Abstract
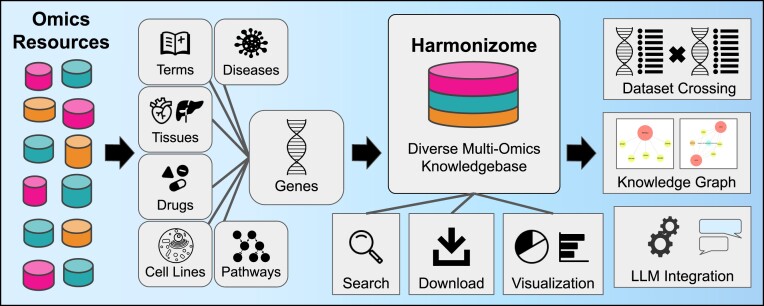
By processing and abstracting diverse omics datasets into associations between genes and their attributes, the Harmonizome database enables researchers to explore and integrate knowledge about human genes from many central omics resources. Here, we introduce Harmonizome 3.0, a significant upgrade to the original Harmonizome database. The upgrade adds 26 datasets that contribute nearly 12 million associations between genes and various attribute types such as cells and tissues, diseases, and pathways. The upgrade has a dataset crossing feature to identify gene modules that are shared across datasets. To further explain significantly high gene set overlap between dataset pairs, a large language model (LLM) composes a paragraph that speculates about the reasons behind the high overlap. The upgrade also adds more data formats and visualization options. Datasets are downloadable as knowledge graph (KG) assertions and visualized with Uniform Manifold Approximation and Projection (UMAP) plots. The KG assertions can be explored via a user interface that visualizes gene–attribute associations as ball-and-stick diagrams. Overall, Harmonizome 3.0 is a rich resource of processed omics datasets that are provided in several AI-ready formats. Harmonizome 3.0 is available at https://maayanlab.cloud/Harmonizome/.
Graphical Abstract
Graphical Abstract.
Introduction
As the volume and diversity of available omics datasets continues to expand, so does the need for platforms capable of integrating and harmonizing them. Many such platforms exist, but most are limited in their breadth and depth. Although omics datasets are well-structured, their integration is not trivial. One critical step for data integration is the need to unify identifiers and align ontologies. One solution to facilitate harmonization is to unify datasets by converting genes, proteins, transcripts, variants and other related entities into the same gene identifiers. For example, GeneCards (1) is one of the leading platforms that collects information about genes and proteins from many sources. Information about annotated and predicted human genes is consolidated into gene landing pages that present associations with pathways, phenotypes, expression levels and many other aspects. However, GeneCards is a commercial product, which limits reuse and integration into other tools. There is no free and open application programming interface (API), and the database and datasets are not available for download. Another leading platform is UniProt (2). UniProt focuses specifically on proteins with emphasis on the amino acid and DNA sequence, as well as the three dimensional structure of each protein. Protein information is mainly from literature curation. However, UniProt is also cross-referenced with 185 other databases that provide, for example, information about protein–protein interactions and disease associations. Pharos (3) is another resource that provides aggregated knowledge about human genes and proteins. The Pharos platform, created for the NIH Common Fund program Illuminating the Druggable Genome (IDG), has aggregated data from 83 resources with a focus on druggable targets from three protein families: G-protein coupled receptors, ion channels and protein kinases. The NCBI Gene database (4) also integrates knowledge about genes for many organisms with a focus on sequence and nomenclature. It has information about protein-protein interactions, pathway and phenotype associations, and gene expression data gathered from 22 resources. Other relevant human gene-centric knowledgebases include Model organism Aggregated Resources for Rare Variant ExpLoration (MARRVEL) (5), which integrates data from multiple similar sources with a focus on human genetics and genetic variants. Wikipedia and WikiGenes (6) also have dedicated gene pages with well-organized information in free text and tabular formats, for example, links to protein structure from the Protein Data Bank (PDB) (7), information about pathways, function, protein interactions, clinical significance, chromosomal location and more. The Human Protein Atlas (HPA) (8) is another widely used repository with extensive and well-organized gene pages.
Harmonizome is also an aggregator of omics knowledge about genes and proteins, with some unique features that set it apart. Launched in 2015 (9), the platform contains processed data from 79 diverse online resources. Data from these resources is processed uniformly into 137 unique datasets that contain 83 472 718 associations between 379 691 biological and biomedical attributes and 58 400 mammalian genes. Here, we describe a major upgrade to Harmonizome. The Harmonizome 3.0 update expands the original knowledgebase with new features that include a chatbot interface powered by a large language model (LLM), ability to cross pairs of datasets and form hypotheses about highly significant overlapping gene sets from dataset pairs, and a knowledge graph (KG) representation of the database with a user interface (UI) that visualizes associations as interactive ball-and-stick diagrams.
Materials and methods
Dataset processing and ingestion
Due to the diverse nature of resources ingested into Harmonizome, unique processing scripts were developed for each resource and dataset. For datasets with scored gene–attribute associations, tables were created with genes as the rows and attributes as columns, with each table entry representing the score of an association between a gene and an attribute. A cutoff value was established for each dataset to only keep the top gene–attribute associations. For some datasets, z-scores were computed for each gene by taking the average and standard deviation of each row. These z-scores were used to define gene–attribute associations with different cutoffs for specific datasets. Specifically, if the origin of the dataset measured the expression of a gene or a protein across many conditions, for example, expression levels across human tissues, z-scores were computed. For datasets without gene–attribute association values, associations were kept as an edge list. For these datasets, the relationship between the genes and their attributes are discrete, for example, membership in cell signaling pathways or gene knockout mouse phenotypes.
To ensure data integrity and consistency, we apply few basic rules. First, gene–attribute associations mostly come from primary and reliable experimentally validated resources. We do not take gene–attribute associations from other knowledge aggregators, and do not include any predicted associations from AI/ML applications. However, six datasets in Harmonizome are labeled as ‘predicted’ associations. These datasets include transcription factor binding motifs, predicted protein domains, and predicted targets for microRNAs (miRNAs) based on sequence matching. We also ensure that the database is not overwhelmed by few resources that contribute most of the associations. Importantly, the multi-omics resources must cover the entire genome/proteome/transcriptome/epigenome unbiasedly. We do not include resources that have a pre-defined subset of the genome such as a specific protein family, or data collected by a panel of predefined subset of the coded genome. Finally, the sources only originate from humans, rats, and mice. Data from other model organisms are excluded.
To harmonize gene identifiers, all gene, protein, transcript, and variant identifiers were converted to human NCBI Entrez gene symbols. When appropriate, attributes were mapped to community established ontologies and dictionaries. For example, this was done for chemicals, drugs, anatomical structures, cell types, cell lines, tissues, diseases, phenotypes and more. To map attributes to their most appropriate identifiers in ontologies and dictionaries, we first used those selected by the original datasets. If such mapping was not available, ontology terms and identifiers were selected from community established resources such as PubChem (12) for drugs and small molecules, and Uberon (13) for anatomical structures. The process of mapping attributes to relevant ontology terms varied by dataset. A table of up-to-date and approved gene symbols was obtained from the NCBI Gene database (4). A mapping file was created to convert synonymous gene symbols to matching current gene symbols, as well as to common identifiers for proteins, transcripts, and variants. This file was created using available gene names mapping tools from NCBI (4), Ensembl (10), UniProt (2) and the mouse genome informatics (MGI) (11). To convert mouse gene symbols to their human homologs, we utilized the NCBI Homologene and Ensembl BioMart resources to create a mapping file that maps mouse genes to their homologous human genes. Each dataset is made available for download in various formats together with the processing scripts. All these scripts are publicly available at the Harmonizome dataset landing pages and on GitHub at: https://github.com/MaayanLab/HarmonizomePythonScripts.
Visualization of datasets with UMAP
To visualize Harmonizome datasets as UMAP plots (12), we started with the gene set representation of each dataset. Terms describing each gene set were converted into inverse document frequency (IDF) vectors using the Scikit-learn library (13). Next, the vectors were converted into two-dimensional space with UMAP using the default parameters for most datasets.
Dataset crossing
To create the dataset crossing feature, pairs of gene sets were compared to find the most significant overlaps for each pair of datasets. For each dataset pair, all gene sets from the corresponding gene set libraries were crossed, computing the right-tailed P-value from the Fisher’s exact test. A Jaccard index and the number of overlapping genes for each pair of gene sets are also reported. The gene set pairs are then ordered by ascending P-value, and the top 5000 gene set pairs are kept for each pair of datasets.
Chatbot and hypotheses generations of gene set overlaps with GPT-4o
The Harmonizome 3.0 upgrade has a chatbot interface feature, and a dataset crossing hypothesis generation feature. Both features utilize OpenAI’s API. The chatbot communicates with an OpenAI assistant, which is defined with a specific system prompt to control its behavior. The assistant uses the GPT-4o model with a temperature setting of 0.05 to mostly eliminate randomness. Several functions are provided to the assistant to define the formatting of answers and for querying the Harmonizome database. The dataset crossing hypothesis generation feature uses the chat completion endpoint with a custom system prompt. The GPT-4o model is used with max_tokens = 1024, temperature = 0.0, frequency_penalty = 0.15 and presence_penalty = 0.15. Both features use a shared backend API controller to handle requests and responses from OpenAI.
Knowledge graph user interface
To develop a UI for interacting with the Harmonizome datasets and ball-and-stick diagrams, each Harmonizome dataset was converted into KG assertions. Once the data was converted into assertions, it was ingested into a Neo4j database. Once all the Harmonizome datasets were ingested into Neo4j, we leveraged an original UI that we developed for a separate project (14). The KG UI data visualization of ball-and-stick diagrams is achieved with the Cytoscape.JS library (15). The KG UI receives the results from Cypher queries in JSON format and converts these into nodes and links subnetworks for visualization. The UI was customized to accommodate specific requirements unique to the Harmonizome 3.0 datasets such as icons, headers and footers, and tooltips.
Results
Added datasets and data resources
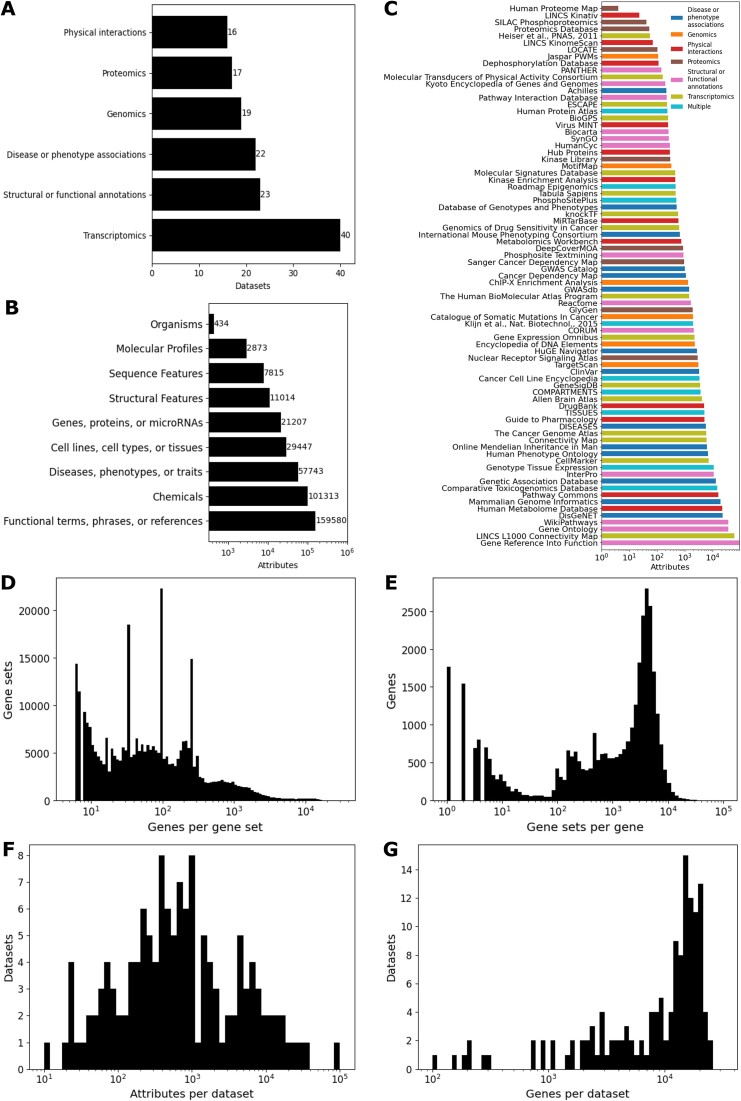
The core Harmonizome 3.0 database currently contains 83 472 718 associations between 58 400 genes and their 379 691 attributes. The attributes are linked to ontologies and dictionaries with additional metadata that describes each one of the 79 resources. The 137 datasets created from the 79 resources in Harmonizome 3.0 can be divided into six categories based on their source type (Figure 1A). These six categories are disease or phenotype associations, genomics, physical interactions, proteomics, structural or functional annotations, and transcriptomics. There are 40 transcriptomics datasets, 23 structural or functional annotation datasets, 22 disease and phenotype association datasets, 19 genomics datasets, 17 proteomics datasets, and 16 physical interaction datasets. Physical interactions include drug–target interactions, substrates for kinases and phosphatases, protein–protein interactions, metabolite–enzyme interactions, microRNAs and their direct targets, and viral–host protein–protein interactions. Similarly, attributes associated with genes can be divided into nine groups (Figure 1B). The Harmonizome database contains 159 580 functional terms, phrases or references; 101 313 chemicals; 57 743 diseases, phenotypes or traits; 29 447 cell lines, cell types or tissues; 21 207 genes, proteins or microRNAs; 11 014 structural features; 7815 sequence features; 2873 molecular profiles; and 434 organisms. The number of attributes contributed by each resource follows a log-normal distribution (Figure 1C). Resources providing structural and functional annotation datasets make up the second largest dataset group but contribute many gene sets in each dataset forming the largest attribute group. The distribution of gene set sizes follows a multimodal distribution with peaks at common gene set length sizes (Figure 1D). These peaks are likely due to arbitrary cutoffs set to limit or normalize set sizes, for example, the number of differentially expressed genes in large perturbation experiments, or basal gene or protein expression in normal tissues and cell types. The number of gene sets that each gene appears in also follows a multimodal distribution (Figure 1E). This could be a mixture of genes that are widely studied, genes that are highly expressed across cells and tissues, genes and proteins that commonly show high variability in their expression, longer and larger genes and proteins. The attributes present in the associations from each dataset follow a log-normal distribution (Figure 1F). This distribution highlights the diversity of the resources in Harmonizome. The gene coverage of each dataset follows a bimodal distribution with a large peak between 10 000 and 25 000 genes (Figure 1G). The is likely because the proteomics and literature-based datasets are likely present at the lower level of coverage, while the genomics and transcriptomics datasets cover all coding genes. Overall, these statistical summaries provide intuition about how knowledge about human genes is distributed across Harmonizome data sources and processed datasets.
Figure 1.
Summary statistics of datasets and attributes in Harmonizome 3.0. (A) Datasets belonging to each group. (B) Gene sets belonging to each attribute group. (C) Gene sets from each resource. (D) Distribution of gene set lengths. (E) Distribution of gene inclusion across gene sets. (F) Distribution of dataset attribute coverage. (G) Distribution of dataset gene coverage.
Updating and expanding the Harmonizome database is crucial for the continued reuse of the most relevant omics resources. Toward this aim, 19 new and 7 updated datasets have been processed and ingested to expand and upgrade Harmonizome (Table 1 and Supplementary Table S1). These datasets contain nearly 12 million gene–attribute associations. The attributes of each of the new datasets fit into one of the following groups: (i) cell line, cell type, or tissue; (ii) chemical; (iii) disease, phenotype, or trait; (iv) functional term, phrase, or reference; or (v) gene, protein, or microRNA. The addition of these new and updated datasets represents ∼132 000 gene sets with an average set size of 74 genes per term. In addition to providing the new and updated processed datasets for download in various formats, several additional features were added to each dataset landing page. Each new and updated dataset can be downloaded as a set of KG assertions, and it is visualized as a Uniform Manifold Approximation and Projection (UMAP) (12) plot. To create the KG serializations, each dataset was organized into a set of node-edge-node relations (triples), where genes/proteins and their attributes constitute the nodes, and the association type became the relationship edges. These triples are provided in Resource Description Framework (RDF), JavaScript Object Notation (JSON), and tab-separated values (TSV) file formats, creating a consistent serialization framework that can be used to construct a gene-centric KG. The UMAP plots visualize each attribute as a point in the UMAP projection (Supplementary Figure S1). Points are colored by automated clustering computed with the Leiden clustering algorithm (16). The IDF vectorized gene set libraries created from each dataset are used to compute the UMAP coordinates and projections. Additionally, 545 new dataset pairs are visualized as hierarchically clustered heatmaps (Supplementary Figure S3). Such new heatmaps visualize the similarity of gene–attribute relationships across pairs of Harmonizome datasets. A summary of features available for each dataset is provided in Supplementary Table S2.
Table 1.
Summary of datasets added to the Harmonizome 3.0 upgrade. Datasets are listed by attribute groups, relevant publication, attributes, genes, and the mean and range of gene set length. Additional attributes such as URLs and other information are provided in Supplementary Table S1
| Name | PMID | Attributes | Genes | Minimum gene set length | Mean gene set length | Maximum gene set length |
|---|---|---|---|---|---|---|
| CellMarker Gene-Cell Type Associations | 36300619 | 7217 | 13 607 | 91 | 9 | 1169 |
| CCLE Cell Line Proteomics | 31978347 | 375 | 8959 | 277 | 326 | 395 |
| DepMap CRISPR Gene Dependency | 34930405 | 1095 | 15 946 | 78 | 637 | 2999 |
| GTEx Tissue Gene Expression Profiles 2023 | 32913098 | 54 | 17 369 | 1000 | 1000 | 1000 |
| GTEx TIssue-Specific Aging Signatures | 135 | 16 047 | 250 | 250 | 250 | |
| HuBMAP Azimuth Cell Type Annotations | 31178118 | 1426 | 3560 | 3 | 10 | 12 |
| MoTrPAC Rat Endurance Exercise Training | 32589957 | 160 | 8833 | 1 | 78 | 4015 |
| Sanger Cancer Dependency Map Cancer Cell Line Proteomics | 35839778 | 949 | 8087 | 93 | 99 | 100 |
| Tabula Sapiens Gene-Cell Associations | 35549404 | 469 | 8184 | 100 | 100 | 100 |
| DeepCoverMOA Drug Mechanisms of Action | 36593396 | 874 | 7750 | 92 | 99 | 100 |
| GlyGen Glycosylated Proteins | 31616925 | 1910 | 2231 | 1 | 11 | 808 |
| LINCS L1000 CMAP Chemical Perturbation Consensus Signatures | 35524556 | 23913 | 12 126 | 1 | 114 | 577 |
| MW Enzyme Metabolite Associations | 26467476 | 734 | 1050 | 1 | 7 | 215 |
| DisGeNET Gene-Disease Associations | 31680165 | 15709 | 15 960 | 1 | 42 | 9666 |
| DisGeNET Gene-Phenotype Associations | 6832 | 14 002 | 1 | 29 | 5592 | |
| IMPC Knockout Mouse Phenotypes | 36305825 | 667 | 6763 | 1 | 55 | 1896 |
| MGI Mouse Phenotype Associations 2023 | 25348401 | 10234 | 12 894 | 1 | 20 | 2016 |
| Gene Ontology Biological Process Annotations 2023 | 30395331 | 12318 | 14 811 | 1 | 16 | 2029 |
| Gene Ontology Cellular Component Annotations 2023 | 926 | 11 089 | 1 | 45 | 5176 | |
| Gene Ontology Molecular Function Annotations 2023 | 3851 | 12 478 | 1 | 13 | 1412 | |
| SynGO Synaptic Gene Annotations | 31171447 | 267 | 1593 | 1 | 15 | 291 |
| Wikipathways PFOCR | 33168034 | 35464 | 13 173 | 3 | 9 | 324 |
| ChEA Transcription Factor Targets 2022 | 31114921 | 757 | 17 962 | 9 | 1211 | 4897 |
| Kinase Library Serine Threonine Kinase Atlas | 36631611 | 303 | 5046 | 100 | 100 | 100 |
| KnockTF Gene Expression Profiles with Transcription Factor Perturbations | 31598675 | 566 | 17 964 | 1 | 94 | 200 |
| LINCS L1000 CRISPR Knockout Consensus Signatures | 35524556 | 5049 | 9551 | 201 | 249 | 250 |
Crossing pairs of Harmonizome datasets
One of the advantages of abstracting and harmonizing omics datasets into the same format is the ability to combine these datasets to identify unexpected relationships. With the goal of uncovering new and surprising attribute–attribute associations from across Harmonizome 3.0 datasets, we have added a dataset pair crossing feature. Using this feature, pairs of gene sets from two selected datasets can be examined for gene set overlap. This enables users to find new significant relationships between attributes from pairs of Harmonizome 3.0 datasets (Figure 2A). After selecting two available datasets from the dropdown menus, a table of the top gene set pairs (up to 5000) sorted by the overlap P-values is displayed. Each row includes the name and size of both gene sets, the Fisher’s exact test P-value, the Jaccard index and the number of shared genes contributing to the overlap. Next to each row, there is also an option to generate a hypothesis about the corresponding pair of gene sets with OpenAI’s GPT-4o LLM. When clicking on this button, the descriptions of the gene sets, the top five enriched terms of the overlapping genes computed using the Fisher’s exact test with Enrichr (17), and the overlapping genes are submitted to the LLM via API. The top five terms are kept from the selected libraries listed below if these enriched terms meet the Benjamini–Hochberg (BH) corrected P-value of < 0.01. The LLM returns an abstract that contains a hypothesis that attempts to explain the reasons for the overlap. To guide the hypothesis, the top five enriched pathways from KEGG (18), knockout mouse phenotypes from International Mouse Phenotyping Consortium (IMPC) (19), biological processes from the Gene Ontology (20) and terms from the GWAS Catalog (21), along with their respective enrichment scores, are provided to the OpenAI API. In addition, the overlapping genes can be examined by clicking the overlap number, and this invokes a window that shows the gene names. There are also options to copy the genes to the clipboard or send them for enrichment analysis with Enrichr (17), Rummagene (22) and RummaGEO (23). Altogether, by identifying significant overlaps between gene sets from different Harmonizome datasets users can generate novel hypotheses.
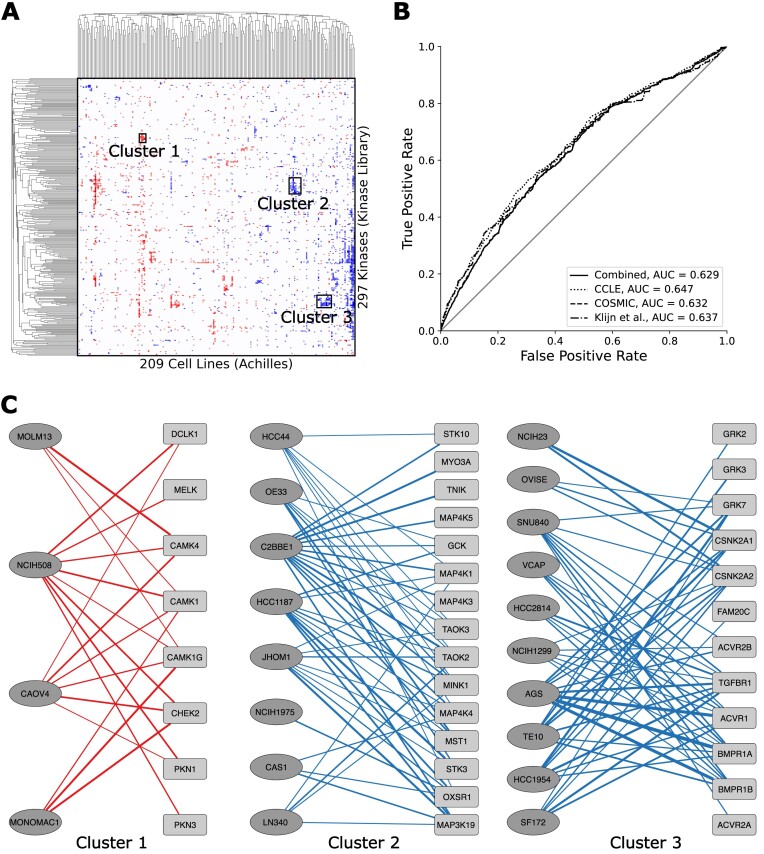
Figure 2.
Crossing the Kinase Library Serine Threonine Kinome Atlas dataset with the Achilles Cell Line Gene Essentiality Profiles dataset. (A) Hierarchically clustered heatmap showing the cosine similarity of kinases and cell lines based on the significance of their overlaps. Three clusters are identified for further examination. (B) Receiver Operating Characteristic (ROC) curves assesses the concordance of the overlapping genes of each crossing with known mutations in cell lines from the COSMIC, CCLE and Klijn et al. datasets from Harmonizome. (C) Subnetworks of the kinases and cell lines identified in each cluster. Red and blue lines indicate increased and decreased cell line fitness, respectively. Line width is proportional to the magnitude of -log(P-value).
Use Case 1: Crossing serine/threonine kinase substrates with cancer cell line knockouts from Achilles
Protein kinases belong to one of the most successful families of drug targets. Their role in controlling cell signaling pathways in cancer, and the ability to selectively target protein kinases with small molecules, provide an alternative to chemotherapy (24). As a result, identifying kinases that are dysregulated in specific cancer subtypes could potentially lead to new personalized targeted therapies. Here, we demonstrate how by using some of the newly added Harmonizome 3.0 datasets, and the newly implemented datasets crossing feature, we can discover specific kinase targets for specific subtypes of cancers. By crossing the Kinase Library Serine Threonine Kinome Atlas (25) with the Achilles Cell Line Gene Essentiality Profiles (26) datasets, we further prioritize protein kinases as targets for specific cell lines (Supplementary Video S1). By examining overlapping proteins that are phosphorylated by specific protein kinases and at the same time lead to decreased cell line fitness following knockdown of the genes that encode these proteins, we can identify regulatory cell signaling pathways that can be targeted in specific cell lines. The most significant overlaps are visualized in a heatmap where red pixels indicate crossings where the kinase substrates overlap with genes that increase cell line fitness when knocked down, while blue pixels indicate overlaps where the substrates cause decreased cell line fitness following knockdown (Figure 2A). In order to benchmark the predictions made by the gene set crossings, we compared the overlapping genes of all crossings that have at least one overlapping gene, ranked by P-value (Fisher’s exact test), against known mutations in these cell lines as reported in CCLE (27), COSMIC (28) and Klijn et al. (29). The cell-line gene mutation profiles are available as processed datasets in Harmonizome (Figure 2B). We observe that the ranked overlapping sets contain genes with known mutations in the respective cell lines. Next, we highlight three clusters of cell lines and kinases to further examine the overlapping genes in these clusters (Figure 2C). In one of these clusters, the kinase substrates that overlap with the knockdowns increases cell line fitness, while the two other highlighted clusters contain kinase substrates that overlap with gene knockdowns that decrease cell line fitness. Within the first cluster, overlaps between two acute myeloid leukemia (AML) derived cell lines and several kinases from the Ca2+/calmodulin-dependent protein kinase (CAMK) family of kinases, and the checkpoint kinase CHEK2, align with previous publications that showed that these kinases are key drivers in AML (30–32). In the third cluster, we identify overlaps between kinases involved in the transforming growth factor beta (TGF-β) bone morphogenetic protein (BMP) signaling pathway, and cell lines derived from gastrointestinal carcinomas. Perturbations of this pathway have previously been linked to the development of esophageal and colorectal cancers (33,34). Overall, the presence of these connections, which have been previously reported in the literature, suggest that other unstudied connections are real and should merit additional investigation. To further validate such hypotheses, the identified kinases could be knocked out, over-expressed, or targeted by available small molecule kinase inhibitors. It should be noted that the crossing analysis identified concentrated phosphorylation reactions between the kinases and the genes that give rise to the kinases' substrates, but the effect of such phosphorylations is unclear. In other words, the kinase activity may induce or inhibit proliferation, it is only apparent that the kinase is likely involved in either of these phenotypes. To determine the directionality, experimental validation is warranted.
Use Case 2: Crossing rat endurance exercise training signatures with knockout mouse phenotypes
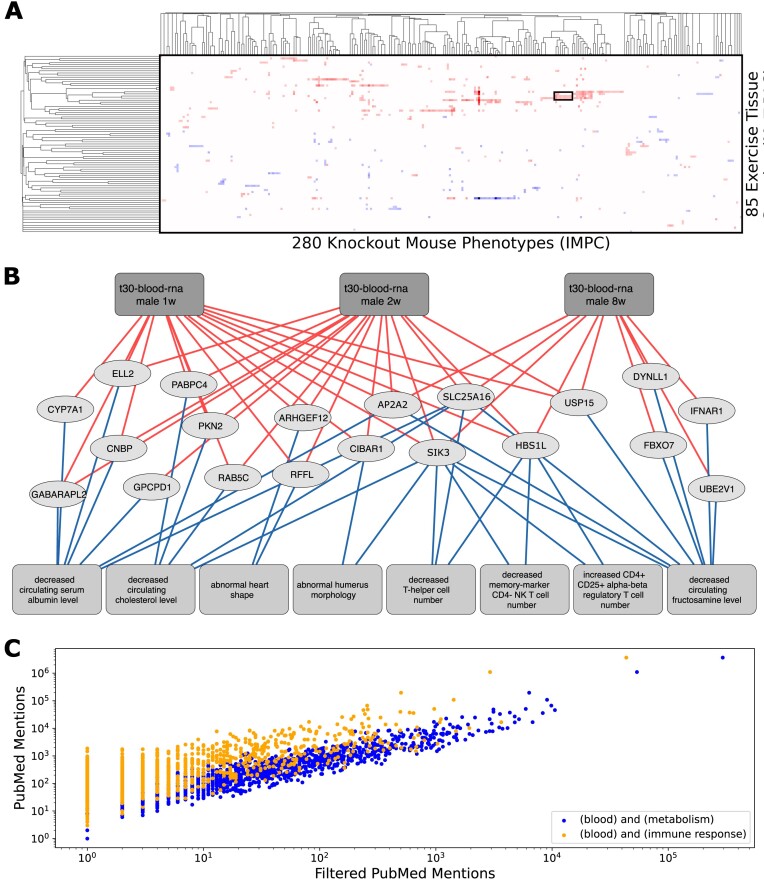
For the second use case, we show how the crossing of gene expression signatures induced by exercise in rats with genes that induce specific phenotypes in mice when knocked out can lead to interesting insights. Specifically, we crossed the Molecular Transducers of Physical Activity Consortium (MoTrPAC) Rat Endurance Exercise Training dataset (35) with the IMPC Knockout Mouse Phenotype dataset (19) (Figure 3A; Supplementary Text S1 and Supplementary Table S3). This approach identifies genes that either increase or decrease in their expression following endurance exercise training in rats that also induce adverse phenotypes when knocked out in mice. In this instance, we investigated a cluster of genes that increase in their expression due to endurance exercise in blood following 1, 2 and 8 weeks of endurance exercise training (Figure 3B). We observe multiple significant connections with metabolic and immune related processes, highlighting specific pathways that are modulated by exercise training. Specifically, several genes involved in metabolic and immune related phenotypes increase in their expression in the blood after prolonged aerobic exercise. Some of these genes have prior evidence to be involve in the process based on surveying the literature. For example, SLC25A16, a solute carrier family mitochondrial protein, is known to be involved in the accumulation of coenzyme-A in the mitochondria, a process essential for lipid metabolism (36). SLC25A16 expression is induced after exercise, and this can be explained by the alteration it causes to cholesterol metabolism and T-cell distribution when it is knocked out in mice. The salt-inducible kinase 3 (SIK3) is also associated with dysregulation of T-cell number when knocked out in mice. Prior research demonstrated that SIK3 is involved in mTOR signaling (37) and this could be related to an increase in protein synthesis needed in response to endurance exercise. Despite their involvement in immune and metabolic processes, both SLC25A16 and SIK3 are relatively understudied (Figure 3C). Their presence in the most significant overlapping set pairs, and the fact that these two genes were identified in the blood after exercise, suggest that they could potentially become biomarkers and potentially targeted to mimic the beneficial effects of endurance exercise training. It would be interesting to see if injecting SLC25A16 and SIK3 recombinant proteins, or targeting SLC25A16 and SIK3 with antibodies, will impact exercising and sedentary rats’ muscle composition and exercise capacity. Alternatively, targeting these two proteins in the same way in mouse models of muscular dystrophies could be tested for potential beneficial effects.
Figure 3.
Crossing the MoTrPAC Rat Endurance Exercise Training and IMPC Knockout Mouse Phenotypes datasets. (A) Hierarchically clustered heatmap showing the cosine similarity between rat endurance exercise training tissue samples and knockout mouse phenotypes based on the significance of their overlap. A cluster is identified for further inspection. (B) Subnetwork of clustered tissue samples, phenotypes and overlapping genes. Lines from the top boxes to the genes indicate genes with increased expression following endurance exercise training, and lines to the boxes at the bottom depict the presence of a phenotype following gene knockout. (C) Scatter plot comparing PubMed mentions for all genes appearing in the top overlaps between exercise tissue samples and knockout phenotypes. The PubMed mentions were derived by querying the PubMed API with the gene symbol, and the mentions were retrieved by querying the gene symbol with the terms ‘blood,’ ‘metabolism’ and ‘immune response’ to evaluate any known associations.
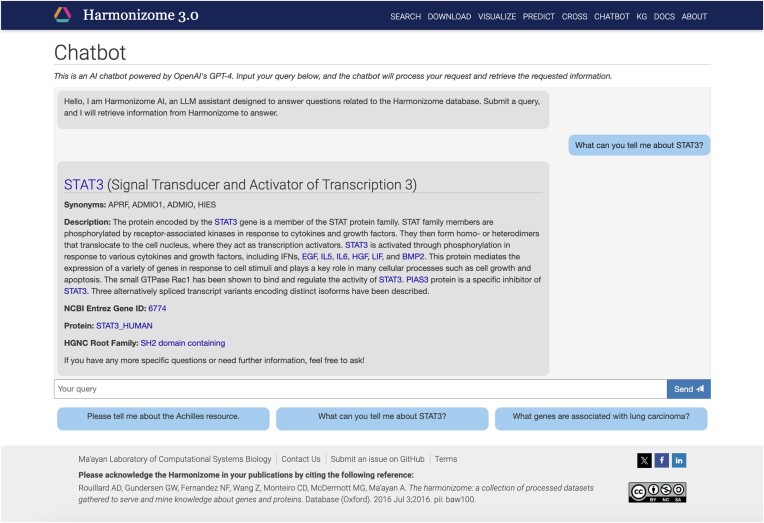
The Harmonizome 3.0 chatbot
LLMs recently emerged as a transforming technology with many applications across domains including biomedicine. With LLMs, it is now possible to develop high-quality interactive chatbots to interface with data using free text queries. Particularly powerful is the ability of LLMs to interact with structured databases to serve knowledge in response to text-based queries from a user (38). Powered the by OpenAI’s GPT-4o model, we have implemented a chatbot for Harmonizome 3.0. This is achieved by defining an Assistant through the OpenAI API. To set up the Assistant, the LLM model is selected, the system prompt that determines its behavior is defined, and the external resources that are available to it are established. To ensure reliability and reproducibility, we have set up a chatbot that limits the scope of accepted inputs to relevant queries, and uses a low temperature setting to reduce the variability of responses. The Harmonizome backend API controller class sends user messages to the Assistant, and then the Assistant constructs and displays messages as responses (Figure 4 and Supplementary Text S2). Besides providing a text-based interface, the chatbot interface also has chips with example queries to direct users about the potential and type of questions that should be composed to receive useful responses. To ensure that the Harmonizome chatbot provides accurate responses, we have implemented several functions that facilitate the chatbot to query the Harmonizome database and only use the resulting information when generating responses. Functions describe actions that the Assistant can select from based on their descriptions. The implemented API controller processes function-calls that are returned from the Harmonizome API. When a function call is received, the information is retrieved from the database and passed back to the Assistant. The Assistant then processes the response and incorporates it into the final output. In this way, we ensure that only relevant but comprehensive data are passed to the Assistant, providing factual and reliable framework to construct responses while limiting the potential for incorrect replies.
Figure 4.
Example from the Harmonizome 3.0 chatbot. The example query ‘What can you tell me about STAT3?’ is selected to query the chatbot for information about the transcription factor STAT3. The query is passed to the OpenAI Assistant through the backend Harmonizome API controller. The Assistant returns a function call to retrieve information about the gene from the Harmonizome database. The controller resolves the function call and returns the relevant metadata to the Assistant as a tool output. The Assistant reformats the gene’s metadata into a human readable format and returns it to the API controller. The controller then renders the chat history for the user, providing the gene name, a description of its functions, NCBI Entrez Gene ID, encoded protein, and protein family. In the rendered response, gene symbols and names of resources and datasets in Harmonizome are hyperlinked to direct users to the respective Harmonizome pages to learn more about the topic.
The Harmonizome 3.0 Knowledge Graph
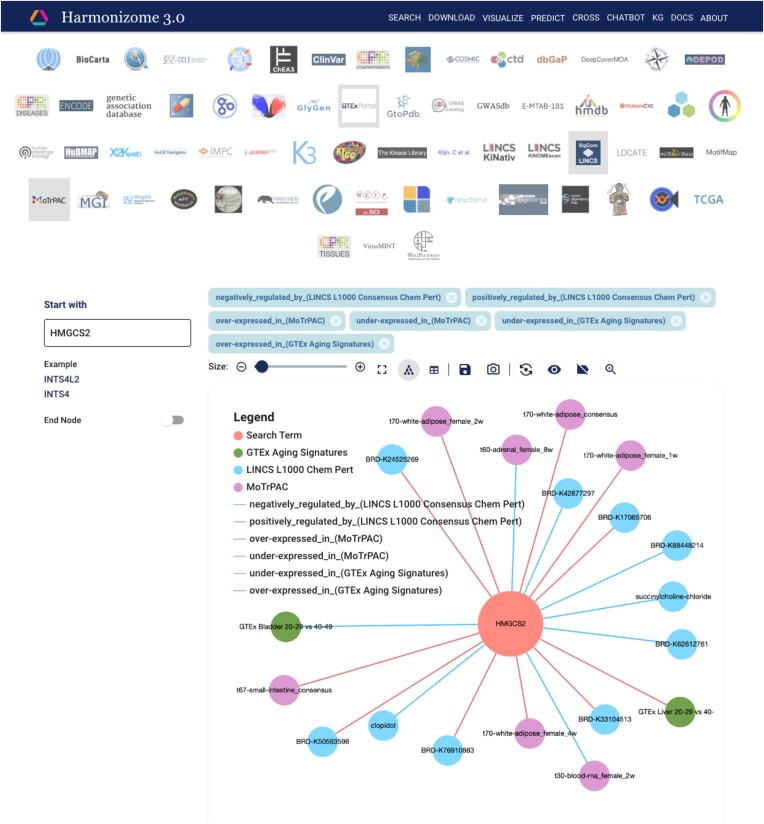
As part of the Harmonizome 3.0 update, we have also developed Harmonizome-KG, a platform to serve the serialized Harmonizome 3.0 datasets as interactive ball-and-stick subnetwork diagrams. To achieve this, we converted all Harmonizome datasets into a format that can be ingested into Neo4j, a commercial KG database. Next, we utilized a customizable web-based UI that we developed for a separate project (14) to query the data in the KG database. The UI enables users to create customized subnetworks originating from one- and two-term searches. Subnetworks can be created by selecting a gene or an attribute, and at most five Harmonizome datasets. Nodes and links from multiple Harmonizome datasets can be added or removed to create customized views. The two-term search requires a start and end node to create subnetworks. The query identifies the shortest paths between the two input terms. To demonstrate the applications of the KG for a specific query, we constructed a subnetwork centered around the gene 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2). HMGCS2 encodes a ketogenesis enzyme that has been linked to negative survival outcomes in mice when deleted (39). To explore drugs that may mimic the effects of exercise and counteract the effects of aging by preserving ketogenesis, we created a subnetwork from HMGCS2 and its associations with aging signatures from the GTEx Tissue-Specific Aging Signatures dataset, tissue samples from the MoTrPAC Rat Endurance Exercise Training dataset, and chemicals from the LINCS L1000 Chemical Perturbation Consensus Signatures dataset (Figure 5). We found several drugs (BRD-K93109423, BRD-K21356734, BRD-K63270352, BRD-K25176380 and BRD-K01910220) that increase the expression of HMGCS2, opposite to its decreased expression in aging bladder tissue and aligning with its increased expression in white adipose tissue following exercise. To reproduce the subnetwork shown in the figure, a tutorial screen capture video was generated (Supplementary Video S2).
Figure 5.
Screenshot from the Harmonizome 3.0 KG. The associations between genes and attributes are visualized as ball-and-stick diagrams. The queried gene (HMGCS2) is at the center of the subnetwork diagram. The attributes associated with HMGCS2 are from each selected Harmonizome 3.0 dataset: GTEx Tissue Specific Aging Signatures, LINCS L1000 CMap Chemical Perturbation Consensus Signatures, and MoTrPAC Rat Endurance Exercise Training. Edges can have positive or negative effects. Associations from other datasets can be selected by clicking and on the resources' icons. The subnetwork can be changed to use force-directed, hierarchical, or geometric layouts. The number of displayed associations can be adjusted using a slider. There are also options to view the subnetworks as a table, save the subnetwork in tabular format, export the subnetworks as an image, and view edge labels, and a figure legend.
Discussion
Harmonizome 3.0 integrates many published omics datasets to provide researchers a rich collection of features and attributes about mammalian genes. It provides uniformly processed datasets in various data structures for data analysis and reuse. Several additional datasets were introduced into the Harmonizome 3.0 release. In addition to new and updated datasets, new methods to interact with and visualize the harmonized and ingested data were implemented. Each Harmonizome dataset can be visualized with a UMAP that displays the embeddings of gene sets from each dataset as points colored by automatically computed clusters. The Harmonizome datasets can be explored by interacting with the datasets via an interactive network visualization. The Harmonizome datasets were converted into a KG representation and stored in a Neo4j database. We have implemented an open-source UI to interact with the contents of this database. Another addition to Harmonizome 3.0 is the dataset-crossing functionality. This feature enables users to find interesting overlaps between pairs of Harmonizome datasets. In addition to discovering unexpected high overlap across dataset pairs, users can generate hypotheses about the reasons behind the high overlap between pairs of datasets using the OpenAI GPT-4o LLM model. Finally, the GPT-4o LLM model was utilized to create the Harmonizome chatbot. The OpenAI Assistant framework was utilized to enable the chatbot by defining the nature of interactions between users and Harmonizome. The chatbot feature provides a new way to interact with Harmonizome using natural language. To ensure chatbot responses are consistent, within a limited scope, and are accurate, the chatbot was given strict instructions to only reply with the data and tools we provided to it. The application of LLMs to form hypotheses based on the dataset crossing feature of Harmonizome attempts to minimize randomness and distorted facts by keeping the temperature low. This also promotes reproducibility. However, it can be argued that such restriction limits the opportunity for the LLM to be more creative. It was recently discussed that as LLMs improve, they also tend to reason well based on false facts (40). This means that hypotheses generated by such LLMs may be convincing but scientifically wrong. LLMs, in general, perform well with creative tasks, but they struggle with separating real fact from fiction.
Overall, by abstracting diverse omics datasets into associations between genes/proteins and their functional attributes, the Harmonizome 3.0 datasets can be seemingly fused. Such data fusion can directly pinpoint to new undiscovered biology. In addition, by concatenating attributes of genes and proteins from many resources, gene/protein functions can be predicted with Machine Learning. For example, we can predict the knockout phenotypes in mice for genes not previously knocked out. Similarly, we can predict pathway membership and Gene Ontology (GO) terms for genes with no annotations. Many other applications that build upon the processed datasets available from Harmonizome 3.0 can be created. In the future, we plan to make several improvements to Harmonizome to further increase its functionality. We will continue processing and integrating new datasets, as well as updating those already in the Harmonizome. We also intend to further incorporate LLMs into various features of Harmonizome. For example, we plan to have an LLM that will interface directly Harmonizome-KG. As mentioned above, the large collection of well annotated diverse knowledge about genes and gene sets in Harmonizome can be used to predict new knowledge about mammalian genes with Machine Learning methods. These predictions can be presented alongside accumulated knowledge from direct evidence. Altogether, these improvements will ensure that Harmonizome will continue to serve as a unique and valuable resource for the biomedical research community.
Supplementary Material
Acknowledgements
Authors contributions: ID and DC: develop and maintain the database backend and frontend, processed the dataset for ingestion, created figures, and wrote the text. JEE and NL developed the knowledge graph component. AM: managed and supervised the project, provided funding, conceptualized the project, and wrote the paper.
Contributor Information
Ido Diamant, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, Box 1603 New York, NY, USA.
Daniel J B Clarke, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, Box 1603 New York, NY, USA.
John Erol Evangelista, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, Box 1603 New York, NY, USA.
Nathania Lingam, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, Box 1603 New York, NY, USA.
Avi Ma’ayan, Department of Pharmacological Sciences, Mount Sinai Center for Bioinformatics, Icahn School of Medicine at Mount Sinai, One Gustave L. Levy Place, Box 1603 New York, NY, USA.
Data availability
The Harmonizome 3.0 database is available at: https://maayanlab.cloud/Harmonizome/. The Harmonizome processed datasets are available in multiple formats at: https://maayanlab.cloud/Harmonizome/download/. The Harmonizome data processing scripts are available at: https://github.com/MaayanLab/HarmonizomePythonScripts. A snapshot of the code of these processing scripts can be access from Zenodo at: https://doi.org/10.5281/zenodo.13971451.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
NIH [R01DK131525, OT2OD036435, OT2OD030160, U24CA264250, U24CA271114, RC2DK131995]. Funding for open access charge: NIH [U24CA224260].
Conflict of interest statement. None declared.
References
- 1. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y.et al.. The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 2. The UniProt Consortium Bateman A., Martin M.-J., Orchard S., Magrane M., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bye-A-Jee H.et al.. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2022; 51:D523–D531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nguyen D.-T., Mathias S., Bologa C., Brunak S., Fernandez N., Gaulton A., Hersey A., Holmes J., Jensen L.J., Karlsson A.et al.. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017; 45:D995–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R.et al.. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015; 43:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J., Al-Ouran R., Hu Y., Kim S.-Y., Wan Y.-W., Wangler M.F., Yamamoto S., Chao H.-T., Comjean A., Mohr S.E.et al.. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet. 2017; 100:843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008; 40:1047–1051. [DOI] [PubMed] [Google Scholar]
- 7. Rose P.W., Prlić A., Bi C., Bluhm W.F., Christie C.H., Dutta S., Green R.K., Goodsell D.S., Westbrook J.D., Woo J.et al.. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015; 43:D345–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thul P.J., Lindskog C.. The human protein atlas: a spatial map of the human proteome. Protein Sci. 2018; 27:233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma’ayan A.. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016; 2016:baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin F.J., Amode M.R., Aneja A., Austine-Orimoloye O., Azov A.G., Barnes I., Becker A., Bennett R., Berry A., Bhai J.et al.. Ensembl 2023. Nucleic Acids Res. 2023; 51:D933–D941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baldarelli R.M., Smith C.L., Ringwald M., Richardson J.E., Bult C.J.. Mouse Genome Informatics Group (2024) Mouse Genome Informatics: an integrated knowledgebase system for the laboratory mouse. Genetics. 227:iyae031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McInnes L., Healy J., Saul N., Großberger L.. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Software. 2018; 3:861. [Google Scholar]
- 13. Saxena P. Ultimate Machine Learning with Scikit-Learn. 2024; London, United Kingdom: Orange Education Pvt. Ltd. [Google Scholar]
- 14. Evangelista J.E., Clarke D.J.B., Xie Z., Marino G.B., Utti V., Jenkins S.L., Ahooyi T.M., Bologa C.G., Yang J.J., Binder J.L.et al.. Toxicology knowledge graph for structural birth defects. Commun. Med. 2023; 3:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Franz M., Lopes C.T., Fong D., Kucera M., Cheung M., Siper M.C., Huck G., Dong Y., Sumer O., Bader G.D.. Cytoscape.js 2023 update: a graph theory library for visualization and analysis. Bioinformatics. 2023; 39:btad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Traag V.A., Waltman L., van Eck N.J.. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019; 9:5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M.. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023; 51:D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Groza T., Gomez F.L., Mashhadi H.H., Muñoz-Fuentes V., Gunes O., Wilson R., Cacheiro P., Frost A., Keskivali-Bond P., Vardal B.et al.. The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. 2023; 51:D1038–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ontology Consortium G., Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L.et al.. The Gene Ontology knowledgebase in 2023. Genetics. 2023; 224:iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J.et al.. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023; 51:D977–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clarke D.J.B., Marino G.B., Deng E.Z., Xie Z., Evangelista J.E., Ma’ayan A.. Rummagene: massive mining of gene sets from supporting materials of biomedical research publications. Commun. Biol. 2024; 7:482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marino G.B., Clarke D.J.B., Deng E.Z., Ma’ayan A.. RummaGEO: automatic mining of human and mouse gene sets from GEO. Patterns. 2024; 5:101072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhullar K.S., Lagarón N.O., McGowan E.M., Parmar I., Jha A., Hubbard B.P., Rupasinghe H.P.V.. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol. Cancer. 2018; 17:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson J.L., Yaron T.M., Huntsman E.M., Kerelsky A., Song J., Regev A., Lin T.-Y., Liberatore K., Cizin D.M., Cohen B.M.et al.. An atlas of substrate specificities for the human serine/threonine kinome. Nature. 2023; 613:759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F., Pantel S.E.et al.. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014; 1:140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D.et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A.et al.. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011; 39:D945–D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J.et al.. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 2015; 33:306–312. [DOI] [PubMed] [Google Scholar]
- 30. Kang X., Cui C., Wang C., Wu G., Chen H., Lu Z., Chen X., Wang L., Huang J., Geng H.et al.. CAMKs support development of acute myeloid leukemia. J. Hematol. Oncol. 2018; 11:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monaco S., Rusciano M.R., Maione A.S., Soprano M., Gomathinayagam R., Todd L.R., Campiglia P., Salzano S., Pastore L., Leggiero E.et al.. A novel crosstalk between calcium/calmodulin kinases II and IV regulates cell proliferation in myeloid leukemia cells. Cell. Signal. 2015; 27:204–214. [DOI] [PubMed] [Google Scholar]
- 32. Didier C., Demur C., Grimal F., Jullien D., Manenti S., Ducommun B.. Evaluation of checkpoint kinase targeting therapy in acute myeloid leukemia with complex karyotype. Cancer Biol. Ther. 2012; 13:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Z., Liu C., Jiang W.G., Ye L.. Deregulated bone morphogenetic proteins and their receptors are associated with disease progression of gastric cancer. Comput. Struct. Biotechnol. J. 2020; 18:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hardwick J.C., Kodach L.L., Offerhaus G.J., van den Brink G.R.. Bone morphogenetic protein signalling in colorectal cancer. Nat. Rev. Cancer. 2008; 8:806–812. [DOI] [PubMed] [Google Scholar]
- 35. Sanford J.A., Nogiec C.D., Lindholm M.E., Adkins J.N., Amar D., Dasari S., Drugan J.K., Fernández F.M., Radom-Aizik S., Schenk S.et al.. Molecular Transducers of Physical Activity Consortium (MoTrPAC): mapping the dynamic responses to exercise. Cell. 2020; 181:1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prohl C., Pelzer W., Diekert K., Kmita H., Bedekovics T., Kispal G., Lill R.. The yeast mitochondrial carrier leu5p and its human homologue graves’ disease protein are required for accumulation of coenzyme A in the matrix. Mol. Cell. Biol. 2001; 21:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Csukasi F., Duran I., Barad M., Barta T., Gudernova I., Trantirek L., Martin J.H., Kuo C.Y., Woods J., Lee H.et al.. The PTH/PTHrP-SIK3 pathway affects skeletogenesis through altered mTOR signaling. Sci. Transl. Med. 2018; 10:eaat9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan S., Luo L., Wang Y., Chen C., Wang J., Wu X.. Unifying large language models and knowledge graphs: a roadmap. IEEE Transactions on Knowledge and Data Engineering. 2023; 36:3580–3599. [Google Scholar]
- 39. Tomita I., Tsuruta H., Yasuda-Yamahara M., Yamahara K., Kuwagata S., Tanaka-Sasaki Y., Chin-Kanasaki M., Fujita Y., Nishi E., Katagiri H.et al.. Ketone bodies: A double-edged sword for mammalian life span. Aging Cell. 2023; 22:e13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou L., Schellaert W., Martínez-Plumed F., Moros-Daval Y., Ferri C., Hernández-Orallo J.. Larger and more instructable language models become less reliable. Nature. 2024; 634:61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Harmonizome 3.0 database is available at: https://maayanlab.cloud/Harmonizome/. The Harmonizome processed datasets are available in multiple formats at: https://maayanlab.cloud/Harmonizome/download/. The Harmonizome data processing scripts are available at: https://github.com/MaayanLab/HarmonizomePythonScripts. A snapshot of the code of these processing scripts can be access from Zenodo at: https://doi.org/10.5281/zenodo.13971451.