ABSTRACT
Accurately predicting individual antidepressant treatment response could expedite the lengthy trial‐and‐error process of finding an effective treatment for major depressive disorder (MDD). We tested and compared machine learning‐based methods that predict individual‐level pharmacotherapeutic treatment response using cortical morphometry from multisite longitudinal cohorts. We conducted an international analysis of pooled data from six sites of the ENIGMA‐MDD consortium (n = 262 MDD patients; age = 36.5 ± 15.3 years; 154 (59%) female; mean response rate = 57%). Treatment response was defined as a ≥ 50% reduction in symptom severity score after 4–12 weeks post‐initiation of antidepressant treatment. Structural MRI was acquired before, or < 14 days after, treatment initiation. The cortex was parcellated using FreeSurfer, from which cortical thickness and surface area were measured. We tested several machine learning pipeline configurations, which varied in (i) the way we presented the cortical data (i.e., average values per region of interest, as a vector containing voxel‐wise cortical thickness and surface area measures, and as cortical thickness and surface area projections), (ii) whether we included clinical data, and the (iii) machine learning model (i.e., gradient boosting, support vector machine, and neural network classifiers) and (iv) cross‐validation methods (i.e., k‐fold and leave‐one‐site‐out) we used. First, we tested if the overall predictive performance of the pipelines was better than chance, with a corrected 10‐fold cross‐validation permutation test. Second, we compared if some machine learning pipeline configurations outperformed others. In an exploratory analysis, we repeated our first analysis in three subpopulations, namely patients (i) from a single site, (ii) with comparable response rates, and (iii) showing the least (first quartile) and the most (fourth quartile) treatment response, which we call the extreme (non‐)responders subpopulation. Finally, we explored the effect of including subcortical volumetric data on model performance. Overall, performance predicting antidepressant treatment response was not significantly better than chance (balanced accuracy = 50.5%; p = 0.66) and did not vary with alternative pipeline configurations. Exploratory analyses revealed that performance across models was only significantly better than chance in the extreme (non‐)responders subpopulation (balanced accuracy = 63.9%, p = 0.001). Including subcortical data did not alter the observed model performance. Cortical structural MRI alone could not reliably predict individual pharmacotherapeutic treatment response in MDD. None of the used machine learning pipeline configurations outperformed the others. In exploratory analyses, we found that predicting response in the extreme (non‐)responders subpopulation was feasible on both cortical data alone and combined with subcortical data, which suggests that specific MDD subpopulations may exhibit response‐related patterns in structural data. Future work may use multimodal data to predict treatment response in MDD.
Keywords: antidepressant treatment response, ENIGMA, machine learning, magnetic resonance imaging, major depressive disorder, mega‐analysis, Radiomics
In this mega‐analysis of cortical structural MRI in 262 individuals with major depression disorder (MDD), pooled from six sites of the ENIGMA‐MDD consortium, we tested various machine‐learning pipeline configurations. We provide compelling evidence that cortical structural MRI alone is not a reliable predictor of individualized pharmacotherapeutic treatment response in MDD.

Summary.
FreeSurfer‐derived cortical thickness and surface area measures showed no predictive value for pharmacotherapeutic treatment response in major depressive disorder in the current sample of the population at large.
Classification performance was not dependent on machine learning pipeline configuration, that is, cortical data representation, the inclusion of clinical data, the machine learning method used, or the cross‐validation scheme used.
Exploratory analyses suggested that response could be predicted from cortical structural data for a specific subpopulation of MDD patients, that is, in the 25% least and most responsive categories.
1. Introduction
Major depressive disorder (MDD) is a highly debilitating psychiatric disorder with a high and growing lifetime prevalence of ~20% (Proudman, Greenberg, and Nellesen 2021). MDD is the second leading contributor to disability, with annual worldwide cost estimated at US$ 1 trillion in lost productivity alone (Bromet et al. 2011; Chodavadia et al. 2023; Greenberg et al. 2021). The first line of treatment often consists of antidepressant treatment because of its established efficacy, and known side‐effects and safety profile (Cipriani et al. 2018; Santarsieri and Schwartz 2015). However, individual response to antidepressant treatment is highly variable among patients, and there remains no validated predictor of individual treatment effect. Therefore, antidepressant treatment planning resorts to a trial‐and‐error approach and initial treatment only achieves significant symptom relief in one‐third of patients (Rush et al. 2009). This means that individuals with MDD are frequently subjected to multiple futile treatments, which prolongs disease burden and risks adverse effects such as further worsening of symptoms and risk of suicide (Zisook et al. 2009). To improve treatment planning and reduce disease burden, early predictors of treatment efficacy are needed.
Neuroimaging techniques, such as magnetic resonance imaging (MRI), have substantially improved our understanding of brain alterations in MDD. For example, large‐scale structural MRI analyses have shown that MDD is associated with patterns of thinner cortical gray matter in the orbitofrontal cortex, anterior and posterior cingulate, and insula and temporal lobes, as well as lower cortical surface area in frontal regions and in primary and higher‐order visual, somatosensory and motor areas compared to healthy volunteers (Schmaal et al. 2017). Furthermore, structural MRI can differentiate treatment‐resistant depression from other forms of MDD (Klok et al. 2019). Although multiple reviews also postulate the predictive value of such biomarkers from structural MRI (Fonseka, MacQueen, and Kennedy 2018; Schrantee, Ruhé, and Reneman 2020), the generally low to moderate effect sizes observed in these studies impede the clinical translatability of these predictors. A promising avenue to enable individual‐level inference is the use of radiomics—the extraction of a large number of features from medical images—and machine learning methods. A recent review of studies that applied deep learning methods to a wide range of features (clinical, demographic, genetic, functional neuroimaging) to predict treatment response in MDD found that they outperform regression models, achieving relatively high area‐under‐the‐curve (AUC) (Squarcina et al. 2021). Therefore, MRI predictors, combined with machine learning or deep learning, may support the search for effective treatment for MDD.
Unfortunately, many studies that aimed to predict treatment response in MDD were restricted by small sample sizes or small training or test sets, with a high risk of overestimating the performance of the predictive models (Cohen et al. 2021; Flint et al. 2021; Sajjadian et al. 2021). Moreover, the variation in techniques and analysis approaches is a major challenge in translating potential predictive biomarkers for clinical application (Schrantee, Ruhé, and Reneman 2020). Collaborative efforts, such as the Enhancing Neuroimaging Genetics through Meta‐Analysis (ENIGMA) MDD consortium, offer a vital solution to overcome this challenge. This global initiative has pooled MRI and demographic data from existing samples around the world from 52 independent sites in 17 countries and six continents. By applying standardized processing, quality control, and analysis procedures to these large‐scale data, ENIGMA‐MDD is addressing some of the core challenges facing prior smaller‐scale studies of MDD (Schrantee, Ruhé, and Reneman 2020). In addition, this approach enables the modeling of individual patient‐level predictors rather than site‐averaged data, which is more potent than traditional meta‐analyses (Harrewijn et al. 2021). Such large‐scale approaches with global representation are crucial for identifying reliable and generalizable brain alterations associated with MDD (Shrout and Rodgers 2018), as previous studies from this consortium have demonstrated (Schmaal et al. 2020; Thompson et al. 2020).
In this study, we tested the hypothesis that machine learning approaches—applied to pre‐treatment cortical structural MRI‐derived measures—can predict pharmacotherapeutic treatment response better than chance. For our secondary analyses, we hypothesized that more advanced predictive modeling pipeline configurations (e.g., a deep learning residual network) would outperform more classical machine learning approaches (e.g., a support vector machine). We compared several machine learning pipeline configurations varying in four aspects: the way we presented the cortical structural data, the inclusion of clinical information, and in the machine learning algorithm and cross‐validation (CV) scheme used (see Figure 1). Together, these modeling variations cover a range of common machine learning approaches discussed in the literature, which allows a thorough investigation of the effect of model configurations on predictive performance. We also conducted exploratory analyses on three subpopulations to further evaluate the modeling configurations. We selected these subpopulations to either increase the homogeneity of the sample (subpopulations I and II) or to improve the homogeneity of the dichotomous outcome labels (subpopulation III). We identified the following subpopulations: (i) participants from a single cohort, (ii) participants from cohorts with comparable mean response rates, and (iii) participants who showed the most and least percentage changes in symptom severity in response to antidepressant treatment.
FIGURE 1.
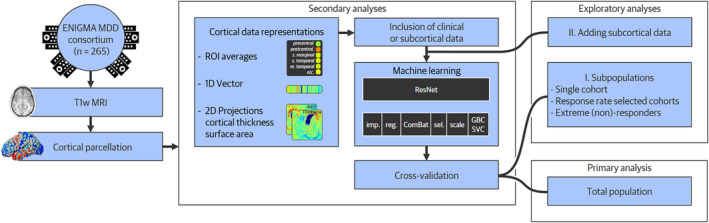
Processing and analysis pipeline. The complete analysis pipeline is presented from left to right, starting with data acquisition and preprocessing steps. Four steps, part of the machine learning pipeline, follow these. We tested various machine learning pipeline configurations for the four steps presented in our secondary analyses. Finally, the model is trained and tested, initially on the full population and subsequently in exploratory analyses on three subpopulations. Finally, we test if adding subcortical data improves predictive performance for our secondary exploratory analysis. ENIGMA MDD, Enhancing Neuroimaging Genetics through Meta‐Analysis Major Depressive Disorder Working Group; GBC, Gradient boosting classifier; imp., imputation; sel, feature selection; SVC, support vector classifier; reg., regressing out confounders; ROI, Region of interest; T1w MRI, T1‐weighted Magnetic Resonance Imaging.
2. Materials and Methods
The analyses described below (with exception of the exploratory analyses) were pre‐registered in an analysis plan before receiving the data. The plan was shared with and approved by the ENIGMA MDD consortium, and is attached to the Supplementary Methods in Data S1. All code related to this work is available through our online repository (Poirot et al. 2023).
2.1. Population
Six international ENIGMA MDD Working Group cohorts contributed data to our analysis (Table 1). All participating sites obtained approval from their institutional review boards and ethics committees and acquired written informed consent from all participants.
TABLE 1.
Demographic, clinical and outcome characteristics across cohorts.
| Characteristic | Total | Cohorts | Subpopulations a | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total (n = 262) | AFFDIS (n = 16) | DEP‐ARREST‐CLIN (n = 57) | Hiroshima cohort (n = 92) | Melbourne (n = 49) | Minnesota (n = 13) | Milano OSR (n = 35) | Response Rate Selected (n = 157) | Extreme (non‐) responders (n = 132) | ||||||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| Age (years) | 36.5 | 15.3 | 44.1 | 14.1 | 34.1 | 12.7 | 43.0 | 11.6 | 19.6 | 3.0 | 15.0 | 2.2 | 51.3 | 9.6 | 35.8 | 14.9 | 34.3 | 14.8 |
| Treatment duration (weeks) | 8.3 | 3.2 | 5.1 | 0.7 | 12.0 | 0.0 | 6.0 | 0.0 | 12.0 | 0.0 | 9.9 | 1.9 | 4.3 | 0.7 | 7.8 | 2.9 | 8.9 | 3.3 |
| Normalized pre‐treatment symptom severity | 0.04 | 0.99 | −0.85 | 1.20 | 0.79 | 0.78 | −0.31 | 0.79 | 0.28 | 0.72 | 0.15 | 0.95 | −0.25 | 1.12 | −0.18 | 0.89 | 0.03 | 0.98 |
| Age at first depressive episode (years) | 30.3 | 14.8 | 0.4 | 15.7 | 0.3 | 11.7 | 0.4 | 0.1 | 15.7 | 2.7 | 11.2 | 2.6 | 36.5 | 11.1 | 31.4 | 15.7 | 27.2 | 13.8 |
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female | 154 | 59 | 5 | 31 | 39 | 68 | 47 | 51 | 33 | 67 | 9 | 69 | 21 | 60 | 85 | 54 | 81 | 61 |
| MDD is recurrent | 160 | 61 | 15 | 94 | 29 | 51 | 46 | 50 | 31 | 63 | 9 | 69 | 30 | 86 | 92 | 59 | 83 | 63 |
| Responds to treatment | 149 | 57 | 6 | 38 | 48 | 84 | 45 | 49 | 22 | 45 | 8 | 62 | 20 | 57 | 73 | 47 | 66 | 50 |
| Uses SSRI | 194 | 74 | 9 | 56 | 0 | 0 | 91 | 99 | 49 | 100 | 13 | 100 | 32 | 91 | 149 | 95 | 91 | 69 |
| Uses SNRI | 68 | 26 | 3 | 19 | 57 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 23 | 3 | 2 | 42 | 32 |
| Uses atypical antidepressant | 16 | 6 | 8 | 50 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 7 | 20 | 9 | 6 | 5 | 4 |
| Scanned after treatment initiation | 137 | 52 | 13 | 81 | 0 | 0 | 86 | 93 | 0 | 0 | 0 | 0 | 27 | 100 | 99 | 63 | 57 | 43 |
| In the response rate selected subpopulation | 157 | 60 | 16 | 100 | 0 | 0 | 92 | 100 | 49 | 100 | 0 | 0 | 0 | 0 | 157 | 100 | 67 | 51 |
| In the extreme (non‐)responders subpopulation | 132 | 50 | 8 | 50 | 36 | 63 | 33 | 36 | 26 | 53 | 11 | 85 | 18 | 51 | 67 | 43 | 132 | 100 |
| Symptom severity scoring method | MADRS | HDRS | HDRS | MADRS | BDI | HDRS | — | — | ||||||||||
Note: Characteristics are shown for the total population, each of the included cohorts, and for the subpopulations used in our exploratory analyses. In the first part of the table, the mean and SD per characteristic are provided; in the second part of the table, the participant numbers and the percentage per characteristic are provided.
Abbreviations: BDI, Beck Depression Inventory; HDRS, Hamilton Depression Rating Scale; MADRS, Montgomery–Åsberg Depression Rating Scale; SNRI, selective serotonin and norepinephrine reuptake inhibitor; MDD, major depressive disorder; SSRI, selective serotonin reuptake inhibitor.
The third subpopulation analyzed, the single cohort subpopulation, was omitted from this table as it consisted of patients from the Hiroshima cohort only.
Inclusion criteria were patients with MDD, for whom pharmacological treatment with any antidepressant was deemed necessary, the presence of both pre‐treatment and follow‐up measurement of symptom severity, and a pre‐treatment structural MRI scan. If patients switched medication, a treatment duration of ≥ 4 weeks with the new medication before the follow‐up measurement was required. Patients were not required to be free of antidepressant medication at the time of scanning. Exclusion criteria consisted of the use of tricyclic antidepressants or quetiapine, a treatment duration of < 4 weeks, and a pre‐treatment structural MRI scan obtained > 14 days after antidepressant treatment initiation. To assess the influence of the treatment duration threshold on our findings, we additionally conducted a sensitivity analysis where we restricted our primary analyses to patients with a treatment duration of ≥ 8 weeks, the results of which can be found in Table S1. In addition, to assess the influence of the used baseline MRI cut‐off values of 14 days, we additionally conducted a sensitivity analysis where we repeated our primary analyses but excluded all participants with a baseline MRI scan obtained > 7 days after treatment initiation in Table S2.
2.2. Treatment Response
We calculated treatment response as a dichotomous outcome, defined as a ≥ 50% reduction in symptom severity score from pre‐treatment to post‐treatment (Rush et al. 2006). Symptom severity scores were measured using one or more of the following scales: the Montgomery Asberg Depression Rating Scale (MADRS), the Hamilton Depression Rating Scale (HDRS), and the Beck Depression Inventory (BDI). When scores were available from more than one scale, we selected clinician‐administered (MADRS, HDRS) over self‐reported (BDI) scales and the MADRS over the HDRS (Carmody et al. 2006). See Table 1 for the scoring method used per cohort. Outcomes were collected between 4 and 12 weeks after the start of treatment, depending on the study design per participating site (Table 1).
2.3. Data Acquisition and Preprocessing
Structural T1‐weighted 3D brain MRI scans were obtained from all six sites and processed according to the ENIGMA protocols (http://enigma.ini.usc.edu/protocols/imaging‐protocols/). We used FreeSurfer software (Fischl 2012) to perform cortical parcellation. These parcellations were visually inspected and statistically evaluated for outliers as part of quality control. Table S3 details the MRI scanners, acquisition parameters, and the FreeSurfer version used per site. For our exploratory analyses, subcortical segmentations were also created with FreeSurfer, from which we used the volumetric measures.
2.4. Machine Learning Pipeline Configurations
The primary goal of this study was to predict pharmacotherapeutic treatment response based on cortical structural MRI‐derived predictors. The primary outcome of predictive performance was balanced accuracy (bAcc), defined as the mean of the sensitivity and specificity. For our secondary aim, we investigated whether predictive performance was affected by the configuration of the machine learning pipeline. To this end, we tested variations in the pipeline configuration in four ways, of which an overview is available in Table S4. First, we created different representations from the cortical structural data derived from FreeSurfer. Second, we tested if including clinical data in the models yielded a predictive performance better than chance. Third, we tested the accuracy of three different machine learning models of varying complexity. Lastly, we applied two different CV schemes. When each of these four pipeline configurations was under investigation, we fixed the other three to isolate the effect of the configuration of interest. Variations were fixed by either taking the average of all available options (for data representations and machine learning models) or by defining a default method (for clinical predictors and CV‐scheme). An overview of these options and the used defaults can be found in Table S4, the motivation of which can be found in the Supplementary Methods in Data S1.
2.4.1. Data Representations
FreeSurfer output consisted of a three‐dimensional mesh of the cortical surface of each hemisphere. Each mesh consists of 163 thousand vertices at which data was available on the cortical thickness, surface area, and Desikan–Killiany‐based (Desikan et al. 2006) gray matter regions of interest (ROI). We processed these data in three ways, resulting in three different cortical data representations (Figure 2). First, we averaged the cortical thickness and the surface area for each of the 34 ROIs per hemisphere, resulting in 136 predictors (a. ROI average). Second, we converted the voxel‐wise cortical surface area and cortical thickness measurements to a single one‐dimensional (1D) vector by downsampling using spatial linear interpolation, resulting in 900 predictors (b. cortical vector). Third, we created two other cortical data representations by projecting the cortical surface thickness (c. cortical thickness projection) and area (d. surface area projection) measurements to two‐dimensional (2D) planes of 64 × 64 pixels using stereographic projection (Su et al. 2013).
FIGURE 2.
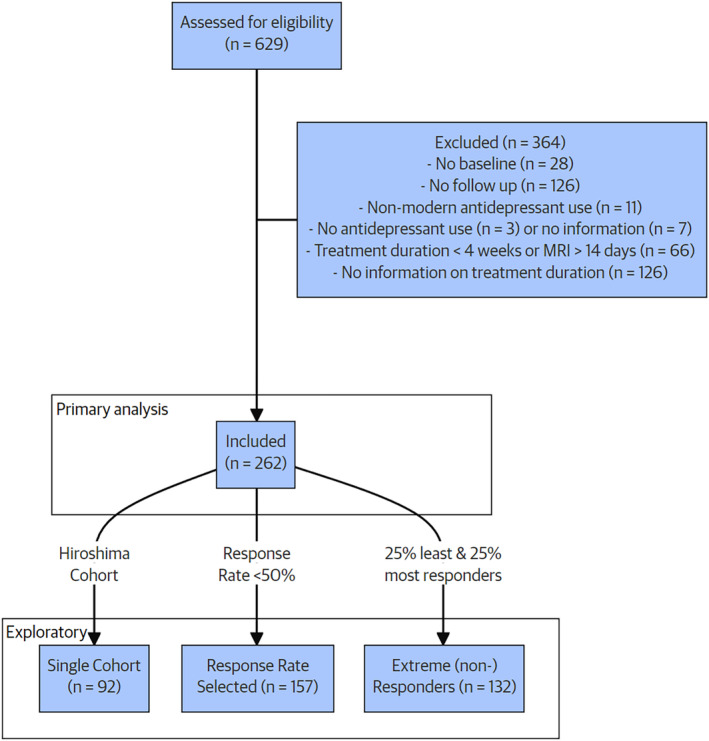
CONSORT flow diagram of patient inclusion.
In addition to cortical data, subcortical volumes generated by FreeSurfer were available for seven subcortical regions. These regions were the nucleus accumbens, amygdala, caudate nucleus, hippocampus, globus pallidus, putamen, and thalamus. Volumes were available for both hemispheres. In addition, total intracerebral volume was available, which was used to normalize subcortical region volumes. Thus a total of 15 additional predictors was included in this exploratory analysis.
2.4.2. Clinical Predictors
Clinical and demographic variables included as additional predictors were age, sex, age at first depressive episode, recurrence of MDD, antidepressant use at the time of scanning, and pre‐treatment symptom severity. When multiple symptom scorings were available, a single preferred score was used. The preferred score was the same as described in Section 2.2: preferring MADRS over HDRS, and HDRS over BDI. Symptom severity scores were normalized for each scoring instrument (Fang et al. 2020) by centering them at zero and scaling them to have a standard deviation of one for each specific scoring method across all cohorts.
2.4.3. Machine Learning Model
To test different machine learning variations, we trained three types of models: (i) a Support Vector Classifier (SVC) implemented in SciKit‐Learn version 1.1.2; (ii) a Gradient Boosting Classifier (GBC) implemented in XGBoost (version 2.1.1) (Pedregosa et al. 2011) with key algorithm hyperparameters optimized using Bayesian optimization implemented in SciKit‐Optimize (version 0.10.2) for 25 iterations (see Supplementary Methods for details in Data S1), and (iii) a deep learning residual network (ResNet) implemented in PyTorch (v.1.13.0). These methods aim to capture both the most common, and more promising methods (Gao, Calhoun, and Sui 2018). The machine learning models were trained in the following setup.
For the SVC and GBC, the machine learning setup consisted of five steps: imputation, harmonization, feature selection, scaling, and classification. Each step in the pipeline was only fitted on data from the training partition to avoid information leakage from the testing partition. First, we imputed the few FreeSurfer values missing due to internal quality control using K‐nearest‐neighbor imputation (K = 5), as SVC does not support missing values. Second, we removed potential confounding effects of age and age2 using linear regression. Third, we used ComBat harmonization (Janssen, Mourao‐Miranda, and Schnack 2018) to mitigate confounding site effects. Covariates used in harmonization were age, age2, sex, and brain volume. Third, feature selection was performed using L1‐based feature selection. Fourth, we scaled feature values using Z‐score scaling to reduce scale sensitivity. Finally, we fitted the estimator.
The ResNet model was an 18‐layer ResNet (He et al. 2016), pre‐trained on ImageNet 1 K version 1 (Deng et al. 2009), with the final fully connected layer swapped with a 512 × 2 fully connected layer. We used the Adam optimization algorithm with a binary cross‐entropy loss implemented in PyTorch (Kingma and Ba 2014). Twenty percent of the training samples were held out as a validation set. Models were trained using a batch size of 32 for a minimum of 20 epochs, after which training was stopped if performance on the validation set ceased to improve for 10 epochs. The model with the lowest loss on the validation set was tested on the test set. Models were trained on GeForce RTC 2090 SUPER (NVIDIA Corporation, Santa Clara, California) for about 5 h.
2.4.4. Cross‐Validation Methods
All machine learning models were trained in one of two CV methods implemented in SciKit‐Learn (v.1.1.2). The first method was outcome‐stratified k‐fold (SKF) CV, for 10 folds. The second method systematically excluded a single cohort from the training set to be used as a test set to assess the generalizability and robustness of our method across cohorts (leave‐one‐site‐out cross‐validation; LSO‐CV). These methods help distinguish between inherent variance and inter‐site variance in model performance.
2.5. Primary and Secondary Analyses
For our primary analysis, we tested if the mean accuracy of the models on the test set was statistically better than chance. Chance was defined as the prevalence of the majority response class, which was determined based on the training set. Whenever we tested accuracy against chance, we used permutation testing implemented in SciPy (v.1.7.3) with 100 permutations. p values were calculated using conservative approximation (Phipson and Smyth 2010; Ernst 2004). For model configurations found to be significantly better than chance, we report classifier feature importance using the coefficients of the SVC and impunity‐based feature importance in the GBC.
In the secondary analyses, we tested if there were significantly different mean performances among the four data representations, the inclusion of clinical predictors, three machine learning models, and two CV methods. For the cortical data representations and machine learning models, we performed a permutational multivariate analysis of variance test (PermANOVA) (Anderson 2005). For the inclusion of clinical data and the CV method, we compared the options using the permutation test mentioned earlier.
2.6. Exploratory Analyses
In the first post hoc exploratory analysis, we tested the accuracy of our machine learning model configurations on subpopulations with increased homogeneity in either the sample or in the dichotomous treatment response labels. For this purpose, we defined three subpopulations for which we repeated all our analyses steps described previously. In the first subpopulation analysis, we limited the sample to the single largest cohort to ascertain if inter‐cohort variance played a role in our prediction outcomes (a. single cohort). This subpopulation consisted of 92 patients from the Hiroshima cohort. Second, we repeated our analyses in cohorts with a mean response rate below 50% since the response rate varied substantially among cohorts (b. response rate selected cohorts). This subpopulation comprised 157 patients from the AFFDIS, Hiroshima, and Melbourne cohorts. Lastly, we created more homogeneous response outcome labels by defining a subpopulation consisting of the extreme subgroups of responders and non‐responders, that is, the 25% of patients showing the lowest percentage changes in depression severity and 25% responding the largest percentage changes to antidepressant treatment (c. extreme (non‐)responders). This subpopulation consisted of 132 patients (roughly equally distributed across cohorts). More demographic information about this subpopulation can be found in Table S5.
Finally, in our secondary exploratory analysis, we repeated our primary analysis but also included subcortical volumetric measures (only available for the ROI average data representation). We compared model performances between models that did or did not include subcortical data with the permutation test (see Supplementary Methods in Data S1 for the methodology used for this exploratory analysis).
3. Results
3.1. Population
Six international ENIGMA MDD Working Group samples contributed data to our analysis (Table 1). We received neuroimaging and clinical data from 629 participants with MDD. Following screening, 364 patients were excluded (for detailed information on the exclusion of patients, see the CONSORT flow diagram in Figure 1). Two of the main reasons for excluding patients were lack of follow‐up information (n = 126) and lack of information on treatment duration (n = 126). A total of 262 patients were included in the analyses. The mean age was 36.5 ± 15.3 years; 154 (59%) were female; the mean response rate was 57%; (Table 1; Figure 1). Two cohorts included adolescents (age < 20 years, Melbourne: 23/49 and Minnesota: 13/13 patients) with a considerably lower average age. Response rates varied substantially across sites (38%–84%) without clear differences between responders and non‐responders in treatment duration (8.7 ± 3.3 vs. 7.8 ± 3.1 weeks) or mean age as non‐responders (35.6 ± 15.0 vs. 37.6 ± 15.6 years, Table S6 for patient characteristics by treatment response). Response rates were high in SNRI users (78%, n = 52/66), as 83% of the SNRI users originated from the site that showed the highest response rate. Baseline clinical symptomatology was not predictive of treatment outcome at follow‐up (bAcc = 52.2% ± 8.5). FreeSurfer data for cortical thickness and surface area projections were available for 258/262 patients.
3.2. Treatment Response Performance
Overall, the performance in predicting antidepressant treatment response in MDD patients, using combined cortical thickness and surface area pre‐treatment from structural MRI data, was not significantly better than chance across machine learning pipeline configurations (bAcc = 50.5%; p = 0.66). These results were not different if we restricted our sample to subjects scanned < 7 days after start of treatment, nor influenced by treatment duration ( Tables S1 and S2).
3.3. Comparative Analyses of Machine Learning Pipeline Configurations
In secondary analyses, we first compared four types of cortical data representations (i.e., ROI average, cortical vector, and cortical thickness and surface area projections). None of the cortical data representations outperformed others (p = 0.10), and none of the cortical data representation performed significantly better than chance. Second, the additional inclusion of clinical data was examined. Performance of models that included clinical data (bAcc = 50.5%) did not outperform models without (bAcc = 51.0%; p = 0.70). In addition, none of these models significantly performed better than chance. Third, we compared three machine learning model types: SVC, GBC, and ResNet classifier. None of the models outperformed others (p = 0.15), and none of the models outperformed chance. Fourth, we assessed CV methods. LSO‐CV performance (bAcc = 52.3%) did not differ significantly from SKF‐CV (bAcc = 50.5%; p = 0.73). A complete overview of all outcomes and bAcc per model configuration is presented in Table 2.
TABLE 2.
Main model performance for all research questions.
| RQ1: Overall performance | Balanced Accuracy | Accuracy | Chance | Different from chance | |||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | p‐value | |
| Full population | 50.5 | 5.9 | 53.6 | 7.2 | 53.2 | 6.8 | 0.657 |
| RQ2‐I: Cortical data representations | 0.101 | ||||||
| a. ROI average | 50.6 | 5.4 | 54.6 | 5.6 | 54.6 | 5.1 | 0.612 |
| b. Cortical vector | 50.9 | 7.7 | 55.1 | 6.6 | 54.9 | 5.7 | 0.630 |
| c. Cortical thickness projection | 50.3 | 5.0 | 48.1 | 6.1 | 46.6 | 6.4 | 0.273 |
| d. Surface area projection | 49.8 | 2.1 | 53.9 | 9.1 | 53.7 | 8.2 | 0.436 |
| RQ2‐II: Adding clinical data | 0.703 | ||||||
| Cortical data only | 51.0 | 5.5 | 54.4 | 6.3 | 54.2 | 5.7 | 0.617 |
| Clinical data added | 50.5 | 5.9 | 53.6 | 7.2 | 53.2 | 6.8 | 0.657 |
| RQ2‐III: Machine learning model | 0.153 | ||||||
| Support vector classifier | 50.5 | 4.3 | 55.9 | 4.3 | 55.8 | 3.8 | 0.545 |
| Gradient boosting classifier | 51.0 | 8.3 | 53.8 | 7.4 | 53.7 | 6.5 | 0.701 |
| ResNet | 50.0 | 3.9 | 51.0 | 8.3 | 50.1 | 8.2 | 0.372 |
| RQ2‐IV: Cross‐validation method | 0.727 | ||||||
| 10‐Fold cross‐validation | 50.5 | 5.9 | 53.6 | 7.2 | 53.2 | 6.8 | 0.657 |
| Leave‐site‐out cross‐validation | 52.3 | 5.5 | 51.5 | 10.4 | 48.7 | 11.5 | 0.235 |
| Exploratory I: Subgroup performance | 0.002 | ||||||
| Single cohort | 49.6 | 7.1 | 48.4 | 8.0 | 47.5 | 10.6 | 0.666 |
| Response rate selected cohorts | 50.1 | 7.2 | 52.1 | 10.0 | 52.1 | 9.3 | 0.732 |
| Extreme (non‐)responders | 63.9 | 10.6 | 63.6 | 8.7 | 52.1 | 10.3 | 0.001 |
| Exploratory II: Adding subcortical data | 0.703 | ||||||
| Cortical data only | 51.6 | 3.7 | 55.3 | 3.8 | 55.4 | 5.7 | 0.587 |
| Subcortical data added | 50.5 | 5.9 | 53.6 | 7.2 | 53.2 | 6.8 | 0.657 |
Note: The balanced accuracy, accuracy, and priori chance are provided for each machine learning pipeline configuration investigated. The p‐value is provided on the right, which illustrates whether a machine learning pipeline configuration outperforms chance. The overarching p‐values for each research question express the probability of significantly different mean performance among (using permutational multivariate ANOVA) or between (permutation test) the configuration variations tested, for example, for RQ2‐II whether the balanced accuracy of “Cortical data only” differs significantly from “Clinical data added,” tested using the permutation test. Significance was inferred when p < 0.05.
Abbreviations: ResNet, deep learning residual network; ROI, region of interest; RQ, research question.
3.4. Exploratory Subpopulation Analyses
We conducted exploratory subpopulation analyses to evaluate the performance of the models in three different subpopulations. Neither the performance of the single cohort subpopulation (bAcc = 49.6%; p = 0.67) nor of the response rate selected subpopulation (bAcc = 50.1%; p = 0.73) outperformed chance across pipeline configurations. In the extreme (non‐)responders subpopulation, our models did perform significantly better than chance (bAcc = 63.9%; p = 0.001). All outcomes per machine learning pipeline configuration for this subpopulation are provided in Table S7. In short, all pipeline configurations we tested for this subpopulation significantly outperformed chance. BAcc did not improve significantly when clinical or subcortical data was added, or when LSO‐CV was applied. Predictors contributing most to this prediction were bilateral increased cortical thickness in the precentral gyri, smaller surface area of the precentral gyri, and larger surface area of the superior frontal gyri (Figure 3). All coefficients are provided in Table S8.
FIGURE 3.
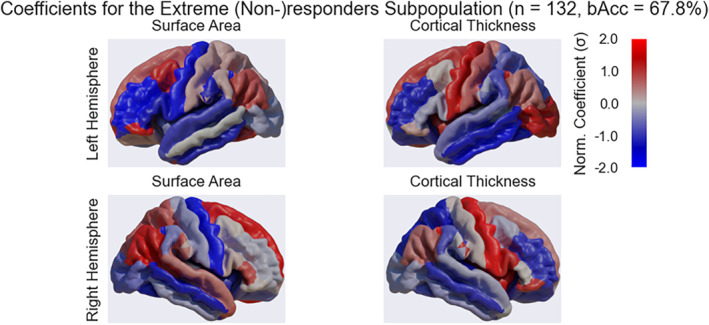
Region of interest relevance for the exploratory analyses. The figure shows the normalized coefficients used by a machine learning model to predict treatment response in the extreme (non‐)responders subpopulation, overlaid on a standard structural brain (cortical data representation: ROI average; model: gradient boosting classifier [GBC]; only cortical data). The sign indicates the direction of the relationship between a positive treatment response and either surface area (left two panels) or cortical thickness (right two panels). Red indicates a positive direction, whilst blue indicates a negative direction. The magnitude (visualized as saturation) of the coefficients indicates the strength of the relationship.
Finally, we tested whether addition of subcortical predictors improved predictive performance. Performance of models including subcortical data (bAcc = 51.6%) was not significantly better than models without subcortical data (bAcc = 50.5; p = 0.31). Again, our models only performed significantly better than chance in the extreme (non‐)responders subpopulation. All outcomes of this analysis, and of alternative classifiers are provided in Tables S9, and S10, respectively.
4. Discussion
The present study aimed to investigate machine learning based classification approaches using pre‐treatment cortical structural MRI‐derived measures to predict antidepressant treatment response. Using the largest sample of cortical structural MRI data to date (Cohen et al. 2021; Lee et al. 2018), our work presents compelling evidence that, using common machine learning approaches, cortical thickness and surface area alone are not viable biomarkers for predicting antidepressant treatment response in individuals with MDD. This finding was independent of machine learning pipeline configurations concerning (I) cortical data representations, (II) inclusion of clinical data, (III) machine learning models, and (IV) cross‐validation methods. Moreover, our exploratory subpopulation analyses highlight the possible viability of employing cortical structural measures as predictive markers for antidepressant treatment response in patients with MDD, elucidating specific patterns linked to the highest and lowest responsive individuals only.
4.1. Overall Prediction of Treatment Response
In contrast to our primary hypothesis, we could not predict treatment response in MDD better than chance level. Although several prior studies highlighted associations between baseline cortical thickness—in, for example, the anterior cingulate cortex (Phillips et al. 2015), supplementary motor area (Wu et al. 2021), and insula (Pimontel et al. 2021)—and pharmacological treatment response or remission in patients with MDD, we were unable to provide support for this framework. However, in line with our findings, another similar study also reported very limited value in using baseline structural MRI to predict pharmacological treatment outcome in MDD (Beliveau et al. 2022). Moreover, a large multi‐center study also reported no differences in pre‐treatment or follow‐up cortical thickness between responders and non‐responders (Suh et al. 2020). These divergent findings underscore the possibly complex relationship between cortical thickness and treatment response in MDD, which is further illustrated by a whole‐brain imaging study demonstrating intricate patterns of cortical and subcortical regions involved in the prediction of remission and residual symptoms in MDD (Costafreda et al. 2009). However, as these previous studies investigated associations with treatment response at the group level and not at the individual level, our findings cannot directly be compared to these previous findings. In addition, although subcortical regions such as the hippocampus have previously been suggested as a strong predictor for antidepressant treatment outcomes (Colle et al. 2018; Fu, Steiner, and Costafreda 2013; Hu et al. 2019; MacQueen et al. 2008), the inclusion of subcortical data in our exploratory analyses did not significantly alter model performance. However, it should be noted that only volumetric measures were available for subcortical data. This prevented the use of more sophisticated machine learning models (e.g., deep learning residual network) for the individualized pharmacotherapeutic treatment response predictions and, therefore, these findings should be interpreted with caution.
4.2. Effect of Model Configurations on Performance
For our second hypothesis, we tested several common machine learning pipeline configurations. Contrary to our expectations, we generally did not find a significant performance difference when including clinical data, nor for more complex data representations and different machine learning models. One outlier was a significant difference in performance for different data representations, but performance here was still not significantly better than chance level. Similarly, other studies suggest that deep learning does not consistently outperform classical machine learning when applied to high‐dimensional data and relatively low sample sizes (Sajjadian et al. 2021; Squarcina et al. 2021).
Our results do not demonstrate an association between baseline clinical symptomatology and treatment outcome. This is in accordance with some (Friedman et al. 2012; Klein, Shankman, and Rose 2008) but in contrast to other (Perlman et al. 2019; Uher et al. 2012) previous works. This heterogeneity in findings is one of the reasons why the on‐going search for additional biomarkers for treatment response remains so relevant to date (Rost, Binder, and Brückl 2023). As prior studies by Rajpurkar et al. (2020) and Poirot et al. (2024) have shown that symptomatological predictors can be integrated with machine learning to improve predictive power, we additionally included clinical symptomatology to our machine learning approach. However, we found that including these predictors did not significantly boost model performance. However, our results are severely limited by the lack of models that performed well, which reduces any effect that can be expected in the comparisons we have made between machine learning models. This also limits the generalizability of these findings to other predictive studies. At the same time, it provides thorough evidence to researchers that cortical structural properties may contain limited information and alternative approaches such as other MRI sequences (Squarcina et al. 2021), multimodal data (Sajjadian et al. 2021) or alternative analysis methods such as normative modeling (Rutherford et al. 2023) may prove more fruitful.
4.3. Exploratory Subpopulation Analyses
Our exploratory analyses suggest that predicting pharmacotherapeutic treatment response on an individual level is only feasible when limiting the sample using a variable that is defined at the outcome level. Here, we selected a subpopulation of patients with MDD exhibiting either the least or the most improvement in clinical symptoms following pharmacological treatment. Therefore, we advise caution in interpreting these findings as we selectively examined a particularly distinct subpopulation with limited generalizability to a clinical population. Our findings seem to suggest the presence of distinct populations within the MDD population, based on the association between cortical structural MRI and treatment outcome. These results appear to indicate the presence of biologically distinct response‐related subpopulations within MDD. Supporting this, the most influential predictors within this subgroup are cortical structural features rather than clinical variables. As response prediction was only feasible for individuals showing the most and least improvement in symptoms, it is possible that antidepressant response in MDD cannot readily be characterized using a continuous spectrum. A similar observation was made in a study by Deserno et al. (2022), who investigated phenotypic clustering based on questionnaire data in a combined sample of attention–deficit hyperactivity disorder and autism spectrum disorder populations. They similarly found only clear clusters of more extreme symptom‐related subgroups, but no clear segregation between the other individuals. However, the extent and underlying mechanisms of potential biological differences in MDD subpopulations remain unclear and further research is needed to elucidate these aspects.
The predictors that appear to contribute most to the prediction of treatment response in the extreme (non‐)responders subpopulation were located in the precentral gyri (higher cortical thickness and smaller surface area) and superior frontal gyri (larger surface area). Interestingly, prior studies have shown that individuals with MDD tend to have lower cortical thickness in the precentral gyrus and lower gray matter volumes compared to healthy volunteers (Xiong et al. 2019; Zhang et al. 2012). Prior studies have also highlighted alterations in gray matter volume (Lai and Wu 2014) and functional connectivity (Yang et al. 2017; Zhu et al. 2022) of the superior frontal gyrus in individuals with MDD compared to healthy controls. A systematic review by (Porta‐Casteràs et al. 2021) also recently reported increased functional connectivity of the superior frontal and middle frontal gyri following electroconvulsive therapy. Taken together, the brain regions contributing most to the model may tentatively be implicated in the pathophysiology of MDD and altered following treatment, although the underlying mechanisms involved are not yet understood.
4.4. Strengths and Limitations
We performed the largest mega‐analysis to date, using data from six cohorts from the ENIGMA MDD consortium, the largest consortium for MDD neuroimaging research (Dinga et al. 2018). The homogeneity of measurement strategies across settings is of paramount importance for prediction research (Luijken et al. 2019). Preprocessing of the structural data was standardized across all participating sites. Crucially, a previous ML study using data from the ENIGMA‐MDD consortium showed that remaining site‐effects may still affect classification outcomes (Belov et al. 2022). Therefore, we used ComBat to remove remaining site‐effects in a step called harmonization. Since site differences that are not modeled explicitly are regressed out as a site‐effect, harmonization potentially reduces the signal‐to‐noise ratio (Orlhac et al. 2022). In our exploratory analyses, we further investigated the effect of heterogeneity in the sample by selecting more homogeneous subpopulations and testing SKF‐CV against LSO‐CV. Model performance for these subpopulations did not increase significantly, and LSO‐performance was comparable to SKF‐CV, suggesting that this heterogeneity is not the main factor contributing to the difficulty of predicting treatment response using cortical MRI predictors.
In our mega‐analysis approach, patients were drawn from various international cohorts. Smaller and more homogeneous samples run the risk of overfitting and reduced out‐of‐sample performance, as noted in previous studies (Bracher‐Smith, Crawford, and Escott‐Price 2021). Notably, reviews by Sajjadian et al. (2021) and Flint et al. (2021) report an inverse correlation between sample size and accuracy, with smaller samples tending to yield higher reported accuracies. Both decreased overfitting and increased homogeneity in the current sample may account for the low performance observed in the present study compared to previous studies reviewed by Squarcina et al. (2021). This discrepancy underscores the importance of sample size and diversity considerations, as well as standardized protocols, to reliably evaluate the performance of machine learning models for personalized medicine.
Our analyses were pre‐registered and approved by the ENIGMA MDD consortium. By pre‐registering our analyses before receiving the data, we avoid inadvertently tuning our analyses to our data (Smith and Ebrahim 2002). However, this preregistration also limited the scope of this work to analyses of cortical structural data in the entire population. To retain a clear distinction between pre‐registered and post hoc analyses, while expanding the predefined scope with additional analyses, we explicitly report non‐preregistered analyses as exploratory.
As an ENIGMA MDD mega‐analysis on the prediction of individual pharmacological treatment response, major strengths of our investigation include the large sample size, the preregistered analyses, and the use of various machine learning pipeline configurations. The sample size specifically, is on the higher end of the field (Sajjadian et al. 2021) and within the estimated range of the number of required samples of 100–300 (Beleites et al. 2013; Luedtke, Sadikova, and Kessler 2019). However, drawbacks remain. We were unable to investigate whether the inclusion of additional MRI modalities contributes to model performance, as this data was not available for the current study. In addition, we included pre‐existing data from international populations, which means that study design and data collection techniques were not standardized across sites. Many factors, including for example the inclusion criteria, the timing of pre‐treatment MRI scans, treatment duration, use of additional medication, timepoints during which response is determined, and study design (naturalistic follow‐up vs. clinical trials), remained unstandardized across the included sites. In addition, further heterogeneity may be introduced by variation in cultural variation in the clinical presentation of depression (Kirmayer 2001) and the clinical assessment of depression symptom severity (e.g., inter‐rater variability and diagnostic tools used per site). It should be acknowledged that these sources of variation may have a considerable effect on model performance and the outcomes of our mega‐analysis. In addition, it is currently unclear how the choices of MRI processing impact performance of prediction models. It is possible that, in this study, possibly clinically relevant features in cortical structural MRI data are omitted by the (pre‐)processing and dimensionality reduction choices made.
4.5. Future Directions
While this study used a single imaging modality to develop personalized predictions of treatment response to antidepressant medication, incorporating multiple predictors across modalities may increase accuracy. A meta‐analysis by Lee et al. (2018) found that combined or multi‐modal predictors performed better than any single modality alone, which was recently corroborated by a similar machine‐learning based prediction study for pharmacological treatment outcome in depression (Poirot et al. 2024). Although accuracy in our study did not improve when adding clinical data, another study that focused solely on clinical and behavioral predictors was able to predict pharmacological treatment response with high accuracy (Zhou et al. 2022). In addition, Poirot et al. (2024) reported that including clinical data to multi‐modal MRI‐based predictors boosted model performance. These results suggest that future research may benefit from using machine learning algorithms incorporating multimodal data to predict pharmacotherapeutic treatment response in individuals with MDD.
Another method that may boost model performance is to obtain early change information (e.g., after 1 or 2 weeks of treatment) in cortical structural MRI‐derived measures for predictions later in treatment, or to consider including multiple time points. A study by Bartlett et al. (2018) observed that the change in anterior cingulate cortical thickness during the first week of treatment predicts treatment response to SSRIs. Harris et al. (2022) also showed that models using MRI predictors obtained both pre‐treatment and 2 weeks after treatment initiation outperformed models using one of these measures. Since this data was unavailable, we were unable to assess the effect of adding the information to the analyses in the current study.
Encouraging findings have also emerged from studies exploring alternative approaches. For example, studies considering heterogeneity in symptom profiles, for example assessed with psychometric network modeling, have shown distinctions in symptom network structure between remitters and non‐remitters following pharmacotherapeutic treatment (van Borkulo et al. 2015). This highlights the potential of these methods for unraveling interindividual variations in depression symptom presentation in relation to treatment outcome. Alternatively, a recent study using normative models of structural MRI measures and brain functional connectivity demonstrated promising results in group difference testing and classification tasks (Rutherford et al. 2023). Although the application of normative modeling for complex classifications such as individual pharmacological response prediction has not been evaluated, further investigations of this method's efficacy may prove fruitful.
5. Conclusion
In this mega‐analysis of cortical structural MRI in 265 individuals with MDD based on data from the ENIGMA‐MDD cohort, we provide compelling evidence that cortical structural MRI alone is not a reliable predictor of individualized pharmacotherapeutic treatment response in MDD. This finding was observed consistently across the machine learning pipeline configurations we employed, which included the majority of predictive methods commonly covered in the literature. Specifically, we varied the cortical data representation, the inclusion of clinical data, machine learning model, and CV scheme. Findings from our exploratory subpopulation analyses, however, suggest the potential of cortical structural measures, alone or combined with subcortical volumetric measures, in predicting antidepressant treatment response for MDD patients, linking distinct patterns to the most and least responsive individuals. To improve the accuracy of personalized treatment response prediction, we suggest further evaluation of alternative approaches, such as integrating multiple imaging modalities.
Conflicts of Interest
M. W. A. Caan is a shareholder of Nico‐lab International Ltd. Dr. H. G. Ruhe received speaking fees from Lundbeck and Janssen, and grants from ZonMW, Hersenstichting, the Dutch ministry of health and an unrestricted educational grant from Janssen. All other authors declare no financial relationships with commercial interests.
Supporting information
Data S1: Supporting Information.
Acknowledgments
This work was done with the ENIGMA major depressive disorder (ENIGMA‐MDD) working group and was supported by the Eurostars funding program (Reference number 113351 DEPREDICT). AS is supported by a Dutch Research Council Veni grant (016.196.153). LS is supported by an National Health and Medical Research Council (NHMRC) Investigator grant 2017962. VB and RGM were supported by the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF: 01 ZX 1507, “PreNeSt – e:Med”) and the University Medical Center Göttingen. LH was funded by the Rubicon award (grant number 452020227) from the Dutch NWO. CC was funded by grants R01 MH129742‐01, R56 AG058854, R01 MH116147. The Minnesota cohort was funded by the National Institute of Mental Health (K23MH090421), the National Alliance for Research on Schizophrenia and Depression, the University of Minnesota Graduate School, the Minnesota Medical Foundation, and the Biotechnology Research Center (P41 RR008079 to the Center for Magnetic Resonance Research), University of Minnesota, and the Deborah E. Powell Center for Women's Health Seed Grant, University of Minnesota. BJH, CGD, AJJ acknowledge that the data collected in Melbourne (Australia) was supported by National Health and Medical Research Council of Australia (NHMRC) Project Grants 1064643 (principal investigator, BJH) and 1024570 (principal investigator, CGD). The DEP‐ARREST‐CLIN cohort was supported by ANR SAMENTA 2012. Funding for the San Raffaele cohort was provided by RF‐2018‐12367249 by the Italian Ministry of Health; A_201779W93T by the Italian Ministry of University and Scientific Research. BCD is supported by a NHMRC CJ Martin Fellowship (app 1161356). The Hiroshima cohort was supported by the Japan Agency for Medical Research and Development (AMED, Grant Number: JP18dm0307002). NJ is supported by the National Institutes of Health, grants R01 MH134004, and R01 MH117601. RGM, NJ, SIT, and PMT are supported in part by U.S. NIH grant R01 MH131806. SIT was supported by R56 AG058854 (The ENIGMA World Aging Center); R01 MH116147 (The ENIGMA Sex Differences Initiative), R01 MH129742‐01 (The ENIGMA Bipolar Initiative). The ENIGMA Working Group acknowledges the NIH Big Data to Knowledge (BD2K) award for foundational support and consortium development (U54EB020403 to Paul M. Thompson). The ENIGMA‐Major Depressive Disorder working group is supported by the National Institute of Mental Health grant numbers MH129832, MH117601 and MH129742. For a complete list of ENIGMA‐related grant support please see: http://enigma.ini.usc.edu/about‐2/funding/.
Funding: This work was supported by Ministry of health, Italy, RF‐2018‐12367249 Ministry of University and Scientific Research, Italy, A_201779W93T. Japan Agency for Medical Research and Development, JP18dm0307002. Biotechnology Research Center, P41 RR008079 Eurostars, 113351. Nederlandse Organisatie voor Wetenschappelijk Onderzoek, Rubicon 452020227, Veni 016.196.153. National Institute of Mental Health, K23MH090421, MH117601, MH129742, MH129832, R01 MH116147, R01 MH129742‐01, R01 MH131806, R01 MH134004. National Health and Medical Research Council, CJ Martin Fellowship 1161356, Investigator grant 1024570, Investigator grant 1064643, Investigator grant 2017962. Bundesministerium für Bildung und Forschung, 01 ZX 1507. National Institute of Aging, R56 AG058854.
Maarten G. Poirot, Daphne E. Boucherie, Henricus G. Ruhe and Liesbeth Reneman are contributed equally to this study.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- Anderson, M. J. 2005. Permutational Multivariate Analysis of Variance. Wuhan, China: Scientific Research Publishing. [Google Scholar]
- Bartlett, E. A. , DeLorenzo C., Sharma P., et al. 2018. “Pretreatment and Early‐Treatment Cortical Thickness Is Associated With SSRI Treatment Response in Major Depressive Disorder.” Neuropsychopharmacology 43, no. 11: 2221–2230. 10.1038/s41386-018-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beleites, C. , Neugebauer U., Bocklitz T., Krafft C., and Popp J.. 2013. “Sample Size Planning for Classification Models.” Analytica Chimica Acta 760: 25–33. 10.1016/j.aca.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Beliveau, V. , Hedeboe E., Fisher P. M., et al. 2022. “Generalizability of Treatment Outcome Prediction in Major Depressive Disorder Using Structural MRI: A NeuroPharm Study.” Neuroimage Clinical 36: 103224. 10.1016/j.nicl.2022.103224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov, V. , Erwin‐Grabner T., Gonul A. S., et al. 2022. “Global Multi‐Site Benchmark Classification of Major Depressive Disorder Using Machine Learning on Cortical and Subcortical Features of 5,365 Participants From the ENIGMA MDD Dataset.” arXiv Preprint arXiv:2206.08122.
- Bracher‐Smith, M. , Crawford K., and Escott‐Price V.. 2021. “Machine Learning for Genetic Prediction of Psychiatric Disorders: A Systematic Review.” Molecular Psychiatry 26, no. 1: 70–79. 10.1038/s41380-020-0825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromet, E. , Andrade L. H., Hwang I., et al. 2011. “Cross‐National Epidemiology of DSM‐IV Major Depressive Episode.” BMC Medicine 9, no. 1: 90. 10.1186/1741-7015-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody, T. J. , Rush A. J., Bernstein I., et al. 2006. “The Montgomery Äsberg and the Hamilton Ratings of Depression: A Comparison of Measures.” European Neuropsychopharmacology 16, no. 8: 601–611. 10.1016/j.euroneuro.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavadia, P. , Teo I., Poremski D., Fung D. S. S., and Finkelstein E. A.. 2023. “Prevalence and Economic Burden of Depression and Anxiety Symptoms Among Singaporean Adults: Results From a 2022 Web Panel.” BMC Psychiatry 23, no. 1: 104. 10.1186/s12888-023-04581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani, A. , Furukawa T. A., Salanti G., et al. 2018. “Comparative Efficacy and Acceptability of 21 Antidepressant Drugs for the Acute Treatment of Adults With Major Depressive Disorder: A Systematic Review and Network Meta‐Analysis.” Lancet 391, no. 10128: 1357–1366. 10.1016/S0140-6736(17)32802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, S. E. , Zantvoord J. B., Wezenberg B. N., Bockting C. L. H., and van Wingen G. A.. 2021. “Magnetic Resonance Imaging for Individual Prediction of Treatment Response in Major Depressive Disorder: A Systematic Review and Meta‐Analysis.” Translational Psychiatry 11, no. 1: 168. 10.1038/s41398-021-01286-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colle, R. , Dupong I., Colliot O., et al. 2018. “Smaller Hippocampal Volumes Predict Lower Antidepressant Response/Remission Rates in Depressed Patients: A Meta‐Analysis.” World Journal of Biological Psychiatry 19, no. 5: 360–367. 10.1080/15622975.2016.1208840. [DOI] [PubMed] [Google Scholar]
- Costafreda, S. G. , Chu C., Ashburner J., and Fu C. H.. 2009. “Prognostic and Diagnostic Potential of the Structural Neuroanatomy of Depression.” PLoS One 4, no. 7: e6353. 10.1371/journal.pone.0006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J. , Dong W., Socher R., Li L.‐J., Li K., and Li F.‐F.. 2009. “ImageNet: A Large‐Scale Hierarchical Image Database.” In 2009 IEEE Conference on Computer Vision and Pattern Recognition, 248–255. New Jersey, United States: IEEE. [Google Scholar]
- Deserno, M. K. , Bathelt J., Groenman A. P., and Geurts H. M.. 2022. “Probing the Overarching Continuum Theory: Data‐Driven Phenotypic Clustering of Children With ASD or ADHD.” European Child & Adolescent Psychiatry 32: 1909–1923. 10.1007/s00787-022-01986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan, R. S. , Segonne F., Fischl B., et al. 2006. “An Automated Labeling System for Subdividing the Human Cerebral Cortex on MRI Scans Into Gyral Based Regions of Interest.” NeuroImage 31, no. 3: 968–980. 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dinga, R. , Marquand A. F., Veltman D. J., et al. 2018. “Predicting the Naturalistic Course of Depression From a Wide Range of Clinical, Psychological, and Biological Data: A Machine Learning Approach.” Translational Psychiatry 8, no. 1: 241. 10.1038/s41398-018-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, M. D. 2004. “Permutation Methods: A Basis for Exact Inference.” Statistical Science 19, no. 4: 676–685. [Google Scholar]
- Fang, Y. , Scott L., Song P., Burmeister M., and Sen S.. 2020. “Genomic Prediction of Depression Risk and Resilience Under Stress.” Nature Human Behaviour 4, no. 1: 111–118. 10.1038/s41562-019-0759-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. 2012. “FreeSurfer.” NeuroImage 62, no. 2: 774–781. 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, C. , Cearns M., Opel N., et al. 2021. “Systematic Misestimation of Machine Learning Performance in Neuroimaging Studies of Depression.” Neuropsychopharmacology 46, no. 8: 1510–1517. 10.1038/s41386-021-01020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseka, T. M. , MacQueen G. M., and Kennedy S. H.. 2018. “Neuroimaging Biomarkers as Predictors of Treatment Outcome in Major Depressive Disorder.” Journal of Affective Disorders 233: 21–35. 10.1016/j.jad.2017.10.049. [DOI] [PubMed] [Google Scholar]
- Friedman, E. S. , Davis L. L., Zisook S., et al. 2012. “Baseline Depression Severity as a Predictor of Single and Combination Antidepressant Treatment Outcome: Results From the CO‐MED Trial.” European Neuropsychopharmacology 22, no. 3: 183–199. 10.1016/j.euroneuro.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C. H. , Steiner H., and Costafreda S. G.. 2013. “Predictive Neural Biomarkers of Clinical Response in Depression: A Meta‐Analysis of Functional and Structural Neuroimaging Studies of Pharmacological and Psychological Therapies.” Neurobiology of Disease 52: 75–83. 10.1016/j.nbd.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Gao, S. , Calhoun V. D., and Sui J.. 2018. “Machine Learning in Major Depression: From Classification to Treatment Outcome Prediction.” CNS Neuroscience & Therapeutics 24, no. 11: 1037–1052. 10.1111/cns.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, P. E. , Fournier A.‐A., Sisitsky T., et al. 2021. “The Economic Burden of Adults With Major Depressive Disorder in the United States (2010 and 2018).” PharmacoEconomics 39, no. 6: 653–665. 10.1007/s40273-021-01019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrewijn, A. , Cardinale E. M., Groenewold N. A., et al. 2021. “Cortical and Subcortical Brain Structure in Generalized Anxiety Disorder: Findings From 28 Research Sites in the ENIGMA‐Anxiety Working Group.” Translational Psychiatry 11, no. 1: 1–15. 10.1038/s41398-021-01622-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, J. K. , Hassel S., Davis A. D., et al. 2022. “Predicting Escitalopram Treatment Response From Pre‐Treatment and Early Response Resting State fMRI in a Multi‐Site Sample: A CAN‐BIND‐1 Report.” NeuroImage Clinical 35: 103120. 10.1016/j.nicl.2022.103120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K. , Zhang X., Ren S., and Sun J.. 2016. “Deep Residual Learning for Image Recognition.” In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778. New Jersey, United States: IEEE. [Google Scholar]
- Hu, X. , Zhang L., Hu X., et al. 2019. “Abnormal Hippocampal Subfields May Be Potential Predictors of Worse Early Response to Antidepressant Treatment in Drug‐Naive Patients With Major Depressive Disorder.” Journal of Magnetic Resonance Imaging 49, no. 6: 1760–1768. 10.1002/jmri.26520. [DOI] [PubMed] [Google Scholar]
- Janssen, R. J. , Mourao‐Miranda J., and Schnack H. G.. 2018. “Making Individual Prognoses in Psychiatry Using Neuroimaging and Machine Learning.” Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 3, no. 9: 798–808. 10.1016/j.bpsc.2018.04.004. [DOI] [PubMed] [Google Scholar]
- Kingma, D. P. , and Ba J.. 2014. “Adam: A Method for Stochastic Optimization.” arXiv Preprint arXiv:1412.6980 .
- Kirmayer, L. J. 2001. “Cultural Variations in the Clinical Presentation of Depression and Anxiety: Implications for Diagnosis and Treatment.” Journal of Clinical Psychiatry 62: 22–30. [PubMed] [Google Scholar]
- Klein, D. N. , Shankman S. A., and Rose S.. 2008. “Dysthymic Disorder and Double Depression: Prediction of 10‐Year Course Trajectories and Outcomes.” Journal of Psychiatric Research 42, no. 5: 408–415. 10.1016/j.jpsychires.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok, M. P. C. , van Eijndhoven P. F., Argyelan M., Schene A. H., and Tendolkar I.. 2019. “Structural Brain Characteristics in Treatment‐Resistant Depression: Review of Magnetic Resonance Imaging Studies.” BJPsych Open 5, no. 5: e76. 10.1192/bjo.2019.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.‐H. , and Wu Y.‐T.. 2014. “Frontal‐Insula Gray Matter Deficits in First‐Episode Medication‐naïve Patients With Major Depressive Disorder.” Journal of Affective Disorders 160: 74–79. 10.1016/j.jad.2013.12.036. [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Ragguett R. M., Mansur R. B., et al. 2018. “Applications of Machine Learning Algorithms to Predict Therapeutic Outcomes in Depression: A Meta‐Analysis and Systematic Review.” Journal of Affective Disorders 241, no. March: 519–532. 10.1016/j.jad.2018.08.073. [DOI] [PubMed] [Google Scholar]
- Luedtke, A. , Sadikova E., and Kessler R. C.. 2019. “Sample Size Requirements for Multivariate Models to Predict Between‐Patient Differences in Best Treatments of Major Depressive Disorder.” Clinical Psychological Science 7, no. 3: 445–461. 10.1177/2167702618815466. [DOI] [Google Scholar]
- Luijken, K. , Groenwold R. H. H., Van Calster B., Steyerberg E. W., and van Smeden M.. 2019. “Impact of Predictor Measurement Heterogeneity Across Settings on the Performance of Prediction Models: A Measurement Error Perspective.” Statistics in Medicine 38, no. 18: 3444–3459. 10.1002/sim.8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen, G. M. , Yucel K., Taylor V. H., Macdonald K., and Joffe R.. 2008. “Posterior Hippocampal Volumes are Associated With Remission Rates in Patients With Major Depressive Disorder.” Biological Psychiatry 64, no. 10: 880–883. 10.1016/j.biopsych.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Orlhac, F. , Eertink J. J., Cottereau A. S., et al. 2022. “A Guide to ComBat Harmonization of Imaging Biomarkers in Multicenter Studies.” Journal of Nuclear Medicine 63, no. 2: 172–179. 10.2967/jnumed.121.262464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa, F. , Varoquaux G., Gramfort A., et al. 2011. “Scikit‐Learn: Machine Learning in Python.” Journal of Machine Learning Research 12: 2825–2830. [Google Scholar]
- Perlman, K. , Benrimoh D., Israel S., et al. 2019. “A Systematic Meta‐Review of Predictors of Antidepressant Treatment Outcome in Major Depressive Disorder.” Journal of Affective Disorders 243: 503–515. 10.1016/j.jad.2018.09.067. [DOI] [PubMed] [Google Scholar]
- Phillips, J. L. , Batten L. A., Tremblay P., Aldosary F., and Blier P.. 2015. “A Prospective, Longitudinal Study of the Effect of Remission on Cortical Thickness and Hippocampal Volume in Patients With Treatment‐Resistant Depression.” International Journal of Neuropsychopharmacology 18, no. 8: pyv037. 10.1093/ijnp/pyv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson, B. , and Smyth G. K.. 2010. “Permutation P‐Values Should Never Be Zero: Calculating Exact P‐Values When Permutations Are Randomly Drawn.” Statistical Applications in Genetics and Molecular Biology 9: 39. 10.2202/1544-6115.1585. [DOI] [PubMed] [Google Scholar]
- Pimontel, M. A. , Solomonov N., Oberlin L., et al. 2021. “Cortical Thickness of the Salience Network and Change in Apathy Following Antidepressant Treatment for Late‐Life Depression.” American journal of geriatric psychiatry 29, no. 3: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirot, M. G. , Ruhe H. G., Mutsaerts H. M. M., et al. 2024. “Treatment Response Prediction in Major Depressive Disorder Using Multimodal MRI and Clinical Data: Secondary Analysis of a Randomized Clinical Trial.” American Journal of Psychiatry 181, no. 3: 223–233. 10.1176/appi.ajp.20230206. [DOI] [PubMed] [Google Scholar]
- Poirot, M. , Boucherie D., Reneman L., and Schrantee A.. 2023. “Antidepressant Treatment Response Prediction Using Cortical Structural Magnetic Resonance Imaging: A Mega‐Analysis on the ENIGMA MDD Working Group.” https://github.com/DEPREDICT/ENIGMA/,version=0.1.
- Porta‐Casteràs, D. , Cano M., Camprodon J. A., et al. 2021. “A Multimetric Systematic Review of fMRI Findings in Patients With MDD Receiving ECT.” Progress in Neuro‐Psychopharmacology and Biological Psychiatry 108: 110178. 10.1016/j.pnpbp.2020.110178. [DOI] [PubMed] [Google Scholar]
- Proudman, D. , Greenberg P., and Nellesen D.. 2021. “The Growing Burden of Major Depressive Disorders (MDD): Implications for Researchers and Policy Makers.” PharmacoEconomics 39, no. 6: 619–625. 10.1007/s40273-021-01040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurkar, P. , Yang J., Dass N., et al. 2020. “Evaluation of a Machine Learning Model Based on Pretreatment Symptoms and Electroencephalographic Features to Predict Outcomes of Antidepressant Treatment in Adults With Depression: A Prespecified Secondary Analysis of a Randomized Clinical Trial.” JAMA Network Open 3, no. 6: e206653. 10.1001/jamanetworkopen.2020.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, N. , Binder E. B., and Brückl T. M.. 2023. “Predicting Treatment Outcome in Depression: An Introduction Into Current Concepts and Challenges.” European Archives of Psychiatry and Clinical Neuroscience 273, no. 1: 113–127. 10.1007/s00406-022-01418-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush, A. J. , Warden D., Wisniewski S. R., et al. 2009. “STAR*D Revising Conventional Wisdom.” CNS Drugs 23, no. 8: 627–647. [DOI] [PubMed] [Google Scholar]
- Rush, A. J. , Trivedi M. H., Wisniewski S. R., et al. 2006. “Acute and Longer‐Term Outcomes in Depressed Outpatients Requiring One or Several Treatment Steps: A STAR*D Report.” American Journal of Psychiatry 163: 1905–1917. [DOI] [PubMed] [Google Scholar]
- Rutherford, S. , Barkema P., Tso I. F., et al. 2023. “Evidence for embracing normative modeling.” eLife 12: e85082. 10.7554/elife.85082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjadian, M. , Lam R. W., Milev R., et al. 2021. “Machine Learning in the Prediction of Depression Treatment Outcomes: A Systematic Review and Meta‐Analysis.” Psychological Medicine 51, no. 16: 2742–2751. 10.1017/S0033291721003871. [DOI] [PubMed] [Google Scholar]
- Santarsieri, D. , and Schwartz T. L.. 2015. “Antidepressant Efficacy and Side‐Effect Burden: A Quick Guide for Clinicians.” Drugs in Context 4: 212290. 10.7573/dic.212290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Hibar D. P., Sämann P. G., et al. 2017. “Cortical Abnormalities in Adults and Adolescents With Major Depression Based on Brain Scans From 20 Cohorts Worldwide in the ENIGMA Major Depressive Disorder Working Group.” Molecular Psychiatry 22, no. 6: 900–909. 10.1038/mp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Pozzi E., Ho C. T., et al. 2020. “ENIGMA MDD: Seven Years of Global Neuroimaging Studies of Major Depression Through Worldwide Data Sharing.” Translational Psychiatry 10, no. 1: 172. 10.1038/s41398-020-0842-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrantee, A. , Ruhé H. G., and Reneman L.. 2020. “Psychoradiological Biomarkers for Psychopharmaceutical Effects.” Neuroimaging Clinics of North America 30, no. 1: 53–63. 10.1016/j.nic.2019.09.006. [DOI] [PubMed] [Google Scholar]
- Shrout, P. E. , and Rodgers J. L.. 2018. “Psychology, Science, and Knowledge Construction: Broadening Perspectives From the Replication Crisis.” Annual Review of Psychology 69: 487–510. 10.1146/annurev-psych-122216-011845. [DOI] [PubMed] [Google Scholar]
- Smith, G. D. , and Ebrahim S.. 2002. “Data Dredging, Bias, or Confounding.” British Medical Journal 352: 1437–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarcina, L. , Villa F. M., Nobile M., Grisan E., and Brambilla P.. 2021. “Deep Learning for the Prediction of Treatment Response in Depression.” Journal of Affective Disorders 281: 618–622. 10.1016/j.jad.2020.11.104. [DOI] [PubMed] [Google Scholar]
- Su, Z. , Zeng W., Shi R., Wang Y., Sun J., and Gu X.. 2013. Area Preserving Brain Mapping. Paper presented at The 2013 IEEE Conference on Computer Vision and Pattern Recognition.
- Suh, J. S. , Minuzzi L., Raamana P. R., et al. 2020. “An Investigation of Cortical Thickness and Antidepressant Response in Major Depressive Disorder: A CAN‐BIND Study Report.” NeuroImage Clinical 25: 102178. 10.1016/j.nicl.2020.102178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. M. , Jahanshad N., Ching C. R. K., et al. 2020. “ENIGMA and Global Neuroscience: A Decade of Large‐Scale Studies of the Brain in Health and Disease Across More Than 40 Countries.” Translational Psychiatry 10, no. 1: 100. 10.1038/s41398-020-0705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uher, R. , Perlis R. H., Henigsberg N., et al. 2012. “Depression Symptom Dimensions as Predictors of Antidepressant Treatment Outcome: Replicable Evidence for Interest‐Activity Symptoms.” Psychological Medicine 42, no. 5: 967–980. 10.1017/s0033291711001905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Borkulo, C. , Boschloo L., Borsboom D., Penninx B. W. J. H., Waldorp L. J., and Schoevers R. A.. 2015. “Association of Symptom Network Structure With the Course of Depression.” JAMA Psychiatry 72, no. 12: 1219–1226. 10.1001/jamapsychiatry.2015.2079. [DOI] [PubMed] [Google Scholar]
- Wu, P. , Zhang A., Sun N., et al. 2021. “Cortical Thickness Predicts Response Following 2 Weeks of SSRI Regimen in First‐Episode, Drug‐Naive Major Depressive Disorder: An MRI Study.” Frontiers in Psychiatry 12: 751756. 10.3389/fpsyt.2021.751756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Y. , Dong D., Cheng C., et al. 2019. “State‐Independent and Dependent Structural Alterations in Limbic‐Cortical Regions in Patients With Current and Remitted Depression.” Journal of Affective Disorders 258: 1–10. [DOI] [PubMed] [Google Scholar]
- Yang, X.‐H. , Tian K., Wang D.‐F., et al. 2017. “Anhedonia Correlates With Abnormal Functional Connectivity of the Superior Temporal Gyrus and the Caudate Nucleus in Patients With First‐Episode Drug‐Naive Major Depressive Disorder.” Journal of Affective Disorders 218: 284–290. 10.1016/j.jad.2017.04.053. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Yao S., Zhu X., Wang X., Zhu X., and Zhong M.. 2012. “Gray Matter Volume Abnormalities in Individuals With Cognitive Vulnerability to Depression: A Voxel‐Based Morphometry Study.” Journal of Affective Disorders 136: 443–452. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Zhang Z., Wang C., et al. 2022. “Predictors of 4‐Week Antidepressant Outcome in Patients With First‐Episode Major Depressive Disorder: An ROC Curve Analysis.” Journal of Affective Disorders 304: 59–65. 10.1016/j.jad.2022.02.029. [DOI] [PubMed] [Google Scholar]
- Zhu, Z. , Wang Y., Lau W. K. W., et al. 2022. “Hyperconnectivity Between the Posterior Cingulate and Middle Frontal and Temporal Gyrus in Depression: Based on Functional Connectivity Meta‐Analyses.” Brain Imaging and Behavior 16, no. 4: 1538–1551. 10.1007/s11682-022-00628-7. [DOI] [PubMed] [Google Scholar]
- Zisook, S. , Trivedi M. H., Warden D., et al. 2009. “Clinical Correlates of the Worsening or Emergence of Suicidal Ideation During SSRI Treatment of Depression: An Examination of Citalopram in the STAR⁎D Study.” Journal of Affective Disorders 117, no. 1: 63–73. 10.1016/j.jad.2009.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: Supporting Information.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


