Abstract
The detection of norm deviations is fundamental to clinical decision making and impacts our ability to diagnose and treat diseases effectively. Current normative modeling approaches rely on generic comparisons and quantify deviations in relation to the population average. However, generic models interpolate subtle nuances and risk the loss of critical information, thereby compromising effective personalization of health care strategies. To acknowledge the substantial heterogeneity among patients and support the paradigm shift of precision medicine, we introduce Nearest Neighbor Normativity (N3), which is a strategy to refine normativity evaluations in diverse and heterogeneous clinical study populations. We address current methodological shortcomings by accommodating several equally normative population prototypes, comparing individuals from multiple perspectives and designing specifically tailored control groups. Applied to brain structure in 36,896 individuals, the N3 framework provides empirical evidence for its utility and significantly outperforms traditional methods in the detection of pathological alterations. Our results underscore N3’s potential for individual assessments in medical practice, where normativity is not merely a benchmark, but a dynamic tool that adapts to the intricacies of personalized patient care.
Keywords: Normative Modeling, Precision Medicine, Diversity, Density-Estimation
1. Introduction
Normativity, as a conceptual framework, holds profound implications for medical practice [1, 2]. Normative reference values underlie the standards, norms, and criteria that guide the physiological assessments in clinical practice. By relying on these reference norms, clinicians are able to identify deviations from expected physiological states and detect pathological conditions that require medical intervention. The quantification of normativity, i.e., the degree of alignment with expected reference norm values, is therefore essential for clinical decision making and moderates our ability to diagnose and treat diseases effectively. With the advent of precision medicine, the necessity to tailor medical interventions based on individual physiological nuances, as well as genetic, environmental, and lifestyle factors, has become ever more pronounced. Precision medicine highlights the considerable heterogeneity among patients and emphasizes the uniqueness of physiological states and individual healthcare needs [3, 4]. In order to enable the personalization of medical interventions, it is thus not merely an academic exercise but a practical necessity to understand and redefine reference values and normativity definitions in large and heterogeneous datasets.
In neuroimaging, parsing the large inter-individual variability of brain structure and function has been a major endeavor of the past decades. The aim is to ground diagnosis and treatment of neurological and psychiatric diseases on an understanding of disease mechanisms and neurobiological alterations associated with psychopathological symptoms [5, 6]. Yet, the variability in brain structural patterns and disease trajectories highlights the diversity among individuals and underscores the complexity and individuality in brain structure, disease progression, and neurodegenerative processes. While the traditional reliance on case–control studies fails to account for the heterogeneity observed among individuals and across different disease phenotypes, normative modeling has been successfully applied to interpret brain structures in several medical domains [7].
Normative modeling uses statistical distributions to quantify normativity relative to the population average and the typical variance around it [8, 9]. In these models, clinical variables, such as gray matter tissue density, are joined with clinical covariates—such as age, gender or body mass index— to be processed within a single analytical framework. However, while comprehensive in its ability to provide context, these general, typically univariate, models can mask finer pathological details critical for nuanced clinical insights [10]. Moreover, diversity is often methodologically simplified by relying on a central tendency (e.g., the mean). Evaluating all data in relation to a single reference point, the mean, interpolates natural variability, neglects the uniqueness of physiological manifestations and may overlook nuanced inter-individual deviations and anomalies. In addition, it inherently excludes the possibility of multiple, equally viable and healthy normative states. This risks the loss of crucial information and compromises the accuracy of personalized normativity assessments, affects their applicability and effectiveness for personalized healthcare.
Here, we address these methodological shortcomings and propose a novel normativity framework which we call Nearest Neighbor Normativity (N3). The N3 framework advances normativity estimations in large and heterogeneous datasets by not only acknowledging but also embracing the diversity inherent in study populations. We use density estimation techniques to enable refined normativity evaluations and accommodate multiple possible normative population prototypes. Moreover, we rely on multiple, specifically tailored subpopulations and leverage multiple comparative angles to create a multi-facetted individual normativity profile. The N3 framework parses the large inter-individual variability in patient data and enables a refined contextualization of individual patient data, moving closer to the ideals of precision medicine.
We provide evidence for the value of the N3 framework by developing a novel normativity marker for brain structure. It is based on four key strategies.
1. Multi-Prototype Normativity.
We reformulate the normativity estimation problem from “What is the population average in healthy individuals and how much does it vary?” to “How common is this observation in a representative reference sample?”. Underlying is the assumption that low regional sample density - i.e. few similar samples - indicates rareness. Methodologically, this can be expressed with straightforward and distribution-free local density estimation techniques such as the Nearest Neighbor Algorithm9. The Nearest Neighbor Algorithm offers a nuanced evaluation of the relation of individuals to each other and expands the normativity definition from a single prototype (the population average) to several possible normative prototypes (i.e., several clusters of high local sample density). We hypothesize that this shift in perspective provides a more nuanced understanding of inter-individual variability, thereby improving the clinical relevance of normativity estimates. In the example of brain structure, we allow several equally normative prototypical brain structures per age group and overlapping brain structural prototypes across several (neighboring) age groups. This is motivated by the many factors impacting neurodegenerative processes, including genetic, lifestyle and environmental factors, which inevitably results in individual progression rates of brain structural aging effects, and thus in shared normative prototypes in neighboring age groups.
2. Tailored control groups.
Second, to enable the detection of subtle deviations often overlooked in broader models, we propose normativity assessments in specifically tailored control groups. Instead of comparing to the available data collective as a whole, these tailored control groups can be designed to accentuate specific normative nuances and elevate the sensitivity of the analytical models. In the context of brain structure, we avoid population-wide comparisons and compare brain structures in relation to a representative sample of the same sex and chronological age. By stratifying our control groups according to age and chronological age, we remove non-specific brain structural variance and enable refined comparisons within more homogeneous subgroups. As described above, we hypothesize that this narrower comparison facilitates the detection of subtle individual norm deviations.
3. Individual normativity profile.
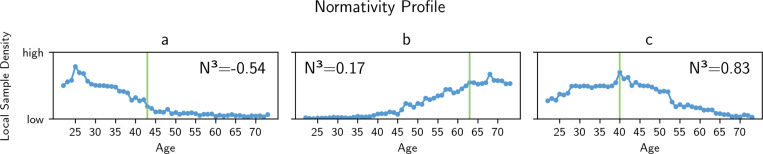
Third, we introduce global context to the normativity assessments and join multiple comparative normativity evaluations per individual into a so-called normativity profile. Such an approach looks at an individual from multiple meaningful angles or viewpoints, culminating in what we refer to as a multi-perspective normativity profile. This profile offers a comprehensive summary of an individual’s alignment with different, not mutually exclusive, subpopulations. It blends a broad overview with subgroup-specific details, thereby contextualizing individual nuances from a holistic, yet granular, perspective. We hypothesize that, compared to a single normativity estimation, such a multi-perspective normativity profile may reveal additional information about an individual’s health status. In the context of brain structure, we utilize the manifold of age-group specific models to evaluate brain structure from different viewpoints along the age continuum. This method assesses an individual’s alignment with different norms seen along the age continuum and positions it within the spectrum of aging effect (see Figure 2). Consequently, a very normative brain structure exhibits high local sample density within its own age group and shows decreasing alignment within other age groups (see Figure 2c). Alternatively, an individual brain structure might align with the aging effects seen further along the aging continuum, resembling older brain structures (see Figure 2b), or younger brain structures (see Figure 2a).
Fig. 2.
Individual brain structural normativity profiles of three exemplary individuals of the training sample (see Methods 1 and 2). The normativity profiles show the alignment of an individual brain structure with brain structure seen in reference samples of different age groups (blue). The alignment, i.e. its normativity, is measured using density estimation techniques, allowing several equally normative prototypical brain structures per age group and overlapping brain structural prototypes across several (neighboring) age groups. Chronological age is depicted in green. a) An individual’s brain structure aligns with younger brain structures, indicating fewer aging effects as commonly seen in same-aged individuals b) An individual brain structure aligns with older brain structures, indicating premature neurodegeneration processes. c) A brain structure exhibits high alignment within its own age group and shows deprecating alignment within other age groups.
4. Meta Normativity.
Finally, to synthesize the comprehensive data captured in an individual’s normativity profile into a singular, actionable metric, we conduct a final normativity estimation. Here, the normativity profile itself is subject to normativity estimation (meta-normativity). In the case of brain structure, we evaluate the normativity profile with respect to age groups. The final normativity marker, which we abbreviate with N3, therefore expresses how common a normativity profile is for a specific age group. We hypothesize that this second layer of normativity estimation will further increase clinical utility. Moreover, it can be adapted to diverse clinical inquiries, e.g., expressing the commonness of a normativity profile for patients with high treatment response or adverse side effects.
In this work, we benchmark the efficacy of the N3 framework relative to conventional normative modeling approaches. We provide evidence that the N3 approach is able to interpret clinical information effectively, and finds individual nuances and norm deviations related to disease in large and heterogeneous data.
2. Results
All normative models are trained with neuroimaging data from T1-weighted MRI scans of 29,883 individuals of a large population-based study (see Methods 2). Our analysis focuses on gray matter (GM), white matter (WM), hyperintense white matter (WMH), total intracranial volume (TIV) and cerebrospinal fluid (CSF) volumes. These global measures provide a comprehensive overview of brain structure[11]. We use these broad aggregates of complex physiological features to appropriately represent typical clinical measurements, and verify the detection of individually nuanced norm deviations. Particularly, we test the ability of different normative modeling approaches to derive meaningful disease indicators from these global parameters. We employ instances of Mild Cognitive Impairment (MCI), Alzheimer’s Disease (AD) and Frontotemporal Dementia (FTD) as model diseases to represent brain structural alterations and different pathological states.
We evaluate the N3 marker efficacy against conventional normative modeling approaches. Using classical normative modeling [7, 8], we derive two normativity scores, the first being the sum of the absolute z-scores (NM-S), the second counting the number of z-scores whose magnitude deviates beyond a threshold of ±1.96 (NM-C). We also benchmark our approach against the Brain Age paradigm, which utilizes a machine learning model to predict chronological age from brain structural data[12, 13]. Deviations between predicted and actual age, referred to as the Brain Age Gap (BAG) indicate neurodegenerative alterations (for details please refer to Methods Section 1, 3 and 4)
Applying the normative models to 7,013 individuals with varying levels of neurodegeneration (see Methods 2), we validate the ability of each normative marker to differentiate between healthy inter-individual variability and (early) pathological states of neurodegeneration. We thereby compare the efficacy of the different normative modeling approaches in identifying individual pathological norm deviations
2.1. Increased statistical explanatory power in distinguishing neurodegenerative diseases
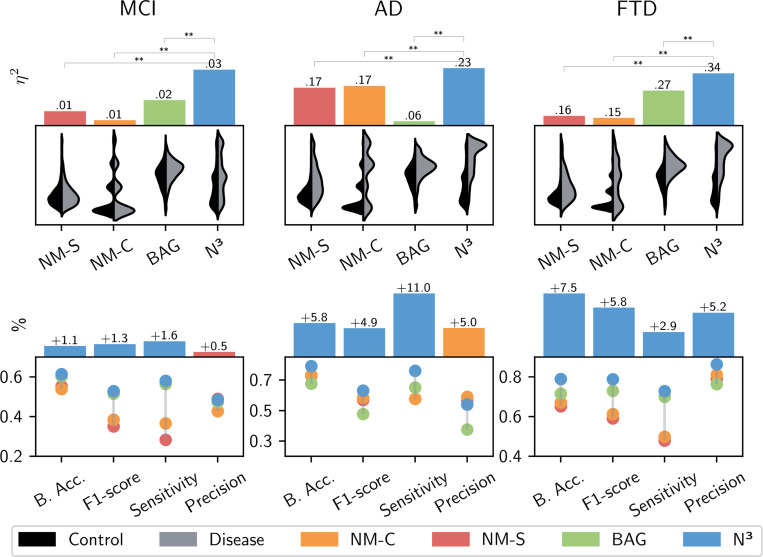
First, we assessed the statistical power of each normativity marker, specifically examining the extent to which the marker detects neurodegenerative alterations in group-level analyses. We calculated the effect size (partial eta squared, ) for the classification of healthy individuals from those affected by disease (MCI, AD or FTD, respectively; see Methods 5.5). Post-hoc comparisons then enabled us to evaluate which normativity marker was able to provide the most statistical power. The N3 marker consistently showed higher discriminative ability across all neurodegenerative conditions compared to other markers used in the study (see Figure 3 and Table 1).
Fig. 3.
Top: The top panel shows the results of the statistical analyses. Statistical effect sizes (partial eta squared - ) are given for the different normativity markers (N3 - our approach, NM-S - the sum of the absolute z-scores, NM-C - the number of z-scores whose magnitude deviates beyond a threshold of ±1.96, and the BAG - Brain Age Gap). We evaluate each normative modeling approach’s ability to parse inter-individual variability and detect pathological alterations. For each marker, we test the ability to differentiate between controls and diseased individuals in group-level analyses, using neurodegeneration as representative model disease. Results are given for Mild Cognitive Impairment (MCI), Alzheimer’s Disease (AD), and Frontotemporal Dementia (FTD), respectively. Post-hoc comparisons of the effect sizes revealed larger explained variance of our N3 marker in all neurodegenerative conditions. The level of significance in the differences between the of N3 and of the other normativity markers is indicated above. Significance was confirmed through permutation testing using 1000 random class assignments. The distribution plots below show each normativity marker’s value distributions for healthy controls (black) and diseased individuals (gray). Bottom: We use machine learning to evaluate the expressiveness of each normativity marker on a single-subject level. The N3 maker demonstrated increased accuracy in predicting the occurrence of neurodegenerative diseases for individual patients. We show the different normativity marker’s performance metrics [balanced accuracy (B.Acc), F1-Score, Recall and Precision] and the performance advantage of the best normativity marker in relation to the second best marker in percentage (above).
Table 1.
Overview of the results achieved in statistical and machine learning analyses. To quantify the expressiveness of the different methodological approaches, we evaluate the different normative markers’ ability in distinguishing between normative inter-individual variability and pathological alterations. We report the effect size , representing the amount of variance explained by each of the different normativity markers in statistical group comparisons. We compare N3 - our approach, NM-S - the sum of the absolute z-scores, NM-C - the number of z-scores whose magnitude deviates beyond a threshold of ±1.96, and the BAG - Brain Age Gap for Mild Cognitive Impairment (MCI), Alzheimer’s Disease (AD), and Frontotemporal Dementia (FTD), respectively. Moreover, we report the F-statistic, reflecting the relation of the marker variance between cognitive unimpaired and diseased individuals in relation to the respective intra-group variance, further indicating its ability to identify pathology in group-level analyses. All F-statistics and effect sizes are significant (p<0.001). The performance results of the machine learning analyses are given, where the normativity markers are used to predict the occurrence of the neurodegenerative diseases in individual cases. The metrics provide insights into each marker’s clinical utility, and overall efficacy in handling inter-individual variability and pathological variations across different neurodegenerative conditions on a single subject level. Highest performance is indicated in bold. We see that the N3 brain structural normativity marker shows relative superiority in relation to the other normativity markers, indicating the approach’s efficacy in processing inter-individual variability and delineating potential anomalies.
| Marker | F-statistic | Effect size [95% CI] | Balanced Accuracy | F1-score | Sensitivity | Precision |
|---|---|---|---|---|---|---|
| Mild Cognitive Impairment (MCI) | ||||||
|
| ||||||
| NM-C | F(1,4565) = 41 | 0.009 [0.004, 0.016] | 0.539 ± 0.010 | 0.385 ± 0.057 | 0.367 ± 0.090 | 0.427 ± 0.028 |
| NM-S | F(1,4565) = 58 | 0.013 [0.007, 0.021] | 0.553 ± 0.013 | 0.352 ± 0.044 | 0.284 ± 0.070 | 0.490 ± 0.044 |
| BAG | F(1,4565) = 78 | 0.017 [0.011, 0.026] | 0.603 ± 0.011 | 0.516 ± 0.014 | 0.566 ± 0.030 | 0.475 ± 0.016 |
| N3 | F(1,4565) = 135 | 0.029 [0.019, 0.039] | 0.614 ± 0.011 | 0.529 ± 0.013 | 0.582 ± 0.023 | 0.485 ± 0.014 |
|
| ||||||
| Alzheimer’s Disease (AD) | ||||||
|
| ||||||
| NM-C | F(1,3709) = 762 | 0.174 [0.148, 0.206] | 0.733 ± 0.020 | 0.583 ± 0.027 | 0.578 ± 0.047 | 0.591 ± 0.010 |
| NM-S | F(1,3709) = 732 | 0.168 [0.136, 0.201] | 0.727 ± 0.023 | 0.570 ± 0.031 | 0.578 ± 0.057 | 0.567 ± 0.022 |
| BAG | F(1,3709) = 226 | 0.059 [0.042, 0.075] | 0.676 ± 0.023 | 0.477 ± 0.025 | 0.651 ± 0.054 | 0.376 ± 0.014 |
| N3 | F(1,3709) = 1091 | 0.232 [0.206, 0.263] | 0.791 ± 0.020 | 0.632 ± 0.020 | 0.761 ± 0.049 | 0.541 ± 0.010 |
|
| ||||||
| Frontotemporal Dementia (FTD) | ||||||
|
| ||||||
| NM-C | F(1,580) = 112 | 0.163 [0.116, 0.211] | 0.671 ± 0.028 | 0.613 ± 0.043 | 0.499 ± 0.063 | 0.812 ± 0.073 |
| NM-S | F(1,580) = 117 | 0.169 [0.126, 0.217] | 0.653 ± 0.042 | 0.592 ± 0.034 | 0.479 ± 0.047 | 0.790 ± 0.097 |
| BAG | F(1,580) = 177 | 0.235 [0.166, 0.301] | 0.715 ± 0.076 | 0.731 ± 0.073 | 0.700 ± 0.073 | 0.765 ± 0.077 |
| N3 | F(1,580) = 325 | 0.341 [0.290, 0.435] | 0.790 ± 0.063 | 0.789 ± 0.059 | 0.729 ± 0.063 | 0.864 ± 0.080 |
For AD, the N3 marker showed the largest effect size (), signifying that approximately 26% of the total variability in the data could be attributed to differences in the N3 marker levels between the AD group and controls. In the context of FTD, all markers demonstrated large effect sizes, while the N3 stood out with an effect size of . The results for Mild Cognitive Impairment (MCI) differed, as all markers showed generally lower explanatory power. Nonetheless, the N3 marker displayed a relative advantage, with an effect size of , compared to for the Brain Age Gap (BAG) and for the normative modeling scores. Overall, the results suggest N3’s enhanced capability of discerning the subtle and complex neurostructural alterations associated with different stages of neurodegeneration in group level analysis.
2.2. Improved personalized predictions
Second, we conduct machine learning analyses to evaluate each normativity marker’s utility in predicting the occurrence of a neurodegenerative disease. Machine learning models transcend conventional statistical models by handling multivariate and non-linear relationships and shifting the focus from group average comparisons to predictions on an individual level[14]. We estimate how well the different normativity markers predict the existence of pathological neurodegenerative states in unseen individuals. To do so, we employ cross-validation strategies, which systematically tests each marker against new, unseen data to verify the accuracy, robustness, and generalizability of the models. Such validation is imperative to ensure reliability when these markers are applied in clinical environments [15]. The performance of the ML models is quantitatively evaluated using metrics such as sensitivity, precision, balanced accuracy, and the F1-score —each providing a different lens through which to assess clinical utility. Balanced accuracy provides a holistic view, ensuring that both the presence and absence of disease are accurately identified. Sensitivity is particularly critical in a clinical setting as it measures the model’s capability to capture as many diseased patients as possible, thus effectively measuring a marker’s utility as a screening tool. Complementary precision ensures that the majority of patients identified by the model as having a disease truly have the disease. The F1-score is crucial for its balance of precision and sensitivity—a vital feature to avoid unnecessary interventions or over-treatment or unnecessary expensive screening programs.
The findings, as presented in Figure 3 and Table 1, elucidate the efficacy of the N3 marker across various neurodegenerative disorders. In the specific cases of AD and FTD, the N3 marker demonstrated notable improvements in balanced accuracy scores—surpassing the second-best markers by 5.8% for AD and 7.5% for FTD. However, in alignment with the small effect sizes observed in statistical analysis, the efficacy of all markers notably declined in predicting the presence of MCI from the given variables. Here, the N3 reached an 1.1% improvement to the next best marker, the BAG. With regard to the F1-scores, the N3 marker achieved the highest performance in all neurodegenerative diseases, demonstrating its adeptness at balancing sensitivity and precision in detecting disease cases. While N3’s precision for MCI was 0.5% behind the normative modeling marker (NM-S) and by 5.1% in AD (NM-C), it was superior by 5.2% for FTD compared to the second best result (NM-C). Moreover, the N3 marker displayed superior sensitivity rates in all conditions, highlighting its sensitivity in identifying (subtle) neurodegenerative patterns. Given the overlap to normative aging patterns and the individuality in disease manifestations, particularly in MCI, this is a notable performance increase and indicates the N3 approach’s utility in decoding sparse associations. Overall, N3’s relative superiority over other markers emphasizes its efficacy in differentiating inter-individual variability from pathological variations in unseen individuals. The results provide evidence for the expressiveness of the proposed N3 normative modeling approach, indicating its ability to parse inter-individual heterogeneity effectively to evaluate individual measurements intricately within the broader landscape of diverse medical data.
2.3. Stability and Robustness of the N3 marker
The calculation of the N3 marker relies on local density estimation. As such it is highly dependent on the composition of the reference sample. Therefore, we investigate how changes to the sample composition and sample size affect the stability of the N3 model. We retrained N3 models with downsampled subsets of varying size, thereby mimicking smaller studies and different study participants. We then apply the different normativity models and predict normativity on an external dataset. Particularly, we evaluate if predictions remain consistent across different sample sizes and sample compositions. We quantify the stability of the normativity estimates by calculating the Intraclass Correlation Coefficient (ICC) 18 (see Methods Section 5). Results are visualized in Figure 4. We see that random samples of 200 individuals and above show consistently high stability (ICC of 0.75 and above). Moreover, the ICC converges to excellent levels (0.9 and above) in larger sample sizes, starting at 300 participants. While the results are calculated for the use case of brain structural normativity estimation, they are a first indication density-estimation based normative models can be realized by dividing larger samples into subgroups of a few hundred samples and above.
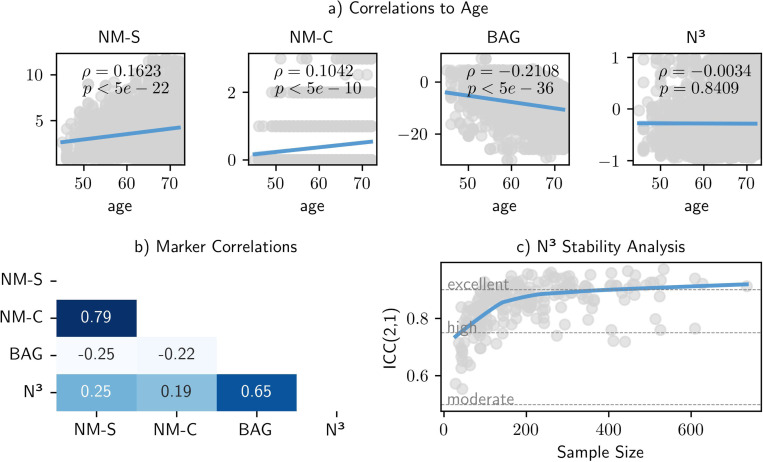
Fig. 4.
Our evaluations revealed high robustness and consistency of the N3 framework. a) We explored the age bias across different brain structural normativity markers in a healthy reference sample. In contrast to the other normativity estimation approaches, the N3 marker showed no significant association to age, which allows a consistent interpretability across different age groups. b) Additionally, we calculated the correlation matrix among markers, which emphasize the distinctiveness and complementarity of the N3 marker. c) We tested the impact of sample size and sample composition on the reliability of the N3 marker through intraclass correlation coefficients. To do so we repeatedly downsampled the training data to a random subset, mimicking smaller samples and different sample compositions. We see that the N3 marker exhibits high stability (ICC of 0.75 and above) starting from small sample sizes of around 100 individuals and converges to excellent stability (ICC of 0.9 and above) in sample sizes of three hundred individuals and above.
Furthermore, it is essential for normativity estimations to remain consistent and interpretable along the aging continuum, i.e., across different age groups, to avoid age biases that could complicate both research and clinical interpretations. An analysis of the age correlation of the N3 marker (presented in Figure 4a) indicates its stability over the age range, showing no significant association to age. In comparison, traditional normative models show a significant but smaller correlation to age (, p<0.001). This is a contrast to the Brain Age Gap (BAG), which exhibits a moderate age bias (, p<0.001), even after bias correcting adjustments are made, (see Methods Section 3).
In terms of inter-marker relationships (detailed in Figure 4), the correlation analysis shows generally weak associations () among the various markers. Two exceptions were noted: a strong correlation () between the two normative modeling markers — expected due to their derivation from the same normative models — and a moderate to strong correlation () between the BAG and the N3 marker. The correlations indicate underlying differences in what these markers are measuring about brain structural normativity, suggesting a potential for a combined utility in clinical settings.
3. Discussion
We have introduced the N3 framework, which extends existing normative modeling approaches by accommodating several normative population prototypes and evaluating individuals from multiple comparative angles. We applied it to brain structure, which resulted in an informative biomarker assessing aging effects from multiple perspectives along the aging continuum. Notably, the N3 framework provides holistic context while at the same time refining individual assessments by benchmarking against a specifically tailored reference sample. In this context, individual normativity profiles were compared to a reference group of same-aged individuals, facilitating the detection of fine-granular norm deviations. We provided evidence that the strategic alterations of the N3 framework yield increased expressiveness and enabled superior differentiation between natural inter-individual variability and pathological alterations. In comparison to commonly used normativity scores and the widely referenced Brain Age approach, the N3 marker showed increased efficacy in identifying pathological neurodegenerative brain structural changes.
Notably, our evaluations are based on only five variables reflecting global brain structure volumes. As such, they are broad aggregates of complex physiological features and represent the character of many clinical measurements. In our application, the N3 approach has demonstrated its ability to effectively decode the relevant information contained in these limited neurobiological variables and was able to extract meaningful insights.
We developed the N3 approach in alignment to the goals of precision medicine. As diversity and scale of datasets increase, we need to reevaluate how population norms are derived, applied, and interpreted in clinical practice [16–19]. A refined modelation of reference values and population norms enhances our understanding of normative variability in diverse populations and fosters the detection of individual pathological alterations [20–24]. The N3 framework embraces the complexity in patient data, contextualizes it against heterogeneous population standards and parses the diversity into an interpretable and actionable metric.
Our approach accommodates the multivariate nature of brain structures [25] and aligns with other modern understandings of heterogeneity, such as the concept of neurotypicality [26–28]. Traditionally seen as a uniform standard, brain architectures are now understood to encompass a spectrum of neurological function and structures, reflecting the rich diversity of the human brain. Moreover, our findings resonate with recent work by Yang et al., where the authors found a range of multiple, co-occuring patterns of brain aging [29]. Their research underscores the significant inter-individual and also intra-individual variability, underscoring the complexity and uniqueness of individual neurodegenerative processes beyond population averages.
Limitations of our proposed N3 framework include its reliance on larger sample sizes, a factor not always feasible in clinical studies where resource efficiency dictates smaller study populations. To maximize statistical power and mitigate the confounding effects of clinical covariates, the heterogeneity in these smaller studies is often restricted, which inadvertently limits their generalizability and applicability of outcomes across the heterogeneous population [30, 31]. In our evaluations, the N3 marker exhibited high stability in samples of a few hundred individuals, indicating substantial robustness in moderately-sized research study populations. Moreover, the N3 marker showed consistency across age groups, i.e., no correlation to age, which means that its interpretation is consistent across individuals from different age groups and facilitates its interpretability in statistical analyses. Next to the overall sample size, the N3 framework depends on the coherence and precision of defined control groups. Without carefully stratified and representative control groups, fragmented and inconsistent normative assessments may ensue. Here, it is crucial that clinical knowledge is used to design comprehensive stratification strategies that capture relevant sources of heterogeneity and enable refined normativity estimations. Within the control groups, the framework’s effectiveness relies on the choice of a density estimation algorithm. In our application, the Nearest Neighbor Algorithm depends on the k parameter, which defines the number of neighbors considered in the estimation of the local sample density. In our approach, limiting the number of neighbors to 10% with an upper bound to 15 prevented overly broad comparisons while maintaining sufficient robustness across all control groups. In general, the underlying algorithm can be customized for different scenarios, or adapted to accommodate different medical data modalities, e.g., by using custom distance metrics or dimensionality reduction techniques [32, 33].
The interpretation and contextualization of individual brain structures holds significant potential for various domains. For example, a reliable biomarker for brain structural normativity is eagerly sought in neuropsychiatric research. Here, biomarkers hold promises to enable comprehensive assessments of neurostructural alterations to better understand the etiology and pathogenesis of different disease phenotypes [10, 14, 34]. In general, a valid and robust neurostructural biomarker would allow us to measure the impact of environmental factors, treatment options and neuroinflammatory processes to understand disease mechanics and optimize individual disease management strategies [35–37]. In the realm of neurodegenerative diseases, the ability to detect brain structural alterations early is of critical clinical relevance, as it has been shown that structural changes in the brain can manifest well before clinical symptoms become apparent [38, 39]. Furthermore, evidence supports the presence of multiple underlying neuropathological processes [40, 41], underscoring the methodological importance for models accommodating multiple disease prototypes. Here, a reliable brain structural screening tool could be attached to routine MRI scans to promote early disease interception and facilitate timely interventions that may prevent or delay disease progression [42–44]. To this end, we intend to extend our approach to process scans of different MRI tissue contrasts and evaluate different deep-learning based embeddings to optimize information gain. Moreover, we intend to investigate the resulting marker’s relation to genetic risk factors [29, 45–47].
As the critical role of individual norm deviations resonates through every facet of personalized medicine, we aim to refine and expand our normativity estimation approach to medical domains beyond brain structure. To illustrate this, consider some exemplary applications. In the context of diabetes, our N3 approach might enable fine-grained analysis of normative glucose tolerance levels. By considering factors such as age, insulin sensitivity, lifestyle habits or ethnicity, the identification of nuances relevant to achieve optimal glycemic control might be facilitated [48–51]. In renal function assessment, particularly in conditions like chronic kidney disease, the N3 approach could aid in evaluating individual glomerular filtration rate patterns. By establishing normative trajectories of GFR, deviations from expected patterns could be identified early on [52, 53]. Finally, in the management of hypertension, the N3 approach could be employed to establish normative trajectories of blood pressure. Here, it could help to identify individual pattern deviations, adjusted for factors such as age, sex, body mass index, ethnicity, and lifestyle habits, that signal an elevated risk of cardiovascular events [54, 55].
In general, we believe that the N3 framework holds promise for dynamically generated ad-hoc normativity assessments in the clinical routine, guided by the expertise of healthcare professionals and adeptly adjusted to meet the individual needs of various clinical scenarios. This forward-thinking application of the N3 framework could assist individual assessments in medical practice, where normativity is not merely a benchmark, but a dynamic tool that adapts to the intricacies of personalized patient care.
4. Conclusion
This approach that we call Nearest Neighbor Normativity (N3) interprets individual patient data in reference to a particularly matched sample, accommodates diverse population norms, and analyzes several different perspectives of normativity. Thereby, it holds significant promise for personalized healthcare. It can be applied across various medical domains to contextualize individual patient data in large and heterogeneous datasets. As we continue to refine and validate our normativity estimation approach, it is our belief that the insights gained will be invaluable for shaping normativity assessments and contribute to more personalized patient care and improved clinical outcomes.
5. Methods
5.1. N3 algorithm
The N3 approach is based on local density estimation in tailored control groups. To establish a normative reference for the local density seen in a representative sample, we here use the simple and intuitive Nearest-Neighbor algorithm [33, 56].
5.1.1. Local density estimation in tailored control groups
Let be a control group of dataset and be the set of control groups, where control groups are allowed to overlap. Each control group contains samples , which are characterized by features .
As a first step, we normalize the features in each control group , so that their value lies in [0,1].
| (1) |
where represents feature of the sample in the control group , and are all values of feature in the control group . Each sample is thus represented as a feature vector of normalized features . To estimate the local sample density around a particular point in , we define a subset such that it contains the points which are the closest to . Distance is measured using the Euclidean distance. We define as the set of all distances from to points in . After sorting the points in into a tuple , where , the nearest neighbors are the first elements.
Next, we quantify the local sample density of as the inverse of the sum of the distance to its nearest neighbors in control group .
| (2) |
For each individual in each of the control groups containing samples, respectively, we calculate the local sample densities as described above.
| (3) |
To ensure comparability between the different control groups, we divide the local densities by the control-group specific median.
| (4) |
As a result we have a set of normalized local sample density estimations for all of the control groups .
We introduce context to the local sample density estimations and analyze their distribution across all control groups. Due to its flexibility in accommodating various distributive shapes, we use the exponentiated Weibull distribution [57]. The distribution is fitted on all normalized local sample density estimation in . Using the fitted distribution, we derive the likelihood of a normalized local sample density estimation.
| (5) |
where is the normalized local density value of sample in control group , is the exponentiation parameter, and is the shape parameter of the non-exponentiated Weibull law.
We use the fitted distribution to convert all local sample density estimations into measures of likelihood. To keep as much information as possible, we add a sign to , which indicates in which direction a sample is deviating from the median. In this context, samples whose local sample density is smaller than the medium, receive a negative value, while samples whose local sample density is larger than the medium, have a positive value.
| (6) |
Finally, to foster intuitive interpretation, we scale the signed likelihood to an interval of [–1, 1], where −1 indicates lowest sample density found and 1 indicates maximal sample density found.
| (7) |
The final value is a normativity estimation on how common the sample appears within a particular control group , measured by its local sample density .
5.1.2. Normativity Profile
To create a normativity profile for an individual sample , several normativity estimations in different, not mutually exclusive, control groups can be combined, evaluating the commonness of an individual measurement from multiple meaningful angles or viewpoints.
| (8) |
5.1.3. Meta Normativity
To synthesize the comprehensive information entailed in an individual normativity profile into a single, actionable metric, we conduct a second layer of normativity estimation (meta-normativity).
Basis to this is the first layer of normativity estimation, in which the local density estimation algorithm described in section 5.1.1 is applied to medical data of a population or study sample. In this step, the local sample density estimation is based on the medical data features. Using the algorithm outputs, a normativity profile can be generated for each individual. The normativity profile expresses how common the medical observations are in relation to the samples contained in each control group.
In the second layer of normativity estimation, we use the normativity profile as input data and repeat the local sample density estimation approach. Now, the local density estimation algorithm is using the normativity measures of as features. Thereby, we measure the commonness of a normativity profile in relation to other normativity profiles seen a particular reference population. This can either be done globally (on all normativity profiles of the sample), or again in in tailored control groups (evaluating the commmonnness of a normativity profile with respect to a particular sample subpopulation). The output of this meta-normativity estimation is the return value of the N3 algorithm, what we call the N3 marker.
| (9) |
5.1.4. Training vs. Inference Phase
The N3 algorithm is trained using a normative reference sample . There are two subsequent layers of local density estimation. The first layer operates on the algorithm’s input data. During the process, scaling parameters for the input features, as well as the median local sample density are derived and persisted per control group, respectively. Also, the parameters of the fitted probability density function and the final scaling function are persisted. Afterwards, all samples in undergo the normativity evaluations and are expressed in individual normativity profiles (see Equation 8).
Using the resulting normativity profiles of the normative reference sample as input, a second layer of normativity estimation is applied. This time, the inidivudal normativity profiles are subject to local sample density estimation . Again, the scaling parameters as well as the median local sample density are persisted per control group, respectively. Control groups may now be different than those in the first stage. Finally, another probability density function is fitted, this time on the local sample densities of . Again, the fitting parameters of as well as those of the scaling function are persisted.
During inference time, a novel sample is evaluated in relation to the controls groups of training sample . For each control group, the feature values of are scaled according to the parameters persisted during training, and the nearest neighbors of are determined, respectively. We calculate in relation to samples seen in . After applying the first layer of local sample density estimation, several normativity evaluations in different control groups are summarized in a normativity profile . In the second step, the normativity profile is evaluated in relation to the normativity profiles seen in the reference sample , using the parameters persisted during the second stage of training. The final output is derived by
5.1.5. Application to Brain Structure
In our application to brain structure, we stratify the training sample by sex and age, resulting into 100 control groups containing same-aged females or males (22 to 72 years), respectively. Each sample is characterized by 5 different features, namely the brain structural volumes (GM, WM, WMH, CSF, TIV) of each individual. To mitigate different sample sizes of different age groups, we join either the lower, the upper, or both neighboring age groups of underrepresented age groups, so that the sample size per age group approximates the median sample size available per sex. We set the k parameter to 10% of the control group sample size, but limit its upper bound to 15 to prevent too broad comparisons . Applying the N3 algorithm, we then first evaluate the commonness of an individual brain structure in comparison to all available age groups of the same sex. The result are normativity profiles, indicating the alignment of the brain structure in relation to the reference samples seen across the aging continuum. In the next step, we use all normativity profiles (across genders) and evaluate their normativity in relation to other representative samples of the same chronological age. The final N3 marker indicates how common a brain structural normativity profile is in the chronological age group of the individual.
5.2. Materials
Neuroimaging data from six different studies were provided by the respective consortia. Our study includes data from the German National Cohort (NAKO)[58–60], the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [61], the Münster-Marburg Affective Disorder Cohort (MACS) [62], the Australian Imaging, Biomarker Lifestyle Study of Aging (AIBL) [63], the Frontotemporal Lobar Degeneration Neuroimaging Initiative (NIFD), and the Open Access Series of Imaging Studies 3 (OASIS3) [64, 65]. We give a short overview of our approach to integrate these resources in our analyses, before we introduce each study population in detail below.
5.2.1. Training and Test Data
In general, if more than one measurement was available per participant, we restrict each study’s dataset to the first (baseline) measurement of the participant. Exclusion criteria were applied based on age; participants younger than 22 years or older than 72 were omitted from the study, due to insufficient sample sizes in the normative reference sample. All neuroimaging data utilized in this study were T1-weighted MRI scans from these baseline measurements. These images underwent preprocessing using the standard software CAT12 (version: cjp_v0008, spm12 build v7771; cat12 build r1720) default parameters. In short, images were bias-corrected, tissue classified, and normalized to MNI-space using linear and non-linear transformations. Subsequently, the derived GM, WM, WMH, CSF, and TIV volumes were extracted.
Training Data
The training data for fitting models of the different normative modeling approaches comprised 30,047 samples from the population-based NAKO cohort (for details see below). We exclude age groups below 22 years and above 72 years due to small sample sizes (n < 100), which restricts the final sample to 29,883. We then fit the models of the different normative model approaches using this large and diverse sample.
Test Data
To investigate each normativity marker’s effectiveness in identifying brain structural anomalies and (early) signs of neurodegeneration, additional data involving 5,857 participants were utilized, sourced from ADNI, AIBL, OASIS and NIFD datasets (for details see section 5.2.2). The collective samples include cognitively unimpaired individuals as well as those diagnosed with Mild Cognitive Impairment, Alzheimer’s Disease and Frontotemporal Dementia.
Data for Stability Analysis
Finally, to evaluate the robustness of the N3 brain structural normativity assessments, we use artificially downsampled subgroups of the NAKO study for training. Validation subsets included n=835 healthy control participants from the MACS study which predominantly comprises younger and middle-aged adults, and an additional n=1073 healthy older adults from the ADNI study to span a wider age demographic (see Methods section 5.5).
5.2.2. Study Populations
German National Cohort (NAKO)
The German National Cohort is a population-based longitudinal study initiated in 2014 aiming to investigate the risk factors for major chronic diseases in 200,000 persons living in Germany. It contains high-quality neuroimaging data from participants spanning a broad age range. In this study, we utilize the participants’ 3.0-Tesla T1w-MPRAGE MRI scans (voxel size 1×1×1 mm3, repetition time/echo time=2300/2.98, flip angle=9°) [58–60].
Alzheimer’s Disease Neuroimaging Initiative (ADNI)
ADNI is a major multicenter study started in 2003, designed to develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease. The ADNI was launched as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of neurodegeneration. We included 1.5 and 3.0-Tesla T1w-MPRAGE MRI scans adhering to the ADNI sequence protocol (voxel size 1.3×1×1 mm3, repetition time/echo time=2300/2.98, flip angle=8°, for scanner specific details please see https://adni.loni.usc.edu/data-samples/adni-data/neuroimaging/mri/mri-scanner-protocols/)
Australian Imaging, Biomarker & Lifestyle Study of Aging (AIBL)
AIBL is an Australian study launched in 2006 focusing on understanding the pathways to Alzheimer’s disease. The cohort includes participants diagnosed with Alzheimer’s disease, mild cognitive impairment, and cognitively unimpaired elderly participants, providing insights into the aging process and the development of neurodegenerative diseases. AIBL study methodology has been reported previously 65. MRI scans were performed using a 3D MPRAGE image (voxel size 1.2×1×1 mm3, repetition time/echo time=2300/2.98, flip angle=8°)[63].
NIFD Dataset
The Frontotemporal Lobar Degeneration Neuroimaging Initiative (FTLDNI) was funded through the National Institute of Aging, and started in 2010. The primary goals of FTLDNI were to identify neuroimaging modalities and methods of analysis for tracking frontotemporal lobar degeneration (FTLD) and to assess the value of imaging versus other biomarkers in diagnostic roles. The Principal Investigator of NIFD was Dr. Howard Rosen, MD at the University of California, San Francisco. We use the provided 3D MPRAGE T1-weighted images (voxel size 1×1×1 mm3, repetition time/echo time=2300/2.9, matrix = 240 × 256 × 160) The data are the result of collaborative efforts at three sites in North America. For up-to-date information on participation and protocol, please visit http://memory.ucsf.edu/research/studies/nifd
Open Access Series of Imaging Studies 3 (OASIS3)
OASIS3 serves as a comprehensive digital repository for MRI brain data that supports longitudinal studies of normal aging and cognitive decline [64, 65]. The project is distinguished by its wide age range of participants, providing diverse datasets that enhance the understanding of late-life brain diseases alongside physiological aging processes. We include 3D MPRAGE T1-weighted images (voxel size 1.0 or 1.2×1×1 mm3, repetition time/echo time=2300/2.95 or 2400/3.16 (depending on the scanner), flip angle=9°, FoV=240 or 256mm)
Marburg-Münster Affective Disorder Cohort Study (MACS)
The MACS cohort is part of the DFG-funded research group FOR2107 cohort, researching the etiology and progression of affective disorders [62]. The goal is to integrate and understand the clinical and neurobiological effects of genetisc and environmental factors, and their complex interactions. Participants received financial compensation and gave written informed consent. We use the T1-weighted neuroimaging scans of n=835 healthy control participants to evaluate stability of the N3 models. Images were in Marburg (MR) or Münster (MS) (voxel size 1×1×1 mm3, repetition time/echo time=MR: 1900, MS: 2130/MR: 2.26, MS: 2.28, flip angle=8°, FoV = 256 mm, matrix = 256 × 256, slice thickness = 1 mm)
5.3. Brain Age Model
In the Brain Age paradigm, the brain structure is evaluated with respect to aging effects seen in a healthy reference sample. This is realized by means of a machine learning model trained to predict chronological age from brain structure. The deviation between chronological and predicted age is referred to as the Brain Age Gap (BAG). While a small BAG is considered normative and age-appropriate, a larger positive or negative BAG symbolizes premature or delayed neurostructural degeneration, respectively. The resulting normativity estimation, i.e. the BAG values, have been associated with numerous neurological and psychiatric conditions [13, 35]. For comparison with N3, we train a Brain Age Model using the Python library photonai [66]. We use 90% of the available normative dataset for model training. We use a Support Vector Machine (SVM), for which we optimize the C and gamma parameters in the nested-cross-validation procedure (k=10 outer folds and two randomly shuffled inner folds with a test size of 0.1). The best model achieves an average MAE of 5.43. Finally, we use the remaining 10% of the normative training data to train a linear age bias correction as described in Peng et al. [67]. For the evaluation of unseen samples, we use the Brain Age SVM model to predict age and apply the age correction model, before we calculate the difference between the chronological and predicted age, the BAG.
5.4. Normative Modeling
We calculate normative models on the training data using the Predictive Clinical Neuroscience toolkit as described in Rutherford et al. [9]. To train the models, we normalize GM, WM, WMH, CSF by Total Intracranial Volume (TIV) and fit Bayesian Linear Regression models with default parameters. Subsequently, z-scores for each of the variables are derived, which we aggregate into two normative modeling markers: one being the sum of the absolute z-scores, the second counting the number of absolute z-scores > 1.96.
5.5. Statistical Analysis
A Type III Sum of Squares ANOVA was performed using an ordinary least squares (OLS) model to assess the discriminative and explanatory power of each normativity marker in distinguishing patients from controls. The model was adjusted for potential confounders, including age, age squared (to mitigate non-linear effects), sex and scanner. Partial eta squared () was used to quantify effect size, providing an estimate of how much variance in disease progression could be explained by each normativity marker, alongside a 95% confidence interval.
We evaluate and rank the different normativity markers by post-hoc comparisons of their effect size. To test the observed marker differences for statistical significance, we calculate the ANOVA for each marker with 1000 random permutations. To determine the p value of the marker differences, we evaluate the actual difference between the of our marker N3 and the another marker, with those found in the 1000 random permutations.
To assess each normativity marker’s consistency across age groups, an analysis of age bias was conducted using Spearman’s rank correlation to evaluate the correlation between the normativity estimation values and age in healthy controls.
To assess stability of the N3 models, the Intraclass Correlation Coefficient (ICC) model (2,1) was applied. For this purpose, we used the NAKO sample to train the normativity models, which were downsampled to mimic smaller study populations. Particularly, we divide the training set in k=[10, 5, 3, 2] non-overlapping parts of equal size, train normativity models within each of these subsets, and use external test data to ensure the stability of the normativity estimates. The stability of the normativity estimates was tested using data from the ADNI and MACS cohort, (see Methods section 5.2.1). To ensure validity of the test, we use only age groups with more than 500 samples available from the training sample and more than 20 samples in the test samples.
All statistical analyses were implemented in Python using the scipy, statsmodels and pingouin libraries.
5.6. Machine Learning Analysis
The effectiveness of aging markers in classifying neurodegenerative diseases was further explored through machine learning techniques. We assessed various performance metrics including balanced accuracy, recall, precision, and F1-score. Our analytical pipeline employed the open-source Python framework photonai [66]. The analysis involved nested cross-validation to robustly estimate model performance and avoid overfitting, using k=5 outer folds and k=10 inner folds, each fold stratified to entail a balanced proportion of samples from the diseased class. Hyperparameter optimization was performed via Grid Search to fine-tune the support vector machine (SVM) parameters C and gamma. The machine learning pipeline included steps for z-normalization and balanced sampling (random under-sampling techniques) to address class imbalance within the training data. We measure balanced accuracy, recall, precision and f1 score of each of the normativity markers in the classification of neurodegenerative diseases.
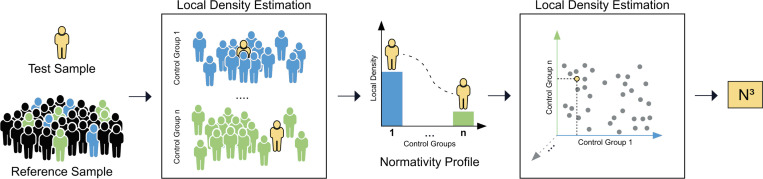
Fig. 1.
Our proposed N3 framework entails methodological innovations that refine normativity assessments in large and diverse medical datasets. We refrain from comparisons to a single normative tendency such as the population average. Instead, we propose to quantify normativity assessment with local density estimation algorithms, which effectively embraces diversity and acknowledges the possibility of multiple, equally viable health states in the population. Moreover, we propose to use several carefully tailored control groups to promote the detection of subtle and nuanced anomalies that may escape broader comparative models. On top of that, we introduce global context to the normativity assessments and join multiple comparative normativity estimations per individual into a so-called normativity profile. This normativity profile acts as a holistic representation of a patient’s health status and provides a multifaceted contextualization to the complex and heterogeneous nature of medical observations. Finally, we convert the normativity profile into a singular, actionable metric, which we call N3. It synthesizes the accumulated information of prior steps and can be adapted to a variety of clinical inquiries. For example, the final N3 normativity assessment can be fine-tuned to express normativity in relation to specific clinical outcomes, such as alignment with normativity profiles in patients who exhibit high treatment responses. The N3 approach is universally applicable, and we see great potential that its application will advance normativity assessments and contribute to personalized patient care.
Table 2.
Study Data Summary
| Study | Group | N Included | Mean Age | Sex |
|---|---|---|---|---|
|
| ||||
| ADNI | HC | 1073 | 68.36 ± 3.3 | 634 females (59.09%) |
| MCI | 1529 | 66.71 ± 4.25 | 729 females (47.67%) | |
| AD | 588 | 67.2 ± 4.65 | 291 females (49.48%) | |
|
| ||||
| AIBL | HC | 368 | 68.00 ± 2.77 | 217 females (58.97%) |
| MCI | 78 | 68.05 ± 3.54 | 33 females (42.31%) | |
| AD | 28 | 66.89 ± 4.44 | 16 females (57.14%) | |
|
| ||||
| OASIS3 | HC | 1643 | 63.36 ± 6.85 | 1028 females (62.57%) |
| MCI | 63 | 66.67 ± 4.85 | 37 females (58.73%) | |
| AD | 228 | 66.54 ± 4.94 | 97 females (42.54%) | |
|
| ||||
| NIFD | HC | 263 | 62.71 ± 6.41 | 148 females (56.27%) |
| FTD | 317 | 63.26 ± 5.66 | 120 females (37.85%) | |
|
| ||||
| MACS | HC | 835 | 35.71 ± 12.6 | 528 females (63.23%) |
|
| ||||
| NAKO | HC | 29883 | 48.45 ± 12.09 | 13201 females (44.18%) |
Acknowledgements.
This work was funded by the German Research Foundation (DFG grants HA7070/2-2, HA7070/3, and HA7070/4 to T.H.), the Interdisciplinary Center for Clinical Research (IZKF) of the medical faculty of Münster (grants Dan3/012/17 to U.D. and MzH 3/020/20 to T.H.) and the IMF research instrument of the medical faculty of Münster (grant LE 1 1 24 09 to R.Leenings). X. Jiang was supported by the Deutsche Forschungsgemeinschaft (DFG) under Grant CRC 1450—431460824.
This project was conducted with data from the German National Cohort (NAKO) (www.nako.de). The NAKO is funded by the Federal Ministry of Education and Research (BMBF) [project funding reference numbers: 01ER1301A/B/C, 01ER1511D, 01ER1801A/B/C/D and 01ER2301A/B/C], federal states of Germany and the Helmholtz Association, the participating universities and the institutes of the Leibniz Association. We thank all participants who took part in the NAKO study and the staff of this research initiative.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12–2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research Development, LLC.; Johnson Johnson Pharmaceutical Research Development LLC.; Lumosity; Lundbeck; Merck Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data was in part provided by OASIS-3: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50AG00561, P30NS09857781, P01AG026276, P01AG003991, R01AG043434, UL1TR000448, R01EB009352. AV-45 doses were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
Data collection and sharing for this project was funded by the Frontotemporal Lobar Degeneration Neuroimaging Initiative (National Institutes of Health Grant R01 AG032306). The study is coordinated through the University of California, San Francisco, Memory and Aging Center. FTLDNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California The investigators at NIFD/FTLDNI contributed to the design and implementation of FTLDNI and/or provided data, but did not participate in analysis or writing of this report (unless otherwise listed). The FTLDNI investigators included the following individuals: Howard Rosen; University of California, San Francisco (PI) Bradford C. Dickerson; Harvard Medical School and Massachusetts General Hospital Kimoko Domoto-Reilly; University of Washington School of Medicine David Knopman; Mayo Clinic, Rochester Bradley F. Boeve; Mayo Clinic Rochester Adam L. Boxer; University of California, San Francisco John Kornak; University of California, San Francisco Bruce L. Miller; University of California, San Francisco William W. Seeley; University of California, San Francisco Maria-Luisa Gorno-Tempini; University of California, San Francisco Scott McGinnis; University of California, San Francisco Maria Luisa Mandelli; University of California, San Francisco
Footnotes
Declarations
Code Availability
Code to realize the normativity estimation calculations within the N3 framework is written in the Python programming language and is provided as an open-source resource to the scientific community on Github (link tbd).
Supplementary information.
Data Availability
Data were obtained from the German National Cohort (NAKO), the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the Open Access Series of Imaging Studies 3 (OASIS3), the Frontotemporal Lobar Degeneration Neuroimaging Initiative (NIFD, and the Australian Imaging, Biomarker Lifestyle Study of Aging (AIBL). Data of the MACS study are not publicly available. All other data are available upon request via the access management systems of the respective studies. (NAKO: nako.de/forschung, ADNI, AIBL, NIFD: ida.loni.usc.edu, OASIS3: sites.wustl.edu/oasisbrains)
References
- [1].Canguilhem G. On the Normal and the Pathological 171–179 (1978).
- [2].Debru C. The Concept of Normativity from Philosophy to Medicine: An Overview. Medicine Studies 3, 1–7 (2011). [Google Scholar]
- [3].Collins F. S. & Harold V. A New Initiative on Precision Medicine. New England Journal of Medicine 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Khoury M. J., Iademarco M. F. & Riley W. T. Precision Public Health for the Era of Precision Medicine. American Journal of Preventive Medicine 50, 398–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stephan K. E. et al. Charting the Landscape of Priority Problems in Psychiatry, Part 1: Classification and Diagnosis. The Lancet Psychiatry 3, 77–83 (2016). [DOI] [PubMed] [Google Scholar]
- [6].Bzdok D. & Meyer-Lindenberg A. Machine Learning for Precision Psychiatry: Opportunities and Challenges. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 3, 223–230 (2017). [DOI] [PubMed] [Google Scholar]
- [7].Rutherford S. et al. Evidence for embracing normative modeling. eLife 12, e85082 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marquand A. F., Rezek I., Buitelaar J. & Beckmann C. F. Understanding Heterogeneity in Clinical Cohorts Using Normative Models: Beyond Case-Control Studies. Biological Psychiatry 80, 552–561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rutherford S. et al. The normative modeling framework for computational psychiatry. Nature Protocols 17, 1711–1734 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Segal A. et al. Embracing variability in the search for biological mechanisms of psychiatric illness. Trends in Cognitive Sciences (2024).
- [11].Bethlehem R. A. I. et al. Brain charts for the human lifespan. Nature 604, 525–533 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Franke K. & Gaser C. Ten Years of BrainAGE as a Neuroimaging Biomarker of Brain Aging: What Insights Have We Gained? Frontiers in Neurology 10, 789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Baecker L., Garcia-Dias R., Vieira S., Scarpazza C. & Mechelli A. Machine learning for brain age prediction: Introduction to methods and clinical applications. EBioMedicine 72, 103600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bzdok D., Altman N. & Krzywinski M. Statistics versus machine learning. Nature Methods 15, 233–234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Poldrack R. A., Huckins G. & Varoquaux G. Establishment of Best Practices for Evidence for Prediction. JAMA Psychiatry 77, 534–540 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oh S. S. et al. Diversity in Clinical and Biomedical Research: A Promise Yet to Be Fulfilled. PLoS Medicine 12, e1001918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hood L., Lovejoy J. C. & Price N. D. Integrating big data and actionable health coaching to optimize wellness. BMC Medicine 13, 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bodicoat D. H. et al. Promoting inclusion in clinical trials—a rapid review of the literature and recommendations for action. Trials 22, 880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Elhussein A. et al. A framework for sharing of clinical and genetic data for precision medicine applications. Nature Medicine 1–12 (2024).
- [20].Belle A. et al. Big Data Analytics in Healthcare. BioMed Research International 2015, 370194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ghassemi M. et al. A Review of Challenges and Opportunities in Machine Learning for Health. AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science 2020, 191–200 (2020). [PMC free article] [PubMed] [Google Scholar]
- [22].Xuan Y. et al. Standardization and harmonization of distributed multi-center proteotype analysis supporting precision medicine studies. Nature Communications 11, 5248 (2020). [Google Scholar]
- [23].Froelicher D. et al. Truly privacy-preserving federated analytics for precision medicine with multiparty homomorphic encryption. Nature Communications 12, 5910 (2021). [Google Scholar]
- [24].Zhang Y. & Chung Y. Nonparametric estimation of linear personalized diagnostics rules via efficient grid algorithm. Statistics in Medicine 43, 1354–1371 (2024). [DOI] [PubMed] [Google Scholar]
- [25].Tozzi L. et al. Personalized brain circuit scores identify clinically distinct biotypes in depression and anxiety. Nature Medicine 30, 2076–2087 (2024). [Google Scholar]
- [26].Blume H. Neurodiversity - On the neurological underpinnings of geekdom. The Atlantic (1998). URL https://www.theatlantic.com/magazine/archive/1998/09/neurodiversity/305909/.
- [27].Ortega F. The Cerebral Subject and the Challenge of Neurodiversity. BioSocieties 4, 425–445 (2009). [Google Scholar]
- [28].Dwyer P. The Neurodiversity Approach(es): What Are They and What Do They Mean for Researchers? Human Development 66, 73–92 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yang Z. et al. Brain aging patterns in a large and diverse cohort of 49,482 individuals. Nature Medicine 1–12 (2024).
- [30].Tan Y. Y. et al. Comparing clinical trial population representativeness to real-world populations: an external validity analysis encompassing 43895 trials and 5685738 individuals across 989 unique drugs and 286 conditions in England. The Lancet Healthy Longevity 3, e674–e689 (2022). [DOI] [PubMed] [Google Scholar]
- [31].Spall H. G. C. V., Toren A., Kiss A. & Fowler R. A. Eligibility Criteria of Randomized Controlled Trials Published in High-Impact General Medical Journals: A Systematic Sampling Review. JAMA 297, 1233–1240 (2007). [DOI] [PubMed] [Google Scholar]
- [32].Abid A., Zhang M. J., Bagaria V. K. & Zou J. Exploring patterns enriched in a dataset with contrastive principal component analysis. Nature Communications 9, 2134 (2018). [Google Scholar]
- [33].Halder R. K., Uddin M. N., Uddin M. A., Aryal S. & Khraisat A. Enhancing K-nearest neighbor algorithm: a comprehensive review and performance analysis of modifications. Journal of Big Data 11, 113 (2024). [Google Scholar]
- [34].Meehan A. J. et al. Clinical Prediction Models in Psychiatry: A Systematic Review of Two Decades of Progress and Challenges. Molecular Psychiatry 1–9 (2022).
- [35].Cole J. H. Multimodality neuroimaging brain-age in UK biobank: relationship to biomedical, lifestyle, and cognitive factors. Neurobiology of Aging 92, 34–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wrigglesworth J. et al. Factors associated with brain ageing - a systematic review. BMC Neurology 21, 312 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gafson A. R. et al. Neurofilaments: neurobiological foundations for biomarker applications. Brain 143, 1975–1998 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Braak H. & Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathologica 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- [39].Wilson D. M. et al. Hallmarks of neurodegenerative diseases. Cell 186, 693–714 (2023). [DOI] [PubMed] [Google Scholar]
- [40].Dong A. et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain 140, 735–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Skampardoni I. et al. Genetic and Clinical Correlates of AI-Based Brain Aging Patterns in Cognitively Unimpaired Individuals. JAMA Psychiatry 81, 456–467 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Finch C. E. & Crimmins E. M. Inflammatory Exposure and Historical Changes in Human Life-Spans. Science 305, 1736–1739 (2004). [DOI] [PubMed] [Google Scholar]
- [43].Gillman M. W. Developmental Origins of Health and Disease. The New England Journal of Medicine 353, 1848–1850 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dehnel T. The European Dementia Prevention Initiative. The Lancet Neurology 12, 227–228 (2013). [DOI] [PubMed] [Google Scholar]
- [45].Yu M., Sporns O. & Saykin A. J. The human connectome in Alzheimer disease — relationship to biomarkers and genetics. Nature Reviews Neurology 17, 545–563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Prabhakar C. et al. ViT-AE++: Improving Vision Transformer Autoencoder for Self-supervised Medical Image Representations. arXiv (2023).
- [47].Shen L. & Thompson P. M. Brain Imaging Genomics: Integrated Analysis and Machine Learning. Proceedings of the IEEE 108, 125–162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Misra S. Precision health could mitigate clinical biases that impact care. Nature Medicine 30, 1804–1804 (2024). [Google Scholar]
- [49].Charalampopoulos D. et al. Exploring Variation in Glycemic Control Across and Within Eight High-Income Countries: A Cross-sectional Analysis of 64,666 Children and Adolescents With Type 1 Diabetes. Diabetes Care 41, 1180–1187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bermingham K. M. et al. Effects of a personalized nutrition program on cardiometabolic health: a randomized controlled trial. Nature Medicine 30, 1888–1897 (2024). [Google Scholar]
- [51].Leslie R. D. et al. Understanding diabetes heterogeneity: key steps towards precision medicine in diabetes. The Lancet Diabetes & Endocrinology 11, 848–860 (2023). [DOI] [PubMed] [Google Scholar]
- [52].Delanaye P. et al. CKD: A Call for an Age-Adapted Definition. Journal of the American Society of Nephrology 30, 1785–1805 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wetzels J., Kiemeney L., Swinkels D., Willems H. & Heijer M. Age- and gender-specific reference values of estimated GFR in Caucasians: The Nijmegen Biomedical Study. Kidney International 72, 632–637 (2007). [DOI] [PubMed] [Google Scholar]
- [54].Cassano P. A., Segal M. R., Vokonas P. S. & Weiss S. T. Body fat distribution, blood pressure, and hypertension A prospective cohort study of men in the normative aging study. Annals of Epidemiology 1, 33–48 (1990). [DOI] [PubMed] [Google Scholar]
- [55].Rosner B., Prineas R., Loggie J. & Daniels S. Blood pressure nomograms for children and adolescents, by height, sex, and age, in the United States. The Journal of Pediatrics 123, 871–886 (1993). [DOI] [PubMed] [Google Scholar]
- [56].Cover T. & Hart P. Nearest neighbor pattern classification. IEEE Transactions on Information Theory 13, 21–27 (1967). [Google Scholar]
- [57].Mudholkar G. & Srivastava D. Exponentiated Weibull family for analyzing bathtub failure-rate data. IEEE Transactions on Reliability 42, 299–302 (1993). [Google Scholar]
- [58].Peters A. et al. Framework and baseline examination of the German National Cohort (NAKO). European Journal of Epidemiology 37, 1107–1124 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Consortium G. N. C. G. The German National Cohort: aims, study design and organization. European Journal of Epidemiology 29, 371–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bamberg F. et al. Whole-Body MR Imaging in the German National Cohort: Rationale, Design, and Technical Background. Radiology 277, 206–220 (2015). [DOI] [PubMed] [Google Scholar]
- [61].Petersen R. C. et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI) Clinical characterization. Neurology 74, 201–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Vogelbacher C. et al. The Marburg-Münster Affective Disorders Cohort Study (MACS): A Quality Assurance Protocol for MR Neuroimaging Data. NeuroImage 172, 450–460 (2018). [DOI] [PubMed] [Google Scholar]
- [63].Fowler C. et al. Fifteen Years of the Australian Imaging, Biomarkers and Lifestyle (AIBL) Study: Progress and Observations from 2,359 Older Adults Spanning the Spectrum from Cognitive Normality to Alzheimer’s Disease. Journal of Alzheimer’s Disease Reports 5, 443–468 (2021). [Google Scholar]
- [64].Marcus D. S. et al. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. Journal of Cognitive Neuroscience 19, 1498–1507 (2007). [DOI] [PubMed] [Google Scholar]
- [65].LaMontagne P. J. et al. OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. medRxiv 2019.12.13.19014902 (2019).
- [66].Leenings R. et al. PHOTONAI—A Python API for Rapid Machine Learning Model Development. PLoS ONE 16, e0254062 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Peng H., Gong W., Beckmann C. F., Vedaldi A. & Smith S. M. Accurate brain age prediction with lightweight deep neural networks. Medical Image Analysis 68, 101871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data were obtained from the German National Cohort (NAKO), the Alzheimer’s Disease Neuroimaging Initiative (ADNI), the Open Access Series of Imaging Studies 3 (OASIS3), the Frontotemporal Lobar Degeneration Neuroimaging Initiative (NIFD, and the Australian Imaging, Biomarker Lifestyle Study of Aging (AIBL). Data of the MACS study are not publicly available. All other data are available upon request via the access management systems of the respective studies. (NAKO: nako.de/forschung, ADNI, AIBL, NIFD: ida.loni.usc.edu, OASIS3: sites.wustl.edu/oasisbrains)