Summary
The placenta is vital to maternal and child health but often overlooked in pregnancy studies. Addressing the need for a more accessible and cost-effective method of placental assessment, our study introduces a computational tool designed for the analysis of placental photographs. Leveraging images and pathology reports collected from sites in the United States and Uganda over a 12-year period, we developed a cross-modal contrastive learning algorithm consisting of pre-alignment, distillation, and retrieval modules. Moreover, the proposed robustness evaluation protocol enables statistical assessment of performance improvements, provides deeper insight into the impact of different features on predictions, and offers practical guidance for its application in a variety of settings. Through extensive experimentation, our tool demonstrates an average area under the receiver operating characteristic curve score of over 82% in both internal and external validations, which underscores the potential of our tool to enhance clinical care across diverse environments.
Keywords: contrastive learning, cross-modal, knowledge distillation, placenta analysis, vision and language
Graphical abstract
Highlights
-
•
Unified placenta image encoder excels in placenta analysis
-
•
A large, diverse dataset enhances the model’s accuracy and robustness
-
•
Robustness evaluation shows the model’s resilience to various imaging conditions
-
•
Cross-national validation confirms consistent performance across populations
The bigger picture
The placenta plays a vital role in the health of both mother and baby during pregnancy, but it is often not thoroughly examined at birth, especially in resource-limited settings. This gap can lead to missed opportunities to detect critical conditions. Neonatal sepsis—a life-threatening infection—affects millions of newborns globally, particularly where early detection is challenging because of limited medical resources. This research introduces a powerful tool that enables quick and accessible placental assessment using just a photograph, potentially reducing the risk of undetected issues such as infection or placental abnormalities.
Adaptable into mobile applications, this innovation promises greater accessibility in both high- and low-resource settings. With further refinement, it has the potential to transform neonatal and maternal care by enabling early, personalized interventions that prevent severe health outcomes and improve the lives of mothers and infants worldwide.
The placenta, vital for maternal and neonatal health, presents assessment challenges due to limited accessibility and expertise. This study presents a novel approach utilizing placenta photos spanning a 12-year period. Employing a unified model, it integrates cross-modal training between photos and pathology reports for diverse tasks. A proposed robustness evaluation protocol ensures method reliability. Validation across diverse populations underscores its potential for widespread clinical applicability.
Introduction
The placenta serves as a significant indicator of both pregnancy events and the health of the mother and baby.1,2,3,4,5,6,7,8,9,10,11,12,13 However, even in a high-resource country like the United States, only approximately 20% of placentas undergo pathology examinations,14,15 and placental data are often overlooked in pregnancy research.16 The underutilization of placental pathology is primarily due to time, cost, expertise, and facility requirements, even in resource-abundant areas.17 In low- and middle-income countries (LMICs), the incidence of adverse maternal and newborn outcomes is higher, but resources are typically lacking to conduct placental pathology.18,19 Therefore, enhancing the accessibility of placental assessment to pathologists, clinicians, and researchers is crucial.20,21 Immediate placental assessment at birth is expected to significantly aid clinical decisions.
Existing automatic approaches in research often require expensive equipment and time (e.g., MRI,22,23 computed tomography,24,25 or histological images21) and are not suitable for immediate assessment after birth. Assessing abnormalities in the delivered placenta has substantial value by revealing events in pregnancy and labor that could impact clinical care for the postpartum mother and newborn. As an example, chorioamnionitis due to infection in the placenta may indicate a subclinical infection in the newborn. Histologic diagnosis of chorioamnionitis currently takes days17 and therefore is not used to guide the immediate clinical care of the newborn. Due to the increased risk of early-onset neonatal sepsis,26 antimicrobial agents are used even if the neonate appears well27 before obtaining a diagnosis from placental pathology. A tool that could accurately estimate a diagnosis of chorioamnionitis (before a full pathology exam) and perhaps, more importantly, the absence of chorioamnionitis for the well-appearing newborn would help to initiate treatment for newborns most at risk of infection and appropriately limit antimicrobial use for those at low risk.
Recent efforts in placenta analysis have primarily focused on segmentation28,29,30 and classification31,32,33,34,35,36,37 using histopathological, ultrasound, or MRI images. Previous studies that utilized photographic images—a low-cost and immediate tool—to evaluate placental characteristics38,39,40,41 and to perform placental diagnoses42,43 required a separate model for each of these tasks due to the lack of a unified method. Moreover, the clinical outcomes that can be inferred from these placental diagnoses are often overlooked. A better utilization of the available data should explore the connection between the visual placental feature and the textual description from the pathology report independent of the downstream task. Additionally, using one model for multiple tasks would greatly save computational resources and improve the deployability of the resulting model. To fully leverage the information available in pathology reports and to train a unified placental feature encoder, our preceding work44,45 introduced a vision-and-language contrastive learning (VLC) approach for placenta analysis. Similarly, in this work, we aim to further enhance the VLC approach in placental analysis toward robust deployment under various settings. VLC approaches46,47 have garnered significant interest, particularly following the success of contrastive language and image pre-training (CLIP).47 Research in this area has focused on improving VLC methodologies through innovations in model architectures,48,49 visual representations,50,51,52 textual representations,45,53 and loss functions54,55 as well as sampling strategies,56,57 training strategies,58 and classifier performance.59 Another significant line of research60,61,62 aims to improve the efficiency of VLC techniques. In the context of medical applications, a recent survey63 provides a comprehensive overview of popular methods. Representative strategies have been proposed to enhance local feature alignment,64,65,66,67 introduce auxiliary reconstruction tasks,68,69 and incorporate external prior knowledge.70,71,72
Building upon our unified pre-training methods,44,45 this research seeks to harness the simplicity and affordability of digital photography combined with the proposed cross-modal pre-training techniques to develop a unified model capable of comprehensive and immediate placental assessment using photographic images. Our contributions are as follows. (1) Introduction of a cross-modal contrastive learning technique designed to enhance both the performance and robustness of the placenta analysis model. (2) Development of three key modules: a cross-modal pre-alignment module for improved alignment between images and pathology reports using external image data, a cross-modal distillation module that leverages external textual information to capture nuanced relationships between placental features, and a cross-modal retrieval module for matching textual and visual features, fostering robust representation learning. (3) Creation of a robustness evaluation protocol tailored for placenta photographs to assess model robustness, facilitate explainability, and generate application guidelines. (4) Expansion of the training dataset 3-fold and broadening of the evaluation dataset to include a diverse, multinational collection, enhancing the model’s applicability and performance in LMICs.
Results
Dataset
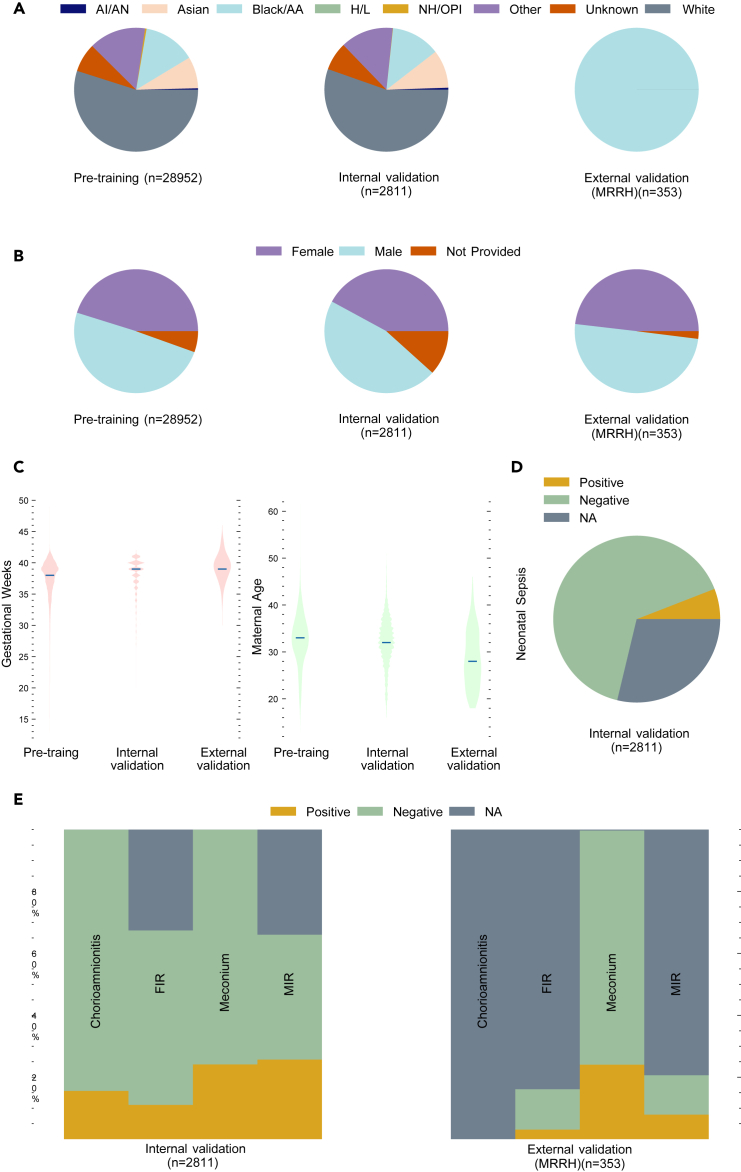
The characteristics of the collected dataset are categorized in Figure 1. The “not applicable” (NA) category was excluded from the results. The primary dataset was collected in the pathology department at Northwestern Memorial Hospital (NMH) (Chicago, IL, USA) between January 1, 2010 and December 31, 2022, following the placenta imaging protocol.73 Photographs were taken using a dedicated pathology specimen photography system (Macropath, Milestone Medical, Kalamazoo, MI, USA) with an integrated, fixed camera and built-in lighting to reduce technical variability. Pathologists generated the pathology reports based on histological findings and widely adopted definitions.74 The refined dataset from NMH comprises 31,763 fetal-side placenta images, each accompanied by a pathology report. We selected 2,811 image-report pairs from the year 2017 for internal validation and the rest for pre-training.
Figure 1.
The characteristics of the primary dataset and the external validation dataset
(A) Distribution of self-reported race.
(B) Distribution of infant sex.
(C) Distribution of gestational age and maternal age.
(D) Distribution of the neonatal sepsis label from the tuning and validation set.
(E) Distribution of placental feature labels.
Each placenta from the primary dataset has one image and one pathology report, while placentas from the external validation dataset have a median (25%–75% percentile) of 4 (3–5) images and one pathology report. The NA category represents instances where information could not be derived due to missing data. AI/AN, American Indian or Alaska Native; Black/AA, Black or African American; H/L, Hispanic or Latino; NH/OPI, Native Hawaiian or other Pacific Islander; NMH, Northwestern Memorial Hospital; MRRH, Mbarara Regional Referral Hospital; FIR, fetal inflammatory response; MIR, maternal inflammatory response; NA, not applicable.
Additionally, we identified 166 cases where the neonate was diagnosed with sepsis and 1,837 potential negative cases from the internal validation set. Furthermore, the external validation set was collected at the Mbarara Regional Referral Hospital (MRRH) (Mbarara, Uganda) between December 1, 2019 and November 30, 2023, under the Placentas, Antibodies, and Child Outcomes study using a Fujifilm FinePix XP130 digital camera. The imaging protocol is included in Data S1. The pathology reports were generated using the same method as described for the primary dataset. We obtained 353 placenta and pathology report pairs from MRRH for external validation. Following our preliminary research,44,45 the AI-based placental assessment and examination (AI-PLAX) algorithm42 was used to mask the background of each image in the NMH dataset. Additionally, the automatic and interactive segment anything model (AI-SAM) algorithm75 was used to mask the background of each image in the MRRH dataset.
For the internal validation set, we manually checked the images to ensure that the placenta was complete and that its visibility was unobscured. We first labeled each image according to the pathology report on four placental feature identification tasks outlined in previous work42,43; namely, meconium-laden macrophages in the amnion or chorion (hereafter referred to as meconium), fetal inflammatory response (FIR), maternal inflammatory response (MIR), and chorioamnionitis. Different levels or stages exist for some lesions. We labeled images as positive for meconium and chorioamnionitis regardless of the reported severity. For FIR and MIR, an image was labeled as negative if the report either lacked relevant information or explicitly indicated a negative diagnosis. Conversely, an image was labeled as positive if the report identified the placenta as stage 2 or higher. To enhance the model’s ability to differentiate significant cases, images were excluded if their associated stage was 1.
A clinical outcome, neonatal sepsis, relates to placental features from the pathology report and is the most immediate cause of neonatal deaths in LMICs.19,76 We retrieved images of cases with neonatal sepsis based on diagnoses made by treating physicians using clinical criteria from infant charts. We selected negative samples from the fine-tuning dataset that were free from FIR, MIR, and chorioamnionitis—the placental features related to sepsis and placental cause of death77—to minimize the possibility of having false negative samples. Evaluating the model’s performance on such a task can infer its prediction capability on clinical outcomes that are related to placental features but not in the pathology report.
We used all positive examples for each task and uniformly sampled a comparable number of negative samples from the internal validation set to perform linear evaluation. The error range computation was based on random divisions of these data, with a 50:50 split for tuning and evaluating the linear classifier.
For the external validation set, we identified three placental feature identification tasks (namely, meconium, FIR, and MIR) and produced the classification labels using the same method. We excluded samples lacking corresponding pathology reports.
Model design
In general, we address two primary tasks: pre-training and downstream classification. Formally, in the pre-training phase, our objective is to learn a function using another function so that, for any given pair of inputs and a similarity function , we have and . The objective function is defined as
| (Equation 1) |
where τ is the temperature in the contrastive loss function, used to control the strength of contrastive learning.
In the downstream classification task, our goal is to learn a function using the learned function for each task so that, for a pair of input ,
| (Equation 2) |
which can be achieved by using a linear classifier.
Given that one of the objectives of placenta pathology reporting is to identify clinically significant findings and make diagnoses, an effective placenta photo analysis model needs to achieve comparable performance on both placental features identified in pathology reports and related clinical findings. To achieve these goals, we adopt the well-established pre-training and fine-tuning paradigm using contrastive learning techniques and propose the placenta feature encoder through CLIP (PlacentaCLIP).
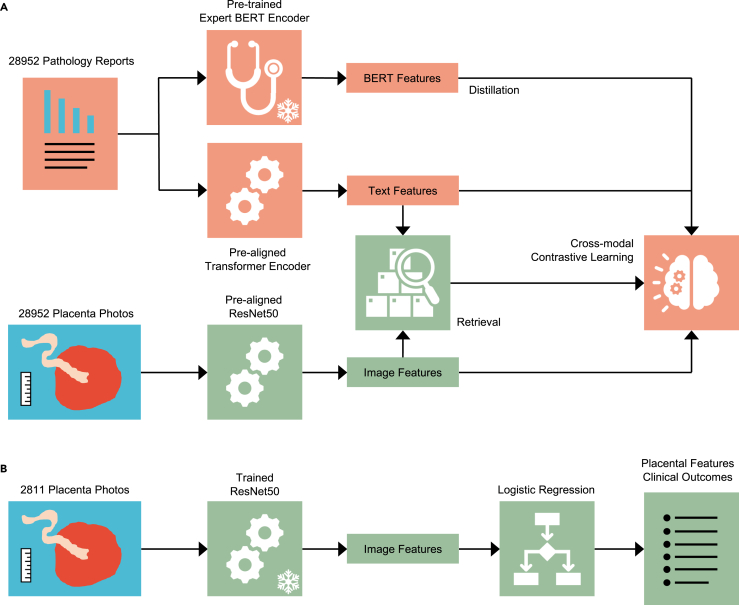
During the pre-training stage (Figure 2A), we train a generalizable fetal-side placenta image encoder that is task agnostic. In the fine-tuning stage (Figure 2B), we train a simple classifier (logistic regression) using the encoded image features for each task. PlacentaCLIP builds on our preliminary work44,45 by introducing cross-modal pre-alignment, distillation, and retrieval strategies. The cross-modal pre-alignment technique is designed to improve performance by pre-aligning the encoders, a ResNet-based78 image encoder, and a transformer-based79 text encoder, with a large collection of natural image-text pairs.47 This reduces the model’s dependency on extensive placenta-specific data. The cross-modal distillation module distills intra-placental feature reasoning capabilities from a language model80 trained on a large medical text corpus into the image encoder to enhance performance. Moreover, the cross-modal retrieval module improves robustness by retrieving the image regions relevant to the textual features for more effective image-text alignment. More details are provided in the methods section.
Figure 2.
An overview of the pre-training and fine-tuning paradigm and the cross-modal contrastive learning algorithm PlacentaCLIP
(A) The pre-training stage, where a frozen pre-trained BERT encoder and a trainable transformer text encoder were used to encode the text from pathology reports, while a trainable ResNet50 was used to encode image features. The proposed cross-modal contrastive learning algorithm guides this training stage. BERT, bidirectional encoder representations from transformers.
(B) The fine-tuning stage, where the frozen ResNet50, trained in the pre-training stage, was used to extract image features, and logistic regression was applied to these features to predict the placental features or clinical outcomes. The 2,811-image fine-tuning dataset was randomly split into training and validation sets.
Robustness evaluation design
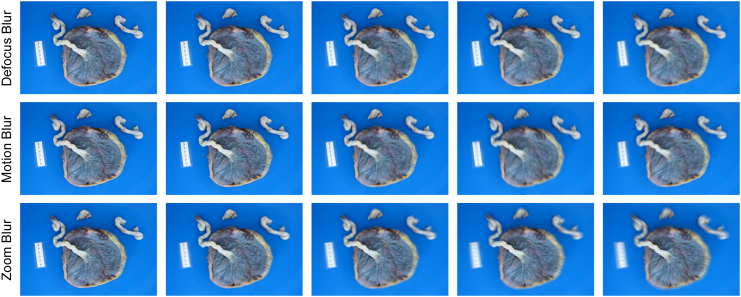
Placenta photographs are often subject to various non-ideal conditions that can affect their quality,81 making it challenging for a modern analysis models to interpret them accurately. While a previous study43 has tackled similar problems, our research offers a more comprehensive analysis. To evaluate the robustness of placenta analysis models in practical environments, it was necessary to account for these common variations and understand their impact on model performance. We developed a robustness evaluation dataset that includes common artifacts specific to placenta photographs, including motion blur, blood stains, and lighting variations. This evaluation provided valuable insights into the model’s strengths and weaknesses, identifying areas for further enhancement. The evaluation protocol focused on three key objectives: first, to assess the robustness of the proposed modules (i.e., module evaluation); second, to identify potential correlations between tasks and image features (i.e., model explainability); and third, to offer guidance on optimizing photo-taking procedures in real-world applications (i.e., application guideline). The first two objectives contribute to model design and evaluation, while the third directly informs clinical practice. More details are provided under Robustness evaluation dataset generation.
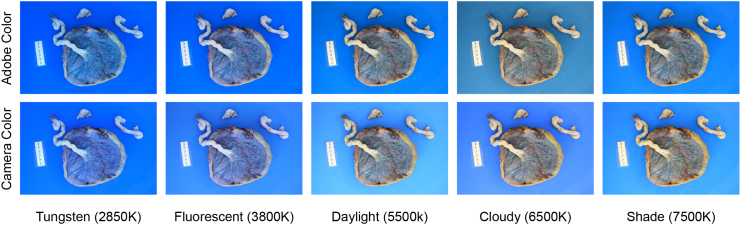
To evaluate the model’s performance under different artifacts, we intentionally introduced these variations or artifacts into the internal validation set. Unlike previous work,82 we focused on creating a list of placenta-specific common corruptions based on our experiences and understanding of placenta photo-taking procedures, aiming to more accurately simulate real-world settings. We divided the corruptions into three groups: image artifacts (blood, glare, JPEG compression, and shadow), image blur (defocus, motion, and zoom), and exposure artifacts (brightness, contrast, and saturation). Each corruption was assigned five levels, with each level representing a different degree of severity. It was important to set realistic corruption strengths to ensure that our evaluation of the model’s robustness was relevant to practical placental analysis. First, we chose levels that maintained placental visibility across all corrupted images. Then, through consultation with a pathologist, we determined the highest corruption level at which placental features remained discernible. These levels were standardized to level 3 (of 5), and the other levels were adjusted accordingly. Additionally, we included common white balance inaccuracies (e.g., tungsten, fluorescent, daylight, cloudy, and shade), simulated using white balance augmentation.83 Further details and examples are provided in the methods section.
Model performance and robustness evaluation
The performance of the linear classifiers was compared and quantified using the area under the receiver operating characteristic curve (AUC), mean average precision (mAP), and 84 to ensure that the threshold for positive predictions did not affect the scores. We measured robustness by observing the performance drop when introducing image artifacts.
Overall performance
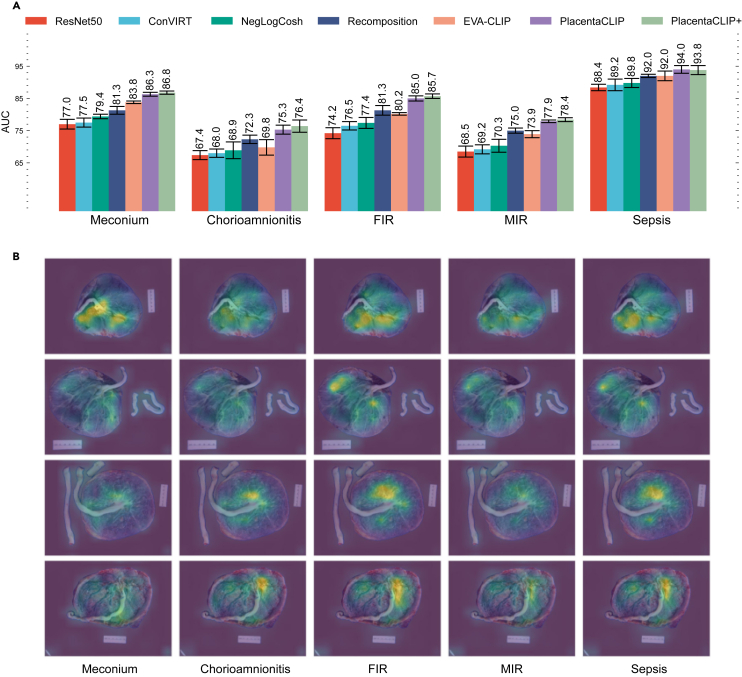
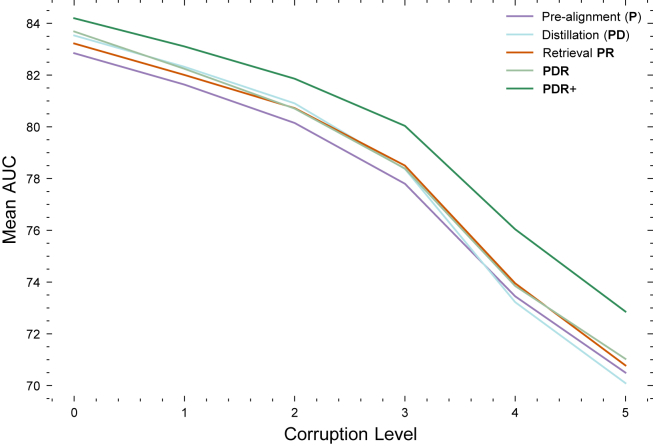
PlacentaCLIP was trained using 10,193 image-text pairs from 2014 to 2018, in alignment with previous work.44,45 PlacentaCLIP+ was trained on 28,952 image-text pairs from 2010 to 2022 to demonstrate its full capability and scalability. The results in Figure 3A indicate that PlacentaCLIP achieves state-of-the-art AUC. Additionally, the AUC improvements from incorporating additional data (PlacentaCLIP+) highlight the scalability of our proposed method. Figure 3B illustrates how the cross-modal retrieval module aids the pre-training process. We visualize the attention weights from each text query to the image features to demonstrate the changes in the feature space. Including stage 1 for FIR and MIR increased the variance of the model performance, but the performance was similar. This result is provided in Table S1.
Figure 3.
Average AUC for four placental feature identification tasks and one clinical outcome prediction task on the primary dataset and visualization of the cross-modal retrieval module
(A) The AUC and the corresponding standard deviation from five random splits. Results of ResNet50, ConVIRT,46 and NegLogCosh are taken from Pan et al.44 Results of recomposition are taken from Pan et al.45 The result for EVA-CLIP58 is from the “EVA02_CLIP_B_psz16_s8B″ model, tuned on our pre-training data. The error bars represent the standard deviations computed from five random splits.
(B) The attention weights from the cross-modal retrieval module during the pre-training stage. Different features are retrieved to assist the image encoder pre-training based on query text for better image-text alignment. In the illustration, the full name of each task is used as the text query to retrieve the visual features, except for sepsis, where the concatenation of FIR, MIR, and chorioamnionitis is used as the query. The actual process uses part of the report as the text query. The ground-truth labels for the images from top to bottom are as follows: row 1: −, −, −, −, −; row 2: +, −, −, −, −; row 3: −, −, 1, 1, −; row 4: +, −, 1, 1, −. −: negative; +: positive; 1: stage 1. FIR, fetal inflammatory response; MIR, maternal inflammatory response; PlacentaCLIP+, PlacentaCLIP trained with additional data.
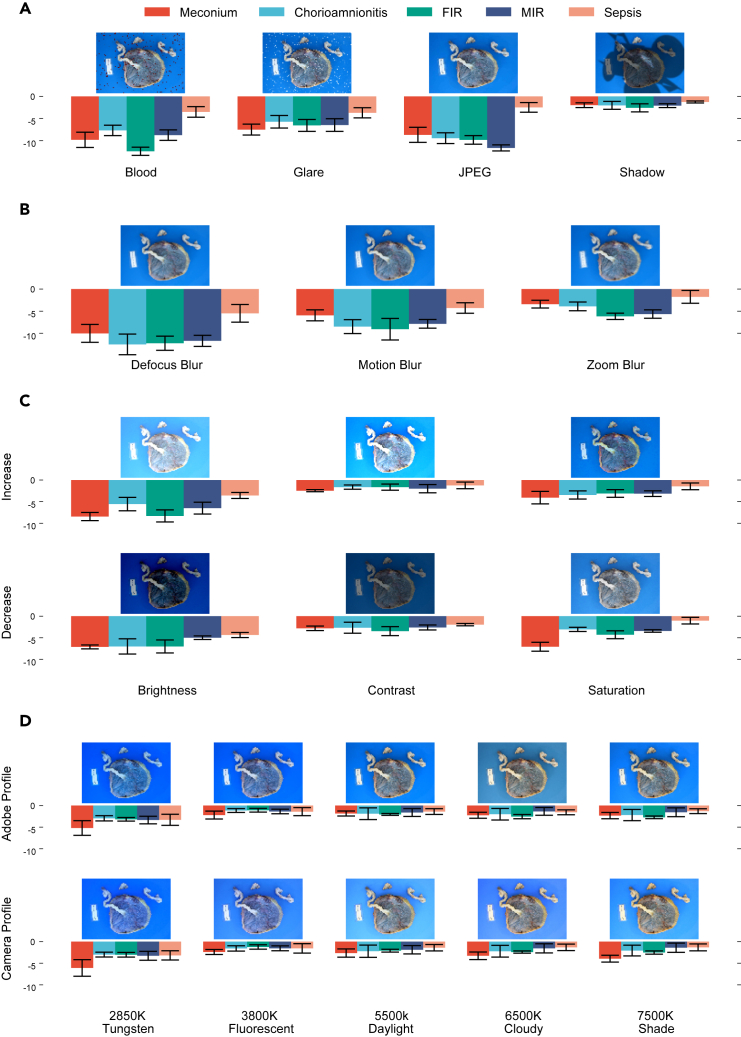
Application guideline from robustness evaluation
As shown in Figure 4, the introduction of any form of corruption adversely affected the model’s performance. Despite significant corruption, factors such as shadows, zoom blur, contrast, and saturation changes exerted a relatively minor impact on performance. These results offered new insights into the photo-taking process, challenging general assumptions. Notably, commonly used lossy image compression techniques (e.g., JPEG) significantly degraded the model’s performance, even though the input size () of the model was smaller than images produced by most smartphones. Consequently, users are advised to avoid lossy compression and instead opt for smaller image sizes if storage space is a concern. Additionally, glare had a more detrimental effect on the model’s performance than shadows, suggesting that shielding light sources to reduce glare could improve performance. The model also showed greater sensitivity to brightness changes compared to contrast and saturation, implying that adjusting contrast and saturation could be a viable method to compensate for brightness issues. Each aspect of robustness was assessed in isolation to avoid complex factorial comparisons and to facilitate the generation of application guidelines. Initial analysis on the combined aspects are included in Tables S2–S6, and original statistical results are shown in Table S8; however, no additional insights were gained due to the complexity of analyzing the large number of comparisons.
Figure 4.
The average AUC performance drop of PlacentaCLIP+ from using the original images to corrupted images on each task identified in the primary dataset
The AUC drop (y axis) is computed by subtracting the AUC of PlacentaCLIP+ on the original images from the AUC on corrupted images, averaged across all corruption levels for each random split. Error bars represent the standard deviations computed using five random splits.
(A) Performance under different image artifacts.
(B) Performance under different types of image blurring.
(C) Performance under different exposure artifacts.
(D) Performance under different WB inaccuracies.
FIR, fetal inflammatory response; MIR, maternal inflammatory response.
As shown in Figure 4D, the model’s performance degradation under white balance inaccuracies was moderate and consistent, except when the white balance preset used was extremely inaccurate (e.g., tungsten 2850K). Moreover, when the white balance preset aligned closely with the actual lighting conditions (e.g., between fluorescent and daylight), the model demonstrated its best performance. This indicated that the model possesses a degree of adaptability to various white balance inaccuracies. Nevertheless, users are advised to verify the white balance setting for optimal performance or use the camera’s automatic setting to secure reasonable performance.
Model explainability from robustness evaluation
Figure 4 illustrates that each task exhibits distinct levels of robustness against various artifact. Sepsis prediction demonstrated the highest overall robustness, likely because the model depends on multiple placental features to predict sepsis risk, which weakens the effect of individual artifacts. Meconium was particularly sensitive to color alterations (e.g., saturation changes in Figure 4C and extreme white balance inaccuracies in Figure 4D), whereas MIR was more susceptible to the loss of textural details due to JPEG compression, as shown in Figure 4A. Moreover, blur significantly compromised performance by removing textural details. As depicted in Figure 4B, tasks like FIR, MIR, and chorioamnionitis were less robust to blur compared to meconium, underscoring their reliance on textural details. Zoom blur, which affected images non-uniformly, impaired textural features in the outer part of the image, particularly where the umbilical cord is present, more than the center. The lesser impact of zoom blur compared to uniform blur suggests a stronger reliance on the features in the center placenta disk than on the cord for all tasks. Notably, while the relative performance between other tasks stayed consistent across all three types of blur, chorioamnionitis exhibited a much lower performance drop with zoom blur, suggesting greater robustness to blur in the cord region. This finding was further supported by the relative performance change between tasks under varying brightness levels. Increasing brightness (or overexposure) removed information more quickly from brighter regions, while decreasing brightness (or underexposure) similarly affected darker regions. Chorioamnionitis was more adversely affected by decreased brightness than increased brightness, suggesting that the model relies more on darker regions (e.g., the disk) than the brighter regions (e.g., the cord) for predicting this condition. This result partially aligns with pathological examination, where MIR and chorioamnionitis are found in the disk region, while FIR at stage 2 or higher is primarily found in the cord.
Module contribution to performance and robustness
To further investigate the differential behavior of the proposed modules, we analyzed their performance across varying corruption levels as part of the robustness evaluation protocol. Figure 5 demonstrates how each module responded to changes in corruption severity. Specifically, applying the cross-modal distillation module () yielded superior AUC scores at lower corruption levels (levels 0–2) compared to the retrieval module (). Conversely, at higher corruption levels (levels 3–5), the retrieval module outperformed the distillation module in terms of AUC. This variation in performance suggests that the retrieval module primarily enhances robustness, while the distillation module boosts performance. Importantly, the two modules are complementary, as their combined use () resulted in higher AUC scores. Finally, these modules demonstrated scalability, with the inclusion of additional data () improving both performance and robustness.
Figure 5.
Module AUC performance at varying corruption levels
At lower corruption levels (below level 3), the distillation module outperformed the retrieval module. As the corruption level increased, the retrieval module showed better performance. Adding the distillation module on top of the retrieval module did not further improve robustness (i.e., the performance of PDR and PR converged as the corruption level increased). These results validate our design; distillation enhances performance, while retrieval improves robustness.
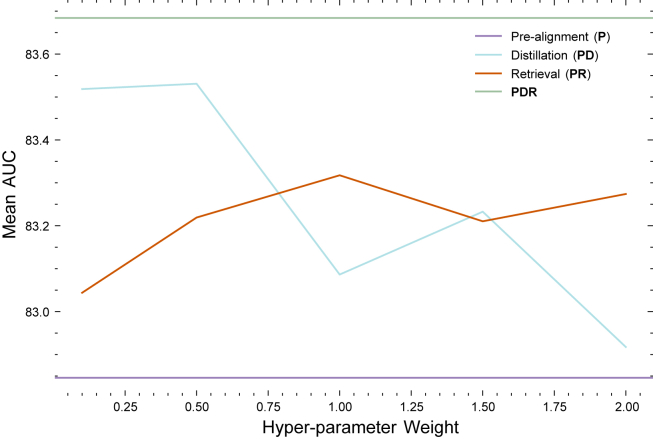
Module hyperparameter evaluation
To analyze the effects of the hyperparameter for each proposed module, we trained the model with each module individually across a range of parameter settings, as shown in Figure 6. The x axis represents the hyperparameter values regulating the strength of each loss function, while the y axis depicts the corresponding model performance. The line represents the performance of the final model with all modules and additional data incorporated, and the line indicates the performance of the baseline pre-aligned model without any proposed modules. It is observed that the performance of the model with individual modules falls between these two lines for all parameter settings experimented.
Figure 6.
The AUC scores obtained by applying different hyperparameters to the pre-training modules on a subset of the primary dataset
Particularly noteworthy is the trend observed in the distillation line. We noted a general decline in performance as the weight of the loss increased. This aligns with our expectations; introducing a distillation loss from a pre-trained model enhances reasoning capabilities, but an excessively high weight assigned to this loss can lead to a considerable domain shift from the pre-trained language model. This shift can adversely affect model performance. If the weight for the distillation loss is set too high, then it effectively reduces to a distillation-only loss, as seen in our previous work.44 Thus, it is preferable to assign a lower weight for this module. In contrast, the cross-modal retrieval module exhibits a different pattern; model performance improves and then plateaus as the weight of this loss increases. This trend, opposite to that of distillation, is anticipated, since this module is linked to trainable parameters and does not introduce domain shift. Increasing this loss introduces additional priors, but beyond a certain point, it adds no further benefit. Consequently, further performance improvement are not anticipated after a certain threshold, making it advisable to select a more conservative weight for this module.
Statistical analysis
We contrasted each module’s performance on the primary dataset and under the robustness evaluation protocol to assess whether they met their intended design rationales. Furthermore, to identify potential biases within the model, we examined its performance in relation to the demographic information present in the primary dataset. We used a paired t test with a significance level of 0.05 applied to the AUC scores. This involved treating the five classifiers trained on different splits as individual subjects. The t test was conducted as a within-subjects test, considering that all other configurations remained identical except for the test variables. Additionally, we applied the Benjamini-Hochberg procedure85 to all multiple tests to control the false discovery rate. All results are presented in the following sections.
Module contribution to performance and robustness
Performance was measured using the internal validation set, while robustness was assessed using the robustness evaluation set (i.e., internal evaluation with introduced artifacts). The use of the cross-modal retrieval module () significantly strengthened robustness, as indicated in rows 1 and 2 of Table 1, though its impact on performance enhancement was comparatively modest. Conversely, rows 2 and 3 demonstrate that the use of the distillation module () led to a significant uplift in performance but had a limited affect on robustness. These results are consistent with our model designs. Additionally, the enhancements in accuracy and robustness provided by the proposed modules were complementary (). When both modules were applied (row 6), there was a significant improvement in both accuracy and robustness. Last, the proposed modules demonstrated scalability, as the addition of extra data () into the training set consistently resulted in significant gains in both accuracy and robustness (rows 7–10). These findings align with the robustness evaluation results from Module contribution to performance and robustness.
Table 1.
Pairwise t test results applied to the AUC between applying set A of modules and set B of modules on both the primary dataset and the robustness evaluation
| A | B | DoF | Performance |
Robustness |
|||
|---|---|---|---|---|---|---|---|
| T score | p-Corr | T score | p-Corr | ||||
| 1 | PD | PD | 4.0 | 1.351 | 0.248 | 3.880 | 0.030 |
| 2 | P | P | 4.0 | 2.874 | 0.057 | 3.452 | 0.037 |
| 3 | PR | PR | 4.0 | 4.010 | 0.023 | 0.970 | 0.385 |
| 4 | P | P | 4.0 | 4.569 | 0.017 | 1.435 | 0.250 |
| 5 | P | P | 4.0 | 2.718 | 0.059 | −1.811 | 0.181 |
| 6 | P | P | 4.0 | 20.550 | 0.000 | 4.031 | 0.030 |
| 7 | P+ | P | 4.0 | 16.077 | 0.000 | 12.114 | 0.001 |
| 8 | PD+ | PD | 4.0 | 5.631 | 0.010 | 9.442 | 0.002 |
| 9 | PR+ | PR | 4.0 | 7.930 | 0.003 | 13.286 | 0.001 |
| 10 | PDR+ | PDR | 4.0 | 8.237 | 0.003 | 11.273 | 0.001 |
The performance of all modules is reported using the same image encoder. Cross-modal retrieval improved robustness, while cross-modal distillation enhanced the performance of the image encoder. Performance was measured using the average AUC score for each set of modules on the original primary dataset. Robustness was measured using the average AUC score for each set of modules on the corrupted primary dataset (robustness evaluation protocol). P represents the use of pre-aligned encoders, D indicates cross-modal distillation, and R refers to cross-modal retrieval. PDR+ denotes the use of additional data in conjunction with all modules. A bar is placed above the differing module. DoF, degree of freedom; p-corr, corrected p value.
Bias assessment
In the bias assessment, our aim was to identify any potential biases in model performance across different demographic groups. Statistical analysis was conducted on the main dataset for various demographic categories.
We first analyzed the mean AUC across all tasks, considering race as the within-subject factor and each model trained on a different random split as the subject. The t test results, presented in Table 2, indicated no statistically significant differences in model performance among the known racial groups. Only the performance for the “unknown” racial category was notably better than that for the “White” group. Additional analysis (Table S7) revealed that the disparity in class distribution for the sepsis classification task contributed to this performance difference.
Table 2.
Pairwise t test comparing the AUC between demographic groups A and B
| A | B | T | DoF | p-Corr |
|---|---|---|---|---|
| Black/African American | American Indian/Alaska Native | 0.885 | 4.0 | 0.502 |
| Black/African American | White | 0.832 | 4.0 | 0.502 |
| White | American Indian/Alaska Native | 0.737 | 4.0 | 0.502 |
| Asian | American Indian/Alaska Native | 1.329 | 4.0 | 0.358 |
| Asian | Black/African American | 1.735 | 4.0 | 0.338 |
| Asian | White | 3.387 | 4.0 | 0.103 |
| Other | American Indian/Alaska Native | 1.303 | 4.0 | 0.358 |
| Other | Asian | 0.754 | 4.0 | 0.502 |
| Other | Black/African American | 2.037 | 4.0 | 0.278 |
| Other | White | 4.049 | 4.0 | 0.077 |
| Unknown | American Indian/Alaska Native | 1.588 | 4.0 | 0.351 |
| Unknown | Asian | 2.737 | 4.0 | 0.156 |
| Unknown | Black/African American | 4.737 | 4.0 | 0.068 |
| Unknown | other | 1.391 | 4.0 | 0.358 |
| Unknown | White | 9.690 | 4.0 | 0.010 |
Performance was measured using the average AUC score for each set of modules on the original primary dataset. DoF, degree of freedom; p-Corr, corrected p value.
Next, the mean AUC across all tasks was compared using maternal age groups (in years) as defined in the obstetric care consensus,86 with each model trained on a different random split as the subject. As shown in Table 3, the model demonstrated improved performance in the age group of 45 and above. There was no clear biological explanation for this. However, the increase in average age from pre-training to the internal validation set, as shown in Figure 1C, may have contributed to this performance difference.
Table 3.
Pairwise t test comparing the AUC of PlacentaCLIP+ using images from maternal age groups A and B
| A | B | T | DoF | p-Corr |
|---|---|---|---|---|
| 45 ≤ MA | 35 ≤ MA < 40 | 6.936 | 4.0 | 0.007 |
| 45 ≤ MA | 40 ≤ MA < 45 | 4.460 | 4.0 | 0.022 |
| 45 ≤ MA | MA < 35 | 7.205 | 4.0 | 0.007 |
| 35 ≤ MA < 40 | 40 ≤ MA < 45 | 1.465 | 4.0 | 0.217 |
| MA < 35 | 35 ≤ MA < 40 | 2.417 | 4.0 | 0.110 |
| MA < 35 | 40 ≤ MA < 45 | 2.126 | 4.0 | 0.121 |
Performance was measured using the average AUC score for each set of modules on the original primary dataset. MA, maternal age; DoF, degree of freedom; p-Corr, corrected p value.
Additionally, the analysis involved comparing the mean AUC across all tasks with gestational age groups (in weeks), as outlined in the committee opinion,87 as the within-subject factor. The results revealed no statistically significant differences in model performance across the different gestational age groups.
Finally, we assessed the mean AUC across all tasks, using fetal sex as the within-subject factor, with each model trained on a different random split as the subject. The analysis yielded a t score of 1.069 and a p value of 0.345, indicating no statistically significant difference in model performance based on fetal sex.
External validation
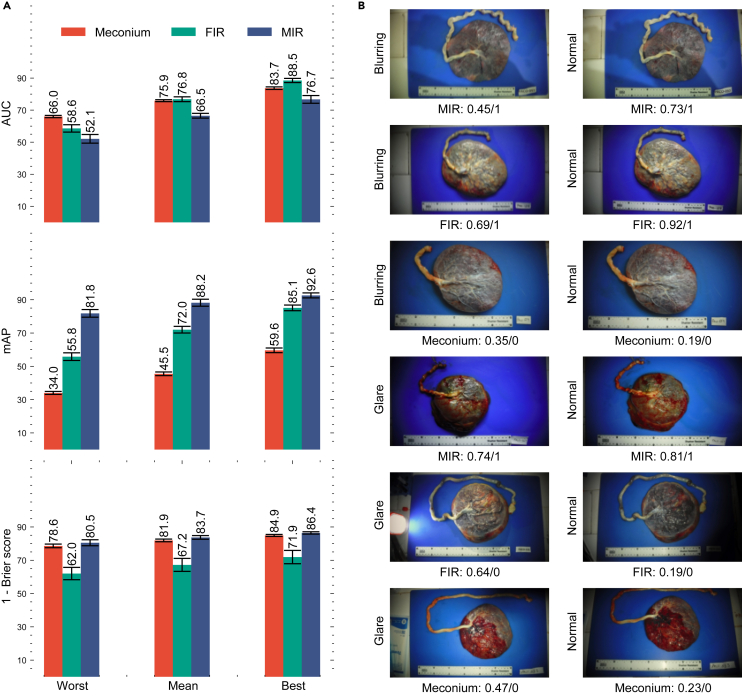
To evaluate the model’s performance in practical application scenarios, where users may capture placental images under diverse conditions, we conducted tests using the dataset from MRRH. The distinctiveness of this dataset lies in its provision of multiple images, varying in quality, for each placenta. For our analysis, we computed the best, worst, and mean performance metrics. The best and worst performances were determined by selecting the images where the model achieved the highest and lowest effectiveness for each placenta, respectively. Meanwhile, the mean performance was calculated by averaging the predicted probabilities across all images for each placenta. As shown in Figure 7A, the mean performance on the MRRH dataset, which we regard as the most representative metric, was satisfactory but lower than expected. Apart from the domain shift from the training data, this reduced performance is attributable to variations in image quality, as evidenced by the substantial gap between the best and worst results and the variations in Figure 7B. Thus, it is reasonable to anticipate that the model’s real-world performance in various clinical settings, on good-quality images, would fall between the mean and the best-observed results.
Figure 7.
Performance of PlacentaCLIP+ on three placental feature identification tasks in the external validation set from MRRH and some qualitative examples of performance variation
(A) The performance for the three identified tasks using the models trained on the NMH dataset. Worst: the metrics were generated by selecting an image for each case where PlacentaCLIP+ performed the worst. Mean: the metrics were generated by averaging the probabilities predicted by PlacentaCLIP+ over all the images for each case. Best: the metrics were generated by selecting the image for each case there PlacentaCLIP+ performed the best. AUC, area under the receiver operating characteristic curve; mAP, mean average precision.
(B) Example performance and image quality variation. The examples on the left are more affected by the identified artifacts than those on the right. The reported model performance under each image is presented in the form of a task: prediction/ground truth.
Discussion
This study advances placenta analysis by introducing new data, models, and evaluation techniques. The integration results in a unified placenta analysis model with promising capabilities for placental feature identification and neonatal sepsis prediction, validated through our robustness evaluation protocol and a cross-national dataset. Unlike previous methods,47,64,65,66,67,68,69 our approach synchronizes both internal and external representations. Moreover, our method extracts external knowledge from pre-trained language models without human intervention, whereas other methods70,71,72 often rely on expert input. This distinction improves the generalizability of our approach, particularly in scenarios where expert knowledge is scarce or difficult to obtain. Moreover, this study examines robustness across different application settings, a critical concern in medical imaging. Our robustness analysis clarifies the factors that influence performance, providing insights for future model design. Finally, our findings offer guidance for clinical photography, identifying significant factors such as glare, JPEG compression, and blur, while noting lesser impacts from contrast, shadow, and minor white balance variations.
Clinical implementation and global health implications
In the United States and most birth settings around the world, the placenta undergoes only a brief visual examination after delivery.14,15 Clinicians often receive minimal training on what to look for in the placenta,17 focusing primarily on obvious signs such as incomplete sections that may indicate retained placenta. Generally, only a small proportion of placentas—around 20% in the United States—are sent for a full pathological examination, which takes 2–4 days to complete14,15; the remainder are discarded. In low-resource settings, such as Uganda, pathology departments may lack the capacity to examine placentas entirely, or such examinations are performed rarely, potentially missing crucial information about the pregnancy that could influence health outcomes.26,27 Another issue is that hospital protocols for identifying which placentas should undergo pathological examination are often ineffective, with clinicians either unaware of College of American Pathologists guidelines or their own institutional guidelines, leading to the selection of placentas that do not provide the most critical information.20 This results in wasted resources and missed opportunities to examine placentas with significant clinical relevance.
We aim to further refine the PlacentaCLIP+ algorithm to eventually integrate it into a mobile app that clinicians worldwide could use at the bedside for real-time, clinically relevant diagnoses concerning the placenta and maternal and neonatal health immediately after birth.42,44,45 This would augment the clinician’s placental expertise, allowing the model to identify important abnormalities, such as incomplete placentas or signs of infection.42 Due to its ease of use, the app could be beneficial in any delivery setting worldwide, with the potential to significantly reduce morbidity and mortality. For example, by enabling the early identification of undetected incomplete placenta or infection risks, providers could intervene more quickly to reduce hemorrhage and sepsis rates. In high-resource settings, PlacentaCLIP+ could be used to triage placentas for full histopathological examination. In this workflow, obstetric providers would photograph the placenta in the delivery room and use the findings, along with their clinical judgment, to determine whether to submit the placenta for further examination. This process could increase the proportion of clinically relevant placentas sent to pathology and enable prioritization for rapid examination.
If we can sufficiently refine PlacentaCLIP+ to provide strong predictive value within an app, then we would evaluate its feasibility with the target end-users—clinicians working across various settings. Its similarity to existing smartphone applications that use photographs, such as those for submitting bank checks or receipts, should ease adoption and simplify training (i.e., minimal training would be needed for clinicians to take a photograph of the placenta). Point-of-care use would be straightforward, and integration with electronic health records could be achieved through the Health Level 7 system.88 While introducing the app might add to the birth workflow, we anticipate that the tool and process could be streamlined to take minimal time (approximately 5 min) due to its simplicity and similarity to existing smartphone applications. Ultimately, it could save time and resources by reducing the incidence of poor health outcomes at birth.
PlacentaCLIP+ could also be valuable in research on pregnancy, birth, and childhood, providing a cost-effective way to include placental pathology and improve our understanding of long-term maternal and child health outcomes. A substantial body of evidence demonstrates links between placental pathologies/features and pregnancy complications,8,9,10,11,89,90 risk of recurrence,12,91 risks to the future health of the mother13,92 and child,93,94,95 and adverse long-term offspring health outcomes.7,96,97,98,99,100,101,102
Privacy and security concerns related to PlacentaCLIP+ would be minimal, as the algorithm analyzes images of a discarded organ. However, if clinical or demographic data were incorporated into the app, then additional measures would be required to ensure patient privacy and data security. PlacentaCLIP+ is part of our broader PlacentaVision project103 aimed at enhancing the timely diagnosis of maternal, placental, and neonatal conditions that could affect health outcomes. Our findings suggest that bias across different ethnic and racial groups presents a relatively low risk. We are actively expanding our datasets from various sites to improve the algorithm’s generalizability. PlacentaCLIP+ likely meets the definition of software as a medical device104 by the US Food and Drug Administration (FDA). If new FDA regulations are implemented, then clearance would be required before its use in the United States. However, given our focus on global health, we anticipate that the algorithm will be of significant value worldwide before we pursue FDA clearance.
Limitations of the study
Despite its advancements, the model still exhibits a performance decline under the robustness evaluation protocol, suggesting potential areas for further enhancement. Ideally, the model’s performance should mirror the consistency of a pathologist’s ability to interpret visual cues under varying conditions. This gap in robustness compared to human experts may be attributed to biases in the training data and limitations in the model architecture, where images are of uniform quality and captured under consistent lighting conditions. To address this, increasing the diversity of the training dataset or introducing additional regularization into the model architecture could potentially enhance its robustness.
While the proposed robustness evaluation protocol is comprehensive, it inevitably has limited scope due to practical constraints and the complexity of real-world scenarios. In the future, a more dynamic and adaptive protocol, similar in concept to Autoaugment,105 could be developed to generate meaningful combinations of aspects for a more thorough evaluation and explanation of a model’s capabilities. Additionally, while the external validation dataset, which comprises images of varying quality, offers valuable insights into real-world model performance, it lacks sufficient diversity across demographic groups and devices. Another limitation of our study is its exclusive focus on the fetal side of the placenta without incorporating additional clinical data and considering the maternal side. Given the established correlations between clinical data and placental outcomes106 as well as the relevance of maternal-side placental features to health outcomes,77 expanding the analysis to both sides of the placenta and incorporating clinical data is a critical next step.
Conclusion
In conclusion, this study presents a comprehensive analysis and three enhancements to a placenta analysis model. With proper photo-taking techniques,73 the model’s ability to accurately analyze placental images captured under various conditions, as indicated by the robustness evaluation protocol and the external validation results, makes it particularly suitable for environments where access to high-quality imaging equipment and expert medical personnel is limited. By leveraging commonly available devices like smartphones and tablets for image capture, this model can bridge the gap in placental assessment in regions where traditional pathology resources are scarce or non-existent. The potential of this model to improve neonatal care in low-resource environments, where such advancements are most urgently needed, is particularly promising. This holds significant implications for advancing equitable and accessible maternal-fetal healthcare on a global scale with the potential to transform neonatal care in LMICs.
Methods
Ethics statement
This work was conducted under Penn State single institutional review board (IRB) approval (STUDY00020697). The primary data collection was conducted under Northwestern IRB approval (STU00207700 and STU00215628). The external validation data collection was conducted under MUST REC MUREC 1/7 and Mass General Brigham IRB (2019P003248).
Module design and motivation
Cross-modal pre-alignment using natural images
In our previous method,44 we trained the image encoder () using an unmodifiable (“frozen”) pre-trained text encoder (), similar to the ConVIRT method.46 To enhance the robustness and generalizability of the model, we incorporated the NegLogCosh similarity and used sub-features. This approach was viewed as a form of distilling knowledge from the text encoder to the image encoder. However, in the context of placenta analysis, a frozen text encoder pre-trained on other tasks may not adapt to the specific demands of this domain without fine-tuning. A frozen text encoder does not learn the information specific to placenta features during training, which can limit both the accuracy and generalizability of the model. Moreover, pathology reports are highly structured, which may not suffice for training a language model without compromising its reasoning capability. Therefore, directly aligning trained text features with untrained image features may not be reasonable, as the text and image encoders may not be calibrated to a common conceptual space. Overcoming this misalignment would require a large corpus of image-text paired training data, which is prohibitively costly for placenta images and pathology reports. To navigate these constraints, in our work, we used a cost-effective dataset of natural image-text pairs sourced from the internet as the initial training data to pre-align the encoders and used our placenta data to shift the encoders to our specific placenta domains. To conserve computational resources, we started with the CLIP47 ResNet50 and transformer models, which have already been trained on 400 million image-text pairs. Through continued training of the aligned encoders, we were able to adapt the text encoder to the demands of placenta analysis tasks and more effectively guide the image encoder. This approach addresses the limitations inherent in using a frozen pre-trained text encoder and allows for simultaneous training of both encoders while preserving the specific knowledge of placenta features learned during training.
Cross-modal distillation
In our prior research, we used a pre-trained bidirectional encoder representations from transformers (BERT) model80,107 as the text encoder for encoding pathology reports, leveraging its language understanding and reasoning capabilities. BERT is a language model that is capable of understanding the relationships between words in a given text, which makes it particularly useful for tasks that involve language understanding and reasoning. For example, a trained BERT model can recognize that the presence of meconium staining may affect the diagnosis of other placenta features, such as inflammation responses and chorioamnionitis. This capability comes from BERT’s extensive training on diverse text data, including medical documents that discuss the relationships between various terms. However, neither the pathology report nor the placenta image contains all the necessary information to model these relationships. Therefore, the text encoder only serves to encode the pathology report, and it is not designed to reason about these relationships, as suggested by Shen et al.108 To address this limitation, we distill the knowledge from the pre-trained BERT by guiding the contrastive loss. Specifically, we split the text encoder into the alignment text encoder and the reasoning text encoder , which allows us to obtain and . We applied the feature recomposition45 technique on to reduce feature suppression. Then, we modified (Equation 1) into
| (Equation 3) |
where is a hyperparemeter. The first objective aligns the image features with the alignment text feature to ensure that we have a text encoder that co-evolves with the image encoder as the training progresses; the second objective aligns the image features with the frozen BERT encoder to ensure that the reasoning capability of the text encoder is retained.
By distilling knowledge from the BERT encoder, we aim to improve the reasoning capabilities of the model and use the captured relationships between placental features that are not explicitly present in the pathology report or placenta image. This approach allows us to leverage the BERT’s robust contextual modeling capabilities and improve the model’s accuracy in placenta analysis tasks.
Cross-modal retrieval
The pathology report can vary in length depending on the number of placental features identified by the pathologist. The CLIP text encoder, however, is designed to accept a maximum of 77 text tokens, which means that we inevitably have to truncate the reports to fit the encoder. This truncation potentially results in information loss and may impair the model’s ability to accurately encode the relationships between the words within the report. Additionally, CLIP generally performs better with shorter textual inputs, as it was trained on brief text segments. To address the issue, we previously randomly rearranged the placental features at each iteration to ensure that all features were covered in the training stage. Nonetheless, this approach of rearrangement and truncation may inadvertently generate false positive feature matches, where different text features are matched to the same image feature at each iteration or where features from non-relevant parts of the images are used in the matching process, which can cause the model to learn spurious relationships. False positive samples can have a negative impact on the model’s performance and accuracy, as they can lead to robustness issues or poor generalization to new inputs.
To address the issue of false positives, we propose a cross-modal retrieval method. Traditional pooling layers use average pooling or its variants, which are based solely on the image modality, to obtain a feature vector from the feature map. Our method, however, necessitates aligning textual features with specific regions of the feature map based on the textual content. For example, if a pathology report mentions the presence of meconium staining, then we should not expect the model to match the textual feature related to meconium with the region of the feature map that corresponds to the umbilical cord. Our cross-modal retrieval module is designed to enhance the alignment between textual and visual features in a way that minimizes false positives and improves the model’s performance. Formally, let V be the image feature map and u the corresponding textual feature. We obtain the textual query , the image key , and the image value as follows:
| (Equation 4) |
where LN is the layer-norm layer and Ws are the learnable weights. Then, we obtain the image feature based on the query text as and perform a final projection and residual connection following Gorti et al.109 to obtain , where FC is a fully connected layer. Then, we update Equation 3 to
| (Equation 5) |
where the λs are hyperparameters.
By enhancing the alignment between textual and visual features, we can reduce the number of false positive samples and guide the model to learn more meaningful relationships between placental features and pathology reports, thereby improving its robustness.
Image pre-processing
We applied the AI-PLAX algorithm42 to the primary dataset to mask out the background of each image, aligning with methodologies used in previous research.
We employed our AI-SAM algorithm,75 trained using the dataset described in AI-PLAX,42 to mask the background of each image in the external validation dataset. The preference for AI-SAM over AI-PLAX in segmentation tasks stems from AI-PLAX’s limited robustness to domain shifts43 and AI-SAM’s capability for interactive modifications, advantageous in application settings.
All images were resized to pixels to preserve all content. For pre-training augmentation, we applied random adjustments of brightness and contrast by up to 20%, saturation and hue shifts by up to 5%, and random rotation by up to 180°. For fine-tuning or validations, we used no augmentation.
Pathology report pre-processing
We used a simple pre-processing procedure for the pathology reports. The reports were split by anomalies and stored as a set. Irrelevant text, such as standard descriptions and information about the pathologist, was removed using keyword matching. When training our PlacentaCLIP model, we performed bootstrap sampling45 from the set and concatenated the sampled items into complete sentences.
Model implementation
The image encoder was a ResNet50,78 and the text encoder was a transformer model.79 The BERT model80 used for cross-modal distillation was trained in a self-supervised manner107 on the MEDLINE/PubMed corpus.110
Pre-training stage
Our model and training code were written in Python 3.10.6 and PyTorch 1.11.0. Pre-training was conducted for 30 epochs with a batch size of 64. We utilized the PyTorch implementation of the AdamW optimizer111 with default settings combined with a cosine learning rate scheduler. The initial learning rate was set to , with a weight decay of 0.2 and 10% warm-up steps.
Fine-tuning stage
Evaluations were performed using scikit-learn 1.0.2 and pingouin 0.5.4. Fine-tuning was conducted by encoding the images using the pre-trained image encoder and training a logistic regression model for each task using the scikit-learn package. We trained the model using five nearly balanced random splits. An example is shown in Table 4.
Table 4.
An example random split of the fine-tuning dataset
| Meconium (n = 1,400) | FIR (n = 669) | MIR (n = 1,419) | Chorioamnionitis (n = 886) | Sepsis (n = 340) | |
|---|---|---|---|---|---|
| Fine-tuning | 700/1,400 (50.0%) | 334/669 (49.9%) | 709/1,419 (50.0%) | 443/886 (50.0%) | 170/340 (50.0%) |
| Positive | 349/700 (49.9%) | 167/334 (50.0%) | 345/709 (48.7%) | 224/443 (50.6%) | 86/170 (50.6%) |
| Negative | 351/700 (50.1%) | 167/334 (50.0%) | 364/709 (51.3%) | 219/443 (49.4%) | 84/170 (49.4%) |
| Evaluation | 700/1,400 (50.0%) | 335/669 (50.1%) | 710/1,419 (50.0%) | 443/886 (50.0%) | 170/340 (50.0%) |
| Positive | 330/700 (47.1%) | 145/335 (43.3%) | 378/710 (53.2%) | 215/443 (48.5%) | 80/170 (47.1%) |
| Negative | 370/700 (52.9%) | 190/335 (56.7%) | 332/710 (46.8%) | 228/443 (51.5%) | 90/170 (52.9%) |
The other four random splits are similar in number of each cases. FIR, fetal inflammatory response; MIR, maternal inflammatory response.
Robustness evaluation dataset generation
Common image artifacts in placenta photos
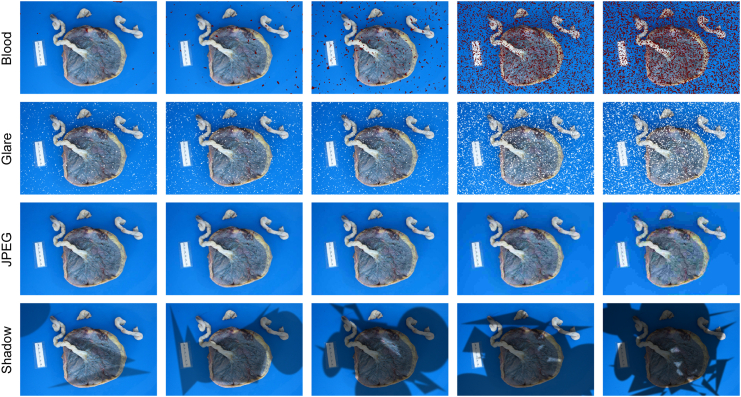
Various artifacts can be introduced during placenta photo capture, potentially negatively affecting the accurate identification of placental features. In this study, we considered four common artifact types: blood stains, glare, JPEG compression, and shadow, as shown in Figure 8. Blood stains are often present on the placenta and can obscure important features. These stains are usually dark red with irregular shapes, making them difficult to differentiate from actual features. Glare, caused by direct or reflected bright light sources, is common on reflective surfaces such as the placenta and can distort features, complicating identification. Many photo-taking devices, such as mobile phones, use JPEG compression to reduce file sizes, which can remove high-frequency or detailed information. While this generally does not affect standard object recognition tasks, it can be detrimental to identifying fine placental features. Shadows are another common challenge, appearing when the light source is behind the camera or when other objects cause uneven lighting across the placenta. Computationally, we simulated blood stains with randomly placed dark red spatters, glare with randomly placed white spatters, and shadows using a combination of randomly generated polygons and ellipses with blurred edges.
Figure 8.
Examples of common image artifacts in placenta photos
The images from left to right are in the order of increasing corruption level.
Common image blurring in placenta photos
Accurate diagnosis of placental features relies on observing low-level patterns present in the placenta photographs. However, when an image is blurry, these critical details are frequently distorted or lost, posing challenges to the diagnostic process. In practical settings, blurry placenta photographs are common, arising from a multitude of factors. To evaluate the robustness of the model, we introduced common types of blur, as shown in Figure 9, which include defocus, motion, and zoom blur. Defocus blur can result from an improperly focused lens when adjusting the camera position to accommodate the placenta’s size. Motion blur may occur if the photographs are captured while the camera is still in motion. In addition, altering the camera’s proximity to the placenta or using the lens’ zoom function to adjust the placenta’s apparent size in the image without pausing to refocus can cause zoom blur.
Figure 9.
Examples of common image blur in placenta photos
The images from left to right are in the order of increasing corruption level.
Common exposure artifacts in placenta photos
The brightness of an image refers to the perceived luminance of an image. It can be affected by both ambient lighting conditions and specific camera settings. For example, a lower exposure setting yields a darker image, while a higher exposure setting produces a brighter one. Similarly, contrast refers to the range of differentiation between the darkest and lightest parts within an image. This can be affected by the lighting conditions at the time of capture and the camera settings. For example, a photograph taken under bright sunlight generally shows a higher contrast than one taken for the same subject on an overcast day. Finally, saturation refers to the intensity and vividness of colors in an image. This can again be affected by lighting conditions and camera settings. For example, a subject photographed under brighter lighting conditions tends to show higher color vibrancy.
To evaluate the robustness of the placenta analysis model against alterations in brightness, contrast, and saturation, we systematically manipulated these attributes in the placenta photographs. This was achieved through adjusting the brightness, contrast, and saturation levels in the original images using image processing techniques, resulting in a set of images with varying levels of these factors as shown in Figure 10.
Figure 10.
Examples of common exposure artifacts in placenta photos
The images from left to right are in the order of increasing corruption level.
Common white balance inaccuracies in placenta photos
White balance (WB) is the process of adjusting the color temperature of a photograph to eliminate color casts and accurately represent the colors in the image. The WB setting can drastically affect the appearance and diagnostic quality of a photo. When the color temperature of the light source does not match the camera’s WB setting, the photo shows a color cast that distorts the original colors of the objects. For instance, an image taken under incandescent lighting might acquire a warm hue. WB is important in placenta analysis because the color of certain features in a placenta photo can be a key factor in accurate diagnosis. To evaluate the robustness of the model under different WB settings, we need to account for different color temperature preset options.
Various methods exist for adjusting WB, including using preset options, manual adjustment, or automatic correction. Modern digital cameras usually offer WB presets that cover common light sources, such as daylight, cloudy, tungsten light, and flash photography. Nonetheless, these presets are not always accurate and may require manual adjustment. Incorrect WB can also occur if it is not properly set or if lighting conditions change during a photo session (e.g., from sunny to overcast or from natural light to artificial light). To simulate the effects of incorrect WB on placenta analysis, we adopted the method proposed by Afifi and Brown,83 which alters each placenta image to five different WB presets using two color profiles. This technique allowed us to evaluate the robustness of the model under different color casts commonly encountered in real-world scenarios. We followed the same approach and modified each placenta image to five different WB presets using two color profiles, as shown in Figure 11.
Figure 11.
Examples of common WB inaccuracies in placenta photos
The images from left to right are in the order of increasing color temperature presets.
Resource availability
Lead contact
Requests for information and resources used in this article should be addressed to the lead contact, James Wang (jwang@ist.psu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Data collected in this study, including de-identified patient information, will be accessible upon reasonable request and subject to IRB approval. Access to the data requires the submission and approval of IRB protocols at both the originating and requesting institutions along with the execution of data use agreements between the institutions. Currently, the data are owned by individual sites and shared with Penn State through established data use agreements.
Our source code is available on GitHub (https://github.com/ymp5078/PlacentaCLIP)112 and has been archived on Zenodo.113 Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (NIH) under award R01EB030130. Patient data used for training the algorithm were collected by Northwestern Memorial Hospital independent of the NIH’s financial support. The external validation data came from a study supported by the National Institute of Child Health and Human Development of the NIH under awards R01HD112302 and K23AI138856 and the Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene Postdoctoral Fellowship (ASTMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or ASTMH. This work used cluster computers at the National Center for Supercomputing Applications and the Pittsburgh Supercomputing Center through an allocation from the Advanced Cyberinfrastructure Coordination Ecosystem: Services & Support (ACCESS) program, which is supported by NSF grants 2138259, 2138286, 2138307, 2137603, and 2138296. The work also used the Extreme Science and Engineering Discovery Environment (XSEDE) under National Science Foundation grant ACI-1548562.
Author contributions
Y.P., J.A.G., A.D.G., and J.Z.W. contributed to the conceptualization and design of the experiments. Y.P. was responsible for methodology, software, formal analysis, visualization, and drafting of the manuscript. M.M., J.A.G., J.N., L.M.B., D.J.R., C.K.C., R.E.W., and A.D.G. contributed to data curation and the IRB protocol. A.D.G., J.Z.W., and J.A.G. acquired funding for the study. J.Z.W. secured high-performance computing resources. All authors contributed to the review and editing of the manuscript. Y.P. and J.Z.W. directly accessed and verified the raw data, and all authors had access to the data and had final responsibility for the decision to submit for publication.
Declaration of interests
J.Z.W., A.D.G., and J.A.G. are named inventors on US patent 11,244,450, “Systems and Methods Utilizing Artificial Intelligence for Placental Assessment and Examination.” It is assigned to The Penn State Research Foundation and Northwestern University. These interests do not influence the integrity of the research, and all efforts have been made to ensure that the research was conducted and presented in an unbiased manner.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used ChatGPT-4o in order to improve the readability of the manuscript. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the published article.
Published: November 19, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2024.101097.
Contributor Information
Alison D. Gernand, Email: adg14@psu.edu.
James Z. Wang, Email: jwang@ist.psu.edu.
Supplemental information
References
- 1.Fitzgerald E., Shen M., Yong H.E.J., Wang Z., Pokhvisneva I., Patel S., O’Toole N., Chan S.-Y., Chong Y.S., Chen H., et al. Hofbauer cell function in the term placenta associates with adult cardiovascular and depressive outcomes. Nat. Commun. 2023;14:7120. doi: 10.1038/s41467-023-42300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ursini G., Punzi G., Chen Q., Marenco S., Robinson J.F., Porcelli A., Hamilton E.G., Mitjans M., Maddalena G., Begemann M., et al. Convergence of placenta biology and genetic risk for schizophrenia. Nat. Med. 2018;24:792–801. doi: 10.1038/s41591-018-0021-y. [DOI] [PubMed] [Google Scholar]
- 3.Reis A.S., Barboza R., Murillo O., Barateiro A., Peixoto E.P.M., Lima F.A., Gomes V.M., Dombrowski J.G., Leal V.N.C., Araujo F., et al. Inflammasome activation and IL-1 signaling during placental malaria induce poor pregnancy outcomes. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thornburg K.L., Marshall N. The placenta is the center of the chronic disease universe. Am. J. Obstet. Gynecol. 2015;213:S14–S20. doi: 10.1016/j.ajog.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker D.J.P., Thornburg K.L. Placental programming of chronic diseases, cancer and lifespan: a review. Placenta. 2013;34:841–845. doi: 10.1016/j.placenta.2013.07.063. [DOI] [PubMed] [Google Scholar]
- 6.Barker D.J.P., Gelow J., Thornburg K., Osmond C., Kajantie E., Eriksson J.G. The early origins of chronic heart failure: impaired placental growth and initiation of insulin resistance in childhood. Eur. J. Heart Fail. 2010;12:819–825. doi: 10.1093/eurjhf/hfq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson J.G., Kajantie E., Thornburg K.L., Osmond C., Barker D.J.P. Mother’s body size and placental size predict coronary heart disease in men. Eur. Heart J. 2011;32:2297–2303. doi: 10.1093/eurheartj/ehr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hutcheon J.A., McNamara H., Platt R.W., Benjamin A., Kramer M.S. Placental weight for gestational age and adverse perinatal outcomes. Obstet. Gynecol. 2012;119:1251–1258. doi: 10.1097/AOG.0b013e318253d3df. [DOI] [PubMed] [Google Scholar]
- 9.Lema G., Mremi A., Amsi P., Pyuza J.J., Alloyce J.P., Mchome B., Mlay P. Placental pathology and maternal factors associated with stillbirth: An institutional based case-control study in northern tanzania. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou S.K., Keating S., Kolomietz E., Shannon P. Diagnostic utility of pathological investigations in late gestation stillbirth: a cohort study. Pediatr. Dev. Pathol. 2020;23:96–106. doi: 10.1177/1093526619860353. [DOI] [PubMed] [Google Scholar]
- 11.Levy M., Alberti D., Kovo M., Schreiber L., Volpert E., Koren L., Bar J., Weiner E. Placental pathology in pregnancies complicated by fetal growth restriction: recurrence vs. new onset. Arch. Gynecol. Obstet. 2020;301:1397–1404. doi: 10.1007/s00404-020-05546-x. [DOI] [PubMed] [Google Scholar]
- 12.Hauspurg A., Redman E.K., Assibey-Mensah V., Tony Parks W., Jeyabalan A., Roberts J.M., Catov J.M. Placental findings in non-hypertensive term pregnancies and association with future adverse pregnancy outcomes: a cohort study. Placenta. 2018;74:14–19. doi: 10.1016/j.placenta.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Holzman C.B., Senagore P., Xu J., Dunietz G.L., Strutz K.L., Tian Y., Bullen B.L., Eagle M., Catov J.M. Maternal risk of hypertension 7–15 years after pregnancy: clues from the placenta. BJOG An Int. J. Obstet. Gynaecol. 2021;128:827–836. doi: 10.1111/1471-0528.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtin W.M., Krauss S., Metlay L.A., Katzman P.J. Pathologic examination of the placenta and observed practice. Obstet. Gynecol. 2007;109:35–41. doi: 10.1097/01.AOG.0000437385.88715.4a. [DOI] [PubMed] [Google Scholar]
- 15.Spencer M.K., Khong T.Y. Conformity to guidelines for pathologic examination of the placenta: rates of submission and listing of clinical indications. Arch. Pathol. Lab Med. 2003;127:205–207. doi: 10.5858/2003-127-205-CTGFPE. [DOI] [PubMed] [Google Scholar]
- 16.Taylor L.A., Gallagher K., Ott K.A., Gernand A.D. How often is the placenta included in human pregnancy research? a rapid systematic review of the literature. Gates Open Res. 2021;5:38. doi: 10.12688/gatesopenres.13215.1. [DOI] [Google Scholar]
- 17.Khong T.Y., Mooney E.E., Nikkels P.G., Morgan T.K., Gordijn S.J. Springer; 2019. Pathology of the Placenta: A Practical Guide. [DOI] [Google Scholar]
- 18.Brizuela V., Cuesta C., Bartolelli G., Abdosh A.A., Abou Malham S., Assarag B., Castro Banegas R., Díaz V., El-Kak F., El Sheikh M., et al. Availability of facility resources and services and infection-related maternal outcomes in the who global maternal sepsis study: a cross-sectional study. Lancet Global Health. 2021;9:e1252–e1261. doi: 10.1016/S2214-109X(21)00248-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milton R., Gillespie D., Dyer C., Taiyari K., Carvalho M.J., Thomson K., Sands K., Portal E.A.R., Hood K., Ferreira A., et al. Neonatal sepsis and mortality in low-income and middle-income countries from a facility-based birth cohort: an international multisite prospective observational study. Lancet Global Health. 2022;10:e661–e672. doi: 10.1016/S2214-109X(22)00043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redline R.W., Roberts D.J., Parast M.M., Ernst L.M., Morgan T.K., Greene M.F., Gyamfi-Bannerman C., Louis J.M., Maltepe E., Mestan K.K., et al. Placental pathology is necessary to understand common pregnancy complications and achieve an improved taxonomy of obstetrical disease. Am. J. Obstet. Gynecol. 2023;228:187–202. doi: 10.1016/j.ajog.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanea C., Džigurski J., Rukins V., Dodi O., Siigur S., Salumäe L., Meir K., Parks W.T., Hochner-Celnikier D., Fraser A., et al. Mapping cell-to-tissue graphs across human placenta histology whole slide images using deep learning with happy. Nat. Commun. 2024;15:2710. doi: 10.1038/s41467-024-46986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutter J., Harteveld A.A., Jackson L.H., Franklin S., Bos C., van Osch M.J.P., O’Muircheartaigh J., Ho A., Chappell L., Hajnal J.V., et al. Perfusion and apparent oxygenation in the human placenta (PERFOX) Magn. Reson. Med. 2020;83:549–560. doi: 10.1002/mrm.27950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saini B.S., Darby J.R.T., Marini D., Portnoy S., Lock M.C., Yin Soo J., Holman S.L., Perumal S.R., Wald R.M., Windrim R., et al. An mri approach to assess placental function in healthy humans and sheep. J. Physiol. 2021;599:2573–2602. doi: 10.1113/JP281002. [DOI] [PubMed] [Google Scholar]
- 24.Shchegolev A.I., Tumanova U.N., Lyapin V.M., Kozlova A.V., Bychenko V.G., Sukhikh G.T. Complex method of CT and morphological examination of placental angioarchitechtonics. Bull. Exp. Biol. Med. 2020;169:405–411. doi: 10.1007/s10517-020-04897-4. [DOI] [PubMed] [Google Scholar]
- 25.Aughwane R., Schaaf C., Hutchinson J.C., Virasami A., Zuluaga M.A., Sebire N., Arthurs O.J., Vercauteren T., Ourselin S., Melbourne A., David A.L. Micro-CT and histological investigation of the spatial pattern of feto-placental vascular density. Placenta. 2019;88:36–43. doi: 10.1016/j.placenta.2019.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck C., Gallagher K., Taylor L.A., Goldstein J.A., Mithal L.B., Gernand A.D. Chorioamnionitis and risk for maternal and neonatal sepsis: a systematic review and meta-analysis. Obstet. Gynecol. 2021;137:1007–1022. doi: 10.1097/AOG.0000000000004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins R.D., Saade G., Polin R.A., Grobman W.A., Buhimschi I.A., Watterberg K., Silver R.M., Raju T.N.K., Chorioamnionitis Workshop Participants Evaluation and management of women and newborns with a maternal diagnosis of chorioamnionitis: summary of a workshop. Obstet. Gynecol. 2016;127:426–436. doi: 10.1097/AOG.0000000000001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Specktor-Fadida B., Link-Sourani D., Ferster-Kveller S., Ben-Sira L., Miller E., Ben-Bashat D., Joskowicz L. Placental and Preterm Image Analysis. Springer; 2021. A bootstrap self-training method for sequence transfer: state-of-the-art placenta segmentation in fetal MRI Proc. Uncertainty for Safe Utilization of Machine Learning in Medical Imaging, and Perinatal Imaging; pp. 189–199. [DOI] [Google Scholar]
- 29.Pietsch M., Ho A., Bardanzellu A., Zeidan A.M.A., Chappell L.C., Hajnal J.V., Rutherford M., Hutter J. APPLAUSE: Automatic Prediction of PLAcental health via U-net Segmentation and statistical Evaluation. Med. Image Anal. 2021;72:102145. doi: 10.1016/j.media.2021.102145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Li Y.-Z., Lai Q.-Q., Li S.-T., Huang J. RU-Net: An improved U-Net placenta segmentation network based on ResNet. Comput. Methods Progr. Biomed. 2022;227:1–7. doi: 10.1016/j.cmpb.2022.107206. [DOI] [PubMed] [Google Scholar]
- 31.Asadpour V., Puttock E.J., Getahun D., Fassett M.J., Xie F. Automated placental abruption identification using semantic segmentation, quantitative features, SVM, ensemble and multi-path CNN. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khodaee A., Grynspan D., Bainbridge S., Ukwatta E., Chan A.D. Proc. IEEE Instrum. Meas. Technol. Conf. (IEEE) 2022. Automatic placental distal villous hypoplasia scoring using a deep convolutional neural network regression model; pp. 1–5. [DOI] [Google Scholar]
- 33.Dormer J.D., Villordon M., Shahedi M., Leitch K., Do Q.N., Xi Y., Lewis M.A., Madhuranthakam A.J., Herrera C.L., Spong C.Y., et al. CascadeNet for hysterectomy prediction in pregnant women due to placenta accreta spectrum. Proc. SPIE. 2022;12032:120320N. doi: 10.1117/12.2611580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobadersany P., Cooper L.A.D., Goldstein J.A. GestAltNet: Aggregation and attention to improve deep learning of gestational age from placental whole-slide images. Lab. Invest. 2021;101:942–951. doi: 10.1038/s41374-021-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun H., Jiao J., Ren Y., Guo Y., Wang Y. Multimodal fusion model for classifying placenta ultrasound imaging in pregnancies with hypertension disorders. Pregnancy Hypertens. 2023;31:46–53. doi: 10.1016/j.preghy.2022.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Ye Z., Xuan R., Ouyang M., Wang Y., Xu J., Jin W. Prediction of placenta accreta spectrum by combining deep learning and radiomics using T2WI: A multicenter study. Abdom. Radiol. 2022;47:4205–4218. doi: 10.1007/s00261-022-03673-4. [DOI] [PubMed] [Google Scholar]
- 37.Gupta K., Balyan K., Lamba B., Puri M., Sengupta D., Kumar M. Ultrasound placental image texture analysis using artificial intelligence to predict hypertension in pregnancy. J. Matern. Fetal Neonatal Med. 2022;35:5587–5594. doi: 10.1080/14767058.2021.1887847. [DOI] [PubMed] [Google Scholar]
- 38.Yampolsky M., Salafia C.M., Shlakhter O., Haas D., Eucker B., Thorp J. Modeling the variability of shapes of a human placenta. Placenta. 2008;29:790–797. doi: 10.1016/j.placenta.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haeussner E., Schmitz C., Von Koch F., Frank H.-G. Birth weight correlates with size but not shape of the normal human placenta. Placenta. 2013;34:574–582. doi: 10.1016/j.placenta.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Ernst L.M., Minturn L., Huang M.H., Curry E., Su E.J. Gross patterns of umbilical cord coiling: correlations with placental histology and stillbirth. Placenta. 2013;34:583–588. doi: 10.1016/j.placenta.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Horikoshi Y., Yaguchi C., Furuta-Isomura N., Itoh T., Kawai K., Oda T., Matsumoto M., Kohmura-Kobayashi Y., Tamura N., Uchida T., et al. Gross appearance of the fetal membrane on the placental surface is associated with histological chorioamnionitis and neonatal respiratory disorders. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Zhang Z., Wu C., Davaasuren D., Goldstein J.A., Gernand A.D., Wang J.Z. AI-PLAX: AI-based placental assessment and examination using photos. Comput. Med. Imag. Graph. 2020;84 doi: 10.1016/j.compmedimag.2020.101744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z., Davaasuren D., Wu C., Goldstein J.A., Gernand A.D., Wang J.Z. Multi-region saliency-aware learning for cross-domain placenta image segmentation. Pattern Recogn. Lett. 2020;140:165–171. doi: 10.1016/j.patrec.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan Y., Gernand A.D., Goldstein J.A., Mithal L., Mwinyelle D., Wang J.Z. Proc. Int. Conf. Med. Image Comput. Comput. Assist. Interv. Springer; 2022. Vision-language contrastive learning approach to robust automatic placenta analysis using photographic images; pp. 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan Y., Cai T., Mehta M., Gernand A.D., Goldstein J.A., Mithal L., Mwinyelle D., Gallagher K., Wang J.Z. Proc. Int. Conf. Med. Image Comput. Comput. Assist. Interv. Springer; 2023. Enhancing automatic placenta analysis through distributional feature recomposition in vision-language contrastive learning; pp. 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y., Jiang H., Miura Y., Manning C.D., Langlotz C.P. Mach. Learn. for Healthc. Conf. PMLR; 2022. Contrastive learning of medical visual representations from paired images and text; pp. 2–25.https://openreview.net/forum?id=T4gXBOXoIUr [Google Scholar]
- 47.Radford A., Kim J.W., Hallacy C., Ramesh A., Goh G., Agarwal S., Sastry G., Askell A., Mishkin P., Clark J., et al. Proc. Int. Conf. Mach. Learn. (PMLR) 2021. Learning transferable visual models from natural language supervision; pp. 8748–8763.https://proceedings.mlr.press/v139/radford21a [Google Scholar]
- 48.Wen K., Xia J., Huang Y., Li L., Xu J., Shao J. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. IEEE; 2021. COOKIE: Contrastive cross-modal knowledge sharing pre-training for vision-language representation; pp. 2208–2217. [DOI] [Google Scholar]
- 49.Bakkali S., Ming Z., Coustaty M., Rusiñol M., Terrades O.R. VLCDoC: Vision-language contrastive pre-training model for cross-modal document classification. Pattern Recogn. 2023;139:1–11. doi: 10.1016/j.patcog.2023.109419. [DOI] [Google Scholar]
- 50.Dong X., Bao J., Zheng Y., Zhang T., Chen D., Yang H., Zeng M., Zhang W., Yuan L., Chen D., et al. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2023. MaskCLIP: Masked self-distillation advances contrastive language-image pretraining; pp. 10995–11005. [DOI] [Google Scholar]
- 51.Zhang P., Li X., Hu X., Yang J., Zhang L., Wang L., Choi Y., Gao J. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2021. Vinvl: Revisiting visual representations in vision-language models; pp. 5579–5588. [DOI] [Google Scholar]
- 52.Sun Z., Fang Y., Wu T., Zhang P., Zang Y., Kong S., Xiong Y., Lin D., Wang J. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2024. Alpha-CLIP: A clip model focusing on wherever you want; pp. 13019–13029.https://openaccess.thecvf.com/content/CVPR2024/html/Sun_Alpha-CLIP_A_CLIP_Model_Focusing_on_Wherever_You_Want_CVPR_2024_paper.html [Google Scholar]
- 53.Boecking B., Usuyama N., Bannur S., Castro D.C., Schwaighofer A., Hyland S., Wetscherek M., Naumann T., Nori A., Alvarez-Valle J., et al. Proc. Eur. Conf. Comput. Vis. Springer; 2022. Making the Most of Text Semantics to Improve Biomedical Vision–Language Processing; pp. 1–21. [DOI] [Google Scholar]
- 54.Li T., Fan L., Yuan Y., He H., Tian Y., Feris R., Indyk P., Katabi D. Proc. IEEE Comput. Soc. Winter Conf. on Applications of Comput. Vis. (IEEE) 2023. Addressing feature suppression in unsupervised visual representations; pp. 1411–1420. [DOI] [Google Scholar]
- 55.Zhai X., Mustafa B., Kolesnikov A., Beyer L. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. (IEEE) 2023. Sigmoid loss for language image pre-training; pp. 11975–11986. [DOI] [Google Scholar]
- 56.Cui Q., Zhou B., Guo Y., Yin W., Wu H., Yoshie O., Chen Y. Proc. Eur. Conf. Comput. Vis. Springer; 2022. Contrastive vision-language pre-training with limited resources; pp. 236–253. [DOI] [Google Scholar]
- 57.Jia C., Yang Y., Xia Y., Chen Y.-T., Parekh Z., Pham H., Le Q., Sung Y.-H., Li Z., Duerig T. Proc. Int. Conf. Mach. Learn. (PMLR) 2021. Scaling up visual and vision-language representation learning with noisy text supervision; pp. 4904–4916.https://proceedings.mlr.press/v139/jia21b [Google Scholar]
- 58.Sun Q., Fang Y., Wu L., Wang X., Cao Y. EVA-CLIP: Improved training techniques for clip at scale. arXiv. 2023 doi: 10.48550/arXiv.2303.15389. Preprint at. [DOI] [Google Scholar]
- 59.Wu W., Sun Z., Song Y., Wang J., Ouyang W. Transferring vision-language models for visual recognition: A classifier perspective. Int. J. Comput. Vis. 2024;132:392–409. doi: 10.1007/s11263-023-01876-w. [DOI] [Google Scholar]
- 60.Vasu P.K.A., Pouransari H., Faghri F., Vemulapalli R., Tuzel O. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2024. MobileCLIP: Fast image-text models through multi-modal reinforced training; pp. 15963–15974.https://openaccess.thecvf.com/content/CVPR2024/html/Vasu_MobileCLIP_Fast_Image-Text_Models_through_Multi-Modal_Reinforced_Training_CVPR_2024_paper.html [Google Scholar]
- 61.Li R., Kim D., Bhanu B., Kuo W. RECLIP: Resource-efficient CLIP by training with small images. Trans. Mach. Learn. Res. 2023 https://openreview.net/forum?id=Ufc5cWhHko [Google Scholar]
- 62.Chen Y., Qi X., Wang J., Zhang L. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2023. DisCo-CLIP: A distributed contrastive loss for memory efficient clip training; pp. 22648–22657. [DOI] [Google Scholar]
- 63.Zhao Z., Liu Y., Wu H., Li Y., Wang S., Teng L., Liu D., Li X., Cui Z., Wang Q., et al. CLIP in medical imaging: A comprehensive survey. arXiv. 2023 doi: 10.48550/arXiv.2312.07353. Preprint at. [DOI] [Google Scholar]
- 64.Salari S., Rasoulian A., Rivaz H., Xiao Y. Proc. Int. Conf. Med. Image Comput. Comput. Assist. Interv. Springer; 2023. Towards multi-modal anatomical landmark detection for ultrasound-guided brain tumor resection with contrastive learning; pp. 668–678. [DOI] [Google Scholar]
- 65.Park S., Lee E.S., Shin K.S., Lee J.E., Ye J.C. Self-supervised multi-modal training from uncurated images and reports enables monitoring AI in radiology. Med. Image Anal. 2024;91 doi: 10.1016/j.media.2023.103021. [DOI] [PubMed] [Google Scholar]
- 66.Huang S.-C., Shen L., Lungren M.P., Yeung S. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. (IEEE) 2021. GLoRIA: A multimodal global-local representation learning framework for label-efficient medical image recognition; pp. 3922–3931. [DOI] [Google Scholar]
- 67.Müller P., Kaissis G., Zou C., Rueckert D. Eur. Conf. on Comput. Vis. Springer; 2022. Joint learning of localized representations from medical images and reports; pp. 685–701. [DOI] [Google Scholar]
- 68.Cheng P., Lin L., Lyu J., Huang Y., Luo W., Tang X. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. (IEEE) 2023. PRIOR: Prototype representation joint learning from medical images and reports; pp. 21361–21371. [DOI] [Google Scholar]
- 69.Zhang K., Yang Y., Yu J., Jiang H., Fan J., Huang Q., Han W. Multi-task paired masking with alignment modeling for medical vision-language pre-training. IEEE Trans. Multimed. 2024;26:4706–4721. doi: 10.1109/TMM.2023.3325965. [DOI] [Google Scholar]
- 70.Zhang X., Wu C., Zhang Y., Xie W., Wang Y. Knowledge-enhanced visual-language pre-training on chest radiology images. Nat. Commun. 2023;14:4542. doi: 10.1038/s41467-023-40260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z., Liu C., Zhang S., Dou Q. Proc. Int. Conf. Med. Image Comput. Comput. Assist. Interv. Springer; 2023. Foundation model for endoscopy video analysis via large-scale self-supervised pre-train; pp. 101–111. [DOI] [Google Scholar]
- 72.Wu C., Zhang X., Zhang Y., Wang Y., Xie W. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. (IEEE) 2023. Medklip: Medical knowledge enhanced language-image pre-training; pp. 21372–21383. [DOI] [Google Scholar]
- 73.Gernand A.D., Goldstein J., Wang J.Z., Gallagher K., Walker R.E. OSF; 2023. Placental Imaging Protocol. [DOI] [Google Scholar]
- 74.Khong T.Y., Mooney E.E., Ariel I., Balmus N.C.M., Boyd T.K., Brundler M.-A., Derricott H., Evans M.J., Faye-Petersen O.M., Gillan J.E., et al. Sampling and definitions of placental lesions: Amsterdam placental workshop group consensus statement. Arch. Pathol. Lab Med. 2016;140:698–713. doi: 10.5858/arpa.2015-0225-CC. [DOI] [PubMed] [Google Scholar]
- 75.Pan Y., Zhang S., Gernand A.D., Goldstein J.A., Wang J.Z. AI-SAM: Automatic and interactive segment anything model. arXiv. 2023 doi: 10.48550/arXiv.2312.03119. Preprint at. [DOI] [Google Scholar]
- 76.Madrid L., Alemu A., Seale A.C., Oundo J., Tesfaye T., Marami D., Yigzaw H., Ibrahim A., Degefa K., Dufera T., et al. Causes of stillbirth and death among children younger than 5 years in eastern hararghe, ethiopia: a population-based post-mortem study. Lancet Global Health. 2023;11:e1032–e1040. doi: 10.1016/S2214-109X(23)00211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dhaded S.M., Saleem S., Goudar S.S., Tikmani S.S., Hwang K., Guruprasad G., Aradhya G.H., Kusagur V.B., Patil L.G.C., Yogeshkumar S., et al. The causes of preterm neonatal deaths in india and pakistan (PURPOSe): a prospective cohort study. Lancet Global Health. 2022;10:e1575–e1581. doi: 10.1016/S2214-109X(22)00384-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He K., Zhang X., Ren S., Sun J. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 79.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017;30:6000–6010. doi: 10.5555/3295222.3295349. [DOI] [Google Scholar]
- 80.Kenton J.D.M.-W.C., Toutanova L.K. Proc. NAACL-HLT. ACL; 2019. BERT: Pre-training of deep bidirectional transformers for language understanding; pp. 4171–4186. [DOI] [Google Scholar]
- 81.Camm E., Wong G., Pan Y., Wang J., Goldstein J., Arcot A., Murphy C., Hansji H., Mangwiro Y., Saffery R., et al. Assessment of an AI-based tool for population-wide collection of placental morphological data. Eur. J. Obstet. Gynecol. Reprod. Biol. 2024;299:110–117. doi: 10.1016/j.ejogrb.2024.05.043. [DOI] [PubMed] [Google Scholar]
- 82.Hendrycks D., Dietterich T. Proc. Int. Conf. Learn. Represent. 2018. Benchmarking neural network robustness to common corruptions and perturbations.https://openreview.net/forum?id=HJz6tiCqYm&hl=es [Google Scholar]
- 83.Afifi M., Brown M. Proc. IEEE Comput. Soc. Int. Conf. Comput. Vis. (IEEE) 2019. What else can fool deep learning? addressing color constancy errors on deep neural network performance; pp. 243–252. [DOI] [Google Scholar]
- 84.Brier G.W. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 1950;78:1–3. doi: 10.1175/1520-0493(1950)078<0001:VOFEIT>2.0.CO;2. [DOI] [Google Scholar]
- 85.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc.: Ser. Bibliogr. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 86.American College of Obstetricians and Gynecologists’ Committee on Clinical Consensus-Obstetrics. Gantt A., Society for Maternal-Fetal Medicine. Metz T.D., Kuller J.A., Louis J.M., Society for Maternal-Fetal Medicine. Turrentine M.A., American College of Obstetricians and Gynecologists. Cahill A.G. Obstetric care consensus# 11, pregnancy at age 35 years or older. Am. J. Obstet. Gynecol. 2023;228:B25–B40. doi: 10.1016/j.ajog.2022.07.022. [DOI] [PubMed] [Google Scholar]
- 87.American College of Obstetricians and Gynecologists ACOG committee opinion No 579: Definition of term pregnancy. Obstet. Gynecol. 2013;122:1139–1140. doi: 10.1097/01.AOG.0000437385.88715.4a. [DOI] [PubMed] [Google Scholar]
- 88.Health Level Seven International. Health level seven international. https://www.hl7.org/.
- 89.Alwasel S.H., Harrath A.-H., Aljarallah J.S., Abotalib Z., Osmond C., Al Omar S.Y., Thornburg K., Barker D.J.P. The velocity of fetal growth is associated with the breadth of the placental surface, but not with the length. Am. J. Hum. Biol. 2013;25:534–537. doi: 10.1002/ajhb.22405. [DOI] [PubMed] [Google Scholar]
- 90.Parks W.T. Placental hypoxia: the lesions of maternal malperfusion. Semin. Perinatol. 2015;39:9–19. doi: 10.1053/j.semperi.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 91.Weiner E., Mizrachi Y., Grinstein E., Feldstein O., Rymer-Haskel N., Juravel E., Schreiber L., Bar J., Kovo M. The role of placental histopathological lesions in predicting recurrence of preeclampsia. Prenat. Diagn. 2016;36:953–960. doi: 10.1002/pd.4918. [DOI] [PubMed] [Google Scholar]
- 92.Parks W.T., Catov J.M. The placenta as a window to maternal vascular health. Obstet. Gynecol. Clin. 2020;47:17–28. doi: 10.1016/j.ogc.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 93.Chisholm K.M., Heerema-McKenney A., Tian L., Rajani A.K., Saria S., Koller D., Penn A.A. Correlation of preterm infant illness severity with placental histology. Placenta. 2016;39:61–69. doi: 10.1016/j.placenta.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlatterer S.D., Murnick J., Jacobs M., White L., Donofrio M.T., Limperopoulos C. Placental pathology and neuroimaging correlates in neonates with congenital heart disease. Sci. Rep. 2019;9:4137. doi: 10.1038/s41598-019-40894-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barker D., Osmond C., Grant S., Thornburg K.L., Cooper C., Ring S., Davey-Smith G. Maternal cotyledons at birth predict blood pressure in childhood. Placenta. 2013;34:672–675. doi: 10.1016/j.placenta.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barker D.J.P., Osmond C., Thornburg K.L., Kajantie E., Eriksson J.G. The shape of the placental surface at birth and colorectal cancer in later life. Am. J. Hum. Biol. 2013;25:566–568. doi: 10.1002/ajhb.22409. [DOI] [PubMed] [Google Scholar]
- 97.Barker D.J.P., Osmond C., Forsén T.J., Thornburg K.L., Kajantie E., Eriksson J.G. Foetal and childhood growth and asthma in adult life. Acta Paediatr. 2013;102:732–738. doi: 10.1016/j.placenta.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 98.Eriksson J.G., Kajantie E., Phillips D.I.W., Osmond C., Thornburg K.L., Barker D.J.P. The developmental origins of chronic rheumatic heart disease. Am. J. Hum. Biol. 2013;25:655–658. doi: 10.1002/ajhb.22425. [DOI] [PubMed] [Google Scholar]
- 99.Eriksson J.G., Gelow J., Thornburg K.L., Osmond C., Laakso M., Uusitupa M., Lindi V., Kajantie E., Barker D.J.P. Long-term effects of placental growth on overweight and body composition. Int. J. Paediatr. 2012;2012 doi: 10.1155/2012/324185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barker D.J.P., Osmond C., Thornburg K.L., Kajantie E., Eriksson J.G. The intrauterine origins of hodgkin’s lymphoma. Cancer Epidemiol. 2013;37:321–323. doi: 10.1016/j.canep.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 101.Barker D.J.P., Larsen G., Osmond C., Thornburg K.L., Kajantie E., Eriksson J.G. The placental origins of sudden cardiac death. Int. J. Epidemiol. 2012;41:1394–1399. doi: 10.1093/ije/dys116. [DOI] [PubMed] [Google Scholar]
- 102.Barker D.J.P., Thornburg K.L., Osmond C., Kajantie E., Eriksson J.G. The surface area of the placenta and hypertension in the offspring in later life. Int. J. Dev. Biol. 2010;54:525–530. doi: 10.1387/ijdb.082760db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.PlacentaVision. PlacentaVision project. http://www.placentavision.com/.
- 104.U.S. Food and Drug Administration. How to determine if your product is a medical device. https://www.fda.gov/medical-devices/classify-your-medical-device/how-determine-if-your-product-medical-device.
- 105.Cubuk E.D., Zoph B., Mane D., Vasudevan V., Le Q.V. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2019. AutoAugment: Learning augmentation strategies from data; pp. 113–123. [DOI] [Google Scholar]
- 106.De Francesco D., Reiss J.D., Roger J., Tang A.S., Chang A.L., Becker M., Phongpreecha T., Espinosa C., Morin S., Berson E., et al. Data-driven longitudinal characterization of neonatal health and morbidity. Sci. Transl. Med. 2023;15 doi: 10.1126/scitranslmed.adc9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Google. BERT experts. https://www.kaggle.com/models/google/experts-bert/frameworks/tensorFlow2/variations/pubmed/versions/2.
- 108.Shen S., Li L.H., Tan H., Bansal M., Rohrbach A., Chang K.-W., Yao Z., Keutzer K. Proc. Int. Conf. Learn. Represent. 2021. How much can CLIP benefit vision-and-language tasks?https://openreview.net/forum?id=zf_Ll3HZWgy [Google Scholar]
- 109.Gorti S.K., Vouitsis N., Ma J., Golestan K., Volkovs M., Garg A., Yu G. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. (IEEE) 2022. X-pool: Cross-modal language-video attention for text-video retrieval; pp. 5006–5015. [DOI] [Google Scholar]
- 110.National Institutes of Health Medline/PubMed corpus. https://www.nlm.nih.gov/databases/download/pubmed_medline.html
- 111.Loshchilov I., Hutter F. Proc. Int. Conf. Learn. Represent. 2018. Decoupled weight decay regularization.https://openreview.net/forum?id=Bkg6RiCqY7 [Google Scholar]
- 112.Pan Y. GitHub Repository; 2024. PlacentaCLIP Training and Evaluation.https://github.com/ymp5078/PlacentaCLIP [Google Scholar]
- 113.Pan Y. Zenodo; 2024. PlacentaCLIP Training and Evaluation. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data collected in this study, including de-identified patient information, will be accessible upon reasonable request and subject to IRB approval. Access to the data requires the submission and approval of IRB protocols at both the originating and requesting institutions along with the execution of data use agreements between the institutions. Currently, the data are owned by individual sites and shared with Penn State through established data use agreements.
Our source code is available on GitHub (https://github.com/ymp5078/PlacentaCLIP)112 and has been archived on Zenodo.113 Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.