Abstract
Laparoscopic exploration (LE) is crucial for diagnosing intra-abdominal metastasis (IAM) in advanced gastric cancer (GC). However, overlooking single, tiny, and occult IAM lesions during LE can severely affect the treatment and prognosis due to surgeons’ visual misinterpretations. To address this, we developed the artificial intelligence laparoscopic exploration system (AiLES) to recognize IAM lesions with various metastatic extents and locations. The AiLES was developed based on a dataset consisting of 5111 frames from 100 videos, using 4130 frames for model development and 981 frames for evaluation. The AiLES achieved a Dice score of 0.76 and a recognition speed of 11 frames per second, demonstrating robust performance in different metastatic extents (0.74–0.76) and locations (0.63–0.90). Furthermore, AiLES performed comparably to novice surgeons in IAM recognition and excelled in recognizing tiny and occult lesions. Our results demonstrate that the implementation of AiLES could enhance accurate tumor staging and assist individualized treatment decisions.
Subject terms: Endoscopy, Gastric cancer, Metastasis, Surgical oncology
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed malignant tumor worldwide and the fourth primary cause of cancer-related deaths1,2, often due to its advanced stage at the time of diagnosis. Intra-abdominal metastasis (IAM) is the primary mode of distant metastasis of GC, and 14% of GC patients were diagnosed with IAM at their initial presentation3,4. Once IAM occurs, the initial therapeutic management of patients shifts from curative resection surgery toward systemic treatment with a median survival of 3–21 months5,6. Therefore, accurate tumor staging, especially for distant metastasis staging, is crucial for effective treatment decision-making and long-term prognosis5. The staging of GC patients is predominantly conducted using enhanced computed tomography (CT) scanning, while PET/CT is also employed for diagnosing suspected metastases7. However, studies have demonstrated that CT scanning exhibits limited sensitivity in accurate tumor staging and metastasis detection8,9, especially IAM. Laparoscopic exploration (LE) is inexpensive and minimally invasive, allowing systematic examination of the abdominal cavity under direct vision10. Prospective clinical trials have shown that 0.9–1.7% of GC patients had undetected metastasis preoperatively while discovered during the surgery11–14. Due to its critical role in tumor staging and treatment decision-making, LE is recommended by several international guidelines for advanced GC7,15,16.
As the first phase of laparoscopic cancer surgery, LE is often overlooked as surgeons prioritize performing lymph node dissection and reconstruction. However, the accuracy of LE for detecting IAM depends on the surgeon’s experience and disease extent5. This procedure is typically performed by junior surgeons under the supervision of senior surgeons; however, junior surgeons may lack sufficient experience and skills17. Moreover, metastatic lesions can be variable in their location, shape, and size18, making them particularly challenging to detect, especially the single and tiny implantation metastases on the peritoneum. These kinds of lesions are prone to be missed by surgeons due to the limitations of light and tissue color perception within the surgical field19, which may result in the omission of IAM, leading to incorrect tumor staging and unnecessary gastrectomy. This impedes making the optimal comprehensive treatment strategy intraoperatively, such as cytoreductive surgery (CRS)20, hyperthermic intraperitoneal chemotherapy (HIPEC)21, or systemic chemotherapy22. Therefore, an emerging solution is to develop an intraoperative artificial intelligence (AI) system to recognize metastatic lesions, improving detection accuracy and reducing the risk of omission.
Computer vision (CV) is the computerized analysis of digital images, aiming to automate human visual capabilities, often utilizing machine learning methods, especially deep learning23. CV has demonstrated potential in assisting surgeons during minimally invasive surgery, such as surgical instrument identification24, key anatomical structures segmentation25, phase recognition26, safety zone detection27, and skill assessment28. However, these studies primarily focus on laparoscopic cholecystectomy or colorectal surgery, with few studies on complex GC surgery, particularly the phase of LE. Semantic segmentation, as a type of algorithm in CV, has the potential to enable precise pixel-level segmentation of IAM lesions, thereby facilitating real-time accurate recognition of metastasis29. Thus, it is essential to integrate an AI algorithm into laparoscopic GC surgery to assist surgeons in recognizing IAM lesions.
The objective of the present study was to develop an artificial intelligence laparoscopic exploration system (AiLES) using the semantic segmentation algorithm for the automatic recognition of IAM lesions during LE for GC, to assist intraoperative tumor staging and treatment decision-making.
Results
Study design and dataset
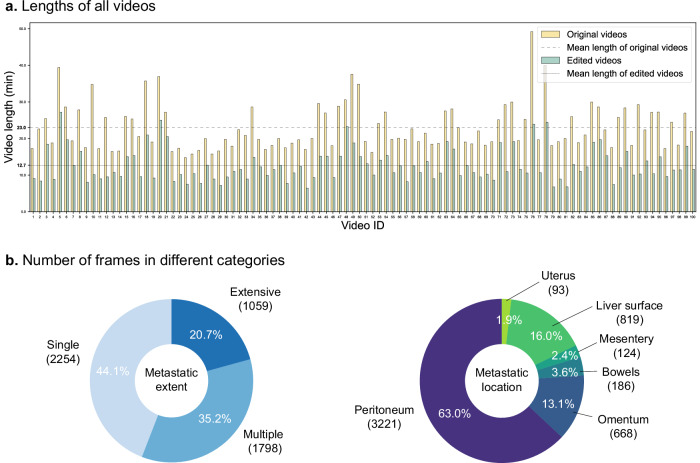
The study workflow is shown in Fig. 1. The technical characteristics and descriptions of the LE are detailed in Table 1. Patient and dataset characteristics are displayed in Supplementary Table 1. There were 46 males and 54 females in 100 GC patients. The mean age was 62.4 (±6.5) years, and the mean Body Mass Index (BMI) was 24.2 (±4.1) kg/m2. The lengths of the original and edited videos in the dataset are displayed in Fig. 2a. The average lengths of the original videos and edited videos were 23.0 ( ± 4.1) min and 12.7 ( ± 1.8) min, respectively. As shown in Fig. 2b, a total of 5111 frames were divided into three categories according to the extent of metastasis: (1) single metastases (2254 frames); (2) multiple metastases (1798 frames); (3) extensive metastases (1059 frames); and divided into six categories according to the location of metastasis: (1) peritoneum (3221 frames); (2) omentum (668 frames); (3) bowels (186 frames); (4) mesentery (124 frames); (5) liver surface (819 frames); (6) uterus (93 frames). Notably, the metastatic location “bowels” included small bowel (154 frames, 3.0% of the dataset) and large bowel (32 frames, 0.6% of the dataset). Due to their small proportions, we did not categorize frames of different bowel segments as separate groups in this study. For model development, the dataset was divided into a development set with 4130 frames and an independent test set with 981 frames at the patient level. The development set is then further divided into a training set (3304 frames) and a validation set (826 frames).
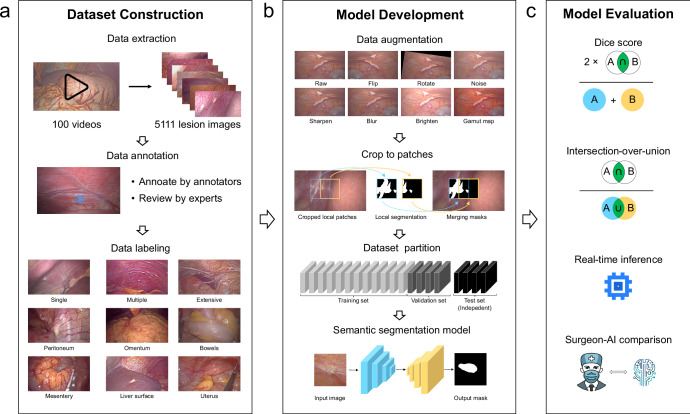
Fig. 1. Workflow of this study.
a Dataset construction: videos of LE were converted to images for dataset construction. All frames from the dataset were annotated by annotators and reviewed by the expert surgeon; after data annotation, each lesion image was labeled according to metastatic extents and locations. b Model development: the dataset was preprocessed with data augmentation and cropping patches for modeling. c Model evaluation: the performance of the models was evaluated using the Dice score, intersection-over-union (IOU), inference speed, and surgeon-AI comparison.
Table 1.
The technical characteristics and detailed descriptions of the LE duration for gastric cancer
| No. | Step | Description | Content |
|---|---|---|---|
| 1 | Trocar insertion |
Start: insert a trocar End: check whether there are puncture injuries |
Insert a 12-mm trocar at the umbilical level and insert a laparoscope into the abdominal cavity. Establish two operating poles on the left (or right) abdomen and plug them into two 5-mm trocars for inserting instruments. Then check for any bruising or puncture injuries to the bowels and confirm if there is any damage. |
| 2 | Exploration of the anterior abdominal wall and the surface of abdominal viscera |
Start: exploration of bilateral diaphragmatic dome End: exploration of the surface of the ascending colon |
The exploration areas include: (1) bilateral diaphragmatic dome, ligamentum teres hepatis, and falciform ligament; (2) the anterior abdominal wall; (3) the diaphragmatic surface of the liver lobe; (4) the surface of the transverse colon and the great omentum; (5) the inferior abdominal wall; (6) the left side of abdominal wall, the left paracolic sulcus and the surface of descending colon; and (7) the right side of abdominal wall, the right paracolic sulcus and the surface of ascending colon. |
| 3 | Exploration of the pelvic cavity and the surface of abdominal viscera |
Start: exploration of bilateral fossa iliaca End: exploration of the sigmoid colon and upper rectum |
The exploration areas include: (1) bilateral fossa iliaca, bilateral accessories, and uterus surface (for females); (2) pelvic floor and peritoneal reflection; (3) sigmoid colon and upper rectum. |
| 4 | Exploration of mesentery and the small bowel |
Start: exploration of the mesocolon transversum End: exploration of the surface of small bowel |
The exploration areas include: (1) the descending mesocolon, transverse mesocolon, ascending mesocolon, and their roots; (2) the Treitz ligament; (3) the mesentery of the small bowel and its root; (4) the surface of the small bowel. |
| 5 | Exploration of the stomach, adjacent structures, and omental bursa |
Start: exploration of the anterior gastric wall and the greater curvature End: exploration of the recessus of hepatorenalis |
The exploration areas include: (1) the serosa of the anterior gastric wall and the greater curvature; (2) the lesser curvature and the lesser omentum; (3) cardia and pylorus; (4) Ligamentum duodenum; (5) posterior gastric wall; (6) omental bursa (if tumor located in the posterior gastric wall); (7) recessus of hepatorenalis. |
| 6 | Peritoneal cytology |
Start: transfuse saline solution into the abdominal cavity End: suck and collect the peritoneal lavage |
Transfuse 200 ml saline solution into hepatorenal recess, splenic recess, bilateral paracolic sulcus, and pelvic floor, respectively. Suck and collect the peritoneal lavage. |
| 7 | Suspicious lesion resection with biopsy |
Start: resect the suspicious lesion End: use electrocautery for hemostasis |
Use laparoscopic scissors or harmonic to resect the lesion if there are any suspicious tumor deposits. Then use electrocautery to perform hemostasis. Send the specimens to biopsy. |
| 8 | Closure of the abdominal incisions |
Start: suture peritoneal muscular sheath layers End: complete the LE |
Suture the peritoneal and muscular sheath layers of the portholes and complete the LE. |
| 9 | Others |
Start: clean the laparoscopic lens, idle or any extra-abdominal procedures End: any one of Steps 1–8 begins |
The steps of lens cleaning, extra-abdominal procedures, or idle are defined as others. The idle step is defined as hold-on time for the surgeon to change the surgical tools, and adjust the angle of scope. The extra-abdominal procedures include removing the resected lesions, inserting instruments, and so on. |
The sequence of exploration from step 2 to step 5 is flexible and can be adjusted based on the surgeon’s experience.
Fig. 2. Videos and frames in the study dataset.
a Lengths of all videos. Original videos include clips of all LE steps (trocar insertion, intra-abdominal exploration, peritoneal cytology, resection of suspicious lesions with biopsy, closure of abdominal incisions, and others). Edited videos focus only on clips of intra-abdominal exploration. b Number of frames in different categories including metastatic extents and locations.
Annotation consistency of IAM
The intra- and inter-annotator consistencies between the expert surgeon (H.C.) and novice surgeons (L.G. and H.B.C.) on the randomly selected subset of 50 frames were displayed in Table 2. For intra-annotator consistency of the novice surgeons, the mean Dice score was 0.94 (±0.03), with the Dice scores for different lesion locations and metastatic extents all exceeding 0.90. For the same-level inter-annotator consistency, the mean Dice score was 0.91 (±0.05), with the lowest consistency observed in annotating lesions with extensive metastasis, achieving a Dice score of 0.88 (±0.05). For inter-annotator consistency of novice and expert surgeons, the mean Dice score was 0.89 (±0.06), with the lowest consistency observed in annotating lesions with extensive metastasis, achieving a Dice score of 0.86 (±0.05). All the annotation consistencies are satisfactory with all the Dice scores greater than 0.85.
Table 2.
Intra-annotator consistency and inter-annotator consistency between novice surgeon annotators and expert surgeon annotators
| Category | Intra-annotator (Novice) | Inter-annotator (Novice–Novice) | Inter-annotator (Expert–Novice) | |||
|---|---|---|---|---|---|---|
| Dice score | IOU | Dice score | IOU | Dice score | IOU | |
| Overall | 0.94 (0.03) | 0.89 (0.04) | 0.91 (0.05) | 0.84 (0.09) | 0.89 (0.06) | 0.81 (0.09) |
| Metastatic extent | ||||||
| Single | 0.95 (0.02) | 0.91 (0.03) | 0.92 (0.06) | 0.85 (0.09) | 0.90 (0.06) | 0.83 (0.09) |
| Multiple | 0.94 (0.02) | 0.88 (0.04) | 0.92 (0.04) | 0.86 (0.07) | 0.89 (0.05) | 0.81 (0.09) |
| Extensive | 0.92 (0.03) | 0.86 (0.06) | 0.88 (0.05) | 0.79 (0.08) | 0.86 (0.05) | 0.76 (0.08) |
| Metastatic location | ||||||
| Peritoneum | 0.93 (0.03) | 0.88 (0.05) | 0.91 (0.06) | 0.84 (0.10) | 0.89 (0.06) | 0.81 (0.10) |
| Single | 0.95 (0.02) | 0.90 (0.03) | 0.91 (0.09) | 0.84 (0.13) | 0.90 (0.08) | 0.82 (0.12) |
| Multiple | 0.94 (0.02) | 0.88 (0.04) | 0.93 (0.03) | 0.88 (0.05) | 0.91 (0.05) | 0.84 (0.07) |
| Extensive | 0.92 (0.03) | 0.86 (0.06) | 0.88 (0.05) | 0.79 (0.08) | 0.86 (0.05) | 0.76 (0.08) |
| Omentum | 0.94 (0.01) | 0.88 (0.02) | 0.92 (0.05) | 0.88 (0.07) | 0.88 (0.05) | 0.80 (0.08) |
| Bowels | 0.95 (0.02) | 0.90 (0.04) | 0.90 (0.03) | 0.82 (0.04) | 0.88 (0.04) | 0.79 (0.06) |
| Mesentery | 0.95 (0.03) | 0.90 (0.05) | 0.95 (0.02) | 0.91 (0.04) | 0.93 (0.05) | 0.87 (0.09) |
| Liver surface | 0.96 (0.01) | 0.92 (0.02) | 0.93 (0.04) | 0.88 (0.07) | 0.91 (0.04) | 0.84 (0.07) |
| Uterus | 0.96 (0.02) | 0.92 (0.04) | 0.89 (0.03) | 0.80 (0.05) | 0.88 (0.04) | 0.78 (0.06) |
Data were expressed in mean (±standard deviation, SD).
IOU intersection-over-union.
Comparison of AI models for IAM recognition on test set
The performance of developed IAM recognition AI models is presented in Table 3. The AiLES demonstrated the best performance among all the models, achieving a Dice score (same as F1 score) of 0.76 (±0.17). AiLES also demonstrated optimal performance across various evaluation metrics, including intersection-over-union (IOU) of 0.61 (±0.19), recall (same as sensitivity) of 0.73 (±0.21), specificity of 0.99 (±0.01), the accuracy of 0.99 (±0.01), the precision of 0.79 (±0.16), mean Average Precision at 50% IOU (mAP@50) of 0.65 and the similarity indices (Table 3 and Supplementary Table 2) including structural similarity index measure (SSIM) of 0.99 (±0.21) and Hausdorff distance (HD) of 67.88 (±40.82). In contrast, the first universal image segmentation foundation model, Segment Anything Model (SAM), achieved poor Dice scores of 0.14 (±0.30), 0.29 (±0.32), and 0.02 (±0.10) for automatic full segmentation, one-box-prompt segmentation and one-point-prompt segmentation, respectively. Nevertheless, it is worth highlighting that the fine-tuned Medical SAM Adapter (MSA) showed a remarkable enhancement in performance compared to the original SAM, with an improved Dice score of 0.63 (±0.31). The performance of the DeeplabV3+ model was comparable to MSA with a Dice score of 0.67 (±0.14). Additionally, the AiLES has the fastest real-time inference speed at a rate of 11 frames per second (fps), whereas the DeeplabV3+ model achieves 7 fps and SAM/MSA detects at 2 fps. The detailed inference speed testing results are shown in Supplementary Table 3.
Table 3.
Performance metrics of different IAM recognition AI models
| Model | Dice score (F1 score) | IOU | Recall (sensitivity) | Specificity | Accuracy | Precision | mAP@50 | Inference speed (fps) |
|---|---|---|---|---|---|---|---|---|
| SAM-Anything | 0.14 (0.30) | 0.07 (0.28) | 0.51 (0.27) | 0.95 (0.19) | 0.94 (0.19) | 0.08 (0.33) | 0.16 | 2 |
| SAM-box | 0.29 (0.32) | 0.17 (0.31) | 0.63 (0.26) | 0.98 (0.06) | 0.97 (0.06) | 0.19 (0.37) | 0.41 | 2 |
| SAM-point | 0.02 (0.10) | 0.01 (0.07) | 0.26 (0.40) | 0.77 (0.27) | 0.76 (0.27) | 0.01 (0.14) | 0.01 | 2 |
| MSA | 0.63 (0.31) | 0.46 (0.29) | 0.59 (0.28) | 0.94 (0.20) | 0.88 (0.19) | 0.67 (0.34) | 0.51 | 2 |
| DeeplabV3+ | 0.67 (0.14) | 0.50 (0.13) | 0.61 (0.06) | 0.93 (0.19) | 0.85 (0.16) | 0.75 (0.23) | 0.59 | 7 |
| AiLES | 0.76 (0.17) | 0.61 (0.19) | 0.73 (0.21) | 0.99 (0.01) | 0.99 (0.01) | 0.79 (0.16) | 0.65 | 11 |
Data were expressed in mean (±standard deviation, SD). In this study, the Dice score and F1 score have the same values, and recall is also referred to as sensitivity.
IAM intra-abdominal metastasis, AI artificial intelligence, IOU intersection-over-union, mAP@50 mean average precision at 50% IOU, fps frame per second, SAM segment anything model, MSA medical SAM adapter, AiLES artificial intelligence laparoscopic exploration system.
Model performance of AiLES for IAM recognition under different conditions
For tiny lesions, AiLES demonstrated superior segmentation performance with a Dice score of 0.87 (±0.17) (Supplementary Fig. 1). For single lesions located on the peritoneum (Table 4), which are typically occult and easily overlooked, AiLES also achieved excellent recognition performance, with a Dice score of 0.90 (±0.08). For lesions with different extents of metastasis (Table 4), AiLES achieved good segmentation performance with Dice scores of 0.76 (±0.18), 0.74 (±0.16), and 0.76 (±0.14) for single, multiple, and extensive metastases, respectively. For lesions with different locations (Table 4), the Dice scores of AiLES were: 0.78 (±0.15), 0.65 (±0.20), 0.67 (±0.08), 0.80 (±0.06), 0.65 (±0.16), and 0.93 (±0.07) for IAM on the peritoneum, omentum, bowels, mesentery, liver surface and uterus. Additionally, there were two cases showcasing the capability of the AiLES for IAM recognition during laparoscopic surgery in Supplementary Movie 1.
Table 4.
Summary of performance metrics of AiLES for IAM recognition
| Category | Dice score (F1 score) | IOU | Recall (sensitivity) | Specificity | Accuracy | Precision |
|---|---|---|---|---|---|---|
| Test dataset | 0.76 (0.17) | 0.61 (0.19) | 0.73 (0.21) | 0.99 (0.01) | 0.99 (0.01) | 0.79 (0.16) |
| Metastatic extent | ||||||
| Single | 0.76 (0.18) | 0.62 (0.21) | 0.69 (0.21) | 0.99 (0.01) | 0.99 (0.01) | 0.86 (0.16) |
| Multiple | 0.74 (0.16) | 0.59 (0.18) | 0.73 (0.21) | 0.99 (0.01) | 0.99 (0.01) | 0.75 (0.16) |
| Extensive | 0.76 (0.14) | 0.61 (0.15) | 0.75 (0.20) | 0.99 (0.01) | 0.99 (0.01) | 0.77 (0.12) |
| Metastatic location | ||||||
| Peritoneum | 0.78 (0.15) | 0.64 (0.18) | 0.80 (0.19) | 0.99 (0.01) | 0.99 (0.01) | 0.77 (0.15) |
| Single | 0.90 (0.08) | 0.73 (0.12) | 0.90 (0.08) | 0.99 (0.01) | 0.99 (0.01) | 0.90 (0.12) |
| Multiple | 0.78 (0.14) | 0.64 (0.16) | 0.82 (0.18) | 0.99 (0.01) | 0.99 (0.01) | 0.75 (0.15) |
| Extensive | 0.74 (0.15) | 0.59 (0.16) | 0.75 (0.21) | 0.99 (0.01) | 0.99 (0.01) | 0.73 (0.12) |
| Omentum | 0.65 (0.20) | 0.49 (0.21) | 0.59 (0.24) | 0.99 (0.01) | 0.99 (0.01) | 0.74 (0.17) |
| Bowels | 0.67 (0.08) | 0.50 (0.09) | 0.64 (0.12) | 0.99 (0.01) | 0.99 (0.01) | 0.69 (0.18) |
| Mesentery | 0.80 (0.06) | 0.67 (0.07) | 0.75 (0.09) | 0.99 (0.01) | 0.98 (0.01) | 0.86 (0.10) |
| Liver surface | 0.65 (0.16) | 0.48 (0.16) | 0.50 (0.18) | 0.99 (0.01) | 0.99 (0.01) | 0.92 (0.12) |
| Uterus | 0.93 (0.07) | 0.86 (0.10) | 0.95 (0.05) | 0.99 (0.01) | 0.99 (0.01) | 0.91 (0.11) |
Data were expressed in mean (±standard deviation, SD). In this study, the Dice score and F1 score have the same values, and recall is also referred to as sensitivity.
AiLES artificial intelligence laparoscopic exploration system, IAM intra-abdominal metastasis, IOU intersection-over-union.
Comparison of IAM recognition between AiLES and surgeon
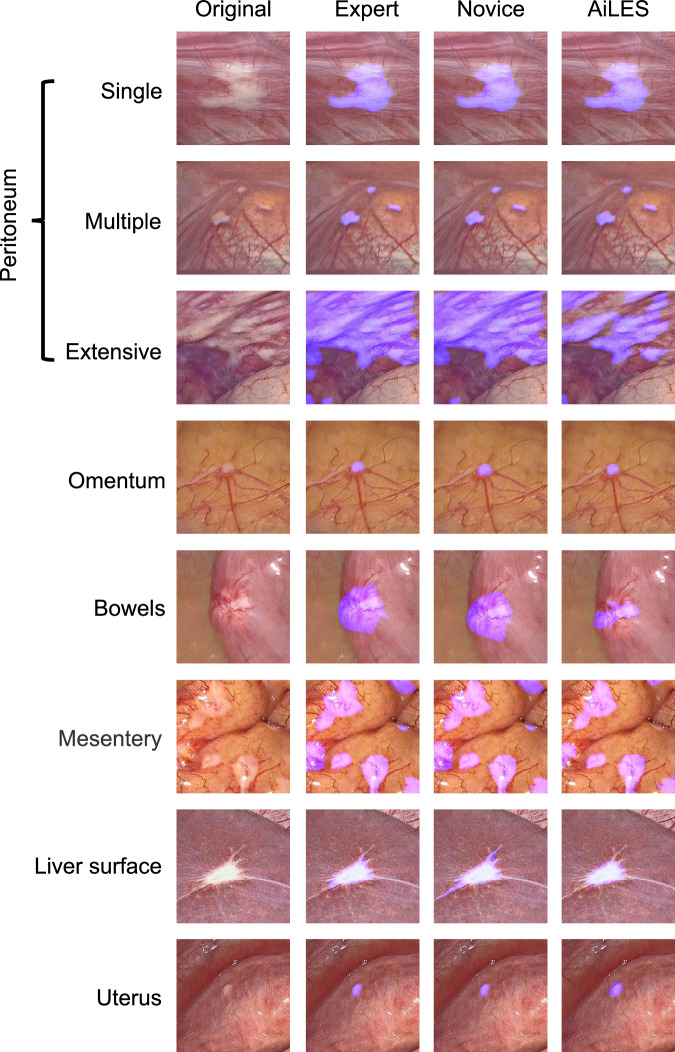
Figure 3 displays eight different scenes of IAM according to the metastatic extents (single, multiple, extensive) and locations (peritoneum, omentum, bowels, mesentery, liver surface, and uterus). The IAM recognition performance of the AiLES, expert surgeons, and novice surgeons is visualized and compared in Fig. 3. To assess the clinical utility of the AiLES, Fig. 4 showcases six key frames from LE to illustrate instances where the novice surgeon overlooked IAM, contrasted with the developed AiLES’s accurate recognition. It is inspiring that the AiLES could effectively detect the tiny, isolated, and occult IAM lesions, which were difficult for novice surgeons.
Fig. 3. Examples of AiLES recognition performance.
These examples displayed the visualization effect of IAM segmented by the AiLES, expert surgeon, and novice surgeon in different scenes according to metastatic extents on the peritoneum and various metastatic locations. To improve the clarity of the lesion segmentation results, all images were resized to 512 × 512 pixels. AiLES: artificial intelligence laparoscopic exploration system.
Fig. 4. Case presentation for clinical utility assessment of the AiLES.
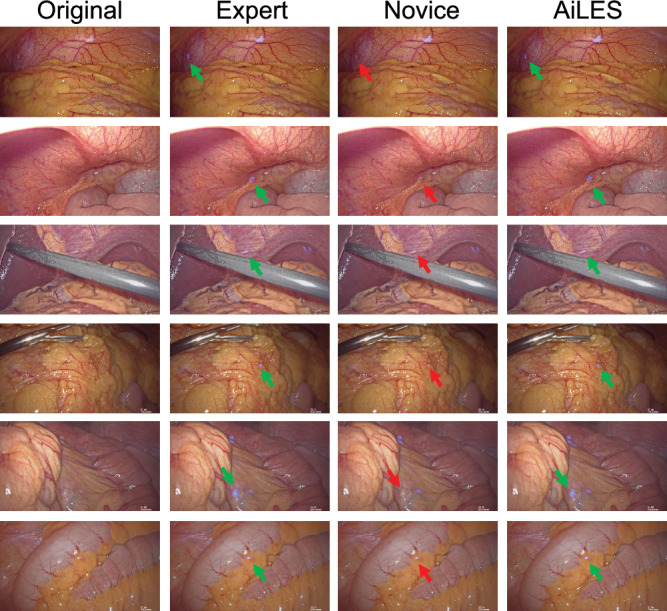
The metastatic lesions in these cases were overlooked by the novice surgeon (red arrow) but detected by both the AiLES (green arrow) and expert surgeon. AiLES: artificial intelligence laparoscopic exploration system.
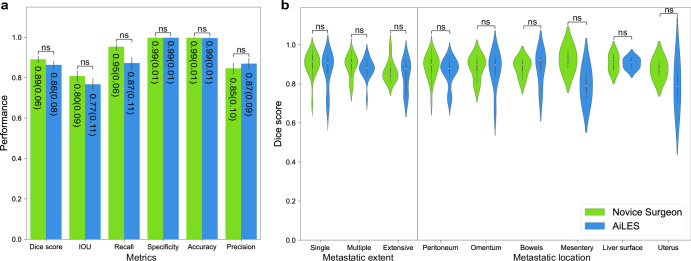
Furthermore, we compared the recognition performance between the AiLES and novice surgeons in randomly selected 50 frames from the test set. As Fig. 5a demonstrated, the AiLES’s performance metrics including Dice score (same as F1 score), IOU, recall (same as sensitivity), specificity, accuracy, and precision were non-inferior to those of the novice surgeons (P > 0.05). Figure 5b further illustrated that AiLES’s segmentation performance across different metastatic extents and locations was also non-inferior to that of novice surgeons. This indicated that the AiLES had initially reached a level comparable to the novice surgeon in recognizing IAM lesions’ shapes and boundaries.
Fig. 5. The comparison of performance metrics between novice surgeons and AiLES.
a Illustrations of performance metrics including Dice score (same as F1 score), intersection-over-union (IOU), sensitivity (same as recall), specificity, accuracy, and precision. b Violin plot illustrations of Dice score for different metastatic extents and locations. AiLES: artificial intelligence laparoscopic exploration system; ns: no significance (P > 0.05).
Discussion
AI technologies, particularly CV, are increasingly utilized in minimally invasive surgery, with prior applications focused on identifying surgical instruments, phases, and anatomical structures to minimize risks and enhance skill assessment23. However, limited research has focused on intraoperative diagnosis and treatment decision-making, particularly for GC patients with IAM30,31. Accurate LE is as crucial as safe surgical resection for the treatment and prognosis of GC patients. Yet, surgeon experience may still lead to misjudgments and omissions of IAM lesions in this phase. This study is the first to apply the real-time semantic segmentation algorithm during the LE phase of GC surgery, achieving satisfactory recognition and segmentation of IAM lesions with a Dice score of 0.76, an IOU of 0.61, and an inference speed of 11 fps. Dice score, IOU, recall, specificity, accuracy, and precision are the common metrics used for evaluating model performance in segmentation tasks. To further compare model performance comprehensively, we utilized mAP@50 to evaluate the model’s ability to accurately segment and distinguish different recognition object classes within the IAM dataset. Additionally, we employed SSIM and HD to assess structural similarity and point set similarity between predictions and ground truth to evaluate model performance. AiLES, based on RF-Net, outperformed SAM, MSA, and DeeplabV3+ across these metrics. RF-Net was originally developed using an ultrasound breast cancer dataset similar to our IAM dataset, making it more suitable for the segmentation task in our study. Notably, AiLES achieved Dice scores of 0.90, 0.87, and 0.80 for the commonly overlooked single peritoneal lesions, tiny lesions, and mesenteric lesions, respectively. The model matches the accuracy of novice surgeons in segmenting IAM lesions, while also effectively recognizing lesions that novice surgeons failed to recognize (Figs. 4 and 5).
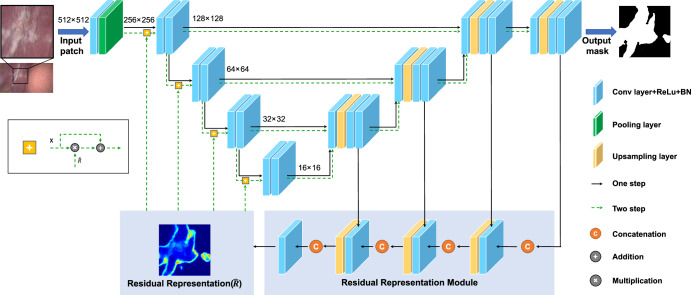
For AI studies aimed at clinical applications, it is most important to choose the appropriate model. Our findings indicated that, compared to other models, the AiLES based on residual feedback network (RF-Net) model performed best in identifying IAM lesions, achieving a Dice score of 0.76. This is primarily because RF-Net was developed based on the breast cancer ultrasound images, which were similar to the IAM lesions in our study, exhibiting significant variability in shape, size, and extent (Supplementary Fig. 2)32. AiLES employed RF-Net’s core module, the residual representation module, to learn the ambiguous boundaries and complex areas of lesions, thereby enhancing segmentation performance (Fig. 6). The DeeplabV3+ model is widely utilized in surgical AI studies for recognizing anatomical structures and surgical instruments33,34. However, it only achieved a Dice score of 0.67 in our study, which was lower than the satisfactory performance (Dice score ≥ 0.75) reported in previous studies for anatomy and instrument segmentation33,34. This is mainly due to the irregular shapes, locations, and extent of the IAM lesions, which are significantly different from more regular anatomical structures and surgical instruments. Additionally, SAM is the first universal image segmentation foundation model35, which was released during the design phase of our study. However, it had not been thoroughly explored in segmenting medical images36, particularly surgical images, and demonstrated poor performance in different segmentation modes including automatic, one-point, and one-box for IAM lesions, with Dice scores of 0.14, 0.02, and 0.29, respectively. We thought the unique characteristics of IAM lesion images were significantly different from natural images, leading to the unsatisfactory performance of SAM which was trained by natural images35. After fine-tuning with the MSA trained on our dataset37, the MSA demonstrated a notable improvement over the original SAM in segmenting IAM, with a Dice score of 0.63. Although the SAM and MSA were unspecific for our study, we think our results provide valuable data support and references for applying SAM in surgical images. As the IAM dataset was original and unique, there were no available studies or algorithms for references and no gold standard model for our dataset. Based on the above factors, we included and evaluated the current mainstream AI models for IAM segmentation, offering a reference workflow for future research on surgical image segmentation. Moreover, the evaluation of different models for real-time intraoperative deployment showed inference speeds of 2 fps for SAM and MSA, 7 fps for DeeplabV3+, and 11 fps for AiLES. According to previous surgical AI studies, inference speeds for surgical image segmentation typically range from 6 fps to 14 fps33,38,39, indicating that the inference speed of AiLES in our study is applicable and acceptable. In the future, the real-time inference speed of AiLES could be further improved by utilizing deep learning acceleration techniques such as TensorRT33.
Fig. 6. The model architecture of the AiLES for IAM segmentation.
ResNet-34 is adopted as the Encoder path with pre-trained parameters. This network includes two steps: The first step (black arrows) is used to generate the initial segmentation results and learn the residual representation of missing/ambiguous boundaries and confusing regions. The second step is used to (green dotted arrows) feed the residual representation into the encoder path and generate more precise segmentation results. AiLES: artificial intelligence laparoscopic exploration system. IAM: intra-abdominal metastasis.
The clinical practicability of the AI model depends not only on the model performance, but also on the appropriate annotation approach for constructing a high-quality dataset40. Currently, there are several common annotation approaches for computer vision image analysis, including point, bounding box, and polygon. Our study focuses on recognizing and segmenting IAM lesions, which have irregular shapes, extents, and boundaries. For IAM lesion annotation, the point approach only marks the main location of the lesion and fails to capture sufficient visual features, especially for extensive lesions; the bounding-box approach may include partial normal areas, which may cause the model to misidentify normal tissues as lesions, increasing false positives and leading to inaccurate results. In contrast, the polygon approach can accurately outline the boundaries of lesions, regardless of their shape and extent, similar to how surgeons recognize lesions (Supplementary Fig. 3). Therefore, we selected the polygon approach for annotating IAM lesions. Finally, we constructed a high-quality IAM dataset with 5111 frames from 100 LE videos, with a similar video-to-frame ratio to that of several excellent surgical AI studies that used the same annotation approach as ours (Supplementary Table 4)25,34,41–43.
In clinical practice, peritoneal metastasis is the predominant type of IAM in GC patients. While multiple and extensive peritoneal metastases are readily identifiable, single peritoneal metastatic lesions may be prone to being overlooked due to their tiny size, occult location, or rapid movement of the camera during LE31. The JCOG0501 trial showed that the incidence of omitted IAM lesions in the omentum and mesentery ranges between 1.9% and 5.1%, not less than the 5.1% incidence of peritoneal metastases44. Also, studies have shown a 10.6–20.0% probability of missing IAM lesions during LE45,46, which could potentially lead to the under-staging of stage IV GC patients. In addition, the REGATTA trial suggests that initial gastrectomy may not be an appropriate treatment option for patients with IAM because of poor prognosis47. Therefore, patients might receive inappropriate gastrectomy due to the underestimation of the tumor stage caused by IAM omission, severely affecting their survival and prognosis. The AiLES excels in segmenting single peritoneal lesions (Dice score 0.90), tiny lesions (Dice score 0.87), and mesenteric lesions (Dice score 0.80), and shows robust performance across different locations (Dice scores 0.62–0.93), effectively identifying and segmenting lesions often missed by humans, thereby significantly enhancing the accuracy of tumor staging (Supplementary Movie 1).
Individualized conversion therapy is crucial for stage IV GC patients, who often exhibit tumor heterogeneity and complex metastases48. The calculation of the peritoneal cancer index (PCI) using LE offers a comprehensive evaluation of the extent and pattern of tumor dissemination in the abdominal cavity20, serving as critical information for determining an optimal individualized treatment strategy for stage IV GC patients. Current research and clinical guidelines advocate for the incorporation of PCI scores into treatment decision-making processes: patients with low PCI scores are recommended to receive CRS and HIPEC for potential survival benefits49; patients with high PCI scores are considered to receive systemic chemotherapy to manage symptoms and improve quality of life50. All the above research results confirm the importance of accurate PCI assessment. It’s well known that the PCI assessment must take into account the metastatic conditions on the parietal peritoneum, visceral peritoneum, and the surfaces of various organs. Recently, Thomas et al. developed a computer-assisted staging laparoscopy (CASL) system that employs object detection and classification algorithms to detect and distinguish between benign and malignant oligometastatic lesions31. The CASL is currently limited to analyzing static images of single lesions in the peritoneum and liver surface. In contrast, our AiLES could recognize the IAM lesions with different metastatic extents (single, multiple, and extensive) and metastatic locations (peritoneum, omentum, bowels, mesentery, liver surface, and uterus) in real-time (Supplementary Movie 1). Consequently, the AiLES could serve as a valuable complement to the CASL system, compensating for limitations and illustrating the tremendous potential of AI algorithms to assist intraoperative PCI assessment for GC patients. However, our AiLES is not yet capable of recognizing specific organs or abdominal regions where IAM lesions are located, nor can it measure lesion size during surgery, which are also fundamental elements for AI-based automatic PCI assessment. AI for IAM recognition and automatic PCI assessment are not only different clinical studies, but also different CV tasks. It should be emphasized that, IAM recognition in our study is an important foundation and first step toward achieving automatic PCI assessment in the future.
In the clinical applications of AI, researchers are primarily concerned with how AI compares to the proficiency of professional physicians23. In studies involving object detection and classification algorithms, AI and physician performance can be evaluated through simple ‘fast/slow’ and ‘yes/no’ evaluations25,31. However, these evaluation methods fall insufficient for semantic segmentation tasks33, where performance differences between humans and AI are less quantifiable. To address this, we utilized two approaches to assess the comparison between surgeons and AI. Firstly, using expert-annotated segmentations as the ground truth, we assessed the segmentation performance of AI and novice surgeons on metastatic lesions with varying extents and locations, finding no significant difference. This suggests that AI performs at a level comparable to that of novice surgeons in lesion identification. Subsequently, we selected images of tiny, occult lesions for identification by AI and a novice surgeon. In several cases, AI successfully identified metastatic lesions missed by the novice surgeon. Since we used the IAM annotations reviewed by expert surgeons as the gold standard for model training and development, the segmentation performance of AiLES can’t surpass that of the expert surgeons, that’s the reason why we didn’t compare AiLES with experts. Furthermore, LE is typically performed by novice surgeons under the supervision of senior surgeons, and novice surgeons may lack experience in recognizing IAM and omitting occult lesions in clinical practice. Therefore, there may be sufficient clinical interest and demand to develop an AI system to assist novice surgeons in recognizing IAM lesions. As expected, the surgeon-AI comparison demonstrated that AiLES had comparable performance to novice surgeons and outperformed them in the recognition of tiny and easily overlooked lesions. The assistance of AiLES is akin to collaborative judgment and decision-making between two surgeons, thereby shortening the learning curve for recognizing IAM and reducing the omission risk.
LE is recommended by several international guidelines for advanced GC in clinical practice7,15,16, which suggests that AiLES holds significant promise for IAM recognition during actual surgeries. Since AiLES is developed based on actual surgery videos, it can be integrated into laparoscopic devices. Our plan is to display visual results of AiLES recognition on a separate screen, alongside the laparoscopic screen51,52. This visual system will assist surgeons in performing accurate intraoperative tumor staging, avoiding unnecessary gastrectomy, and reducing related healthcare costs, without changing their established surgical routines. The AiLES has not only potential clinical applications in workflow, but also practical value in surgical training for residents, fellows, and young attending surgeons. Preoperatively, young surgeons can observe both the original and AI-assisted lesion visualization videos to enhance their lesion recognition skills, rapidly improving their abilities and reinforcing their memory of various lesion shapes and extents. Intraoperatively, AiLES can assist in identifying tiny, single, and occult IAM lesions and avoid incorrect tumor staging. Postoperatively, AiLES allows young surgeons to comprehensively review lesion images from surgery videos, helping them identify weaknesses and make targeted improvements. Finally, it is important to emphasize that our AI model for IAM recognition can assist surgeons but can’t replace surgeons in making treatment decisions. Automated treatment decision-making is a different clinical challenge and multimodal AI task. In surgical practice, even if AiLES’s performance improves in the future, final intraoperative treatment decisions should be made by surgeons.
This study has several limitations. Firstly, the current dataset exhibits an imbalance in the distribution of IAM lesions across different locations, such as the omentum, bowels, mesentery, and uterus. Especially, the dataset did not include frames of ovarian implantation metastasis and lacked sufficient frames of metastasis located on small and large bowels53. It’s necessary to expand the IAM dataset with a more balanced distribution of various metastatic locations. Secondly, all videos in our dataset were collected from GC surgeries, which may limit the generalization of AiLES to other types of abdominal tumor surgeries. To address this issue, we will collect image data from surgery videos with IAM in different cancers (e.g., colorectal cancer, liver cancer, and pancreatic cancer). Thirdly, it is a single-center retrospective study, requiring further multi-center and multi-device validation to strengthen the robustness and applicability of AiLES.
In conclusion, this study effectively showcases the utilization of a real-time semantic segmentation AI model during the LE phase of laparoscopic GC surgery, leading to an enhancement in the recognition of IAM lesions. The AiLES demonstrate comparable performance to novice surgeons and outperform them in the identification of tiny and easily overlooked lesions, thereby potentially reducing the risk of inappropriate treatment decisions due to under-staging. The results emphasize the significance of incorporating AI technologies within surgery to enhance accurate tumor staging and assist individualized treatment decision-making. In the future, we will improve our model’s generalization and robustness by expanding the IAM dataset with comprehensive metastatic locations from various abdominal cancer videos. Moreover, it is imperative to carry out multi-center and multi-device validation for the clinical implementation of AiLES.
Methods
Study design
Videos of the LE for GC patients were used to develop and validate the AI model to recognize IAM except for the primary tumor lesion. The metastatic lesions could be classified into different categories based on their locations, including peritoneum, omentum, bowels, mesentery, liver surface, and uterus. Additionally, metastatic lesions could also be categorized according to the extent of metastasis within the field of view, including single, multiple, and extensive metastases (Supplementary Note 1)18. Metastatic lesions are further defined as “tiny lesions” if they meet the following criteria: (1) lesions with a diameter ≤ 0.5 cm, meanwhile, similar to or smaller than the tip of the surgical instruments, such as laparoscopic gripper (Supplementary Fig. 4); (2) presence of only a single lesion in the scene. The detailed technical characteristics and descriptions of the LE in GC surgery were described in Table 1, which reflects both the clinical experiences at our center and the published “Four-Step Procedure”46. All metastatic lesions were confirmed for their consistency through LE and pathologic biopsy. Ethical approval (Approval No. NFEC-2023-392) was provided by the Institutional Review Board of Nanfang Hospital, Southern Medical University, and informed consent was waived due to its retrospective design and the anonymization of patient data. This study was conducted in accordance with the TRIPOD + AI statement (Supplementary Table 5)54.
Dataset
All videos of laparoscopic GC surgery confirmed as IAM at initial exploration were collected from April 2017 to September 2022 in Nanfang Hospital, which is the initiating unit of the Chinese Laparoscopic Gastrointestinal Surgery Study (CLASS) Group. All duplicate videos were removed. Videos were included for the first-time LE, without perioperative chemotherapy or radiotherapy. All videos were transformed into MP4 format, with a display resolution of 1920 × 1080 pixels and a frame rate of 25 fps. The intra-abdominal exploration of various regions and suspicious IAM lesions is the most important step for LE. We cut out clips involving trocar insertion, peritoneal cytology, lesion resection with biopsy, closure of the abdominal wall incision, and other steps, retaining only the clips of intra-abdominal exploration for study video dataset construction. The expert surgeons (H.C., J.Y., H.D., and G.L.) used ffmpeg 4.2 software (www.ffmpeg.org) to manually extract and select frames from edited videos. The inclusion criteria of frame selection were as follows: (1) IAM lesions are located in the peritoneum, omentum, mesentery, bowel surface, liver surface, uterus, and adnexa, excluding the primary gastric tumor, perigastric lymph nodes, and nearby invaded structures; (2) if the lesion is observed over a period of time, experts select frames from different angles and distances, then remove any similar ones; (3) the frames are clear, without artifacts, glare, smoke, or bloodstains that could interfere with IAM lesion recognition; and (4) pathological data is reviewed to confirm that the IAM lesions are metastatic through biopsy. All frames were modified to have an aspect ratio of 16:9. The specific process for constructing the dataset can be seen in Fig. 1a.
Annotations
The annotation group consisted of four expert surgeons (laparoscopic gastric surgery experience ≥ 300 procedures) and four medical annotators (including two novice surgeons). All expert surgeons are core members of the CLASS group. The medical annotators (J.Z. and C.N.) are students who have completed undergraduate medical courses, including anatomy and surgery, and have watched at least 30 laparoscopic GC surgery videos, covering both LE and gastrectomy. They must be familiar with the features of IAM lesions. The novice surgeons (H.C. and L.G.), in addition to meeting the qualifications of a medical annotator, have completed the rotation of the surgical department, served as an assistant in surgical procedures, and participated in no more than 20 laparoscopic GC surgeries. All annotators received training including learning the LE process, observing actual laparoscopic surgeries, identifying IAM lesions, and using the annotation tool. The medical annotators used the Labelme image polygonal annotation tool to annotate the IAM lesions. For single IAM lesions, annotators outlined their boundaries to create accurate segmentation masks. For multiple IAM lesions within the same frame, each lesion was annotated separately. For extensive IAM lesions, particularly large and confluent ones, annotators outlined the boundaries of each visible lesion and annotated any surrounding normal structures or tissues using the polygonal method, excluding these normal structures or tissues during mask generation to ensure accurate annotation. After completing the annotations, we used the OpenCV module to generate binary masks by converting the outlined boundaries into segmented areas (Supplementary Note 2 and Supplementary Fig. 5). Subsequently, all annotations were reviewed by expert surgeons to ensure the final accuracy of the IAM annotations. To assess the consistency of the annotators, we evaluated three kinds of consistencies: intra-annotator consistency for the same annotator across different times, inter-annotator consistency among annotators at the same level, and inter-annotator consistency among annotators at different levels. To compute the consistencies mentioned above, a randomly selected subset of 50 frames from the whole dataset was annotated twice at different times, with a seven-day interval between each annotation. The intra- and inter-annotator consistencies were expressed in terms of Dice scores and IOU55, which were further explained in the subsection “Model evaluation and statistical analysis ”.
Data preprocessing and postprocessing
In the data preprocessing step, all frames underwent preprocessing for normalization to minimize variations stemming from different imaging systems, uneven lighting, and motion blur due to the rapid movement of laparoscopic cameras. Data augmentation is used to expand the dataset and improve the robustness of model training including flipping, rotating, noising, sharpening, blurring, brightening, and gamut mapping. In addition, to segment high-resolution (1920 × 1080 pixels) laparoscopic frames without losing image information, we evenly divided these high-resolution frames into multiple 512 × 512 pixels patches using a 256 pixels step. All patches were input in the model mentioned in the “Semantic segmentation model” subsection to obtain prediction masks. Finally, we merged the patch segmentation results to obtain the final high-resolution segmentation masks in the postprocessing step.
Semantic segmentation model
The dataset of IAM lesions was randomly divided on a patient-level basis into a development set (used for model training and validation) and a test set, with an 8:2 ratio. Since the heterogeneity of breast cancer lesions is similar to that of the IAM lesions in our study, exhibiting significant variability in shape and extent. The RF-Net originally developed for segmenting the breast cancer ultrasound image was selected to construct the AiLES32. Figure 6 shows the network architecture of AiLES, which is based on the U-net. In addition, to identify the most appropriate semantic segmentation model for our task of interest, various state-of-the-art models including the DeeplabV3+ network and segmentation anything model (SAM) are tested. The DeeplabV3+ neural network architecture with an Xception backbone is adopted because it has been widely used in surgical AI studies for identifying anatomical structures and surgical instruments25,33,34. The SAM is the first universal image segmentation foundation model in the field of CV35, analogous to ChatGPT in natural language processing, which has strong segmentation capabilities based on prompts (e.g., boxes, and points). Furthermore, a medical SAM adapter (MSA) was employed to improve the performance of the original SAM due to the lack of optimization in medical images37. The process of data preprocessing and model development (Supplementary Note 3) is displayed in Fig. 1b. All codes were implemented in Python and Pytorch.
A workstation was used for the development and evaluation of AI models, which was equipped with 2 NVIDIA RTX 3090 GPUs with 24GB GPU memory, 86GRAM, and Intel(R) Xeon(R) Platinum8157 CPU@3.60 GHz, using Ubuntu 20.04. Specifically, during model training, both GPUs were used with a batch size of 32. For inference speed testing, since surgical videos are processed frame by frame, the batch size was adjusted to 1 with an input image resolution of 1920 × 1080. We utilized only one GPU when testing the inference speed to simulate the limited computational resources of the AI-assisted module for laparoscopy in the operation room. It is important to note that the reported inference speed of AiLES reflects the total time for image loading, model inference, and prediction result visualization.
Model evaluation and statistical analysis
The primary evaluation metrics used in this study were the Dice score and IOU, which are commonly employed in CV to quantify the overlap between the predicted segmented object area by the AI model and the actual area of the object annotated by expert surgeon, which was called ground truth (GT). The Dice score and IOU are calculated as follows: , . In this study, A denotes the segmentation predicted by the AI algorithm, while B refers to the manually annotated reference segmentation. The symbol “∩” denotes the intersection between A and B, while the symbol “∪” represents the union between A and B. A value closer to 1 for either the Dice score or IOU signifies a higher degree of overlap between predicted and GT areas, reflecting a perfect match at 1 and no overlap at 0. The secondary evaluation metrics (Supplementary Note 4) derived from true positive (TP), true negative (TN), false positive (FP) and false negative (FN) included recall (, same as sensitivity), specificity (), accuracy (), precision (), and F1 score (, same as Dice score). Additionally, the mAP@50 and similarity indices (including SSIM and HD) are used to provide a more comprehensive evaluation of different models. Furthermore, the inference time of models is tested to assess their real-time recognition speed. The model evaluation process is displayed in Fig. 1c. To assess statistical significance (P < 0.05), we utilized a two-sided Student’s t-test to compare the performance of AiLES with that of the surgeons. All statistical analyses were conducted using the Python (v3.9).
Supplementary information
Acknowledgements
This study was supported by the Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (grant no. pdjh2024a086) and the National Natural Science Foundation of China (grant no.82203712).
Author contributions
G.L., J.Y., and H.D. conceived and designed this study. H.C. defined the study dataset and designed the workflow. L.G., Z.F., and H.B.C. conducted model development and evaluation. L.G., H.B.C., J.Z., and C.N. annotated the study dataset. H.C., G.L., J.Y., and H.D. reviewed the annotation. L.G., C.C., and Y.Q. performed the data analysis and visualization. Q.D. revised the manuscript and contributed to model development and statistical analysis. Y.H. provided clinical insights and analysis for discussion. All authors discussed the results and contributed to the final manuscript.
Data availability
The IAM dataset generated and analyzed in the present study is not publicly available due to ethical regulations on confidentiality and privacy, but is available on reasonable request from the corresponding author.
Code availability
The AI models developed in this study include RF-Net, DeeplabV3+, SAM, and MSA. The underlying codes for all models are available on GitHub. The links are as follows: RF-Net is hosted at https://github.com/mniwk/RF-Net. DeeplabV3+ is hosted at https://github.com/VainF/DeepLabV3Plus-Pytorch; SAM is hosted at https://github.com/facebookresearch/segment-anything. MSA is hosted at https://github.com/KidsWithTokens/Medical-SAM-Adapter. In addition, the custom code of AiLES is hosted at https://github.com/CalvinSMU/AiLES.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hao Chen, Longfei Gou, Zhiwen Fang.
Contributor Information
Haijun Deng, Email: denghj@i.smu.edu.cn.
Jiang Yu, Email: balbc@163.com.
Guoxin Li, Email: gzliguoxin@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-024-01372-6.
References
- 1.Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. & Lordick, F. Gastric cancer. Lancet396, 635–648 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Cortés-Guiral, D. et al. Primary and metastatic peritoneal surface malignancies. Nat. Rev. Dis. Prim.7, 91 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Thomassen, I. et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int J. Cancer134, 622–628 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Schena, C. A. et al. The role of staging laparoscopy for gastric cancer patients: current evidence and future perspectives. Cancers15, 3425 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coccolini, F. et al. Intraperitoneal chemotherapy in advanced gastric cancer. Meta-analysis of randomized trials. Eur. J. Surg. Oncol.40, 12–26 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Ajani, J. A. et al. Gastric cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw.20, 167–192 (2022). [DOI] [PubMed] [Google Scholar]
- 8.Sarela, A. I., Lefkowitz, R., Brennan, M. F. & Karpeh, M. S. Selection of patients with gastric adenocarcinoma for laparoscopic staging. Am. J. Surg.191, 134–138 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Karanicolas, P. J. et al. Staging laparoscopy in the management of gastric cancer: a population-based analysis. J. Am. Coll. Surg.213, 644–651 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Borgstein, A. B. J., Keywani, K., Eshuis, W. J., van Berge Henegouwen, M. I. & Gisbertz, S. S. Staging laparoscopy in patients with advanced gastric cancer: a single center cohort study. Eur. J. Surg. Oncol.48, 362–369 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Hu, Y. et al. Morbidity and mortality of laparoscopic versus open D2 distal gastrectomy for advanced gastric cancer: a randomized controlled trial. J. Clin. Oncol.34, 1350–1357 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Liu, F. et al. Morbidity and mortality of laparoscopic vs open total gastrectomy for clinical stage I gastric cancer: the CLASS02 multicenter randomized clinical trial. JAMA Oncol.6, 1590–1597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, W. et al. Decreased morbidity of laparoscopic distal gastrectomy compared with open distal gastrectomy for stage I gastric cancer: short-term outcomes from a multicenter randomized controlled trial (KLASS-01). Ann. Surg.263, 28–35 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Lee, H.-J. et al. Short-term outcomes of a multicenter randomized controlled trial comparing laparoscopic distal gastrectomy with D2 lymphadenectomy to open distal gastrectomy for locally advanced gastric cancer (KLASS-02-RCT). Ann. Surg.270, 983–991 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Lordick, F. et al. Gastric cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol.33, 1005–1020 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Japanese Gastric Cancer Association. Japanese gastric cancer treatment guidelines 2021 (6th edition). Gastric Cancer26, 1–25 (2023). [DOI] [PMC free article] [PubMed]
- 17.Kim, C. Y. et al. Learning curve for gastric cancer surgery based on actual survival. Gastric Cancer19, 631–638 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Schnelldorfer, T. et al. Can we accurately identify peritoneal metastases based on their appearance? An assessment of the current practice of intraoperative gastrointestinal cancer staging. Ann. Surg. Oncol.26, 1795–1804 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Suliburk, J. W. et al. Analysis of human performance deficiencies associated with surgical adverse events. JAMA Netw. Open2, e198067 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquet, P. & Sugarbaker, P. H. Clinical research methodologies in diagnosis and staging of patients with peritoneal carcinomatosis. Cancer Treat. Res.82, 359–374 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Rau, B. et al. Effect of hyperthermic intraperitoneal chemotherapy on cytoreductive surgery in gastric cancer with synchronous peritoneal metastases: the phase III GASTRIPEC-I trial. J. Clin. Oncol.42, 146–156 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan, W.-L., He, Y. & Xu, R.-H. Gastric cancer treatment: recent progress and future perspectives. J. Hematol. Oncol.16, 57 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mascagni, P. et al. Computer vision in surgery: from potential to clinical value. NPJ Digit Med.5, 163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitaguchi, D. et al. Development and validation of a model for laparoscopic colorectal surgical instrument recognition using convolutional neural network-based instance segmentation and videos of laparoscopic procedures. JAMA Netw. Open5, e2226265 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima, S. et al. Deep-learning-based semantic segmentation of autonomic nerves from laparoscopic images of colorectal surgery: an experimental pilot study. Int J. Surg.109, 813–820 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng, K. et al. Artificial intelligence-based automated laparoscopic cholecystectomy surgical phase recognition and analysis. Surg. Endosc.36, 3160–3168 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Mascagni, P. et al. Artificial intelligence for surgical safety: automatic assessment of the critical view of safety in laparoscopic cholecystectomy using deep learning. Ann. Surg.275, 955–961 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Igaki, T. et al. Automatic surgical skill assessment system based on concordance of standardized surgical field development using artificial intelligence. JAMA Surg.158, e231131 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, S. et al. Semantic instance segmentation with discriminative deep supervision for medical images. Med Image Anal.82, 102626 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Wei, G.-X., Zhou, Y.-W., Li, Z.-P. & Qiu, M. Application of artificial intelligence in the diagnosis, treatment, and recurrence prediction of peritoneal carcinomatosis. Heliyon10, e29249 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnelldorfer T., Castro J., Goldar-Najafi A., Liu L. Development of a deep learning system for intra-operative identification of cancer metastases. Ann. Surg. 280, 1006–1013 (2024). [DOI] [PubMed]
- 32.Wang, K. et al. (Springer International Publishing, 2021).
- 33.Kolbinger, F. R. et al. Anatomy segmentation in laparoscopic surgery: comparison of machine learning and human expertise - an experimental study. Int J. Surg.109, 2962–2974 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebigbo, A. et al. Vessel and tissue recognition during third-space endoscopy using a deep learning algorithm. Gut.71, 2388–2390 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander, K. et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) 4015–4026 (ICCV, 2023).
- 36.Mazurowski, M. A. et al. Segment anything model for medical image analysis: an experimental study. Med. Image Anal.89, 102918 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, J. et al. Medical SAM adapter: adapting segment anything model for medical image segmentation. Preprint at 10.48550/arXiv.2304.12620 (2023).
- 38.Kawamura, M. et al. Development of an artificial intelligence system for real-time intraoperative assessment of the critical view of safety in laparoscopic cholecystectomy. Surg. Endosc.37, 8755–8763 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Kitaguchi, D. et al. Real-time vascular anatomical image navigation for laparoscopic surgery: experimental study. Surg. Endosc.36, 6105–6112 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Carstens, M. et al. The dresden surgical anatomy dataset for abdominal organ segmentation in surgical data science. Sci. Data10, 3 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitaguchi, D. et al. Artificial intelligence for the recognition of key anatomical structures in laparoscopic colorectal surgery. Br. J. Surg.110, 1355–1358 (2023). [DOI] [PubMed] [Google Scholar]
- 42.den Boer, R. B. et al. Deep learning-based recognition of key anatomical structures during robot-assisted minimally invasive esophagectomy. Surg. Endosc.37, 5164–5175 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura, T. et al. Precise highlighting of the pancreas by semantic segmentation during robot-assisted gastrectomy: visual assistance with artificial intelligence for surgeons. Gastric Cancer27, 869–875 (2024). [DOI] [PubMed] [Google Scholar]
- 44.Hato, S. et al. Effectiveness and limitations of staging laparoscopy for peritoneal metastases in advanced gastric cancer from the results of JCOG0501: a randomized trial of gastrectomy with or without neoadjuvant chemotherapy for type 4 or large type 3 gastric cancer. J. Clin. Oncol.35, 9–9 (2017). [Google Scholar]
- 45.Irino, T. et al. Diagnostic staging laparoscopy in gastric cancer: a prospective cohort at a cancer institute in Japan. Surg. Endosc.32, 268–275 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Liu, K. et al. “Four-Step Procedure” of laparoscopic exploration for gastric cancer in West China Hospital: a retrospective observational analysis from a high-volume Institution in China. Surg. Endosc.33, 1674–1682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujitani, K. et al. Gastrectomy plus chemotherapy versus chemotherapy alone for advanced gastric cancer with a single non-curable factor (REGATTA): a phase 3, randomised controlled trial. Lancet Oncol.17, 309–318 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Hu, C., Terashima, M. & Cheng, X. Conversion therapy for stage IV gastric cancer. Sci. Bull.68, 653–656 (2023). [DOI] [PubMed] [Google Scholar]
- 49.Lin, T. et al. Laparoscopic cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for gastric cancer with intraoperative detection of limited peritoneal metastasis: a phase II study of CLASS-05 trial. Gastroenterol. Rep.12, goae001 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chicago Consensus Working Group. The Chicago Consensus on peritoneal surface malignancies: management of gastric metastases. Cancer126, 2541–2546 (2020). [DOI] [PubMed]
- 51.Mascagni, P. et al. Early-stage clinical evaluation of real-time artificial intelligence assistance for laparoscopic cholecystectomy. Br. J. Surg.111, znad353 (2024). [DOI] [PubMed] [Google Scholar]
- 52.Cao, J. et al. Intelligent surgical workflow recognition for endoscopic submucosal dissection with real-time animal study. Nat. Commun.14, 6676 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsushita, H., Watanabe, K. & Wakatsuki, A. Metastatic gastric cancer to the female genital tract. Mol. Clin. Oncol.5, 495–499 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins, G. S. et al. TRIPOD + AI statement: updated guidance for reporting clinical prediction models that use regression or machine learning methods. BMJ385, e078378 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin, K. et al. FIVES: a fundus image dataset for artificial intelligence based vessel segmentation. Sci. Data9, 475 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The IAM dataset generated and analyzed in the present study is not publicly available due to ethical regulations on confidentiality and privacy, but is available on reasonable request from the corresponding author.
The AI models developed in this study include RF-Net, DeeplabV3+, SAM, and MSA. The underlying codes for all models are available on GitHub. The links are as follows: RF-Net is hosted at https://github.com/mniwk/RF-Net. DeeplabV3+ is hosted at https://github.com/VainF/DeepLabV3Plus-Pytorch; SAM is hosted at https://github.com/facebookresearch/segment-anything. MSA is hosted at https://github.com/KidsWithTokens/Medical-SAM-Adapter. In addition, the custom code of AiLES is hosted at https://github.com/CalvinSMU/AiLES.