% | 2.9% ± 1.6% | 0.950 ± 0.013 | 94.0% ± 0.9% | 4.0% ± 0.5% | 2.0% ± 1.2% | 3.8% ± 2.0% |
| 60% | 0.944 ± 0.013 | 93.7% ± 1.5% | 4.8% ± 0.9% | 1.5% ± 0.7% | 4.5% ± 1.8% | 0.941 ± 0.019 | 93.0% ± 3.0% | 5.1% ± 1.9% | 1.9% ± 1.1% | 4.3% ± 1.6% |
| 65% | 0.937 ± 0.006 | 92.7% ± 1.4% | 5.4% ± 1.3% | 1.9% ± 0.5% | 4.7% ± 1.0% | 0.932 ± 0.012 | 92.3% ± 2.0% | 5.7% ± 1.5% | 2.0% ± 0.7% | 5.0% ± 0.3% |
| 70% | 0.886 ± 0.033 | 86.6% ± 4.3% | 10.6% ± 3.8% | 2.8% ± 0.9% | 10.7% ± 4.8% | 0.890 ± 0.027 | 88.8% ± 2.2% | 9.1% ± 1.6% | 2.1% ± 1.1% | 10.6% ± 4.1% |
NR noise rate, ACC label accuracy, CMR correct modification rate, EMR error modification rate, MR miss rate, FLR false labeling rate.
RETFound classification performance
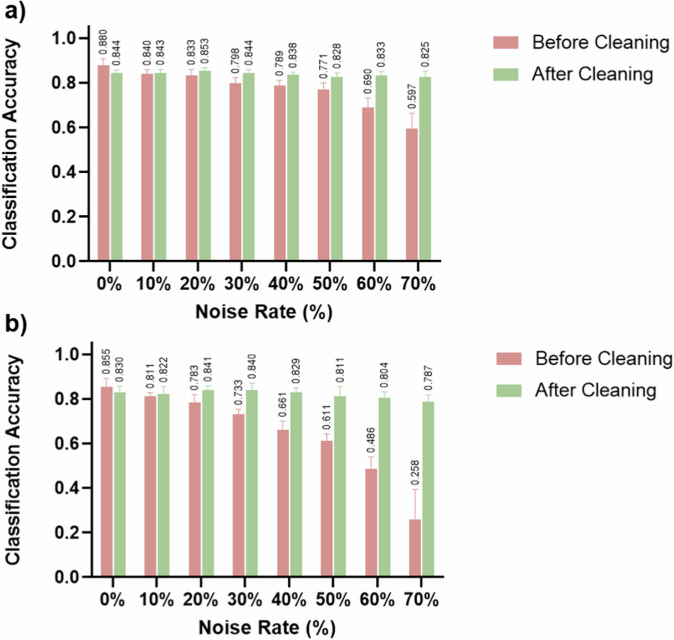
Noise in datasets resulted in decreased classification accuracy of the RETFound model (Fig. 4). The noisier dataset, the lower the classification accuracy. Even a 10% noise rate resulted in a 4.0 and 4.4% decrease in classification accuracy in CFP and OCT, respectively. Except in the clean datasets (0% noise rate), the classification accuracy of the RETFound improved using the datasets after six iterations of label cleaning compared to that before cleaning, with the classifying accuracies increased by 0.3 to 52.9%, with all p values <0.05 except in 10% noise rate. The noisier datasets, the more improvement in classification accuracy after label cleaning. The classification accuracy achieved using the cleansed datasets was comparable to that of the noise-free dataset in both CFP and OCT. This consistent trend was also observed across precision, recall, and F1-score (Supplementary Figs. 2, 3, 4).
Fig. 4. Classification accuracy of RETFound finetuning before and after cleaning.
The original datasets with noisy labels and the ones after 6-iterations of label cleaning were used to fine-tune the RETFound model and subsequently test on the hold-out testing sets, respectively. The classification accuracies in both CFP (a) and OCT (b) demonstrated notable enhancements, with improvements becoming more evident as the noise rates increased. Except in the clean datasets (0% noise rate), the classification accuracy of the RETFound improved using the dataset after cleaning compared to that before cleaning.
External validation on public datasets
The results of external validation on EyePACS and APTOS-2019 have demonstrated that Cleanlab is capable of significantly improving both label accuracies and dataset quality scores in real-world unbalanced settings. A substantial collection of 14,358 CFPs from EyePACS and 3178 CFPs from APTOS-2019 underwent multi-grader re-annotation to refine the datasets. The detailed statistics of the re-annotation and distribution of label noise within these datasets are presented in Supplementary Table 1. As illustrated in Supplementary Table 2, after six iterations, the label accuracies increased from 0.916 to 0.929 for EyePACS, and from 0.906 to 0.928 for APTOS-2019. Additionally, the DQSs improved from 0.9547 to 0.9950 for EyePACS and from 0.9755 to 0.9987 for APTOS-2019. Following the DQS-guided strategy, during the third round of cleaning for EyePACS, the DQS decreased, so the final label accuracy from the previous round was used, which was 0.930 in the final. In contrast, after three rounds of cleaning, the DQS of APTOS-2019 exceeded the threshold of 0.9965, allowing cleaning to be halted in the third round, resulting in a final label accuracy of 0.924. These results further confirm the effectiveness and generalizability of this approach.
In cross-validation using EyePACS and APTOS-2019 with updated consensus labels as ground truth and fine-tuned on RETFound, a marked increase in AUC after cleaning was noted, with the values increasing from 0.972 ± 0.008 to 0.979 ± 0.004 for EyePACS and from 0.785 ± 0.019 to 0.810 ± 0.021 for APTOS-2019 (both with p = 0.036), as illustrated in Supplementary Table 3. A modest rise in classification accuracy was also observed; while this did not achieve statistical significance for EyePACS (p = 0.105), it did for APTOS-2019 (P = 0.006). Additionally, we observed consistent upward trends in precisions, recalls, and F1-scores across both datasets. This trend of a slight enhancement after label cleaning aligns with our findings in the private CFP datasets with a 10% noise level.
Furthermore, to demonstrate the effectiveness of Cleanlab, two advanced projects in the field of unstructured data noise, namely Docta and Fastdup, were included for comparison. We have conducted label cleaning on the EyePACS and APTOS-2019 datasets using the same methodology. After six iterations of label cleaning and data updating, Cleanlab achieved an increase in label accuracies from 0.916 to 0.929 on the EyePACS dataset and from 0.906 to 0.928 on the APTOS-2019 dataset, as illustrated in Supplementary Fig. 5. In contrast, Docta’s performance significantly dropped to 0.836 on EyePACS but increased to 0.912 on APTOS-2019. Fastdup reached a plateau after two rounds, with accuracy decreasing to 0.899 on EyePACS and 0.901 on APTOS-2019. These results suggest that Cleanlab demonstrates a superiority over other state-of-the-art methods.
Discussion
In this study, we found that Cleanlab can efficiently correct the label errors automatically for both multi-category CFP and OCT datasets, with a low risk of missed or mis-correction. Dataset cleaning using Cleanlab can improve the performance of multi-category classification of subsequent models. Furthermore, we showed that the DQS can be used to predict the accuracy of the label, and we set up a strategy using DQS to guide the start and stop of the iteration. Finally, we demonstrated the effectiveness of Cleanlab through external validation on two public datasets.
Label noise had a substantial impact on the model’s performance. Although deep learning may have some robustness against label noise22,23, our study revealed that 10% of the noise labels resulted in a 3.6 and 4.5% decrease in classification accuracy for CFP and OCT, respectively, which deteriorated further as the noise proportion increased. This finding is consistent with the previous report that only a 6% increase in label noise on ImageNet can seriously affect the rankings of resnet-18 and resnet-50, highlighting the crucial importance of label cleaning15.
Label noise is a kind of uncertainty in the field of machine learning. There are two types of uncertainty: aleatoric (data) and epistemic (model). In our previous study, we addressed the challenge of epistemic uncertainty by developing an uncertainty-inspired open set learning model, which can quantify the uncertainty and use it to detect out-of-distribution data, including images with atypical features of retinal disease, non-target categories not included in training and non-CFP images24. In the current study, we resolve the challenge of aleatoric uncertainty using automatic confident learning, which is based on the principles of pruning noisy data, counting with probabilistic thresholds to estimate noise, and ranking examples to train with confidence.
Data cleaning can be performed manually or automatically. Manual re-labeling is very laborious and time-consuming and cannot resolve the problem of graders’ subjectivity17. Cleanlab can automatically identify the data with higher uncertainty in labels and resolve the problem of labor and subjectivity. The identified images with label issues can be managed using various strategies: discarding, manual correction, and automated correction. The images with labeling issues can be removed from the datasets, and the classification models can be retrained15. However, due to the scarcity of medical data, the images identified with labeling issues can be reinspected manually in an interactive manner. While this approach aims to conserve resources, it still requires workforce and time if there is a high proportion of noisy data. Therefore, we directly adopted the suggested label generated by Cleanlab to replace the original label during each round of label cleaning, thereby maximizing the utilization of each valuable medical image and minimizing the need for a workforce. Our results illustrated that cleaned datasets are comparable to initial noise-free datasets in CFP and only mildly inferior in OCT for classification accuracy of the downstream RETFound model, even for the datasets with noise rates up to 70%. These results confirmed the efficiency of our approach, fully automatic multi-iteration data cleaning. This approach is successful in both CFP and OCT datasets, suggesting its efficiency is not dependent on the modality of images.
We also accessed the limitations and risks of automated multi-iteration data cleaning. There may be 0.5–2.8% of missed detection and 0.4–10.6% of error modification rate in all label errors (Tables 1, 2). Consequently, achieving a completely clean dataset is not feasible with this approach. One notable risk involves misidentifying actual labels as label issues and subsequently mis-modifying them. Our observations reveal that label accuracy diminishes under two conditions: initial cleaning of noise-free datasets and repeated excessive cleaning of noisy datasets. For instance, in noise-free datasets, label accuracy may decrease from 100% to approximately 99.3 or 99.4% after six iterations of cleaning. Conversely, datasets initially containing 5 or 10% noise may experience an initial accuracy increase with cleaning iterations, followed by a subsequent decrease. Throughout, Cleanlab consistently exhibited DQS with minimal flagged label errors in both situations. Ambiguous cases with overlapping category representations likely contribute to higher rates of false labeling compared to correct modifications of pseudo-labels, thereby impacting label accuracy. To mitigate this issue, we propose using DQS as a guide for determining when to initiate and cease label cleaning, given its strong correlation with label accuracy. Alternatively, in cases where few label errors are flagged, manual confirmation based on the ranked label scores of each image can be employed. This iterative human-machine feedback loop ensures efficient validation of Cleanlab’s suggested changes, maintaining high label accuracy without excessive time or effort.
To our knowledge, this study is the first to investigate the efficiency and risk of automatic data cleaning in retinal images. Our fully automatic approach further minimizes the need for manual re-inspection. This method was applied in two different modalities of images, suggesting its potential applicability to other medical specialties or imaging modalities. CFP and OCT are the most widely used screening methods in the context of fundus diseases despite having completely different imaging principles and effects. Notably, the flexibility of Cleanlab, which is not tightly bound to specific data modalities or models, holds great promise and has garnered interest from clinical professionals21. This indicates that it is more data-centric than strongly model-dependent, showcasing its robustness and generalization capabilities.
The application of automated label cleaning in medicine holds substantial promise. The code-free or simple implementation nature makes this method accessible to medical practitioners who do not have profound knowledge of coding. By reducing annotation workload and improving efficiency, medical practitioners can benefit significantly from the ability to detect and rectify label errors in a timely and automated manner. This enhances dataset quality and improves model performance, ultimately advancing the development and clinical implementation of artificial intelligence in healthcare. This method can be applied to both public and private datasets to optimize the datasets for deep learning algorithm training. It also has the potential to identify misdiagnosis in private datasets and prevent medical risk.
It is imperative to acknowledge the limitations of our study. Firstly, the lesions of categories included were typical, representative, and distinctive to some extent, yet they do not fully encapsulate the intricate clinical landscape. Therefore, a multi-label, multi-class study instead of a single-label one would be more appropriate. Secondly, we only included images obtained from a single piece of equipment. Further investigation is necessary to assess the applicability of our methodology across multiple devices. Thirdly, the category distribution within the dataset was balanced and did not align with the intricacies of clinical epidemiology. Hence, an evaluation of our approach using real-world, large-scale clinical datasets is warranted. Finally, the included datasets were relatively small. A more comprehensive evaluation would be possible with larger datasets. In future studies, we would intend to expand our research to incorporate more extensive datasets.
In conclusion, the utilization of Cleanlab in the context of CFP and OCT emerges as an efficacious and low-risk method in the realm of label error detection and correction, which would further improve the performance of the classification model trained with these datasets. The DQS-guided strategy may help to prevent the risk of over-cleaning.
Methods
Dataset construction and pseudo-label strategy
All image data were gathered from the Joint Shantou International Eye Center (JSIEC). This study was approved by the Ethics Committee of JSIEC following the principles of the Helsinki Declaration. It was conducted retrospectively, and all data were deidentified to ensure the utmost protection of patient privacy without the need for patient’s informed consent forms.
The CFPs were captured using a TRC-NW8 Mydriatic Retinal Camera (Topcon, Japan), with images at a resolution of 3046*2574. A total of eight distinct classifications were compiled for label cleaning. These classifications encompassed age-related macular degeneration (AMD), central serous chorioretinopathy (CSCR), diabetic retinopathy (DR), normal fundus(N), pathological myopia (PM), retinal detachment (RD), retinitis pigmentosa (RP), and retinal vein occlusion (RVO). The OCTs were acquired using the Cirrus HD-OCT 5000 (Zeiss, Germany), with images at a resolution of 1389*926. A collection of seven disease classifications was amassed, including AMD, CSCR, DR, epiretinal membrane (ERM), N, PM, and RVO.
To ensure sample diversity, one image per patient was randomly selected for inclusion in this study. Initially, 2296 CFPs and 1340 OCTs in total were collected following protocols detailed in Supplementary Table 4. Each image underwent independent annotation by two ophthalmologists in a blinded manner, with only data having consistent labels included in the study. As a result, 33 inconsistent CFPs (1.44% of the total CFPs) and 24 inconsistent OCTs (1.79% of the total OCTs) were excluded to ensure the integrity of the data. Within the consensus-labeled images, the CFP experimental set for data cleaning were structured to include eight disease categories, with each category containing 200 images from 200 unique patients. Similarly, the OCT experimental sets were composed of seven disease categories, each with 140 B-scans from 140 distinct patients. To benchmark the classification accuracy of the RETFound model, we additionally curated two separate hold-out datasets, consisting of 663 CFPs and 336 OCTs sourced from a unique cohort distinct from the experimental sets. These datasets were noise-free and specifically reserved for comparative testing, providing a robust framework for evaluating the model’s performance. A comprehensive data distribution is provided in Supplementary Table 5. Subsequently, all included images were resized to (224, 224) and paired with their corresponding single-labels.
To evaluate the efficacy of Cleanlab in detecting label issues in CFP and OCT images, we employed a proportional pseudo-label strategy, as illustrated in Fig. 1. Initially, a random selection of 5% of images from each category was evenly redistributed among the remaining classes. This approach allowed for the creation of 15 noisy gradients for both CFP and OCT, ranging from noise-free (0% noise rate) to very noisy (70% noise rate) in a progressive manner. To mitigate the potential sampling errors, we performed triple sampling at each step of adding 5% noise, resulting in the formation of 45 sub-datasets in total.
Data cleaning using cleanlab
Cleanlab excels at detecting common real-world issues such as label errors, outliers, and near duplicates. To address the intricate problem of label noise, we implemented Cleanlab, which uses Confidence Learning principles to infer and correct label errors. Cleanlab can estimate the probability of each label’s correctness and identifies potentially incorrect labels by using cross-validation to compute out-of-sample predicted class probabilities and using trained models to generate feature embeddings (numeric vector representations) for all the image data, subsequently updating the label information within the dataset. The code was downloaded from Cleanlab’s GitHub repository (https://github.com/cleanlab/cleanlab) and implemented on the public platform PyTorch.
In our study, we utilized Cleanlab iteratively to inspect the dataset and rectify mislabeled errors. The parameter settings were configured based on the default settings of the framework, employing the Swin Transformer model trained on varied noisy groups. The Swin Transformer, known for its ability to handle long-range dependencies in images through self-attention mechanisms, was utilized to compute predicted class probabilities and extract feature embeddings. Specifically, the hyperparameters were as follows: the batch size was set to 32, the learning rate was 0.0001, the number of epochs was 10, the number of folds was 5, the patience level was set to 2, the optimization algorithm used was Adam, and the loss function utilized was the cross-entropy loss.
After completing model training, the predicted class probabilities and feature embeddings were loaded into Datalab, a component of Cleanlab. This tool was used to inspect the dataset for potential label issues. A comprehensive audit of the overall dataset quality score (DQS) was conducted to assess the database’s reliability regarding incorrect labeling, with a higher DQS indicating a cleaner dataset. Additionally, Datalab can flag suspicious images—those with a high likelihood of being mislabeled—quantified by a numeric label score ranging from 0 to 1. A lower label score suggests a lower label quality and a greater likelihood of mislabeling. Moreover, Datalab can suggest an alternative label, known as the predicted category, which Cleanlab deems more suitable for the data point than the original label. It corresponds to the predicted class probability of the most likely class in these multi-class datasets.
Multi-iteration cleaning and DQS-guided cleaning
At first, six sequential rounds of unsupervised label cleaning, data updating, and model retraining were conducted using Cleanlab without manual confirmation. The means and standard deviations of the label accuracies and DQSs of the triple sampling were calculated after each cleaning iteration.
In real-world implementation, the label may be unreliable, so the label accuracy or noise rate is not available. To optimize the number of iterations, we developed a DQS-guided strategy. Linear regression was used to investigate the relationship between label accuracy and DQS. Furthermore, we defined a threshold of label accuracy = 0.98 to categorize datasets into two classes: those requiring further cleaning (label accuracy <0.98) and those that are sufficiently clean and do not require additional cleaning (label accuracy ≥ 0.98). This binary classification, predicated on the defined threshold, facilitated the conversion of the continuous label accuracy metric into a binary format amenable for receiver operating characteristic (ROC) curve analysis. This approach enabled us to explore the capacity of dataset quality score (DQS) to predict label accuracy and to ascertain the DQS threshold that would trigger the initiation of the label-cleaning process. The criteria for terminating the label cleaning iteration were set when the DQS decreased or remained unchanged after cleaning, and the dataset before this iteration was used. The numbers of iterations were recorded for each dataset with different noise rates.
RETFound finetuning and classification accuracy
To investigate the effect of dataset noise on model performance, we employed the pre-trained RETFound foundation model for transfer learning25. We fine-tuned models on datasets before and after label cleaning separately using PyTorch and Nvidia Geforce RTX 2080ti GPU (11 G), and compared their best-predicting accuracy on the same hold-out testing set. Adam was chosen as the optimizer, with an initial learning rate and weight decay both set to 0.0001. The number of epochs was set to 10, while the batch size was set to 8. The images were randomly divided into training and validation sets at an 8:2 ratio, so the validation sets may also contain noisy labels. Models with higher validation accuracy were saved and further tested on the same hold-out testing set, which are free of noise.
The best testing accuracies before and after cleaning were recorded and compared. To mitigate sampling errors that may arise from dataset allocating and noise distribution, the above steps were then repeated three times using different random seeds, and their means and standard deviations of evaluation indicators were calculated and compared using a two-sided Wilcoxon signed rank test, depending on the distribution of data.
External validation on public datasets
Furthermore, we conducted external validation on two public datasets, EyePACS and APTOS-2019, which were initially considered noisy. The original labels for these datasets were annotated by a single individual, resulting in numerous subjective errors. To improve label accuracy, previous research organized a group of doctors to re-annotate EyePACS and open-source detailed image labels17. The process of data preparation for the EyePACS dataset was illustrated in Supplementary Fig. 6. The original datasets comprising 88,702 were obtained from Kaggle, while 57,213 were relabeled17. The diabetic retinopathy was classified into DR grades 0–4, while other diagnoses as “others” and used a majority-voting principle to establish ground truth. After excluding 3450 single-annotation images, 53,763 were retained. Out of these, 2898 images could not reach a consensus, and 36,508 were predominantly classified as “others”, leading to 14,358 images with consensus DR grades 0–4 in final. For our study, we invited four ophthalmologists to thoroughly re-annotate APTOS-2019 according to the International Clinical Diabetic Retinopathy Disease Severity Scale. Adhering to the majority-voting consensus approach, only labels with over 75% agreement among annotators were accepted as the definitive annotations for the images. This criterion allowed us to include 3178 out of the total 3662 images in our analysis. Diabetic retinopathy was further classified as either non-referable DR (grades 0–1) or referable DR (grades 2–4).
Following the application of Cleanlab, we documented the label accuracy and dataset quality score after each cleaning cycle for both datasets. Consistent with our approach to the private dataset, we employed both the Six-Iteration strategy and the DQS-guided strategy. For the DQS-guided strategy, we applied a threshold derived from the experimental set of our private CFP data, setting the DQS cutoff at 0.9965, as shown in Fig. 3 c.
To assess the impact of label cleaning on model classification performance, we conducted cross-validation using the EyePACS dataset before and after label cleaning, fine-tuning the RETFound model on these datasets, and then testing the model using the APTOS-2019’s consensus labels. Similarly, we also fine-tuned RETFound on APTOS-2019 and tested with EyePACS’s consensus labels.
To further illustrate the effectiveness of the Cleanlab-based approach over existing methods, we compared Cleanlab’s performance with two leading projects addressing: Docta26 and Fastdup. Like the approach using Cleanlab, multi-iterations were conducted without manual confirmation, while label accuracies were calculated after each cleaning iteration.
Evaluation indicator
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
| 6 |
| 7 |
| 8 |
| 9 |
Label accuracy was defined as the proportion of images in the dataset that have been correctly labeled, which reflects the overall results and be calculated in accordance with formula (1). Actual labels refer to the true, verified labels for the images, which may be obtained through expert annotation and consensus labeling, serving as the benchmark for evaluating the accuracy of the predicted labels generated by different approaches. Correct labels in formulae (1) are defined as the labels within the dataset that are consistent with the actual labels. To comprehensively assess the impact and potential risks associated with label error detection, we employed formulae (2–5) to calculate the miss rate, correct modification rate, error modification rate, and false labeling rate, where label issues in formulae (2) indicates the examples, whose given label is estimated to be incorrect and flagged by Cleanlab.
Furthermore, to evaluate the impact of label cleaning on model performance, we computed classification accuracy, precision, recall and F1-score employing formulae (6-9). The TP, FP, TN, and FN stand for true positive, false positive, true negative, and false negative, respectively.
Supplementary information
Acknowledgements
This research is supported by the Internal research project of Joint Shantou International Eye Center (21-003 to T.L.), the National Key R&D Program of China (2018 YFA0701700 to H.C.), Shantou Science and Technology Program (190917085269835 to H.C.), Department of Education of Guangdong Province (2024ZDZX2024 to H.C.).
Author contributions
T.L.: conceptualization, methodology, data collection, and writing the original draft. M.W.: software, experimental deployment, and writing the original draft. A.L. and X.M.: image annotation, reviewing, and editing. H.L.: data collection, image annotation, and reviewing. Y.T.: Reviewing and editing. H.C.: conceptualization, supervision, clinical assessment and curation, clinical support, reviewing, and editing.
Data availability
Data sets supporting the findings of this study are not publicly available due to the confidentiality policy of the Chinese National Health Council and institutional patient privacy regulations. However, they are available from the corresponding authors upon request. For replication of the findings and/or further academic and AI-related research activities, data may be requested from the corresponding author H.C. (drchenhaoyu@gmail.com), and any requests will be responded to within 10 working days. Two public datasets are available as follows. EyePACS: http://www.eyepacs.com/data-analysis. Corresponding multi-labels are available from supplementary material to the article17. APTOS-2019: https://www.kaggle.com/datasets/mariaherrerot/aptos2019?select=train_1.csv.
Code availability
Codes for label cleaning are available at https://github.com/cleanlab/cleanlab. Codes for RETFound finetuning and testing are available at https://github.com/LooKing9218/RetClean. Codes for Docta are available at https://github.com/Docta-ai/docta. Codes for Fastdup are available at https://github.com/visual-layer/fastdup.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tian Lin, Meng Wang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-024-01424-x.
References
- 1.Steinmetz, J. D. et al. Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Glob. Health9, 144–160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu, H. et al. Economic evaluation of combined population-based screening for multiple blindness-causing eye diseases in China: a cost-effectiveness analysis. Lancet Glob. Health11, 456–465 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Quellec, G. et al. Automatic detection of rare pathologies in fundus photographs using few-shot learning. Med Image Anal.61, 101660 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Yim, J. et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat. Med.26, 892–899 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Ruamviboonsuk, P. et al. Real-time diabetic retinopathy screening by deep learning in a multisite national screening programme: a prospective interventional cohort study. Lancet Digit Health4, 235–244 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Moraes, G. et al. Quantitative analysis of OCT for neovascular age-related macular degeneration using deep learning. Ophthalmology128, 693–705 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, C. Y. et al. Big data in ophthalmology. Asia Pac. J. Ophthalmol.9, 291–298 (2020). [DOI] [PubMed]
- 8.Dong, L. et al. Artificial intelligence for screening of multiple retinal and optic nerve diseases. JAMA Netw. Open5, e229960 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cen, L. P. et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat. Commun.12, 4828 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhardt, M. et al. Active label cleaning for improved dataset quality under resource constraints. Nat. Commun.13, 1161 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majkowska, A. et al. Chest radiograph interpretation with deep learning models: assessment with radiologist-adjudicated reference standards and population-adjusted evaluation. Radiology294, 421–431 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Xiao, T., Xia, T., Yang, Y., Huang, C. & Wang X. Learning from massive noisy labeled data for image classification. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2691-2699 (2015).
- 13.Song, H., Kim, M. & Lee, J. G. SELFIE: refurbishing unclean samples for robust deep learning. In International Conference on Machine Learning. 5907–5915 (2019).
- 14.Lee, K. H., He, X., Zhang, L. & Yang, L. CleanNet: transfer learning for scalable image classifier training with label noise. In Proc. IEEE conference on computer vision and pattern recognition. 5447–5456 (2018).
- 15.Northcutt, C. G., Athalye, A. & Mueller J. Pervasive label errors in test sets destabilize machine learning benchmarks. Preprint at arXiv:2103.14749 (2021).
- 16.Gao, M. et al. Bayesian statistics-guided label refurbishment mechanism: Mitigating label noise in medical image classification. Medical Physics49.9, 5899–5913 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Ju, L. et al. Improving medical images classification with label noise using dual-uncertainty estimation. IEEE Trans. Med. Imaging41, 1533–1546 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Arpit, D. et al. A closer look at memorization in deep networks. Int. Conf. Mach. Learn. 233–242 (2017).
- 19.Li, J., Zhang, M., Xu, K., Dickerson, J. & Ba, J. How does a neural network’s architecture impact its robustness to noisy labels. Adv. Neural Inf. Process. Syst.34, 9788–9803 (2021). [Google Scholar]
- 20.Han, B. et al. A survey of label-noise representation learning: Past, present and future. Preprint at arXiv:2011.04406 (2020).
- 21.Northcutt, C., Jiang, L. & Chuang, I. Confident learning: estimating uncertainty in dataset labels. J. Artif. Intell. Res.70, 1373–1411 (2021). [Google Scholar]
- 22.Zhou, Y. et al. A foundation model for generalizable disease detection from retinal images. Nature622, 156–163 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, W. R. et al. Understanding generalization through visualizations. Preprint at arXiv: 1906.03291 (2019).
- 24.Rolnick, D., Veit, A., Belongie, S. & Shavit, N. Deep learning is robust to massive label noise. Preprint at arXiv:1705.10694 (2017).
- 25.Wang, M. et al. Uncertainty-inspired open set learning for retinal anomaly identification. Nat. Commun.14, 6757 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu, Z., Wang, J., Cheng, H. & Liu, Y. Unmasking and improving data credibility: A study with datasets for training harmless language models. Preprint at arXiv:2311.11202 (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets supporting the findings of this study are not publicly available due to the confidentiality policy of the Chinese National Health Council and institutional patient privacy regulations. However, they are available from the corresponding authors upon request. For replication of the findings and/or further academic and AI-related research activities, data may be requested from the corresponding author H.C. (drchenhaoyu@gmail.com), and any requests will be responded to within 10 working days. Two public datasets are available as follows. EyePACS: http://www.eyepacs.com/data-analysis. Corresponding multi-labels are available from supplementary material to the article17. APTOS-2019: https://www.kaggle.com/datasets/mariaherrerot/aptos2019?select=train_1.csv.
Codes for label cleaning are available at https://github.com/cleanlab/cleanlab. Codes for RETFound finetuning and testing are available at https://github.com/LooKing9218/RetClean. Codes for Docta are available at https://github.com/Docta-ai/docta. Codes for Fastdup are available at https://github.com/visual-layer/fastdup.