Abstract
While sleep is important, our understanding of its molecular mechanisms is limited. Over the last two decades, protein kinases including Ca2+/calmodulin-dependent protein kinase II (CaMKII) α and β have been implicated in sleep regulation. Of all the known mouse genetic mutants, the biggest changes in sleep is reported to be observed in adult mice with sgRNAs for Camk2b injected into their embryos: sleep is reduced by approximately 120 min (mins) over 24 h (hrs). We have reexamined the sleep phenotype in mice with either Camk2a or Camk2b gene knocked-out by conventional gene targetting. While the basal sleep is reduced in Camk2a knockout mice, it remains unaltered in Camk2b mutants. Knockout of either Camk2a or Camk2b reduces sleep rebound after deprivation, indicating their roles in sleep homeostasis. These results indicate the involvement of CaMKIIα in both basal sleep and sleep homeostasis while CaMKIIβ is mainly required physiologically for sleep homeostasis, serving as a stimulus for rigorous studies in the future.

Subject terms: Molecular neuroscience, Behavioural genetics
Sleep analysis in stable transgenic mouse strains reveals the involvement of CaMKIIα in basal sleep and homeostatic sleep rebound, and that of CaMKIIβ in homeostatic sleep rebound but not in basal sleep.
Introduction
Genetics has been very helpful for our understanding of molecular mechanisms of sleep regulation. Genetic mutants of mice have been particularly useful for revealing roles of specific genes involved in regulating sleep. The most famous mouse mutants with sleep phenotypes are perhaps mutants for orexin and its receptor1,2. They are required for maintaining wakefulness during daytime and loss of function (LOF) mutant mice (or dogs and humans) show characteristic narcoplepsy with cataplexy. Another well-known mouse gene is Sik3, whose gain of function (GOF) mutation was discovered to increase sleep3–5. We have found reduced daytime rapid eye movement (REM) sleep in SIK3 LOF mutants6, consistent with the sleep increase phenotype of SIK3 GOF mutant mice3. However, the extents of sleep reduction in LOF mutants of either orexin or SIK3 were moderate: approximately 30 min (mins) per 24 h (hrs)6.
Other protein kinases implicated in sleep include: protein kinase A7–9, the extracellular signal-regulated kinase10–12, the adenosine monophosphate-activated protein kinase13–15, calmodulin-dependent kinase II (CaMKII) α and β16–18, c-Jun N-terminal kinase19, and the liver kinase B (LKB1)20. The most prominent changes in sleep was reported in Camk2b knockout mice17: a reduction of more than 120 min in 24 h. This change is bigger than changes in all other known mouse mutants1–6,9,12,18,20–33.
It thus seemed that Camk2b knockout mice are the most important mutant mice for further studies of sleep regulation. We constructed the embryonic knockout mice and tested the sleep behavior after backcross, yet could not validate the result from the functional quick screen reported previously17,18.
Results
Genetic knockout of Camk2a and Camk2b by conventional means
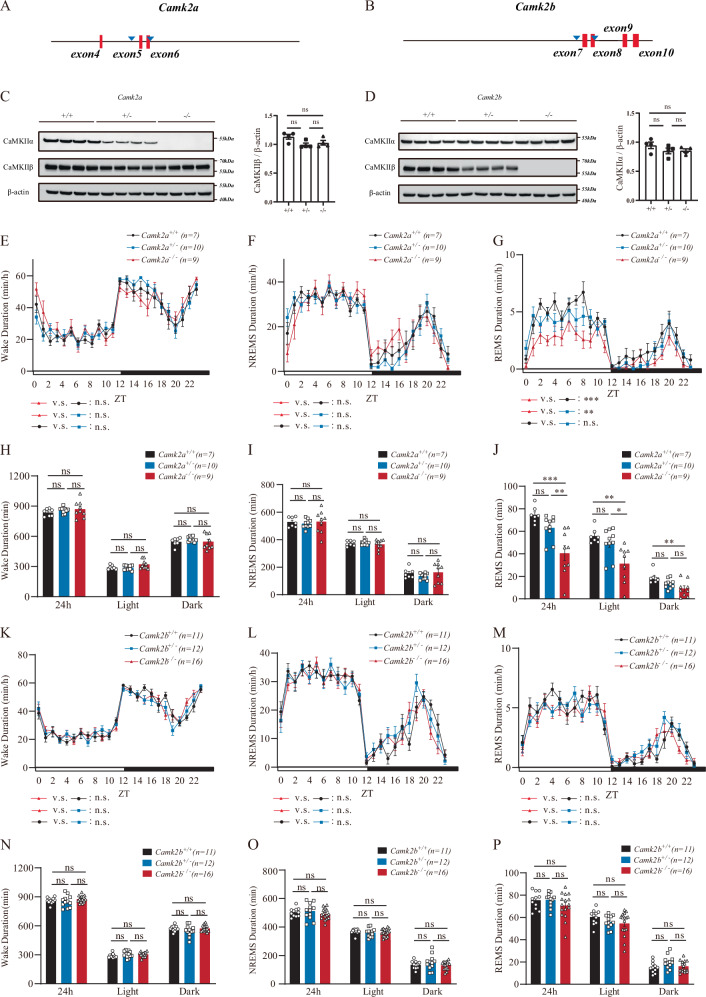
In our efforts to study protein phosphorylation in sleep, we generated Camk2a and Camk2b knockout mice (Fig. 1A, B). While the previous paper reporting sleep reduction used embryonic injection of single guide (sg) RNAs to create LOF Camk2a and Camk2b mutants for functional quick screens17,18, we used sgRNAs for gene targetting to create genetically transmissible mutants and crossed them for at least six generations before functional testing. We deleted exons 5 and 6 of Camk2a which encoded amino acid residues (aa) 92 to 137, with an additional frameshift mutation of CCU (Pro) to CUG (Leu), resulting in truncation thereafter (Fig. 1A). The shifted frame covered all aa sequences targetted for deletion in the previous paper17. We deleted exons 7 and 8 of Camk2b encoding aa 139 to 200, with an additional frameshift mutation from GGGGUG (Gly-Val) to GGGUGA (Gly-*), which also covered all sequences deleted in the the previous paper17. Western blot analysis with antibodies recognizing CaMKIIα and CaMKIIβ proteins confirmed the effectiveness of Camk2a and Camk2b gene targeting in heterozygous mutant mice and homozygous mutant mice. (Fig. 1C, D) And there’s no compensation in either CaMKIIα or CaMKIIβ knockout mice by the other isoform. (Fig. 1C, D). As behavioral controls, we tested Camk2a and Camk2b knockout mice in Morris water maze and confirmed that they were defective (Supplementary Fig. S1A, B).
Fig. 1. Generation of Camk2a and Camk2b Mutant Mice and Their Sleep Phenotypes.
A A schematic diagram illustrating the strategy using a pair of sgRNAs to mediate the deletion of exons 5 and 6 in Camk2a with sgRNAs CTGGATCACGAAGACCCCTG and ACCTGAAGGTGAGTAACCCT. B A schematic diagram for the deletion of exons 7 and 8 in Camk2b with sgRNAs TTGCAGTCACCTATGTCACG and TGTGTGAGGGGAAACACCTG. C Western blot analysis of CaMKIIα and CaMKIIβ from the brain lysates of mice with the genotypes of Camk2a−/−, Camk2a+/− and Camk2a+/+ (4 mice for each genotype shown here). Grayscale values of each genotype were analyzed. D Western blot analysis of CaMKIIα and CaMKIIβ from the brain lysates of mice with the genotypes of Camk2b−/−, Camk2b+/− and Camk2b+/+ (4 mice for each genotype shown here). Grayscale values of each genotype were analyzed. E–J Analysis of sleep in Camk2a+/+ (n = 7, black curve and bar), Camk2a+/− (n = 10, blue curve and bar) and Camk2a−/− (n = 9, red curve and bar) mice. Profiles showing wake time of each hour in min/hr. E profiles of NREM sleep (F), or profiles of REM sleep (G). The x axis shows zeitgeber time (ZT) with the white box indicating light phase (or daytime) and black box dark phase (or nighttime). Twenty four hours, daytime and nighttime durations of wake (H), NREM sleep (I) or REM sleep (J) shown in mins. K–P Sleep of Camk2b+/+ (n = 11, black curve and bar), Camk2b+/− (n = 12, blue curve and bar) and Camk2b−/− (n = 16, red curve and bar) mice. Profiles of wake time (K), NREM sleep (L) or REM sleep (M). Twenty four hours, daytime and nighttime durations of wake (N), NREM sleep (O) and REM sleep (P). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001; mean ± standard error of the mean (mean ± SEM). Two-way ANOVA (E, F, G, K, L, M); One-way ANOVA (C, D, H, I, J, N, O, P).
No significant change in basal sleep observed for Camk2b−/− mice
We then analyzed the sleep phenotypes in mutant mice according to procedures we have published previously6,20. In Camk2a knockout mice, while no significant change was observed in total wake duration (Fig. 1E and H, Supplementary Fig. S2A and S2B), daytime or nighttime non-REM (NREM) sleep (Fig. 1F and I, Supplementary Fig. S2E and S2F), REM sleep was found to be reduced in daytime for 25.5 min and nighttime for 9.7 min (Fig. 1G and J, Table 1, Supplementary Fig. S2I and S2J). These were not very different from the total of approximately 50 min over 24 h reported previously.
Table 1.
Total time spent in different sleep-wake states of Camk2a knockout male mice
| Total time | 24 hours (Mean ± SEM, min) | Light phase (Mean ± SEM, min) | Dark phase (Mean ± SEM, min) | |
|---|---|---|---|---|
| Sleep | Camk2a+/+ | 603.0 ± 13.9 | 428.6 ± 7.9 | 174.4 ± 15.4 |
| Camk2a+/− | 579.1 ± 9.1 | 430.7 ± 7.2 | 148.4 ± 8.1 | |
| Camk2a−/− | 570.3 ± 27.0 | 397.6 ± 14.7 | 172.6 ± 24.5 | |
| NREM | Camk2a+/+ | 528.3 ± 13.7 | 372.8 ± 7.4 | 155.5 ± 8.2 |
| Camk2a+/− | 516.0 ± 10.4 | 380.9 ± 5.7 | 135.1 ± 7.3 | |
| Camk2a−/− | 529.7 ± 26.9 | 366.3 ± 11.5 | 163.4 ± 23.7 | |
| REM | Camk2a+/+ | 74.7 ± 2.7 | 55.8 ± 2.6 | 18.9 ± 2.0 |
| Camk2a+/− | 63.1 ± 3.3 | 49.8 ± 4.0 | 13.3 ± 1.3 | |
| Camk2a−/− | 40.5 ± 6.6 | 31.3 ± 5.3 | 9.2 ± 2.4 | |
| Wake | Camk2a+/+ | 837.0 ± 13.9 | 291.4 ± 7.9 | 545.6 ± 15.4 |
| Camk2a+/− | 860.9 ± 9.1 | 289.3 ± 7.2 | 571.6 ± 8.1 | |
| Camk2a−/− | 869.7 ± 27.0 | 322.4 ± 14.7 | 547.4 ± 24.5 |
However, when we analyzed the sleep phenotype of Camk2b knockout mice (Fig. 1K–P, Table 2, and Supplementary Fig. S2C, S2D, S2G, S2H, S2K, S2L), no significant change in sleep was observed: from total wake duration, to total sleep, daytime or nighttime NREM sleep or REM sleep.
Table 2.
Total time spent in different sleep-wake states of Camk2b knockout male mice
| Total time | 24 hours (Mean ± SEM, min) | Light phase (Mean ± SEM, min) | Dark phase (Mean ± SEM, min) | |
|---|---|---|---|---|
| Sleep | Camk2b+/+ | 578.4 ± 9.7 | 429.4 ± 6.3 | 149.0 ± 9.0 |
| Camk2b+/− | 587.3 ± 17.4 | 416.5 ± 9.6 | 170.8 ± 16.5 | |
| Camk2b−/− | 562.8 ± 8.5 | 415.6 ± 6.0 | 147.2 ± 7.9 | |
| NREM | Camk2b+/+ | 503.1 ± 9.1 | 369.2 ± 5.6 | 133.9 ± 8.2 |
| Camk2b+/− | 511.6 ± 16.4 | 360.6 ± 8.7 | 151.0 ± 15.0 | |
| Camk2b−/− | 492.2 ± 9.5 | 361.1 ± 6.7 | 131.1 ± 7.2 | |
| REM | Camk2b+/+ | 75.4 ± 2.3 | 60.3 ± 2.0 | 15.1 ± 1.5 |
| Camk2b+/− | 75.7 ± 2.8 | 55.9 ± 1.7 | 19.8 ± 1.7 | |
| Camk2b−/− | 70.6 ± 2.8 | 54.5 ± 2.7 | 16.0 ± 1.2 | |
| Wake | Camk2b+/+ | 861.6 ± 9.7 | 290.6 ± 6.3 | 571.0 ± 9.0 |
| Camk2b+/− | 852.7 ± 17.4 | 303.5 ± 9.6 | 549.2 ± 16.5 | |
| Camk2b−/− | 877.2 ± 8.5 | 304.4 ± 6.0 | 572.8 ± 7.9 |
Requirement of Camk2a and Camk2b in sleep homeostasis
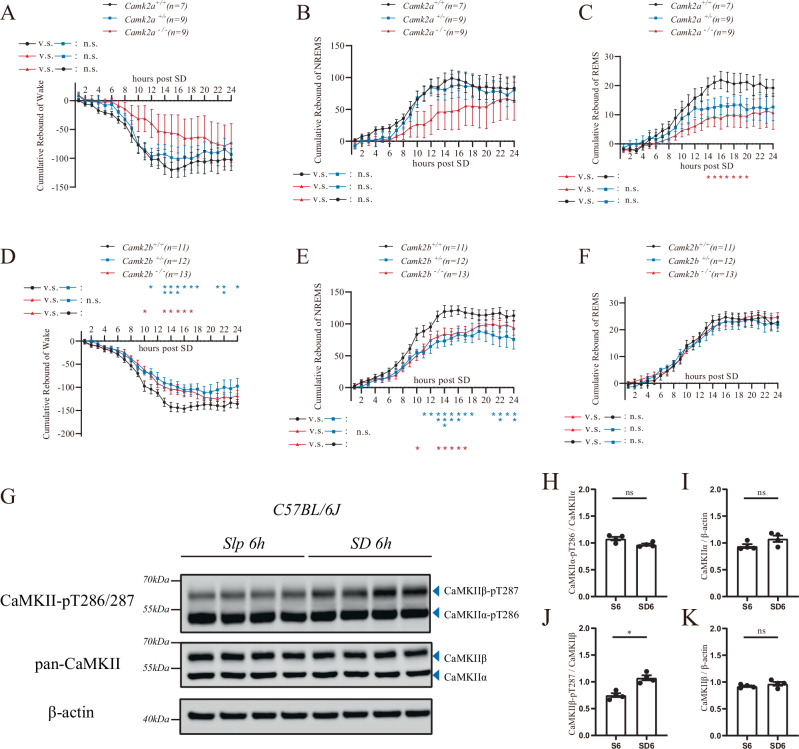
We also conducted sleep deprivation (SD) experiment and analyzed the rebound after 6 h of SD in both mutant mice. In Camk2a knockout mice, there were no significant alterations in the NREM rebound or wake reduction (Fig. 2A, B). A slight decrease in REM rebound was found after 14 to 20 h; however, no significant differences were observed 24 h post-SD (Fig. 2C). In Camk2b knockout mice, although cumulative NREM rebound and wake reduction were found to decrease slightly after 13 to 17 h, there were no significant differences in sleep rebound 24 h after SD comparing to wild-type mice (Fig. 2D, E). And no differences in cumulative REM rebound were detected (Fig. 2F).
Fig. 2. Involvement of CaMKIIα and CaMKIIβ in homeostatic sleep rebound.
A–C Cumulative wake reduction, and NREM, REM rebound of Camk2a+/+ (n = 7, black curve), Camk2a+/− (n = 9, blue curve) and Camk2a−/− (n = 9, red curve) after 6 h of SD. D–F Cumulative wake reduction, and NREM, REM rebound of Camk2b+/+ (n = 11, black curve), Camk2b+/− (n = 12, blue curve) and Camk2b−/− (n = 13, red curve) after 6 h of SD. G Western blot analysis of pan-CaMKII and CaMKII-pT286/287 from the brain lysates of C57BL/6 J mice with (SD 6 h) and without 6 h of SD (Slp 6 h). (4 mice for each group). H–K The grayscale value ratio CaMKIIα-pT286/CaMKIIα (H), CaMKIIα/β-actin (I), CaMKIIβ-pT287/CaMKIIβ (J) and CaMKIIβ/β-actin (K) were compared between Slp 6 h (S6) and SD 6 h (SD6). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; mean ± standard error of the mean (mean ± SEM). Two-way ANOVA (A–F). Mann–Whitney test (H–K).
We then analyzed the NREM delta power density of the mutants before and after SD. The NREM delta power during NREM sleep was significantly increased after SD in Camk2a+/+, Camk2b+/+, Camk2a+/− and Camk2b+/− mice (Supplementary Fig. S3A–D), while no remarkable change in NREM delta power density was observed in Camk2a−/− and Camk2b−/− mice (Supplementary Fig. S3E and S3F). The result suggested that in Camk2a−/− and Camk2b−/− mice, the homeostatic rebound in delta power after SD was attenuated, which was consistent with their reduction in homeostatic rebound sleep duration.
We further examined whether CaMKIIα and CaMKIIβ activities were regulated by SD. Autophosphorylation at CaMKIIα-T286 and CaMKIIβ-T287 are known to be indicative of their activities34. Immunoblot analyses showed that, while protein levels of CaMKIIα and CaMKIIβ were not significantly changed after a 6 h of SD (Fig. 2G, I and K), CaMKIIβ-pT287 phosphorylation was significantly increased (p = 0.0286) (Fig. 2J) but CaMKIIα-T286 phosphorylation was not significantly changed (Fig. 2H). At the present, it is unclear whether the absence of observed changes in CaMKIIα-T286 phosphorylation was due to the insensitivity of the antibody or the low percentage of phosphorylated CaMKIIα-T286 in the total amount of CaMKIIα in the brain, or actual no change in CaMKIIα phosphorylation.
Discussion
Our results have shown the involvement of CaMKIIα in basal sleep and homeostatic sleep rebound, and that of CaMKIIβ in homeostatic sleep rebound. We have not replicated the previous finding of a role for CaMKIIβ in basal sleep regulation. Both CaMKIIα and CaMKIIβ mutant mice did show learning deficits as expected. Our work also emphasize the importance of using traditional gene targetting and genetically crossing mice for several generations after the initial targetting.
There can be multiple explanations for the discrepancies between the previous results17 and our present results. As a quantitative trait, sleep is not only sensitive to the genetic background but also the environmental factors such as ambient temperature35, chow36, social conditions37 and environmental enrichment38; Differences in recording devices and surgery might also influence the phenotype. However, it would be unfriendly to the field if moderate discrepancies in such factors caused a difference in almost 2 h in basal sleep.
Another explanation would be the possibility that, while the previous study had used mutants quickly generated by embryonic injection of sgRNAs and functionally tested in the same generation, we have used the traditional, reliable and commonly accepted method of generating stable mutants which were functionally tested after a few generations. While the quick method has the advantage of time-saving, ours is the reliable method used by the vast majority of users.
Methods
Mouse stocks
WT C57 BL/6J mice (8 to 10 weeks old) were purchased from Beijing Vital River Laboratories Technology Co., Ltd. or Laboratory animal resource center in Chinese Institute for Brain Research. CaMKIIα and CaMKIIβ knock out mice were constructed with CRISPR-Cas9 gene targeting technology. For Camk2a, the gRNA sequences were designed as 5′-CTGGATCACGAAGACCCCTG-3′ and 5′-ACCTGAAGGTGAGTAACCCT-3′ to delete the 5th and 6th exons. For Camk2b, the gRNA sequences were designed as 5′-TCTAGGACCTCCATGTTGGG-3′ and 5′- TAAGACCTGTGTGTGAGAGG-3′ to delete the 7th and 8th exons. A mixture of Cas9-expressing mRNA and sgRNAs was injected into fertilized eggs through electroporation and the eggs were then transplanted into the womb of foster mothers. F0 and F1 mice were genotyped through PCR to make sure the presence of recombination. The genotyping primers for Camk2a−/− were 5′-AGGGGACAGAGAGGGGTAAG-3′ and 5′-ACAGGGCAGCCTCAGTCTAA-3′. For Camk2b−/−, primers were 5′-CAGGCCTACGATGGAGATGT-3′ and 5′-ATCCTGGTGTCCACTTGCTC-3’. Mutant lines were back-crossed to C57 BL/6J for at least 5 generations to exclude possible off-targeting.
Mouse housing
All experimental procedures were performed in accordance with the guidelines and were approved by the Animal Care and Use Committee of Chinese Institute for Brain Research, Beijing. We have complied with all relevant ethical regulations for animal use. Mice were maintained on a C57 BL/6J background. Mice were housed under a 12 h: 12 h light/dark cycle and controlled temperature and humidity conditions. Food and water were delivered ad libitum. Mice used in all experiments were 10–14 weeks old.
EEG and EMG recording and analysis
EEG and EMG data recording and analysis were performed as our previous study20. EEG and EMG data at basal sleep conditions were recorded for 2 consecutive days, with a sample frequency of 200 Hz and epoch length of 4 s. EEG and EMG data were initially processed using AccuSleep39 and then were manual correction in SleepSign. EEG and EMG signals were classified into Wake (fast and low amplitude EEG, high amplitude and variable EMG), NREM (high amplitude and 1–4 Hz dominant frequency EEG, low EMG tonus) and REM (low amplitude and 6–9 Hz frequency EEG, complete silent of EMG). The state episode was defined as at least three continuous and unitary state epochs. Epoch contained movement artifacts were included in sleep duration analysis but excluded from the subsequent power spectrum analysis. For power spectrum analysis, EEG was subjected to fast Fourier transform analysis (FFT). Power spectra represents the mean ratio of each 0.25 Hz to total 0–25 Hz of EEG signals during 24 h baseline condition. The power density of NREMS represents the ratio of delta power density (1–4 Hz) to total power (0–25 Hz) in each hr. Cumulative rebound represented cumulative changes of time in post-SD compared with relative ZT under the baseline condition. The difference before and after sleep deprivation are calculated by comparing the NREMS delta density after sleep deprivation with the baseline at the equivalent ZT point from ZT6-ZT12. The NREMS delta density in the six-hour after SD was compared with the baseline for each mouse.
Morris water maze
Spatial learning and memory were assessed using the hidden-platform version of the Morris water maze40,41. A circular tank (150 cm in diameter) was filled with opaque water (21 ± 2 °C) to a depth of 35 cm to make the platform invisible. The tank was divided into four quadrants, with the escape platform consistently positioned 1 cm below the water surface in the center of the SE quadrant. Data collection was facilitated by a digital camera connected to an image tracking system. The mice were given 1 min of free swimming on the first day. Subsequently, the mice were brought to the water maze room 1 h early for acclimatization before each daily session. Over 6 consecutive days, the mice performed four trials per day with an intertrial interval of 30 min. The mice started each trial facing the tank wall, and the starting position was randomized. A maximum of 60 s was allowed for the mice to locate the platform. After reaching the platform within 60 s, the mice were allowed to remain on it for 10 s. In cases where the mice failed to locate the platform within 60 s, they were gently guided to the platform and allowed to stay on it for 15 s. The time taken to reach the platform was analyzed.
Sleep deprivation
After 2 consecutive days of EEG and EMG signals recording, mice were introduced into new cages at ZT0. Mice were gently handled or touched to keep them awake for 6 h of sleep deprivation, before being returned to the recording cage for another 24 h of recording.
Brain lysate preparation and immunoblotting
Mouse whole brains were quickly dissected at indicated ZT, rinsed with PBS and immediately homogenized in a glass tissue homogenizer with 4 ml of lysis buffer (50 mM Tris, 1.0% Triton X-100, pH8.0, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxychilate) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche) freshly before usage. The homogenates were incubated on ice over 20 min and then centrifuged at 12,000 rpm for 20 min. The supernatant was collected and diluted four times for immunoblotting.
Western blotting was performed according to standard procedures. Antibodies were used at the optimal concentration according to the manufacturer’s instructions. Antibodies used in this study included anti-β-actin antibody (ab213262, Abcam); anti-CaMKIIα (#11945, Cell Signaling), anti-CaMKIIβ anitibody (ab34703, Abcam); anti-phospho-CaMKII (Thr286) (#12716, Cell Signaling), anti-CaMKII (#4436, Cell Signaling). Grayscale values were calculated with ImageJ.
Statistics and reproducibility
All statistical analyses were performed using GraphPad Prism 9.0. One-way ANOVA was used to compare differences among more than three groups, followed by Tukey’s multiple comparisons tests. Kruskal-Wallis tests were used for non-parameters tests. Two-way ANOVA was used to compare the differences between different groups with different treatments, followed by Tukey’s multiple comparisons tests. Two-way ANOVA with repeated measurements (Two-way RM ANOVA) was used when the same individuals were measured on the same outcome variable more than once, followed by Tukey’s multiple comparisons test. Data are presented as mean ± SEM. In all cases, p values more than 0.05 were considered not significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
We are grateful to the National Center for Protein Sciences at Peking University for assistance with the Morris Water Maze test.
Author contributions
Weiwen Yang, Jingyi Shi and Jingqun Yang performed the research and collected the data; Weiwen Yang, Jingyi Shi and Chenggang Li analyzed the data; Jianjun Yu and Juan Huang designed, mutated and housed the transgenic mice; Weiwen Yang and Jingyi Shi wrote and revised the manuscript; Yi Rao designed and supervised the research.
Peer review
Peer review information
Communications Biology thanks Paul Shaw, Thomas Kilduff and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Benjamin Bessieres.
Data availability
All data behind the graphs in this paper are presented in the Supplementary Data file. Uncropped gel images and the gray value of the bands are provided in Supplementary Information. Detailed information supporting the results and conclusions of this paper are available upon reasonable request from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ethics approval
All experimental procedures were performed in accordance with the guidelines and were approved by the Animal Care and Use Committee of Chinese Institute for Brain Research, Beijing.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07449-y.
References
- 1.Chemelli, R. M. et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell98, 437–451 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Lin, L. et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell98, 365–376 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Funato, H. et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature539, 378–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda, T. et al. A single phosphorylation site of SIK3 regulates daily sleep amounts and sleep need in mice. Proc. Natl. Acad. Sci. USA115, 10458–10463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park, M. et al. Loss of the conserved PKA sites of SIK1 and SIK2 increases sleep need. Sci. Rep.10, 8676 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, Y. et al. Sleep need, the key regulator of sleep homeostasis, is indicated and controlled by phosphorylation of threonine 221 in salt-inducible kinase 3. Genetics225, iyad136 (2023). [DOI] [PubMed]
- 7.Lelkes, Z., Alföldi, P., Erdos, A. & Benedek, G. Rolipram, an antidepressant that increases the availability of cAMP, transiently enhances wakefulness in rats. Pharm. Biochem. Behav.60, 835–839 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Hendricks, J. C. et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat. Neurosci.4, 1108–1115 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Hellman, K., Hernandez, P., Park, A. & Abel, T. Genetic evidence for a role for protein kinase A in the maintenance of sleep and thalamocortical oscillations. Sleep33, 19–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foltenyi, K., Greenspan, R. J. & Newport, J. W. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci.10, 1160–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Vanderheyden, W. M., Gerstner, J. R., Tanenhaus, A., Yin, J. C. & Shaw, P. J. ERK phosphorylation regulates sleep and plasticity in Drosophila. PLoS ONE8, e81554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikhail, C., Vaucher, A., Jimenez, S. & Tafti, M. ERK signaling pathway regulates sleep duration through activity-induced gene expression during wakefulness. Sci. Signal.10, eaai9219 (2017). [DOI] [PubMed]
- 13.Chikahisa, S., Fujiki, N., Kitaoka, K., Shimizu, N. & Séi, H. Central AMPK contributes to sleep homeostasis in mice. Neuropharmacology57, 369–374 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Nagy, S. et al. AMPK signaling linked to the schizophrenia-associated 1q21.1 deletion is required for neuronal and sleep maintenance. PLoS Genet14, e1007623 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yurgel, M. E. et al. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biol.17, e2006409 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta, S., O’Malley, M. W. & Patterson, E. H. Calcium/calmodulin kinase II in the pedunculopontine tegmental nucleus modulates the initiation and maintenance of wakefulness. J. Neurosci.31, 17007–17016 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatsuki, F. et al. Involvement of Ca(2+)-dependent hyperpolarization in sleep duration in mammals. Neuron90, 70–85 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Tone, D. et al. Distinct phosphorylation states of mammalian CaMKIIβ control the induction and maintenance of sleep. PLoS Biol.20, e3001813 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahama, K. et al. Pan-neuronal knockdown of the c-Jun N-terminal Kinase (JNK) results in a reduction in sleep and longevity in Drosophila. Biochem Biophys. Res. Commun.417, 807–811 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Liu, Z. et al. LKB1 is physiologically required for sleep from Drosophila melanogaster to the Mus musculus. Genetics221, iyac082 (2022). [DOI] [PMC free article] [PubMed]
- 21.Parmentier, R. et al. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J. Neurosci.22, 7695–7711 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popa, D. et al. Contribution of 5-HT2 receptor subtypes to sleep-wakefulness and respiratory control, and functional adaptations in knock-out mice lacking 5-HT2A receptors. J. Neurosci.25, 11231–11238 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Z. L. et al. Adenosine A2A, but not A1, receptors mediate the arousal effect of caffeine. Nat. Neurosci.8, 858–859 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Anaclet, C. et al. Orexin/hypocretin and histamine: distinct roles in the control of wakefulness demonstrated using knock-out mouse models. J. Neurosci.29, 14423–14438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorness, T. E., Kelly, C. L., Gao, T., Poffenberger, V. & Greene, R. W. Control and function of the homeostatic sleep response by adenosine A1 receptors. J. Neurosci.29, 1267–1276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, Y. et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science325, 866–870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu, W. M. et al. Essential role of dopamine D2 receptor in the maintenance of wakefulness, but not in homeostatic regulation of sleep, in mice. J. Neurosci.30, 4382–4389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, X., Yan, H., Luo, Y., Huang, Z. & Rao, Y. Thermoregulation-independent regulation of sleep by serotonin revealed in mice defective in serotonin synthesis. Mol. Pharm.93, 657–664 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kashiwagi, M. et al. Widely distributed neurotensinergic neurons in the brainstem regulate NREM sleep in mice. Curr. Biol.30, 1002–1010.e1004 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Ma, Y. et al. Galanin neurons unite sleep homeostasis and α2-adrenergic sedation. Curr. Biol.29, 3315–3322.e3313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexandre, C. et al. Early life blockade of 5-hydroxytryptamine 1A receptors normalizes sleep and depression-like behavior in adult knock-out mice lacking the serotonin transporter. J. Neurosci.26, 5554–5564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, S. J. et al. Kinase signalling in excitatory neurons regulates sleep quantity and depth. Nature612, 512–518 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Zhou, R. et al. A signalling pathway for transcriptional regulation of sleep amount in mice. Nature612, 519–527 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Yasuda, R., Hayashi, Y. & Hell, J. W. CaMKII: a central molecular organizer of synaptic plasticity, learning and memory. Nat. Rev. Neurosci.23, 666–682 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Lo Martire, V., Silvani, A., Bastianini, S., Berteotti, C. & Zoccoli, G. Effects of ambient temperature on sleep and cardiovascular regulation in mice: the role of hypocretin/orexin neurons. PLoS ONE7, e47032 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perron, I. J., Pack, A. I. & Veasey, S. Diet/energy balance affect sleep and wakefulness independent of body weight. Sleep38, 1893–1903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sotelo, M. I. et al. Neurophysiological and behavioral synchronization in group-living and sleeping mice. Curr. Biol.34, 132–146.e135 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, Y. M. et al. Long-term environmental enrichment overcomes depression, learning, and memory impairment in elderly CD-1 mice with maternal sleep deprivation exposure. Front. Aging Neurosci.15, 1177250 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barger, Z., Frye, C. G., Liu, D., Dan, Y. & Bouchard, K. E. Robust, automated sleep scoring by a compact neural network with distributional shift correction. PLoS ONE14, e0224642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva, A. J., Paylor, R., Wehner, J. M. & Tonegawa, S. Impaired spatial learning in alpha-calcium-calmodulin kinase II mutant mice. Science257, 206–211 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Vorhees, C. V. & Williams, M. T. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat. Protoc.1, 848–858 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
All data behind the graphs in this paper are presented in the Supplementary Data file. Uncropped gel images and the gray value of the bands are provided in Supplementary Information. Detailed information supporting the results and conclusions of this paper are available upon reasonable request from the corresponding author.