Abstract
Coordination between growth and division is a fundamental feature of cells. In many syncytia, cell growth must couple with multiple nuclear divisions in one cytoplasm. In the fungus, Ashbya gossypii, cell-cycle progression and hyphal elongation require condensates formed by the protein Whi3 in complex with distinct mRNA species. We hypothesized the condensates may act through local translation regulation and find that Whi3 target mRNAs show distinct spatial biases in translation in vivo. Whi3-RNA condensates can both promote and repress RNA translation in an RNA- and condensate size-dependent manner in vitro. Interestingly, we observe a sub-condensate enrichment of translation that is tunable by RNA valency and protein phospho-state. Together, these data suggest that Whi3 condensates generate a continuum of translation states, resulting in asynchronous nuclear divisions coordinated with growth. This local regulation requires a minimal complement of molecular components at the nano scale to support global coordination at the cell scale.
Introduction
Large cells such as unicellular slime molds, filamentous fungi, alga, skeletal muscles and neurons occupy spatial scales at which diffusion becomes limiting for establishing patterning at distal sites1. These cells face a challenge of mounting local responses such as growth and chemotaxis in coordination with global cues from the environment or developmental programs. This two-fold challenge is exemplified in the growth of the multinucleate filamentous fungus Ashbya gossypii, in which the rate of nuclear division is coupled to the rate of hyphal elongation and branching morphogenesis to maintain a constant nucleus-to-cytoplasm ratio. However, nuclear divisions, hyphal elongation and branching all require precisely regulated biochemical programs at different locations within the cell that are often 100s of microns apart. Thus, cells like Ashbya need to maintain and regulate local cell cycles and polarized growth but do so in a way that results in global coordination of these activities across the whole cell. How such simultaneous global- and local biochemical control is achieved in these large cells is not well understood.
Biomolecular condensates can both locally control biochemistry and be responsive to signaling, serving as an attractive mechanism for coupling global cell states to a local process2–4. At the cell scale, condensates assemble and disassemble in response to internal or external cell cues5–9. This control manifests through post-translational modifications or direct environmental sensitivities that result in altered molecular interactions. The strength and specificity of interactions amongst components within the dense phase can in turn impact its composition and physiochemical properties such as pore size, charge and pH10–13. Thus, the fact that the fundamental molecular driving forces that regulate condensation are also sensitive to signaling make condensates responsive to cell-scale cues.
RNA-protein (RNP) condensates have long been appreciated as regulators of RNA translation. Stress granules and P-bodies are enriched for non-translating RNAs14,15. During embryogenesis, P granules can localize and regulate RNAs for specification and patterning of the germline16,17 and FXR1 granules show selective translation activation during spermiogenesis18. A series of recent papers focused on the ability of Drosophila germline granules to locally promote translation of specific RNAs, indicating that the condensate environment can enhance RNA translation19,20. RNP granules are also key passengers for long-range transport in neurons where the mRNAs generated in the nucleus are packaged and moved to distal sites for translation in response to stimulation and other cues3,21,22. Notably, across these varied contexts, RNP condensates must toggle between states where mRNAs are repressed and translated. Thus, an underappreciated property of mRNA-enriched condensates is the ability to both promote or repress translation of resident RNAs. However, the mechanisms controlling this kind of translation switch remain cryptic and whether this dual role in repression and activation of translation is a ubiquitous feature of mRNA condensates is unclear. In general, much effort in the condensate field has focused on binary descriptions of functions where the dense phase either activates or inhibits processes. There are likely many contexts, however, where condensates act as a rheostat, serving multiple and nuanced regulatory roles depending on the needs of the cell.
Our lab previously reported that the RNA-binding protein Whi3 forms cytoplasmic condensates that drive clustering of RNAs in distinct patterns in the cytosol of the fungus Ashbya gossypii. Whi3’s targets include the G1 cyclin mRNA CLN3 which is clustered near nuclei, and the formin mRNA BNI1 which is enriched at hyphal tips23,24. Perturbing Whi3’s glutamine-rich region (QRR) that drives condensation or Whi3’s ability to bind mRNA, both decrease mRNA clustering in the cell. In these mutants, not only do the mRNAs lose their spatial patterning, but this dramatically affects growth phenotypes such as nuclear division asynchrony and branching23,24. Here, we set out to understand the molecular basis underlying Whi3 condensate function aside from mRNA localization.
We began with the unexpected observation that in growing Ashbya hyphae, growth speed and nuclear asynchrony are both correlated with spatial heterogeneity in Whi3 condensates. These data suggested that condensate function may be temporally dynamic and spatially variable in these cells. Additionally, the Whi3 interactome is enriched for regulators of translation and RNA metabolism. We therefore used three orthogonal approaches to test the hypothesis that Whi3 regulates the translation of its target mRNAs in space. We first used a FISH-based strategy to probe spatial variation in ribosome association with Whi3 target mRNAs in cells. This revealed that CLN3 and BNI1 both show distinct patterns of spatial enrichment and repression of translation. We then measured the impact of condensates on bulk translation of mRNAs in cell lysates and observed that RNA identity, number of Whi3 binding sites, condensate size and Whi3 concentration can all modulate the translation of mRNAs. Finally, we used an imaging-based approach involving a real-time translation readout in condensates and observed a striking pattern of intra-condensate positional bias in translation. Together, these lines of evidence point strongly to the ability of Whi3-RNA condensates to both activate and repress protein translation by integrating RNA sequence information along with signaling cues. We propose that translation state-tuning allows Whi3 condensates to serve as responsive hubs that regulate local protein abundance while integrating various cellular cues to coordinate cell-scale processes.
Results
Whi3 condensates vary in appearance throughout the cell
We previously reported that Whi3 forms spatially distinct condensates that localize G1 cyclin mRNA CLN3 near nuclei, and formin mRNA BNI1 to hyphal tips23,24. To visualize how Whi3 condensate localization is correlated with cell growth we performed 3D time-lapse fluorescence imaging of Whi3:tdTomato using lattice light sheet microscopy. We observed a surprising negative correlation between proximity of Whi3 condensates to the hyphal tip and the hyphal elongation rate. Hyphae without a visible Whi3 punctum near the tip showed faster elongation compared to those with a punctum (Figure 1A, B).
Figure 1. Whi3 condensate variability at hyphal tips and around nuclei is correlated with variability in hyphal elongation and cell cycle state.
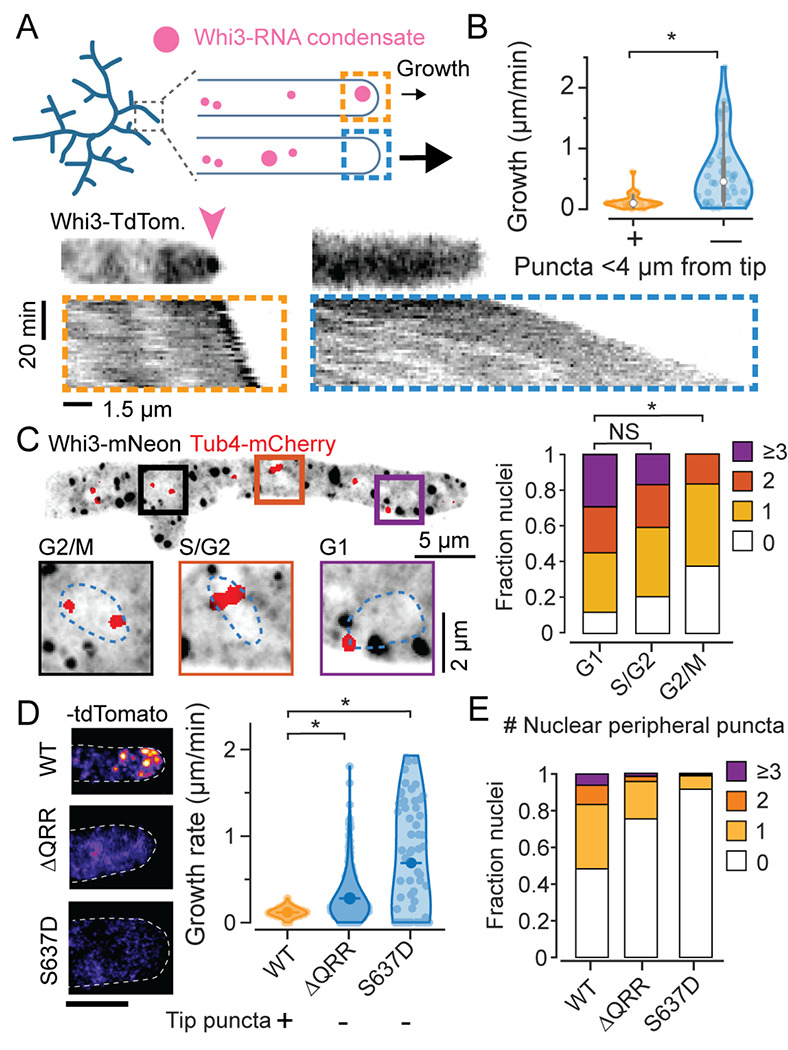
A. Schematic illustration of Whi3 condensates and tip elongation in Ashbya gossypii (top) and representative timelapse kymographs of hypha expressing Whi3:tdTomato (bottom). Kymographs were created from MIPs of 3D lightsheet timelapses. Orange box shows an example of a hypha with a prominent visible condensate at tip, marked by a pink arow, and the blue box shows an example of a hypha without a large tip-proximal condensate. Scale bar x: 1.5 μm, time: 20 min, origin on top left.
B. Instantaneous hyphal growth extracted from a total of 21 movies, split into 40 clips with or without tip proximal condensate (<4 μm from tip) each. * = p<0.01 using KS test.
C. Representative MIP of Tub4-mCherry expressing plasmidic Whi3-mNeon, scale bar 5 μm. Insets shows G1, S/G2 and G2/M nuclei with variable numbers of nuclear-peripheral Whi3 puncta, scale bar 2 μm. Stacked histograms quantifying relative nuclear proportions having varying peri-nuclear Whi3 puncta counts pooled from 3 biological replicates from a total of 238 G1, 88 S/G2, 24 G2/M phase nuclei. * = p<0.01 using KS test.
D. Representative images of WT Whi3:mNeon, non-condensate-forming whi3-QRR:tdTomato and whi3 S637D:tdTomato Ashbya hyphae (left) and hyphal elongation rates (right) measured using ConA pulse labeling. N>40 hyphae from >10 cells per strain. Scale bar 5 μm.
E. Stacked histograms quantifying relative nuclear proportions having varying peri-nuclear Whi3 puncta counts, from WT Whi3-tdTomato, n=180, whi3(ΔQRR):tdTomato, n=148, whi3(S637D):tdTomato, n=194.
We next used high-resolution confocal reconstructions of Whi3:mNeon/Tub4-mCherry cells to determine the distribution of Whi3 puncta around nuclei, where they were previously shown to associate with the ER membrane25. The periphery of nuclei, defined as the region 500 nm around to the edge of a segmented nuclear boundary, showed considerable variability in the number and size of Whi3 puncta (representative image, insets and histogram in Figure 1C). Whi3 condensates also showed a clear cell-cycle-biased perinuclear localization, based on spindle pole appearance which was visualized using Tub4-mCherry26. Nuclei with two well separated spindle poles, representing late G2/M phases had fewer peri-nuclear condensates relative to G1 and S/G2 nuclei. Taken together, these data demonstrate that Whi3 condensates show high variability both in the vicinity of nuclei and at hyphal tips, which is associated with nuclear division asynchrony and variable hyphal elongation rates, respectively. We next examined if phenotypic variability was dependent on Whi3 condensation.
We measured hyphal elongation rates for hyphae in mutants that were previously reported to decrease condensation including a mutant lacking the Whi3 glutamine rich region (QRR) that drives condensation, and a phosphomimic of the conserved PKA phosphosite S6372,23,27. We confirmed previous observations of a strong reduction in tip condensates in the two mutant strains (Figure 1D, left) and found that both strains with decreased tip condensates showed significantly faster hyphal elongation rates (Figure 1D, right) than wildtype. These data suggest that the condensed state of Whi3 limits tip elongation rate.
We also quantified the distribution of perinuclear puncta. In Whi3-tdTomato cells, 48% of nuclei do not have a detectable Whi3 punctum, 35% of nuclei have a clear punctum in proximity, whereas ~15% of the nuclei showed >2 puncta (N=180 nuclei) (Figure 1E), demonstrating substantial variability in relative proportions of nuclei with proximal Whi3 puncta. However, whi3(ΔQRR) and whi3(S637D), both showed decreased overall condensate number and size, resulting in a dramatic reduction in the fraction of nuclei with a proximal condensate. Thus, these mutations abrogate condensate-associated function and decrease inter-nuclear condensate variability. This is consistent with the increase in nuclear division synchrony phenotype observed in these mutants2,24.
These data combined prompted us to consider how Whi3 condensates, aside from simply localizing target mRNAs, can mediate biochemical programs such as the cell-cycle and polarized growth.
Whi3 protein interacts with regulators of mRNA and translation
To determine molecular functions for Whi3 condensates, we identified the proteins that associate with Whi3 using an affinity purification protocol to enrich and isolate Whi3-condensates. Whi3 condensates proved difficult to isolate intact using standard IP-protocols, prompting us to design an alternative membrane-association affinity purification approach. Briefly, lipid-coated beads functionalized with Ni-NTA lipids were coated in recombinantly expressed 6xHis-Whi3. These beads, when incubated in Ashbya lysate (Figure 2A), recruit endogenous Whi3 which is evident in the enrichment of Whi3-tdTomato puncta on the beads as well as associated nucleic acids and proteins (Figure 2B). Bead-enriched proteins were analyzed by mass spectrometry (Figure 2A,C,D). Gene ontology analysis on identified proteins showed an enrichment translation regulators and RNA decay factors (Figure 2C; Figure S1A; Table S1). Notably, a majority of Whi3-protein interactions are RNA-dependent (Figure 2C,D; Figure S1A; 160 enriched proteins identified in non-treated lysates, 6 proteins identified RNAse A treated lysates). Thus, Whi3 is well positioned to function as a post-transcriptional regulator.
Figure 2. Whi3 condensates interact with regulators of translation and RNA metabolism.
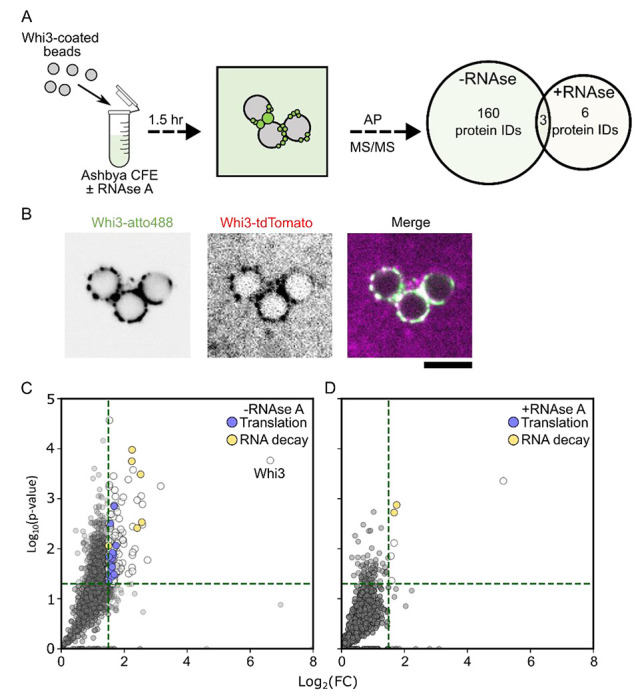
A. Whi3-assemblies can be recovered and purified from Ashbya cell-free extracts. Proteins identified are expressed as a Venn diagram indicating the number of enriched proteins for each treatment group. The central overlap indicates the number of unique protein IDs shared between the treatments.
B. Microspheres coated with recombinantly expressed Whi3 recruit endogenously expressed Whi3-tdTomato from Ashbya lysates.
C. Protein IDs corresponding to mass spectrometry identification plotted as a function of fold-change (FC) over the control condition (microspheres without recombinant Whi3). Green lines indicate the criteria for significance. Translation regulators are indicated in light-blue, and factors involved in RNA decay are shown in orange. Results are from three independently prepared Ashbya lysates (see methods).
D. Same as C for RNAse A treated lysates.
Whi3 target mRNAs show spatially variable translation
To examine the role of Whi3 condensates in translation regulation in vivo we performed polysome profiling on WT and whi3(ΔQRR). We reasoned that if Whi3 condensates primarily activated or repressed translation of its targets, the absence of condensates would result in a prominent difference in polysome association of Whi3 targets CLN3 and BNI1. While both RNAs showed strong polysome association (fractions 7-11), only CLN3 showed a detectable loss of a non-translating population in whi3(ΔQRR) relative to WT (fractions 4-5) (Figure S1B). This result suggests that while Whi3 condensates may differentially impact translation of its targets, condensate state does not completely reshape polysome association of these mRNAs in bulk. A limitation of polysome profiling is that it may not capture small subpopulations of transcripts or spatial variation of translation, which may be critically relevant for these transcripts that show strongly biased localizations. We therefore next tested the hypothesis that Whi3 targets show spatial variation in translation in cells.
To determine if Whi3 condensates locally regulate translation in vivo, we turned to the recently reported “tri-probe” proximity ligation (PL)-based strategy to detect ribosome associations with endogenous mRNAs28. Briefly, four and five sets of “padlock” and “primer” DNA probes tiling CLN3 and BNI1 respectively were used in conjunction with five Ag18s rRNA splint probes. The splint probes allowed ligation of proximal padlock probes by T4 ligase, creating a circular ssDNA template for rolling circle amplification (RCA) by Phi29 DNA polymerase. Rolling circle amplicons were detected via AlexaFluor647- or AlexaFluor555- labeled readout probes (Figure 3A). The method was successfully adapted to Ashbya, and we observed bright, punctate PL-RCA signal that robustly colocalized with smFISH spots. On average, >50% of CLN3 smFISH spots were colocalized with PL-RCA signals, whereas >80% of PL-RCA puncta overlapped with an smFISH spot, suggesting high specificity of detection (Figure 3B). We validated the sensitivity of this approach by perturbing translation rates pharmacologically with translation elongation inhibitors G418 or initiation inhibitor rapamycin which drastically decreased the number and brightness of PL-RCA puncta (Figure 3C). We conclude that PL-RCA puncta (henceforth “ribo”) represent not just ribosome proximity but active translation in Ashbya.
Figure 3. Whi3 target mRNAs show spatially biased translation in Ashbya.
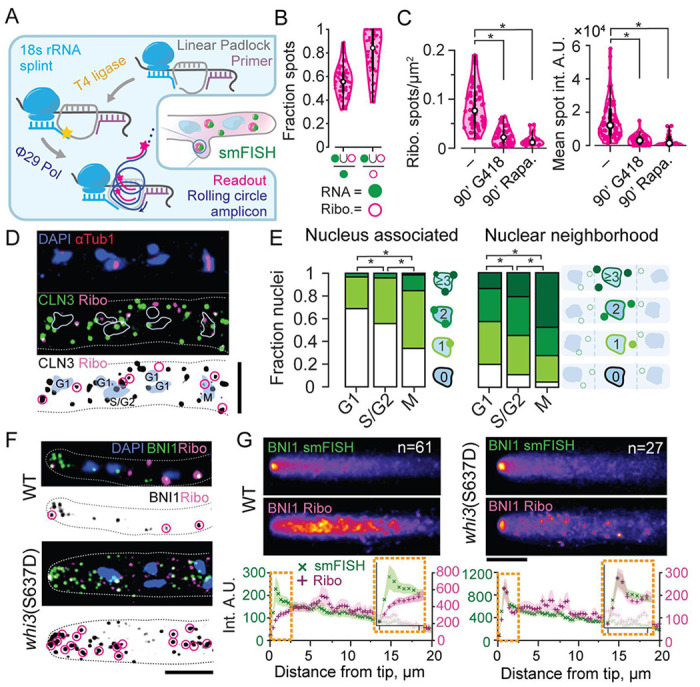
A. Schematic of tri-probe PL-RCA strategy for detection of ribosome association using FISH in Ashbya.
B. Most PL-RCA amplicons for CLN3 show specific colocalization with CLN3 smFISH spots, representing a majority of RNA spots. Spot masks in the smFISH (green filled circle) and RCA readout (magenta hollow circle) channels obtained using a semi-automated Otsu threshold were used to determine the fraction of colocalized spots with respect to counts of smFISH and PL-RCA spots per hypha, n=62 cells.
C. PL-RCA amplicons represent true translation. The PL-RCA spot density per hyphal mask and mean brightness both significantly decreased (KS test, p<0.01), upon 90 minutes of 200 μg/mL G418 (44 hyphae, 3757 RNA, 957 PL-RCA spots) or 90 minutes of 10 μg/mL rapamycin (17 hyphae, 348 RNA, 791 PL-RCA spots). Drugs were added to 10 mL of 14 h old cultures just prior to fixation.
D. Representative image of CLN3 PL-RCA showing nuclear proximal translation. Blue, Hoechst-stained nuclei; red, AlexaFluor488 labeled secondary, for anti-Tub1 antibody; green, CLN3 smFISH using TAMRA-labeled probes from Stellaris; magenta, CLN3-PL-RCA amplicons detected using AlexaFluor647-labeled readout probe. Scale bar 1 μm.
E. Cell cycle stage correlations with counts of PL-RCA at the nuclear-periphery (left) or nuclear-neighborhood (right). *=p<0.01, KS test on average spot count per nucleus.
F. Representative images of BNI1 smFISH (green) and PL-RCA (magenta) in WT and whi3(S637D). Scale bar 5 μm. G. Mean intensity projections of BNI1 smFISH (top) and PL-RCA for WT and whi3(S637D). Mean intensity and SEM at points sampled at 0.365 μm interval along the length of the hyphae aligned to the tip. Hyphal counts for each strain are in inset. Orange boxes show insets of the profiles 2.5 μm closest to the hyphal tip. Scale bar 5 μm.
We first applied this method to study spatial variation in translation of CLN3 mRNA, which was previously reported to form clusters near nuclei in a Whi3-dependent manner23. We observed a striking pattern of nuclear-periphery-associated CLN3-ribo spots (Figure 3D). Based on data in Figure 1C, we next determined the correlation between nuclear-peripheral CLN3 translation and nuclear cell-cycle state. To do this, we combined CLN3 PL-RCA with immunofluorescence for tubulin. We quantified the number of CLN3-ribo puncta on the nuclear periphery and in the cytoplasmic neighborhood of nuclei, defined manually using the midpoint of the shortest line between two nuclei. When binned by the cell cycle state, the counts showed a striking correlation with cell cycle state: M-phase nuclei showed the greatest number of nuclear-peripheral as well as nuclear proximal CLN3 translation spots, followed by S/G2, then G1, which showed the fewest spots (Figure 3E). This anti-correlation with G1 phase was unexpected, given that CLN3 encodes a putative G1 cyclin, but may be consistent with mutant phenotypes of cln3Δ showing delays at G2/M 29. We note that while the overall proportions are reminiscent of the cell-cycle-associated variation in Whi3 condensates (Figure 1C), bleaching and fixation artifacts limited in our ability to visualize Whi3 in this assay. Importantly, these data show that the Whi3 target mRNA CLN3 is locally translated in specific windows of the cell cycle, indicating another layer of regulation aside from simply localization.
We next investigated the sites of translation of BNI1, which was previously reported to cluster at hyphal tips in a Whi3-binding-dependent manner24. While smFISH revealed the expected clustering of mRNAs at tips, we saw low levels of translation at these sites. Importantly, the tip of growing hyphae is crowded with secretory vesicles, which may lead to this compartment being generally inhibitory to translation. However, this turned out not to be the case, as whi3(S637D), which has rapid tip growth and limited Whi3 puncta (Figure 1D; right), showed both BNI1 mRNA clusters and enriched translation of those mRNAs at the hyphal tip (Figure 3F). We quantified this tip-specific, strain-dependent translation difference using mean projections of translation signals of hyphae, with plots aligned to hyphal tips. Intensity profiles revealed a depletion of tip translation compared to the rest of the hypha in WT, and an enrichment at the tip in whi3(S637D) (Figure 3G). We conclude that BNI1 can show variable translation at hyphal tips but is predominantly repressed at tips in WT cells. Thus, Whi3 target mRNAs show spatially and temporally variable translation, with clear spatially biased zones of translation activation and repression.
Whi3 condensates differentially regulate translation of distinct targets in vitro
We next turned to an in vitro luciferase reporter assay to assess how Whi3 condensates modulate target mRNA translation30. To encompass the majority of the predicted Whi3 recognition motifs (4x and 3x UGCAU, for CLN3 and BNI1, respectively) in the reporters we placed the 5’ UTR of CLN3, and the 5’UTR and first 1500 nts of the CDS of BNI1 upstream of Nanoluciferase (Nluc) (Figure 4A,C). Whi3 and reporter RNAs were allowed to mix prior to the addition of Rabbit Reticulocyte (RR) lysate, and nano-luciferase activity was measured as a dose response of Whi3 concentrations in end point measurements. We chose two Whi3 concentrations that reflect concentrations of Whi3 measured in the dilute phase of the Ashbya cytoplasm (50 nM31), and two concentrations that resulted in the formation of Whi3 condensates in vitro (Figure 4A–D). We observe little change in luciferase activity for either CLN3-Nluc or BNI1-Nluc at dilute concentrations of 50 or 100 nM Whi3 compared to no-Whi3 reference reactions (Figure 4A,C). To our surprise, at Whi3 concentrations that promote condensate formation, we saw a biphasic response in BNI1-Nluc translation: 1 μM Whi3 protein showed >2-fold enhancement of translation while 3 μM showed strong repression (Figure 4A,B). In contrast, at multiple condensate-forming concentrations of Whi3, translation of CLN3-Nluc showed monotonic Whi3 dose-dependent repression, reaching extreme levels of >100-fold repression at 3 μM (Figure 4C,D). Notably, this dose-dependent CLN3-Nluc repression was detectable at concentrations in which visible condensates did not form (Figure S2A), suggesting that Whi3 impacts different target RNAs distinctly, with different density-dependent effects on translation.
Figure 4. Condensate size and RNAs specify translation.
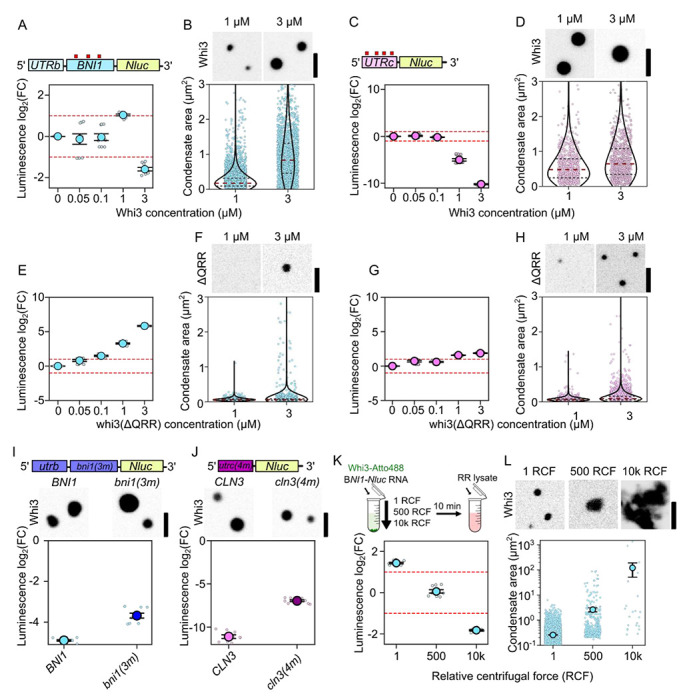
A. Translation of BNI1-Nluc reporter RNA as a function of Whi3 concentration. Luminescence values are displayed as log2(FC) over no-Whi3 conditions. Measurements are taken from three separate RR lysates with three technical replicates per lysate (see Methods).
B. Fluorescence images of Whi3 condensates. Measurements of condensate area of BNI1-Nluc-containing condensates in either 1 μM (n=5201) or 3 μM (n=1890) Whi3 conditions. Individual dots indicate a single condensate. Measurements were taken from reactions prepared with three different RR lysates. Scale bar = 2.5 μm.
C. Translation of CLN3-Nluc reporter RNA as a function of Whi3 concentration.
D. Fluorescence images of Whi3 condensate. Condensate area of CLN3-Nluc-containing condensates in either 1 μM (n=792) or 3 μM (n=900) Whi3 conditions. Scale bar = 2.5 μm.
E. Translation of BNI1-Nluc reporter RNA as a function of Whi3(ΔQRR) concentration.
F. Representative images of whi3(ΔQRR) condensates and areas with BNI1-NLuc. Measurements of condensate area of BNI1-Nluc-containing condensates in either 1 μM (n=785) or 3 μM (n=1718) whi3(ΔQRR) conditions.
G. Translation of CLN3-Nluc reporter RNA as a function of Whi3(ΔQRR) concentration.
H. Fluorescence images of whi3(ΔQRR) condensates. Measurements of condensate area of CLN3-Nluc condensates in either 1 μM (n=1310) or 3 μM (n=3985) whi3(ΔQRR) conditions. Scale bar = 2.5 μm.
I. Representative images of Whi3 condensates formed with either BNI1 or bni1(3m). Translation of BNI1-Nluc or bni1(3m) reporter RNA in 3 μM Whi3 conditions.
J. Representative images of Whi3 condensates formed with either CLN3 or cln3(4m). Translation of CLN3-Nluc or cln3(4m) reporter RNA in 3 μM Whi3 conditions.
K. Luminescence of BNI1-Nluc in reactions containing 1 μM Whi3 left to form condensates on the bench top (1 RCF) or centrifuged at 500 or 10000 RCF prior to their addition to RR lysate. Luminescence values are shown as log2(FC) over RNA-only conditions spun at 1, 500, or 10000 RCF respectively.
L. Fluorescence images of Whi3 condensates left to form on the bench top (1 rcf) or under 500 or 10000 rcf. Scale bar = 2.5 μm. Condensate area of BNI1-Nluc condensates 1 μM Whi3 conditions left to incubate on the bench top (n=5201), at 500 rcf (n=389), or 10000 rcf μM (n=23). Individual dots indicate a single condensate. Measurements were taken from reactions prepared with three different RR lysates.
To test the impact of the dense phase properties on translation regulation, we measured translation in the presence of whi3(ΔQRR), as the Q-rich region controls Whi3 oligomerization and condensate properties 23,24,32. Presence of whi3(ΔQRR) in the 1-phase regime resulted in modest increases in RNA translation for both reporter RNAs (Figure 4E,G). Higher concentrations of whi3(ΔQRR) generate smaller condensates for both CLN3- and BNI1-Nluc when compared to similar concentrations of WT Whi3, as anticipated, (Figure 4B,D compared to 4F,H) and resulted in pronounced enhancement of translation for both RNAs relative to the no protein control (Figure 4E,G). The RNA-specific regulation and translation repression is therefore sensitive to the presence of the QRR.
RNA-binding is another critical determinant of condensation and dense phase characteristics31,33,34, so we next tested the impact of eliminating the Whi3 recognition motifs on CLN3 and BNI1 on translation regulation. We created nano-luciferase reporter mRNAs bni1(3m)-Nluc and cln3(4m)-Nluc that preserved nucleotide composition but scrambled the Whi3 recognition motifs. At 3 μM WT Whi3, both RNAs still formed detectable condensates (Figure 4I,J). However, these RNAs showed decreased translation repression relative to RNAs with intact binding sites, when normalized to their respective no-Whi3 conditions (Figure 4 I,J). We conclude that translation repression arises primarily from the dense phase, but the magnitude of repression is tunable by RNA sequence, binding sites for Whi3, and Whi3 protein-protein interactions .
Taken together, Whi3 shows distinct RNA-specific translation outcomes that are influenced by interactions that drive condensation. Condensate-promoting compositions are in general more repressive than single phase compositions except for a window of Whi3 concentrations that selectively and strongly enhance BNI1 translation. The removal of the QRR completely reverses translation repression in both the 1-phase and 2-phase regimes in both RNAs, and removal of Whi3 recognition motifs from target RNAs decreases the magnitude of repression in the 2-phase regime.
Condensate size is inversely correlated with translation repression
The different lengths, structures, and Whi3-binding site valency of these two RNAs makes direct comparison of their translational outputs challenging. However, reaction conditions that caused the greatest degree of translational repression for both BNI1-Nluc and CLN3-Nluc, were associated with large condensates. We thus wondered whether condensate size may influence the translational output (Figure 4A–H). To test this idea, we sought to directly perturb condensate size without changing RNA and protein concentrations. We chose the Whi3 concentration that resulted in the highest enhancement of translation for BNI1-Nluc (1 μM Whi3, Figure 4A), and subjected the protein-RNA mixture to low- or moderate-speed centrifugation (500 and 10,000 RCF)33, prior to the addition of RR lysate (Figure 4K; schematic). Increasing centrifugation force resulted in progressively larger condensates (Figure 4L) and as predicted, condensate size was negatively correlated with luciferase activity (Figure 4K). Thus, artificially altering condensate size is sufficient to reverse translation enhancement and can even achieve strong repression. Since this centrifugation is not expected to shift the dense or dilute phase concentrations, we suggest this shift in translation outcome originates within the arrangement of the dense phase.
This result is consistent with a model in which Whi3 condensates sequester target mRNAs and repress their translation in a condensate size-dependent manner. We conclude that the positive and negative regulation of BNI1 translation could arise from condensate size-mediated repression.
Whi3-condensate size impacts interface-associated translation
The apparent role of the condensed state of Whi3 in regulating translation led us to next investigate the location of translation relative to the dense phase. To do this, we measured translation of Whi3-associated mRNAs using a fluorescence approach based on the MoonTag epitope arrays35. We generated reporters with mRNA regions engineered upstream of a CDS encoding 24x repeats of the gp41 peptide (“MoonTags”). MoonTag peptides are expected to bind fluorescently labeled gp41 nanobody with high affinity as they emerge from ribosomes, resulting in an accumulation of fluorescence reporting translation elongation (Figure 5A). We adapted this system for RR lysates, and generated BNI1- and CLN3-MoonTag reporter mRNAs (Figure 5C,D). As in the luciferase assays, we pre-mixed 3 μM Whi3 and MoonTag reporter mRNAs to allow for the formation of sufficiently large Whi3 condensates, and then supplemented the reaction with RR lysate and performed sequential 2-color fluorescence imaging of Whi3 and the gp41 nanobody (Figure 5B). It was immediately apparent that condensates formed with both RNA reporters showed frequent association with punctate MoonTag nanobody signal, but unexpectedly this signal was not homogeneous across the condensate volume (Figure 5C,D). Radial intensity profiles of the MoonTag signal aligned to the condensate boundary (located at x = 0.1) showed significant enrichment of the MoonTag signal within the condensate relative to the dilute phase, but a dramatic enrichment at the periphery (Figure 5C,D).
Figure 5. Condensate interfaces are sites for translation.
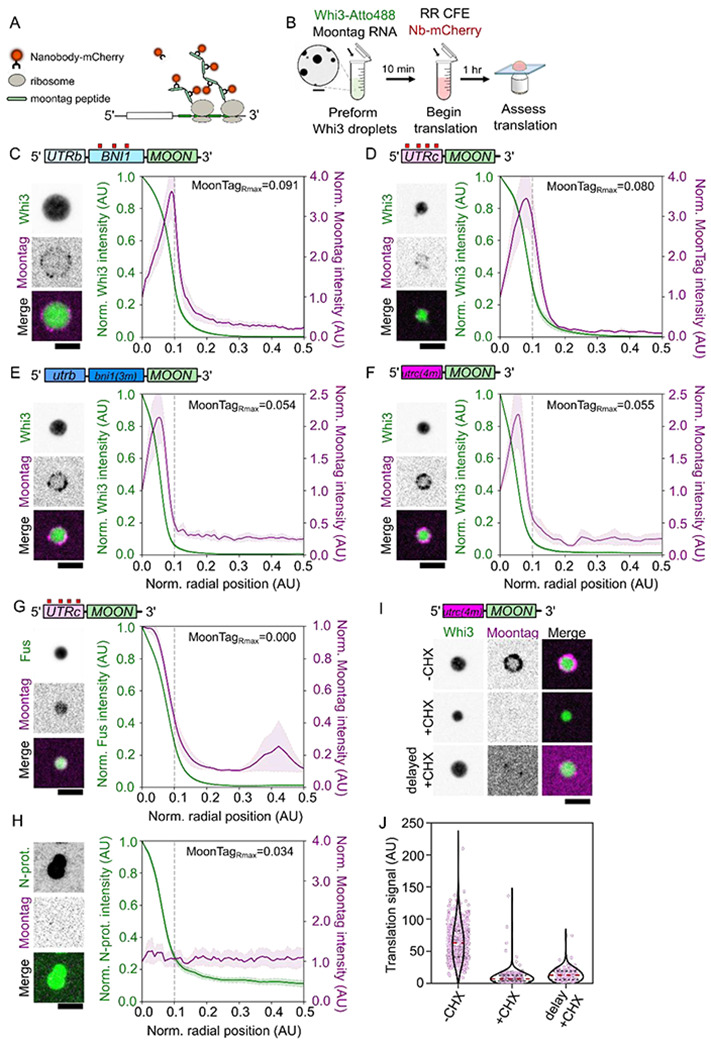
A. Design of MoonTag reporter mRNA.
B. Schematic of MoonTag reaction in cell extracts.
C. Fluorescence image of a Whi3 condensate formed with BNI1-Moon showing translation of mRNAs on the surface. Radial intensity profile of Whi3 condensates formed with BNI1-Moon, showing fluorescence intensity as a function of radial position in the condensate for Whi3 or MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 123 condensates ± SEM. Scale bar = 2.5 μm.
D. Fluorescence images of Whi3 condensate formed with CLN3-Moon showing translation of mRNAs on the surface. Radial intensity profile of Whi3 condensates formed with CLN3-Moon, showing fluorescence intensity as a function of radial position in the condensate for Whi3 or MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 42 condensates ± SEM. Scale bar = 2.5 μm.
E. Fluorescence images of Whi3 condensates formed with bni1(3m)-Moon showing translation of mRNAs at the condensate interface. Radial intensity profile of Whi3 condensates formed with bni1(3m)-Moon, showing fluorescence intensity as a function of radial position in the condensate for Whi3 and MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 127 condensates ± SEM. Scale bar = 2.5 μm.
F. Fluorescence images of Whi3 condensates formed with cln3(4m)-Moon showing translation of mRNAs at the condensate interface. Radial intensity profile of Whi3 condensates formed with cln3(4m)-Moon, showing fluorescence intensity as a function of radial position in the condensate for Whi3 and MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 147 condensates ± SEM. Scale bar = 2.5 μm.
G. Fluorescence images of Fus condensates formed with CLN3-Moon showing translation of mRNAs within the Fus condensate. Radial intensity profile of Fus condensates formed with CLN3-Moon, showing fluorescence intensity as a function of radial position in the condensate for Fus or MoonTag Nb-mCherry. Scale bar = 2.5 μm. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 265 condensates ± SEM.
H. Fluorescence images of N-protein condensates formed with CLN3-Moon showing no translation of mRNAs within or at the surface of the N-protein condensate. Radial intensity profile of N-protein condensates formed with CLN3-Moon, showing fluorescence intensity as a function of radial position in the condensate for N-protein or MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 162 condensates ± SEM. Scale bar = 2.5 μm.
I. Fluorescence images showing translation of cln3(4m)-Moon at Whi3 condensates in RR lysates where translation is intact (−CHX), supplemented with cycloheximide (+CHX), or in lysates that have been allowed to translate CLN3-Moon mRNAs prior to either the addition of cycloheximide or Whi3 droplets (delayed +CHX). Scale bar = 2.5 μm.
J. Quantification of average translation signal per condensate in translation competent lysates (−CHX; n=460), lysates supplemented with cycloheximide (+CHX; n=253), or lysates that have first been allowed to translate CLN3(0xWBS)-Moon prior to the addition of either cycloheximide or Whi3 droplets (delayed +CHX; n=127). Measurements were taken from reactions prepared with three different RR lysates.
We were curious about the significance of condensate-associated MoonTag events, since 3 μM Whi3 is largely repressive at the bulk scale in the luciferase experiments, and further dissected this phenomenon. We repeated the experiment with bni(3m)- and cln3(4m)-MoonTag reporters. Again, we observed an annular localization of the MoonTag signal (Figure 5E,F). However, these signals were significantly brighter (Figure S3A,B), and the peak of the signal showed a shift towards the interior of the condensate (Figure 5 E–F), reflecting an increase in translation, consistent with the trends of the luciferase assay (Figure 4I,J). The overall trends of translation within condensates thus reflect the overall trend of the luciferase translation results: a major portion of the mRNAs are repressed within the condensate for all RNAs, however the binding site mutant RNAs show a reduction in repression relative to WT. These changes in RNA valency are also associated with a change in the sub-condensate localization of mRNA translation. Notably, even condensates that appear repressive for translation in a bulk assay support some translation at their surface.
We confirmed this apparent spatial bias in MoonTag signal by performing a series of controls. First, we established that labeled RNAs themselves showed a uniform localization profile across these condensates, suggesting that MoonTag translation did not reflect a biased RNA distribution (Figure S3C). Second, we probed the accessibility of the condensate interior with fluorescently labeled dextran to determine if it excluded ribosome-sized particles. We did not see any exclusion of dextran, suggesting that the bias in translation was not reflective of a physical exclusion of translation machinery (Figure S3D). Next, we asked if the MoonTag signal represents recruitment of nanobody-bound, MoonTag peptides from the dilute phase. We preincubated RR lysate with 5 nM cln3(4m)-MoonTag and 500 nM Nb-mCherry without Whi3, allowing these RNAs to translate for 1 h. These lysates were treated with cycloheximide for 1 h before the addition of Whi3-condensates and an additional 500nM Nb-mCherry. We saw a near-complete loss of MoonTag signal associated with condensates, comparable to a complete cycloheximide-arrest of translation (Figure 5I,J). These results demonstrate that free nanobody or epitopes by themselves are not enriched within condensates, and any MoonTag signal in the condensate represents active translation rather than recruitment of paused elongation events from the dilute phase.
Having established that condensates are indeed able to support translation even in a regime that appeared repressive in a bulk assay, we revisited the inverse correlation between condensate size and translation. At the level of individual condensates, the average translation per unit surface area at the interface showed a low correlation with the size of the condensate for both BNI1 and CLN3 (Figure S4A,B,D,E). However, the peak of the radial MoonTag profile, is closer to the center of the condensate in smaller condensates (Figure S4C,F). The translation enrichment towards the center of smaller condensates does not necessarily reflect a widening in the zone of translation enrichment (Figure S4G) but is likely the consequence of the condensate-interface constituting a greater proportion of the total volume in smaller condensate. We conclude that condensate-associated translation shows a distinct sub-condensate localization that is size-dependent and independent of the specific RNA.
Taken together, these results suggest that RNAs within Whi3 condensates are accessible for translation, however a variable zone, close to the condensate interface is particularly permissive for translation. The ability for Whi3 condensates to increase (1 μM Whi3; Figure 4A) or decrease (3 μM Whi3; Figure 4A) translation depending on the size of condensates may emerge from changes in the relative surface area/volume rather than simply switching translation states across dense/dilute phases. Since Whi3 associates with populations of translating and repressed RNAs, it is conceivable both actively translating and repressed mRNAs may be contained within the same Whi3 assembly. An alternate explanation for these results is that larger condensates arise as a consequence of enriching translationally repressed mRNAs, as has been postulated for stress granules36. We tested the latter hypothesis by measuring the condensate size distribution upon translation arrest. We only see a modest change in average condensate size associated with CHX-treated RR lysates (Figure S3E) changing from 0.681 and 1.02 μm2 for non-treated and CHX-treated lysates, respectively. This small size change corresponds to a >50-fold drop in condensate associated translation suggesting large-sized condensates are not simply reservoirs of repressed mRNAs in the assay (Figure 5J).
Regardless of the causality between condensate size and translation repression, the enrichment of translation at the interface of Whi3 condensates shows striking similarity with recent work in Drosophila germ granules, where the translation of Nanos in the germplasm occurs at the interface or surface of germ granules19,20. To test if surface-enhanced translation is a general feature of condensate-associated translation, we mixed our CLN3-Moon reporter RNA with two other well described RNA-binding proteins (RBPs), Fus and N-protein (Figure 5G,H). To our surprise, Fus condensates showed strong internal translation signal, while N-protein condensates completely repressed the translation of the reporter RNA (Figure 5G,H). These results indicate that the interface-enriched translation we observe for Whi3 condensates is specific to Whi3 and are tuned in part by the specific RNA-protein and protein-protein interactions therein.
Whi3 PKA mutant eliminates translation-state switching
We next evaluated how this model for Whi3 condensate translation regulation played out in the PKA phosphomimic S637D mutants that show prominent cell and condensate phenotypes (Figure 1C). Concentrations of whi3(S637D) in the single-phase regime resulted in no changes in translation for either BNI1-Nluc or CLN3-Nluc over the no-Whi3 reference (Figure 6A,C). At higher concentrations, whi3(S637D) forms condensates with both BNI1- and CLN3-Nluc mRNAs (Figure 6B,D). However, we saw a significant departure from WT Whi3 condensate translation behavior: whi3(S637D) condensates formed with BNI1 now show only an enhancement in translation of these mRNAs, while CLN3-containing condensates no longer show significant repression of translation (Figure 6 A,C). Notably, whi3(S637D) condensates formed with either CLN3 or BNI1 led to the formation of smaller condensates when compared to similar concentrations of WT Whi3, suggesting this phospho-state alters the assembly of Whi3 condensates (Figure 6B,D). Considering this PKA phospho-site lies within the RRM, we tested if this translation phenotype observed for CLN3 was related to a difference in RNA binding affinity of the mutant. We did not observe a significant difference in RNA interaction Kd between WT and whi3(S637D) for CLN3 (154 and 147 nM respectively), relative to the measurable difference in affinity between WT Whi3 and a full length CLN3 mutant with no Whi3 binding sites (345 nM) (Figure S5A).
Figure 6. PTM state of Whi3 tunes translational outcome of condensate-associated RNAs.
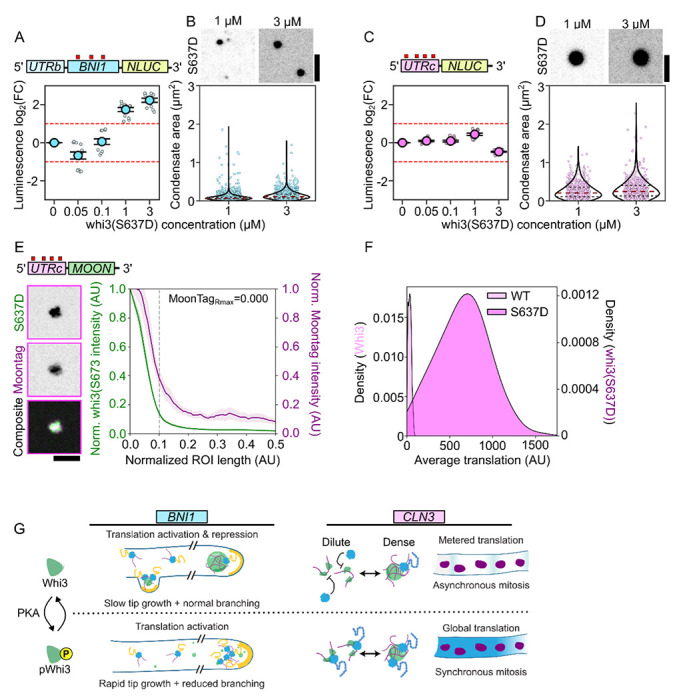
A. Translation of BNI1-Nluc reporter RNA as a function of whi3(S637D) concentration. Luminescence values are displayed as log2(FC) over RNA-only conditions. Measurements are taken from at least three separate RR lysates with three technical replicates per lysate (see Methods).
B. Fluorescence images of whi3(S637D) condensates formed with BNI1-Nluc. Measurements of condensate area of BNI1-Nluc condensates in either 1 μM (n=3449) or 3 μM (n=2020) whi3(S637D) conditions. Individual dots indicate a single condensate. Measurements were taken from reactions prepared with three different RR lysates. Scale bar = 2.5 μm.
C. Translation of CLN3-Nluc reporter RNA as a function of whi3(S637D) concentration. Luminescence values are displayed as log2(FC) over RNA-only conditions. Measurements are taken from at least three separate RR lysates with three technical replicates per lysate (see Methods).
D. Fluorescence images of whi3(S637D) condensates formed with CLN3-Nluc. Measurements of condensate area of CLN3-Nluc condensates in either 1 μM (n=593) or 3 μM (n=607) whi3(S637D) conditions. Individual dots indicate a single condensate. Measurements were taken from reactions prepared with three different RR lysates. Scale bar = 2.5 μm.
E. Fluorescence images of whi3(S637D) condensates formed with CLN3-Moon showing translation of mRNAs within whi3(S637D) condensates. Radial intensity profile of whi3(S637D) condensates formed with CLN3-Moon, showing fluorescence intensity as a function of radial position in the condensate for whi3(S637D) or MoonTag Nb-mCherry. The condensate periphery, grey line, is approximately 0.1 AU from the center of the condensate. The radial position associated with the maximum MoonTag intensity recorded, MoonTagRmax, is shown. Trace shows the average intensity of 163 condensates ± SEM. Scale bar = 2.5 μm.
F. Kernel probability density estimate of the average translation signal associated with condensates formed with CLN3-Nluc and either Whi3 (n=299) or whi3(S637D) (n=369).
G. Schematic of translation state-switching model underlying normal hyphal growth and asynchrony, tuned by phospho-signaling.
Next, we looked at translation at the sub-condensate scale associated with whi3(S637D). In the conditions we tested with 3 μM Whi3(S673D) and 5 nM RNA, we were unable to reliably form condensates with the BNI1-MOON reporter RNA. In contrast, CLN3-MOON readily formed small condensates that showed internal enrichment for MoonTag signal (Figure 6E). These results further reinforce our previous conclusions regarding condensate size control: the degree of repression is inversely related to condensate size, and in smaller condensates the zone of translation occupies a greater proportion of the interior of the condensate. Additionally, these condensates were associated with a substantial increase in translation signal when compared to WT Whi3 condensates (Figure 6F). It is likely that this ~10-fold increase in translation per whi3(S637D) condensate over WT Whi3 condensates (Figure 6F; light pink) contributes to the net loss of translation repression observed in our whi3(S637D)/CLN3-Nluc reactions (Figure 6C). Despite these increases in translation per condensate, CLN3-Nluc reactions never reach translation enhancement as is observed for BNI1, in agreement with RNA-dependent translation outcomes. Intriguingly, this single point mutant can completely shift the regulation of both RNAs, leading only to the monotonic enhancement of translation for BNI1 and the loss of translation repression for CLN3. This is reflected in the cell-scale phenotype of S637D where a loss of spatial variation in CLN3 and BNI1 translation results in nuclear division synchrony as well as loss of branching and coordinated hyphal extension (Figure 6G). Thus, beyond repression, the ability of Whi3 to regulate translation state-switching in response to growth cues appears critical to coordinated cell growth.
Discussion
These observations emphasize two distinct scales at which Whi3-RNA condensates show spatial variability in protein translation: at the sub-micron scale within condensates, and at the ~10-100 micron scale across cells (Figure 7). At the submicron scale, we observe a striking translation-permissive zone within Whi3 condensates that is biased towards the condensate interface. The radial position of this zone, however, is tunable by the valency of the RNA and Whi3-phosphostate. These condensates, rather than toggling translation on/off, show a continuum of regulatory states observable both in bulk and at the level of individual condensates in vitro. The most surprising finding at this scale is the exquisite specificity that the RNAs themselves bring to these condensates: CLN3 5’UTR shows consistent repression that is reversed by Whi3 phosphostate and binding site abundance; BNI1 shows more a complex biphasic response to Whi3 concentration. While we tested the contribution of Whi3 binding site abundance to translation regulation, we predict other factors such as RNA length, structure, motif position and clustering and 5’UTR features may all influence this specificity.
Figure 7. Whi3 condensates coordinate hyphal growth by dynamically switching translation states in response to multiple cues to tune mRNA-specific local translation.
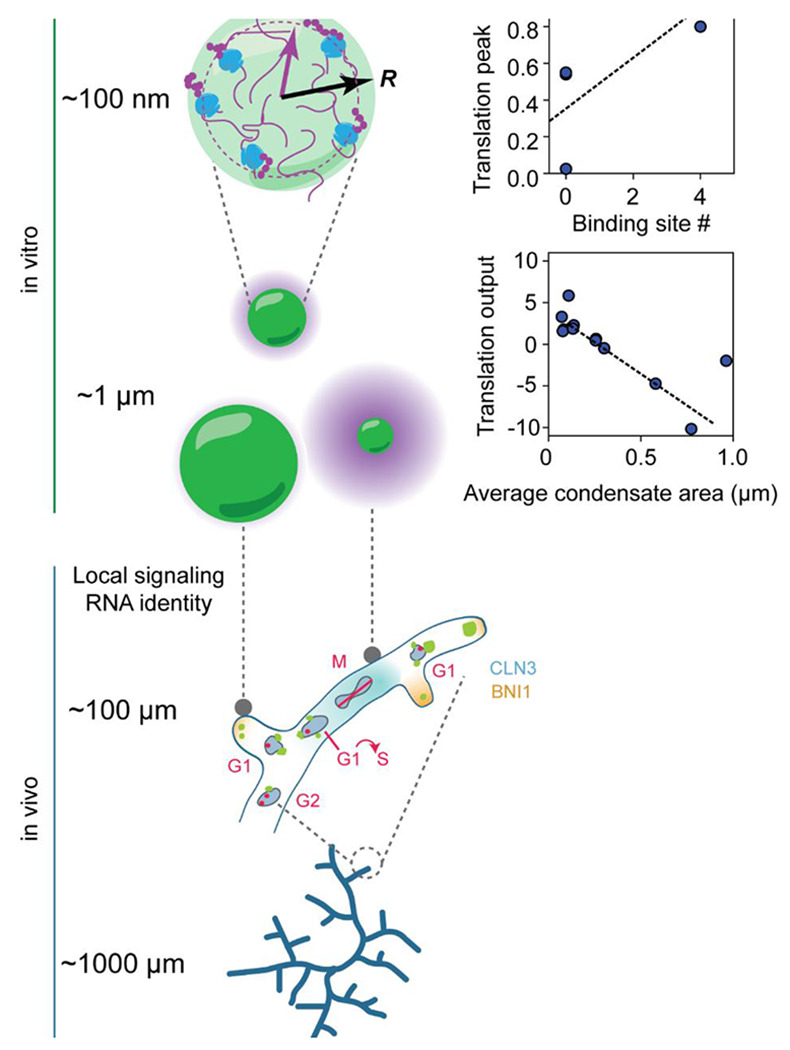
Top to bottom: RNA identity, binding-site number and Whi3 phospho-state shape protein translation at condensates and specify the location of sub-compartment enrichment. This results in a continuum of translation states that is additionally modulated by condensate size in vitro. This continuum is also reflected in the spatial variation in Whi3-target translation at the cell scale in vivo. In addition to spatial variation in translation, the ability to both repress and activate translation underlies normal nuclear division asynchrony and branching phenotypes. This paradigm underscores how a single protein such as Whi3 can differentially regulate RNAs whose encoded protein requirements are vastly different but whose local translation needs to nevertheless be tightly coordinated.
The strongest RNA-independent predictor of translation outcome at the micron scale appears to be condensate size. Across all the Whi3 and RNA mutants and across a range of protein/RNA concentrations we test, the magnitude of bulk translation repression increases with condensate size, providing a compelling rationale for the tight size control seen in vivo25. Intriguingly, we also see condensate size and number variation in vivo that is associated with translation phenotypes: the largest Whi3 condensates appear at hyphal tips and are correlated with slower growth and lower BNI1 translation at these sites, whereas M-phase nuclei are associated with both fewer condensates and highest CLN3 translation. Thus size-dependent condensate function regulation may be a critical feature of condensate biology, as attested by the diverse biological systems in which it is starting to be reported37,38.
Another regime where bulk translation is repressed is in the dilute phase, where CLN3 with WT Whi3 is modestly repressed. We propose that these repressive regimes are critical for the overall regulatory circuit: both Cln3 and Bni1 proteins are likely to be potent at low concentrations, when dosed in at the appropriate subcellular location. Consequently, bulk translation levels measured in vitro may not be the most relevant parameter in vivo: for instance, large condensates show the highest repression at the bulk level, but also show varying levels of interface-enriched translation, and these may be sufficient for shaping local Bni1 activity at the tip. In contrast, repression of Cln3 in the dilute phase may help accentuate local bursts of protein synthesis at certain cell-cycle states, allowing for enhanced spatial heterogeneity in composition.
Correspondingly, at the ~100 micron scale we observe well-defined spatial biases in translation in Ashbya. Cln3 shows an unexpected bias for translation at the periphery and neighborhoods of non-G1 phase nuclei, and substantial repression around G1 nuclei. How does the observed spatial variation impact nuclear division asynchrony? We previously reported that Whi3 CDK phosphomutants show increased nuclear division synchrony potentially due to being potentially regulated normally by CDK signaling2. This may result in “out of phase coupling” wherein Whi3 mediates nuclear division asynchrony by regulating cell-cycle-dependent CLN3 translation, such that nuclei that are out-of-phase with G1 nuclei can promote G1-exit of neighbors. Alternatively, Cln3 may show non G1-associated functions in Ashbya that explain the G2/M phase enrichment in translation. Irrespective of the mechanism of Cln3-translation mediated asynchrony, we propose that local variation in Cln3 translation emerges out of dynamic switching between a repressive and translation-permissive Whi3 states.
BNI1 also shows an unexpected spatial bias in translation and is predominantly repressed at hyphal tips where the transcript is enriched. Taking live cell growth observations and condensate size observations together, we propose that large Whi3 condensates associated with slower hyphal growth are also repressive for translation in the cell. Notably, this allows Bni1 transcripts to be positioned, poised for translation, at the tip, but otherwise repressed, consistent with the low demand for fresh protein at these sites. This repression is sensitive to signaling, and the Whi3 PKA phosphomimic dramatically reverses repression both in vitro and in vivo. However, translation-state switching is also important for Bni1: a constitutively “active” mutant of Whi3 that allows high tip-associated translation results in abnormal growth morphology, and poor branching2. Thus, Whi3 serving dual roles in activation and inhibition of repression is critical for function in growth.
How these two RNAs are handled so differently by Whi3 remains unclear. However, specific perturbations to valency in both RNAs result in predictable translation regulation outcomes, suggesting that RNA valency is one RNA-specific tunable knob for translation regulation. Such valency-dependent organization of molecules within condensates has been observed in other phase-separated systems39, suggesting that general condensate organizing principles are co-opted for translation regulation in this system.
Thus Whi3’s ability to appropriately tune the translation of its targets relies on its ability to switch between a repressive and an activating state. Failure to switch states, as in whi3(QRR) and whi3(S637D) results in highly abnormal growth phenotypes, including abnormal nuclear density, nuclear division synchrony, irregular branching interval and premature tip-splitting 2,23,24. Surprisingly, we rarely see these defects in WT cells, suggesting that nuclear division phenotypes are normally tightly co-regulated with branching and hyphal growth. This raises the intriguing possibility that by regulating local Cln3 and Bni1 abundance, Whi3 may serve the role of a coupler that is able to co-regulate nuclear division and hyphal growth in Ashbya’s shared cytoplasm to maintain coordinated growth.
This work expands our understanding of how complex post-transcriptional regulation underlying growth can arise from a minimal network of RNA-protein interactions and highlights how mesoscale condensate properties can tune biological function by switching between regulatory states.
Supplementary Material
Acknowledgements
We thank Qiang Chen and Christopher Nicchitta for generous help with designing the polysome profiling experiments. We thank Christine Roden and Wilton Snead for their generous gift of N-protein and Fus, respectively. We thank Laura Herring and her team at the Michael Hooker Metabolomics and Proteomics Core located at the University of North Carolina at Chapel Hill.
Funding:
This work was supported Air Force Office of Scientific Research (grant FA9550-20-1-0241) to ASG, NIH grant 7R01GM081506-13 to ASG, Duke School of Medicine International Chancellor’s Scholarship, NIH F32 1F32GM147989 to APJ and F32 GM151858-02 to ZMG.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Data and materials are available upon request.
References
- 1.Dill K. A., Ghosh K. & Schmit J. D. Physical limits of cells and proteomes. Proc Natl Acad Sci U S A 108, 17876–17882, doi: 10.1073/pnas.1114477108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerbich T. M. et al. Phosphoregulation provides specificity to biomolecular condensates in the cell cycle and cell polarity. J Cell Biol 219, doi: 10.1083/jcb.201910021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T. H. et al. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829, doi: 10.1126/science.aax4240 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Sasazawa M., Tomares D. T., Childers W. S. & Saurabh S. Biomolecular condensates as stress sensors and modulators of bacterial signaling. PLoS Pathog 20, e1012413, doi: 10.1371/journal.ppat.1012413 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wippich F. et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805, doi: 10.1016/j.cell.2013.01.033 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Jalihal A. P. et al. Multivalent Proteins Rapidly and Reversibly Phase-Separate upon Osmotic Cell Volume Change. Mol Cell 79, 978–990.e975, doi: 10.1016/j.molcel.2020.08.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iserman C. et al. Condensation of Ded1p Promotes a Translational Switch from Housekeeping to Stress Protein Production. Cell 181, 818–831.e819, doi: 10.1016/j.cell.2020.04.009 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang M. et al. The intrinsically disordered region from PP2C phosphatases functions as a conserved CO(2) sensor. Nat Cell Biol 24, 1029–1037, doi: 10.1038/s41556-022-00936-6 (2022). [DOI] [PubMed] [Google Scholar]
- 9.Chen D. et al. Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol Cell 82, 3015–3029.e3016, doi: 10.1016/j.molcel.2022.05.026 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Mittag T. & Pappu R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol Cell 82, 2201–2214, doi: 10.1016/j.molcel.2022.05.018 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jülicher F. & Weber C. A. Droplet Physics and Intracellular Phase Separation. Annual Review of Condensed Matter Physics 15, 237–261, doi: 10.1146/annurev-conmatphys-031720-032917 (2024). [DOI] [Google Scholar]
- 12.King M. R. et al. Macromolecular condensation organizes nucleolar sub-phases to set up a pH gradient. Cell 187, 1889–1906.e1824, doi: 10.1016/j.cell.2024.02.029 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Dai Y. et al. Biomolecular condensates regulate cellular electrochemical equilibria. Cell 187, 5951–5966.e5918, doi: 10.1016/j.cell.2024.08.018 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker C. J. & Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4, a012286, doi: 10.1101/cshperspect.a012286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ripin N., Macedo de Vasconcelos L., Ugay D. A. & Parker R. DDX6 modulates P-body and stress granule assembly, composition, and docking. J Cell Biol 223, doi: 10.1083/jcb.202306022 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brangwynne C. P. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732, doi: 10.1126/science.1172046 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Lee C. S. et al. Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. Elife 9, doi: 10.7554/eLife.52896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang J. Y. et al. LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs. Science 377, eabj6647, doi: 10.1126/science.abj6647 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Chen R., Stainier W., Dufourt J., Lagha M. & Lehmann R. Direct observation of translational activation by a ribonucleoprotein granule. Nat Cell Biol 26, 1322–1335, doi: 10.1038/s41556-024-01452-5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramat A., Haidar A., Garret C. & Simonelig M. Spatial organization of translation and translational repression in two phases of germ granules. Nat Commun 15, 8020, doi: 10.1038/s41467-024-52346-x (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alami N. H. et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81, 536–543, doi: 10.1016/j.neuron.2013.12.018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Cabau C. et al. Kinetic stabilization of translation-repression condensates by a neuron-specific microexon. bioRxiv, 2023.2003.2019.532587, doi: 10.1101/2023.03.19.532587 (2024). [DOI] [Google Scholar]
- 23.Lee C. et al. Protein aggregation behavior regulates cyclin transcript localization and cell-cycle control. Dev Cell 25, 572–584, doi: 10.1016/j.devcel.2013.05.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C., Occhipinti P. & Gladfelter A. S. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J Cell Biol 208, 533–544, doi: 10.1083/jcb.201407105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snead W. T. et al. Membrane surfaces regulate assembly of ribonucleoprotein condensates. Nat Cell Biol 24, 461–470, doi: 10.1038/s41556-022-00882-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gladfelter A. S., Hungerbuehler A. K. & Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J Cell Biol 172, 347–362, doi: 10.1083/jcb.200507003 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizunuma M. et al. Ras/cAMP-dependent protein kinase (PKA) regulates multiple aspects of cellular events by phosphorylating the Whi3 cell cycle regulator in budding yeast. J Biol Chem 288, 10558–10566, doi: 10.1074/jbc.M112.402214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng H. et al. Spatially resolved single-cell translatomics at molecular resolution. Science 380, eadd3067, doi: 10.1126/science.add3067 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hungerbuehler A. K., Philippsen P. & Gladfelter A. S. Limited functional redundancy and oscillation of cyclins in multinucleated Ashbya gossypii fungal cells. Eukaryot Cell 6, 473–486, doi: 10.1128/ec.00273-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.England C. G., Ehlerding E. B. & Cai W. NanoLuc: A Small Luciferase Is Brightening Up the Field of Bioluminescence. Bioconjug Chem 27, 1175–1187, doi: 10.1021/acs.bioconjchem.6b00112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H. et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell 60, 220–230, doi: 10.1016/j.molcel.2015.09.017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seim I. et al. Dilute phase oligomerization can oppose phase separation and modulate material properties of a ribonucleoprotein condensate. Proc Natl Acad Sci U S A 119, e2120799119, doi: 10.1073/pnas.2120799119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boeynaems S. et al. Spontaneous driving forces give rise to protein-RNA condensates with coexisting phases and complex material properties. Proc Natl Acad Sci U S A 116, 7889–7898, doi: 10.1073/pnas.1821038116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langdon E. M. et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927, doi: 10.1126/science.aar7432 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boersma S. et al. Multi-Color Single-Molecule Imaging Uncovers Extensive Heterogeneity in mRNA Decoding. Cell 178, 458–472.e419, doi: 10.1016/j.cell.2019.05.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khong A. et al. The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820.e805, doi: 10.1016/j.molcel.2017.10.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gil-Garcia M. et al. Local environment in biomolecular condensates modulates enzymatic activity across length scales. Nat Commun 15, 3322, doi: 10.1038/s41467-024-47435-w (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chattaraj A. & Shakhnovich E. I. Multi-condensate state as a functional strategy to optimize the cell signaling output. bioRxiv, doi: 10.1101/2024.01.14.575571 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Burgos I., Espinosa J. R., Joseph J. A. & Collepardo-Guevara R. Valency and Binding Affinity Variations Can Regulate the Multilayered Organization of Protein Condensates with Many Components. Biomolecules 11, doi: 10.3390/biom11020278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


