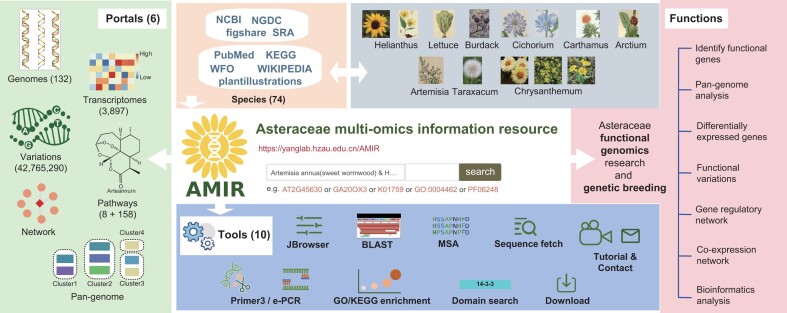
Abstract
As the largest family of dicotyledon, the Asteraceae family comprises a variety of economically important crops, ornamental plants and numerous medicinal herbs. Advancements in genomics and transcriptomic have revolutionized research in Asteraceae species, generating extensive omics data that necessitate an efficient platform for data integration and analysis. However, existing databases face challenges in mining genes with specific functions and supporting cross-species studies. To address these gaps, we introduce the Asteraceae Multi-omics Information Resource (AMIR; https://yanglab.hzau.edu.cn/AMIR/), a multi-omics hub for the Asteraceae plant community. AMIR integrates diverse omics data from 74 species, encompassing 132 genomes, 4 408 432 genes annotated across seven different perspectives, 3897 transcriptome sequencing samples spanning 131 organs, tissues and stimuli, 42 765 290 unique variants and 15 662 metabolites genes. Leveraging these data, AMIR establishes the first pan-genome, comparative genomics and transcriptome system for the Asteraceae family. Furthermore, AMIR offers user-friendly tools designed to facilitate extensive customized bioinformatics analyses. Two case studies demonstrate AMIR’s capability to provide rapid, reproducible and reliable analysis results. In summary, by integrating multi-omics data of Asteraceae species and developing powerful analytical tools, AMIR significantly advances functional genomics research and contributes to breeding practices of Asteraceae.
Graphical Abstract
Graphical Abstract.
Introduction
The Asteraceae family, accounting for 7% of flowering plant diversity with approximately 1600–1700 genera and over 26 000 species (1–4), holds paramount economic and ecological significance. It includes well-known oilseed crops such as sunflowers, vegetables such as lettuce, chicory and artichoke, ornamental plants such as chrysanthemum, and various weeds and invasive species (1–3). Additionally, Asteraceae plants are widely used in traditional medicine and valued for their production of diverse secondary metabolites (5,6).
Recent advancements in genomics and transcriptomics have revolutionized research in Asteraceae species, facilitating the identification of functional genes, crop genetic breeding and pharmaceutical discovery (7–12). Several Asteraceae species, including sunflower, lettuce, cultivated chrysanthemum, chicory, endive, great burdock and yacon, have been decoded (7–9,12), providing valuable references for genetic information mining. However, genomic studies of Asteraceae plants still face challenges due to scarcity of publicly accessible gene annotations, lack of comparative genomics/transcriptomic platforms, and difficulties in managing and analyzing multi-omics data. Platforms such as LettuceGDB, SGD (https://sunflowergenome.org/), the burdock database and Chrysanthemum Genome Database (http://210.22.121.250:8880/) have been developed to integrate and utilize these data (13,14). For example, LettuceGDB integrates multi-omics data and germplasm resources, and provides several tools for lettuce (13). SGD contains tools such as JBrowse, BLAST and gene expression browser, and allows the users to download genomic, variations and transcriptomes data of sunflower. The burdock multi-omics database integrates mitochondrial, chloroplast, and nuclear genome, transcriptome and SRAP fingerprints of different varieties of burdock (14). The Chrysanthemum Genome Database was launched in April 2023, and contains genomes, transcriptomes, comparative genomics and various bioinformatics analysis tools. While these existing databases address specific species, they struggle with efficient gene mining and cross-species studies. Therefore, there is a critical need for an integrative Asteraceae omics database that offers comprehensive annotations, data integrity and user-friendly tools to support fundamental research and breeding practices across the Asteraceae family.
To address these challenges, we introduce the Asteraceae Multi-omics Information Resource (AMIR) platform. AMIR integrates various data types, including genomes, gene functional annotations, homolog relationships, transcriptome and variations for 74 Asteraceae species. It provides the first pan-genome and comparative genomics platform for Asteraceae species, along with a comprehensive transcriptome system comprising over 3897 RNA sequencing (RNA-seq) libraries. AMIR also features user-friendly tools, including JBrowser, BLAST, Multi-alignment viewer, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (GO/KEGG) enrichment, Primer3 and e-PCR, enabling extensive customized bioinformatics analyses. By integrating multi-omics data of Asteraceae species and offering powerful analytical tools, AMIR aims to advance functional genomics research and enhance breeding practices of Asteraceae.
Materials and methods
Data collection
To encompass a broad range of Asteraceae species, we compiled 132 genome assembly datasets from 74 species sourced primarily from the National Center for Biotechnology Information (NCBI) (15), National Genomics Data Center (16) and European Bioinformatics Institute (17). These include well-known species such as sunflower (7), lettuce (9), cultivated Chrysanthemum (10) and Artemisia annua (18), with 76 genomes assembled at the chromosome level. Additionally, we collected 3897 RNA-seq libraries for 44 species from NCBI’s SRA database, which were utilized for genetic variation analysis and co-expression network construction.
Gene annotation pipeline
Gene annotation was performed for 24 Asteraceae species lacking gene annotation data using Braker3 (19). This tool integrates ab initio gene prediction, homology-based protein prediction using sequences from Arabidopsis thaliana (TAIR10) (https://www.arabidopsis.org/), sunflower (HanXRQr2.0-SUNRISE) (7), cultivated Chrysanthemum (Chrysanthemum_x_morifolium_Ramat_Zszgv0) (10) and lettuce (Lsat_Salinas_v11) (9), and RNA-seq data with a mapping rate exceeding 70% across diverse tissues and treatments (Supplementary Table S1) (20). The completeness of protein-coding annotations was evaluated using BUSCO v4.0.5 (21), and results are detailed in Supplementary Table S2. For ease of access, a uniform gene identifier (AMID) was assigned to genes in general feature format (GFF) files across 68 collected and predicted genomes.
Functional annotation of protein-coding genes
To enhance genome annotation completeness, we conducted unified functional annotation for each genome. Homologous genes from Asteraceae species were identified using a sequence similarity-based approach previously described (22). Specifically, protein sequences from genes in Arabidopsis and sunflower (7) were used as queries. BLASTP (2.10.0+) searches were performed against protein sequences of each Asteraceae species under the conditions of E-value <1e−5 and identity >50% (22). Subsequently, all protein sequences from Asteraceae species were reciprocally searched against protein sequences from Arabidopsis and sunflower. For protein sequences matching those from Arabidopsis and sunflower, only the best alignment sequence was kept. Based on the results from these searches, homologous genes in Asteraceae species were extracted.
Transcription factors (TFs) in 68 Asteraceae genomes were predicted using PlantTFDB v5.0 (23) and iTAK (24). Amino acid sequences were uploaded to PlantTFDB and iTAK database for analysis. Gene families in the Asteraceae genomes were identified based on corresponding gene family members in Arabidopsis (https://www.arabidopsis.org). To identify conserved motifs, protein sequences from Asteraceae species were subjected to HMMER models using HMMER v3.2.1 hmmsearch (25) with an E-value threshold of 1e−5. Protein sequences from each pair of genomes were compared using Diamond v.0.9.14.115 (26). Subsequently, gene collinearity was detected using the McScan (Python version) (27). Visualization of collinearity results for specific species and regions was achieved using ShinySyn (28) to facilitate user access.
Ortholog groups among the Asteraceae genomes
To analyze ortholog groups among Asteraceae genomes, we first constructed a distance-based phylogenetic tree using JolyTree with default parameters (29). The tree was saved in Newick format and visualized with the ggtree R package (30). We selected genomes from 43 Asteraceae species to establish a robust pan-genome based on two criteria: (i) BUSCO completeness scores >80% (31) and (ii) the inclusion of RNA-seq data for gene structure prediction. Drawing from pan-genome construction approaches in Bambusoideae (32), rice (20), wild grape (33) and poplar (34), we developed a gene-based pan-genome for the Asteraceae (33). Protein sequences from genes in 43 Asteraceae genomes were collected and analyzed using OrthoFinder v2.5.4 (35) with default parameters to identify ortholog groups, which are clusters of genes. These ortholog groups include three categories: core, dispensable and species-specific gene clusters. Core gene clusters are genes present in at least 39 genomes (>90% genomes) (36), species-specific gene clusters occur only in one species and the remaining gene clusters found in 2–38 species are classified as dispensable gene clusters.
RNA-seq analysis and gene regulatory network construction
After removing adapter sequences and low-quality reads using fastp (v.0.23.0) (37), clean reads were aligned to the reference genome of each species using Hisat2 (v.2.1.2) (38) with default parameters. Expression quantification was performed using StringTie (v.1.3.5) (39) with default settings, and gene expression levels were normalized using transcripts per kilobase million reads. Raw gene expression counts were generated using featureCounts (v1.6.4) (40). Differential gene expression analysis was conducted using the Bioconductor package DESeq2 (41) in R (v4.3.1). Genes with a log2-transformed fold change ≥1 or ≤−1 and a false discovery rate ≤0.05 were considered differentially expressed genes (DEGs). Tissue-specific gene expression was assessed using transcriptomes from various developmental. Batch effects were removed using the ComBat (42) package in R (v4.3.1). To measure tissue specificity, expression specificity metric (Tau) (43) and specificity measure (SPM) (44) were calculated. A co-expression network was constructed by calculating the Pearson correlation coefficient (PCC) of pairwise gene expression levels with gene pairs having a PCC > 0.8 retained to form the network. Furthermore, TF regulatory networks for 54 species were predicted using the regulatory prediction function in PlantRegMap (45), based on the regulatory network of Arabidopsis.
Genome variation calling
Due to the complexity and large size of Asteraceae genomes, population-level resequencing studies are limited (10,46). Nonetheless, transcriptome-based variant identification has emerged as a cost-effective alternative and has been successfully applied in several species, such as lettuce, maize and rice (47–49). In this study, we adopted a workflow previously used for variant identification using RNA-seq data (47–49). Initially, high-quality clean reads were aligned to the reference genome using STAR with default parameters (50). The alignment outputs were subsequently converted and sorted using Samtools (v1.13) (51). The mapped reads were further processed for sorting, read group addition and duplicate marking with Sambamba (v0.8.2) (52). After that, GATK’s SplitNCigarReads was utilized to split ‘N’ cigar reads (53). GATK HaplotypeCaller was then employed to generate Genomic Variant Call Format (GVCF) files for each sample. These individual GVCF files were combined using CombineGVCFs and GenotypeGVCFs to produce a unified Variant Call Format (VCF) file. The raw VCF file was filtered using GATK’s VariantFiltration to remove low-quality single-nucleotide polymorphisms (SNPs), applying criteria of ‘QUAL < 30.0 || MQ < 50.0 || QD < 2’. SNPs and Insertions and Deletions (InDels) with minor allele frequencies <0.01 or missing rates >0.1 were discarded using VCFtools (v.0.1.16) (54). Finally, genetic variant annotation and effect prediction were carried out using SnpEff (5.0d) (55).
Database implementation
AMIR was implemented using ThinkPHP framework (v.5.0.24) (https://www.thinkphp.cn/) for its backend. The core JavaScript libraries utilized include Vue.js as the main framework (https://vuejs.org), vis.js for network visualization (https://visjs.org), plotly.js (https://plotly.com), echarts (https://echarts.apache.org), D3.js (https://d3js.org), igv.js (56) and BlasterJS (57) for interactive charts. The system operates on an Apache2 Web server (v.2.4.53) and employs MySQL (v.8.0.29) as its database engine. The database is publicly accessible online without requiring registration and has been optimized for Chrome (recommended), Opera, Firefox, Windows Edge and macOS Safari. Gene and protein sequence alignments were performed using NCBI BLAST (v.2.13.0+) (58) and MAFFT (v7.490) with the ‘--maxiterate 1000’ parameter (59). Genome sequences, gene models, variations and gene expression levels were displayed using JBrowse 2 (60). Web BLAST functionality was powered by Sequencesever (61), while primer design was facilitated by Primer3 (62). Gene set enrichment analysis was conducted using the R package fgsea (63).
Database content and usage
Overview of AMIR
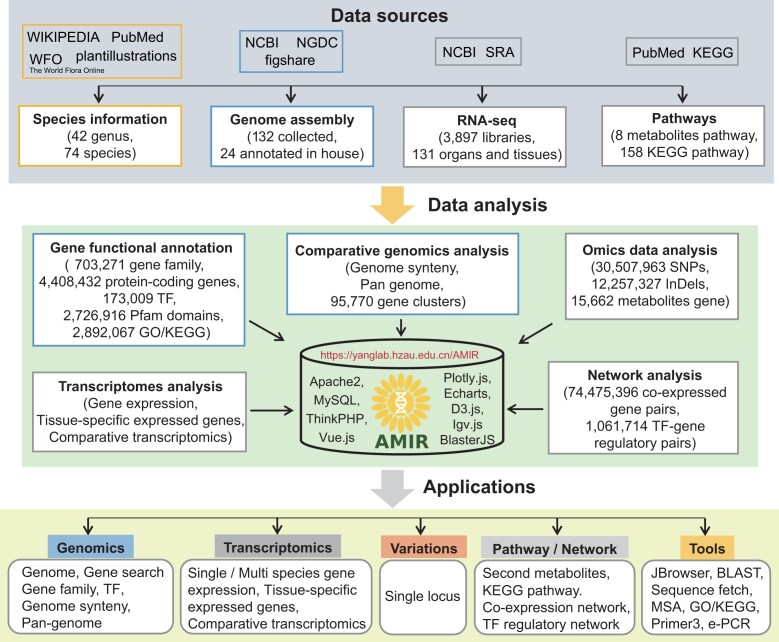
AMIR is dedicated to developing a comprehensive and user-friendly multi-omics data platform for the Asteraceae family. To this end, we have constructed eight specialized omics modules focusing on genes, including species, genomes, transcriptomes, variome, networks, pathways, tools, downloads and help (Figure 1). In the genomics module, AMIR encompasses 132 genome assembly data for 74 Asteraceae species to date (Supplementary Table S2). To address the scarcity of publicly available gene structure annotations, we predicted gene structure for 24 species using genomes at or above the scaffold level (Supplementary Table S2). We then performed comprehensive and integrated functional annotation on 4 408 432 protein-coding genes, identifying 2 892 067 GO/KEGG terms (64), 2 726 916 Pfam domains (65), 158 KEGG pathways (66), 173 009 TFs and 703 271 gene families (Figure 1). For construction of the Asteraceae pan-genome, the most frequently used reference genome from each species was selected, resulting in 95 770 gene clusters. AMIR incorporates 3897 RNA-seq datasets covering 44 species, 55 tissues and 76 stress treatments (Supplementary Table S1). Genetics variations were identified from transcriptome data, revealing a total of 30 507 963 SNPs and 12 257 327 InDels.
Figure 1.
Construction pipelines of AMIR, depicting data collection, data processing and database implementation. The upper section represents data sources. The middle section depicts the data analysis outcomes. The lower section illustrates data storage and the corresponding functional modules accessible in AMIR.
Gene search with unified identifier (AMID)
To facilitate cross-species research, AMIR has implemented a standardized naming system (AMID) for 68 genomes. AMID provides details on species, genome versions, chromosomes and gene order (Supplementary Figure S1A). Within AMIR, researchers can search for genes using multiple methods, such as AMID, homologous in Arabidopsis and sunflower, gene symbols, GO, KEGG, EC and PFAM identifier. For example, by selecting Artemisia argyi and entering the gene name ‘MEE23’, ‘AT2G34790’, ‘AargV1_Chr01g0000021’ or KEGG number ‘K22395’, users can access the same gene's information (Supplementary Figure S1B). Additionally, initiating a search from the homepage or the top right corner of the navigation bar directs users to a global search page, where users can easily access detailed results for the target gene across relevant functional modules by clicking corresponding buttons (Supplementary Figure S1C).
Genome module
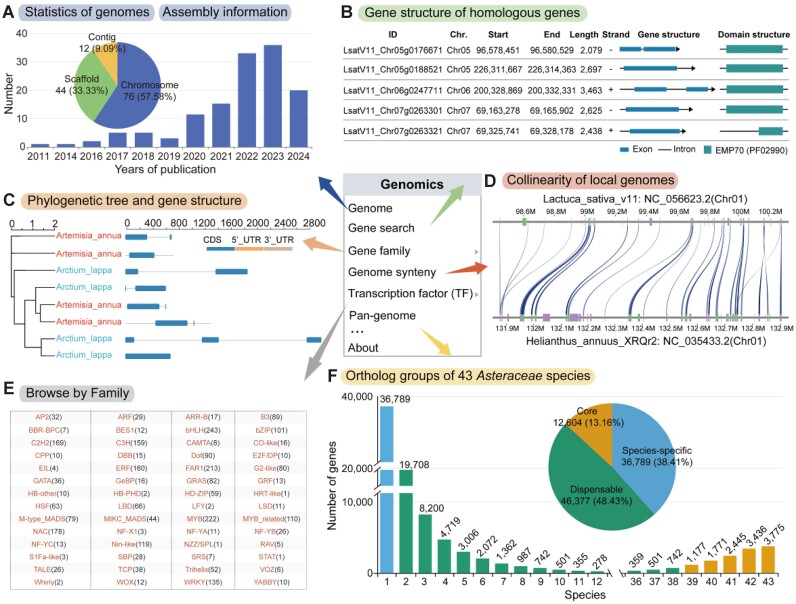
The genomics module is structured into eight key sections: genome, gene search, gene family, genome synteny, TFs, pan-genome and genome variations. The genome page displays assembly information and data sources for 132 Asteraceae genomes (Figure 2A). In the gene search section, users can select a species and search for genes by ID and position (Figure 2B), with results providing comprehensive annotations, including homologous in Arabidopsis/sunflower, gene position and functional annotations. Moreover, the page integrates a phylogenetic tree and visual representations of gene structures and protein domains based on homologous genes in Arabidopsis (Figure 2B). The gene family module is divided into gene family overview and phylogenetic sections. The overview presents detailed information on 181 gene families across 54 species, while the phylogenetic section illustrates evolutionary relationships within each gene family across different species (Figure 2C). Users can customize phylogenetic trees by selecting a varying number of species. Genome synteny section allows pairwise comparison of genomes from 53 species (Figure 2D), enabling users to explore collinearity relationships across chromosomes, identifying conserved regions and functional genes. The homolog search allows for the quick identification of homologous genes using gene IDs (Supplementary Figure S2A). It also provides flexibility in selecting genomes from different species, effectively identifying homologs and paralogs in the chosen genomes (Supplementary Figure S2B).
Figure 2.
Functionality and utilization of the ‘Genomics’ section in AMIR. (A) Assembly information for 132 genomes within the Asteraceae family. (B) Gene search results, including genomic location, gene structure, functional annotation and homologous genes in Arabidopsis and sunflower. (C) Gene family phylogenetic analysis across different species. (D) Genome synteny pages displaying collinearity of chromosomes in target species. (E) TF page providing annotated information on TFs from various Asteraceae species. (F) Ortholog groups page presenting homologous gene clusters among 43 species, enabling exploration of gene presence or absence across species.
TFs play a pivotal role as key regulatory elements in various biological processes, including plant growth, development and responses to external environments (67). The TF module utilizes iTAK (24) and PlantTFDB (23) online tools to identify TFs in 54 species. iTAK detected a total of 70 TF families and 176 072 TF genes, while PlantTFDB identified 58 TF families and 173 009 TF genes. Users have the option to select a specific TF family or input an AMID to access TF information for a particular species (Figure 2E). The super-pangenome offers a comprehensive view of genomic variation within a genus, facilitating the identification of conserved and adaptive genes, and presenting significant opportunities for crop improvement (68). Drawing from pan-genome constructions in Bambusoideae (32), rice (20), wild grape (33), Brassica oleracea (69) and poplar (34), we developed the gene-based pan-genome of Asteraceae (33). Ortholog groups were identified using OrthoFinder (35), resulting in 95 770 gene clusters. Among these clusters, 12 604 (13.16%) are core gene clusters present in at least 39 genomes, 46 377 (48.43%) are dispensable gene clusters found in 2–38 species, and the remaining 36 789 (38.41%) represent species-specific gene clusters (Figure 2F). Within the pan-genome page, users can explore the presence/absence patterns of each gene across 43 species and examine gene conservation categories (core, dispensable and species-specific) (Figure 2F).
Transcriptome module
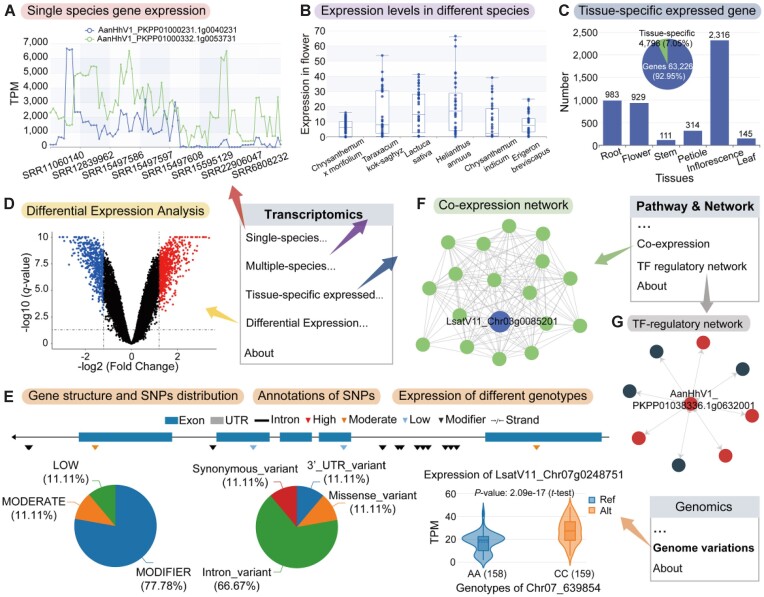
The ‘Transcriptomics’ module, compiled from 3897 transcriptome datasets across 44 species, is divided into four sections: ‘Single-species gene expression profiles’; ‘Multi-species gene expression profiles’; ‘Tissue-specific expressed genes’; and ‘Differential expression analysis’. In the ‘Single-species gene expression profile’ section, users can explore the expression profiles of genes or gene sets of a specific species across diverse samples (Figure 3A). Leveraging RNA-seq data from the same tissues and treatments across different species (Supplementary Table S3), we have developed multi-species gene expression profiles, enabling users to compare gene expression patterns across different species (Figure 3B). Tissue-specific expressed genes, responsible for conferring distinct morphological structures or physiological functions to various tissue or cell types (44), are identified and presented on a dedicated page using statistical measures like Tau and SPM (Supplementary Table S4). Users can search for tissue-specific expressed genes across various species on this page (Figure 3C). Additionally, by clicking the ‘Expression Profile’ button, users can navigate to the single-species gene expression profile page to explore the expression patterns of chosen genes. The ‘Differential expression analysis’ section compiles data from 53 transcriptome projects spanning 23 species and covering 25 different treatment conditions (Supplementary Table S5). On this page, users can search for DEGs under various conditions (Figure 3D and Supplementary Table S6). By selecting specific DEGs, users can transition to other functional pages, such as conducting GO/KEGG enrichment analysis or querying expression profiles for these genes, thereby enhancing the platform's exploratory capabilities.
Figure 3.
Functionality and utilization of the ‘Transcriptomic’, ‘Genome variation’ and ‘Pathway & Network’ sections in AMIR. (A) Single-species expression module providing gene or gene set expression profiles for a specific species. (B) Multi-species expression profile page displaying comparisons of expression levels in the same tissue across different species. (C) Tissue-specific expression page allowing users to query and retrieve information on genes specifically expressed in tissues across multiple species. (D) Differential expression analysis page offering data on DEGs under various conditions, with links to the GO/KEGG enrichment analysis. (E) Genome variations module allows users to search for SNP distributions and functional annotations on genes and facilitates comparisons of expression levels between different genotypes. (F) Co-expression network visualizes queried genes and co-expressed genes. (G) TF regulatory network features the queried TF gene as the central dot, with surrounding dots representing genes regulated by the TF, and directional arrows indicating the regulation.
Genome variation module
The identification of SNPs is essential for unraveling the regulatory mechanisms underlying phenotypic or expression variation. These SNPs are predominantly derived from whole-genome resequencing data. However, population resequencing can be expensive, particularly for the intricate and large-size genomes in Asteraceae (10,46). Alternatively, transcriptome sequencing presents a more cost-effective option for those genomes, enabling the identification of SNPs within transcribed regions (70). In some species, transcriptome-based variant identification has led to the identification of expression quantitative trait locus (eQTLs) and, subsequently, the revelation of genes associated with key traits (47–49).
To understand the impact of genetic variation on gene expression in Asteraceae species, we selected 2392 high-quality RNA-seq datasets for variant identification (Supplementary Table S7). Across 12 species, a total of 30 507 963 SNPs and 12 257 327 InDels were identified. These genetic variations formed the basis of the genome variations module. In the SNP mode of the single-locus page, distinct genetic variant loci are visually displayed below the gene structure diagram, allowing users to select various loci for detailed information and assess whether the locus is significantly associated with gene expression (Figure 3E). Furthermore, to facilitate user exploration of the relationship between haplotypes composed of multiple genetic variants and gene expression, we developed the ‘Haplotype’ mode. Users can select different haplotypes to obtain correlations with gene expression.
Pathway and gene regulatory network
The discovery of new functional genes through cross-species homologs has been widely applied in several important species, such as A. thaliana, rice and sorghum (71–73). To identify candidate genes associated with the synthesis of secondary metabolites in the Asteraceae, we compiled eight validated pathways. These pathways include the synthesis of terpenoids (74–76), costunolide (77), artemisinin (78,79), chicoric acids (80,81) and cyanidin (82). These secondary metabolites are either unique to the Asteraceae or distinguished by their high content and widespread distribution. Key genes from multiple species, such as A. thaliana, A. annua, sunflower and chicory, contribute to these pathways. Candidate genes from other species were identified through homologous relationships, and eight interactive scalable vector graphics were designed to investigate these candidate genes (Supplementary Figure S3). Users can hover over key enzyme nodes in the metabolic pathways to reveals basic information and click for detailed information about the candidate genes in each species. Additionally, we collected and expanded 158 pathways from the KEGG database (66), resulting in the creation of the KEGG pathway page. Users can access detailed pathways for their species of interest by selecting the pathway map ID (83). Yellow nodes within these maps can be hovered over to reveal associated functional genes. Furthermore, the KEGG Pathway tool includes an AMID-based search function, facilitating the discovery of pathways containing target genes. Users can select the species of interest and then explore pathway details by clicking on the respective map ID. Yellow nodes can also be hovered over to identify functional gene locations. Users can also directly query the pathways involving specific genes using the search function.
We constructed a co-expression module using 74 475 396 co-expression pairs from 31 species. This module allows users to input genes or gene sets to generate a gene co-expression network. Moreover, users can adjust parameters such as search depth and threshold to access varying levels of the co-expression network (Figure 3F). Additionally, we established a transcriptional regulatory network for 54 Asteraceae species based on binding motifs from PlantTFDB. Users have the flexibility to select from three modes to retrieve regulations according to their requirements. In the TF-regulation network, the ‘TF’ mode facilitates the retrieval of downstream targets of input TFs, the ‘Target’ mode retrieves upstream regulators of input genes and the ‘Gene’ mode uncovers internal regulations among input genes (Figure 3G). Overall, the ‘Network’ module provides researchers with a comprehensive platform to explore gene functions and potential regulatory patterns.
Tools and download
To improve user convenience and accessibility, we integrated nine bioinformatics tools into the Tools portal in AMIR (Supplementary Figure S4A). These tools encompass a range of functions, including JBrowser, BLAST, Multi-alignment viewer, Sequence fetch, GO and KEGG enrichment, Primer3, e-PCR and Domain search. Notably, the ‘JBrowser’ tool incorporates the 68 Asteraceae genome and GFF3 annotations, enabling users to visually explore genomic sequence and gene information within specific regions (Supplementary Figure S4B). With the ‘BLAST tool, users can input query nucleotide or protein sequences for homo comparisons within or across species, facilitating functional analysis (Supplementary Figure S4C). The ‘Multi Alignment Viewer’ allows users to perform multiple sequence alignments to identify conserved regions across sequences (Supplementary Figure S4D). Moreover, the ‘Sequence fetch’ tool enables users to retrieve reference sequences, including genome sequence, coding sequence (CDS) and protein sequence, for customized analysis (Supplementary Figure S4E). For functional prediction of gene sets, the ‘GO/KEGG enrichment’ tools offer enrichment analysis capabilities with downloadable results and visualizing of enrichment pathway bubble diagrams (Supplementary Figure S4F). Modules such as ‘Primer3’ and ‘e-PCR’ aid users in quickly selecting optimal primer pairs for genes or nucleotide sequences to facilitate downstream experiments (Supplementary Figure S4G and H). Lastly, the ‘Domain search’ tool facilitates the identification of protein domains, providing insights into their functional roles. In summary, the development of the Tools in AMIR supports functional gene discovery, comparative genomic studies and molecular experiments within Asteraceae species.
To enhance user-customized analysis and facilitate data mining, we organized and categorized the data in AMIR, offering convenient downloads for four types of omics data. The genomic data include comprehensive details such as version information, assembly quality metrics and gene counts for 132 Asteraceae genomes; the transcriptomic data offer information on sources and sampling details for 3897 transcript datasets from 44 species; the variation data encompass variant information across 12 species, providing access to downloading VCF files of variants on different chromosomes; and metabolomics data feature 15 662 genes associated with the synthesis of various secondary metabolites.
Case study illustrating the utility of AMIR
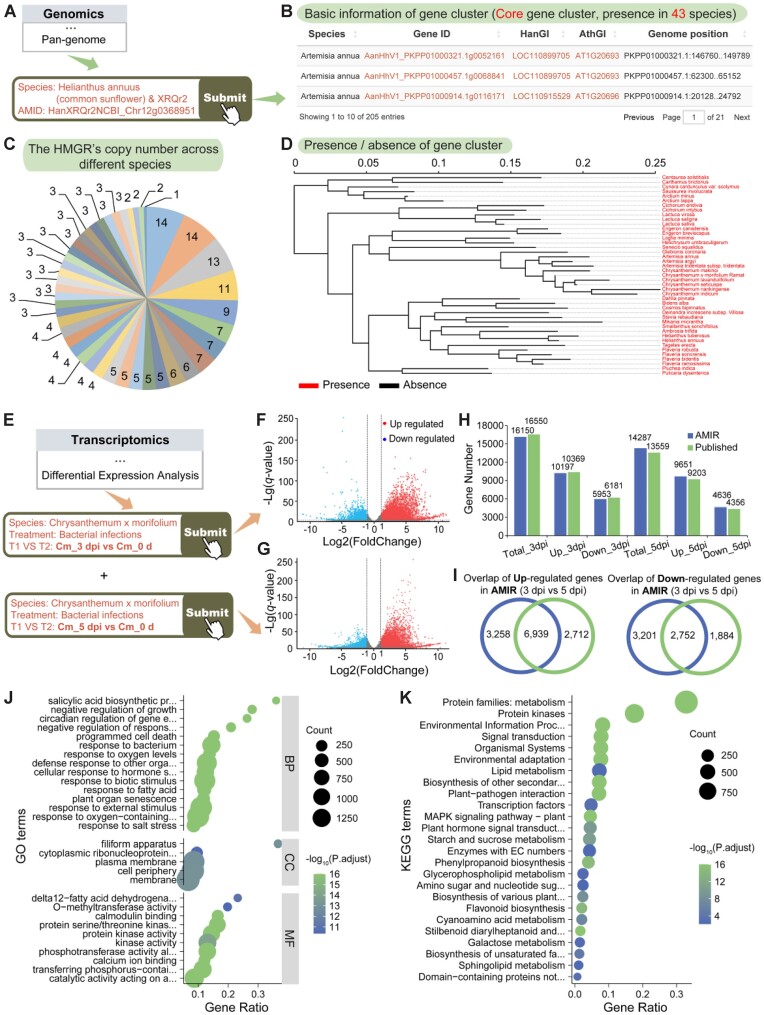
To illustrate the capabilities of AMIR, we present two case studies that confirm the conservation of the HMGR gene and explore DEGs induced by Alternaria sp. infection. Mevalonic acid (MVA) is the most critical precursor in the biosynthetic pathway of terpenoids in plants (84), and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), the key rate-limiting enzyme in the MVA pathway, is widely present in Asteraceae (18,85). Using the ‘Pan-genome’ module, we verified the conservation of the HMGR gene in Asteraceae. Specifically, LOC110894660 (https://www.ncbi.nlm.nih.gov/gene/110894660) corresponds to the HMG1/2-like protein in sunflower, with AMIR search ID HanXRQr2NCBI_Chr12g0368951. By selecting Helianthus annuus (common sunflower) and XRQr2 in the species selection box and entering HanXRQr2NCBI_Chr12g0368951 in AMIR’s search box (Figure 4A), we obtained results including homologous genes, conservation analysis and a phylogenetic tree of HMGR homologs within Asteraceae (Figure 4B–D). These findings confirm the presence of HMGR gene across all 43 species in the pan-genome construction, indicating its status as a core gene in Asteraceae (Figure 4D). Additionally, the gene's copy number varies from 1 to 14 across different species (Figure 4C), underscoring its conservation and highlighting the utility of the ‘Pan-genome’ module.
Figure 4.
Case study illustrating the utility of AMIR. (A) Process of searching for HMGR genes in the ‘Pan-genome’ module. (B) Pan-genome search results, including conservation of the queried gene, its distribution across different Asteraceae species and detailed gene information. (C) Number of homologous genes in each species. (D) Presence or absence of the queried gene in Asteraceae species. (E) Process of searching for DEGs in the ‘Differential expression analysis’ module. (F) Volcano plot of DEGs in Chrysanthemum morifolium leaves after 3 and 5 days of Alternaria sp. infection. (G) Comparison of the number of DEGs in AMIR with those reported in the original study. (H) Continuously upregulated genes after 3 and 5 days of Alternaria sp. infection. (I) Continuously downregulated genes after 3 and 5 days of Alternaria sp. infection. (J) GO enrichment analysis of continuously upregulated genes after 3 and 5 days of Alternaria sp. infection. (K) KEGG enrichment analysis of continuously upregulated genes after 3 and 5 days of Alternaria sp. infection.
AMIR’s ‘Differential expression analysis’ module includes data from project PRJNA448499, which involves transcriptomes of C. morifolium leaves at various time points inoculated with Alternaria sp. (86). Here, we will conduct a differential expression analysis to demonstrate AMIR’s robust data analysis capabilities. First, in the species and treatment selection menu of the module, Chrysanthemum × morifolium and Zszgv0 and Bacterial infections are chosen. Then, Cm_3 dpi versus Cm_0d and Cm_5 dpi versus Cm_0d are selected in the comparison box (Figure 4E). The search results include volcano plots of DEGs, detailed information on these genes and buttons for GO/KEGG enrichment analysis (Figure 4F and G). The results show that the number of DEGs observed after 3 and 5 days of Alternaria sp. infection are 16 150 and 14 287, respectively, closely aligning with the original study’s reported figures of 16 550 and 13 559 (86) (Figure 4H). Moreover, persistent upregulated and downregulated gene counts at Cm_3 dpi and Cm_5 dpi are 6939 and 2752, respectively, slightly surpassing previously reported numbers of 5952 and 2435 (Figure 4I). Clicking the GO enrichment button reveals predominant enrichment in terms such as ‘salicylic acid biosynthetic process’, ‘response to bacterium’ and ‘protein kinase activity’ (Figure 4J). Furthermore, KEGG analysis highlights significant enrichment in the ‘signal transduction’, ‘plant–pathogen interaction’, ‘phenylpropanoid biosynthesis’ and ‘flavonoid biosynthesis’ categories (Figure 4K), consistent with the original study’s findings (86). Additionally, comparing KEGG enriched terms based on gene hits between AMIR and the original study shows high consistency (Kendall’s coefficient of concordance = 0.81, P-value = 9.06e−09). These results underscore AMIR’s capability to quickly deliver reproducible and reliable analysis outcomes, including the identification of conserved genes, DEGs and differential pathways, without necessitating raw data downloads or complex bioinformatics procedures.
Discussion and future perspectives
With the rapid development of sequencing technology, numerous species in Asteraceae have been sequenced. However, publicly available gene annotations for most Asteraceae genomes have been limited, impeding the discovery of functional genes. Addressing this gap, we collected and annotated genomes from 74 species, totaling 132. Using 11 475 543 annotation data, we established the AMIR. AMIR stands as the first platform to systematically integrate, analyze and store multi-omics data for the Asteraceae, offering the most comprehensive collections of genomes, transcriptomes and variations available to date. This resource empowers users to conduct comparative, evolutionary and functional genomic studies effectively.
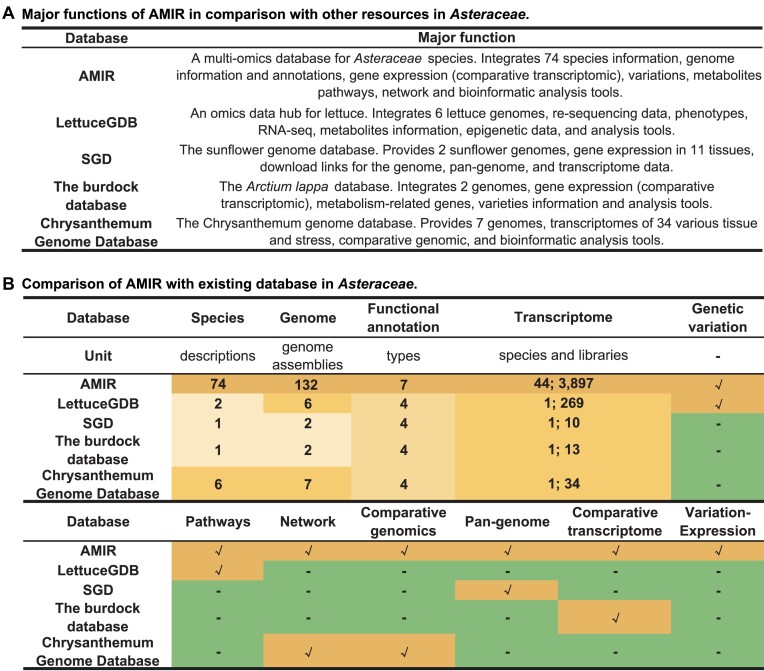
Compared to existing data resources in the Asteraceae such as LettuceGDB, SGD, the burdock database and Chrysanthemum Genome Database, AMIR provides enhanced features (Figure 5A): (i) it allows retrieval of annotation data for individual genes or gene sets by using gene IDs, GO/KEGG/Pfam identifiers or TF names, significantly enhancing the discovery of functional genes; (ii) it introduces the first super-pangenome and comparative genomics platform for the Asteraceae, enabling rapid exploration of gene conservation and function; (iii) it includes a comparative transcriptomics system to identify genes expressed specifically in certain tissues and to mine DEGs related to key traits or biological processes; and (iv) AMIR enables retrieval of SNPs/InDels in important genes or genomic regions, along with their frequency, haplotypes and correlation with gene expression, thereby uncovering variations and genes linked to important traits (Figure 5B).
Figure 5.
Comparison of AMIR with existing database in Asteraceae. (A) Major functions of AMIR in comparison with other resources in Asteraceae. (B) Comparison of main functions and data volumes between AMIR and other databases, the values representing data volume and tick marks indicating the presence of specific functional modules.
Moreover, AMIR integrates nine commonly used bioinformatics analysis tools for constructing gene regulatory network, performing functional enrichment analysis and identifying potential functional genes and metabolic pathways. We validate AMIR’s capabilities through two case studies: one confirming the conservation of the HMGR gene and the other exploring the DEGs induced by Alternaria sp. infection. These studies illustrate AMIR’s integration of extensive, high-quality data and robust analytical workflows. These resources and tools will facilitate the discovery of evolutionarily conserved genes and pathways specific to certain species or associated with distinct environments and essential traits. In summary, this multi-omics database marks a significant advancement in Asteraceae genomic research, providing a foundational resource for molecular breeding and drug discovery efforts in Asteraceae plants.
Currently, AMIR houses a variety of data types, including species information, genomes and transcriptomes, as well as variations and co-expression data derived from transcriptome processing and analysis. As genomic research on Asteraceae progresses and the volume of whole genome sequencing data and epigenetic data (such as methylation, histone modifications, Assay for Targeting Accessible-Chromatin with high-throughout sequencing (ATAC-seq) and Chromatin Immunoprecipitation sequencing (ChIP-seq)) expands, we are committed to continuously enriching AMIR with new types of omics information. Through regular updates and maintenance, we hope that AMIR can serve as the central hub for functional research in the Asteraceae family, driving advancements in molecular breeding, drug discovery and landscaping.
Supplementary Material
Acknowledgements
We thank the bioinformatics computing platform of the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, managed by Hao Liu.
Author contributions: Q.-Y.Y. and Z.M. designed the project. Q.-Y.Y. and D.L. managed and coordinated the project. R.D., X.H., C.L. and L.H. collected datasets. C.L., R.D., X.H. and D.L. performed the bioinformatics analysis. X.H., R.D., C.L., D.L., L.H., X.C., H.M., J.L. and L.Z. constructed the database. D.L., C.L., R.D., X.H. and L.H. wrote the manuscript. Q.-Y.Y. and Z.M. directed the project.
Contributor Information
Dongxu Liu, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Chengfang Luo, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Rui Dai, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Xiaoyan Huang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Xiang Chen, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Lin He, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; College of Plant Science & Technology, Huazhong Agricultural University, Wuhan 430070, China.
Hongxia Mao, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Jiawei Li, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Linna Zhang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Qing-Yong Yang, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Hubei Key Laboratory of Agricultural Bioinformatics and Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Yazhouwan National Laboratory, Sanya 572025, China.
Zhinan Mei, National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops, Huazhong Agricultural University, Wuhan 430070, China; College of Plant Science & Technology, Huazhong Agricultural University, Wuhan 430070, China.
Data availability
AMIR is accessible for free at https://yanglab.hzau.edu.cn/AMIR. All associated datasets can be downloaded via the ‘Download’ module of the database.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Key R&D Program of China [2022YFC3502200 and 2022YFF1001400]; National Natural Science Foundation of China [32322061 and 32070559]; Seed Industry High-quality Development Project Of Hubei Province [HBZY2023B00503]; Fundamental Research Funds for the Central University HZAU [2662023XXPY001].
Conflict of interest statement. None declared.
References
- 1. Zhang C., Zhang T., Luebert F., Xiang Y., Huang C.-H., Hu Y., Rees M., Frohlich M.W., Qi J., Weigend M.et al.. Asterid phylogenomics/phylotranscriptomics uncover morphological evolutionary histories and support phylogenetic placement for numerous whole-genome duplications. Mol. Biol. Evol. 2020; 37:3188–3210. [DOI] [PubMed] [Google Scholar]
- 2. Zhang C., Huang C.H., Liu M., Hu Y., Panero J.L., Luebert F., Gao T., Ma H.. Phylotranscriptomic insights into Asteraceae diversity, polyploidy, and morphological innovation. J. Integr. Plant Biol. 2021; 63:1273–1293. [DOI] [PubMed] [Google Scholar]
- 3. Zhang G., Yang J., Zhang C., Jiao B., Panero J.L., Cai J., Zhang Z.-R., Gao L.-M., Gao T., Ma H.. Nuclear phylogenomics of Asteraceae with increased sampling provides new insights into convergent morphological and molecular evolution. Plant Commun. 2024; 5:100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christenhusz M.J.M., Byng J.W.. The number of known plants species in the world and its annual increase. Phytotaxa. 2016; 261:201–217. [Google Scholar]
- 5. Medeiros-Neves B., Teixeira H.F., von Poser G.L.. The genus pterocaulon (Asteraceae)—A review on traditional medicinal uses, chemical constituents and biological properties. J. Ethnopharmacol. 2018; 224:451–464. [DOI] [PubMed] [Google Scholar]
- 6. Toyang N.J., Verpoorte R.. A review of the medicinal potentials of plants of the genus Vernonia (Asteraceae). J. Ethnopharmacol. 2013; 146:681–723. [DOI] [PubMed] [Google Scholar]
- 7. Badouin H., Gouzy J., Grassa C.J., Murat F., Staton S.E., Cottret L., Lelandais-Brière C., Owens G.L., Carrère S., Mayjonade B.et al.. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017; 546:148–152. [DOI] [PubMed] [Google Scholar]
- 8. Fan W., Wang S., Wang H., Wang A., Jiang F., Liu H., Zhao H., Xu D., Zhang Y.. The genomes of chicory, endive, great burdock and yacon provide insights into Asteraceae palaeo-polyploidization history and plant inulin production. Mol. Ecol. Resour. 2022; 22:3124–3140. [DOI] [PubMed] [Google Scholar]
- 9. Reyes-Chin-Wo S., Wang Z., Yang X., Kozik A., Arikit S., Song C., Xia L., Froenicke L., Lavelle D.O., Truco M.-J.et al.. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017; 8:14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song A., Su J., Wang H., Zhang Z., Zhang X., Van de Peer Y., Chen F., Fang W., Guan Z., Zhang F.et al.. Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat. Commun. 2023; 14:2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wei T., van Treuren R., Liu X., Zhang Z., Chen J., Liu Y., Dong S., Sun P., Yang T., Lan T.et al.. Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat. Genet. 2021; 53:752–760. [DOI] [PubMed] [Google Scholar]
- 12. Wen X., Li J., Wang L., Lu C., Gao Q., Xu P., Pu Y., Zhang Q., Hong Y., Hong L.et al.. The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic. Res. 2022; 9:uhab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou W., Yang T., Zeng L., Chen J., Wang Y., Guo X., You L., Liu Y., Du W., Yang F.et al.. LettuceDB: an integrated multi-omics database for cultivated lettuce. Database. 2024; 2024:baae018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song Y., Yang Y., Xu L., Bian C., Xing Y., Xue H., Hou W., Men W., Dou D., Kang T.. The burdock database: a multi-omic database for Arctium lappa, a food and medicinal plant. BMC Plant Biol. 2023; 23:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Farrell C.M., Feldgarden M., Fine A.M., Funk K.et al.. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023; 51:D29–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xue Y., Bao Y., Zhang Z., Zhao W., Xiao J., He S., Zhang G., Li Y., Zhao G., Chen R.et al.. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2023. Nucleic Acids Res. 2023; 51:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thakur M., Buniello A., Brooksbank C., Gurwitz K.T., Hall M., Hartley M., Hulcoop D.G., Leach A.R., Marques D., Martin M.et al.. EMBL’s European Bioinformatics Institute (EMBL-EBI) in 2023. Nucleic Acids Res. 2024; 52:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen Q., Zhang L., Liao Z., Wang S., Yan T., Shi P., Liu M., Fu X., Pan Q., Wang Y.et al.. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant. 2018; 11:776–788. [DOI] [PubMed] [Google Scholar]
- 19. Gabriel L., Brůna T., Hoff K.J., Ebel M., Lomsadze A., Borodovsky M., Stanke M.. BRAKER3: fully automated genome annotation using RNA-seq and protein evidence with GeneMark-ETP, AUGUSTUS, and TSEBRA. Genome Res. 2024; 34:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shang L., Li X., He H., Yuan Q., Song Y., Wei Z., Lin H., Hu M., Zhao F., Zhang C.et al.. A super pan-genomic landscape of rice. Cell Res. 2022; 32:878–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M.. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015; 31:3210–3212. [DOI] [PubMed] [Google Scholar]
- 22. Jia Q., Brown R., Kollner T.G., Fu J., Chen X., Wong G.K., Gershenzon J., Peters R.J., Chen F.. Origin and early evolution of the plant terpene synthase family. Proc. Natl Acad. Sci. U.S.A. 2022; 119:e2100361119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jin J., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J., Gao G.. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017; 45:D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Y., Jiao C., Sun H., Rosli H.G., Pombo M.A., Zhang P., Banf M., Dai X., Martin G.B., Giovannoni J.J.et al.. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant. 2016; 9:1667–1670. [DOI] [PubMed] [Google Scholar]
- 25. Finn R.D., Clements J., Eddy S.R.. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2014; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H.et al.. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012; 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xiao Z., Lam H.-M., Marschall T.. ShinySyn: a Shiny/R application for the interactive visualization and integration of macro- and micro-synteny data. Bioinformatics. 2022; 38:4406–4408. [DOI] [PubMed] [Google Scholar]
- 29. Criscuolo A. A fast alignment-free bioinformatics procedure to infer accurate distance-based phylogenetic trees from genome assemblies. Res. Ideas Outcomes. 2019; 5:e36178. [Google Scholar]
- 30. Yu G., Lam T.T., Zhu H., Guan Y.. Two methods for mapping and visualizing associated data on phylogeny using Ggtree. Mol. Biol. Evol. 2018; 35:3041–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raghavan V., Kraft L., Mesny F., Rigerte L.. A simple guide to de novo transcriptome assembly and annotation. Brief. Bioinform. 2022; 23:bbab563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu Y.-L., Gao S.-Y., Jin G., Zhou M.-Y., Gao Q., Guo C., Yang Y.-Z., Niu L.-Z., Xia E., Guo Z.-H.et al.. BambooBase: a comprehensive database of bamboo omics and systematics. Mol. Plant. 2024; 17:682–685. [DOI] [PubMed] [Google Scholar]
- 33. Cochetel N., Minio A., Guarracino A., Garcia J.F., Figueroa-Balderas R., Massonnet M., Kasuga T., Londo J.P., Garrison E., Gaut B.S.et al.. A super-pangenome of the North American wild grape species. Genome Biol. 2023; 24:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi T., Zhang X., Hou Y., Jia C., Dan X., Zhang Y., Jiang Y., Lai Q., Feng J., Feng J.et al.. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant. 2024; 17:725–746. [DOI] [PubMed] [Google Scholar]
- 35. Emms D.M., Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song J.M., Guan Z., Hu J., Guo C., Yang Z., Wang S., Liu D., Wang B., Lu S., Zhou R.et al.. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants. 2020; 6:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 41. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y., Parmigiani G., Johnson W.E.. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020; 2:lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kryuchkova-Mostacci N., Robinson-Rechavi M.. A benchmark of gene expression tissue-specificity metrics. Brief Bioinform. 2017; 18:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiao S.-J., Zhang C., Zou Q., Ji Z.-L.. TiSGeD: a database for tissue-specific genes. Bioinformatics. 2010; 26:1273–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tian F., Yang D.C., Meng Y.Q., Jin J., Gao G.. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020; 48:D1104–D1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song C., Liu Y., Song A., Dong G., Zhao H., Sun W., Ramakrishnan S., Wang Y., Wang S., Li T.et al.. The Chrysanthemum nankingense genome provides insights into the evolution and diversification of chrysanthemum flowers and medicinal traits. Mol. Plant. 2018; 11:1482–1491. [DOI] [PubMed] [Google Scholar]
- 47. Zhang L., Su W., Tao R., Zhang W., Chen J., Wu P., Yan C., Jia Y., Larkin R.M., Lavelle D.et al.. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat. Commun. 2017; 8:2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu S., Li C., Wang H., Wang S., Yang S., Liu X., Yan J., Li B., Beatty M., Zastrow-Hayes G.et al.. Mapping regulatory variants controlling gene expression in drought response and tolerance in maize. Genome Biol. 2020; 21:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu C., Zhu X., Zhang J., Shen M., Chen K., Fu X., Ma L., Liu X., Zhou C., Zhou D.X.et al.. eQTLs play critical roles in regulating gene expression and identifying key regulators in rice. Plant Biotechnol. J. 2022; 20:2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. 1000 Genome Project Data Processing Subgroup Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P.. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015; 31:2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.et al.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T.et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: sNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson J.T., Thorvaldsdottir H., Turner D., Mesirov J.P.. igv.Js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics. 2023; 39:btac830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Blanco-Míguez A., Fdez-Riverola F., Sánchez B., Lourenço A.. BlasterJS: a novel interactive JavaScript visualisation component for BLAST alignment results. PLoS One. 2018; 13:e0205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y.et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Katoh K., Kuma K., Toh H., Miyata T.. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005; 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Diesh C., Stevens G.J., Xie P., De J., Martinez T., Hershberg E.A., Leung A., Guo E., Dider S., Zhang J.et al.. JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biol. 2023; 24:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Priyam A., Woodcroft B.J., Rai V., Moghul I., Munagala A., Ter F., Chowdhary H., Pieniak I., Maynard L.J., Gibbins M.A.et al.. Sequenceserver: a modern graphical user interface for custom BLAST databases. Mol. Biol. Evol. 2019; 36:2922–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kõressaar T., Lepamets M., Kaplinski L., Raime K., Andreson R., Remm M., Hancock J.. Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics. 2018; 34:1937–1938. [DOI] [PubMed] [Google Scholar]
- 63. Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A.. Fast gene set enrichment analysis. 2021; bioRxiv doi:01 February 2021, preprint: not peer reviewed 10.1101/060012. [DOI]
- 64. Gene Ontology Consortium, Aleksander S.A., Balhoff J., Carbon S., Cherry J.M., Drabkin H.J., Ebert D., Feuermann M., Gaudet P., Harris N.L.et al.. The Gene Ontology knowledgebase in 2023. Genetics. 2023; 224:iyad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kanehisa M., Furumichi M., Sato Y., Kawashima M., Ishiguro-Watanabe M.. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023; 51:D587–D592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chowdhary A.A., Mishra S., Mehrotra S., Upadhyay S.K., Bagal D., Srivastava V.. Srivastava V., Mishra S., Mehrotra S., Upadhyay S.K.. Plant Transcription Factors. 2023; Amsterdam, The Netherlands: Elsevier; 3–20. [Google Scholar]
- 68. Khan A.W., Garg V., Roorkiwal M., Golicz A.A., Edwards D., Varshney R.K.. Super-pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020; 25:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li X., Wang Y., Cai C., Ji J., Han F., Zhang L., Chen S., Zhang L., Yang Y., Tang Q.et al.. Large-scale gene expression alterations introduced by structural variation drive morphotype diversification in Brassica oleracea. Nat. Genet. 2024; 56:517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lopez-Maestre H., Brinza L., Marchet C., Kielbassa J., Bastien S., Boutigny M., Monnin D., Filali A.E., Carareto C.M., Vieira C.et al.. SNP calling from RNA-seq data without a reference genome: identification, quantification, differential analysis and impact on the protein sequence. Nucleic Acids Res. 2016; 44:e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Armstead I., Donnison I., Aubry S., Harper J., Hörtensteiner S., James C., Mani J., Moffet M., Ougham H., Roberts L.et al.. Cross-species identification of Mendel's I locus. Science. 2007; 315:73–73. [DOI] [PubMed] [Google Scholar]
- 72. Duan G., Wu G., Chen X., Tian D., Li Z., Sun Y., Du Z., Hao L., Song S., Gao Y.et al.. HGD: an integrated homologous gene database across multiple species. Nucleic Acids Res. 2023; 51:D994–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hartmann A., Berkowitz O., Whelan J., Narsai R.. Cross-species transcriptomic analyses reveals common and opposite responses in Arabidopsis, rice and barley following oxidative stress and hormone treatment. BMC Plant Biol. 2022; 22:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Phillips M., Leon P., Boronat A., Rodríguez-Concepción M.. The plastidial MEP pathway: unified nomenclature and resources. Trends Plant Sci. 2008; 13:619–623. [DOI] [PubMed] [Google Scholar]
- 75. Shimada T.L., Shimada T., Okazaki Y., Higashi Y., Saito K., Kuwata K., Oyama K., Kato M., Ueda H., Nakano A.et al.. HIGH STEROL ESTER 1 is a key factor in plant sterol homeostasis. Nat. Plants. 2019; 5:1154–1166. [DOI] [PubMed] [Google Scholar]
- 76. Chen F., Tholl D., D’Auria J.C., Farooq A., Pichersky E., Gershenzon J.. Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell. 2003; 15:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de Kraker J.-W., Franssen M.C.R., Joerink M., de Groot A., Bouwmeester H.J.. Biosynthesis of costunolide, dihydrocostunolide, and leucodin. Demonstration of cytochrome P450-catalyzed formation of the lactone ring present in sesquiterpene lactones of chicory. Plant Physiol. 2002; 129:257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D.et al.. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013; 496:528–532. [DOI] [PubMed] [Google Scholar]
- 79. Ro D.-K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M., Ho K.A., Eachus R.A., Ham T.S., Kirby J.et al.. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006; 440:940–943. [DOI] [PubMed] [Google Scholar]
- 80. Clifford M.N., Jaganath I.B., Ludwig I.A., Crozier A.. Chlorogenic acids and the acyl-quinic acids: discovery, biosynthesis, bioavailability and bioactivity. Nat. Prod. Rep. 2017; 34:1391–1421. [DOI] [PubMed] [Google Scholar]
- 81. Fu R., Zhang P., Jin G., Wang L., Qi S., Cao Y., Martin C., Zhang Y.. Versatility in acyltransferase activity completes chicoric acid biosynthesis in purple coneflower. Nat. Commun. 2021; 12:1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006; 57:761–780. [DOI] [PubMed] [Google Scholar]
- 83. Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000; 28:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lu S., Zhou C., Guo X., Du Z., Cheng Y., Wang Z., He X.. Enhancing fluxes through the mevalonate pathway in Saccharomyces cerevisiae by engineering the HMGR and β-alanine metabolism. Microb. Biotechnol. 2022; 15:2292–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Majdi M., Abdollahi M.R., Maroufi A.. Parthenolide accumulation and expression of genes related to parthenolide biosynthesis affected by exogenous application of methyl jasmonate and salicylic acid in Tanacetum parthenium. Plant Cell Rep. 2015; 34:1909–1918. [DOI] [PubMed] [Google Scholar]
- 86. Zhao X., Song L., Jiang L., Zhu Y., Gao Q., Wang D., Xie J., Lv M., Liu P., Li M.. The integration of transcriptomic and transgenic analyses reveals the involvement of the SA response pathway in the defense of chrysanthemum against the necrotrophic fungus Alternaria sp. Hortic. Res. 2020; 7:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
AMIR is accessible for free at https://yanglab.hzau.edu.cn/AMIR. All associated datasets can be downloaded via the ‘Download’ module of the database.