Summary
Background
Pancreatic cancer is highly aggressive and has a low survival rate primarily due to late-stage diagnosis and the lack of effective early detection methods. We introduce here a novel, noninvasive urinary extracellular vesicle miRNA-based assay for the detection of pancreatic cancer from early to late stages.
Methods
From September 2019 to July 2023, Urine samples were collected from patients with pancreatic cancer (n = 153) from five distinct sites (Hokuto Hospital, Kawasaki Medical School Hospital, National Cancer Center Hospital, Kagoshima University Hospital, and Kumagaya General Hospital) and non-cancer participants (n = 309) from two separate sites (Hokuto Hospital and Omiya City Clinic). The main inclusion criteria included a diagnosis of pancreatic cancer based on pathological or imaging examination, while multiple primary cancers were excluded. Extracellular vesicles were enriched using a polymer-based precipitation method, and miRNAs were comprehensively analyzed by small RNA sequencing. A machine learning model for pancreatic cancer detection was developed using a training dataset (n = 315) consisting of 99 pancreatic cancer participants (of which 33 were early-stage [I/IIA]) and 216 non-cancer participants, and validated with a test dataset (n = 147) consisting of 54 pancreatic cancer participants (of which 9 were early-stage [I/IIA]) and 93 non-cancer participants.
Findings
This method showed consistent performance, with areas under the receiver operating characteristic curves of 0.972 (95% confidence interval [CI], 0.928–0.996) and 0.963 (95% CI, 0.932–0.988) in the training and test sets, respectively. The sensitivities for pancreatic cancer detection were 93.9% (95% CI, 87.5%–97.3%) and 77.8% (95% CI, 64.9%–87.3%) overall and 97.0% (95% CI, 83.9%–99.8%) and 77.8% (95% CI, 44.2%–95.9%) for stage I/IIA pancreatic cancer, respectively. The specificities were 91.7% (95% CI, 87.1%–94.7%) and 95.7% (95% CI, 89.4%–98.5%), respectively. We also evaluated the sensitivity of CA19-9 for pancreatic cancer detection in 140 patients with pancreatic cancer, and it was 37.5% (95% CI, 23.5%–53.8%) for stages I/IIA pancreatic cancer. Performance in early-stage cancer detection was significantly higher for miRNA-based pancreatic cancer detection. Functional enrichment analysis of pancreatic cancer-associated urinary miRNAs revealed that the urinary miRNA signature reflects miRNA patterns of the pancreatic cancer tissue itself as well as those of the tumor microenvironment.
Interpretation
Urinary extracellular vesicle miRNAs may reflect signals from both tumor cells and their microenvironment, offering a unique opportunity for detection of pancreatic cancer from early to late stages. While this study has a limitation due to the relatively small sample size, our approach has the potential to contribute to treatment outcomes through population screening. Our primary goal is to make this assay more accessible to a broader population, particularly in areas with limited hospital access where cancer is often detected at a late stage, leveraging the advantage of using urine samples that can be collected at home.
Funding
This research was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Number JP24he2302007 and Craif Inc.
Keywords: Pancreatic cancer, Liquid biopsy, Micro RNA, Machine learning, Urinary assay
Research in context.
Evidence before this study
Cancer antigen 19-9 (CA19-9) is a clinically relevant biomarker of pancreatic cancer, although the sensitivity is stage-dependent and tends to be lower in the early stages. Detection of pancreatic cancer associated circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) is insufficient in sensitivity at early stages when concentrations are low because of their tumor origin. The role of miRNA in pancreatic cancer is well-established in the scientific literature. A PubMed search (March 13, 2024) for research articles with the terms “miRNA” and “pancreatic cancer” (in its various forms) revealed 3274 publications, including 554 review articles. Adding “exosome” and/or “extracellular vesicle” to the search narrowed the results to 289 publications, with 69 being review articles. Notably, only one research article and three review articles specifically investigated the role of urinary exosomal miRNA in pancreatic cancer. The research article employed miRNA microarray to identify differentially expressed miRNAs in urinary exosomes of pancreatic ductal adenocarcinoma patients. However, no study has yet utilized next-generation sequencing (NGS) to profile urinary exosomal miRNA in pancreatic cancer. Moreover, although miRNAs were noted as potential cancer biomarkers, blood miRNA profiles were affected by pre-analytical processes, which poses a challenge for practical application.
Added value of this study
We developed and validated a noninvasive urinary extracellular vesicle miRNA-based assay using small RNA-seq for pancreatic cancer detection. Enriching urinary extracellular vesicles have enabled efficient small RNA sequencing to improve the low concentration of miRNAs in urine. Functional enrichment analysis of pancreatic cancer-associated urinary miRNA and tumor-derived organoid analysis suggested that miRNAs secreted not only from tumor cells but also from the tumor microenvironment contributed to our urinary miRNA-based detection of pancreatic cancer, although pancreatic tumors are thought to be small in the early stages. Detecting pancreatic cancer in early stages could significantly improve the management of pancreatic cancer by triaging patients to earlier diagnostic testing, potentially resulting in diagnosis at a curative stage.
Implications of all the available evidence
The miRNA-based pancreatic cancer detection was superior to CA19-9 in terms of performance, especially in the early stage, despite the limitation of a relatively small sample size. This study has the potential of urinary liquid biopsy and may contribute to the overall outcome of pancreatic cancer treatment through its application in both population screening and the management of patients with increased pancreatic cancer risk.
Introduction
Pancreatic cancer, with its highly aggressive nature and often late-stage diagnosis, is one of the deadliest malignancies worldwide, with a 5-year survival rate of only 12%,1 because of the late-stage diagnosis of the majority of patients owing to the lack of screening tools and the difficulty of early detection; moreover, indications for curative surgery are limited to early-stage cancers, and < 20% of patients can benefit from it.2 Earlier detection of pancreatic cancer when it is still localized in the Surveillance, Epidemiology, and End Results (SEER) stage can improve the 5-year survival rate by up to 44%.1 Cancer antigen 19-9 (CA19-9) is a clinically relevant biomarker of pancreatic cancer (sensitivity: 70–90%), although the sensitivity is stage-dependent and tends to be lower in the early stages.3,4 Thus, there is a crucial need for innovative approaches that can enable early intervention and improve patient outcomes.
Liquid biopsy, a minimally invasive or noninvasive diagnostic technique, is a promising approach in cancer research5 that harnesses the circulatory potential of molecular markers in bodily fluids. Several blood-based liquid biopsy tests developed in recent years have demonstrated efficacy in detecting pancreatic cancer associated circulating tumor cells (CTC) and circulating tumor DNA (ctDNA).6,7 However, owing to the low concentrations and tumor-origin nature of CTCs and ctDNAs, the level of early sensitivity is insufficient to improve prognosis dramatically.
MicroRNAs (miRNA), which are small non-coding RNAs with an average length of 22 nucleotides that are involved in post-transcriptional gene regulation, have garnered attention for their stability in biofluids and their implications in cancer pathogenesis.8 Circulating miRNAs are potential cancer biomarkers, with demonstrable usefulness in detecting early-stage cancers, including pancreatic cancer.9, 10, 11 Nonetheless, blood miRNA profiles are affected by pre-analytical processes, including the type of anticoagulant used in plasma preparation, sample storage, and sampling, which poses a challenge for practical application.12
Urine, an easily accessible biofluid, has immense potential as a noninvasive medium for population screening, and urinary miRNA-based assays constitute a potential tool for detection of cancers13 and pancreatic adenocarcinoma.14,15 Furthermore, research on the relative stability of human urinary miRNAs under various storage conditions supports their potential use as urinary biomarkers.16 However, given the low concentration of urinary miRNAs, and the high ratio of tRNAs compared to those in the blood, efficient recovery of miRNAs using next-generation small RNA sequencing (small RNA-seq) platforms is difficult.17 To overcome the issue, enrichment of extracellular vesicles containing various molecules, including miRNAs,18 from urine may enable efficient isolation and measurement of miRNAs. Moreover, exosomes, which are extracellular vesicle subtypes, have emerged as important biomarkers of cancer because of their involvement in cancer progression and metastasis.19
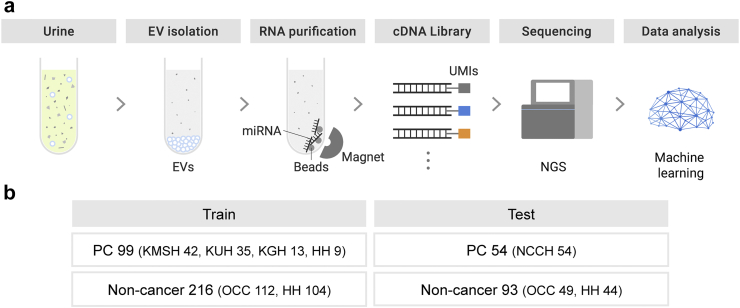
In this study, we developed and validated a noninvasive urinary extracellular vesicle miRNA-based assay using small RNA-seq for the detection of pancreatic cancer across all stages (Fig. 1). This study aimed to assess the potential of urinary liquid biopsy and miRNA markers for the detection of pancreatic cancer.
Fig. 1.
Schematic overview of noninvasive urinary extracellular vesicle miRNA-based liquid biopsy. (a) Workflow of noninvasive urinary extracellular vesicle miRNA-based liquid biopsy with a next-generation small RNA sequencing platform for the detection of pancreatic cancer. (b) Samples included in the training and test sets. Patients with pancreatic cancer from five distinct sites and non-cancer participants from two separate sites were enrolled. Abbreviations: EVs, extracellular vesicles; UMIs, unique molecular identifier; NGS, next-generation sequencing; PC, pancreatic cancer; HH, Hokuto Hospital; KMSH, Kawasaki Medical School Hospital; NCCH, National Cancer Center Hospital; KUH, Kagoshima University Hospital; KGH, Kumagaya General Hospital; OCC, Omiya City Clinic.
Methods
Ethics
This study was approved by the Institutional Review Boards of Hokuto Hospital (No. 1051), Kawasaki Medical School Hospital (No. 2020-1016 and 2021-0598), National Cancer Center Hospital (NCCH, No. 2021-308), Kagoshima University Hospital (No. 200149疫), Kumagaya General Hospital (No. 54), and Craif (No. IEF-20220516). Written informed consent was obtained from all participants.
Study design and cohorts
This case–control study was performed using urine obtained from participants without cancer (controls) and with pancreatic cancer (cases). From September 2019 to July 2023, patients with pancreatic cancer from five distinct sites (Hokuto Hospital, Kawasaki Medical School Hospital, NCCH, Kagoshima University Hospital, and Kumagaya General Hospital) and non-cancer participants from two separate sites (Hokuto Hospital and Omiya City Clinic) were enrolled. The inclusion criteria included a diagnosis of pancreatic cancer based on pathological or imaging examination, whereas individuals with multiple primary cancers were excluded from the study. According to 2020 cancer statistics from the National Cancer Center of Japan, the average age of individuals with pancreatic cancer is 73 years for men and 75 years for women, with similar incidence rates between the genders. Since urine samples were collected from hospitals nationwide, regional influences are minimal, ensuring the cohort represents pancreatic cancer cases in Japan. The majority of the pancreatic cancer participants had pancreatic ductal adenocarcinoma (PDAC), which is consistent with the PDAC incidence of > 90% of all pancreatic cancer cases.20
In this study, stage I/IIA cases of TMN classification by the Union for International Cancer Control (UICC) were defined as early-stage pancreatic cancer group, and stage IIB/III/IV cases were defined as late-stage pancreatic cancer group. For non-cancer participants, the inclusion criteria were confirmed through health checkups, such as comprehensive medical examinations, and those with a history of any cancer within five years were excluded.
The collected cases, through a review of medical records, were divided into training and test sets to develop and validate an miRNA-based pancreatic cancer-detection algorithm. To validate the model's performance in an independent cohort, pancreatic cancer cases from NCCH were exclusively collected for the test set. Non-cancer participants were randomly assigned to the training and test sets in the same proportion as in the pancreatic cancer group. Consequently, 462 participants (pancreatic cancer, n = 153; non-cancer, n = 309) were enrolled. Of these, 315 (pancreatic cancer, n = 99; non-cancer, n = 216) and 147 (pancreatic cancer, n = 54; non-cancer, n = 93) were included in the training and test sets, respectively (Fig. 1, Table 1). Of 153 patients with pancreatic cancer, 18 were diagnosed using imaging.
Table 1.
Patient demographic characteristics.
| Dataset |
Train |
p-value | Test |
p-value | ||
|---|---|---|---|---|---|---|
| Disease | PC (n = 99) | NC (n = 216) | PC (n = 54) | NC (n = 93) | ||
| Age | ||||||
| Median (IQR) | 74.0 (65.5–79.5) | 60.0 (34.0–65.0) | <0.0001 | 67.5 (59.5–73.8) | 60.0 (35.0–65.0) | <0.0001 |
| BMI (kg/m2) | ||||||
| Median (IQR) | 21.6 (19.4–24.4) | 22.6 (20.7–25.2) | 0.013 | 22.1 (19.6–24.4) | 23.2 (21.0–25.2) | 0.032 |
| NA | 0 | 2 | – | 0 | 1 | – |
| Sex | ||||||
| Female | 36 (36.4%) | 105 (48.6%) | 0.056 | 26 (48.1%) | 42 (45.2%) | 0.86 |
| Male | 63 (63.6%) | 111 (51.4%) | 28 (51.9%) | 51 (54.8%) | ||
| Smoking history | ||||||
| Current use | 12 (12.1%) | 29 (13.4%) | 0.024 | 7 (13.0%) | 24 (25.8%) | 0.0049 |
| Never used | 50 (50.5%) | 140 (64.8%) | 21 (38.9%) | 47 (50.5%) | ||
| Past used | 33 (33.3%) | 43 (19.9%) | 26 (48.1%) | 21 (22.6%) | ||
| NA | 4 (4.0%) | 4 (1.9%) | – | 0 (0.0%) | 1 (1.1%) | – |
| Alcohol | ||||||
| No | 55 (55.6%) | 104 (48.1%) | 0.21 | 30 (55.6%) | 44 (47.3%) | 0.50 |
| Yes | 41 (41.4%) | 109 (50.5%) | 24 (44.4%) | 47 (50.5%) | ||
| NA | 3 (3.0%) | 3 (1.4%) | – | 0 (0.0%) | 2 (2.2%) | – |
| Histology | ||||||
| Duct adenocarcinoma, NOS | 83 (83.8%) | – | – | 52 (96.3%) | – | – |
| Adenosquamous carcinoma NOS | – | – | – | 2 (3.7%) | – | – |
| Carcinoma, undifferentiated, NOS | 1 (1.0%) | – | – | – | – | – |
| Intraductal papillary mucinous carcinoma | 1 (1.0%) | – | – | – | – | – |
| NA | 14 (14.1%) | 216 (100.0%) | – | 0 (0.0%) | 93 (100.0%) | – |
| Stage | ||||||
| I | 24 (24.2%) | – | – | 8 (14.8%) | – | – |
| IIA | 9 (9.1%) | – | – | 1 (1.9%) | – | – |
| IIB | 30 (30.3%) | – | – | 0 (0.0%) | – | – |
| III | 14 (14.1%) | – | – | 20 (37.0%) | – | – |
| IV | 17 (17.2%) | – | – | 25 (46.3%) | – | – |
| NA | 5 (5.1%) | 216 (100.0%) | – | 0 (0.0%) | 93 (100.0%) | – |
NA, not available; PC, Pancreatic cancer; NC, Non-cancer; BMI, Body Mass Index.
Urine collection, extracellular vesicle isolation from urine and RNA extraction
Urine samples from Hokuto Hospital, Kawasaki Medical School Hospital, NCCH, Kagoshima University Hospital, and Kumagaya General Hospital were collected at the participants' convenience during their visit to the clinic and stored at −80 °C, whereas those from Omiya City Clinic were stored at −20 °C, and were transported to the analysis facility where all samples were stored at −80 °C until use.
Urine samples (∼3.5 mL) were centrifuged at 2000×g for 30 min at 4 °C to sediment the cells and cellular debris. The supernatant (3 mL) was mixed (1:1) with the Total Exosome Isolation Reagent (from urine) (Thermo Fisher Scientific, Waltham, MA, USA) and incubated at room temperature for 1 h with shaking (100 rpm). Extracellular vesicles were spun down by centrifugation at 3000×g for 1 h at 4 °C. The supernatant was discarded, and the pellet resuspended in 300 μL phosphate-buffered saline (PBS). The extracellular vesicle solution was stored at 4 °C until use. Total RNA was extracted from the extracellular vesicle solution using the MagMAX mirVana Total RNA Isolation Kit on a KingFisher Apex System (both from Thermo Fisher Scientific) following the manufacturer's instructions. RNA extraction was eluted in 30-μL elution buffer. The solution was concentrated using a centrifugal concentrator (Eppendorf, Hamburg, Germany) at 60 °C for 30–45 min and eluted in 5 μL nuclease-free water (Thermo Fisher Scientific). Extracted RNA was stored at −80 °C until use.
To compare miRNA yield (miRNA mapping rate) with and without extracellular vesicle isolation, miRNAs were also extracted from 300 μL of cell-free urine (cfUrine) without extracellular vesicle isolation using the same protocol. All 69 donors were included in this study, consisting of 59 pancreatic cancer and 10 non-cancer cases, randomly selected from available samples.
Small RNA library construction and sequencing analysis
Libraries were prepared from 5 μL RNA using the QIAseq miRNA Library Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Library concentrations were measured using Qubit™ dsDNA HS Assay Kit on a Qubit Flex Fluorometer (both from Thermo Fisher Scientific) and stored at −20 °C until further processing. Libraries were pooled at equimolar concentrations and sequenced on a NextSeq 550 System (Illumina, San Diego, CA, USA) with single-end reads of 75 nucleotides according to the manufacturer's instructions.
Raw sequencing reads were initially processed using UMI-tools,21 which involved the extraction of the initial 5′-end bases, indicative of miRNA, which were positioned anterior to the 3′-adaptor sequence. Concurrently, 12-bp UMI succeeding the 3′-adaptor sequence were integrated into the read identifiers to facilitate subsequent analyses. Following this step, reads longer than 19 bp were aligned to the human miRNA reference provided by miRge 3.022 using Bowtie v1.2.3,23 allowing one mismatch within a 25-bp seed region and no reverse complement mapping. Reads that shared an identical UMI sequence and were aligned to the same miRNA reference were collapsed, and their counts were recorded.
The miRNA yield was evaluated using the current assay. The UMI duplication rate was calculated for each sample as the UMI-unique count divided by the total mapped count averaged over all miRNAs. As miRNA yield depends on sequencing depth, it was normalized to a target depth x = 4 × 106 as miRNA Yield = (x/A)/[1 + (x/B)] where parameters A and B were defined as A = T/m and B = T/(U-1) with total read T, total miRNA read m, and UMI duplication rate U.
Machine learning analysis
The miRNA count profile dataset was divided into the training and testing datasets. Prior to the machine learning analysis, the training set underwent several preprocessing steps. First, the miRNA features commonly detected within the training set were extracted. Features with at least θ counts detected for more than r% of the samples were included (θ = 3 and r = 80% were used). The miRNA count profile was normalized using the median ratio method in the training set, wherein a reference profile was calculated as the geometric mean of the miRNA count profile in the training set. It was used to normalize both training and test sets. The normalized profile was transformed according to log2 (1 + x).
A support vector classifier (SVC) with a linear kernel was fitted to develop a predictive model for pancreatic cancer. The normalized miRNA profile was subjected to a standard scaling transformation before being input into the model. The hyperparameters were tuned according to the maximization of the area under the curve (AUC) of the receiver operating characteristic (ROC) curve evaluated by five-fold cross-validation using Optuna.24 To obtain prediction results in the test set, the classifier model with the selected hyperparameters was fitted to the training set and adopted in the test set. The prediction score was obtained on the scale of [0, 1] according to a sigmoid-like transformation using the Platt scaling method. The optimal threshold was determined from the stacked out-of-fold prediction scores through cross-validation in the training set according to the Youden index method, which maximizes the sum of sensitivity and specificity. To address potential bias from the participant age range, an age-matched subset of the whole dataset was selected according to a frequency matching with 5-year intervals, and an SVC model was fitted and evaluated by a five-fold cross-validation in the same way as described above.
Differential expression analysis and functional enrichment analysis
Differential expression analysis (DEA) was performed using DESeq225 to obtain differentially expressed miRNAs (DEM) according to fold-changes and p-values with the Benjamini–Hochberg adjustment. Before DEA, miRNA features commonly detected across the training set were extracted similarly as in the machine learning preprocessing, albeit with different parameter values (θ = 3 and r = 50% relaxed for DEM discovery). DEMs in the urinary dataset were called according to an adjusted p-value < 0.05 and an absolute log2 fold change > 0.5.
For functional enrichment analysis, the miRNA Enrichment Analysis and Annotation Tool 2.126 was used to perform an overrepresentation analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases using miRTarBase. A Benjamini–Hochberg adjusted p-value < 0.05 was considered statistically significant.
CA19-9 measurement
For 140 pancreatic cancer samples, plasma CA19-9 levels were measured using a chemiluminescent immunoassay at the medical facility at which each patient was diagnosed. According to the standard clinical threshold value for pancreatic cancer detection, samples with CA19-9 > 37.0 U/mL were classified as positive.
Generation of pancreatic organoids and collection of culture supernatant
A pathologist selected tumor and normal tissues, which were washed and shredded to 1–2 mm pieces. These pieces were treated with 50 μg/μL Liberase (Roche, Basel, Switzerland) for 1 h, then centrifuged at 200×g for 5 min at 4 °C. Pellets were resuspended in 50% Matrigel Basement Membrane Matrix (Corning, Corning, NY, USA) and seeded into each well of a 24-well plate (Corning). After solidifying, 500 μL organoid culture medium was added. It was composed of Advanced DMEM/F12 (Thermo), 10 mM HEPES (Sigma-Aldrich, St. Louis, MO, USA), GlutaMax supplement (Thermo), N2 and B27 supplements (Thermo), 10 nM gastrin (Sigma-Aldrich), 1.25 mM N-Acetyl-Cysteine (Sigma-Aldrich), 10 mM Nicotinamide (Sigma-Aldrich), 0.5 μM A83-01 (Sigma-Aldrich), 10 μM SB202190 (Sigma-Aldrich), 100 ng/mL EGF (PeproTech, Cranbury, NJ, USA), 1 ng/mL FGF2 (PeproTech), 100 ng/mL FGF10 (PeproTech), Nogin (PeproTech), 1 μM Prostaglandin E2 (Sigma-Aldrich), 10% Afamin/Wnt3a CM (MBL, Tokyo, Japan), 10% R-Spondin 1 conditioned medium, 10 μM Y27632 (Sigma-Aldrich), antibiotic/antimycotic (Thermo), and Primocin (Invivogen, San Diego, CA, USA). Clinical sequencing confirmed the organoids contained the same gene variants as the tumor tissue, with no variants in normal tissue. After sufficient organoid growth, the medium was replaced with a conditioned medium-free medium to collect the supernatant. After 48-h incubation, the supernatant was collected, centrifuged at 2000×g for 10 min at 4 °C, filtered through a 0.22-μm filter (Merck, Rahway, NJ, USA), and stored at −80 °C.
Statistics
For hypothesis testing in two-group comparisons, the chi-square test was used for categorical variables, and the Mann–Whitney U and Wilcoxon signed-rank sum tests were used for continuous variables in unmatched and matched samples, respectively. The non-parametric approach was adopted as we observed skewed distributions in the variables of interest. A multiple regression analysis was performed by employing a generalized linear model with a logit link function, with cancer status (pancreatic cancer or not) values, age, Body Mass Index (BMI), sex, alcohol, and smoking history as the explanatory variables and the binary outcome of the model as the target variable. The categorical variables were converted into one-hot vectors. Any missing values were reported and were removed from the statistical analyses. Unless otherwise mentioned, p < 0.05 was considered statistically significant. Confidence intervals were estimated using the Wilson's score interval for proportions and the bootstrap method for other numerical metrics, such as AUC.
Role of the funding source
AMED had no role in the study design, the collection/analysis/interpretation of data, the writing of the report, or the decision to submit the paper for publication. Craif Inc. played a role in the study design, data collection/analysis/interpretation of data, and the writing of the report. A pharmaceutical company or other agency has not paid us to write this article. All authors were not precluded from accessing data in the study and accept responsibility for the publication.
Results
Urinary extracellular vesicle-derived miRNA-assay
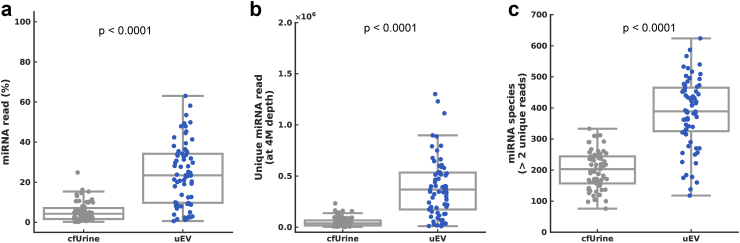
To increase the miRNA concentration, urinary extracellular vesicles were precipitated and enriched before RNA extraction (Fig. 1). Compared to cfUrine-derived miRNA without extracellular vesicle isolation (when RNA is extracted from the same volume of urine as the extracellular vesicle solution), the mapping rate of miRNAs significantly increased in urinary extracellular vesicle-derived miRNA (Wilcoxon signed-rank sum test, p < 0.0001) from 5.22% ± 4.72% to 24.5% ± 15.6% (mean ± SD; Fig. 2a). The count of unique miRNA reads was eight times higher when the miRNA was extracted from extracellular vesicles at the same sequencing depth (Wilcoxon signed-rank sum test, p < 0.0001; Fig. 2b), and hence the number of detected microRNA species (> 2 unique miRNA reads) was improved from 200.7 ± 59.3 to 385.9 ± 111.7 (Wilcoxon signed-rank sum test, p < 0.0001; Fig. 2c). We evaluated extracellular vesicles using nanoparticle tracking analysis (NTA) with urine samples. We demonstrated that our precipitation method for extracellular vesicle extraction provides sufficient extracellular vesicle yields of 1010 particles/mL from just 3 mL of urine (Supplementary Fig. S1).
Fig. 2.
Improved miRNA yield by extracellular vesicle enrichment. miRNAs were extracted from urinary extracellular vesicle (uEV) and cell-free urine (cfUrine) for 69 donors, and their miRNA yields were compared. (a) Percentage of miRNA read in total read. (b) Unique miRNA reads at 4M depth. (c) Number of miRNA species detected (unique miRNA reads > 2).
Participant demographics and baseline characteristics
The clinical characteristics of the cohort, including height, weight, age, sex, smoking history, and drinking history are shown in Table 1. Statistical analysis of the clinical variables showed that, in both the training and test sets, pancreatic cancer participants tended to be older than non-cancer participants (Mann–Whitney U test, p < 0.0001 and p < 0.0001 in the training and test sets, respectively) and had a lower body mass index (Mann–Whitney U test, p = 0.013 and p = 0.032). The pancreatic cancer group had fewer current smokers and more past smokers (chi-square test, p = 0.024 and p = 0.0049, respectively). Other baseline characteristics were comparable between the groups. In the cancer group of the training set, 33.3% (33/99) of patients had stage I/IIA cancer, whereas only 16.7% (9/54) had stage I/IIA cancer in the test set (Table 1).
Development and testing of a miRNA-based pancreatic cancer-detection algorithms
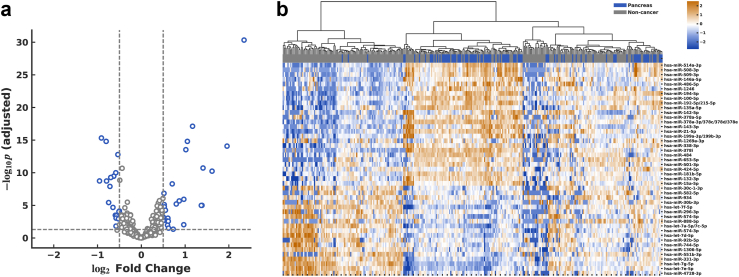
The miRNA profiles of pancreatic cancer and non-cancer participants were compared in the training cohort. DEA revealed 45 DEMs, including 26 up-regulated and 19 down-regulated miRNAs in pancreatic cancer within the training set (Fig. 3a and Supplementary Table S1). An miRNA expression heatmap showed that most pancreatic cancer patients clustered closely (Fig. 3b).
Fig. 3.
Differential expression analysis. (a) Volcano plot in train set with 306 miRNAs. 26 miRNAs were up-regulated, and 19 miRNAs were down-regulated. (b) Heatmap showing expression pattern of urinary miRNAs. Log2-transformed and normalized miRNA expression values were transformed to z-scores for each of 45 differentially expressed miRNAs (DEMs) and values > 2.5 or < −2.5 were rounded for visualization. miRNA features were shown in rows and samples in columns. Hierarchical clustering was performed for rows by Ward method with Euclidean metric, and the dendrogram was shown. Pancreatic cancer and non-cancer were labeled in blue and gray, respectively, in the first row.
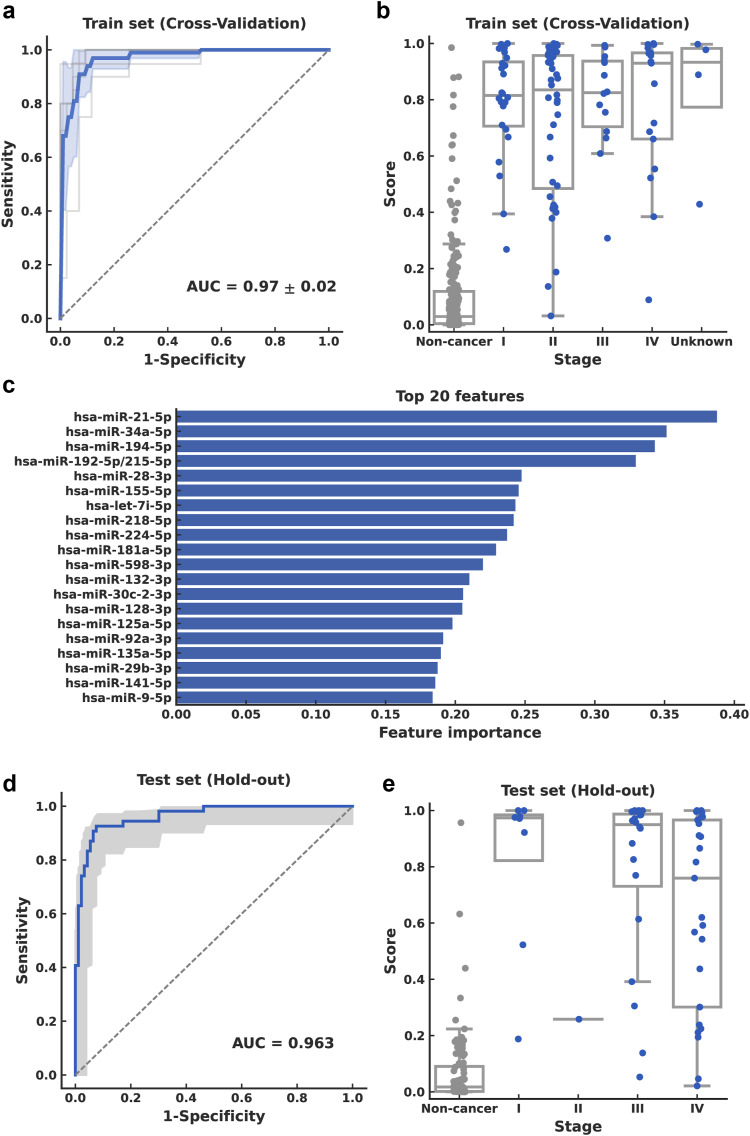
A binary prediction algorithm was constructed using a support vector classifier with normalized miRNA profiles with 183 miRNAs commonly detected in the training set as input features. The performance of the model was evaluated via a five-fold cross-validation, and the best model was selected according to hyperparameter tuning; the AUC of the ROC curve was 0.972 (95% confidence interval [CI], 0.928–0.996; Fig. 4a). The prediction score across stages in the training set showed that the predictive model allowed the detection of early stages, namely, stages I and II (Fig. 4b). The miRNAs with the highest contributions to the constructed algorithm were hsa-miR-21-5p, hsa-miR-34a-5p, and hsa-miR-194-5p (Fig. 4c). The threshold value (0.308) was determined from the training set according to Youden's Index. The overall test sensitivity of this five-fold cross-validation analysis was 93.9% (95% CI, 87.5%–97.3%), with early-stage (I/IIA) sensitivity of 97.0% (95% CI, 83.9%–99.8%) and specificity of 91.7% (95% CI, 87.1%–94.7%; Table 2). When repeating five times with different sample-fold assignments to assess the reproducibility of five-fold cross-validation, AUC (SD = 0.026), sensitivity (SD = 5.58%) and specificity (SD = 6.28%) were consistent.
Fig. 4.
Performance of urinary miRNA-based model for pancreatic cancer prediction. Performance of the support vector classifier model in the training set evaluated via cross-validation (a–c) and in the test set (d, e). (a) Receiver operating characteristic (ROC) curve from the training set. Mean of five-fold cross-validation is shown as the solid blue line, with standard deviation in blue shading. (b) Prediction scores were obtained via cross-validation for all stages in the training set. (c) Feature importance of the model for top 20 features. Absolute values of the model coefficients were used as feature importance. (d) ROC curve in the test set. The shaded region indicates the 95% confidence interval. (e) Prediction scores across stages in the test set.
Table 2.
Sensitivity and specificity in the training and validation data sets and CA19-9.
| Urinary miRNA-based model |
CA19-9 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N |
AUC | Youden index |
95% specificity target |
N |
>37.0 U/mL |
||||||
| PC | NC | Specificity | Sensitivity | Specificity | Sensitivity | PC | NC | Specificity | Sensitivity | ||
| Train | |||||||||||
| All | 99 | 216 | 0.972 (0.928–0.996) | 91.7% (87.1%–94.7%) | 93.9% (87.5%–97.3%) | 94.9% (91.0%–97.3%) | 83.8% (75.3–90.0%) | – | – | – | – |
| Stage I/IIA | 33 | 0.987 (0.955–1.00) | 97.0% (83.9%–99.8%) | 90.9% (76.1%–97.5%) | – | – | – | – | |||
| Test | |||||||||||
| All | 54 | 93 | 0.963 (0.932–0.988) | 95.7% (89.4%–98.5%) | 77.8% (64.9%–87.3%) | 97.8% (92.6%–99.6%) | 74.1% (60.3%–84.5%) | – | – | – | – |
| Stage I/IIA | 9 | 0.983 (0.955–1.00) | 77.8% (44.2%–95.9%) | 77.8% (44.2%–95.9%) | – | – | – | – | |||
| All | |||||||||||
| All | 153 | 309 | 0.963 (0.946–0.978) | 92.9% (89.4%–95.4%) | 88.2% (82.1%–92.6%) | 95.8% (92.9%–97.6%) | 80.4% (73.3%–86.0%) | 140 | 0 | – | 63.6% (55.0%–71.2%) |
| Stage I/IIA | 42 | 0.980 (0.965–0.992) | 92.9% (81.2%–98.0%) | 88.1% (74.7%–95.2%) | 40 | 0 | – | 37.5% (23.5%–53.8%) | |||
PC, pancreatic cancer; NC, non-cancer.
Summary of specificity, overall sensitivity, and early-stage sensitivity in urinary miRNA-based pancreatic cancer-detection algorithm and Cancer antigen 19-9 (CA19-9). For the former, two distinct values of threshold according to the Youden Index and target specificity at 95%, and the performance was evaluated in the train set via cross-validation (Train), in the test set (Test), and in the total data set combining the train and test sets (All). For CA19-9, sensitivity was obtained according to the cutoff value of 37.0 U/mL, and specificity was not obtained because of the lack of CA19-9 values in non-cancer subjects.
The overall performance in the test set, as measured by the AUC of the ROC curve, was 0.963 (95% CI, 0.932–0.988; Fig. 4d) and 0.983 (95% CI, 0.955–1.00) when limited to Stage I/IIA (Table 2). The prediction scores across stages in the test set showed that the predictive model could detect pancreatic cancer at all stages. However, the number of participants with early-stage pancreatic cancer was small (Fig. 4e). A sensitivity of 77.8% (95% CI, 44.2%–95.9%) was observed for early-stage disease, with an overall sensitivity of 77.8% (95% CI, 64.9%–87.3%) and specificity of 95.7% (95% CI, 89.4%–98.5%; Table 2).
In the training set, the median age of the pancreatic cancer group was 14 years higher than that of the non-cancer group. However, the cancer prediction score was not affected by age (Supplementary Fig. S2). Furthermore, when a model was developed for an age-matched dataset (n = 182), a good performance was obtained: AUC = 0.93 (five-fold cross-validation, SD = 0.04) (Supplementary Fig. S3). The cancer prediction scores for sex, body mass index, alcohol consumption, and smoking history were examined; however, no particular trend was observed except smoking history (Supplementary Fig. S2). In the current smoker subgroup, a slight decrease in scores was observed in pancreatic cancer, leading to a lower sensitivity while its specificity remained comparable. A multiple regression analysis against the prediction score showed that only the cancer status (pancreatic cancer or non-cancer) was statistically significant (p < 0.0001). At the same time, the other five variables were not (Supplementary Table S3).
Performance comparison with CA19-9 biomarker
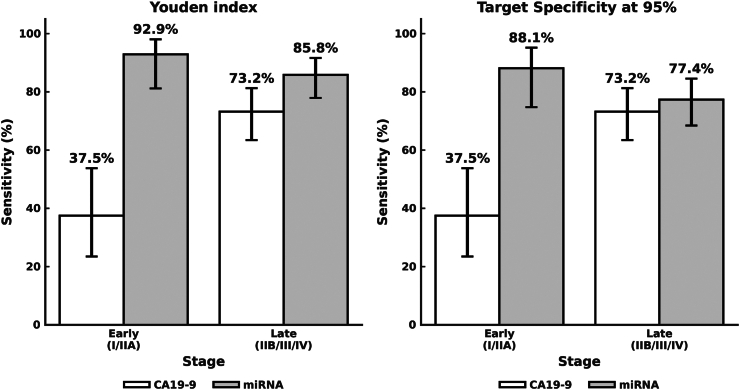
We compared the performance of miRNA-based pancreatic cancer-detection algorithms with CA19-9 detection. A total of 140 pancreatic cancers had CA19-9 measurements, enabling validation of pancreatic cancer-detection performance by stage. CA19-9 tended to show poorer predictive abilities in the early stages, and its overall sensitivity was 63.6% (95% CI, 55.0%–71.2%), and the sensitivity was 37.5% (95% CI, 23.5%–53.8%) for stage I/IIA (Fig. 5, Table 2). The sensitivity was calculated for cases with multiple cutoff values for the miRNA-based pancreatic cancer-detection algorithm. Because the number of early-stage cases was limited to the test set, we evaluated the performance from the total dataset by combining the cross-validation results in the training set and the hold-out results in the test set. When the cutoff value with the best balance between sensitivity and specificity was set based on the Youden Index, the specificity was 92.9% (95% CI, 89.4%–95.4%), the overall sensitivity was 88.2% (95% CI, 82.1%–92.6%), and the sensitivity was 92.9% (95% CI, 81.2%–98.0%) for stage I/IIA. Using the cutoff value (0.484) when fixed at 95.5% as the expected specificity for CA19-9 based on previous reports,7 the overall sensitivity of the miRNA-based algorithm for pancreatic cancer detection was 80.4% (95% CI, 73.3%–86.0%), with 88.1% (95% CI, 74.7%–95.2%) for stage I/IIA (Fig. 5, Table 2). The pancreatic cancer-detection performance in early-stage cancer was significantly higher in the miRNA-based pancreatic cancer detection (chi-square test, p < 0.0001).
Fig. 5.
Performance of urinary miRNA-based model compared to CA19-9. Sensitivity in early-stage (Stage I/IIA) and late-stage (Stage IIB/III/IV) pancreatic cancer are shown for the urinary miRNA-based model and Cancer antigen 19-9 (CA19-9). For the former, sensitivity was calculated for two distinct cutoff values according to the Youden Index and specificity target at 95%. Error bars indicate the 95% confidence intervals.
Functional enrichment analysis of pancreatic cancer-associated urinary miRNA and comparison of DEMs from urine and tumor-derived organoids
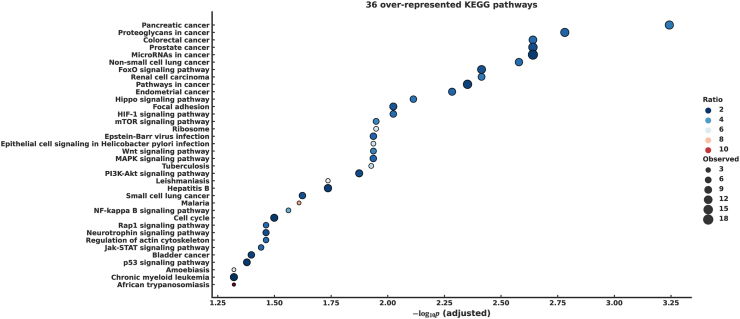
Functional enrichment analysis was performed using miEAA to investigate how 45 DEMs, whose expression varied in the pancreatic cancer group, were related to the pathogenesis of pancreatic cancer. Specifically, we focused on the KEGG pathway and the GO through miRTarBase. From the KEGG pathway, we identified 36 significantly overrepresented pathways associated with 45 urinary DEMs (Fig. 6, Supplementary Table S2a), which included cancer-associated pathways, such as pancreatic cancer, microRNAs in cancer, and pathways in cancer. Moreover, we found several signaling pathways associated with pancreatic cancer development and progression, including PI3K-Akt, MAPK, Jak-STAT, mTOR, P53, TGF-beta, and Wnt. We selected the 100 most strongly over-enriched GO terms (Supplementary Table S2b). We identified several biological processes associated with stroma, such as fibroblast growth factor binding (GO0017134), macrophage activation involved in the immune response (GO0002281), and interleukin-1 receptor binding (GO0005149).
Fig. 6.
Overrepresented KEGG pathways of DEMs in urine. Babble plot showing 36 overrepresented the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in vertical axis against -log10 p (adjusted) in the horizontal axis with the color showing the ratio of observed and expected hits, and the babble size showing the observed hits.
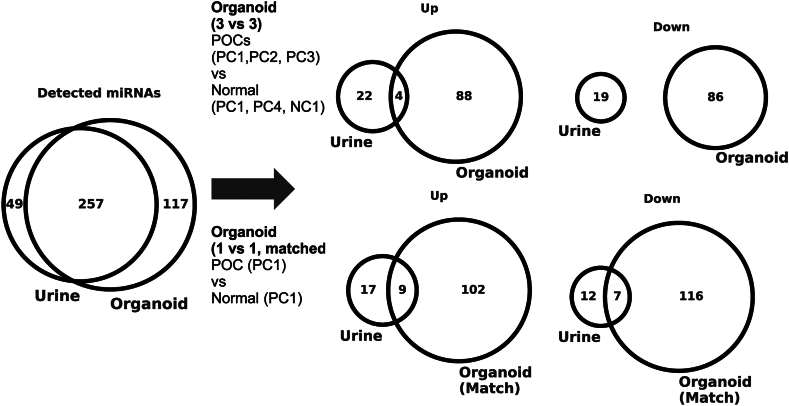
To assess the origin of urinary DEMs, we used tumor-derived organoids to compare the profiles of tumor cell-secreted miRNAs with the urinary miRNA profiles (Supplementary Fig. S4a). We established three pancreatic cancer organoids from patients with pancreatic cancer. As a control group, normal tissues were collected from patients with pancreatic cancer, and a non-cancer patient with common bile duct stones, and three normal tissue organoids were established (Supplementary Fig. S4b). We constructed a pair of pancreatic cancer and normal tissue organoids from the same participant (PC1). Of the 45 urine DEMs (up-regulated 26; down-regulated 19), 33 miRNAs (up-regulated 21; downregulated 12) were detected in the organoids. In one tumor and normal pair (PC1) comparison, approximately 35% (up, 9/21; down, 7/12) of the urinary DEMs matched the organoid DEMs (absolute log2 fold-change > 0.5; Fig. 7, Supplementary Table S4). Approximately 15% (up: 4/21, down: 0/12) of the urinary DEMs were consistent with the organoid DEMs in each of the three tumors compared to normal tissues (absolute log2 fold-change > 0.5; Fig. 7, Supplementary Table S4).
Fig. 7.
Organoid analysis and pathway enrichment analysis. Venn diagrams showing overlaps in urinary DEMs and organoid DEMs. (Left) Overlap of miRNAs detected in urine and in organoids with the thresholds (θ = 3 and r = 50%). (Top right) Overlap of urinary and organoid DEMs with three tumors and three normal organoids. (Bottom right) Overlap of urinary and organoid DEMs with one donor-matched pair of tumor and normal organoids.
Discussion
Pancreatic cancer is one of the deadliest cancers, with an approximately 10% 5-year survival rate.1 A major contributing factor for the poor prognosis is the difficulty in early diagnosis, when surgery is effective and often curative. We developed a noninvasive urinary extracellular vesicle miRNA-based assay for pancreatic cancer detection. A low concentration of miRNAs in urine, which is lower than that in plasma, and a high tRNA fraction17 result in significant challenges in detecting miRNA signatures by small RNA sequencing compared to a clinical assay. To improve the low concentrations of miRNAs, urinary extracellular vesicles were precipitated and enriched before RNA extraction, enabling efficient small RNA-seq with a five-fold higher mapping rate than conventional urinary miRNAs (Fig. 2). Furthermore, this miRNA mapping rate was higher than that previously reported for urinary miRNA sequencing.17 This method showed consistent performance with AUC = 0.972 and 0.963 in the training set (n = 315) and test set (n = 147), respectively, and the sensitivities of pancreatic cancer detection were 93.9% and 77.8% overall and 97.0% and 77.8%, respectively, in early-stage pancreatic cancer. The specificities were 91.7% and 95.7%, respectively (Table 2). It should be noted that the sample size for early-stage pancreatic cancer in the test dataset was relatively small. Nevertheless, our modeling approach with L2 regularization effectively mitigated overfitting and demonstrated generalizability, as indicated by the saturation in the learning curve (Supplementary Fig. S5), confirming that the sample size was sufficient for developing a robust machine learning prediction model. We conducted a sensitivity analysis and found that the performance was consistent between pancreatic cancer cases diagnosed via imaging and those diagnosed through pathology (Supplementary Fig. S2). Since some pancreatic cancer participants may have jaundice, we checked if urinary bilirubin affects the miRNA assay. Although only seven pancreatic cancer participants were bilirubin-positive, all tested positive in the assay (Supplementary Materials).
Through subsequent analyses, we confirmed the validity of the differentially expressed DEMs. We performed DEA on the test set and found a high correlation with the training set results (Supplementary Fig. S6). Additionally, we validated these findings using qPCR on 30 pancreatic cancer and 30 non-cancer cases from the training set, targeting 15 miRNA probes. Three of the eight up-regulated miRNAs from RNA-seq exhibited consistent log2 fold changes above 0.5 in qPCR (Supplementary Fig. S7), demonstrating strong agreement between NGS and qPCR results. While a panel of 183 miRNAs achieves high performance, reducing it to 43 miRNAs maintains performance compared to a target value of AUC = 0.95 (Supplementary Fig. S8). However, further reduction would likely lead to a significant decrease in its performance. Unlike qPCR, using NGS allows us to maintain a larger number of miRNAs, resulting in a robust panel for clinical utility.
CA19-9 is a biomarker routinely used for pancreatic cancer detection but unsuitable for early detection because of its low sensitivity in early-stage pancreatic cancer. Although this study could not directly compare specificity because CA19-9 values in non-cancer participants were unavailable, we evaluated the sensitivity in 140 pancreatic cancer participants. CA19-9 tended to show higher values as the stage progressed, with an overall sensitivity of 63.6% and a sensitivity of 37.5% for stages I/IIA (Table 2). A recent study of CA19-9 performance showed a sensitivity of 57.6% and specificity of 95.5% for the early stages.6 The miRNA-based pancreatic cancer-detection algorithm from this study was superior to CA19-9 in terms of performance, especially in detecting early-stage pancreatic cancer (sensitivity of 88.1% and specificity of 95.8%; Fig. 5, Table 2). Next, we compare the performance and reliability of our urinary miRNA assay with the currently prevalent blood-based miRNA assays. A meta-analysis of blood-based miRNA assays reported an AUC of 0.81, with a sensitivity of 0.79 and a specificity of 0.74 for early-stage pancreatic cancer,27 indicating that our urinary miRNA assay performs comparably well. Furthermore, DEMs identified in this study share common features with previously reported blood and/or tissue miRNAs associated with pancreatic cancer including miR-21, miR-181b, miR-1246, miR-143, miR-132, and the let-7 family.
Functional enrichment analysis showed that urinary DEMs' target genes were associated with tumor development and progression and signaling pathways involving cells comprising the tumor microenvironment, such as fibroblasts and macrophages (Fig. 6, Supplementary Table S2). These results suggest that the urinary miRNA signature reflects the miRNA patterns of the pancreatic tumor tissue and contributes to those secreted from cells in the tumor microenvironment. This may be reasonable given that pancreatic cancer tissue tends to be formed primarily by stromal tissue, and the amount of stromal tissue is often relatively high in pancreatic tumors.28 Our urinary miRNA assay demonstrates high sensitivity for detecting pancreatic cancer from stage I, although this sensitivity does not correlate with progression to later stages. The early occurrence of microenvironmental changes in cancer development likely explains why our miRNA assay detects these variations, contributing to its stage-independent detection capability.
To further assess the origin of urinary DEMs, we used tumor-derived organoids to compare the profiles of tumor cell-secreted miRNAs with urinary miRNA profiles (Fig. 7, Supplementary Material). Tissue collection was not performed in this study due to surgical and treatment considerations. Moreover, analyzing miRNAs from pancreatic cancer tissue itself presents the challenge of distinguishing between secreted and intracellular miRNAs. Since biofluid miRNAs are likely secreted, direct comparisons with tumor-derived miRNAs may not be effective. Organoids are three-dimensional (3D) cell aggregates derived in vitro from the primary tissue and maintain the corresponding parent tumor's histological features, gene expression, and mutation profile.29 They are composed of cells of epithelial origin and do not include stromal or immune cells in the tumor microenvironment. Extracellular vesicle-encapsulated and -free miRNAs secreted from organoids are detected in organoid culture supernatants.30 Our results showed that urinary DEMs partly overlapped with organoid-derived DEMs, suggesting that tumor cells partially contributed to the urinary miRNA signature. Overlapping miRNAs show promise as tissue-specific urinary biomarkers. We developed classifiers based on each miRNA, with their AUCs detailed in Supplementary Table S4. The limited number of organoid samples restricts their effectiveness in forming a robust miRNA panel. Moreover, reducing miRNAs lowers performance, making our current approach more reliable for accurate results. We could only use three tumor tissues from rare cases where preoperative chemotherapy was not required, significantly limiting our sample size. Taken together, functional enrichment and organoid analyses suggested that not only miRNAs secreted from tumor cells but also from the tumor microenvironment contributed to our urinary miRNA-based detection of pancreatic cancer across all stage, although pancreatic tumors are thought to be small in the early stages.
We evaluated the accuracy requirements for early detection of pancreatic cancer and the clinical utility of our urinary miRNA assay. Given the low prevalence of pancreatic cancer in Japan in 2020 (0.056% for those over 40 years old), early detection tests must have very high specificity and high sensitivity. The Japanese Ministry of Health, Labour and Welfare (MHLW) has set positive predictive values (PPV) thresholds in 2023 at 2.5%, 3.0%, and 4.1% for gastric cancer, colorectal cancer, and lung cancer, respectively. For pancreatic cancer, a PPV of 1% or higher is realistic. Our assay, with 77.8% sensitivity and 97.8% specificity, achieves a PPV of 1.94% and a negative predictive value (NPV) of 99.99%, meeting these criteria. We expect the urinary miRNA assay to aid in determining the need for further examinations like computed tomography (CT) or magnetic resonance imaging (MRI), similar to pancreatic cancer biomarkers (e.g., CA19-9) and trans-abdominal ultrasound (TAUS). TAUS shows 88% sensitivity and 94% specificity for overall pancreatic cancer detection,31 indicating that our assay has comparable or even superior detection capability. To further improve the cost-benefit balance, the next effective step would be to develop a pancreatic cancer screening test targeting high-risk groups with higher prevalence rates. Our primary goal is to make these tests accessible to a broader population, particularly in areas with limited hospital access where cancer is often detected late. Unlike blood samples, urine samples can be collected at home.
The present study had several limitations. (1) The pancreatic cancer cases in the test set were sourced from a different institution and kept entirely independent from the training samples to provide a more rigorous assessment of the assay's performance. Therefore, the distributions of the stage and other background factors did not exactly match, and many of the cases assigned to the test set were at a late stage. The assay showed a reasonably high pancreatic cancer-detection sensitivity for early-stage pancreatic cancer in the test set. However, it was slightly lower than that in the training set, and its confidence interval was higher due to the limited number of early-stage cases in the test set (Fig. 4, Table 2). (2) Our urinary miRNA assay is subject to potential confounding factors. The pancreatic cancer group was approximately 14 years older than the non-cancer group. However, the prediction outcomes of the algorithm were not influenced by age. These differences were unexpected to significantly affect the performance of the assay, which was expected to show reasonable results in all age groups (Supplementary Fig. S2) and was confirmed by the high performance obtained by a model developed for the age-matched dataset (Supplementary Fig. S3). The prediction outcome was slightly influenced by smoking history, where the current smoker subgroup had a lower sensitivity while its specificity remained comparable with the other subgroups. Further investigations will be needed to evaluate its impact on the assay's performance. Moreover, the number of participating facilities was limited. (3) This study recruited participants retrospectively; therefore, PPV and NPV were not evaluated. While this design facilitated the rapid collection of pancreatic cancer individuals for proof-of-concept exploration, it presents a limitation regarding the applicability of our model to other populations, particularly high-risk groups including those with chronic pancreatitis, and precancerous neoplasms such as mucinous cystic neoplasms (MCN) and intraductal papillary mucinous neoplasms (IPMN). Therefore, prospective screening studies involving these high-risk populations will be essential for successful translation of the current assay into clinical practice. (4) It also remains unclear whether our model is sensitive to other types of cancer, such as gastrointestinal cancers beyond pancreatic cancer (e.g., stomach or colorectal cancer) or urologic cancers (e.g., kidney or prostate cancer). Further studies are necessary to explore this. (5) This cohort study was limited to Japanese patients. Genetic background may affect miRNA expression,32 further studies are needed to confirm the performance in different racial/ethnic cohorts. (6) Regarding the pre-analytical conditions, the storage temperature for urine samples from participants without cancer was −20 °C at Omiya City Clinic, while all other cohorts stored their samples at −80 °C. We have compared the model's performance for the non-cancer samples stored at −20 °C and −80 °C. The specificity performance according to the threshold values was comparable both in the train and test sets (Supplementary Fig. S9). We simplified the urine collection process by not specifying timing or instructions, yet our model still achieved a high AUC in distinguishing non-cancer from pancreatic cancer cases. Importantly, the test minimizes participant burden while maintaining high assay performance. Given the lack of established standards for miRNA-based screening tests, it is crucial to define best practices for pre-analytical/analytical conditions and quality control, and conduct a large-scale prospective study to overcome these limitations and establish the clinical value of this assay.
In conclusion, we developed and validated a noninvasive urinary extracellular vesicle miRNA-based assay using small RNA-seq that requires only 3 mL of urine. This assay, which can efficiently detect pancreatic cancer from early to late stages with high sensitivity in seemingly healthy participants, may contribute to the overall outcome of pancreatic cancer treatment by improving the rate of detection through population screening and the management of patients with increased risk of pancreatic cancer.
Contributors
The following are the role of each author to this study; Conceptualization: MM, YI, KH, HN, and YK; Data curation: SB, KY, HK, MY, SK, MO, TS, YSI, TY, HY, YA, and YK; Formal analysis: HY; Funding acquisition: KY, MM, YI, and YK; Investigation: SB, HY, YA, and YK; Methodology: HY; Project administration: MM, YI, and YK; Resources: SB, TaK, SH, TN, TA, RO, KY, ToK, HK, HO, MY, JK, JI, KK, SK, MO, TS, YSI, TY, and YK; Software: HY; Supervision: SH, KY, JK, JI, MM, YI, HN, and YK; Validation: HY and YA; Visualization: HY, HN, and YK; Writing-original draft: SB, HY, YA, MM, KH, HN, and YK. All authors reviewed and provided their final approval and agreed to be accountable for all aspects of this study. HY, YI, and YK accessed and verified the data.
Data sharing statement
Data, code supporting the findings of this study, and the study protocol are available from the corresponding author upon reasonable request.
Declaration of interests
AMED supported KY, YI and YK. YK is an advisor and has stock options of Craif Inc. MM and YI are board members and shareholders of Craif Inc. HY and YA are employees and have stock options of Craif Inc. Other authors have no conflicts of interest.
Acknowledgements
We want to thank all our participants and the Omiya City Clinic for contributing to sample collection from healthy individuals.
During the preparation of this work we used Chat GPT4o in order to improve language and readability. After using this tool/service, we reviewed and edited the content as needed and take full responsibility for the content of the publication.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2024.102936.
Contributor Information
Hiroshi Nishihara, Email: hnishihara1971@keio.jp.
Yasutaka Kato, Email: taka.katooo@keio.jp.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Kleeff J., Korc M., Apte M., et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 3.Meng Q., Shi S., Liang C., et al. Diagnostic and prognostic value of carcinoembryonic antigen in pancreatic cancer: a systematic review and meta-analysis. Onco Targets Ther. 2017;10:4591–4598. doi: 10.2147/OTT.S145708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duffy M.J., Sturgeon C., Lamerz R., et al. Tumor markers in pancreatic cancer: a European Group on Tumor Markers (EGTM) status report. Ann Oncol. 2010;21:441–447. doi: 10.1093/annonc/mdp332. [DOI] [PubMed] [Google Scholar]
- 5.Wang K., Wang X., Pan Q., Zhao B. Liquid biopsy techniques and pancreatic cancer: diagnosis, monitoring, and evaluation. Mol Cancer. 2023;22:167. doi: 10.1186/s12943-023-01870-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haan D., Bergamaschi A., Friedl V., et al. Epigenomic blood-based early detection of pancreatic cancer employing cell-free DNA. Clin Gastroenterol Hepatol. 2023;21:1802–1809. doi: 10.1016/j.cgh.2023.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Chen J., Wang H., Zhou L., Liu Z., Tan X. A combination of circulating tumor cells and CA199 improves the diagnosis of pancreatic cancer. J Clin Lab Anal. 2022;36 doi: 10.1002/jcla.24341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura K., Zhu Z., Roy S., et al. An exosome-based transcriptomic signature for noninvasive, early detection of patients with pancreatic ductal adenocarcinoma: a multicenter cohort study. Gastroenterology. 2022;163:1252–1266.e2. doi: 10.1053/j.gastro.2022.06.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyhan A.A. Circulating microRNAs as potential biomarkers in pancreatic cancer-advances and challenges. Int J Mol Sci. 2023;24 doi: 10.3390/ijms241713340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J., Gao G., Ge Y., et al. Development of a serum-based MicroRNA signature for early detection of pancreatic cancer: a multicenter cohort study. Dig Dis Sci. 2024;69(4):1263–1273. doi: 10.1007/s10620-024-08338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki K., Yamaguchi T., Kohda M., et al. Establishment of preanalytical conditions for microRNA profile analysis of clinical plasma samples. PLoS One. 2022;17 doi: 10.1371/journal.pone.0278927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitano Y., Aoki K., Ohka F., et al. Urinary microRNA-based diagnostic model for central nervous system tumors using nanowire scaffolds. ACS Appl Mater Interfaces. 2021;13:17316–17329. doi: 10.1021/acsami.1c01754. [DOI] [PubMed] [Google Scholar]
- 14.Debernardi S., Massat N.J., Radon T.P., et al. Noninvasive urinary miRNA biomarkers for early detection of pancreatic adenocarcinoma. Am J Cancer Res. 2015;5:3455–3466. [PMC free article] [PubMed] [Google Scholar]
- 15.Ishige F., Hoshino I., Iwatate Y., et al. MIR1246 in body fluids as a biomarker for pancreatic cancer. Sci Rep. 2020;10:8723. doi: 10.1038/s41598-020-65695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mall C., Rocke D.M., Durbin-Johnson B., Weiss R.H. Stability of miRNA in human urine supports its biomarker potential. Biomark Med. 2013;7:623–631. doi: 10.2217/bmm.13.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan S., Yeri A., Cheah P.S., et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 19.Tamura T., Yoshioka Y., Sakamoto S., Ichikawa T., Ochiya T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell Vesicles Circ Nucl Acids. 2021;2:148–174. doi: 10.20517/evcna.2021.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng J., Bai L. Pancreatic cancer: current situation and challenges. Gastroenterol Hepatol Lett. 2020;2:1–3. [Google Scholar]
- 21.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil A.H., Halushka M.K. miRge3.0: a comprehensive microRNA and tRF sequencing analysis pipeline. NAR Genom Bioinform. 2021;3 doi: 10.1093/nargab/lqab068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiba T., Sano S., Yanase T., Ohta T., Koyama M. 2019. Optuna: a next-generation hyperparameter optimization framework. [Google Scholar]
- 25.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aparicio-Puerta E., Hirsch P., Schmartz G.P., Kern F., Fehlmann T., Keller A. miEAA 2023: updates, new functional microRNA sets and improved enrichment visualizations. Nucleic Acids Res. 2023;51:W319–W325. doi: 10.1093/nar/gkad392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng C., Wang J., Gao W., et al. Meta-analysis of the diagnostic performance of circulating MicroRNAs for pancreatic cancer. Int J Med Sci. 2021;18:660–671. doi: 10.7150/ijms.52706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hosein A.N., Brekken R.A., Maitra A. Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat Rev Gastroenterol Hepatol. 2020;17(8):487–505. doi: 10.1038/s41575-020-0300-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacob F., Salinas R.D., Zhang D.Y., et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szvicsek Z., Oszvald Á., Szabó L., et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell Mol Life Sci. 2019;76:2463–2476. doi: 10.1007/s00018-019-03052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toft J., Hadden W.J., Laurence J.M., et al. Imaging modalities in the diagnosis of pancreatic adenocarcinoma: a systematic review and meta-analysis of sensitivity, specificity and diagnostic accuracy. Eur J Radiol. 2017;92:17–23. doi: 10.1016/j.ejrad.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Rawlings-Goss R.A., Campbell M.C., Tishkoff S.A. Global population-specific variation in miRNA associated with cancer risk and clinical biomarkers. BMC Med Genomics. 2014;7:53. doi: 10.1186/1755-8794-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.