Abstract
Objectives
Develop risk-adapted conditional biopsy pathways utilizing MRI in combination with prostate-specific antigen (PSA) density (PSAD) and the ratio of free to total PSA (f/tPSA), respectively, to enhance the detection of clinically significant prostate cancer (csPCa) while minimizing ‘negative’ biopsies in low-risk patients.
Methods
The Prostate Imaging Reporting and Data System (PI-RADS) category, PSAD, f/tPSA and biopsy-pathology of 1018 patients were collected retrospectively. Subsequently, PSAD and f/tPSA were divided into four intervals, which were then combined with the MRI findings to construct two risk stratification matrix tables. Six biopsy decision pathways were established: three clinical pathways based solely on PSAD and f/tPSA, and three MRI-combined pathways incorporating both PI-RADS and PSA-derived indicators. The biopsy and clinically insignificant PCa (ciPCa) avoidance, csPCa detection rate, and ‘negative’ biopsies proportion were assessed. Decision curve analysis (DCA) was employed to evaluate the net benefit associated with each pathway.
Results
When reporting PI-RADS 1 - 2, PSAD ≥ 0.20 ng/ml/cm3 or f/tPSA ≤ 0.10 were found to be useful for patient stratification. When reporting PI-RADS 3, PSAD ≥ 0.10 − 0.15 ng/ml/cm3 and f/tPSA ≤ 0.16 − 0.25 were helpful in distinguishing the risk of csPCa. The three MRI-combined pathways showed higher csPCa detection rates (94% to 96%) than the three clinical pathways (85% to 91%); ‘MRI + PSAD + f/tPSA’ demonstrated a high csPCa detection rate of 94% while maintaining the maximum biopsy avoidance and lowest ‘negative’ biopsy proportion of 40% and 25%, respectively. The DCA showed significantly higher net benefits for three MRI-combined pathways compared to all clinical pathways.
Conclusions
The integration of MRI and PSA-derived indicators enables effective patient risk stratification, thereby providing valuable decision-making pathways to enhance the management of csPCa while minimizing ‘negative’ biopsies.
Keywords: Prostate cancer, magnetic resonance imaging, prostate-specific antigen density, the ratio of free to total prostate-specific antigen, biopsy indicator
Introduction
The global incidence of prostate cancer (PCa) has reached 1.5 million new cases in 2022, with a corresponding mortality rate of 397,000 deaths. This positions PCa as the second most prevalent cancer worldwide and the fifth leading cause of male cancer-related fatalities [1]. Prostate-specific antigen (PSA) serves as a primary screening indicator for PCa; the PSA test, however, is susceptible to interference from non-neoplastic conditions such as benign prostatic hyperplasia and inflammation [2]. The positive predictive value (PPV) of PSA ≥ 3 ng/ml as an biopsy indication is notably limited (approximately 16%) and infiluenced by patient age and other factors [3], thus necessitating an accurate procedure to minimize overevaluation. Multi-parameter MRI (mpMRI), serving as a non-invasive triage tool, not only enables the detection of clinically significant PCa (csPCa), but also provides localized staging and guidance for biopsies, thus establishing its reliability in pre-biopsy risk assessment [4, 5]. Compared to systematic biopsies, MRI-based triage pathways can decrease false-positive results effectively and minimize the detection of low-grade tumours by approximately 30%, while maintaining or even improving the identification of clinically significant tumours at a robust level [6, 7].
The MRI pathway is implemented based on the Prostate Imaging Reporting and Data System (PI-RADS). However, the PI-RADS category solely represents the likelihood of csPCa and does not directly inform disease decision-making and management. Particularly when reported as PI-RADS 3, the utility of MRI in determining biopsy procedure is limited due to uncertainty regarding csPCa presence, with a probability range of 10% to 30% [8]. The study conducted by Dominik et al. revealed that both PSA density (PSAD) and PI-RADS independently serve as risk factors for accurately predicting csPCa [9]. Furthermore, the combination of these two factors can effectively reduce benign biopsies and the detection of low-grade cancers in patients with PI-RADS 3 or higher [10]. The utilization of PSAD < 0.15 ng/ml/cm3 enhanced the negative predictive value (NPV) of MRI for csPCa in biopsy-naive men from 83% to 90%. Conversely, when PSAD ≥ 0.20 ng/ml/cm3, the csPCa risk in patients with negative MRI findings escalated to as high as 27% to 40% [11–13]. The updated European Association of Urology (EAU) guideline [14] recommends employing the risk stratification matrix table developed by Schoots et al. which incorporates PI-RADS categories and PSAD, as a decision-making tool for determining whether a biopsy should be performed, thereby mitigating the impact of MRI threshold setting on diagnostic outcomes [15].
Due to the limited accuracy and specificity of PSA as a standalone test, studies have demonstrated an inverse correlation between free PSA (fPSA) levels and the risk of PCa [16, 17]. Consequently, the utilization of the ratio of free to total PSA (f/tPSA) has been proposed to enhance risk stratification when PSA falls within the range of 4 to 10 ug/ml. Evaluation of f/tPSA has indicated that unnecessary biopsies could be reduced by up to 50% among men with PSA in the grey area [18]. Additionally, Ferraro et al. have demonstrated that the integration of f/tPSA into a nomogram model can effectively predict high-grade tumour with International Society of Urology Pathology (ISUP) grade ≥ 3 [19]. However, there is limited literature on the combined use of f/tPSA and MRI, and its potential value in optimizing biopsy indications remains unconfirmed. Additionally, it should be noted that Schoots et al.’s risk decision matrix is a theoretical prediction tool based on data analysis and has not been implemented in real clinical practice. Therefore, this study aims to externally validate the risk matrix while exploring the added benefit of combining f/tPSA with MRI to enhance biopsy strategies.
Materials and methods
The present study was conducted in accordance with the Declaration of Helsinki and granted approval by the institutional review board of Zhejiang Cancer Hospital (IRB-2024-432). Because the study was retrospective in nature and all image data were de-identified to ensure patient confidentiality, the requirement for written informed consent exempted by ethics committee of Zhejiang Cancer Hospital.
Study cohort
The clinical and imaging data of consecutive patients who underwent prostate MRI and ‘targeted + systematic’ biopsies were retrospectively collected from January 2021 to June 2024 at three centres: Zhejiang Province Cancer Hospital, Wuhu City Second People’s Hospital, and the First Affiliated Hospital of Soochow University. The following exclusion criteria were applied: (1) incomplete clinical data (e.g. missing tPSA, fPSA, or f/tPSA); (2) poor image quality (gas or motion artifacts) or incomplete images affecting PI-RADS assessment; (3) the absence of pathological results or presence of PCa from non-acinar cell origin; (4) completion of the sampling procedure before MRI or previous intervention like endocrine therapy and radiotherapy. Ultimately, 1018 eligible patients were included in the study.
Prostate MRI technique and PI-RADS interpretation
The mpMRI was conducted using a 3.0-T MRI scanner equipped with pelvic phased array coils. Essential sequences include axial, sagittal, and coronal T2-weighted imaging (T2WI), high-b-value diffusion-weighted imaging (DWI) with corresponding apparent diffusion coefficient (ADC) maps, and dynamic contrast enhancement (DCE) sequences. The parameters for each sequence are presented in Table 1.
Table 1.
Standardized mpMRI scanning protocols of three centres.
| Parameter | Institution 1 |
Institution 2 |
Institution 3 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| T2WI | DWI | DEC-MRI | T2WI | DWI | DEC-MRI | T2WI | DWI | DEC-MRI | |
| TR (ms) | 5639 | 4000 | 4.2 | 4569 | 3997 | 3.2 | 4000 | 4000 | 3.22 |
| TE (ms) | 125 | 50 | 0 | 91 | 70 | 1.53 | 89 | 56 | 1.18 |
| Thickness (mm) | 3 | 3 | 3 | 4 | 4 | 4 | 3 | 3 | 3 |
| Interslice gap (mm) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Matrix size | 320 × 280 | 184 × 184 | 256 × 256 | 320 × 320 | 128 × 128 | 224 × 224 | 320 × 256 | 256 × 256 | 256 × 256 |
| FOV (mm × mm) | 200 × 200 | 203 × 203 | 300 × 270 | 200 × 200 | 200 × 200 | 250 × 250 | 640 × 640 | 160 × 296 | 512 × 512 |
| NSA | 2 | 2 | 1 | 2 | 3 | 1 | 3 | 1 | |
| b factor (s/mm2) | 0, 800, 1500 | 0, 800, 2000 | 100, 800, 2000 | ||||||
TR Repetition time, TE Time echo, FOV Field of view, T2WI T2-Weighted Imaging, DWI, diffusion weighted imaging DCE dynamic contrast-enhanced.
Re-evaluation of PI-RADS categories was conducted by two radiologists, each with 3 and 5 years of experience in PI-RADS reporting, respectively, adhering to the PI-RADS v2.1 standard [20]. Clinical information and pathological results were concealed from them. Where both the peripheral zone (PZ) and transition zone (TZ) were involved, the lesion was considered to originate from the area exceeding 60%, and adhering to the assessment principle specific to this region. For patients presenting with multiple lesions simultaneously, the patient’s PI-RADS categories were determined based on the lesion with the highest suspicion category. When there was a discrepancy, a consensus was reached through discussion among another senior radiologist (with 10 years of experience in prostate MRI interpretation) and the two radiologists, who jointly determined the appropriate PI-RADS category.
The anterior and posterior diameter (AP), superior and inferior diameter (SI), and transverse diameter (T) of the prostate were measured by another radiologist, and the average of two measurements was recorded to document the results. Prostate volume (PV) was calculated according to the ellipsoid formula (PV = AP * SI * T * 0.52). PSAD was obtained by dividing the total PSA by PV.
Biopsy and histopathology
Prostate biopsies were performed by experienced sonographers under transrectal ultrasound (TRUS) guidance, accurately recording each sample region to match MRI images. All patients underwent a systematic biopsy with 10 – 12 cores through the perineum, while 2 – 3 targeted cores were conducted for PI-RADS 3 or higher lesions using either TRUS-MRI fusion or cognitive fusion techniques.
Biopsy cores were independently reviewed by a pathologist possessing professional qualifications (with 8 years of experience in prostate pathology reporting) to determine the Gleason score (GS) and ISUP grade. Tumours classified as ISUP grade 2 or higher are considered csPCa. Clinical insignificant PCa (ciPCa) is defined as ISUP grade 1 with a GS of 6 or less [21].
Decision-making pathway for biopsy
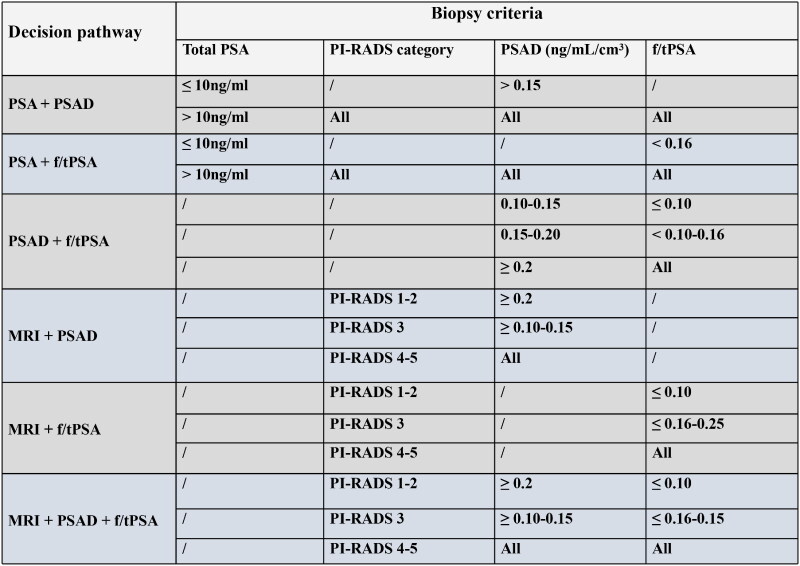
PSAD was categorized into 4 risk levels: low risk (< 0.10 ng/ml/cm3), intermediate-low risk (0.10 − 0.15 ng/ml/cm3), intermediate-high risk (0.15 − 0.20 ng/ml/cm3), and high risk (> 0.20 ng/ml/cm3). f/tPSA was also divided into four intervals: extremely low (≤ 0.10), low (0.10 − 0.16), slightly low (0.16 − 0.25), and normal (≥ 0.25). We analysed and compared the following six risk stratification methods to identify patients require biopsy (Figure 1): ‘PSA + PSAD’, ‘PSA + f/tPSA’, ‘PSAD + f/tPSA’, ‘MRI + PSAD’, ‘MRI + f/tPSA’, and ‘MRI + PSAD + f/tPSA’.
Figure 1.
Six pathways for biopsy decision making.
PSA: Prostate-specific antigen; PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen; PI-RADS: Prostate Imaging Reporting and Data System
Statistical analysis
The statistical software employed SPSS (version 25.0, Chicago, IL, USA) and R language (version 4.2.1, R Foundation for Statistical Computing, Vienna, Austria). After confirming inconformity with normal distribution characteristics, continuous variables were expressed as median (interquartile range [IQR]). Categorical variables were reported as frequencies and percentages. Biopsy avoidance rates, ciPCa avoidance rates, and csPCa detection rates were calculated and compared using the McNemar test. Decision curve analysis (DCA) was utilized to evaluate the clinical ‘net benefit’ of the six decision pathways. DCA was determined by subtracting the detrimental impact of ‘negative’ biopsies (false positives) from the advantageous outcome of detecting csPCa (true positives) at a specific threshold probability, with the following formula (N represents the total number of subjects, pt denotes the threshold probability):
Results
Baseline characteristics
The study enrolled a total of 1,018 patients, with a median age of 70 (IQR 64–76) years, a median tPSA of 10.77 (IQR 7.11 − 18.25) ng/ml, a median PSAD of 0.21 (IQR 0.12 − 0.39) ng/ml/cm3 and a median f/tPSA of 0.14 (IQR 0.10 − 0.19). Among the patients, 39% (396/1018) were reported as PI-RADS 1–2, 20% (199/1018) were PI-RADS 3, and 41% (413/1018) were PI-RADS 4–5. 54% (549/1018) patients observed benign biopsy results, whereas 9% (91/1018) were diagnosed with ciPCa, and the remaining 37% (378/1018) were confirmed with csPCa (Table 2).
Table 2.
The baseline characteristics of the study cohort.
| Characteristics | Values |
|---|---|
| Age, median (IQR), years | 70 (64,76) |
| fPSA, median (IQR), ng/ml | 1.5 (0.9, 2.7) |
| tPSA, median (IQR), ng/ml | 10.8 (7.1, 18.3) |
| f/tPSA, median (IQR) | 0.14 (0.10, 0.19) |
| Prostate volume, median (IQR), cm3 | 51.05 (35.54, 74.14) |
| PSAD, median (IQR), ng/ml/cm3 | 0.21 (0.12, 0.39) |
| Prostate zone, n (%) | |
| PZ | 353 (34%) |
| TZ | 573 (56%) |
| PZ+TZ | 92 (9%) |
| PI-RADS, n (%) | |
| 1 | 28 (3%) |
| 2 | 368 (36%) |
| 3 | 199 (20%) |
| 4 | 205 (20%) |
| 5 | 218 (21%) |
| Biopsy-ISUP grade, n (%) | |
| No PCa | 549 (54%) |
| ISUP 1 (GS 3 + 3) | 91 (9%) |
| ISUP 2 (GS 3 + 4) | 87 (8%) |
| ISUP 3 (GS 4 + 3) | 109 (11%) |
| ISUP 4 (GS 8) | 83 (8%) |
| ISUP 5 (GS 9 ∼ 10) | 99 (10%) |
IQR: interquartile range; fPSA: free prostate-specific antigen; tPSA: total prostate-specific antigen; PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen; PZ: peripheral zone; TZ: transition zone; PI-RADS: Prostate Imaging Reporting and Data System; ISUP: International Society of Urology Pathology; GS: Gleason score.
Distribution of PCa within PSAD and f/tPSA risk intervals
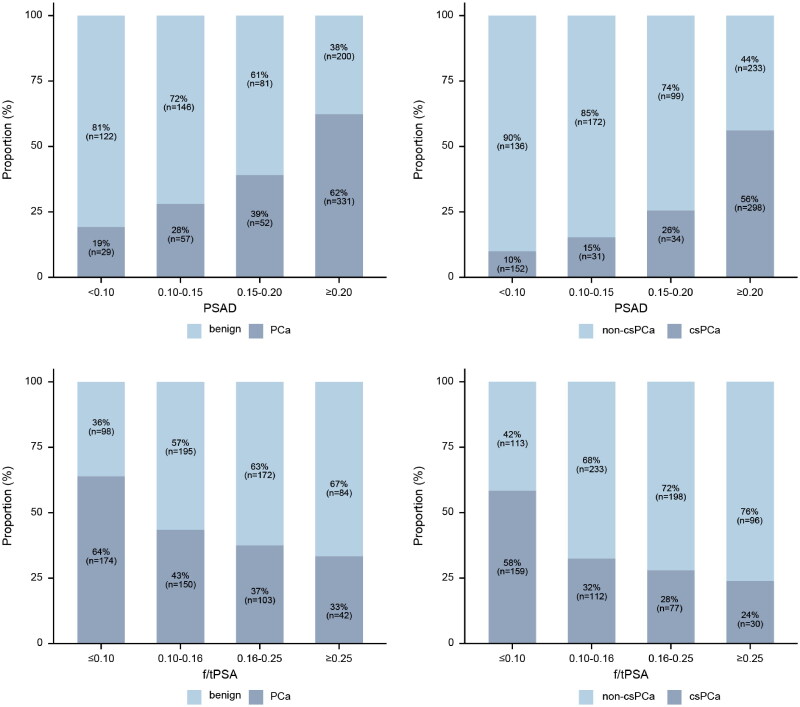
The proportions of PCa and csPCa in low, intermediate-low, intermediate-high, and high intervals of PSAD showed an increasing trend (Table 3, Figure 2), with 62% (331/531) of PCa and 56% (298/531) of csPCa in the high interval. The proportions of PCa and csPCa in extremely low, low, intermediate-low, and normal intervals of f/tPSA showed a decreasing trend (Table 4, Figure 2), with 64.0% (174/272) of PCa and 58% (159/272) of csPCa in the high interval.
Table 3.
The observed proportion of csPCa in relation to PI-RADS categories and PSAD levels.
| PSAD (ng/ml/cm3) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI-RADS | PCa prevalence | csPCa prevalence | <0.10 low |
0.10-0.15 Intermediate-low |
0.15-0.20 Intermediate-high |
≥0.20 high |
||||||||
| Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa |
|||
| 1-2 | 12% 48/396 |
7% 29/396 |
89% 79/89 |
4% 4/89 |
7% 6/89 |
91% 97/107 |
5% 6/107 |
4% 4/107 |
88% 59/67 |
7% 5/67 |
5% 3/67 |
85% 113/133 |
3% 4/133 |
12% 16/133 |
| 3 | 27% 54/199 |
13% 26/199 |
87% 34/39 |
10% 4/39 |
3% 1/39 |
70% 36/51 |
18% 9/51 |
12% 6/51 |
54% 12/22 |
23% 5/22 |
23% 5/22 |
72% 63/87 |
12% 10/87 |
16% 14/87 |
| 4-5 | 87% 367/423 |
76% 323/423 |
39% 9/23 |
26% 6/23 |
35% 8/23 |
29% 13/45 |
24% 11/45 |
47% 21/45 |
23% 10/44 |
18% 8/44 |
59% 26/44 |
8% 24/311 |
6% 19/311 |
86% 268/311 |
| All | 46% 469/1018 |
37% 378/1018 |
81% 122/151 |
9% 14/151 |
10% 15/151 |
72% 146/203 |
13% 26/203 |
15% 31/203 |
61% 81/133 |
13% 18/133 |
26% 34/133 |
38% 200/531 |
6% 33/531 |
56% 298/531 |
| Very low | 0%-5% csPCa (below population risk) | No biopsy | ||||||||||||
| low | 5%-10% csPCa (acceptable risk) | No biopsy | ||||||||||||
| Intermediate-low | 10%-20% csPCa | Consider biopsy | ||||||||||||
| Intermediate-high | 20%-30% csPCa | Highly consider biopsy | ||||||||||||
| High | 30%-40% csPCa | Perform biopsy | ||||||||||||
| Very high | > 40% csPca | Perform biopsy | ||||||||||||
PCa: Prostate cancer; csPCa: clinically significant prostate cancer; PSAD: Prostate-specific antigen density; PI-RADS: Prostate Imaging Reporting and Data System; ISUP: International Society of Urology Pathology.
Figure 2.
The distribution of prostate cancer (PCa) (left) and clinically significant prostate cancer (csPCa) (right) within each PSAD and f/tPSA intervals.
PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen
Table 4.
The observed proportion of csPCa in relation to PI-RADS categories and f/tPSA levels.
| f/tPSA |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI-RADS | PCa prevalence | csPCa prevalence | ≤ 0.10 Extremely low |
0.10-0.16 Low |
0.16-0.25 Intermediate-low |
≥ 0.25 Normal |
|||||||||
| Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa | Benign | ISUP 1 PCa | ISUP ≥2 PCa | ||||
| 1-2 | 12% 48/396 |
7% 29/396 |
82% 62/75 |
3% 2/75 |
15% 11/75 |
88% 125/142 |
6% 8/142 |
6% 9/142 |
92% 114/124 |
5% 6/124 |
3% 4/124 |
86% 47/55 |
5% 3/55 |
9% 5/55 |
|
| 3 | 27% 54/199 |
13% 26/199 |
69% 25/36 |
11% 4/36 |
20% 7/36 |
72% 51/71 |
18% 13/71 |
10% 7/71 |
69% 43/62 |
15% 9/62 |
16% 10/62 |
87% 26/30 |
7% 2/30 |
6% 2/30 |
|
| 4-5 | 87% 367/423 |
76% 323/423 |
7% 11/161 |
5% 9/161 |
88% 141/161 |
14% 19/132 |
13% 17/132 |
73% 96/132 |
17% 15/89 |
12% 11/89 |
71% 63/89 |
27% 11/41 |
17% 7/41 |
56% 23/41 |
|
| All | 46% 469/1018 |
37% 378/1018 |
36% 98/272 |
5% 15/272 |
59% 159/272 |
57% 195/345 |
11.0% 38/345 |
32% 112/345 |
63% 172/275 |
9% 26/275 |
28% 77/275 |
67% 84/126 |
9% 12/126 |
24% 30/128 |
|
| Very low | 0%-5% csPCa (below population risk) | No biopsy | |||||||||||||
| low | 5%-10% csPCa (acceptable risk) | No biopsy | |||||||||||||
| Intermediate-low | 10%-20% csPCa | Consider biopsy | |||||||||||||
| Intermediate-high | 20%-30% csPCa | Highly consider biopsy | |||||||||||||
| High | 30%-40% csPCa | Perform biopsy | |||||||||||||
| Very high | > 40% csPca | Perform biopsy | |||||||||||||
PCa: Prostate cancer; csPCa: clinically significant prostate cancer; f/tPSA: ratio of free to total prostate-specific antigen; PI-RADS: Prostate Imaging Reporting and Data System; ISUP: International Society of Urology Pathology.
Risk stratification of csPCa based on PI-RADS and PSA-derived indicators
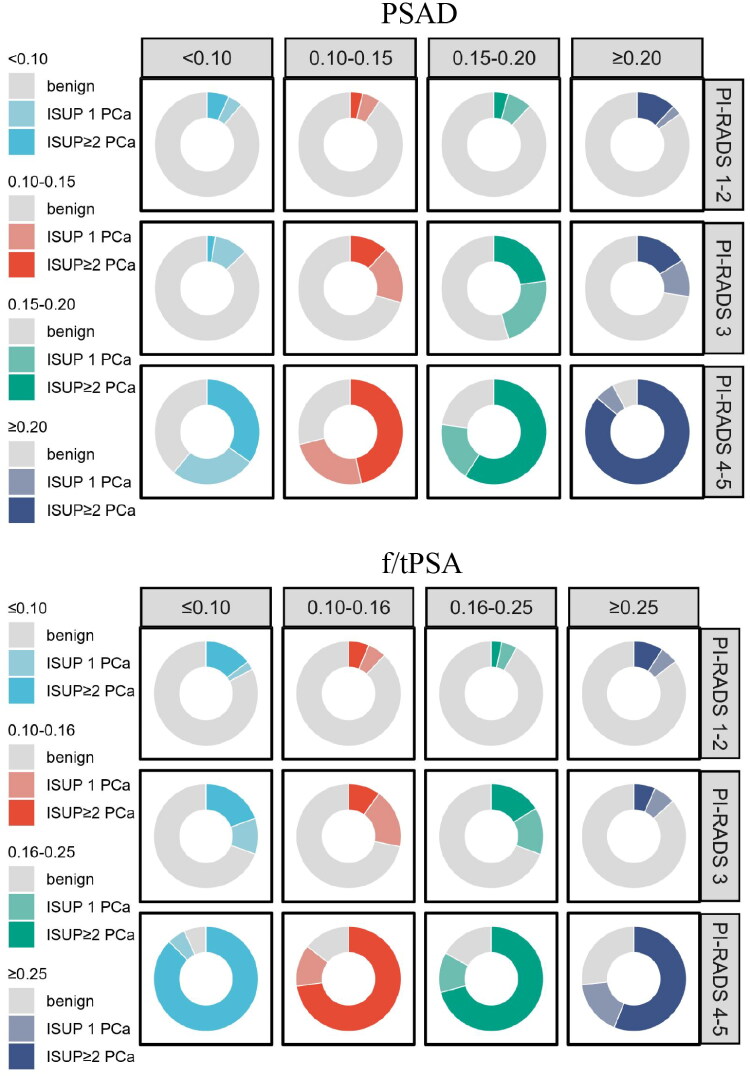
The PI-RADS category was divided into three levels, which were integrated with the risk intervals of PSAD and f/tPSA to establish twelve risk subgroups, formulating two sets of risk-adapted biopsy decision tables (Tables 3 and 4, Figure 3). When the combinations of PI-RADS category and PSAD were ‘PI-RADS 1 - 2 + PSAD ≥ 0.20 ng/ml/cm3’, ‘PI-RADS 3 + PSAD ≥ 0.10 − 0.15 ng/ml/cm3’, and ‘PI-RADS 4 - 5 + any PSAD’, the observed risks of csPCa exceeded 10%, with rates of 12%, 12%−23%, and 35%−86%, respectively. When the combinations of PI-RADS category and f/tPSA were ‘PI-RADS 1–2 + f/tPSA ≤ 0.10’, ‘PI-RADS 3 + f/tPSA ≤ 0.16 − 0.25’, and ‘PI-RADS 4–5 + any f/tPSA’, the observed risk of csPCa exceeded the acceptable level, with rates of 15%, 10%−19%, and 56%−88%, respectively. The observed csPCa risks in remaining combinations of PI-RADS category and PSAD (or f/tPSA) were below 10%.
Figure 3.
Risk distribution map of csPCa for each subgroup categorized by PI-RADS and PSA-derived indicators. The above diagram illustrates the integration of PI-RADS with PSAD, while the below diagram demonstrates the integration of PI-RADS with f/tPSA.
PCa: Prostate cancer; csPCa: clinically significant prostate cancer; PSA: Prostate-specific antigen; PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen; PI-RADS: Prostate Imaging Reporting and Data System; ISUP: International Society of Urology Pathology
Pathological comparison of six decision-making pathways
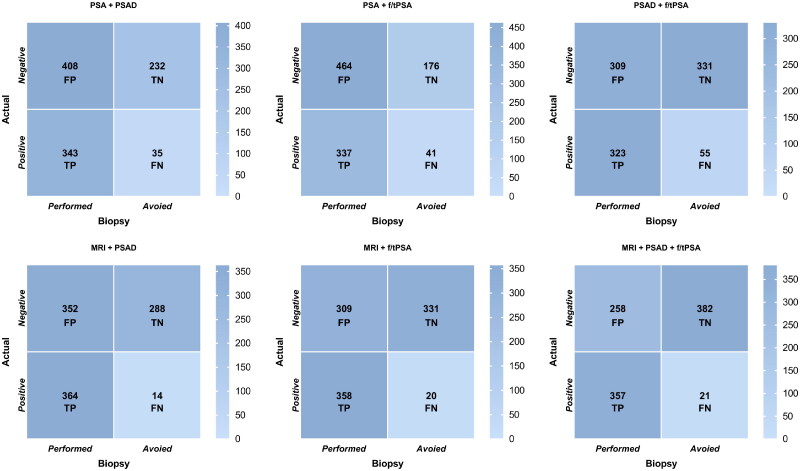
Table 5 and Figure 4 summarizes the biopsy performance, ciPCa and csPCa detection of the six pathways.
Table 5.
The outcomes of the six decision-making pathways.
| Pathway | Biopsies (N = 1018) |
ciPCa (N = 91) |
csPCa (N = 378) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Performed % (n/N) |
Avoied % (n/N) |
P | Detected % (n/N) |
Avoied % (n/N) |
P | Detected % (n/N) |
Missed % (n/N) |
P | |
| PSA + PSAD | 74% (751/1018) |
26% (267/1018) |
– | 64% (58/91) |
36% (33/91) |
– | 91% (343/378) |
9% (35//378) |
– |
| PSA + f/tPSA | 79% (801/1018) |
21% (217/1018) |
<0.001 | 72% (66/91) |
28% (25/91) |
0.008 | 89% (337/378) |
11% (41/378) |
0.03 |
| PSAD + f/tPSA | 62% (632/1018) |
38% (386/1018) |
<0.001 | 49% (45/91) |
51% (46/91) |
<0.001 | 85% (323/378) |
15% (55/378) |
<0.001 |
| MRI + PSAD | 70% (716/1018) |
30% (302/1018) |
<0.001 | 79% (72/91) |
21% (19/91) |
<0.001 | 96% (364/378) |
4% (14/378) |
<0.001 |
| MRI+ f/tPSA | 66% (667/1018) |
34% (351/1018) |
<0.001 | 79% (72/91) |
21% (19/91) |
<0.001 | 95% (358/378) |
5% (20/378) |
<0.001 |
| MRI + PSAD + f/tPSA | 60% (615/1018) |
40% (403/1018) |
<0.001 | 75% (68/91) |
25% (23/91) |
0.002 | 94% (357/378) |
6% (21/378) |
<0.001 |
ciPCa: clinically insignificant prostate cancer; csPCa: clinically significant prostate cancer; PSA: Prostate-specific antigen; PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen.
The McNemar test was employed to compare the rates.
Figure 4.
The confusion matrix evaluates the performance of the six decision pathways in avoiding biopsies, detecting csPCa, and controlling ‘negative’ biopsies. TP represents the number of detected csPCa, FP represents the number of ‘negative’ biopsies, FN represents the number of missed csPCa, and TN represents the number of avoidable biopsies that were confirmed as negative.
csPCa: Clinically significant prostate cancer; TP: True Positive; FP: False Positive; FN: False Negative; TN: True Negative; PSA: Prostate-specific antigen; PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen
‘PSA + PSAD’
This method serves as a widely accepted biopsy reference standard and was utilized as the control group with alternative methods. In this pathway, 26% (267/1018) of biopsies and 36% (33/91) of ciPCa were avoided, with a detection rate of 91% (343/378) for csPCa and 40% (408/1018) of patients received ‘negative’ biopsies.
‘PSA + f/tPSA’
Following this pathway, 21% (217/1018) of biopsies and 28% (25/91) of ciPCa were avoided, with a detection rate of 89% (337/378) for csPCa and 46% (464/1018) of patients received ‘negative’ biopsies.
‘PSAD + f/tPSA’
Following this pathway, 38% (386/1018) of biopsies and 51% (46/91) of ciPCa were avoided, with a detection rate of 85% (323/378) for csPCa and 30% (309/1018) of patients received ‘negative’ biopsies.
‘MRI + PSAD’
Following this pathway, 30% (302/1018) of biopsies and 21% (19/91) of ciPCa were avoided, with a detection rate of 96% (364/378) for csPCa and 35% (352/1018) of patients received ‘negative’ biopsies.
‘MRI + f/tPSA’
Following this pathway, 34% (351/1018) of biopsies and 21% (19/91) of ciPCa were avoided, with a detection rate of 95% (358/378) for csPCa and 30% (309/1018) of patients received ‘negative’ biopsies.
‘MRI + PSAD + f/tPSA’
Following this pathway, 40% (403/1018) of biopsies and 25% (23/91) of ciPCa were avoided, with a detection rate of 94% (357/378) for csPCa and 25% (258/1018) of patients received ‘negative’ biopsies.
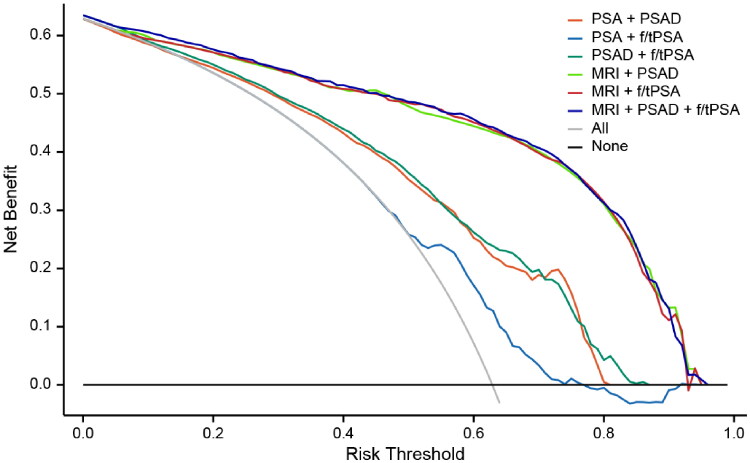
Figure 5 shows the changes in net benefits of the six pathways with the threshold probability. Using f/tPSA (‘PSA + f/tPSA’) alone exhibited the least favourable performance, consistently yielding lower net benefits compared to other pathways across all probability thresholds. The three pathways combined with MRI had significantly higher net benefits than ‘PSA + f/tPSA’ and ‘PSAD + f/tPSA’ in the 4%−92% probability threshold range and those of ‘MRI + PSAD’, ‘MRI + f/tPSA’, and ‘MRI + PSAD + f/tPSA’ were generally comparable.
Figure 5.
The decision curve analysis shows that using f/tPSA alone has the lowest net benefit across all probability threshold ranges. The combined pathways with MRI have higher net benefits than ‘PSA + PSAD’ and ‘PSAD + f/tPSA’ in the probability threshold range of 4%–92%.
PSAD: Prostate-specific antigen density; f/tPSA: ratio of free to total prostate-specific antigen
Discussion
Balancing ‘negative’ biopsies and accurately detecting moderate and high-risk cancers has long been a challenge, particularly when MRI results are inconclusive, which has motivated researchers to explore methods for effectively stratifying patients suspected of having PCa, ensuring that decisions made are in the best interest of the patient [22]. In this study, we replicated Schoots et al.’s biopsy decision table, while also incorporating PI-RADS categories with various f/tPSA intervals to divided patients into distinct risk subgroups. Consistent with their findings, our results also indicated that PSAD at high risk and at least intermediate-low risk have the ability to enhance risk stratification for patients with PI-RADS 1–2 and PI-RADS 3, respectively, warranting consideration for biopsy due to a greater than 10% observed probability of csPCa within these subgroups. Additionally, f/tPSA also demonstrated similar performance in patient stratification. When reported as PI-RADS 1 - 2, f/tPSA ≤ 0.10 may provide valuable guidance for patients considering a biopsy; whereas both PI-RADS 3 and f/tPSA ≥ 0.25 were present, the likelihood of csPCa was very low (6.7%), allowing patients to safely avoid biopsy.
This study also translated the developed risk stratification matrix into clinical practice and compared the diagnostic outcomes of six decision pathways for csPCa. We observed that clinical pathways without incorporating MRI performed suboptimally, with relatively low csPCa detection rates, particularly when relying solely on tPSA and f/tPSA. Although the three clinical pathways avoided the detection of a small amount of ciPCa, the omission of csPCa and high proportion for “negative” biopsies may be more detrimental. The current consensus recognizes MRI as the optimal imaging modality for diagnosing and managing PCa [23]. However, the diagnostic effectiveness of MRI and the potential for reducing biopsies rely heavily on the precise definition of a positive MRI threshold [24]. For instance, setting the threshold at PI-RADS 3 can avoid approximately 30% of biopsies while overlooking around 11% of csPCa. However, raising the threshold to PI-RADS 4 can achieve a biopsy avoidance rate of about 59%, but comes with missing nearly 28% of csPCa [25–27]. Hence, relying solely on PI-RADS categories for biopsy predictions exhibits certain limitations, necessitating the integration of diagnostic tools from different modalities to establish a comprehensive and multi-layered conditional diagnostic pathway.
In this study, the three combinations of MRI and PSA-derived indicators resulted in higher csPCa detection rates compared to the other three clinical pathways, while reduced the need for biopsies. On the DCA chart, the net benefit curves of the three MRI-based pathways consistently surpass those of ‘PSA + PSAD’, ‘PSA + f/tPSA’ and ‘PSAD + f/tPSA’, demonstrating a distinct trend of differentiation. This further validates that combining MRI and PSAD (or f/tPSA) can enhance patients’ benefits. However, the DCA reveals that the overall net yield of ‘MRI + PSAD’, ‘MRI + f/tPSA’ and ‘MRI + PSAD + f/tPSA’ is comparable, albeit with subtle distinctions. Among these combinations, ‘MRI + PSAD’, ‘MRI + f/tPSA’ and ‘MRI + PSAD + f/tPSA’ exhibit nearly identical csPCa detection rates (respectively 96%, 95% and 94%), but ‘MRI + PSAD + f/tPSA’ proved to be more effective in avoiding biopsy, reducing the detection rate of ciPCa, and controlling ‘negative’ biopsy.
Determining the critical value of PSAD necessitates integration with the accuracy of prostate MRI findings and consideration of individual patient factors. A fixed PSAD threshold (e.g. > 0.15 ng/ml/cm3) may not be compatible with all MRI results [28]. The cut-off value of PSAD was not specifically defined in this study. Instead, PSAD was categorized into four intervals and combined with PI-RADS categories to form multiple risk subgroups. The use of different PSAD intervals for patients with PI-RADS 1–2 and PI-RADS 3 can effectively distinguish between those who do not require biopsy, should consider biopsy, and should highly consider biopsy. Compared to a single threshold value, this combination method provides a more diversified dimension for stratification. In addition, the risk-adapted combination is highly flexible and adaptable, allowing for adjustments in the biopsy threshold based on patient preferences and clinician discretion. For instance, considering the reluctance of urologists to perform biopsies on an additional 10 patients just to identify a case of csPCa, Vickers et al. proposed a reasonable threshold probability of 10% [29].
Yim et al. [30]. conducted a follow-up study involving 6727 patients to assess the correlation between f/tPSA and csPCa, found that individuals with a baseline PSA level of 2 ng/mL and an f/tPSA value below 0.10 exhibited a csPCa-free survival rate of 75% after two decades, while those with an f/tPSA range between 0.11 and 0.25 demonstrated an 89% rate, and individuals with an f/tPSA exceeding 0.25 displayed a remarkable rate of 97%. These results highlight the potential improvement in risk stratification capabilities when utilizing a threshold of 0.10 for f/tPSA. The domestic guidelines recommend a reference basis of f/tPSA <0.16 for considering biopsy in patients with total PSA between 4 and 10 ng/ml [17]. Building upon this, the present study categorize f/tPSA into four intervals with ≤0.10, 0.10 − 0.16, 0.16 − 0.25, and ≥0.25.
Our findings revealed a negative correlation between f/tPSA and csPCa risk, with similar stratification effect on PI-RADS 3 as PSAD. Among the six pathways, ‘PSA + f/tPSA’ showed lower dcsPCa detection rates and biopsy avoidance compared to ‘PSA + PSAD’, indicating that f/tPSA alone is not suitable as a primary biopsy criterion. However, ‘PSAD + f/tPSA’ significantly improved biopsy avoidance and reduced the ciPCa detection rate while minimizing missed csPCa; moreover, ‘MRI + PSAD + f/tPSA’ demonstrated the highest number of avoided biopsies while maintaining a high detection rate for csPCa with the lowest proportion of ‘negative’ biopsies among all pathways. These findings suggested that f/tPSA can serve as a secondary indicator providing additional stratification value for patients with low or intermediate–low PSAD and PI-RADS 1–2 or 3 combined with intermediate-low to high PSAD. Additionally, Yim et al. also indicated that f/tPSA holds universal applicability across different ethnic groups; thus suggesting its potential not only for guiding screening but also for offering new options in MRI-based biopsy decision-making pathways. It is worth noting that MRI provides PSAD ‘for free’, whereas f/tPSA involves extra biochemical analysis, which may increase the economic cost.
Our research has several limitations and requires further improvement. Firstly, it is important to acknowledge that this study was conducted retrospectively, without separate analysis of the included multi-centre samples. Secondly, our PI-RADS category was obtained from radiologists with specialized background, the impact of experience on decision-making pathways was not taken into account and needs subsequent application in clinical practice for prospective verification. Thirdly, previous studies have demonstrated that incorporating f/tPSA and PSAD to form a new parameter (f/tPSA/PSAD) is more effective in diagnosing PCa among Italian and Chinese populations compared to using f/tPSA or PSAD alone [31], it will be necessary for further verification. Lastly, although DRE results are crucial considerations for biopsies, they were not included in this study due to lack of DRE administration among most patients.
Conclusions
In summary, our study emphasizes the value of integrating MRI with PSAD or f/tPSA in determining the necessity for biopsy. Additionally, f/tPSA may help optimize the risk stratification within the ‘MRI + PSAD’ pathway and provide personalized biopsy strategies for patients.
Funding Statement
This research was supported by Natural Science Foundation of Zhejiang Province under Grant No.LGF22H220006, Medical Health Science and Technology Project of Zhejiang Province under Grant 2022KY100.
Author contributions
PF Jin: Conceptualization, Methodology, Writing - Original Draft, Visualization XM Wang and ZW Ding: Data collection, Investigation LQ Yang and X Wang: Interpretation, Software, Investigation CY Xu: Data curation, Validation FW Huang: Interpretation, Writing - Review & Editing
All authors have read and approved the final manuscript.
Consent to publication
Authors unanimously concur on the publication.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of data and material
The imaging and clinical data used in this study are not publicly available, because they contain private patient health information. Further enquiries can be directed to the corresponding author.
References
- 1.Bray F, Laversanne M, Sung H, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–263. doi: 10.3322/caac.21834. [DOI] [PubMed] [Google Scholar]
- 2.Nordström T, Adolfsson J, Grönberg H, et al. Repeat prostate-specific antigen tests before prostate biopsy decisions. J Natl Cancer Inst. 2016;108(12):djw165. doi: 10.1093/jnci/djw165. [DOI] [PubMed] [Google Scholar]
- 3.Hugosson J, Månsson M, Wallström J, et al. Prostate cancer screening with PSA and MRI followed by targeted biopsy only. N Engl J Med. 2022;387(23):2126–2137. doi: 10.1056/NEJMoa2209454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stabile A, Giganti F, Rosenkrantz AB, et al. Multiparametric MRI for prostate cancer diagnosis: current status and future directions. Nat Rev Urol. 2020;17(1):41–61. doi: 10.1038/s41585-019-0212-4. [DOI] [PubMed] [Google Scholar]
- 5.Mazzone E, Stabile A, Pellegrino F, et al. Positive predictive value of prostate imaging reporting and data system version 2 for the detection of clinically significant prostate cancer: a systematic review and meta-analysis. Eur Urol Oncol. 2021;4(5):697–713. doi: 10.1016/j.euo.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 6.van Luijtelaar A, Bomers J, Fütterer J.. A comparison of magnetic resonance imaging techniques used to secure biopsies in prostate cancer patients. Expert Rev Anticancer Ther. 2019;19(8):705–716. doi: 10.1080/14737140.2019.1641086. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Kozarek J, Russell R, et al. Diagnostic performance of prostate-specific antigen density for detecting clinically significant prostate cancer in the era of magnetic resonance imaging: a systematic review and meta-analysis. Eur Urol Oncol. 2024;7(2):189–203. doi: 10.1016/j.euo.2023.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Kortenbach KC, Løgager V, Thomsen HS, et al. Comparison of PSA density and lesion volume strategies for selecting men with equivocal PI-RADS 3 lesions on bpMRI for biopsies. Abdom Radiol (NY). 2023;48(2):688–693. doi: 10.1007/s00261-022-03720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deniffel D, Healy GM, Dong X, et al. Avoiding unnecessary biopsy: MRI-based risk models versus a PI-RADS and PSA density strategy for clinically significant prostate cancer. Radiology. 2021;300(2):369–379. doi: 10.1148/radiol.2021204112. [DOI] [PubMed] [Google Scholar]
- 10.Drost FH, Osses DF, Nieboer D, et al. Prostate MRI, with or without MRI-targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst Rev. 2019; Apr 254(4):CD012663. doi: 10.1002/14651858.CD012663.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grönberg H, Eklund M, Picker W, et al. Prostate cancer diagnostics using a combination of the Stockholm3 blood test and multiparametric magnetic resonance imaging. Eur Urol. 2018;74(6):722–728. doi: 10.1016/j.eururo.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Boesen L, Nørgaard N, Løgager V, et al. Prebiopsy biparametric magnetic resonance imaging combined with prostate-specific antigen density in detecting and ruling out Gleason 7-10 prostate cancer in biopsy-naïve men. Eur Urol Oncol. 2019;2(3):311–319. doi: 10.1016/j.euo.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Falagario UG, Martini A, Wajswol E, et al. Avoiding unnecessary magnetic resonance imaging (MRI) and biopsies: negative and positive predictive value of MRI according to prostate-specific antigen density, 4Kscore and risk calculators. Eur Urol Oncol. 2020;3(5):700–704. doi: 10.1016/j.euo.2019.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Cornford P, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG Guidelines on Prostate Cancer-2024 Update. Part I: screening, Diagnosis, and Local Treatment with Curative Intent. Eur Urol. 2024;86(2):148–163. doi: 10.1016/j.eururo.2024.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Schoots IG, Padhani AR.. Risk-adapted biopsy decision based on prostate magnetic resonance imaging and prostate-specific antigen density for enhanced biopsy avoidance in first prostate cancer diagnostic evaluation. BJU Int. 2021;127(2):175–178. doi: 10.1111/bju.15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferraro S, Caruso S, Panteghini M.. Reflex testing of free prostate-specific antigen as effective health care policy. Arch Pathol Lab Med. 2019;143(9):1045–1045. doi: 10.5858/arpa.2019-0117-LE. [DOI] [PubMed] [Google Scholar]
- 17.Gao XD, Miao Q, Zhang JL, et al. Clinical application of free/total PSA ratio in the diagnosis of prostate cancer in men over 50 years of age with total PSA levels of 2.0-25.0 ng ml-1 in Western China. Asian J Androl. 2022;24(2):195–200. doi: 10.4103/aja202182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or = 4.0 ng per milliliter. N Engl J Med. 2004;350(22):2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 19.Ferraro S, Biganzoli D, Rossi RS, et al. Individual risk prediction of high grade prostate cancer based on the combination between total prostate-specific antigen (PSA) and free to total PSA ratio. Clin Chem Lab Med. 2023;61(7):1327–1334. doi: 10.1515/cclm-2023-0008. [DOI] [PubMed] [Google Scholar]
- 20.Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur Urol. 2019;76(3):340–351. doi: 10.1016/j.eururo.2019.02.033. [DOI] [PubMed] [Google Scholar]
- 21.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 22.Hrubá T, Kubas V, Franko M, et al. Precision in prostate cancer detection: integrating prostate-specific antigen density (PSAD) and the Prostate Imaging Reporting and Data System (PI-RADS) to provide additional risk stratification for a more accurate diagnostic decision. Ir J Med Sci. 2024;193(6):2635–2642. doi: 10.1007/s11845-024-03771-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z, Wang K, Kong Z, et al. A multicenter study of artificial intelligence-aided software for detecting visible clinically significant prostate cancer on mpMRI. Insights Imaging. 2023;14(1):72. doi: 10.1186/s13244-023-01421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hectors SJ, Chen C, Chen J, et al. Magnetic resonance imaging radiomics-based machine learning prediction of clinically significant prostate cancer in equivocal PI-RADS 3 lesions. J Magn Reson Imaging. 2021;54(5):1466–1473. doi: 10.1002/jmri.27692. [DOI] [PubMed] [Google Scholar]
- 25.Lim CS, Abreu-Gomez J, Carrion I, et al. Prevalence of Prostate Cancer in PI-RADS Version 2.1 transition zone atypical nodules upgraded by abnormal DWI: correlation with MRI-directed TRUS-guided targeted biopsy. AJR Am J Roentgenol. 2021;216(3):683–690. doi: 10.2214/AJR.20.23932. [DOI] [PubMed] [Google Scholar]
- 26.Brancato V, Aiello M, Basso L, et al. Evaluation of a multiparametric MRI radiomic-based approach for stratification of equivocal PI-RADS 3 and upgraded PI-RADS 4 prostatic lesions. Sci Rep. 2021;11(1):643. doi: 10.1038/s41598-020-80749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang AM, Shumaker LA, Martin KD, et al. Multi-institutional analysis of clinical and imaging risk factors for detecting clinically significant prostate cancer in men with PI-RADS 3 lesions. Cancer. 2022;128(18):3287–3296. doi: 10.1002/cncr.34355. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrino F, Tin AL, Martini A, et al. Prostate-specific antigen density cutoff of 0.15 ng/ml/cc to propose prostate biopsies to patients with negative magnetic resonance imaging: efficient threshold or legacy of the past? Eur Urol Focus. 2023;9(2):291–297. doi: 10.1016/j.euf.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vickers AJ, Van Calster B, Steyerberg EW.. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ. 2016;352:i6. doi: 10.1136/bmj.i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim K, Ma C, Carlsson S, et al. Free PSA and clinically significant and fatal prostate cancer in the PLCO Screening Trial. J Urol. 2023;210(4):630–638. doi: 10.1097/JU.0000000000003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nan LB, Yin XT, Gao JP.. Significant diagnostic value of free-serum PSA (FPSA)/prostate-specific antigen density (PSAD) and (F/T)/PSAD for Prostate Cancer of the Chinese Population in a Single Institution. Med Sci Monit. 2019;25:8345–8351. doi: 10.12659/MSM.916900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The imaging and clinical data used in this study are not publicly available, because they contain private patient health information. Further enquiries can be directed to the corresponding author.