Abstract
Biological data visualization is challenged by the growing complexity of datasets. Traditional single-data plots or simple juxtapositions often fail to fully capture dataset intricacies and interrelations. To address this, we introduce “cross-layout,” a novel visualization paradigm that integrates multiple plot types in a cross-like structure, with a central main plot surrounded by secondary plots for enhanced contextualization and interrelation insights. We also introduce “Marsilea,” a Python-based implementation of cross-layout visualizations, available in both programmatic and web-based interfaces to support users of all experience levels. This paradigm and its implementation offer a customizable, intuitive approach to advance biological data visualization.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-024-03469-3.
Background
Data visualization for biological research and data science faces significant challenges, primarily due to the advancement in technologies that produce data with exponential growth of depth and resolution. These datasets often include numerous features, making it difficult to display and interpret the information effectively. Single-plot visualization often falls short of representing the multi-dimensional nature of data. As a result, researchers have turned to composable visualization that incorporates multiple plot types to convey their findings. Examples include the use of complex heatmaps [1] for genomic data, multiple sequence alignments (MSA) for DNA/protein sequence comparison, upset plots [2] for set intersection, and OncoPrint [3] to display mutation events in patient cohorts. However, the creation of these composable visualizations can be constrained to specific tools with limited customization capability or require a deep understanding of plotting tools to construct a complex layout and integrate various plots. There is currently no standardized approach to creating composable visualizations. As a result, researchers often resort to writing complicated code or through vector graphic editors to create composable visualizations.
Results
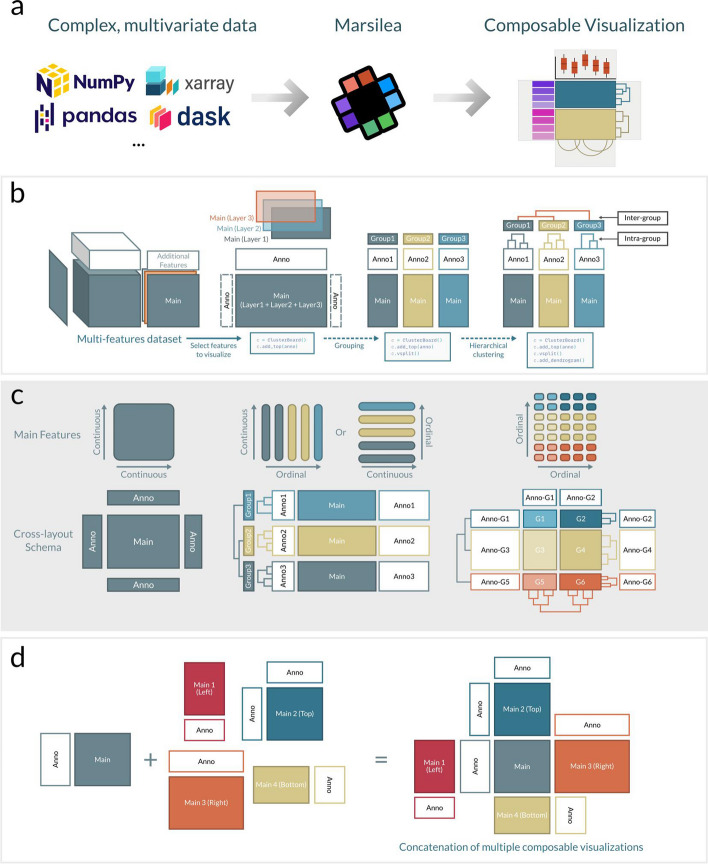
To address contemporary challenges in complex data visualization, we propose a visualization paradigm that introduces a novel and intuitive way to visualize complex datasets in a composable fashion—the cross-layout (Fig. 1a). At its core, it is a central main plot that focuses on representing the key feature of a dataset. Surrounding this central plot are secondary plots, incrementally added to each side. This arrangement forms a cross-like structure, hence the name “cross-layout visualization” (Fig. 1). Each plot, central or secondary, allows the layering of additional features such as text or symbols. These layers add annotations and context, enhancing the understanding of the plots. For datasets with categorical axis, the paradigm allows incorporation of data-driven structure, for example, through hierarchical clustering showcasing similarities within and between data groups, adding a deeper analytical dimension (Fig. 1b–c). Additionally, the paradigm offers versatility through concatenation and recursion: secondary plots can transform into central plots of new cross-layouts that are connected to the initial one, allowing for intricate and detailed visual representations of the data (Fig. 1d). This approach makes complex data more accessible and interpretable and allows the visualization of cross-feature relationships.
Fig. 1.
The novel concept of cross-layout implemented in Marsilea. a Conceptual illustration of transforming different data dimensions into composable visualization. b The assembly process of composable visualization using cross-layout. c Variants of cross-layouts adapted to different input data. d Illustration of the versatility provided by concatenation of multiple composable visualizations
To facilitate the creation and manipulation of composable visualization, we built Marsilea, a Python library designed to create composable visualizations in a declarative way. Marsilea is built with modularity in mind, allowing users to add plot components incrementally as needed. Marsilea offers a diverse range of built-in plot types: four variants of heatmap, statistical plots like line plot, bar plot, violin plot, arc diagrams, text labels, and sequence logos (Additional file 1: Fig. S1). One of Marsilea’s key strengths is its capacity for customization; users can easily adjust the layout and esthetic options, and new plot types can be easily implemented by users when needed. Marsilea boasts compatibility with multiple input formats, from the primitive Python list to NumPy arrays and Pandas DataFrames, ensuring seamless integration into the data analysis pipelines (Fig. 1a).
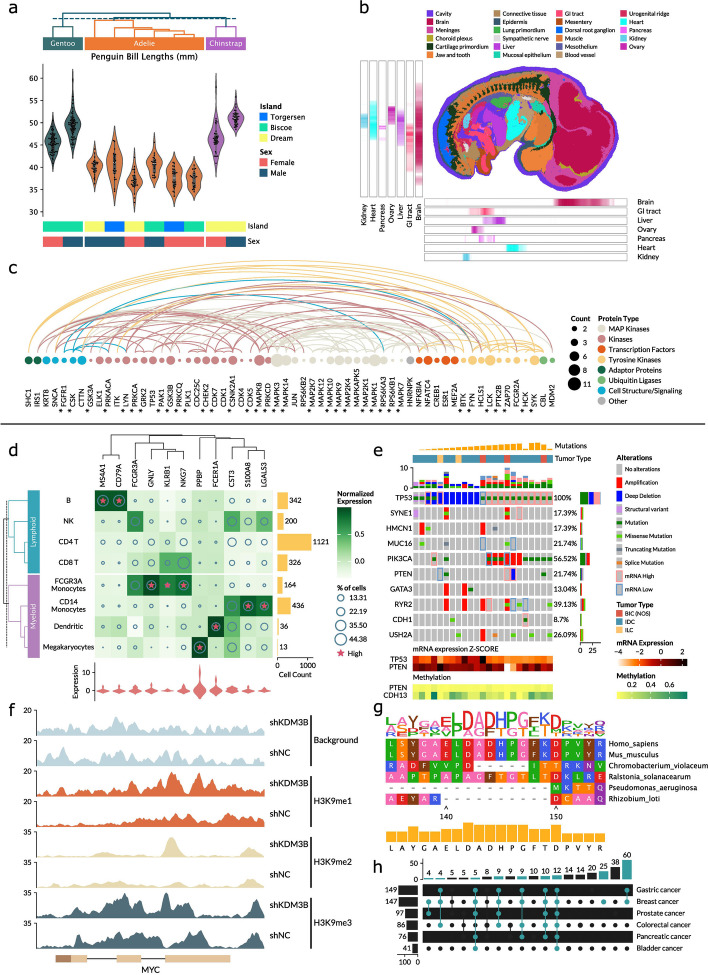
We demonstrate the broad applicability and the intuitive usage of Marsilea by creating different composable visualizations in the domain of Biology. The composable visualization can significantly enhance the single plot visualization. For instance, consider the first example (Fig. 2a), where we employed the Palmer penguin dataset to generate a violin plot exhibiting the bill length of penguins. However, a singular violin plot may not adequately communicate all the pertinent details. We then include supplementary annotations such as text labels or color strips denoting the species, residing island, and sex of the penguins. Moreover, a dendrogram can also be incorporated to indicate the similarity of data distribution. In addition to providing supplementary details, composable visualization can also enhance the clarity of visual data. For example, in the second scenario where we analyzed the 25 cell types of a mouse embryo visualized in 25 colors (Fig. 2b), making it challenging to distinguish less common cell types such as the kidney and pancreas. To remedy this, we can integrate density heatmaps along the x- and y-axes to draw attention to these cells. By doing so, we enable our audience to easily grasp crucial information from our visualization, including those with color vision deficiencies. To showcase the broad application of Marsilea, in the third example, we apply composable visualization in network visualization by presenting an arc diagram to visualize a subset of the protein–protein interaction network through phosphorylation (Fig. 2c).
Fig. 2.
Demonstration of Marsilea’s capabilities to create both novel and existing composable visualization. a Extension of the violine plot with extra annotations in the Palmer Penguin dataset. b Cell map of mouse embryo with visual enhancement of cell density heatmap. c Arc diagram of protein–protein interaction network. d Complex heatmap of single-cell RNA-seq data. e OncoPrint of mutation events in breast cancer patients. f Tracks plot of ChIP-seq data for MYC gene. g Multiple sequence alignment plot for PAH from sites 135 to 155. h Upset plot of shared terms in different cancer pathways
Afterward, we replicated widely used composable visualizations in genomics research to highlight Marsilea’s versatility. (1) Complex heatmap (Fig. 2d): This visualization extends traditional heatmap by incorporating additional data dimensions. Leveraging a well-known single-cell RNA expression dataset PBMC3K, we constructed a complex heatmap. This heatmap effectively displays marker genes across different cell types, organized by cell lineage. (2) OncoPrint (Fig. 2e): Originating from and popularized by cBioPortal, OncoPrint serves as a composable visualization to jointly visualize mutation events and expression profiles in patients. We applied it to breast cancer patients with the expression profile of mRNA and methylation rendered as heatmaps. (3) Tracks plot (Fig. 2f): Commonly employed in the analysis of ChIP-seq or ATAC-seq data. We showcased different ChIP-seq data for the MYC gene under two conditions, facilitating an easy comparison across conditions and targets. (4) Multi-sequence alignment plot (MSA) (Fig. 2g): As a foundational visualization in comparison genomics, MSA enables the examination of DNA or protein sequence variations across species. We presented an MSA for the protein sequence of PAH across six species from sites 135 to 155. The sequence logo positioned on top of the alignment clearly shows the conservation of specific sequences. (5) Upset plot (Fig. 2h): Designed to elucidate the intersection for more than three sets, the Upset plot is useful for complex sets’ data. We employed an Upset plot to show the similarity of terms among different cancer pathways with special emphasis on the interactions with breast cancer in green. Although these examples above can be created by different existing software like ComplexHeatmap [1, 4], cBioPortal [3], ggmsa [5], and Upsetplot [2], they can only create one or a few specific forms of composable visualization. Due to the generalizable approach of the cross-layout paradigm, Marsilea can generate all these plots in an intuitive and unified manner without posing any restriction on customization. Nevertheless, these applications underscore the versatility and efficacy of Marsilea in the creation of advanced composable visualizations in genomics research and its potential application in the field.
Marsilea provides multiple interfaces for users, catering to different user preferences and skill levels. It can be utilized programmatically through a declarative API for those comfortable with coding (Additional file 1: Fig. S2a) or through a no-code, web-based interface, making it accessible to a wider audience beyond just programmers (Additional file 1: Fig. S2b). The current web-based interface (http://marsilea.rendeiro.group) offers extensive customization, albeit not to the level conferred by the infinite customization of the programming interface. To assess the conciseness and simplicity of Marsilea’s code, we evaluated the implementation of the same visualization produced using Marsilea or its low-level counterpart, matplotlib (Additional files). We observed that Marsilea only demands about half the coding effort to produce identical visualizations (Additional file 1: Fig. S3), suggesting it is significantly more user-friendly. Moreover, Marsilea’s layout adjustments are notably precise, requiring only a single line of code to modify plot location, size, and padding (Additional file 1: Fig. S2c), which greatly facilitates going from individual plots to publication-ready figures for researchers.
Conclusions
In summary, the cross-layout is a simple paradigm to create composable visualization with flexibility in composition and customization to unlock infinite possibilities of biological data visualization. With the flexibility and extensibility of Marsilea, we anticipate the application of Marsilea in various data analysis pipelines and research contexts, enabling scientists to create tailored visualizations with ease and in an intuitive manner.
Methods
Benchmarking of Marsilea: we asked three individuals to implement the cooking oils example in Additional file 1: Fig. S3. Their source code files were first automatically formatted using ruff (https://github.com/astral-sh/ruff) with default settings and then measured the number of tokens, lines of code (excluding blank lines and comments), and number of API calls.
Supplementary Information
Additional file 1: Description of the benchmark procedure. Fig. S1: Demonstration of the basic building blocks of Marsilea. Fig. S2: Demonstration of the programmatic, web-based interfaces of Marsilea and the simplicity of making layout adjustments in Marsilea. Fig. S3: The example we benchmarked and the benchmark results.
Acknowledgements
We thank the open-source community efforts of building matplotlib, seaborn, numpy, and pandas, which made this project possible.
Peer review information
Andrew Cosgrove was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. The peer-review history is available in the online version of this article.
Authors’ contribution
YZ proposed the idea of a cross-layout and developed a demo. YZ and ZZ developed the Python package and built the documentation website together. YZ developed the web application for Marsilea. YZ and EC drafted the manuscript together. AFR edited the manuscript and figures. AFR and EC supervised the research.
Funding
This work was supported by the University of Macau [MYRG2020-00100-FHS, MYRG2022-00204-FHS, MYRG-GRG2023-00189-FHS-UMDF]; and Macau Science and Technology Development Fund [0011/2019/AKP, 0137/2020/A3]. YZ and AFR were supported by Angelini Ventures S.p.A. Rome, Italy. This work was supported by the Vienna Science and Technology Fund (WWTF) through project LS23-067.
Data availability
Marsilea is open source and available on GitHub under MIT license (https://github.com/Marsilea-viz/marsilea) [6], and deposited at Zenodo (https://zenodo.org/records/13732108) [7]. A graphical user interface is available from http://marsilea.rendeiro.group. The datasets and the code to recreate the examples in the figure can be found in the GitHub repository. The documentation website is https://marsilea.readthedocs.io/. The penguin dataset is from seaborn [8]. The spatial mapping of mouse embryos at E12.5 is from Mouse Organogenesis Spatiotemporal Transcriptomic Atlas (MOSTA) [9]. The protein-protein interaction data is from CellPhoneDB [10]. The single-cell expression data is a sample dataset from 10X Genomics [11]. The mutation dataset is accessible from cBioportal with the name of Breast Invasive Carcinoma (TCGA, PanCancer Atlas) [12]. The ChIP-seq data is downloaded from Gene Expression Omnibus (GEO) with accession number GSE137105 [13]. The sequence alignment data is from the sample data in the GitHub repository of ggmsa [14]. The pathway dataset of different cancer types is acquired from KEGG [15].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
André F. Rendeiro, Email: arendeiro@cemm.oeaw.ac.at
Edwin Cheung, Email: echeung@um.edu.mo.
References
- 1.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 2.Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph. 2014;20:1983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6: l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding W, Goldberg D, Zhou W. PyComplexHeatmap: a Python package to visualize multimodal genomics data. Imeta. 2023;2. [DOI] [PMC free article] [PubMed]
- 5.Zhou L, Feng T, Xu S, Gao F, Lam TT, Wang Q, et al. ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Brief Bioinform. 2022;23. [DOI] [PubMed]
- 6.Zheng Y, Zheng Z, Rendeiro A, Cheung E. Marsilea: An intuitive generalized paradigm for composable visualization. GitHub. https://github.com/Marsilea-viz/marsilea (2024)
- 7.Zheng Y, Zheng Z, Rendeiro A, Cheung E. Marsilea: An intuitive generalized paradigm for composable visualization. Zenodo. 10.5281/zenodo.13732108 (2024)
- 8.Micheal W. Penguin dataset. Datasets. GitHub. https://github.com/mwaskom/seaborn-data (2024)
- 9.Chen A, Liao S, Cheng M, Ma K, Wu L, Lai Y, et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Datasets. https://db.cngb.org/stomics/mosta/ (2022) [DOI] [PubMed]
- 10.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Datasets. https://www.cellphonedb.org/ (2023) [DOI] [PubMed]
- 11.3k PBMCs from a Healthy Donor. Datasets. https://support.10xgenomics.com/single-cell-gene-expression/datasets/1.1.0/pbmc3k (2016)
- 12.Breast Invasive Carcinoma (TCGA, PanCancer Atlas). Datasets. https://www.cbioportal.org/datasets (2018)
- 13.Wang X, Fan H, Xu C, Jiang G, et al. KDM3B suppresses APL progression by restricting chromatin accessibility and facilitating the ATRA-mediated degradation of PML/RARα. Datasets. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137105 (2020) [DOI] [PMC free article] [PubMed]
- 14.Zhou L, Feng T, Xu S, Gao F, Lam TT, Wang Q, et al. ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Datasets. GitHub. https://github.com/YuLab-SMU/ggmsa (2022) [DOI] [PubMed]
- 15.Kanehisa M. The KEGG database. Datasets. https://www.kegg.jp/kegg/ (2024)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Description of the benchmark procedure. Fig. S1: Demonstration of the basic building blocks of Marsilea. Fig. S2: Demonstration of the programmatic, web-based interfaces of Marsilea and the simplicity of making layout adjustments in Marsilea. Fig. S3: The example we benchmarked and the benchmark results.
Data Availability Statement
Marsilea is open source and available on GitHub under MIT license (https://github.com/Marsilea-viz/marsilea) [6], and deposited at Zenodo (https://zenodo.org/records/13732108) [7]. A graphical user interface is available from http://marsilea.rendeiro.group. The datasets and the code to recreate the examples in the figure can be found in the GitHub repository. The documentation website is https://marsilea.readthedocs.io/. The penguin dataset is from seaborn [8]. The spatial mapping of mouse embryos at E12.5 is from Mouse Organogenesis Spatiotemporal Transcriptomic Atlas (MOSTA) [9]. The protein-protein interaction data is from CellPhoneDB [10]. The single-cell expression data is a sample dataset from 10X Genomics [11]. The mutation dataset is accessible from cBioportal with the name of Breast Invasive Carcinoma (TCGA, PanCancer Atlas) [12]. The ChIP-seq data is downloaded from Gene Expression Omnibus (GEO) with accession number GSE137105 [13]. The sequence alignment data is from the sample data in the GitHub repository of ggmsa [14]. The pathway dataset of different cancer types is acquired from KEGG [15].