ABSTRACT
The number of authorized genetically modified (GM) soybeans has increased worldwide. In Japan, 34 GM soybeans containing single events and their stacked varieties have been approved as food. However, not all approved GM events are commercially cultivated or distributed. In this study, we evaluated domestically distributed samples from the United States (US) and Canada using 17 event-specific detection methods for GM soybeans. Identity-preserved (IP) soybean samples imported from the US and Canada, and non-IP samples from the US in 2021 and 2022 were analyzed. Four GM soybean events consisting of MON89788, A5547–127, MON87708, and DAS-44406 were detected in all lots in the non-IP samples. Furthermore, a single-kernel-based analysis was conducted to determine whether the detected GM soybean events are stacked. The results suggest that DAS-44406 is rapidly increasing, particularly as a single event among GM soybeans.
KEYWORDS: Distribution, event, genetically modified (GM), stacked variety, soybean
Introduction
The global area under genetically modified (GM) crops is continually increasing and reached 206.3 million hectares in 2023.1 However, some consumers still express concerns about the utilization of genetically modified organisms (GMOs). In response, many countries and areas have passed laws requiring food labeling systems to indicate the presence of authorized GM crops. Under such conditions, not only the planted area, but also the number of varieties of GM events has been continuously increased. The main commercially grown GM crops are soybean, maize, cotton, and oilseed rape. In 39 countries and areas, 472 GM crops have been approved for food, feed, or environmental release.2
Soybeans are among the most important crops in Japan. The domestic consumption of soybeans as oil and food exceeds 4 million tons/y, but the self-sufficiency ratio for the crop is only 6%.3 Most of the soybeans consumed in Japan are mainly imported from the United States (US) and Canada where GM soybeans account for over 90% of the soybean cultivation area. In many countries including Japan, GM-labeling of food is mandatory if the GM content exceeds the authorized threshold level of the country. For example, in the European Union, Korea and the US, the threshold levels have been set at 0.9%, 3%, and 5%, respectively.4,5 In Japan, non-GM maize and soybeans are segregated and imported from other countries by an identity-preserved (IP) handling system.6 However, the unintentional commingling of GM products in non-GM materials is inevitable. In such cases, up to 5% unintentional commingling of approved GM maize or soybeans is generally accepted, and foods within this level of GM content can voluntarily be labeled as “handled to prevent commingling of GMO,” while non-GMO labeling is permitted when commingled GM contents are not detectable instead of 5%.7–9
The polymerase chain reaction (PCR) technique is widely used to detect and quantify GM crops in foods and feed. PCR detection methods can be largely classified into event-specific and screening methods. In event-specific detection, a unique sequence at the junction between the plant genome and recombinant DNA is used as the target. Screening methods target commonly conserved elements among many GM events, such as the Cauliflower Mosaic Virus 35S promoter (P35S), the nopaline synthase terminator (TNOS), and the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS).
In the Japanese standard method, qualitative detection methods for processed foods containing GM soybeans targeting MON89788, and P35S screening have been adopted.10 Additionally, quantification methods targeting GTS 40-3-2 (RRS), MON89788, and A2704–12 are used.10 In a previous study, non-IP soybean samples imported to Japan from the US and Brazil in 2017 were analyzed, revealing that the GM events, RRS, MON89788, and A2704–12 constituted the majority of GM events in these samples.11 In subsequent years, the number of approved GM events has been increasing in Japan. By the end of April 2024, 19 single GM soybean events and 15 stacked varieties had been approved.12 There are no surveys for distributed samples in recent years, despite the number of approved GM soybeans, which were not covered in the previous study, increased. In this study, we investigated IP and non-IP soybean samples imported from the US and Canada in 2021 and 2022 using 17 sets of event-specific real-time PCR detection methods. In case of large-scale distribution is revealed of GM event(s) other than the events or stacked varieties covered by the current standard method, it will be necessary to develop a detection method for those event(s).
Materials and Methods
Plant Materials
Non-IP and IP soybean samples from the 2021 and 2022 harvests were obtained from Japanese trading companies. These samples included five different non-IP and IP lots from the US and five different IP lots from Canada. The non-IP samples were imported for oilseeds processing, whereas the IP samples were imported for soy sauce production. Each sample was sourced from a different company, with non-IP samples labeled as N1, N2, N3, N4, and N5, and IP samples as I1, I2, I3, I4, and I5. The sample sizes ranged from 500 to 1500 g. Six certified reference materials (CRMs) in powder form were purchased from Sigma-Aldrich (St. Louis, MO, USA): RRS (Event 40-3-2), DP-305423 (ERM-BF426d), DAS-68416 (ERM-BF432d), DAS-44406 (ERM-BF436b), DAS-81419 (ERM-BF437b), and GMB151 (ERM-BF443b). Eleven CRMs were purchased from the American Oil Chemists’ Society (Urbana, IL): ground seeds of MON89788 (AOCS 0906-B), MON87701 (AOCS 0809-A), MON87705 (AOCS 0210-A), BPS-CV127 (AOCS 0911-C), MON87708 (AOCS 0311-A), MON87769 (AOCS 0809-B), MON87751 (AOCS 0215A), and SYHT0H2 (AOCS 0112A), and DNA extracts of A2704–12 (AOCS 0707-B), A5547–127 (AOCS 0707-C), and FG72 (AOCS 0610-A).
DNA Extraction
Soybean genomic DNAs were extracted using a DNeasy Plant Maxi Kit (Qiagen, Hilden, Germany) and a GM quicker (NIPPON GENE Co., Ltd., Tokyo, Japan) according to the Japanese standard method.10 DNA extraction was performed in duplicate for each sample with the extracted DNA solutions labeled as follows: I1–1 and I1–2 for I1, and N1–1 and N1–2 for N1. The concentration and quality of the extracted DNA solutions were evaluated by measuring ultraviolet absorbance with a NanoDrop One spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The concentration of the genomic DNA was adjusted to 10 ng/μL and used as a template in PCR analysis.
Individual Kernel Detection Analysis
A kernel-based detection analysis was performed, as previously described.11,13,14 In each sample, 48 to 53 soybean kernels were prepared using a grain counter plate (For 100 Soybeans; Fuji Kinzoku, Tokyo, Japan). Each soybean kernel was individually ground using a Multi-beads Shocker (MB601NIHS; Yasui Kikai Co., Osaka, Japan), and DNA extraction from the ground kernels was performed using the GM quicker, following the manufacturer’s instructions.
PCR Analysis
The primers and probes used in this study and the references are listed in Table 1.15–27 Oligonucleotide DNA primers and TaqMan probes were synthesized by FASMAC Co. Ltd. (Kanagawa, Japan) and Thermo Fisher Scientific (Carlsbad, CA), respectively. The probes were labeled with 6-carboxyfluorescein (FAM) and 6-carboxytetramethylrhodamin (TAMRA) at the 5′ and 3′ ends, respectively. The DAS-68416 and FG72 probes were labeled with minor groove binders at the 3′ end, and for the GMB151 and SYHT0H2 probes, black hole quenchers were used instead of TAMRA. The 25-μL reaction mixture contained 5 μL of the template DNA, 12.5 μL of the TaqMan Universal PCR Master Mix (Thermo Fisher Scientific), 0.5 μM primer pair, and 0.2 μM probe. The step-cycle program was set to 2 min at 50°C, 10 min at 95°C, 45 cycles of 30 s at 95°C, and 1 min at 59°C. The assay was repeated twice for each analysis using the ABI PRISM 7900HT (Thermo Fisher Scientific) or the LightCycler 480 (Roche Diagnostics).
Table 1.
Primer and probe sequences.
| Target | Sequences | References | |
|---|---|---|---|
| RRS | RRS 01–5 | CCTTTAGGATTTCAGCATCAGTGG | 15 |
| RRS 01–3 | GACTTGTCGCCGGGAATG | ||
| RRS-Taq | CGCAACCGCCCGCAAATCC | ||
| MON89788 | MON89788-F | TCCCGCTCTAGCGCTTCAAT | 15 |
| MON89788-R | TCGAGCAGGAC CTGCAGAA | ||
| MON89788-P | CTGAAGGCGGGAAACGACAATCTG | ||
| A2704–12 | KVM175 | GCAAAAAAGCGGTTAGCTCCT | 15 |
| SMO001 | ATTCAGGCTGCGCAACTGTT | ||
| TM031 | CGGTCCTCCGATCGCCCTTCC | ||
| A5547–127 | KVM175 | GCAAAAAAGCGGTTAGCTCCT | 15 |
| SMO001 | ATTCAGGCTGCGCAACTGTT | ||
| TM031 | CGGTCCTCCGATCGCCCTTCC | ||
| DP-305423 | DP305-f1 | CGTGTTCTCTTTTTGGCTAGC | 15 |
| DP305-r5 | GTGACCAATGAATACATAACACAAACTA | ||
| DP305-p | TGACACAAATGATTTTCATACAAAAGTCGAGA | ||
| MON87701 | MON87701 1 | CGTTTCCCGCCTTCAGTTTAAA | 16 |
| MON87701 2 | TGGTGATATGAAGATACATGCTTAGCAT | ||
| MON87701 | TCAGTGTTTGACACACACACTAAGCGTGCC | ||
| MON87705 | MON 87,705 | TTCCCGGACATGAAGCCATTTAC | 17 |
| MON 87,705 | ACAACGGTGCCTTGGCCCAAAG | ||
| MON 87,705 | AAGAGACTCAGGGTGTTGTTATCACTGCGG | ||
| MON87769 | MON 87,769 | CATACTCATTGCTGATCCATGTAGATT | 18 |
| MON 87,769 | GCAAGTTGCTCGTGAAGTTTTG | ||
| MON 87,769 probe | CCCGGACATGAAGCCATTTACAATTGAC | ||
| MON87708 | MON87708 | TCATACTCATTGCTGATCCATGTAG | 19 |
| MON87708 | AGAACAAATTAACGAAAAGACAGAACG | ||
| MON 87,708 | TCCCGGACTTTAGCTCAAAATGCATGTA | ||
| CV127 | SE-127-f4 | AACAGAAGTTTCCGTTGAGCTTTAAGAC | 20 |
| SE-127-r2 | CATTCGTAGCTCGGATCGTGTAC | ||
| SE-127-p3 | TTTGGGGAAGCTGTCCCATGCCC | ||
| DAS-68416 | DAS-68416-4_3f5 | GTACATTAAAAACGTCCGCAATGTGT | 21 |
| DAS-68416-4_3r3 | GTTTAAGAATTAGTTCTTACAGTTTATTGTTAG | ||
| DAS-68416-4_3p3 | TTAAGTTGTCTAAGCGTCAATA | ||
| DAS-44406 | DAS-44406-5F | TTATTGTTCTTGTTGTTTCCTCTTTAGG | 22 |
| DAS-44406-5 R | CCTCAATTGCGAGCTTTCTAATTT | ||
| DAS-44406-6-5p1 | ATTCGGACCTCCATGATGACCTTACCGTT | ||
| DAS-81419 | DAS81419-f2 | TCTAGCTATATTTAGCACTTGATATTCAT | 23 |
| DAS81419-r1 | GCTTCAAGATCCCAACTTGCG | ||
| DAS81419-p3 | ATCAACAGGCACCGATGCGCACCG | ||
| FG72 | MAE071 | AGATTTGATCGGGCTGCAGG | 24 |
| SHA097 | GCACGTATTGATGACCGCATTA | ||
| TM325 | AATGTGGTTCATCCGTCTT | ||
| GMB151 | PRIM1040 | TCAAATCAACATGGGTGACTAGAAA | 25 |
| PRIM1041 | CATTGTGCTGAATAGGTTTATAGCTATGAT | ||
| TM1789 | CAGTACTGGGCCCTTGTGGCGCT | ||
| SYHT0H2 | FE08316-F | GGGAATTGGGTACCATGCC | 26 |
| FE08317-R | TGTGTGCCATTGGTTTAGGGT | ||
| FE08318-P | CCAGCATGGCCGTATCCGCAA | ||
| MON87751 | MON 87,751 primer 2 | CTAAATTGCTCTTTGGAGTTTATTTTGTAG | 27 |
| MON 87,751 primer 1 | GGCCTAACTTTTGGTGTGATGATG | ||
| MON 87,751 probe | TGACTGGAGATCTCCAAAGTGAGGGGAAA | ||
| Le1 | Le1n02–5′ | GCCCTCTACTCCACCCCCA | 15 |
| Le1n02–3′ | GCCCATCTGCAAGCCTTTTT | ||
| Le1-Taq | AGCTTCGCCGCTTCCTTCAACTTCAC |
Results
Evaluation of Identity-Preserved-Handled Samples
We examined the commingling (s) of GM soybean in the IP handled samples from the US and Canada produced in 2021 and 2022 by targeting 17 event-specific detection methods. The events tested were listed in Table S1. The results of the US samples are summarized in Table 2. MON89788 was detected in all samples except I1 in 2021, and MON87708 was detected in all samples in 2022 and I4 in 2021, although the obtained Cq values were relatively high across the board. RRS and A5547–127 were detected in I1, I3 and I5 of the samples from 2022, and DAS-44406 was detected in sample I5 from 2022. DNA extraction was repeated twice for each sample, and no other GM events were reproducibly detected from either DNA extraction. In the Canadian samples, MON89788 was detected only in sample I1 from 2021 (Table S2).
Table 2.
List of Cq* values obtained from identity-preserved samples from the United States.
| 2021 |
2022 |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM event | PC | Sample no. |
Sample no. |
||||||||||||||||||
| I1–1 | I1–2 | I2–1 | I2–2 | I3–1 | I3–2 | I4–1 | I4–2 | I5–1 | I5–2 | I1–1 | I1–2 | I2–1 | I2–2 | I3–1 | I3–2 | I4–1 | I4–2 | I5–1 | I5–2 | ||
| RRS | 25.88 | – | – | – | – | 39.53 | – | – | – | – | – | 33.61 | 33.02 | – | – | 34.32 | 33.99 | – | 40.00 | 33.45 | 33.37 |
| MON89788 | 25.02 | – | – | 32.80 | 33.04 | 34.06 | 33.59 | 31.91 | 32.03 | 41.14 | 40.61 | 31.10 | 30.58 | 37.72 | 34.46 | 31.44 | 30.87 | 32.56 | 32.19 | 36.81 | 36.57 |
| A2704–12 | 23.19 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| A5547–127 | 23.80 | – | – | – | – | – | – | – | – | – | – | 33.77 | 33.77 | – | – | 34.52 | 33.94 | – | – | 37.04 | 36.86 |
| DP-305423 | 30.82 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87701 | 24.28 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87705 | 24.51 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87769 | 24.71 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87708 | 24.51 | – | – | – | – | – | – | 32.41 | 32.77 | – | 43.80 | 34.10 | 33.88 | 36.52 | 33.87 | 32.07 | 31.84 | 33.31 | 32.40 | 35.83 | 35.92 |
| CV127 | 24.45 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| DAS-68416 | 29.61 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| DAS-44406 | 26.93 | 39.50 | – | – | – | – | – | – | – | 39.46 | – | – | – | – | – | – | – | – | – | 32.42 | 32.04 |
| DAS-81419 | 25.01 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FG72 | 23.52 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| GMB151 | 25.89 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87751 | 24.72 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| SYHT0H2 | 24.16 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Le1 | 26.76 | 24.22 | 24.11 | 24.13 | 24.11 | 24.24 | 24.25 | 24.14 | 24.17 | 24.32 | 24.22 | 25.11 | 26.19 | 27.26 | 25.04 | 24.56 | 24.30 | 24.29 | 23.11 | 23.33 | 23.07 |
*Cq : quantification cycle for real-time PCR analysis.
To estimate the approximate comingling levels of GM soybeans, we quantified MON89788, which was detected in multiple samples with comparatively low Cq values relative to other events. A quantification method for MON89788 using a standard plasmid has been developed and validated.28,29 The limit of quantification (LOQ) was 0.5% for the quantification method. The contents of MON89788 were near or below the LOQ in all samples (data not shown).
Evaluation of Non-Identity-Preserved Samples
Next, we tested the non-IP samples from the US in a similar manner (Table 3). GM soybeans were detected in all non-IP samples, and MON89788, A5547–127, MON87708, and DAS-44406 were detected in all samples. RRS was detected in samples N1, N3, N4, and N5 from 2021, and in samples N2, N4, and N5 from 2022. A2704–12 and MON87701 were only detected in samples N4 and N3 from 2022, respectively, whereas FG72 was detected in samples N1, N2, N3, and N5 from 2021, and samples N3, N4, and N5 from 2022. For the remaining 9 events, DP-305423, MON87705, MON87769, CV127, DAS-68416, DAS-81419, GMB151, MON87751, and SYHT0H2, no amplification was detected in any sample. Although it was difficult to simply compare the Cq values obtained because of the differences in the performance of each primer and probe, it was generally considered that lower Cq values generally indicated higher copy numbers for the target sequence, suggesting relatively higher comingling. The comingling levels of MON89788 were higher than those of RRS in all samples. RRS, the first generation of glyphosate-resistant soybean, went off-patent in 2015 and has since been gradually phased out from seed stocks.30
Table 3.
List of Cq* values obtained from non-identity-preserved samples in the United States.
| 2021 |
2022 |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM event | Sample no. |
Sample no. |
||||||||||||||||||
| N1–1 | N1–2 | N2–1 | N2–2 | N3–1 | N3–2 | N4–1 | N4–2 | N5–1 | N5–2 | N1–1 | N1–2 | N2–1 | N2–2 | N3–1 | N3–2 | N4–1 | N4–2 | N5–1 | N5–2 | |
| RRS | 31.63 | 31.60 | – | – | 33.03 | 33.45 | 31.95 | 31.89 | 31.68 | 31.37 | – | – | 33.54 | 33.54 | – | – | 35.43 | 34.64 | 34.08 | 33.84 |
| MON89788 | 24.15 | 24.17 | 24.45 | 24.45 | 24.80 | 24.95 | 23.99 | 23.87 | 24.41 | 24.25 | 25.38 | 26.12 | 25.45 | 25.43 | 24.38 | 24.24 | 26.04 | 24.96 | 25.75 | 26.09 |
| A2704–12 | – | – | – | – | – | – | 36.51 | – | – | – | – | – | – | – | – | – | 34.48 | 33.35 | – | – |
| A5547–127 | 26.19 | 26.32 | 25.12 | 25.18 | 25.48 | 25.55 | 25.66 | 25.61 | 26.13 | 25.86 | 26.32 | 27.3 | 26.85 | 26.74 | 26.02 | 25.90 | 27.71 | 26.65 | 27.07 | 27.41 |
| DP-305423 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87701 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 37.55 | 37.63 | – | – | – | – |
| MON87705 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87769 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87708 | 24.70 | 24.86 | 24.74 | 24.85 | 25.24 | 25.41 | 24.33 | 24.29 | 24.97 | 24.83 | 25.72 | 26.74 | 25.85 | 25.79 | 24.73 | 24.70 | 26.21 | 25.29 | 26.18 | 26.33 |
| CV127 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| DAS-68416 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| DAS-44406 | 25.99 | 26.02 | 25.21 | 25.26 | 24.71 | 24.73 | 26.93 | 26.73 | 25.60 | 25.64 | 25.81 | 26.37 | 25.31 | 25.14 | 24.57 | 24.66 | 25.99 | 25.26 | 25.27 | 25.36 |
| DAS-81419 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| FG72 | 27.01 | 27.02 | 28.15 | 28.06 | 30.61 | 30.16 | – | – | 28.84 | 28.62 | – | – | – | – | 33.23 | 33.05 | 37.35 | 35.96 | 34.54 | 34.60 |
| GMB151 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| MON87751 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| SYHT0H2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Le1 | 24.40 | 24.30 | 24.03 | 24.11 | 24.13 | 24.07 | 24.18 | 24.17 | 24.41 | 24.05 | 23.92 | 24.08 | 24.44 | 24.40 | 24.42 | 24.32 | 26.14 | 25.36 | 25.67 | 25.64 |
*Cq : quantification cycle for real-time PCR analysis.
Kernel-Based Detection Analysis for Identification of Stacked Genetically Modified Varieties
To determine whether the detected GM soybean events were stacked in the non-IP samples, we collected 48–53 kernels from each sample in 2021 and 2022 and performed individual kernel detection analyses targeting 4 GM events, MON89788, MON87708, A5547–127, and DAS-44406 (Figure 1). For this analysis, because it was a single kernel, the mixing level would be nearly 100% if the kernel was a GM. We used purified DNA as the template, and it was expected that the Cq values would be approximately the same level as the positive control (23–25) in the case of GMs, and that the Cq value would not be obtained in the case of non-GM kernel. However, during the analysis, several kernels had unexpectedly high Cq values of approximately 35–40. Table S3 shows the Cq values obtained in sample N3. The cause of these high Cq values was unclear, and we speculated the effect of DNA from other kernel sources on the surface when conducting individual kernel detection. Then, we removed the seed coats from the kernels, and DNA was extracted separately from the seed coats and internal seed materials (Figure S1). Fourteen kernels were collected and analyzed. High Cq values were frequently observed in DNA extracted from seed coats, and were rarely detected inside the seeds, even in the same samples (Table S4). These findings indicated that kernels that showed high Cq values were GM negative seeds, which were created when a powder from other GM seeds attached to the surfaces of the tested samples. Based on these results, we set a Cq threshold of 35 or higher to classify seeds as GM-negative.
Figure 1.
Schematic workflow of the GM soybean detection strategy for non-ip samples.
The results of the kernel-based detection analyses are listed in Table 4. In the 2021 samples, the most frequently detected GM soybean variety was the stacked variety MON89788 × MON87708 (42.5%), followed by the single event DAS-44406 (20.4%), the triple stacked variety MON89788 × MON87708 × A5547–127 (20%), the single event A 5547–127 (7.5%), and the triple stacked variety MON89788 × MON87708 × DAS-44406 (5%). MON89788 × MON87708 accounted for the majority and single events of DAS-44406 and A5547–127 were present in all samples. In the 2022 samples, the ratio of DAS-44406 single event increased to 37.5%, followed by MON89788 × MON87708 × A5547–127 (23.4%), MON89788 × MON87708 (10.5%), MON89788 × MON87708 × A5547–127 × DAS-44406 (3.5%) varieties. It appears that nearly all detected MON89788 and MON87708 in these samples were stacked varieties containing both events, although the number of investigated kernels and samples was limited. We previously analyzed the ratios of GM soybean varieties of non-IP samples from 2017 using kernel-based inspections.11 The results of the 2017 samples were added to this study, and changes in the ratios of GM soybean varieties in 2017, 2021, and 2022 were shown in Figure 2. GM soybeans, containing MON89788, were largely detected as single events in 2017, whereas the proportion of stacked varieties containing MON89788 increased in 2021. Multi-stacked varieties containing MON89788 × MON87708, such as MON89788 × MON87708 × A5547–127 further increased by 2022. In contrast, the ratio of single event of DAS-44406 is increasing considerably from 2021 onwards. These trends may reflect an increase in GM soybean varieties containing multiple herbicide-tolerant traits such as MON89788 × MON87708, which confers tolerance to glyphosate and dicamba, and DAS-44406, which confers tolerance to 2, 4-D, glyphosate, and glufosinate.
Table 4.
Summary of individual kernel-based detection analyses from non-identity-preserved samples.
| 2021 |
2022 |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GM variety | Sample |
Sample |
||||||||||||
| N1 | N2 | N3 | N4 | N5 | Total | (%) | N1 | N2 | N3 | N4 | N5 | Total | (%) | |
| MON89788 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0.4 |
| MON87708 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0.4 |
| A5547–127 | 3 | 4 | 5 | 3 | 3 | 18 | 7.5 | 0 | 0 | 0 | 1 | 0 | 1 | 0.4 |
| DAS-44406 | 2 | 12 | 17 | 6 | 12 | 49 | 20.4 | 11 | 15 | 18 | 26 | 26 | 96 | 37.5 |
| MON89788 × MON87708 | 30 | 10 | 16 | 24 | 22 | 102 | 42.5 | 0 | 2 | 14 | 7 | 4 | 27 | 10.5 |
| MON89788 × MON87708 × A5547–125 | 0 | 14 | 10 | 13 | 11 | 48 | 20 | 8 | 12 | 19 | 14 | 7 | 60 | 23.4 |
| MON89788 × MON87708 × DAS-44406 | 9 | 3 | 0 | 0 | 0 | 12 | 5 | 0 | 0 | 0 | 0 | 6 | 6 | 2.3 |
| MON89788 × MON87708 × A5547–27 × DAS-44406 | 1 | 1 | 0 | 0 | 0 | 2 | 0.8 | 0 | 0 | 0 | 0 | 9 | 9 | 3.5 |
| A5547–127 × MON87708 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0.78 |
| DAS-44406 × A5547–127 | 2 | 0 | 0 | 0 | 0 | 2 | 0.8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Others | 1 | 4 | 0 | 2 | 0 | 7 | 2.9 | 32 | 19 | 2 | 0 | 0 | 53 | 20.7 |
Figure 2.
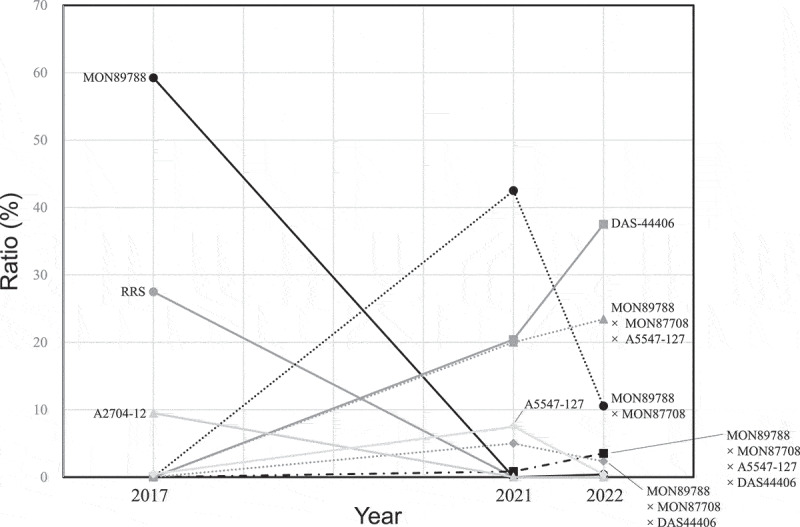
Changes in the ratio of GM soybean varieties from 2017 to 2022. The data in 2017 was cited from Soga et al. Biol Pharm Bull. 43:1259–1266.
Discussion
GM crops, including GM soybeans has been developed continuously in recent years. However, limited information is available on which varieties are primarily cultivated and distributed in the US, which has the largest number of approved GM soybean varieties and the most extensive area of GM crop cultivation. This is particularly relevant for non-IP samples, as these are often intended for export to countries such as Japan, where knowledge of the predominant GM soybean events or stacked varieties in the US cultivation would be useful. In 2017, an investigation into the distribution of non-IP samples from the US reported that over 95% of these samples consisted of single events including RRS, MON89788, and A2704–12.11 In the Japanese standard method, specific detection methods for these 3 events have been adopted,10 and it was sufficient for the labeling system for GM soybean at that time.
More recently, we evaluated maize distribution samples, targeting both IP and non-IP samples like soybean. It was revealed that the GM maize events detected in those samples were adequately covered by the Japanese standard method.31
In this study, we re-investigated soybean samples from the US and Canada domestically distributed in 2021 and 2022. In the single kernel analysis, single events such as RRS and MON89788 had largely been replaced by stacked varieties containing these events. Consequently, it is difficult to determine whether a processed food product contains single events or stacked varieties. Under the Japanese standard method, quantitative detection methods, including kernel-based analysis, are available for seeds but not for processed products. From non-IP samples, it was suggested that certain GM soybeans such as MON87708, DAS-44406, A5547–127, MON87701 and FG72 were not directly detectable with the current method. Among these events, MON87708, DAS-44406, and A5547–127, which were detected in non-IP samples, underwent kernel-based detection analysis. Nearly, all MON87708 were stacked varieties with MON89788, indicating that these stacked GM varieties containing MON87708 were detectable with the MON89788 detection method. If MON87708 were quantified separately from MON89788, it would result in a double quantification of MON89788 and MON87708, suggesting the possibility of overestimating the comingling rate. Therefore, for the stacked varieties containing MON87708, more efficient and accurate results could be achieved by conducting quantitative analysis for MON89788, but not for MON87708. If the prevalence of the single event of MON87708 increases in the future, a specific quantification method for MON87708 will be required. Regular monitoring of distribution samples is therefore important. DAS-44406 and A5547–127 were also detected in all samples. In the case of A5547–127, it tends to contain more stacked varieties than single event (Table 4). A5547–127, as a single event, accounted for 7.5% of the samples from 2021, but this ratio dropped significantly to 0.4% in the samples from 2022. Moreover, A5547–127 contains the P35S sequence, indicating that screening detection targeting P35S is applicable. From these results, a specific detection method for A5547–127 is not currently indispensable. In contrast, DAS-44406 does not contain common sequences such as P35S, TNOS, or EPSPS, which have been used as targets for screening detection. The single event DAS-44406 was detected in all non-IP sample lots and was the second most abundant event in the 2021 samples and was the most abundant in the 2022 samples. Additionally, DAS-44406 was detected in some IP sample lots although the event is not detectable with the current Japanese standard method. Therefore, an event-specific or screening detection method for DAS44406 will be necessary.
Supplementary Material
Acknowledgments
We thank Motoko Funae for experimental support.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Funding Statement
This work was supported by a Grant from the Consumer Affairs Agency of the Government of Japan.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Author Contributions
K.S., T.E., C.T., S.Y., N.S., and R.T., conceived and conceptualized the study. K.S., and R.T., investigated and analyzed the data. Y.H., provided technical support. K.K. provided critical advice. R.T. drafted the manuscript. All authors reviewed the manuscript and approved the final version.
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/21645698.2024.2444048.
References
- 1.AgbioInvestor GM monitor, Global GM Crop Area Review . 2024. https://gm.agbioinvestor.com/.
- 2.GM approved database . https://www.isaaa.org/gmapprovaldatabase/default.asp.
- 3.Ministry of Agriculture, Forestry and Fisheries of Japan . https://www.maff.go.jp/j/tokei/kouhyou/zyukyu/.
- 4.Regulation (EC) No. 1829/2003 of the European parliament and council of Europe. 2003 Sep 22.
- 5.US Department of Agriculture, National Bioengineered Food Disclosure Standard . https://federalregister.gov/d/2018-27283.
- 6.Food Labelling, Consumer Affairs Agency, Government of Japan, Tokyo . 2022. https://www.caa.go.jp/en/policy/foodlabeling.
- 7.Consumer Affairs Agency . Food labelling standards (ordinance of the cabinet office No. 10 of 2015).
- 8.Takabatake R, Egi T, Soga K, Narushima J, Yoshiba S, Shibata N, Nakamura K, Kondo K, Kishine M, Mano J, et al. Development and interlaboratory validation of a novel reproducible qualitative method for GM soybeans using comparative Cq-based analysis for the revised non-gmo labeling system in Japan. Anal Chem. 2022;94(39):13447–54. doi: 10.1021/acs.analchem.2c02447. [DOI] [PubMed] [Google Scholar]
- 9.Soga K, Nakamura K, Egi T, Narushima J, Yoshiba S, Kishine M, Mano J, Kitta K, Takabatake R, Shibata N, et al. Development and validation of a new robust detection method for Low-content DNA using δδcq-based real-Time PCR with optimized standard plasmids as a control sample. Anal Chem. 2022;94(41):14475–83. doi: 10.1021/acs.analchem.2c03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consumer Affairs Agency . https://www.caa.go.jp/policies/policy/food_labeling/food_labeling_act/assets/food_labeling_cms202_210915_01.pdf.
- 11.Soga K, Kimata S, Narushima J, Sato S, Sato E, Mano J, Takabatake R, Kitta K, Kawakami H, Akiyama H, et al. Development and testing of an individual kernel detection system for genetically modified soybean events in non-identity-preserved soybean samples. Biol Pharm Bull. 2020;43(8):1259–66. doi: 10.1248/bpb.b20-00382. [DOI] [PubMed] [Google Scholar]
- 12.Ministry of Health, Labour, and Welfare . https://www.mhlw.go.jp/stf/seisakunitsuite/bunya/kenkou_iryou/shokuhin/bio/idenshi/index.html.
- 13.Akiyama H, Sakata K, Makiyma D, Nakamura K, Teshima R, Nakashima A, Ogawa A, Yamagishi T, Futo S, Oguchi T, et al. Interlaboratory study of DNA extraction from multiple ground samples, multiplex real-time PCR, and multiplex qualitative PCR for individual kernel detection system of genetically modified maize. J AOAC Int. 2011;94(5):1540–47. doi: 10.1093/jaoac/94.5.1540. [DOI] [PubMed] [Google Scholar]
- 14.Akiyama H, Minegishi Y, Makiyama D, Mano J, Sakata K, Nakamura K, Noguchi A, Takabatake R, Futo S, Kondo K, et al. Quantification and identification of genetically modified maize events in non-identity preserved maize samples in 2009 using an individual kernel detection system. J Food Hyg Soc Jpn. 2012;53:157–65. doi: 10.3358/shokueishi.53.157. [DOI] [PubMed] [Google Scholar]
- 15.Mano J, Harada M, Takabatake R, Furui S, Kitta K, Nakamura K, Akiyama H, Teshima R, Noritake H, Hatano S, et al. Comprehensive GMO detection using real-time PCR array: single-laboratory validation. J AOAC Int. 2012;95(2):508–16. doi: 10.5740/jaoacint.11-388. [DOI] [PubMed] [Google Scholar]
- 16.Tsukahara K, Takabatake R, Masubuchi T, Futo S, Minegishi Y, Noguchi A, Kondo K, Nishimaki-Mogami T, Kurashima T, Mano J, et al. Development and evaluation of event-specific quantitative PCR method for genetically modified soybean MON87701. J Food Hyg Soc Jpn. 2016;57:187–92. doi: 10.3358/shokueishi.57.187. [DOI] [PubMed] [Google Scholar]
- 17.Event-specific method for the quantification of soybean MON 87705 using real-time PCR (2012) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/2011-05-20_CRLVL0110%20MON87705_VP.pdf.
- 18.Event-specific method for the quantification of soybean MON87769 using real-time PCR (2012) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/2012-01-27_CRLVL0709%20MON87769_VP.pdf.
- 19.Event-specific method for the quantification of soybean MON87708 using real-time PCR (2013) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-02-11_VM.pdf.
- 20.Event-specific method for the quantification of soybean CV127 using real-time PCR (2011) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/BPS-CV-1279_validated%20Method.pdf.
- 21.Event-specific method for the quantification of soybean DAS-68416-4 using real-time PCR (2014) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-11-10-VM.pdf.
- 22.Event-specific method for the quantification of soybean DAS-44406-6 by real-time PCR (2015) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-01-12VPFinal.pdf.
- 23.Event-specific method for the quantification of soybean DAS-81419-2 by real-time PCR (2015) joint research centre, European commission. https://gmo-.jrc.ec.europa.eu/summaries/EURLVL0313VP_Validated%20method.pdf.
- 24.Event-specific method for the quantification of soybean event FG72 using real-time PCR (2012) joint research centre, European commission. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-04-10%20VP.pdf.
- 25.Event-specific method for the quantification of soybean GMB151 by real-time PCR. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-01-18-VM.pdf.
- 26.Event-specific method for the quantification of soybean SYHT0H2 by real-time PCR. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-04-12-VP.pdf.
- 27.Event-specific method for the quantification of soybean MON 87751 using real-time PCR. https://gmo-crl.jrc.ec.europa.eu/summaries/EURL-VL-03-14-VP-Corrected-Version-1.pdf.
- 28.Takabatake R, Onishi M, Koiwa T, Futo S, Minegishi Y, Akiyama H, Teshima R, Furui S, Kitta K.. Establishment and evaluation of event-specific quantitative PCR method for genetically modified soybean MON89788. J Food Hyg Soc Jpn. 2010;51(5):242–46. doi: 10.3358/shokueishi.51.242. [DOI] [PubMed] [Google Scholar]
- 29.Takabatake R, Onishi M, Koiwa T, Futo S, Minegishi Y, Akiyama H, Teshima R, Kurashima T, Mano J, Furui S, et al. Development and interlaboratory validation of quantitative polymerase chain reaction method for screening analysis of genetically modified soybeans. Biol Pharm Bull. 2013;36(1):131–34. doi: 10.1248/bpb.b12-00766. [DOI] [PubMed] [Google Scholar]
- 30.Nandula VK. Herbicide resistance traits in maize and soybean: current status and future outlook. Plants. 2019;8(9):337. doi: 10.3390/plants8090337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soga K, Taguchi C, Sugino M, Egi T, Narushima J, Yoshiba S, Takabatake R, Kondo K, Shibata N. Investigation of genetically modified maize imported into Japan in 2021/2022 and the applicability of official Japanese methods. J Food Hyg Soc Jpn. 2023;64:218–25. doi: 10.3358/shokueishi.64.218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


