ABSTRACT
Immune cell infiltration into adipose tissue (AT) is a key factor in type 2 diabetes (T2DM). However, research on the impact of fat distribution on immune cells and immune responses in women is still lacking. This study used enrichment, protein–protein interaction network, immune cell infiltration, and correlation analysis to compare the similarities and differences between the transcriptome data of visceral AT (VAT) and subcutprotein-proteinaneous AT (SAT) obtained from the omprehensive database of gene expression in women with non-T2DM and T2DM. DEGs with the same biological function in two types of ATs often exhibited different expression trends. SharedVAT-specific and SAT-specific hub genes were mainly associated with transcription factors, monocyte-macrophage markers, and chemokines, respectively. Immune cells affected by both AT types included monocytes, granulocytes, T and B lymphocytes, and NK cells. VAT affected more immune cells, mainly myeloid cells. Shared hub genes in VAT correlated positively with M1 macrophages, suggesting pro-inflammatory effects, while those in SAT correlated negatively with M1 macrophages and lymphocytes, suggesting anti-inflammatory effects. This study provides a theoretical basis for further understanding the correlation between AT and T2DM in women.
KEYWORDS: Gene Expression Omnibus, visceral fat tissue, subcutaneous fat tissue, type 2 diabetes, immune infiltration analysis
Introduction
Adipose tissue (AT) functions as an endocrine organ that regulates various physiological processes and is associated with dysfunction, metabolic homoeostasis disruption, insulin resistance, and type 2 diabetes mellitus (T2DM) in obese individuals [1]. AT can be categorized into brown AT (BAT) and whiteAT (WAT) based on morphology [2]. WAT constitutes the largest proportion of AT in the body and plays a significant role in the development of cardiovascular disease and its complications. Furthermore, WAT can be divided into two categories based on anatomical location: visceral AT (VAT) and subcutaneous AT (SAT) [3]. Previous studies ha demonstrated significant differences between these two types of AT in terms of cellular and signalling molecule composition, physiological metabolism, clinical manifestations, and disease prognosis [4]. VAT, often referred to as metabolically unhealthy fat, primarily resides in the mesentery and omentum, with its venous blood being directly discharged into the liver via the portal vein. Research indicates that VAT possesses a greater number of blood vessels, an abundant blood supply, stronger neural innervation, and a higher concentration of glucocorticoid and androgen receptors compared to SAT, rendering it more sensitive to catecholamine-induced lipolysis and adrenaline stimulation [5–7]. Elevated levels of cytokines such as tumour necrosis factor alpha (TNF-α), interleukin (IL) -6 , IL-8, and monocyte chemotactic protein-1 (MCP-1), alongside adipokines like matrix metalloproteinase-1 (MMP-1) and plasminogen activator inhibitor-1 (PAI-1), as well as prostaglandin compounds such as T-box transcription factor 2 (TBX2) and prostaglandin E2 (PGE2), which are expressed at higher levels in VAT, can contribute to systemic inflammation, insulin resistance, metabolic syndrome, and other diseases [8,9]. In contrast, SAT is primarily deposited in the hip region, posterior abdominal wall, and anterior abdominal wall [10]. SAT is drained through systemic veins and can store excess free fatty acids (FFAs) and glycerol in the form of triglycerides (TGGs) 10 within adipocytes, thereby forming a neutral metabolic pool [11]. The correlation between SAT and leptin is stronger , while estrogen has a significant effect on the accumulation of SAT [12]. Estrogen deficiency results in an increase in VAT in postmenopausal women [13]. SATis regarded as a metabolically healthy fat, exhibiting a weak association with metabolic diseases, and may even play a protective role [14,15].
AT contains a substantial number of immune cells, including macrophages, natural killer cells, B lymphocytes, and T lymphocytes, whose condition directly influences overall health [16]. The abnormal proliferation of AT can lead to alterations in the immune cell spectrum, resulting in a chronic low-grade inflammatory state that may trigger systemic inflammation, insulin resistance, and metabolic diseases [17]. Research indicates that the types of immune cells present in VAT and SAT differ. Although CD4 and CD8 T cell infiltration occurs in both VAT and SAT, the proportion of pro-inflammatory T helper cells (Th) −1, Th17, and CD8 T cells is higher in VAT compared to SAT [18]. Notably, significant gender differences exist in the number of immune cells within AT, including regulatory T cells (Treg) [19]. AT exhibits gender specificity concerning obesity-related low-grade inflammatory states. A study demonstrated that, in women, SAT volume correlates with the number of monocytes and neutrophils, and the association between certain circulating inflammatory markers, such as IL-6, IL-18BP, TRAILR2, LOX1, and CEACAM8, is stronger than in men [20]. However, there remains a lack of research on the impact of fat distribution on immune cells and immune responses specifically in women.
This study aimed to analyze the transcriptome data of VAT and SAT from women with and without T2DM, obtained from the Gene Expression Omnibus (GEO) database. Bioinformatic analysis was used to examine changes in the spectrum of immune-infiltrating cells and the correlation between key genes and immune cells, with the aim of comparing the differences and similarities in their impact on the pathogenesis of T2DM. From an immunological perspective, this study provides valuable insights into the relationship between AT and T2DM.
Results
Analysis of shared differentially expressed genes between SAT and VAT
The transcriptome data of VAT (dataset GSE29231) and SAT (dataset GSE29226) in women with and without T2DM were analyzed. The results revealed that 796 differentially expressed genes (DEGs) were identified in the VAT of women with T2DM, comprising 690 upregulated and 106 downregulated genes. The SAT of women with T2DM contained 328 DEGs, including 110 upregulated and 218 downregulated genes. Further analysis using the Venn method identified 55 shared DEGs between the AT types, 741 VAT-specific DEGs, and 273 SAT-specific DEGs (Figure 1). Detailed tables are provided in Supplementary File S1.
Figure 1.
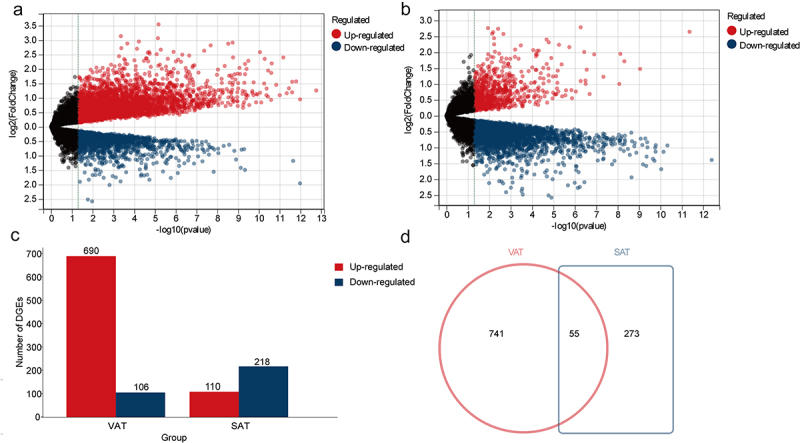
To analyze the DEGs in SAT or VAT gene expression profiles of women with T2DM. (a) The volcanic plot illustrates the distribution of DEGs derived from comparing the gene expression profiles of VAT in women with T2DM (n = 12) versus those without T2DM (n = 12) (GSE 29231). (b) The volcanic plot illustrates the distribution of DEGs derived from comparing the gene expression profiles of women SAT in women with T2DM (n = 12) versus those without T2DM (n = 12) (GSE 29226). (c) The histogram presents the distribution and quantity of DEGs in VAT and SAT, with red indicating upregulated genes and blue indicating downregulated genes; (d) In the Venn diagram, the red circles represent DEGs specific to VAT, the blue rectangles represent DEGs specific to SAT, the overlapping areas indicate shared DEGs (n = 55), while the non-overlapping sections denote VAT-specific DEGs (n = 741) and SAT-specific DEGs (n = 273), respectively.
GO enrichment and KEGG pathway analysis of DEGs
GO enrichment analysis of DEGs with consistent expression trends in the shared DEGs between VAT and SAT revealed associations with response to metal ions, regulation of transcriptions, response to hormones, circadian rhythm, and regulation of inflammatory response (Figure 2a). Conversely, DEGs showing distinct expression trends in the shared DEGs were found to be implicated in regulating body fluid levels, inhibiting cellular catabolic processes, and intracellular signal transduction (Figure 2b). Additionally, the upregulated VAT-specific DEGs were associated with biological processes such as ECM reconstruction processes and transcription (Figure 2c). Conversely, the downregulated VAT-specific DEGs were enriched in regulating of metabolic process (Figure 2d). In SAT, the upregulated SAT-specific DEGs were linked to inflammation and immune-related biological processes (Figure 2e). Finally, the downregulated SAT-specific DEGs were related to ECM reconstruction processes and response to hormones (Figure 2f).
Figure 2.
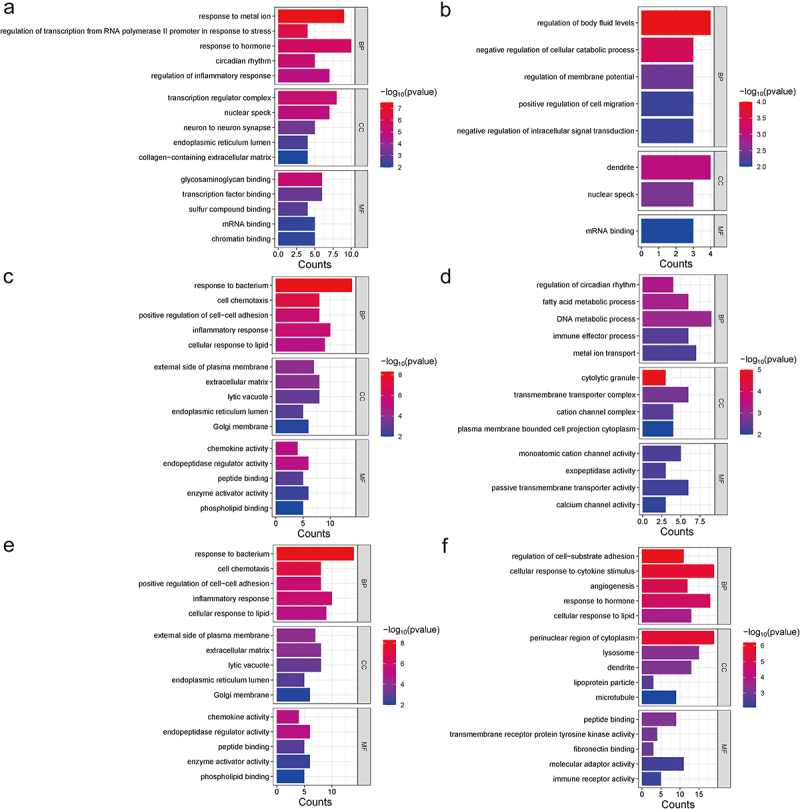
Perform GO enrichment analysis of shared DEGs, VATspecific DEGs, and SAT-specific DEGs in women VAT and SAT gene expression profiles using metascape. Five GO enrichment results were displayed in each term for biological processes (BP), cellular components (CC), and molecular functions (MF) (if enough), with p < 0.01. (a) GO enrichment results of shared DEGs with the same expression trends in VAT and SAT(n = 37). (b) GO enrichment results of shared DEGs with the different expression trends (upregulated in one type and downregulated in the other at type) in VAT and SAT (n = 18). (c, d) GO enrichment analysis of DEGs with VAT-specific DEGs upregulation (n = 642) or downregulation (n = 99). (e, f) GO enrichment analysis of DEGs with SAT-specific DEGs upregulation (n = 78) or downregulation (n = 195).
KEGG pathway analysis revealed that the signal pathways enriched with shared DEGs in VAT and SAT primarily included the IL-17 signaling pathway, Toll-like receptor signaling pathway, parathyroid hormone synthesis, secretion and action, chemokine signalling pathway (Figure 3a). Additionally, the signaling pathways enriched with VAT-specific DEGs were mainly associated with MAPK signaling pathway, PI3K-Akt signaling pathway, IL-17 signaling pathway, oestrogen signaling pathway (Figure 3b). In contrast, the signaling pathways enriched with SAT-specific DEGs mainly involved cytokine-cytokine receptor interaction, Toll-like receptor signaling pathway, cell adhesion molecules, and viral protein interaction with cytokine and cytokine receptors in diabetic complications (Figure 3c).
Figure 3.
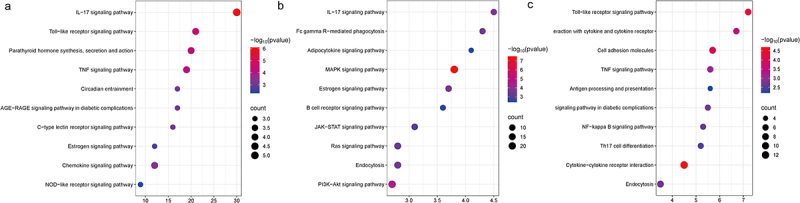
Perform KEGG enrichment analysis of shared DEGs, VAT-specific DEGs, and SAT-specific DEGs in women VAT and SAT gene expression profiles usingMetascape. Each term displays 10 enriched KEGG pathways, with p < 0.01 as the selection criterion. (a) KEGG analysis of shared DEGs(n = 55). (b) KEGG analysis of VAT-specific DEGs (n = 741). (c) KEGG analysis of SAT-specific DEGs (n = 273).
Protein–protein interaction network and hub genes analysis of DEGs
Analysis of the protein–protein interaction network and hub genes revealed that the top 10 shared hub genes in VAT and SAT included composition-related activation protein-1 (FOS, FOSB, JUN, JUNB, and ATF3), early growth response factors (EGR1 and EGR2), ZFP36, NFKBIZ, and BTG2 (Figure 4a). The top 10 VAT-specific hub genes included macrophage-related genes (CD68 and CD163), integrin-related genes (ITGAM and ITGAX), cytokines and chemokines (IL-6, CXCR4, and CCR1), SELL, VAM1, and the downregulated gene GZMB (Figure 4b). The top 10 SAT-specific hub genes were associated with cytokines and chemokines (IL1B, CXCL10, CCL4L2, CCL3L1, CCL3, and CXCL2), as well as CD34, FCGR2A, SOCS3, and NLRP3, with CXCL10 being a crucial gene among the downregulated genes (Figure 4c).
Figure 4.
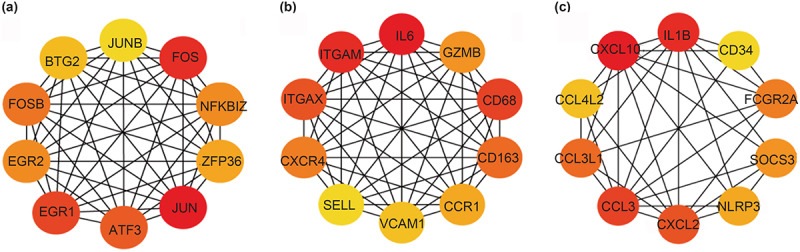
Use STRING to analyze the shared DEGs, VAT-specific DEGs, and SAT-specific DEGs in the women VAT and SAT gene expression profiles, and visualize them using Cytoscape, displaying the top 10 hub genes in each term. (a)The top 10 hub genes in shared DEGs. (b) The top 10 hub genes in VAT-specific DEGs. (c) The top 10 hub genes in SAT-specific DEGs.
Immune cell infiltration and correlation with immune cells analysis
The analysis of the infiltration of 22 types of immune cells reveals that the proportion of macrophages M0, monocytes, and neutrophils in VAT of women with T2DM was significantly higher than that in women without T2DM. Conversely, the proportion of mast cells resting natural killer (NK) cells activated, and T cells gamma-delta is significantly lower in women with T2DM compared to those without T2DM(p < 0.05) (Figure 5a, b). Additionally, in the SAT of women with T2DM, the proportion of B cells memory and mast cells activated was significantly higher than that in women without T2DM, while the proportion of macrophages M0, macrophages M1, and T cells gamma-delta was significantly lower (p < 0.05) (Figure 5c, d).
Figure 5.
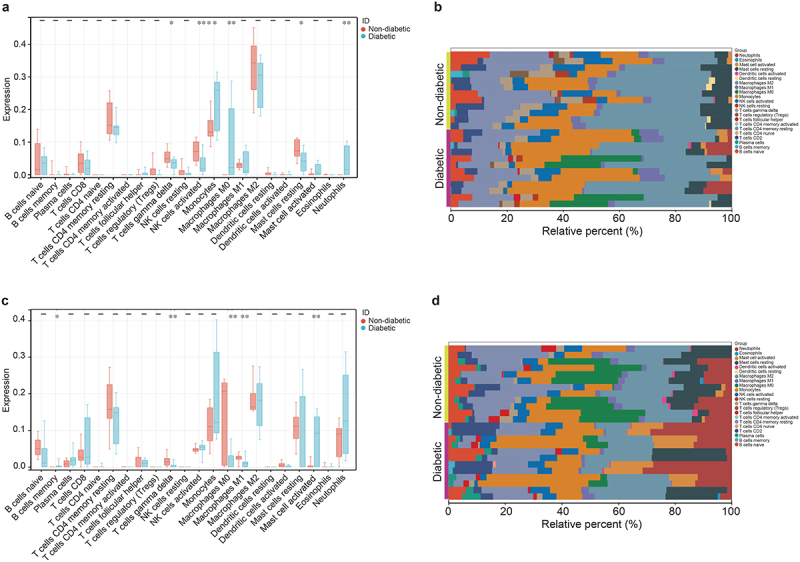
Immunoinfiltration analysis of DEGs in 22 immune cell subtypes in women VAT and SAT using CIBERSORTx. (a, b) block diagram and cumulative histogram show the difference in immune cell infiltration in VAT between diabetes patients (n = 12) and non-diabetes patients (n = 12). (c, d) block diagram and cumulative histogram show the difference in immune cell infiltration in SAT between diabetes patients (n = 12) and non-diabetes patients (n = 12). *p < 0.05 **p < 0.01.
The correlation between shared hub genes and 22 types of immune cells suggested that VAT was primarily positively correlated with monocytes, macrophages M1, and mast cell activated, while strongly negatively correlated with macrophages M2 and mast cells resting. In SAT, there was a strong positive correlation with monocytes, mast cells activated, and neutrophils, and a strong negative correlation with macrophages M1, mast cells resting, B cells naive, and T cells CD4 memory resting. The VAT-specific hub genes increased the correlation between B cells naive, T cells CD4 memory resting, and NK cells activated, although the correlation was mainly negative. Simultaneously, the correlation between mast cells and macrophages was reduced. IL-6 and GZMB showed opposite correlations with other genes, and the SAT-specific hub genes exhibited a strongly correlated immune cell spectrum consistent with the shared hub genes, whereas CD34 and CXCL10 exhibited opposite correlations with other genes (Figure 6).
Figure 6.
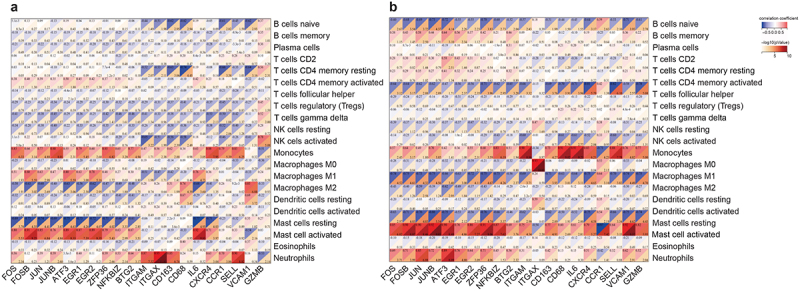
Correlation matrix of infiltration degree of 22 immune cells and hub DEGs in women VAT and SAT using Sangerbox. Red represents a positive correlation, blue represents a negative correlation, and darker colors represent a stronger correlation. (a) Select the top 10 hub genes in shared DEGs and the top 10 hub genes in VAT-specific DEGs to generate a heatmap of their correlation with the degree of immune cell infiltration. (b) Select the top 10 hub genes in shared DEGs and the top 10 hub genes in SAT-specific DEGs to generate a heatmap of their correlation with the degree of immune cell infiltration.
Discussion
T2DM is characterized as a chronic immune-mediated inflammatory ailment. Immune cells, along with their secreted cytokines and chemokines within AT, significantly contribute to the development of T2DM. Notably, SAT and VAT exert distinct functions in the pathogenesis of T2DM. Compared with SAT, VAT is more cellular and vascular, containing more inflammatory factors and immune cells, and has a stronger resistance to insulin [4,7,17,21]. Conversely, SAT exhibits a stronger absorption of circulating free fatty acids and triglycerides. This study compared the biological functions, signaling pathways, key genes, immune-infiltrating cell spectra, and correlation between co-expressed genes and DEGs in VAT and SAT of women with T2DM. This study provides a theoretical basis for further understanding the correlation between AT and T2DM.
This study analyzed the distribution of DEGs in two datasets and found that the VAT of women with T2DM showed a higher number of upregulated DEGs than SAT. Cluster analysis of biological functions revealed that the upregulated genes in VAT were primarily linked to ECM remodeling and alterations in tissue structure, especially concerning the morphology and function of blood vessels and muscles. The ECM serves as a highly dynamic structural framework that is essential for regulating cellular behavior during metabolic homeostasis and the progression of obesity [22]. Increased ECM synthesis creates a physical barrier for glucose transport and reduces insulin delivery and signal transduction in blood vessels, ultimately resulting in insulin resistance [23]. Abnormal accumulation of ECM components and their regulators in AT is also a characteristic feature of T2DM [24]. The biological function of downregulated DEG clusters was mainly related to fatty acid metabolism. Lipids play various roles as signaling molecules, metabolic substrates, and cell membranes [25]. Abnormal lipid metabolism is not only a significant clinical manifestation of T2DM but also a crucial factor in its onset and progression [26]. Notably, the number of downregulated DEGs in SAT was relatively higher than upregulated DEGs. Moreover, the main enriched biological functions were related to ECM reconstruction, consistent with the enrichment of upregulated DEGs in VAT. These findings suggested that ECM plays a significant role in the regulation of T2DM by VAT and SAT. Furthermore, the upregulated DEGs in SAT were mainly associated with immune cell- and factor-related functions, highlighting the presence of chronic low-grade inflammation and changes in immune function in the SAT of patients with T2DM.
Venn analysis of DEGs in the two datasets revealed that 55 shared DEGs were expressed in VAT and SAT. These genes primarily function in adipocyte expansion, immune cell accumulation, angiogenesis, and other processes related to AT reconstruction. The top 10 hub genes among the 55 shared DEGs are primarily transcription factors, including activator protein-1 (AP-1), nuclear transcription factor (NF-κB), and early growth response factor. Transcription factors play crucial roles in various diseases such as cancer, autoimmune/inflammatory diseases, diabetes, and cardiovascular diseases [27]. The activation of NF-κB, AP-1, and EGR can drive the M1 activation state of macrophages, leading to the expression of numerous pro-inflammatory target genes and promoting insulin resistance or T2DM. Activation of these transcription factors is closely associated with the of the IL-17 signaling pathway [28–30]. Ding et al. discovered that AP-1 family genes (FOS, FOSB, ATF3, JUN, and JUNB) are key genes in the WGCNA purple module and are key components in activating mast and Tfh cells in samples of abdominal aortic aneurysm-type perivascular AT, non-dilated perivascular AT, subcutaneous abdominal AT, and omental visceral AT. This study further demonstrated the significant role of the AP-1 family in regulating AT inflammation [31].
The top 10 VAT-specific hub genes were mainly associated with monocyte–macrophage surface markers, including CD68, ITGAX (CD11c), CD163, and ITGAM (CD11b). The expression of CD antigen genes serves as an indicator of inflammation in VAT [32–34]. Additionally, these genes have been implicated in cell adhesion and the promotion of tissue fibrosis. Previous studies have indicated that the chronic inflammatory state in AT may contribute to the excessive synthesis of ECM components and subsequent deposition of fibrotic stroma, which is a characteristic feature of AT in individuals with insulin resistance [35–38]. Importantly, members of the ITGA family (ITGAX and ITGA5) promote adipocyte fibrosis and angiogenesis by activating the PI3k/Akt signaling pathway [39,40]. The PI3k/Akt pathway is one of the main pathways involved in insulin transmission, with functions including the regulation of cell growth, apoptosis, and glucose and lipid metabolism. However, this pathway was not identified among the signaling pathways enriched in SAT. The top 10 SAT-specific hub genes primarily consisted of chemokine-related genes, including CXCL10, CCL4L2, CCL3L1, CCL3, and CXCL2. NLRP3 plays a key role in this process. The interaction between these genes occurs through the TLR/NF-κB signaling pathway [41–43], aligning with the findings of previous KEGG enrichment analysis of DEGs in SAT. Notably, certain substances, such as free fatty acids, saturated fatty acids, and lipopolysaccharides, can be recognized by TLRs through pathogen-associated molecular patterns and danger-associated molecular patterns [44]. This recognition activates the monocyte/macrophage NF-κB pathway, leading to the transcription of the NLRP3 gene and pro-IL-1β [45]. IL-1β, in turn, triggers the production of various cytokines and chemokines, causing the infiltration of macrophages and other immune cells [46]. IL-6 and IL-1β are recognized as the most significant pro-inflammatory factors secreted by adipocytes and macrophages, with elevated IL-6 levels closely linked to the onset of type 2 diabetes mellitus (T2DM) [47]. This study highlights IL-6 as a key central gene in VAT, while IL-1β has been identified as a crucial hub gene in SAT. Research conducted by Wen and Wang on WAT in lean and obese individuals revealed that important hub genes include IL-6 and IL-1β [48]. In a study focusing on healthy overweight and obese subjects, it was found that IL-6 expression, secreted by Th1 and pro-inflammatory macrophages, was significantly higher in VAT than in SAT [18]. However, one study indicated that IL-6 is associated with VAT and deep SAT (dSAT) volume in male obese patients, while IL-6 levels correlate with SAT, SAT hip, and superficial SAT (sSAT) volume in female obese patients [20]. The discrepancies in these research findings can be attributed to factors such as the progression of metabolic diseases (including obesity, insulin resistance, or T2DM), gender, using insulin, and the sampling location within the study population. Nonetheless, these findings underscore the critical roles of these two central genes in different AT depots.
AT contains a large number of immune cells, and the activity of these cells is related to the regulation of adipocytes and systemic metabolic functions [49]. In this study, immune infiltration analysis was conducted to examine the types and distribution patterns of different immune cells in the sample based on the differential expression of marker genes. From the analysis results, it is evident that the differences in immune cells between the two types of AT are mainly associated with bone marrow-derived immune cells such as monocytes, macrophages, granulocytes (neutrophils and mast cells), as well as lymphoid-derived immune cells such as T cells (gamma-delta T cells), B cells, and NK cells. VAT affects the activity of a greater number of immune cells than SAT, and the immune infiltration state is primarily upregulated by myeloid immune cells, including M0 macrophages, monocytes, and neutrophils. An increased level of cellular infiltration indicates a higher degree of inflammation and activation of the innate immune response. Studies have demonstrated that the expansion of AT in obese mice and humans can increase the infiltration of myeloid immune cells [16,50], which may be a crucial mechanism in the development of obesity. According to the results of this study, this infiltration state appears more frequently in VAT, which is also evidence that VAT acts as a ‘bad fat’. Compared with VAT, SAT had a more pronounced effect on memory B cell activity [51]. B cells are acquired immune cells capable of producing specific antibodies, presenting antigens, producing cytokines, and participating in the regulation of AT inflammation and insulin resistance [52]. However, the impact of B1 and B2 cell subtypes in B cells on the development of T2DM differs. B1 cells, found in mouse VAT, have been shown to exacerbate inflammation, insulin resistance, and systemic glucose intolerance, while the introduction of B2 cells into VAT has been found to promote metabolic dysfunction [53]. Further research is required to understand the role of B cell subtypes in SAT. In the present study, the T2DM population exhibited changes in the balance between the two types of mast cells in both AT types. This included a decrease in resting mast cell infiltration and an increase in activated mast cell infiltration. Although statistical significance was not observed in one AT type, potentially owing to variations in data consistency across groups, the trend still indicates a reduction in resting mast cells within AT likely due to their activation. Previous studies have demonstrated that activated mast cells contribute to the development of T2DM through their effects on ECM remodeling (such as blood vessels and fibrosis) and the promotion of inflammatory cell recruitment and proliferation [54]. In line with the observed decrease in mast cell infiltration, the degree of infiltration of gamma-delta T cells in the AT of patients with T2DM also decreased. Taylor et al. demonstrated that hyperglycemia leads to impaired T cell proliferation in the skin of mouse models of obesity and metabolic diseases [55]. Additionally, a long-term ketogenic diet in mice can lead to obesity, impaired metabolic health, and depleted fat-resident gamma-delta T cells [56], which may be responsible for the above results.
This study examined the relationship between shared hub genes found in VAT and SAT and 22 types of immune-infiltrating cells. The findings revealed that most of these genes exhibited a positive correlation with M1 macrophages and a negative correlation with M2 macrophages in VAT. However, in SAT, the correlation was negative for M1 macrophages and lymphocytes, specifically resting B cells and naive CD4 memory cells, and displayed a weak correlation with M2 macrophages. Notably, M1 macrophages are proinflammatory, whereas M2 macrophages are anti-inflammatory [57]. Inflammatory activation by M1-polarized macrophages plays a crucial role in the development of T2DM [58]. B cells can serve as antigen-presenting cells and induce various inflammatory cells to produce cytokines and chemokines, which act as CD4+ T cell co-stimulators [22]. These results confirm that VAT-specific hub genes mainly induce inflammation, whereas SAT-specific hub genes mainly improve inflammation, indicating that the same genes have different interaction mechanisms in different AT types. Whether the difference between the above genes and immune cell infiltration can explain the induction of T2DM in VAT and its protective role in SAT requires further research. Nonetheless, the results of this study offer promising target genes and immune cells for subsequent research endeavors.
By conducting a bioinformatics analysis of VAT and SAT in women patients with T2DM, this study revealed the differences and commonalities between them in terms of biological functions, signaling pathways, key genes, and immune cell infiltration. Although the sample size of this study is limited and lacks experimental verification, these findings still provide a theoretical basis for further exploring the changes of key genes and immune cell infiltration in different AT women’s compartments and their correlation with the occurrence and progress of type 2 diabetes.
Methods
Selection and information introduction of GEO datasets
For the analysis, the GSE29226 and GSE29231 datasets were chosen from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). The selected dataset was the expression profiling array (GPL6947) of the Illumina platform, the non-normalized data was obtained using GenomeStudio data analysis software (Illumina). The data was then normalized by using average normalization. The GSE29226 dataset comprised samples from women’s SAT, whereas the GSE29231 dataset consisted of samples from women’s VAT. The two data sets are composed of 48 samples, including 12 VAT samples and 12 SAT samples from 3 non-diabetes women, 12 VAT samples and 12 SAT samples from 3 diabetes women, with an average age of 57.1 years (range 37 to 58 years). Diabetes patients were treated with sulfonylureas or insulin, no one has ever used pioglitazone, and all received insulin treatment before surgery. Biopsy samples are obtained from tissues exposed and wasted during the surgical process. There was a significant difference in glycosylated hemoglobin (%HbA1c) levels between non-diabetes patients and diabetes patients (p < 0.05), and there was no significant difference in BMI (p > 0.05).
DEG screening and data analysis
GEO2R online analysis software (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to conduct differential expression analysis of the samples from both datasets and to process the data accordingly. DEGs were identified based on the criteria of adjusted p-value <0.05 and |logFC| ≥ 1. Venn analysis was performed using the online software Jvenn (https://jvenn.toulouse.inrae.fr/app/example.html) to identify co-expressed genes and DEGs, which were further analyzed.
GO enrichment and KEGG pathway analysis of DEGs
Metascape online analysis software (https://metascape.org/gp/index.html) was used to conduct GO enrichment analysis of biological processes, cellular components, and molecular functions, as well as KEGG pathway analysis, of DEGs. The critical criteria for selection included a p-value <0.01, a minimum overlap > 3, and a minimum enrichment > 1.5. Five GO enrichment results were displayed in each term for biological processes (BP), cellular components (CC), and molecular functions (MF) (if enough) . For KEGG pathway analysis, the top10 pathways were selected for display.
Protein–protein interaction network and hub gene analysis
The STRING online analysis website (https://cn.string-db.org/) was used to analyze the protein–protein interaction network of the DEGs. Subsequently, the Cytohubba plug-in from Cytoscape 3.7.2 was used to select and visualize the hub genes. Three sets of data were chosen for enrichment analysis: genes of shared DEGs between the two ATs, VAT-specific DEGs, and SAT-specific DEGs. The top 10 hub genes from each dataset were selected for display analysis.
Immune cell infiltration and correlation with immune cells analysis
Obtained the expression matrix, used Sangerbox 3.0 (http://www.sangerbox.com/login.html) to obtain gene names, removed rows with NA values greater than 50%, and used the impute.knn function of the R software package imputes to complete missing values. The CIBERSORTx (https://cibersortx.stanford.edu/) algorithm was utilized to analyze and visualize the infiltration of 22 immune cell subtypes. Box plots and correlation analyses were drawn using the Sangerbox 3.0. Data were processed using JMP version 10.0. As the measurement data did not follow a normal distribution, the Wilcoxon nonparametric test was employed for data comparison. Statistical significance was set at p < 0.05.
Supplementary Material
Acknowledgments
We thank the authors who deposited their microarray datasets, GSE29226 and GSE29231, into the public GEO database.
Funding Statement
This work was supported by the National Clinical Key Specialty Construction Project [grant number: Bingcaishe [2023] 16], the Clinical Medical Research Center for Laboratory Medicine of Xinjiang Production and Construction Corps [grant number: Bingkefa [2023] 12], and the Science and Technology Plan of Xinjiang Production and Construction Corps Hospital [(No.2020002]. The sponsors had no role in the study design; data collection, analysis, or interpretation; writing of the report; or the decision to submit the article for publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Qian Shi is responsible for manuscript writing and data processing. Yongxin Li is responsible for visualizing images. Mengjie Liang is responsible for data statistical analysis. Chunyan Liu and Hefei Zha are responsible for proofreading the manuscript. Xin Zhang and Fuchun Zhang are responsible for manuscript review and editing.
All authors have read and approved the final work.
Data availability statement
The datasets generated and analysed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29231. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29226). Data are available upon request to the corresponding author.
Ethical approval
This study was approved by the Ethics Committee of Xinjiang Production and Construction Corps Hospital (approval number: 20200301).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21623945.2024.2442419
References
- [1].Kusminski CM, Bickel PE, Scherer PE.. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15(9):639–12. doi: 10.1038/nrd.2016.75 [DOI] [PubMed] [Google Scholar]
- [2].Koenen M, Hill MA, Cohen P, et al. Adipose Tissue and Vascular Dysfunction. Circ Res. 2021;128(7):951–968. doi: 10.1161/CIRCRESAHA.121.318093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang CL, Xiao LL, Xu M, et al. Chemerin deficiency regulates adipogenesis is depot different through TIMP1. Genes Dis. 2021;8(5):698–708. doi: 10.1016/j.gendis.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ibrahim MM. Subcutaneous and visceral adipose tissue: structural and functional differences. Obes Rev. 2010;11(1):11–18. doi: 10.1111/j.1467-789X.2009.00623.x [DOI] [PubMed] [Google Scholar]
- [5].Herold J, Kalucka J. Angiogenesis in adipose tissue: the interplay between adipose and endothelial cells. Front Physiol. 2021;11:624903. doi: 10.3389/fphys.2020.624903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lessard J, Tchernof A. Interaction of the glucocorticoid and androgen receptors in adipogenesis. Chem Biol. 2012;19(9):1079–1080. doi: 10.1016/j.chembiol.2012.09.003 [DOI] [PubMed] [Google Scholar]
- [7].Mitchelson KAJ, O’Connell F, O’Sullivan J, et al. Dietary fats, and gastrointestinal cancer risk-potential mechanisms relating to lipid metabolism and inflammation. Metabolites. 2024;14(1):42. doi: 10.3390/metabo14010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qiu Y, Deng X, Sha Y, et al. Visceral fat area, not subcutaneous fat area, is associated with cardiac hemodynamics in type 2 diabetes. Diabetes Metab Syndr Obes. 2020; 13:4413–4422 doi: 10.2147/DMSO.S284420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kahn D, Macias E, Zarini S, et al. Exploring visceral and subcutaneous adipose tissue secretomes in human obesity: implications for metabolic disease. Endocrinology. 2022;163(11):bqac140. doi: 10.1210/endocr/bqac140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chait A, den Hartigh LJ. Adipose tissue distribution, inflammation and its metabolic consequences, including diabetes and cardiovascular disease. Front Cardiovasc Med. 2020;7(22):22. doi: 10.3389/fcvm.2020.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lustig RH, Collier D, Kassotis C, et al. JJ. Obesity I: overview and molecular and biochemical mechanisms. Biochem Pharmacol. 2022;199:115012. doi: 10.1016/j.bcp.2022.115012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee JJ, Britton KA, Pedley A, et al. Adipose tissue depots and their Cross-sectional associations with circulating Biomarkers of metabolic regulation. J Am Heart Assoc. 2016;5(5):e002936. doi: 10.1161/JAHA.115.002936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park YM, Erickson C, Bessesen D, et al. Age- and menopause-related differences in subcutaneous adipose tissue estrogen receptor mRNA expression.Steroids. Steroids. 2017;121:17–21. doi: 10.1016/j.steroids.2017.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jialal I, Devaraj S. Subcutaneous adipose tissue biology in metabolic syndrome. Horm Mol Biol Clin Investig. 2018;33(1):20170074. doi: 10.1515/hmbci-2017-0074 [DOI] [PubMed] [Google Scholar]
- [15].Sam S. Differential effect of subcutaneous abdominal and visceral adipose tissue on cardiometabolic risk. Horm Mol Biol Clin Investig. 2018;33(1):20180014. doi: 10.1515/hmbci-2018-0014 [DOI] [PubMed] [Google Scholar]
- [16].Chung KJ, Nati M, Chavakis T, et al. Innate immune cells in the adipose tissue. Rev Endocr Metab Disord. 2018. Dec;19(4):283–292. doi: 10.1007/s11154-018-9451-6 [DOI] [PubMed] [Google Scholar]
- [17].Michailidou Z, Gomez-Salazar M, Alexaki VI., et al. Innate immune cells in the adipose tissue in health and metabolic disease. J Innate Immun. 2022;14(1):4–30. doi: 10.1159/000515117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McLaughlin T, Liu LF, Lamendola C, et al. T-cell profile in adipose tissue is associated with insulin resistance and systemic inflammation in humans. Arterioscler Thromb Vasc Biol. 2014;34(12):2637–2643. doi: 10.1161/ATVBAHA.114.304636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vasanthakumar A, Chisanga D, Blume J, et al. Kallies A.Sex-specific adipose tissue imprinting of regulatory T cells. Nature. 2020;579(7800):581–585. doi: 10.1038/s41586-020-2040-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van den Munckhof Icl, Bahrar H, Schraa K, et al. Sex-specific association of visceral and subcutaneous adipose tissue volumes with systemic inflammation and innate immune cells in people living with obesity. Int J Obes (Lond). 2024;48(4):523–532. doi: 10.1038/s41366-023-01444-9 [DOI] [PubMed] [Google Scholar]
- [21].Houghton SC, Eliassen H, Tamimi RM, et al. Central adiposity and subsequent risk of breast cancer by menopause status. J Natl Cancer Inst. 2021;113(7):900–908. doi: 10.1093/jnci/djaa197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xia T, Shen Z, Cai J, et al. ColXV aggravates adipocyte apoptosis by facilitating abnormal extracellular matrix remodeling in mice. Int J Mol Sci. 2020;21(3):959. doi: 10.3390/ijms21030959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Williams AS, Kang L, Wasserman DH. The extracellular matrix and insulin resistance. Trends Endocrinol Metab. 2015;26(7):357–366. doi: 10.1016/j.tem.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin D, Chun TH, Kang L. Adipose extracellular matrix remodelling in obesity and insulin resistance. Biochem Pharmacol. 2016;119:8–16. doi: 10.1016/j.bcp.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang Q, Vijayakumar A, Kahn BB. Metabolites as regulators of insulin sensitivity and metabolism. Nat Rev Mol Cell Biol. 2018;19(10):654–672. doi: 10.1038/s41580-018-0044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Savage DB, Petersen KF, Shulman GI. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev. 2007;87(2):507–520. doi: 10.1152/physrev.00024.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vallerga CL, Zhang F, Fowdar J, et al. Analysis of DNA methylation associates the cystine–glutamate antiporter SLC7A11 with risk of Parkinson’s disease. Nat Commun. 2020;11(1):1238. doi: 10.1038/s41467-020-15065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72(1):219–246. doi: 10.1146/annurev-physiol-021909-135846 [DOI] [PubMed] [Google Scholar]
- [29].Schmidt J, Stoffels B, Moore BA, et al. Proinflammatory role of leukocyte-derived Egr-1 in the development of murine postoperative ileus. Gastroenterology. 2008;135(3):926–936.e2. doi: 10.1053/j.gastro.2008.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li XX, R ZJ, Mj M, et al. IL-17 receptor–based signaling and implications for disease. Nat Immunol. 2019;20(12):1594–1602. doi: 10.1038/s41590-019-0514-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ding S, Gan T, Xiang Y, et al. FOS gene associated immune infiltration signature in perivascular adipose tissues of abdominal aortic aneurysm. Gene. 2022;831:146576. doi: 10.1016/j.gene.2022.146576 [DOI] [PubMed] [Google Scholar]
- [32].Fujisaka S, Usui I, Nawaz A, et al. M2 macrophages in metabolism. Diabetol Int. 2016;7(4):342–351. doi: 10.1007/s13340-016-0290-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shi DC, Zhong Z, Xu RC, et al. Association of ITGAX and ITGAM gene polymorphisms with susceptibility to IgA nephropathy. J Hum Genet. 2019;64(9):927–935. doi: 10.1038/s10038-019-0632-2 [DOI] [PubMed] [Google Scholar]
- [34].Kwon EY, Shin SK, Cho YY, et al. Time-course microarrays reveal early activation of the immune transcriptome and adipokine dysregulation leads to fibrosis in visceral adipose depots during diet-induced obesity. BMC Genomics. 2012;13(1):450. doi: 10.1186/1471-2164-13-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Batista ML, Henriques FS, Neves RX, et al. Cachexia-associated adipose tissue morphological rearrangement in gastrointestinal cancer patients. J Cachexia Sarcopenia Muscle. 2016;7(1):37–47. doi: 10.1002/jcsm.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Swendeman SL, Hla T. Lipid mediators, M2 macrophages, and pathological neovascularization. Trends Mol Med. 2018;24(12):977–978. doi: 10.1016/j.molmed.2018.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu XL, Luo TT, Fan ZX, et al. Single cell RNA-seq resolution revealed CCR1+/SELL+/XAF+ CD14 monocytes mediated vascular endothelial cell injuries in Kawasaki disease and COVID-19. Biochim Biophys Acta Mol Basis Dis CCR1. 2023;1869(5):166707. doi: 10.1016/j.bbadis.2023.166707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bourgeois C, Gorwood J, Olivo A, et al. Contribution of adipose tissue to the chronic immune activation and inflammation associated with HIV infection and its treatment. Front Immunol. 2021;12:670566. doi: 10.3389/fimmu.2021.670566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang J, Yang L, Liang F, et al. Integrin alpha x stimulates cancer angiogenesis through PI3K/Akt signaling–mediated VEGFR2/VEGF-A overexpression in blood vessel endothelial cells. J Cell Biochem. 2019;120(2):1807–1818. doi: 10.1002/jcb.27480 [DOI] [PubMed] [Google Scholar]
- [40].Liu YX, Li YZ, Liang JT, et al. The mechanism of leptin on inhibiting fibrosis and promoting browning of white fat by reducing ITGA5 in mice. Int J Mol Sci. 2021;22(22):12353. doi: 10.3390/ijms222212353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Schulthess FT, Paroni F, Sauter NS, et al. CXCL10 impairs β cell function and viability in diabetes through TLR4 signaling. Cell Metab. 2009;9(2):125–139. doi: 10.1016/j.cmet.2009.01.003 [DOI] [PubMed] [Google Scholar]
- [42].Bouma G, Coppens JM, Lam-Tse WK, et al. An increased MRP8/14 expression and adhesion, but a decreased migration towards proinflammatory chemokines of type 1 diabetes monocytes. Clin Exp Immunol. 2005;141(3):509–517. doi: 10.1111/j.1365-2249.2005.02865.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang Y, Li C, Wang ZF, et al. Blocking CXC motif chemokine ligand 2 ameliorates diabetic peripheral neuropathy via inhibiting apoptosis and NLRP3 inflammasome activation. Biol Pharm Bull. 2023;46(5):672–683. doi: 10.1248/bpb.b22-00680 [DOI] [PubMed] [Google Scholar]
- [44].Kim Y, Gromovsky AD, Brown JM, et al. Gamma-tocotrienol attenuates the aberrant lipid mediator production in NLRP3 inflammasome-stimulated macrophages. J Nutr Biochem. 2018;58:169–177. doi: 10.1016/j.jnutbio.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang L, Mosoian A, Schwartz ME, et al. HIV infection modulates il‐1β response to LPS stimulation through a TLR4‐NLRP3 pathway in human liver macrophages. J Leukoc Biol. 2019;105(4):783–795. doi: 10.1002/JLB.4A1018-381R [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Noël A, Xiao R, Perveen Z, et al. Incomplete lung recovery following sub-acute inhalation of combustion-derived ultrafine particles in mice. Part Fibre Toxicol. 2016;13(1):10. doi: 10.1186/s12989-016-0122-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zu Y, Zhao L, Hao L, et al. Browning white adipose tissue using adipose stromal cell-targeted resveratrol-loaded nanoparticles for combating obesity. J Control Release. 2021;333:339–351. doi: 10.1016/j.jconrel.2021.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wen J, Wang LW. Identification of key genes and their association with immune infiltration in adipose tissue of obese patients: a bioinformatic analysis. Adipocyte. 2022;11(1):401–412. doi: 10.1080/21623945.2022.2104512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Frikke-Schmidt H, Zamarron BF, O’Rourke RW, et al. Weight loss independent changes in adipose tissue macrophage and T cell populations after sleeve gastrectomy in mice. Mol Metab. 2017;6(4):317–326. doi: 10.1016/j.molmet.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Todosenko N, Khaziakhmatova O, Malashchenko V, et al. Adipocyte- and monocyte-mediated vicious circle of inflammation and obesity (review of cellular and molecular mechanisms). Int J Mol Sci. 2023;24(15):12259. doi: 10.3390/ijms241512259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cw F, Em S, Benvie A, et al. Exacerbated staphylococcus aureus foot infections in Obese/Diabetic mice are associated with impaired germinal center reactions, Ig Class switching, and humoral immunity. J Immunol. 2018;201(2):560–572. doi: 10.4049/jimmunol.1800253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Winer DA, Winer S, Shen L, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat Med. 2011;17(5):610–617. doi: 10.1038/nm.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kempuraj D, Caraffa A, Ronconi G, et al. Are mast cells important in diabetes? Pol J Pathol. 2016;3(3):199–206. doi: 10.5114/pjp.2016.63770 [DOI] [PubMed] [Google Scholar]
- [54].Taylor KR, Mills RE, Costanzo AE, et al. γδ T cells are reduced and rendered unresponsive by hyperglycemia and chronic TNFα in mouse models of obesity and metabolic disease. PLOS ONE. 2010;5(7):e11422. doi: 10.1371/journal.pone.0011422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goldberg EL, Shchukina I, Asher JL, et al. Ketogenesis activates metabolically protective γδ T cells in visceral adipose tissue. Nat Metab. 2020;2(1):50–61. doi: 10.1038/s42255-019-0160-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shyu KG, Wang BW, Fang WJ, et al. Exosomal MALAT1 derived from High glucose-treated Macrophages up-regulates Resistin expression via miR-150-5p downregulation. Int J Mol Sci. 2022;23(3):1095. doi: 10.3390/ijms23031095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yu M, Duan XH, Cai YJ, et al. Multifunctional nanoregulator reshapes immune microenvironment and enhances immune memory for tumor immunotherapy. Adv Sci (Weinh). 2019;6(16):1900037. doi: 10.1002/advs.201900037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang ZX, Xia QY, Su WX, et al. The commonness in immune infiltration of rheumatoid arthritis and atherosclerosis: screening for central targets via microarray data analysis. Front Immunol. 2022;13:1013531. doi: 10.3389/fimmu.2022.1013531 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29231. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29226). Data are available upon request to the corresponding author.


