Abstract
With the escalating crisis of bacterial multidrug resistance, anti-virulence therapeutic strategies have emerged as a highly promising alternative to conventional antibiotic treatments. Anti-virulence compounds are specifically designed to target virulence factors (VFs), disarming pathogens without affecting bacterial growth and thus reduce the selective pressure for resistance development. However, due to the complexity of bacterial pathogenesis, no anti-virulence small molecules have been approved for clinical use thus far, despite the documentation of hundreds of potential candidates. To provide valuable reference resources for drug design, repurposing, and target selection, the virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/) has systematically collected public data on anti-virulence compounds through extensive literature mining, and further integrated this information with its existing knowledge of bacterial VFs. To date, the VFDB has curated a comprehensive dataset of 902 anti-virulence compounds across 17 superclasses reported by 262 studies worldwide. By cross-linking the current knowledge of bacterial VFs with information on relevant compounds (e.g. classification, chemical structure, molecular targets and mechanisms of action), the VFDB aims to bridge the gap between chemists and microbiologists, providing crucial insights for the development of innovative and effective antibacterial therapies to combat bacterial infections and address antibiotic resistance.
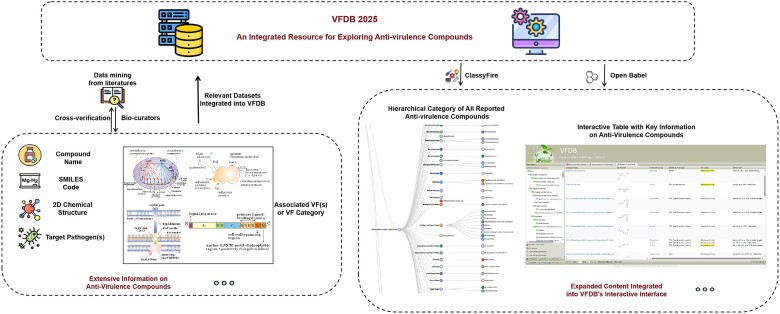
Graphical Abstract
Graphical Abstract.
Introduction
In recent decades, the continuous development of vaccines, anti-infective drugs, and advancements in infection control measures have led to substantial progress in the prevention and treatment of bacterial infections (1,2). However, the rapid emergence of adaptive mutations and the widespread dissemination of antibiotic resistance genes have made infections caused by multidrug-resistant bacterial strains a serious public health threat (3,4). As a result, antimicrobial resistance has emerged an increasingly critical global challenge, with projections suggesting it could cause over 10 million deaths annually by 2050, vastly exceeding the current figure of 700 000 deaths per year (5,6). In addition, conventional antibiotics typically target essential cellular functions of bacteria, such as cell wall, replication, transcription and translation, which not only kill or inhibit bacterial pathogens but also disrupt commensal and beneficial microbiota, often leading to further clinical complications (7). Consequently, there is an urgent need for novel antibacterial strategies to combat drug-resistant bacterial infections.
One emerging strategy is anti-virulence therapy, which focuses on disarming pathogens by suppressing their virulence rather than inhibiting their growth or directly killing them (8). By attenuating virulence systems, anti-virulence drugs are particularly attractive due to their hypothesized ability to exert lower selective pressure for resistance development, as they do not target vital bacterial functions and are less disruptive to the resident microbial flora (9). This approach reduces the severity of clinical manifestations while simultaneously enhancing antibacterial immune responses, positioning anti-virulence therapy as one of the most promising alternatives to traditional antibiotic treatments (10). Anti-virulence compounds can be designed to target various bacterial virulence factors (VFs) by preventing bacterial adhesion (11) or biofilm formation (11), disrupting effector delivery systems (12), blocking quorum sensing (13), and attenuating virulence through global or specific gene expression regulation (14). For instance, a family of bicyclic 2-pyridones, termed pilicides, has been shown to inhibit pilus biogenesis, bacterial adherence, and biofilm formation in uropathogenic bacteria by blocking the chaperone and usher functions (15). Additionally, furan-based compounds inhibit salicylate synthase, the key enzyme in mycobacterial siderophore biosynthesis, thereby disrupting iron acquisition and impairing bacterial growth (16). Another important example is the inhibition of cholesterol-dependent cytolysins, such as pneumolysin and perfringolysin, where small molecule inhibitors block their pore-forming ability and prevent host cell damage (17). With the deepening understanding of bacterial pathogenesis and advancements in computer-aided drug design methods, an increasing number of potential anti-virulence small molecules have been discovered over the past decades (18,19). However, progress in the clinical application of anti-virulence drugs has lagged, with only a limited number of compounds entering clinical trials, while the majority remain in preclinical development stages. Moreover, current research predominantly targets specific virulence factors of individual pathogens, underscoring the need for an in-depth understanding of the interaction dynamics between these agents and pathogenic bacteria to more effectively identify and develop potent new drug candidates.
The virulence factor database (VFDB, http://www.mgc.ac.cn/VFs/) has served as a comprehensive knowledge base and a versatile analysis platform of bacterial VFs for over two decades (20,21). Apart from the VFDB, other databases related to bacterial VFs, such as the PHI-base (22), Victors (23), TADB (24), SecReT4 (25) and SecReT6 (26), primarily focus on host-pathogen interactions or specific categories of VFs. But none of the existing databases provide comprehensive information on anti-virulence compounds. To fill this gap, the VFDB has recently compiled the current knowledge of anti-virulence compounds and integrated it with information on related pathogens and VFs, aiming to bridge the gap between chemists and microbiologists through a comprehensive repository. To date, the VFDB has collected a total of 902 individual anti-virulence compounds of 90 classes from 17 superclasses reported by 262 published studies, covering various bacterial VF categories and major medically significant pathogens. Recent developments in the VFDB will provide valuable insights for the discovery of novel and effective anti-virulence therapies, aimed at mitigating bacterial infections and combating antibiotic resistance.
Database updates
Data collection and database integration
To develop a comprehensive resource for investigating anti-virulence compounds, we retrieved all promising anti-virulence drug candidates and related publications available in the public domain, identifying potential alternatives for the prevention and treatment of critical bacterial infections. Inspired by a previous study (27), we established a comprehensive set of retrieval keywords, including ‘anti-virulence OR antivirulence OR ‘virulence inhibition’ OR ‘virulence inhibitor’ OR ‘virulence factor inhibition’, to systematically search PubMed for relevant literature as an initial step (latest accessed at July 2024). The resulting publications were then reviewed by two independent bio-curators for cross-verification. Please note that in this study, we focus on small molecules, which are relatively easier to develop and manufacture, and therefore exclude studies involving large molecules such as polypeptides and antibodies. We included only original studies on anti-virulence compounds with in vitro or in vivo activity, as well as relevant reports on clinical trials. Currently, we have collected only compounds targeting pathogens from the 32 medically important bacterial genera covered by the VFDB (28). With respect to the enrolled studies, we manually retrieved relevant information from the documents, including the chemical structure of the compound(s), the target pathogen(s) and virulence factors (if reported), the effects on bacterial pathogenesis, and the maximum development stages (e.g. in vitro, in vivo, or clinical trial phases I/II). Particularly, to facilitate storage and management in the backend database, we converted the chemical structure diagrams from the literature into the Simplified Molecular Input Line Entry System (SMILES) strings (29). Next, we performed an associative search in the PubChem database (30) using the unique SMILES string for each collected anti-virulence compound and batch downloaded the expanded chemical information, including canonical and isomeric SMILES codes, molecular formula, standard and synonymous names, and related classifications based on the Medical Subject Headings (MeSH) tree (if available). However, a significant portion of these compounds remains unclassified in the MeSH tree. To address this issue, we utilized the ClassyFire webserver (31), a highly valuable automated chemical taxonomy tool with robust classification capabilities across multiple hierarchical levels (e.g. kingdom, superclass, class and subclass), to systematically categorize the enrolled anti-virulence compounds. Finally, all the aforementioned information were integrated into the backend MySQL database of the VFDB, forming a comprehensive warehouse of anti-virulence compounds.
To enhance global accessibility for browsing and analyzing the dataset of anti-virulence compounds, we designed a dedicated summary page providing an overview of these compounds and integrated related information into the existing webpages of pathogens and VFs. The summary page features a hierarchical classification tree for anti-virulence compounds, allowing users to explore specific drug categories of interest in detail. Upon selecting a specific category, an interactive table displays key information, including the compound name, 2D chemical structure (dynamically generated by the Open Babel software (32) based on the SMILES code), target pathogen(s), associated VF(s) or VF category, and the maximum development phase. Users can click on compound names to access full details or click on the statistics icon at the bottom toolbar to explore pie charts by the related pathogens and VF/compound categories. Additionally, the VFDB incorporates a hierarchical category of all reported anti-virulence compounds for each pathogen into the existing pathogen page, while highlighting representative chemical structures of related anti-virulence compounds on the corresponding VF page, allowing users to click on these categories or compounds to navigate to more detailed information. Taken together, the VFDB seamlessly interlinks the information of anti-virulence compounds with the knowledge of pathogens and VFs, offering a highly interactive and user-friendly platform for users worldwide.
A preliminary digest of anti-virulence compounds summarized in the VFDB
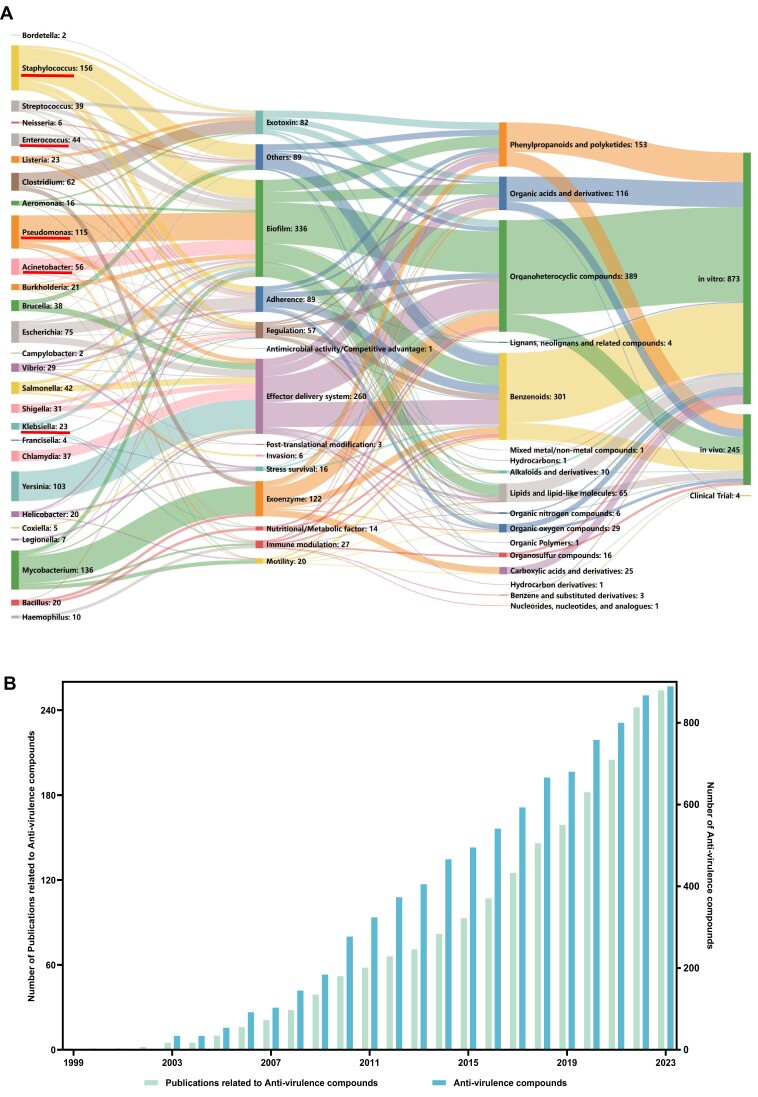
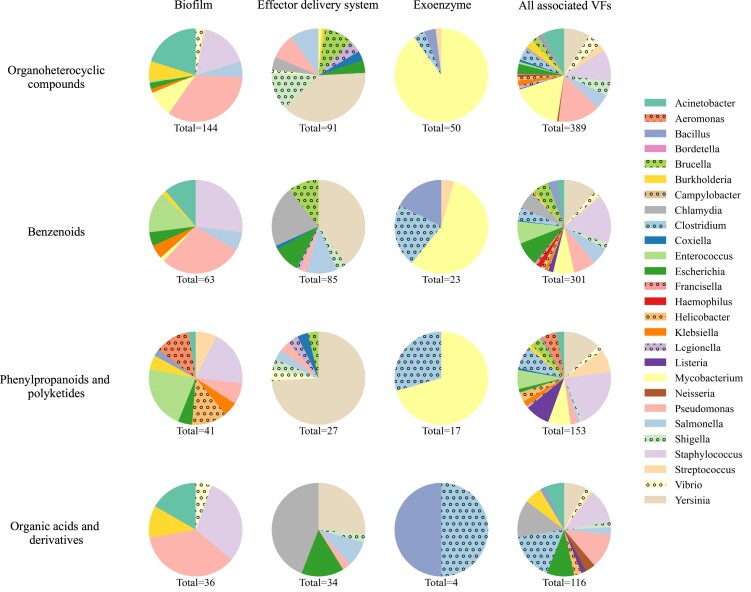
As of September 2024, the anti-virulence compounds collected in the VFDB database are primarily organic compounds, with a significant proportion consisting of organoheterocyclic compounds, benzenoids, phenylpropanoids and polyketides, and organic acids and derivatives (Figure 1A). Despite significant growth in research on anti-virulence small molecules over the past two decades (Figure 1B), the majority of these compounds remain in the preclinical stage, with approximately 78% demonstrating virulence attenuation only in vitro, and only four having progressed to clinical trials. Additionally, about 40% of the anti-virulence compounds we have compiled lack in-depth deciphering of molecular mechanisms, and therefore cannot be linked to any specific target VFs. To better illustrate the possible interactions between anti-virulence compounds and bacterial VFs, we have summarized and grouped the target VF(s) of each compound based on our recently proposed classification scheme VF category (21). Notably, current data indicates that the aforementioned major categories of anti-virulence compounds share similar broad-spectrum antibacterial properties that can target nearly all groups of bacterial VFs (Figure 2). In contrast, some other superclasses of compounds are currently validated against only limited VF categories. For example, carboxylic acids and derivatives are primarily target to exoenzymes thus far.
Figure 1.
Overview of the current resources for the exploration of anti-virulence compounds in the VFDB. (A) A Sankey diagram illustrating the relationships between bacterial genera, target VF categories, superclass of anti-virulence compounds, and the current development stages (from left to right). The number of compounds involved in each node is given after the node name. Note that, since each individual compound can be associated with more than one virulence factor across multiple pathogens, the total number of compounds represented by all nodes (n = 1122) exceeds the total number of anti-virulence compounds in the VFDB (n = 902). Pathogens of the ESKAPE group are highlighted with red underline. (B) Cumulative growth of verified anti-virulence compounds and the related publications over time as collected in the VFDB.
Figure 2.
Characteristics of four major categories of anti-virulence compounds. This figure presents a series of pie charts illustrating the interactions between anti-virulence compounds and virulence factors. Each row corresponds to a specific compound category, while each column represents either a distinct virulence factor category or all related VFs (in the last column). The colors within each pie chart represent various pathogens (keys), and the ‘total’ beneath each chart indicates the total number of compounds of the corresponding category.
As shown in Figure 1A, about two thirds of the currently explored anti-virulence compounds target bacterial VFs involved in biofilm, effector delivery systems and exoenzyme. This focus is logical from the perspective of bacterial pathogenesis. Bacterial biofilms significantly enhance the resistance to host immune system and antibiotics, often contributing to chronic infections. Numerous in vitro and in vivo studies have demonstrated significant inhibition of biofilms by an increasing number of natural compounds and their derivatives, underscoring the importance of identifying novel inhibitors that prevent biofilm formation without promoting drug resistance (33,34). Effector delivery systems, such as type III secretion systems (T3SS), are well-characterized key virulence determinants in many bacterial pathogens. For instance, genetic analysis of Yersinia genomes has revealed the presence of a T3SS, which is crucial for its virulence (35). Consequently, current research on anti-virulence compounds targeting Yersinia has predominantly focused on blocking the T3SS, as illustrated in Figure 1A. In addition, YM155 (sepantronium bromide), which target to Escherichia coli and is currently in phase II trial, has demonstrated strong efficacy in inhibiting the activity of the effector protein NleB1 of the T3SS (36). Exoenzymes are extracellular enzymes functioning outside of the bacterial cells to degrade macromolecules to small soluble molecules, which facilitates the bacterial invasion. Indeed, over 90 currently verified anti-virulence compounds of Mycobacterium target tyrosine phosphatases PtpA or PtpB, which destruct the key components of the host endocytic pathway to arrest phagosome maturation (37). Nonetheless, some other groups of bacterial VFs, such as adherence, exotoxins, regulation and immune modulation factors, also play pivotal roles in bacterial pathogenesis and could be potential targets for future investigations. For example, adherence to host cells is typically the initial step of bacterial infections. Sibofimloc, a novel FimH adherence blocker of E. coli, currently in clinical trials, has shown minimal systemic exposure, good tolerability, and a reduction in several inflammatory biomarkers in its initial study involving patients with active Crohn's disease (38). Therefore, various bacterial VFs remain potential targets for further investigation into anti-virulence compounds.
Among the 32 genera of medically significant bacterial pathogens covered by the current VFDB, 27 genera have associated anti-virulence compounds reported thus far (Figure 1A). As a promising alternative for combatting antibiotic resistance, many current anti-virulence compounds target members of the so-called ESKAPE group (i.e. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp.), five of which are covered by the current VFDB database. ESKAPE pathogens pose critical threats to public health, as they are responsible for the majority of nosocomial infections caused by multidrug-resistant strains worldwide (39). Within the current dataset, biofilm formation and quorum sensing (QS) are primary virulence targets for all members of ESKAPE bacteria (Figure 1A), with biofilm formation being crucial for antibiotic resistance and QS coordinating the expression of virulence-related genes. Nevertheless, the only anti-virulence compound in clinical trials targeting A. baumannii is macrocyclic peptide zosurabalpin, which blocks the lipopolysaccharide transport system and disrupts outer-membrane assembly (40). Actually, different trends of current approaches were observed in different pathogens, primarily based on the respective bacterial pathogenesis. Although current efforts of anti-virulence compounds on Staphylococcus generally diversified in terms of VF category, the validated compounds for Pseudomonas, Yersinia and Mycobacterium predominantly target to biofilm, effector delivery system and exoenzyme, respectively (Figure 1A). Indeed, an in-depth understanding of the bacterial pathogenesis is undoubtedly the foundation for a successful development of anti-virulence therapies.
Discussion
As a novel alternative to antibiotic therapy intended to address the escalating drug resistance crisis, the development of anti-virulence therapies continues to encounter significant challenges. Due to the complexity of bacterial pathogenesis, an in-depth understanding of the molecular mechanisms of virulence factors and their interactions with host organisms is urgently needed. To tackle this, the VFDB has dedicated over two decades to conducting comparative analyses of molecular mechanisms across various bacterial VFs and has proposed a general classification scheme to provide a robust foundation for target design in anti-virulence strategies (20,21,28,41–43). Nevertheless, with most anti-virulence therapies still in the preclinical stage and only four having progressed to clinical trials (36,38,44,45), there is a notable absence of predictive models for determining optimal drug dosing, validated models for forecasting clinical outcomes, and accurate diagnostic tests for identifying pathogens and assessing VF expression - an essential factor given that some virulence factors are pathogen-specific and their expression can be influenced by patient condition and disease state.
Therefore, to accelerate research progress and enhance the efficiency of knowledge discovery in the field of anti-virulence therapy, the VFDB has systematically integrated and rigorously analyzed the current research findings and related data, including the chemical structures, molecular targets, mechanisms of action, and development stages of anti-virulence compounds. By interlinking existing knowledge of relevant bacterial pathogens and VFs in the VFDB, the upgraded resource offers invaluable data references for both chemists and microbiologists, supporting future investigations into novel compounds design, drug repurposing, and strategic target selection.
To provide new perspectives in developing novel and effective antibacterial therapies and address the challenges posed by bacterial infections and antibiotic resistance, the VFDB database will continue to integrate up-to-date knowledge on both bacterial pathogenesis and anti-virulence compounds, maintaining its role as a centralized and comprehensive repository for the global scientific community. In addition, we aim to develop advanced tools for predictive modeling of interactions between bacterial VFs and small molecules, and integrate deep-learning algorithms to predict potential anti-virulence compounds based on existing data. Furthermore, to streamline research on bacterial pathogenicity, we plan to further expand the VFDB datasets and develop new tools capable of identifying bacterial VFs in metagenomic data, thereby broadening the scope and utility of VFDB in future studies.
Contributor Information
Siyu Zhou, NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, P.R. China.
Bo Liu, NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, P.R. China.
Dandan Zheng, NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, P.R. China.
Lihong Chen, NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, P.R. China.
Jian Yang, NHC Key Laboratory of Systems Biology of Pathogens, National Institute of Pathogen Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, P.R. China.
Data availability
The virulence factor database (VFDB) is publicly accessible at http://www.mgc.ac.cn/VFs/.
Funding
National Natural Science Foundation of China [31970635]; National Scientific Data Sharing Platform for Population and Health. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
References
- 1. Cook M.A., Wright G.D.. The past, present, and future of antibiotics. Sci. Transl. Med. 2022; 14:eabo7793. [DOI] [PubMed] [Google Scholar]
- 2. Yeaman M.R., Hennessey J.P. Jr.. Innovative approaches to improve anti-infective vaccine efficacy. Annu. Rev. Pharmacol. Toxicol. 2017; 57:189–222. [DOI] [PubMed] [Google Scholar]
- 3. Davies J., Davies D.. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010; 74:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kunnath A.P., Suodha Suoodh M., Chellappan D.K., Chellian J., Palaniveloo K.. Bacterial persister cells and development of antibiotic resistance in chronic infections: an update. Br. J. Biomed. Sci. 2024; 81:12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399:629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Filipić B., Ušjak D., Rambaher M.H., Oljacic S., Milenković M.T.. Evaluation of novel compounds as anti-bacterial or anti-virulence agents. Front. Cell. Infect. Microbiol. 2024; 14:1370062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng E., Dorjsuren D., Lehman S., Larson C.L., Titus S.A., Sun H., Zakharov A., Rai G., Heinzen R.A., Simeonov A.et al.. A comprehensive phenotypic screening strategy to identify modulators of cargo translocation by the bacterial type IVB secretion system. mBio. 2022; 13:e0024022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clatworthy A.E., Pierson E., Hung D.T.. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007; 3:541–548. [DOI] [PubMed] [Google Scholar]
- 9. Allen R.C., Popat R., Diggle S.P., Brown S.P.. Targeting virulence: can we make evolution-proof drugs?. Nat. Rev. Micro. 2014; 12:300–308. [DOI] [PubMed] [Google Scholar]
- 10. Sheremet A.B., Zigangirova N.A., Zayakin E.S., Luyksaar S.I., Kapotina L.N., Nesterenko L.N., Kobets N.V., Gintsburg A.L.. Small molecule inhibitor of type three secretion system belonging to a class 2,4-disubstituted-4H-[1,3,4]-thiadiazine-5-ones improves survival and decreases bacterial loads in an airway Pseudomonas aeruginosa infection in mice. Biomed. Res. Int. 2018; 2018:5810767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Svensson A., Larsson A., Emtenäs H., Hedenström M., Fex T., Hultgren S.J., Pinkner J.S., Almqvist F., Kihlberg J.. Design and evaluation of pilicides: potential novel antibacterial agents directed against uropathogenic Escherichia coli. ChemBioChem. 2001; 2:915–918. [DOI] [PubMed] [Google Scholar]
- 12. Swietnicki W. Secretory system components as potential prophylactic targets for bacterial pathogens. Biomolecules. 2021; 11:892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Defoirdt T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018; 26:313–328. [DOI] [PubMed] [Google Scholar]
- 14. Trebosc V., Lucchini V., Narwal M., Wicki B., Gartenmann S., Schellhorn B., Schill J., Bourotte M., Frey D., Grünberg J.et al.. Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis. Virulence. 2022; 13:1868–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinkner J.S., Remaut H., Buelens F., Miller E., Aberg V., Pemberton N., Hedenström M., Larsson A., Seed P., Waksman G.et al.. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:17897–17902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mori M., Stelitano G., Cazzaniga G., Gelain A., Tresoldi A., Cocorullo M., Roversi M., Chiarelli L.R., Tomaiuolo M., Delre P.et al.. Targeting siderophore-mediated iron uptake in M. abscessus: a new strategy to limit the virulence of non-tuberculous mycobacteria. Pharmaceutics. 2023; 15:502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aziz U.B.A., Saoud A., Bermudez M., Mieth M., Atef A., Rudolf T., Arkona C., Trenkner T., Böttcher C., Ludwig K.et al.. Targeted small molecule inhibitors blocking the cytolytic effects of pneumolysin and homologous toxins. Nat. Commun. 2024; 15:3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maura D., Ballok A.E., Rahme L.G.. Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 2016; 33:41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang Y., Hawkins B.A., Du J.J., Groundwater P.W., Hibbs D.E., Lai F.. A guide to in silico drug design. Pharmaceutics. 2022; 15:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q.. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005; 33:D325–D328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu B., Zheng D., Zhou S., Chen L., Yang J.. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022; 50:D912–D917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Urban M., Cuzick A., Seager J., Wood V., Rutherford K., Venkatesh S.Y., De Silva N., Martinez M.C., Pedro H., Yates A.D.et al.. PHI-base: the pathogen-host interactions database. Nucleic Acids Res. 2020; 48:D613–D620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sayers S., Li L., Ong E., Deng S., Fu G., Lin Y., Yang B., Zhang S., Fa Z., Zhao B.et al.. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res. 2019; 47:D693–D700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shao Y., Harrison E.M., Bi D., Tai C., He X., Ou H.Y., Rajakumar K., Deng Z.. TADB: a web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011; 39:D606–D611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bi D., Liu L., Tai C., Deng Z., Rajakumar K., Ou H.Y.. SecReT4: a web-based bacterial type IV secretion system resource. Nucleic Acids Res. 2013; 41:D660–D665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J., Guan J., Wang M., Li G., Djordjevic M., Tai C., Wang H., Deng Z., Chen Z., Ou H.Y.. SecReT6 update: a comprehensive resource of bacterial Type VI Secretion Systems. Sci. China Life Sci. 2023; 66:626–634. [DOI] [PubMed] [Google Scholar]
- 27. Dickey S.W., Cheung G.Y.C., Otto M.. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 2017; 16:457–471. [DOI] [PubMed] [Google Scholar]
- 28. Liu B., Zheng D., Jin Q., Chen L., Yang J.. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019; 47:D687–D692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988; 28:31–36. [Google Scholar]
- 30. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A.et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Djoumbou Feunang Y., Eisner R., Knox C., Chepelev L., Hastings J., Owen G., Fahy E., Steinbeck C., Subramanian S., Bolton E.et al.. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J Cheminform. 2016; 8:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open Babel: an open chemical toolbox. J Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jakobsen T.H., van Gennip M., Phipps R.K., Shanmugham M.S., Christensen L.D., Alhede M., Skindersoe M.E., Rasmussen T.B., Friedrich K., Uthe F.et al.. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012; 56:2314–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J.H., Kim Y.G., Shim S.H., Lee J.. Antibiofilm activities of norharmane and its derivatives against Escherichia coli O157:H7 and other bacteria. Phytomedicine. 2017; 36:254–261. [DOI] [PubMed] [Google Scholar]
- 35. Plano G.V., Schesser K.. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol. Res. 2013; 57:237–245. [DOI] [PubMed] [Google Scholar]
- 36. Zhu C., El Qaidi S., McDonald P., Roy A., Hardwidge P.R.. YM155 Inhibits NleB and SseK Arginine Glycosyltransferase Activity. Pathogens. 2021; 10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koul A., Choidas A., Treder M., Tyagi A.K., Drlica K., Singh Y., Ullrich A.. Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 2000; 182:5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reinisch W., Hébuterne X., Buisson A., Schreiber S., Desreumaux P., Primas C., Paillarse J.M., Chevalier G., Bonny C.. Safety, pharmacokinetic, and pharmacodynamic study of sibofimloc, a novel FimH blocker in patients with active Crohn's disease. J. Gastroenterol. Hepatol. 2022; 37:832–840. [DOI] [PubMed] [Google Scholar]
- 39. Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008; 197:1079–1081. [DOI] [PubMed] [Google Scholar]
- 40. Zampaloni C., Mattei P., Bleicher K., Winther L., Thäte C., Bucher C., Adam J.M., Alanine A., Amrein K.E., Baidin V.et al.. A novel antibiotic class targeting the lipopolysaccharide transporter. Nature. 2024; 625:566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang J., Chen L., Sun L., Yu J., Jin Q.. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008; 36:D539–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L., Xiong Z., Sun L., Yang J., Jin Q.. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012; 40:D641–D645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen L., Zheng D., Liu B., Yang J., Jin Q.. VFDB 2016: hierarchical and refined dataset for big data analysis–10 years on. Nucleic Acids Res. 2016; 44:D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zigangirova N.A., Nesterenko L.N., Sheremet A.B., Soloveva A.V., Luyksaar S.I., Zayakin E.S., Balunets D.V., Gintsburg A.L.. Fluorothiazinon, a small-molecular inhibitor of T3SS, suppresses salmonella oral infection in mice. J. Antibiot. (Tokyo). 2021; 74:244–254. [DOI] [PubMed] [Google Scholar]
- 45. Que W., Deng Z., Gao J.. Clinical crusade: zosurabalpin's charge against antibiotic resistance. Trends Mol. Med. 2024; 30:420–422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The virulence factor database (VFDB) is publicly accessible at http://www.mgc.ac.cn/VFs/.