Abstract
Objective
This study aimed to assess the feasibility of the deep learning in generating T2 weighted (T2W) images from diffusion-weighted imaging b0 images.
Materials and methods
This retrospective study included 53 patients who underwent head magnetic resonance imaging between September 1 and September 4, 2023. Each b0 image was matched with a corresponding T2-weighted image. A total of 954 pairs of images were divided into a training set with 763 pairs and a test set with 191 pairs. The Hybrid-Fusion Network (Hi-Net) and pix2pix algorithms were employed to synthesize T2W (sT2W) images from b0 images. The quality of the sT2W images was evaluated using three quantitative indicators: Peak Signal-to-Noise Ratio (PSNR), Structural Similarity (SSIM), and Normalized Mean Squared Error (NMSE). Subsequently, two radiologists were required to determine the authenticity of (s)T2W images and further scored the visual quality of sT2W images in the test set using a five-point Likert scale. The overall quality score, anatomical sharpness, tissue contrast and homogeneity were used to reflect the quality of the images at the level of overall and focal parts.
Results
The indicators of pix2pix algorithm in test set were as follows: PSNR, 20.549±1.916; SSIM, 0.702±0.0864; NMSE, 0.239±0.150. The indicators of Hi-Net algorithm were as follows: PSNR, 20.646 ± 2.194; SSIM, 0.722 ± 0.0955; NMSE, 0.469 ± 0.124. Hi-Net performs better than pix2pix, so the sT2W images obtained by Hi-Net were used for radiologist assessment. The two readers accurately identified the nature of the images at rates of 69.90% and 71.20%, respectively. The synthetic images were falsely identified as real at rates of 57.6% and 57.1%, respectively. The overall quality score, sharpness, tissue contrast, and image homogeneity of the sT2Ws images ranged between 1.63 ± 0.79 and 4.45 ± 0.88. Specifically, the quality of the brain parenchyma, skull and scalp, and middle ear region was superior, while the quality of the orbit and paranasal sinus region was not good enough.
Conclusion
The Hi-Net is able to generate sT2WIs from low-resolution b0 images, with a better performance than pix2pix. It can therefore help identify incidental lesion through providing additional information, and demonstrates the potential to shorten the acquisition time of brain MRI during acute ischemic stroke imaging.
Introduction
Acute Ischemic stroke (AIS), one of the most common diseases in the elderly, accounts for 60% of strokes and has a high clinical mortality and disability rate [1–3]. Rapid and accurate diagnosis is closely related to the prognosis and the subsequent quality of life in AIS patients [4]. Owing to the high sensitivity of Magnetic Resonance (MR) imaging, particularly the Diffusion Weighted (DW) imaging, which is sensitive to the restricted diffusion of free water induced by cytotoxic edema in cerebral infarction areas, it has emerged as the most crucial imaging technique for AIS diagnosis [5,6]. Conventional brain MR scans typically encompass DW, T1 weighted (T1W), T2 weighted (T2W), and T2 FLAIR imaging sequences, all of which are time-consuming to acquire. Given the criticality of time in AIS, encapsulated in the phrase “time is life”, there may be a need for further optimization of these scanning procedures [7,8]. Shortening the acquisition time of brain MR imaging, such as reducing which of above sequences mentioned, will provide an important therapeutic time window for the disease.
Some medical institutions have begun to use a single DW imaging to rule out AIS, but single sequence imaging can lead to a lot of information loss, making it difficult to diagnose other lesions (e.g., hemorrhagic stroke) [9]. DWI scanning requires images with multiple b-values. Typically, the b = 0 sec/mm2 value image (b0 image) should be obtained first, followed by images with high b-value (e.g., 800, 3000 sec/mm2) [10]. Although the b0 image bears visual similarity to the T2W image, it is significantly less sharp than regular T2W images. The synthetic T2W (sT2W) images generated from the b0 images could shorten the acquisition time of brain MRI for stroke evaluation, and in the other hand, provide more information for the clinic to rule out other lesions, such as hemorrhage and otitis media.
Currently, there is burgeoning interest in utilizing deep learning algorithms to generate specific images, with the potential benefits of enabling immediate diagnosis, shortening scanning time, and reducing metabolic stress by minimizing the use of contrast agents [11,12]. Some studies proposed a novel invertible neural network for multimodal image translation [13]. Zhou et al. introduced the Hybrid-Fusion Network (Hi-Net) that comprehensively generates target modality images based on multimodality images, demonstrating advantages in MRI generation [14]. Specifically, the Hi-Net learns the mapping from the source image to the target image through three modules: the modality-specific network learns the representation of the input modality, and the fusion network learns its common latent representation, and then combines the latent representation with hierarchical features of the input modality through synthesis network to synthesize the target image.
Therefore, the aim of this study was to use Hi-Net to generate sT2W images from B0 images to explore the feasibility of shortening the acquisition time of brain MRI and excluding other lesions during the diagnosis of AIS.
Materials and methods
1. Data collection
The Medical Research Ethics Committee of the hospital approved the study, and the requirement for written informed consent was waived. All data were fully anonymized during the analyses. The data for this study were accessed on February 9, 2024. This retrospectively study enrolled the individuals who underwent conventional brain MRI scans in the Hospital between September 1, 2023 and September 4, 2023. Exclusion criteria comprised poor image quality and age less than 18 years. Ultimately, the MRIs of 53 patients were included.
2. Image generation using Hi-Net
2.1 Data preprocessing
To accelerate the convergence of the deep learning network, the input image is first normalized by the min-max normalization method. The calculation formula is as follows:
| (1) |
Where xnew is the normalized image data, xmax is the maximum value of the input image data, and xmin is the minimum value of the input image data.
The performance of the model can be improved by using the data enhancement method. The original image of the training set is randomly rotated at specific angle (90°, 180°, 270°, or 360°). Following horizontal or vertical inversion, the new image is reshaped to 256×256. This process generates eight new images from each original image, thereby expanding the training set eightfold (Fig 1).
Fig 1. Data enhancement flow chart.
2.2 Image calculation
Hi-Net learns the mapping from existing images to target images for specific synthesis tasks. The model consists of three main components: Modality-Specific Network, Multi-Modal Fusion Network and Multi-Modal Synthesis Network [14].
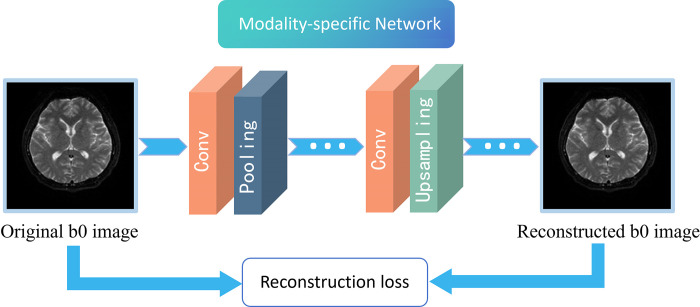
A. Modality-Specific Network. The modality-specific network can capture the unique property information of each modality and exploit the correlation between modalities to improve model learning performance. The loss function for this network is as follows:
| (2) |
where xi represents each individual modality and denotes the reconstructed image of xi, ‖⋅‖1 represents the L1-norm. As shown in Fig 2, the network consists of convolutional layer, pooling layer, upsampling layer, and activation functions, which are used to reconstruct the b0 image.
Fig 2. Workflow of modality-specific network.
It illustrates how the original b0 image is reconstructed through a modality-specific network to capture the characteristic properties of the modality.
B. Multi-Modal Fusion network. The multi-modal fusion network utilizes acquired low-level and high-level features to learn the similarities and differences among different modalities. It connects multiple modules to dynamically weight different fusion strategies, establishing connections for different modalities images.
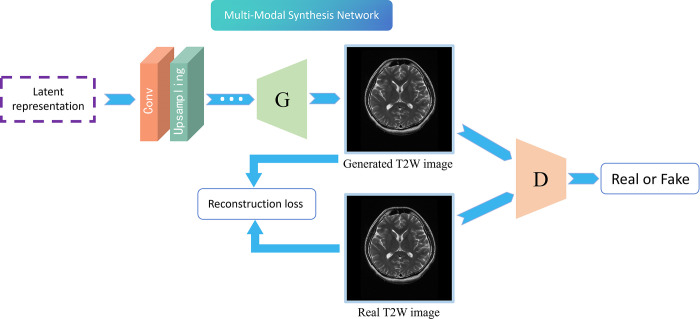
C. Multi-Modal Synthesis Network. The design of multi-modal synthesis network is based on the principle of Generative Adversarial Networks (GAN), which consists of generator and discriminator. The key concept is to carry out continuous adversarial learning between generator and discriminator. The generator attempts to produce a T2W image that confuses the discriminator, while the discriminator endeavors to distinguish the generated T2W images from the real T2W images. Accordingly, the objective function of the generator can be formulated as:
| (3) |
where x represents the real image, G(z) represents the image generated by the G network, D() represents the probability of the D network judging whether the image is real. During the training process, the network D is trained to maximize log(D(x)) and log(1−D(G(z))), and the network G is trained to minimize log(1−D(G(z))), that is, to maximize the loss of D, making the generated samples more realistic.
As shown in Fig 3, during the training, the synthesis network calculating the loss between the generated image and the real image. The model is then trained using the weight of the loss adjusted model. The generated image gradually approaches the real image, rendering the discriminator incapable of distinguishing the generated image and the authentic image.
Fig 3. Workflow of the multi-modal synthesis network.
The multi-modal latent representation generated by the last module in the multi-modal fusion network is synthesized into T2W image through the generator. The generated T2W image and the real T2W image are concurrently transmitted to the discriminator, and the gap between the generated image and the real image is reduced by the loss function.
2.3 Image generation using pix2pix
In order to better verify the effectiveness of the model, an advanced algorithm pix2pix were used to comparison [15]. The pix2pix GAN was the first success to use a conditional GAN to learn the mapping between paired images. It was designed for general purpose image-to-image translation [16].
All networks are trained using an ADAM trainer. A total of 4000 epochs were used to run the model, with an initial learning rate of 0.001. This code is implemented by the PyTorch library.
3. Image evaluation
3.1 Quantitative image evaluation
The T2W image was utilized as the gold standard for the quantitative evaluate the sT2W image generated by the two algorithms. The evaluation metrics included the Peak Signal-to-Noise Ratio (PSNR), Structural Similarity (SSIM), and Normalized Mean Squared Error (NMSE).
The PSNR is generally used as an engineering parameter relating to the maximum signal and background noise. The PSNR is defined by
| (4) |
| (5) |
Where, H, W are the height and width of the image X or Y, MSE represents the mean square error; and n is the number of bits per pixel. The unit of PSNR is usually dB.
The SSIM is a measure of structural similarity between the two images, which is defined as:
| (6) |
Where C1 and C2 are constants to avoid the situation where the denominator is 0. μ represents the mean of image. σ represents the standard noise variance of image. σXY represents the covariance of image X and image Y.
The NMSE is calculated as:
| (7) |
A higher PSNR, higher SSIM and lower NMSE indicate that the quality of the synthesized image is better and closer to the real image.
3.2 Visual assessment
Two readers, Radiologist 1 (a junior radiologist with 5 years of experience) and Radiologist 2 (an attending junior radiologist with 10 years of experience), were required to determine the authenticity of (s)T2W images and further scored the visual quality of sT2W images in the test set using a five-point Likert scale. The relevant visual assessments were divided into two experiments.
In Experiment 1, the 191 pair of original T2W and sT2W images in the test set were randomly numbered as 001–382. Two readers were tasked with independently evaluating the properties of the images (real or synthetic) one by one without knowing the nature of the image.
In Experiment 2, the readers have been told of the authenticity of the T2W images. They were required to further rate the quality of the sT2W images relative to the real images. The quality of overall synthetic image and focal region were both assessed by employing a 5-point Likert scale (5 = very good, 4 = good, 3 = acceptable, 2 = poor, 1 = very poor). Four metrics: overall image quality, sharpness, tissue contrast, and image homogeneity were scored to indicate the overall quality of each sT2w image [15]. Subsequently, three metrics: the sharpness, tissue contrast, and image homogeneity were scored to indicate the quality of focal area. The focal parts included the brain parenchyma, skull and scalp, orbits, middle ear, and paranasal sinuses. This study defined sharpness as the clarity of anatomical structures among the brain parenchyma, cerebrospinal fluid, and skull. Tissue contrast refers to the contrast between the above tissues, and image homogeneity refers to the uniformity and stability of the image.
4. Statistical analysis
All the statistical analysis were performed using SPSS software (version 26; IBM, Armonk, NY, USA). All evaluation values, including quantitative parameters and Likert scores, are expressed as the mean ± standard deviation.
The weighted kappa coefficient was used to evaluate the consistency of the image quality scores between the two observers. The degrees of consistency were as follows: < 0.20, slight; 0.21–0.40, fair; 0.41–0.60, moderate; 0.61–0.80, substantial; and 0.81–1.0, almost perfect.
Results
1. Basic data
The MRI of 53 patients were ultimately included, with each patient contributing 18 pairs of 2D T2W and b0 images. A total of 954 pairs of images were divided into the training set (763 pairs) and the test set (191 pairs).
2. Quantitative assessment of images
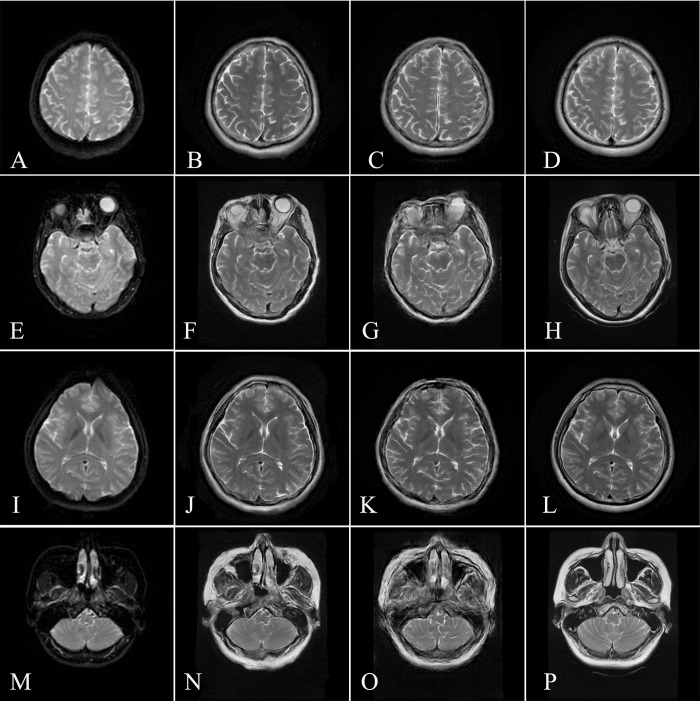
Fig 4 shows a schematic diagram of the images from test set. In the quantitative evaluation of images generated by Hi-Net in the test set, the parameters were as follows: PSNR, 20.646 ± 2.194; SSIM, 0.722 ± 0.0955; NMSE, 0.469 ± 0.124. The parameters of images generated by pix2pix were as follows: PSNR, 20.549 ± 1.916; SSIM, 0.702 ± 0.0864; NMSE, 0.239±0.150. Hi-Net algorithm presented higher PSNR, SSIM and lower NMSE than pix2pix, indicated that Hi-Net performs better.
Fig 4. Representative image in the test set.
The left column shows the DWI b0 image (A/E/I/M), the middle column shows the sT2WI images generated by Hi-Net (B/F/J/N), and pix2pix (C/G/K/O) the respectively, and the right column shows the original T2WI images (D/H/L/P).
3. Visual assessment
3.1 The images generated by Hi-Net were further analyzed
In Experiment 1, Table 1 shows the authenticity judgments by reader 1 and reader 2. The percentages of correct identification of image nature by the two readers were 69.90% (267/382) and 71.20% (272/382), respectively. The percentages of false identification of synthetic image as real were 57.6% (110/192) and 57.1% (109/192), respectively.
Table 1. Judgment of the authenticity of the images by two radiologists.
| Reader 1 | Reader 2 | |||
|---|---|---|---|---|
| True | Synthetic | True | Synthetic | |
| T2W images (n = 191) | 186 | 5 | 190 | 1 |
| Synthetic T2W images by Hi-Net (n = 191) | 110 | 81 | 109 | 82 |
3.2 In Experiment 2, the scores of the sT2W images are shown in Table 2
Table 2. Evaluation of the overall and focal quality of the images.
| Reader 1 | Reader 2 | Kappa | P value | |
|---|---|---|---|---|
| Overall | ||||
| Overall image quality | 3.43 ± 1.23 | 3.51 ± 1.29 | 0.774 | <0.001 |
| Sharpness | 3.82 ± 1.10 | 3.35 ± 1.08 | 0.749 | |
| Tissue contrast | 4.05 ± 1.08 | 3.70 ± 0.99 | 0.498 | |
| Image homogeneity | 3.55 ± 0.97 | 3.50 ± 1.00 | 0.597 | |
| Focal regions | ||||
| Brain parenchyma | ||||
| Sharpness | 4.08 ± 1.02 | 3.87 ± 0.91 | 0.529 | <0.001 |
| Tissue contrast | 4.34 ± 0.92 | 4.17 ± 0.83 | 0.507 | |
| Image homogeneity | 3.93 ± 1.01 | 3.87 ± 0.90 | 0.525 | |
| Skull and scalp | ||||
| Sharpness | 3.89 ± 1.32 | 3.79 ± 1.03 | 0.434 | <0.001 |
| Tissue contrast | 4.07 ± 1.25 | 4.03 ± 0.94 | 0.394 | |
| Image homogeneity | 3.16 ± 1.13 | 3.63 ± 1.01 | 0.424 | |
| Orbit | ||||
| Sharpness | 2.05 ± 0.84 | 2.21 ± 1.12 | 0.610 | <0.001 |
| Tissue contrast | 2.33 ± 1.19 | 2.37 ± 1.31 | 0.662 | |
| Image homogeneity | 1.84 ± 0.92 | 2.21 ± 1.21 | 0.572 | |
| Middle ear | ||||
| Sharpness | 4.00 ± 0.94 | 4.09 ± 0.94 | 0.569 | <0.001 |
| Tissue contrast | 4.45 ± 0.88 | 4.30 ± 0.90 | 0.475 | |
| Image homogeneity | 4.25 ± 0.99 | 4.02 ± 0.93 | 0.445 | |
| Paranasal sinuses | ||||
| Sharpness | 2.05 ± 0.98 | 1.63 ± 0.81 | 0.273 | <0.001 |
| Tissue contrast | 2.19 ± 1.16 | 1.91 ± 1.00 | 0.328 | |
| Image homogeneity | 1.92 ± 1.02 | 1.63 ± 0.79 | 0.270 |
The average kappa coefficient between the two radiologists for evaluating image quality was 0.655 (range, 0.498–0.774; P < 0.001) for the overall evaluation and 0.468 (range, 0.270–0.662; P < 0.001) for the focal regions. The overall image quality, anatomical sharpness, tissue contrast and image homogeneity of sT2W images were rated as good to excellent. The scores for different focal areas suggested that the image quality for the middle ear was very good, for the brain parenchyma, skull, and scalp was good. The image quality for the orbit and paranasal sinus regions was poor to acceptable.
Discussion
To the best of our knowledge, this is the first preliminary exploration proposing the generation of high-resolution T2W images based on DWI b0 images, with the aim of shortening the acquisition time of brain MRI in AIS. We employed the Hi-Net and pix2pix algorithm to synthesize T2W images. Hi-Net algorithm presented higher PSNR, SSIM and lower NMSE than pix2pix, indicated that Hi-Net performs better. When compared with conventional T2W images, sT2W images generated by Hi-Net achieved acceptable or even excellent in terms of quantitative parameters and radiologists’ visual scores.
To generate translated modality images from given modality images is one of the popular directions in deep learning. To minimize the use of contrast agents, Müller-Franzes et al. utilized GANs to explore the synthesis of enhanced images from T1 combined with T2 images or low-contrast agent images [17]. Fujita et al. develop a deep learning algorithm to generate MR angiography images based on 3D synthetic MRI raw data, which may be useful as a screening tool for intracranial aneurysms without requiring additional scanning time [18]. To reduce radiation dosimetry, an increasing number of researchers using deep learning on head and neck MR images to generate corresponding CT images [13,19,20]. Image generation across modalities can aid in simplifying clinical examination procedures and reducing unnecessary expenditures. We hope to use deep learning algorithm to generate sT2W images to avoid T2W image scanning and speed up brain MR imaging acquisition during stroke screening.
Compared with b0 image, the quality of sT2W image by the two algorithms, Hi-Net and pix2pix, is improved. Hi-Net algorithm presented higher PSNR, SSIM and lower NMSE than pix2pix, indicated that Hi-Net performs better. Currently, prevalent image generation algorithms are rooted in the Variational Autoencoder (VAE), PixelRNN/PixelCNN, and GAN series networks. The VAE network, characterized by its robust reasoning properties, is commonly used for simple digital fitting and image completion [21,22]. It can be applied to AI face modification and beauty cameras, but is not good at generating clear images [23]. The PixelRNN/PixelCNN model utilizes the probability chain rule and generates images pixel by pixel. The advantage of this model is its high-quality images, typically widely applied to image inpainting and coloring [24]. However, the speed of image generation by this network is very slow in practical application [24]. GAN produces relatively high quality images, and the dynamic interplay between the generator and the discriminator make it suitable for image enhancement [25]. However, some shortcoming such as training instability, pattern collapse and gradient disappearance limit its application. Therefore, various variants, such as conditional GANs and deep convolutional GANs, have emerged. Deep convolutional GAN is mainly used to improve the visual quality of generated image. The advantage of this approach is that the training is stable [26], but the model needs to readjust the parameters when training different data sets and is prone to model crash. Conditional GAN is mainly used for image completion, style transfer, and image subtitle generation [27,28], it implements conditional image generation by introducing conditional information into generator and discriminator. Pix2pix is based on conditional GAN and performs well on image generation and image conversion tasks [29–31]. Therefore, we choose pix2pix as the comparison algorithm, and finally find that the synthetic image obtained by this algorithm has lower quality than Hi-Net.
The Hi-Net model in this study uses existing modality data as input to synthesize the target modality. The modality-specific network is capable of capturing image feature information from a single modality image, while the fusion network discerns connections between different modalities. Furthermore, the synthetic network employs a generator and a discriminator to synthesize target modality, in conjunction with the correlation between the acquired features and modalities. Ultimately, the cross-domain generation of images is achieved by combining three models.
The visual assessment results from two radiologists indicated that the overall image quality generated by Hi-Net ranged from acceptable to excellent, with the display of the brain parenchyma, skull and scalp, and middle ear region being particularly remarkable, even similar to that of the original T2W images. We also observed that the quality of the regions close to the skull (especially paranasal sinus) and orbit were poorer than that the other regions. This phenomenon may be attributed to the fact that the signal acquisition of the skull base and orbital region is not the focal point in the process of DW image scanning, resulting in blurred or deformed appearance in these regions of the b0 image. Deep learning algorithms can generate images based solely on a limited number of signals. Nevertheless, we believe that, even with the current results, it is feasible to evaluate some crucial areas such as the brain parenchyma and middle ear using sT2W images.
This study has several limitations. Firstly, it constitutes an initial exploration based on a small sample size; multicenter studies with larger sample sizes are necessary to validate the results of this study. Secondly, although our experiments confirmed that the superior quality of the brain parenchyma, middle ear, skull, and scalp regions in the generated synthetic images, the quality of the orbit and paranasal sinus regions still requires further improvement. Subsequent studies will demand additional algorithms and larger sample sizes. Third, only the feasibility of T2W image generation from b0 image was explored, and the feasibility of deep learning to generated other sequence images, such as T1W, T2FLAIR, etc., based on DW images needs to be further studied.
Conclusion
Deep learning algorithms including Hi-Net and pix2pix can generate high-resolution T2W images from low-resolution b0 DW images, and the former shows a higher quantitative index. The sT2W images generated by Hi-Net are acceptable when compared with the real T2W images. They could provide a clearer contrast and offer a significant advantage in revealing craniocerebral anatomy, especially in the middle ear, brain parenchyma, and skull regions. This finding suggests that it is feasible to use deep learning to generate T2W images from b0 images thereby assisting incidental lesion through providing additional information as well as reducing scanning time of brain MR imaging with multi-sequence during the AIS imaging.
Abbreviations
- AIS
Acute Ischemic stroke
- MR
Magnetic Resonance
- DW
Diffusion Weighted
- T1W
T1 weighted
- T2W
T2 weighted
- b0 image
the b = 0 sec/mm2 value image
- sT2W
synthetic T2W
- Hi-Net
Hybrid-Fusion Network
- GAN
Generative Adversarial Networks
- PSNR
Peak Signal-to-Noise Ratio
- SSIM
Structural Similarity
- NMSE
Normalized Mean Squared Error
Data Availability
The datasets generated and/or analysed during the current study are not publicly available due to limitations of ethical approval involving the sensitive patient information and anonymity but are available from the corresponding author or from the ethics committee (contact via efyiec_iit@163.com), on reasonable request.
Funding Statement
This study has received funding by the National Natural Science Foundation of China (No. 91230027). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Yue C, Lin Z, Lu C, Chen H. Efficacy of Monitoring Platelet Function by an Automated PL-12 Analyzer During the Treatment of Acute Cerebral Infarction With Antiplatelet Medicine. Clin Appl Thromb Hemost. 2021;27:10760296211001119. Epub 2021/03/23. doi: 10.1177/10760296211001119 ; PubMed Central PMCID: PMC7989109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;135(10):e146–e603. Epub 20170125. doi: 10.1161/CIR.0000000000000485 ; PubMed Central PMCID: PMC5408160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feigin VL, Brainin M, Norrving B, Martins S, Sacco RL, Hacke W, et al. World Stroke Organization (WSO): Global Stroke Fact Sheet 2022. Int J Stroke. 2022;17(1):18–29. doi: 10.1177/17474930211065917 . [DOI] [PubMed] [Google Scholar]
- 4.Ohtakara K, Arakawa S, Nakao M, Muramatsu H, Suzuki K. Volumetric-Modulated Arc-Based Re-radiosurgery With Simultaneous Reduced-Dose Whole-Brain Irradiation for Local Failures Following Prior Radiosurgery of Brain Oligometastases From Small Cell Lung Cancer. Cureus. 2023;15(8):e44492. Epub 2023/10/04. doi: 10.7759/cureus.44492 ; PubMed Central PMCID: PMC10544458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu W, Yang J, Liu L, Song W, Zhang Z, Xu M, et al. The value of diffusion weighted imaging in predicting the clinical progression of perforator artery cerebral infarction. Neuroimage Clin. 2022;35:103117. Epub 2022/07/26. doi: 10.1016/j.nicl.2022.103117 ; PubMed Central PMCID: PMC9421429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu SC, Yin ZH, Zeng CF, Lin F, Ma L, Zhang Y, et al. Infarction Patterns and Recurrent Adverse Cerebrovascular Events in Moyamoya Disease. Dis Markers. 2022;2022:8255018. Epub 20220329. doi: 10.1155/2022/8255018 ; PubMed Central PMCID: PMC8983186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prabhakaran S, Ruff I, Bernstein RA. Acute stroke intervention: a systematic review. JAMA. 2015;313(14):1451–62. doi: 10.1001/jama.2015.3058 . [DOI] [PubMed] [Google Scholar]
- 8.Lo EH. A new penumbra: transitioning from injury into repair after stroke. Nat Med. 2008;14(5):497–500. doi: 10.1038/nm1735 . [DOI] [PubMed] [Google Scholar]
- 9.Ter Telgte A, Wiegertjes K, Gesierich B, Baskaran BS, Marques JP, Kuijf HJ, et al. Temporal Dynamics of Cortical Microinfarcts in Cerebral Small Vessel Disease. JAMA Neurol. 2020;77(5):643–7. Epub 2020/02/18. doi: 10.1001/jamaneurol.2019.5106 ; PubMed Central PMCID: PMC7042834 Vital GmbH and Pfizer Pharma GmbH outside the submitted work. No other disclosures were reported. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn S, Lee SK. Diffusion tensor imaging: exploring the motor networks and clinical applications. Korean J Radiol. 2011;12(6):651–61. Epub 2011/11/02. doi: 10.3348/kjr.2011.12.6.651 ; PubMed Central PMCID: PMC3194768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenkowicz J, Votta C, Nardini M, Quaranta F, Catucci F, Boldrini L, et al. A deep learning approach to generate synthetic CT in low field MR-guided radiotherapy for lung cases. Radiother Oncol. 2022;176:31–8. Epub 2022/09/06. doi: 10.1016/j.radonc.2022.08.028 . [DOI] [PubMed] [Google Scholar]
- 12.Bahrami A, Karimian A, Arabi H. Comparison of different deep learning architectures for synthetic CT generation from MR images. Phys Med. 2021;90:99–107. Epub 2021/10/02. doi: 10.1016/j.ejmp.2021.09.006 . [DOI] [PubMed] [Google Scholar]
- 13.Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44(4):1408–19. Epub 2017/02/14. doi: 10.1002/mp.12155 . [DOI] [PubMed] [Google Scholar]
- 14.Zhou T, Fu H, Chen G, Shen J, Shao L. Hi-Net: Hybrid-Fusion Network for Multi-Modal MR Image Synthesis. IEEE Trans Med Imaging. 2020;39(9):2772–81. Epub 2020/02/23. doi: 10.1109/TMI.2020.2975344 . [DOI] [PubMed] [Google Scholar]
- 15.Fujioka T, Mori M, Oyama J, Kubota K, Yamaga E, Yashima Y, et al. Investigating the Image Quality and Utility of Synthetic MRI in the Breast. Magn Reson Med Sci. 2021;20(4):431–8. Epub 2021/02/05. doi: 10.2463/mrms.mp.2020-0132 ; PubMed Central PMCID: PMC8922358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Li H, Dillman JR, Parikh NA, He L. Multi-Contrast MRI Image Synthesis Using Switchable Cycle-Consistent Generative Adversarial Networks. Diagnostics (Basel). 2022;12(4). Epub 20220326. doi: 10.3390/diagnostics12040816 ; PubMed Central PMCID: PMC9026507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller-Franzes G, Huck L, Tayebi Arasteh S, Khader F, Han T, Schulz V, et al. Using Machine Learning to Reduce the Need for Contrast Agents in Breast MRI through Synthetic Images. Radiology. 2023;307(3):e222211. Epub 2023/03/22. doi: 10.1148/radiol.222211 . [DOI] [PubMed] [Google Scholar]
- 18.Fujita S, Hagiwara A, Otsuka Y, Hori M, Takei N, Hwang KP, et al. Deep Learning Approach for Generating MRA Images From 3D Quantitative Synthetic MRI Without Additional Scans. Invest Radiol. 2020;55(4):249–56. Epub 2020/01/25. doi: 10.1097/RLI.0000000000000628 . [DOI] [PubMed] [Google Scholar]
- 19.Pan S, Abouei E, Wynne J, Chang CW, Wang T, Qiu RLJ, et al. Synthetic CT generation from MRI using 3D transformer-based denoising diffusion model. Med Phys. 2024;51(4):2538–48. Epub 20231127. doi: 10.1002/mp.16847 ; PubMed Central PMCID: PMC10994752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou L, Ni X, Kong Y, Zeng H, Xu M, Zhou J, et al. Mitigating misalignment in MRI-to-CT synthesis for improved synthetic CT generation: an iterative refinement and knowledge distillation approach. Phys Med Biol. 2023;68(24). Epub 20231212. doi: 10.1088/1361-6560/ad0ddc . [DOI] [PubMed] [Google Scholar]
- 21.Elbattah M, Loughnane C, Guerin JL, Carette R, Cilia F, Dequen G. Variational Autoencoder for Image-Based Augmentation of Eye-Tracking Data. J Imaging. 2021;7(5). Epub 2021/08/31. doi: 10.3390/jimaging7050083 ; PubMed Central PMCID: PMC8321343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cackowski S, Barbier EL, Dojat M, Christen T. ImUnity: A generalizable VAE-GAN solution for multicenter MR image harmonization. Med Image Anal. 2023;88:102799. Epub 20230324. doi: 10.1016/j.media.2023.102799 . [DOI] [PubMed] [Google Scholar]
- 23.Han K, Xiang W. Inference-Reconstruction Variational Autoencoder for Light Field Image Reconstruction. IEEE Trans Image Process. 2022;31:5629–44. Epub 2022/08/23. doi: 10.1109/TIP.2022.3197976 . [DOI] [PubMed] [Google Scholar]
- 24.Sun J, Zhong G, Chen Y, Liu Y, Li T, Huang K. Generative adversarial networks with mixture of t-distributions noise for diverse image generation. Neural Netw. 2020;122:374–81. Epub 20191118. doi: 10.1016/j.neunet.2019.11.003 . [DOI] [PubMed] [Google Scholar]
- 25.Jin R, Li X. Backdoor attack and defense in federated generative adversarial network-based medical image synthesis. Med Image Anal. 2023;90:102965. Epub 2023/10/08. doi: 10.1016/j.media.2023.102965 . [DOI] [PubMed] [Google Scholar]
- 26.Kazeminia S, Baur C, Kuijper A, van Ginneken B, Navab N, Albarqouni S, et al. GANs for medical image analysis. Artif Intell Med. 2020;109:101938. Epub 2020/09/01. doi: 10.1016/j.artmed.2020.101938 . [DOI] [PubMed] [Google Scholar]
- 27.Souibgui MA, Kessentini Y. DE-GAN: A Conditional Generative Adversarial Network for Document Enhancement. IEEE Trans Pattern Anal Mach Intell. 2022;44(3):1180–91. Epub 2020/09/08. doi: 10.1109/TPAMI.2020.3022406 . [DOI] [PubMed] [Google Scholar]
- 28.Wolterink JM, Mukhopadhyay A, Leiner T, Vogl TJ, Bucher AM, Isgum I. Generative Adversarial Networks: A Primer for Radiologists. Radiographics. 2021;41(3):840–57. Epub 20210423. doi: 10.1148/rg.2021200151 . [DOI] [PubMed] [Google Scholar]
- 29.Sun J, Du Y, Li C, Wu TH, Yang B, Mok GSP. Pix2Pix generative adversarial network for low dose myocardial perfusion SPECT denoising. Quant Imaging Med Surg. 2022;12(7):3539–55. doi: 10.21037/qims-21-1042 ; PubMed Central PMCID: PMC9246746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H, Fu X, Abraham S, Jin S, Murphy RF. Improving and evaluating deep learning models of cellular organization. Bioinformatics. 2022;38(23):5299–306. doi: 10.1093/bioinformatics/btac688 ; PubMed Central PMCID: PMC9710556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park KS, Moon JB, Cho SG, Kim J, Song HC. Applying Pix2pix to Translate Hyperemia in Blood Pool Image into Corresponding Increased Bone Uptake in Delayed Image in Three-Phase Bone Scintigraphy. Nucl Med Mol Imaging. 2023;57(2):103–9. Epub 20230103. doi: 10.1007/s13139-022-00786-y ; PubMed Central PMCID: PMC10043061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available due to limitations of ethical approval involving the sensitive patient information and anonymity but are available from the corresponding author or from the ethics committee (contact via efyiec_iit@163.com), on reasonable request.