Abstract
GenBank® (https://www.ncbi.nlm.nih.gov/genbank/) is a comprehensive, public data repository that contains 34 trillion base pairs from over 4.7 billion nucleotide sequences for 581 000 formally described species. Daily data exchange with the European Nucleotide Archive and the DNA Data Bank of Japan ensures worldwide coverage. We summarize the content of the database in 2025 and recent updates such as accelerated processing of influenza sequences and the ability to upload feature tables to Submission Portal for messenger RNA sequences. We provide an overview of the web, application programming and command-line interfaces that allow users to access GenBank data. We also discuss the importance of creating BioProject and BioSample records during submissions, particularly for viruses and metagenomes. Finally, we summarize educational materials and recent community outreach efforts.
Graphical Abstract
Graphical Abstract.
Introduction
GenBank® (1) is a comprehensive public repository of nucleotide sequences and supporting bibliographic and biological annotations built and distributed by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM) at the US National Institutes of Health (NIH). Together with the European Bioinformatics Institute at the European Molecular Biology Lab and the Research Organization of Information and Systems National Institute of Genetics, NLM-NCBI is one of three founding members of the International Nucleotide Sequence Database Collaboration (INSDC) (2). Their sequence data repositories, the European Nucleotide Archive (3), the DNA Data Bank of Japan (4), and GenBank and Sequence Read Archive (SRA; NCBI), share data daily to promote unrestricted, global access to these data. From its inception >40 years ago, GenBank has pioneered the principles of open science and data sharing, and supports these as described by FAIR (findable, accessible, interoperable and reusable) principles (5). As a public archive, GenBank preserves the scientific record, the integrity of that record and the knowledge associated with sequence data that investigators generate over time through reuse of these data. A case in point is the recent COVID-19 pandemic. Before 2020, GenBank contained 3.5 million viral genomes, only 30 000 of which were from coronaviruses. Nonetheless, these sequences formed the basis for and facilitated subsequent genomic analysis of SARS-CoV-2. These data and GenBank’s common data indexing allowed NCBI to rapidly publish a specific coronavirus resource (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=taxid:2697049) to support the public health response. By the end of 2021, GenBank contained 6.8 million viral genomes, of which 2.2 million were from coronaviruses, or about one-third of the viral collection.
GenBank in 2025
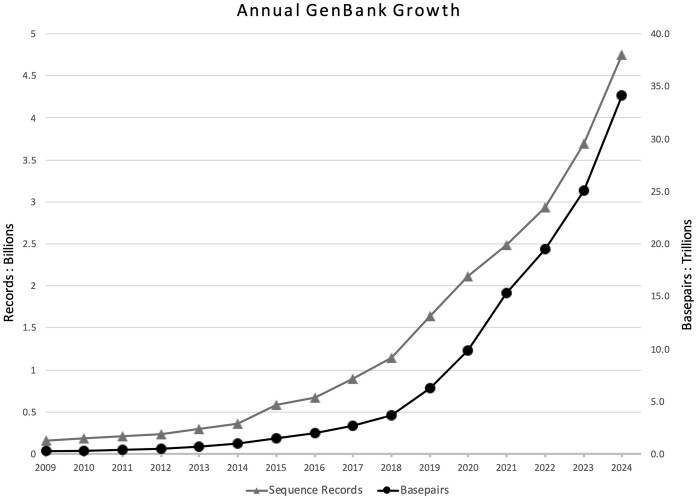
Throughout its history, GenBank has grown continuously (Figure 1) and now contains 34 trillion base pairs from 4.7 billion sequences. GenBank data are partitioned into 21 divisions based on either the source taxonomy or the sequencing strategy used to produce the data. The PAT division contains records provided by patent offices, while the WGS and TSA divisions contain sequences from whole genome shotgun and transcriptome shotgun assembly projects, respectively. Finally, the TLS division contains data from targeted locus studies that focus on, for example, a single gene locus from multiple organisms, such as 16S ribosomal RNA.
Figure 1.
Growth of GenBank recorded in both base pairs (circles) and the number of sequence records (triangles). Each point represents the GenBank release in August of each year, starting with release 173 (August 2009).
GenBank’s exhaustive collection forms the foundation of numerous NCBI products. GenBank is the major source of primary data that NCBI staff use to produce the RefSeq collection (6) and records in NCBI Gene (7). In turn, these GenBank and RefSeq data are the largest sources for NCBI BLAST databases (8), NCBI Datasets (9) and NCBI Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/). The conceptual translations of coding sequences on GenBank records form the largest component (53%) of the Entrez Protein collection. These sequences support the Conserved Domain Database (10) that is a key component of NCBI processes that add functional annotations to genomes. Through these resources, GenBank supports a wide range of scientific activities, including identifying novel organisms, monitoring the geographical distribution of viral strains and understanding the molecular basis of SARS-CoV-2 infections and potential treatments (11).
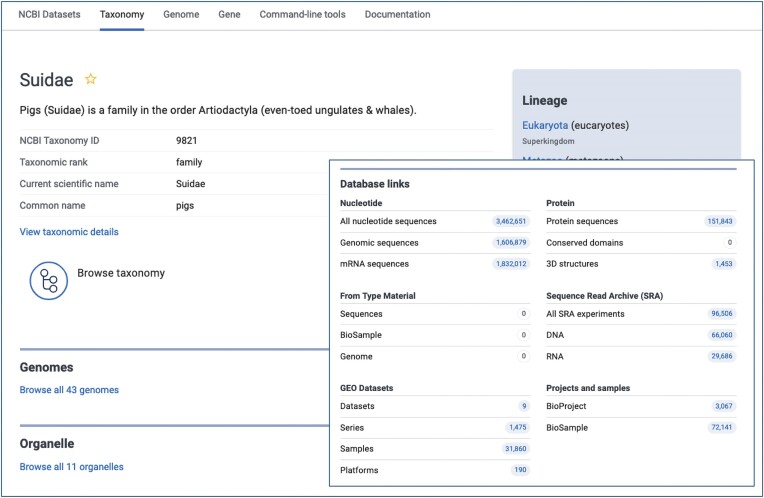
GenBank continues to support community efforts to add value to existing sequence data. While the outputs of next-generation sequencing projects are housed in SRA (12), GenBank collects the valuable annotated assemblies that the community constructs from these raw reads. GenBank also allows investigators to create Third Party Annotation (TPA) records that represent efforts to assemble and/or annotate GenBank data submitted by other researchers (https://www.ncbi.nlm.nih.gov/genbank/tpa). GenBank also supports efforts to increase the accurate reporting and understanding of the Earth’s biodiversity. GenBank currently includes data submitted from 121 countries and contains data for over 581 000 species, an increase of 77 000 species (15%) in the past year. NCBI Datasets now provides a completely redesigned and modern view of the nodes in the GenBank taxonomy tree (Figure 2), providing users an easy way to browse and download data for any of these nodes. Along with INSDC, we continue to encourage submitters to report spatiotemporal metadata to ensure that GenBank data records correctly reflect the geographical distribution of sample collection to support research and public health efforts.
Figure 2.
Taxonomy page in NCBI Datasets for the family Suidae providing easy access to available genomes, nucleotide and protein sequences, SRA data, BioProjects and more, in addition to the other taxonomic nodes in the lineage of Suidae.
In an effort to increase the availability of new influenza sequences to the research community, in 2024 we expedited the release of these sequences, much as we did for SARS-CoV-2 sequences during the global pandemic (13). Submitters of influenza data can now obtain accession numbers and have their data publicly accessible within hours (rather than days) after submission. As part of this process, we subject sequences to several quality checks before annotation, including laboratory adaptor screening and trimming and/or removal of sequences that do not meet minimal thresholds for length and quality. After this screening, features such as coding regions and genes are automatically applied using the FLAN (FLu ANnotation) tool (14). We report problems with annotations and/or sequence data back to the submitter for review. Submissions that pass all these checks meet quality standards and have standardized annotations, making the data more reusable for investigators.
Accessing GenBank
Web interfaces
The most direct way to access GenBank sequences on the NCBI website is through NCBI’s Nucleotide web resource (https://www.ncbi.nlm.nih.gov/nucleotide/). This interface supports text queries that retrieve sequences based on sequence accession numbers or metadata fields such as the title of the record, the source organism, the name of a submitter and many others as described in the help documentation listed on the above website. The Nucleotide web resource also contains sequences originally deposited to partner INSDC data repositories, along with sequences from the RefSeq collection. Users can find conceptual translations of coding regions in GenBank data in NCBI’s Protein resource, while NCBI’s Gene knowledgebase associates information on genes with representative RefSeq and GenBank sequences. The BLAST (8) family of tools (https://blast.ncbi.nlm.nih.gov) offers an alternative method of retrieving sequences based on the sequence data rather than on metadata fields, and so can be a powerful tool for validating annotations and investigating poorly annotated sequences. As GenBank has grown in size, so have the BLAST databases, and so in 2024 we released core_nt, a new default database that increases search efficiency and reduces the redundancy common in search results (https://ncbiinsights.ncbi.nlm.nih.gov/2024/07/18/new-blast-core-nucleotide-database/). Finally, we encourage users interested in acquiring assembled genome data to explore NCBI Datasets (9), a modern interface for browsing and downloading genomes that provides options for website, command-line and application programming interface (API) supported information retrieval.
APIs and command-line tools
NCBI provides the Entrez Programming Utilities (https://eutils.ncbi.nlm.nih.gov) to support programmatic access to all records in the Nucleotide and Protein web resources along with many others. These APIs allow users to submit text queries and retrieve matching sequence records in several formats such as the GenBank flat file, FASTA and XML. Similarly, we offer an API for BLAST (https://blast.ncbi.nlm.nih.gov/doc/blast-help/developerinfo.html), and users can also download BLAST software and databases so that they can perform searches without connecting to NCBI servers (https://blast.ncbi.nlm.nih.gov/doc/blast-help/downloadblastdata.html). NCBI Datasets also provides an API and several command-line tools for accessing genomic datasets (https://www.ncbi.nlm.nih.gov/datasets/docs/v2/download-and-install/).
FTP
NCBI provides bimonthly comprehensive releases of GenBank sequence records in both the traditional flat file format and a structured ASN.1 format by anonymous FTP at ftp.ncbi.nlm.nih.gov/genbank. For release 262 (15 August 2024), there are 5374 files requiring 5.626 TB of uncompressed disk storage. In addition, daily GenBank incremental update files containing new records and those updated since the most recent release are available in flat file format at ftp.ncbi.nlm.nih.gov/genbank/daily-nc/.
Citing GenBank records
As described previously (15), the best way to cite a GenBank record in a publication is to use the full accession.version identifier for the record. For example, AF123456.2 is the accession.version identifier for version 2 of the record AF123456. Using the accession.version identifier in searches is the only way to retrieve the exact version of the record the authors are referencing, since searching the Nucleotide web resource with the unversioned accession (e.g. AF123456) retrieves only the current version of the record. Users can update the record display to the ‘Revision History’ view to review the history of the sequence record. Additional information about accession numbers for GenBank and related INSDC databases is now available online: https://www.ncbi.nlm.nih.gov/genbank/acc_prefix/.
Submitting to GenBank
Submission portal
The NCBI Submission Portal (https://submit.ncbi.nlm.nih.gov) has continued to expand support for eukaryotic nuclear messenger RNA sequences as part of our transition from BankIt to Submission Portal. The interactive wizard now allows submitters to add features, including coding sequence (CDS) and protein annotations, by uploading feature tables. We encourage submitters to explore these new capabilities and to expect additional improvements as we expand the types of submissions accepted for GenBank in Submission Portal.
BioProject and BioSample
This pair of resources, BioProject and BioSample (16), collects data and metadata about submissions, resulting in increased alignment of the data with FAIR principles. BioProject links sequence records from the same research effort but that are in different NCBI repositories (e.g. GenBank and SRA data) not only to one another but also to supporting literature in PubMed and PubMed Central and associated funding sources, providing customers with a single point of access to all the diverse data types available for an initiative. BioSample helps enforce consistency in metadata and accuracy of taxonomic assignments for related sequence records so that data consumers can gather meaningful, descriptive information about the source of the associated sequence data. This is of particular importance in supporting biomedical research and public health, which rely on understanding the provenance of the data to draw conclusions. We highly encourage submitters to create BioProject and BioSample records during their submission, and the Submission Portal interface guides submitters in doing so.
Foreign contamination screening
One of the best ways that submitters can accelerate the release of their data in GenBank is to ensure that their data are free from contaminating sequences from organisms not arising from the intended biological source. We therefore encourage submitters to pre-screen their data using software such as the NCBI Foreign Contamination Screening (FCS) tool (https://github.com/ncbi/fcs), developed as part of the NIH Comparative Genomics Resource (17), and freely available for download and execution on local machines as well as on the Galaxy platform (18). The download includes two tools: FCS-adaptor that detects adaptor and vector contaminants, and FCS-GX (19) that detects sequences from foreign organisms. Detailed instructions are provided on the GitHub site linked above.
Metagenomes
Metagenomes continue to be an increasingly important source of novel sequence data and inform our understanding of biodiversity. Given the common need to assign accurate taxonomic nodes to these data, we encourage submitters to include raw reads in their submissions, along with details about the isolation sources of the biological samples. Access to the underlying reads, in addition to detailed metadata, promotes FAIR principles along with reproducibility of downstream data analyses, increasing the value of the datasets to the community for interpretation and reuse.
Changes to submission support
We expect to make several changes in the coming months that may affect some users. We will retire the Popset database (https://www.ncbi.nlm.nih.gov/popset) in January 2025 (https://ncbiinsights.ncbi.nlm.nih.gov/2024/08/14/popset-to-retire-january-2025/). Sequences from population studies will remain available in the Nucleotide web resource, and we encourage submitters of such sets to create BioProject records to represent them, as this is a robust way for users to access diverse and related data across multiple submissions over the course of a research project. Along with our INSDC partners, we also plan to end support for two types of TPA sequences, experimental and inferential, in January 2025 while continuing support for TPA assembly submissions (https://www.ncbi.nlm.nih.gov/genbank/tpa/). Given recent developments in assembly methods for genomes, we plan to stop accepting AGP files that have been used to specify the assembly of a large sequence from smaller components. If submitters need an alternative to AGP files and have evidence to link adjacent sequences, we advise that they include runs of Ns in FASTA sequences to indicate gaps, where the number of Ns should represent the estimated size of the gap if known (use 100 Ns if the size is unknown). Submitters will be asked to provide information about such gaps during their submission (https://www.ncbi.nlm.nih.gov/genbank/genomesubmit/). Finally, we would note that that the country qualifier for specifying geographical metadata has been replaced by geo_loc_name and will be required on all submitted sequences by the end of 2024 (https://ncbiinsights.ncbi.nlm.nih.gov/2023/12/14/update-genbank-qualifier/).
Supporting the community
GenBank and other NCBI staff coordinate with large sequencing consortia and standards groups and develop a range of outreach efforts (including webinars, conferences, NCBI blog posts and codeathons) throughout the year to provide information about GenBank, the Nucleotide and Protein web resources, BLAST and other services that support researchers, educators, clinicians and public health experts (https://ncbiinsights.ncbi.nlm.nih.gov/ncbi-outreach-events/). These efforts include demonstrating how to access sequence data, use available tools, use NCBI and INSDC data standards, and use NCBI services to submit data to enable reuse by others. For example, as part of ongoing efforts to support submissions globally, in June 2024 we collaborated with the African Centres for Disease Control and Prevention to present a 3-day workshop on sequence data curation. This initial workshop aimed to train staff from public health and research institutions from across Africa in uploading, managing, validating and sharing sequence data from pathogens as part of an effective response to public health threats (https://www.nlm.nih.gov/ncbi/workshops/AfricaCDC_Summer2024/).
GenBank provides several ways for users to learn more about both retrieving and submitting data. Web resources such as Nucleotide, Protein and BLAST offer Quick Start guides, FAQs and detailed documentation from their home pages. NCBI Datasets also provides documentation and online tutorials (https://www.ncbi.nlm.nih.gov/datasets/docs/v2/). We publish regular updates on the NCBI Insights blog (https://ncbiinsights.ncbi.nlm.nih.gov) and the blog’s home page also provides links where users can sign up for our announcement mailing lists. The various Submission Portal wizards and their requirements are described at https://submit.ncbi.nlm.nih.gov/genbank/help, and basic information about GenBank and associated data processing and release policies are described at https://www.ncbi.nlm.nih.gov/genbank/. The NLM Support Center provides additional articles (https://support.nlm.nih.gov/knowledgebase/category/?id=CAT-01240), and email support is available at info@ncbi.nlm.nih.gov.
Mailing address
GenBank, National Center for Biotechnology Information, Building 45, Room 6AN12D-37, 45 Center Drive, Bethesda, MD 20892, USA.
Electronic addresses
www.ncbi.nlm.nih.gov: NCBI home page.
gb-sub@ncbi.nlm.nih.gov: submission of sequence data to GenBank.
update@ncbi.nlm.nih.gov: revisions to, or notification of release of, ‘confidential’ GenBank entries.
info@ncbi.nlm.nih.gov: general information about NCBI resources.
Citing GenBank
If you use the GenBank database in your published research, we ask that this article be cited.
Contributor Information
Eric W Sayers, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Mark Cavanaugh, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Linda Frisse, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kim D Pruitt, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Valerie A Schneider, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Beverly A Underwood, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Linda Yankie, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Ilene Karsch-Mizrachi, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Data availability
GenBank® is freely available at https://www.ncbi.nlm.nih.gov/genbank/.
Funding
This work was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM), National Institutes of Health.
Conflict of interest statement. None declared.
References
- 1. Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2021; 49:D92–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karsch-Mizrachi I., Arita M., Burdett T., Cochrane G., Nakamura Y., Pruitt K.D., Schneider V.A.. The International Nucleotide Sequence Database Collaboration (INSDC). Nucleic Acids Res. 2024; 10.1093/nar/gkae1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yuan D., Ahamed A., Burgin J., Cummins C., Devraj R., Gueye K., Gupta D., Gupta V., Haseeb M., Ihsan M.et al.. The European Nucleotide Archive in 2023. Nucleic Acids Res. 2024; 52:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ara T., Kodama Y., Tokimatsu T., Fukuda A., Kosuge T., Mashima J., Tanizawa Y., Tanjo T., Ogasawara O., Fujisawa T.et al.. DDBJ update in 2023: the MetaboBank for metabolomics data and associated metadata. Nucleic Acids Res. 2024; 52:D67–D71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E.et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goldfarb T., Kodali V.K., Pujar S., Brover V., Robbertse B., Oh D.H., Astashyn A., Ermolaeva O., Farrell C.M., Haddad D.et al.. NCBI RefSeq: reference sequence standards through 25 years of curation and annotation. Nucleic Acids Res. 2024; 10.1093/nar/gkae1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown G.R., Hem V., Katz K.S., Ovetsky M., Wallin C., Ermolaeva O., Tolstoy I., Tatusova T., Pruitt K.D., Maglott D.R.et al.. Gene: a gene-centered information resource at NCBI. Nucleic Acids Res. 2015; 43:D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y.et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Leary N.A., Cox E., Holmes J.B., Anderson W.R., Falk R., Hem V., Tsuchiya M.T.N., Schuler G.D., Zhang X., Torcivia J.et al.. Exploring and retrieving sequence and metadata for species across the tree of life with NCBI datasets. Sci. Data. 2024; 11:732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J., Chitsaz F., Derbyshire M.K., Gonzales N.R., Gwadz M., Lu S., Marchler G.H., Song J.S., Thanki N., Yamashita R.A.et al.. The conserved domain database in 2023. Nucleic Acids Res. 2023; 51:D384–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beyerstedt S., Casaro E.B., Rangel E.B.. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021; 40:905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katz K., Shutov O., Lapoint R., Kimelman M., Brister J.R., O’Sullivan C.. The Sequence Read Archive: a decade more of explosive growth. Nucleic Acids Res. 2022; 50:D387–D390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Underwood B.A., Yankie L., Nawrocki E.P., Palanigobu V., Gotvyanskyy S., Calhoun V.C., Kornbluh M., Smith T.G., Fleischmann L., Sinyakov D.et al.. Rapid automated validation, annotation and publication of SARS-CoV-2 sequences to GenBank. Database (Oxford). 2022; 2022:baac006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao Y., Bolotov P., Dernovoy D., Kiryutin B., Tatusova T.. FLAN: a web server for influenza virus genome annotation. Nucleic Acids Res. 2007; 35:W280–W284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2020; 48:D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T.et al.. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012; 40:D57–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bornstein K., Gryan G., Chang E.S., Marchler-Bauer A., Schneider V.A.. The NIH Comparative Genomics Resource: addressing the promises and challenges of comparative genomics on human health. BMC Genomics. 2023; 24:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galaxy C. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024; 52:W83–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Astashyn A., Tvedte E.S., Sweeney D., Sapojnikov V., Bouk N., Joukov V., Mozes E., Strope P.K., Sylla P.M., Wagner L.et al.. Rapid and sensitive detection of genome contamination at scale with FCS-GX. Genome Biol. 2024; 25:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GenBank® is freely available at https://www.ncbi.nlm.nih.gov/genbank/.