Abstract
Lycium barbarum var. implicatum (2010), is a new variety of L. barbarum in Solanaceae. Here, we sequenced, assembled, and annotated the complete chloroplast (cp) genome of L. barbarum var. implicatum. Results showed that the complete cp genome of L. barbarum var. implicatum was 154,888 bp in length, containing a large single copy (LSC) region of 85,894 bp, a pair of inverted repeats (IR) region of 25,393 bp, and a small single copy (SSC) region of 18,208 bp. Maximum-likelihood (ML) phylogenetic tree elucidated that L. barbarum var. implicatum was sister to L. ruthenicum. Our results provide useful information for future phylogenetic studies in the family of Solanaceae.
Keywords: Complete chloroplast genome, Lycium barbarum var. implicatum, phylogenetic analysis, Solanaceae
1. Introduction
Lycium barbarum var. implicatum T. Y. Chen & Xu L. Jiang in 2010, is a new variety of L. barbarum in Solanaceae, which was found in the Qingshui River tributary of the Yellow River Basin in Ningxia, and the whole plant is characterized by many dense branches, narrow leaves, 2-fid calyx, ovoid or elliptic berries appear purple or white transparent, juicy and sweet (Chen et al. 2012). This species can not only grow well in saline-alkali land and arid land, but also its fruit contains more nutrients, such as amino acids, flavonoids, polyphenols, pigments, polysaccharides, potassium, and trace elements than Lycium barbarum (Zhu et al. 2016). In particular, the higher crude protein content (Liu et al. 2016) can be used in the development of fermented feed and the wolfberry industry. However, studies on the phylogeny of L. barbarum var. implicatum remain limited, and there is little submission for nucleic-acid sequences in NCBI Genbank.
2. Materials and methods
Fresh leaves of L. barbarum var. implicatum were collected from Qingshui River Basin, a branch of the Yellow River, Ningxia Hui Autonomous Region, China (38°57′53.28 N, 106°25′21.32 E, alt. 1069 m), and dried with silica gel. The specimen (No. 2023LBVI001LY) was deposited in the Herbarium of Plant and Plant Physiology Laboratory, Ningxia Technical College of Wine and Desertification Prevention (http://www.nxfszs.cn/, Wangsuo Liu, email: liuwangsuo@sina.com) (Figure 1). Genomic DNAs were extracted by a modified CTAB method (Stefanova et al. 2013). The genome sequencing was performed by Illumina Hiseq 2500 at Biomarker Technologies Corporation. A total of 31,284,518 paired-end raw reads were obtained, and each of which comprised 50-151bp. The sequencing coverage achieved a remarkable 99.9%, effectively capturing the comprehensive information of the L. barbarum var. implicatum chloroplast (cp) genome. By removing low-quality reads, the trimmed reads, which comprised 25,010,894 bp in read bases, were assembled via the program SPAdes 3.15 (Prjibelski et al. 2020), with that of Lycium barbarum (GenBank: MH032560) as the initial reference. The CPGView program (http://www.1kmpg.cn/cpgview/) was conducted to generate the cp genome maps (Liu et al. 2023). A comparison of the junction of LSC, SSC, and IR regions was performed using R (version 4.2.0) to activate the IRscope website (https://irscope.shinyapps.io/irapp/) (Amiryousefi et al. 2018). The sequence of L. barbarum var. implicatum complete cp genome has been submitted to GenBank (accession number OR551485). 18 of the most relevant Lycium cp genomes in Solanaceae were downloaded from the NCBI database, and aligned with four outgroups using MAFFT-7.037 (Katoh and Standley 2013). We constructed a molecular phylogenetic tree based on the already-aligned genomes, a maximum likelihood tree was generated by MEGA-X with the nucleotide model and 1000 bootstrap replicates (Kumar et al. 2018).
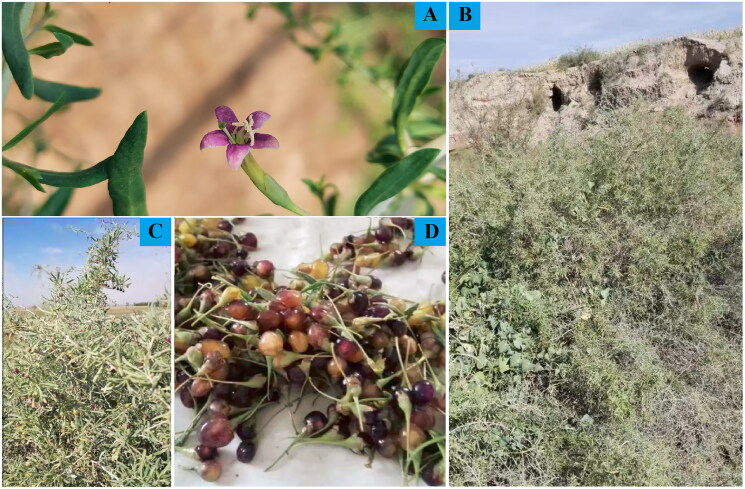
Figure 1.
Morphological characteristics of L. barbarum var. implicatum (A: flower, bifid calyx; B: habitats, extremely arid and water-scarce; C: branches with thorns; D: berries, purple or white transparent). Photographs of L. barbarum var. implicatum were taken by Wangsuo Liu in the Qingshui River Basin (38°57′53.28 N, 106°25′21.32 E).
3. Results
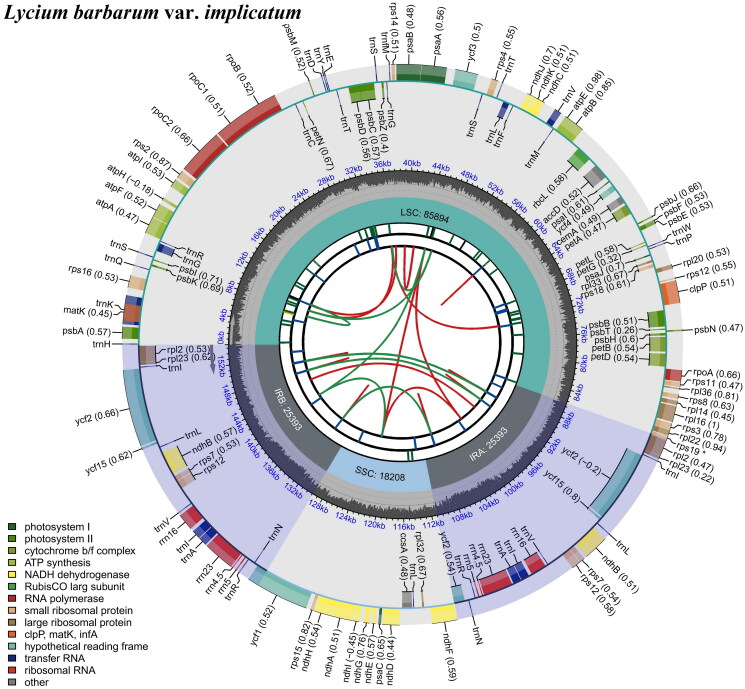
The L. barbarum var. implicatum chloroplast genome is 154,888 bp in length, including a pair of inverted repeats (IRa and IRb) region of 25,393 bp, a large single copy (LSC) region of 85,894 bp, and a small single copy (SSC) region of 18,208 bp (Figure 2). Additionally, the GC content of the L. barbarum var. implicatum cp genome accounted for 37.9%, and the GC content in IR (43.2%) regions was higher than that of LSC (36%) and SSC (32.3%) regions. The results showed that the maximum and average read mapping depths of the assembled genomes were ×8000 and ×4505 (Figure S1), while the structures of cis- and trans-splicing genes are illustrated in Figure S2. As shown in Figure 2, the cp genome of L. barbarum var. implicatum contains 132 genes, including 87 protein-coding genes, 37 tRNA genes, and eight rRNA genes. In the IR region, there were 19 duplicated genes identified including 8 protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf1, ycf15, ycf2), 4 rRNA genes (rrn16S, rrn23S, rrn4.5S, rrn5S), and 7 tRNA genes (trnI-CAU, trnA-UGC, trnL-CAA, trnI-GAU, trnN-GUU, trnV-GAC, trnR-ACG). There were also identified 15 genes with one intron (ndhB, ndhA, petD, petB, atpF, rpl2, rpl16, rps16, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) and 3 genes with two introns (rps12, clpP, ycf3).
Figure 2.
Genomic map of overall features of the cp genome of L. barbarum var. implicatum was visualized using CPGview. This map consists of six tracks arranged from the center outward. The first track represents dispersed repeats, while the red arcs depict direct repeats and the green areas represent palindromic repeats. In the second track, a long tandem repeat is indicated by short blue bars. The third track displays short tandem repeats or microsatellite sequences as colored bars. The fourth track illustrates the regions of small single-copy (SSC), inverted repeat (IRs), and large single-copy (LSC). Additionally, the fifth track showcases the GC content of the cp genome, while in the last track coding genes are categorized based on their functions. Notably, transcription directions for inner and outer genes are depicted as clockwise and anticlockwise, respectively.
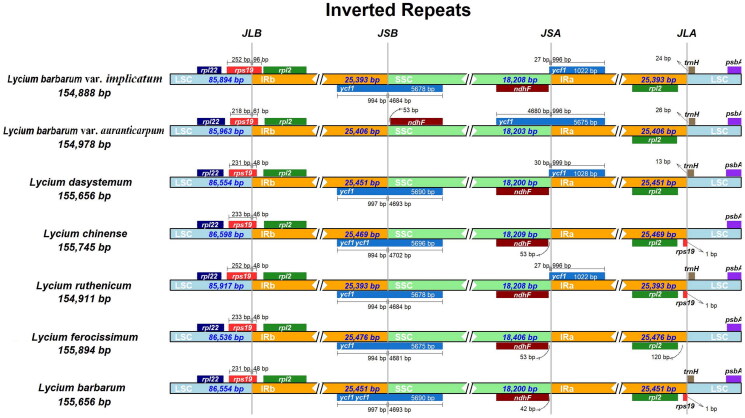
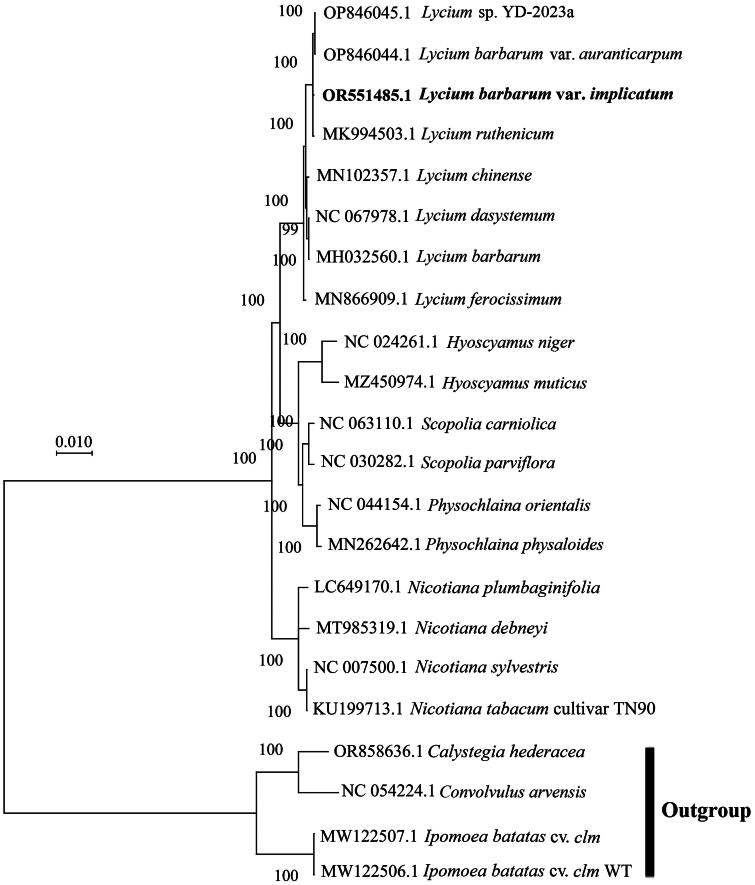
As shown in Figure 3, the IR-LSC and IR-SSC boundaries of L. barbarum var. implicatum were similar to the related species, such as L. dasystemum (NC067978), L. chinense (MN102357), L. ruthenicum (MK994530), L. ferocissimum (MN866909), and L. barbarum (MH032560) except for L. barbarum var. auranticarpum (OP846044). By comprehensive comparison, the boundary of L. barbarum var. implicatum was similar to that of L. ruthenicum, but it is different from that of L. barbarum var. auranticarpum with the same variety. The phylogenetic analysis was performed based on the 22 complete cp genomes (including 4 outgroups) by maximum likelihood (ML), it suggested that L. barbarum var. implicatum was sister to L. ruthenicum with strong support (Figure 4).
Figure 3.
Comparison of borders of LSC, IR, and SSC regions in the complete cp genomes of L. barbarum var. implicatum (OR551485), L. barbarum var. auranticarpum (OP846044), L. dasystemum (NC067978), L. chinense (MN102357) (He et al. 2020), L. ruthenicum (MK994503) (Wang et al. 2019), L. ferocissimum (MN866909) (Li et al. 2020), and L. barbarum (MH032560) (Jia et al. 2018). The junction line between LSC and IRb is abbreviated as JLB; the junction line between SSC and IRb is abbreviated as JSB; the junction line between SSC and IRa is abbreviated as JSA; the junction line between LSC and IRa is abbreviated as JLA.
Figure 4.
Maximum-likelihood (ML) tree generated with 17 complete cp genomes related to L. barbarum var. implicatum and four outgroups. Bootstrap values are shown near the nodes, and the bold font represents the cp genome of L. barbarum var. implicatum in this study. The following sequences, some of which are stored in the NCBI database and most of which remain unpublished, were used: L. dasystemum (NC 067978), Hyoscyamus niger (NC 024261), Scopolia carniolica (NC 063110), S. parviflora (NC 030282), Physochlaina orientalis (NC 044154), Nicotiana sylvestris (NC 007500), Convolvulus arvensis (NC 054224) (Wang et al. 2021), N.tabacum (KU 199713) (Gao et al. 2016), N. plumbaginifolia (LC 649170) (Tong et al. 2022), L. barbarum (MH032560) (Jia et al. 2018), L. ruthenicum (MK994503) (Wang et al. 2019), L. chinense (MN102357) (He et al. 2020), L. ferocissimum (MN866909) (Li et al. 2020), P. physaloides (MN 262642) (Tong et al. 2019), N.debneyi (MT985319) (Zeng et al. 2021), Ipomoea batatas (MW122506, MW122507) (Zou et al. 2021), H. muticus (MZ 450974), Lycium sp. (OP846045), L. barbarum var. auranticarpum (OP846044) and Calystegia hederacea (or 858636).
4. Discussion and conclusions
Our study was the first to report the complete cp genomes of L. barbarum var. implicatum, which is a typical circular form with 154,888 bp in length. A total of 131 genes were found, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, and the GC content accounted for 37.9%. L. barbarum var. implicatum is a new species of Lycium found in Qingshui River Basin, Zhongning County, Ningxia, China (Chen et al. 2012). Early taxonomists identified this species as a variety of Lycium barbarum (Chen et al. 2012), but according to the results of our study, we speculate that it is more closely related to the phylogenetic evolution of L. ruthenicum, and may be its mutant hybrid offspring. In addition, IRs are a conserved region of the cp genome for the plant, and the contraction and expansion of its boundaries often reflect the evolutionary relationship of cp genomes among species (Amiryousefi et al. 2018; Konhar et al. 2019), our study also confirmed the close relationship between L. barbarum var. implicatum and L. ruthenicum. However, according to the unique characteristics of rich flesh and high crude protein content of L. barbarum var. implicatum (Liu et al. 2016), it is in sharp contrast to the characteristics of proanthocyanidins and no flesh of L. ruthenicum (Li et al. 2022). Although Miller (2002) performed a comprehensive systematic phylogenetic analysis of 25 species of Lycium, this species was not included. Therefore, it is necessary to conduct a comprehensive analysis of all Lycium species in the future. The result provides a phylogenetic relationship of the genus Lycium with the other related species, which would provide useful information for the study of plant evolution in Solanaceae and the further study of DNA barcoding.
Supplementary Material
Funding Statement
This study was supported by the Project for High-Level Talent Introduction Research Startup Funding of Hetao College of China [HYRC202401], the Ningxia Higher Education Science Research Project [NYG2022235], National Natural Science Foundation of China [32360327], the Ningxia Natural Science Foundation [2022AAC03641] and the Project for Ecological Environment Innovation Team Building of Hetao College of China.
Ethical approval
The collection and research process of L. barbarum var. implicatum was carried out in strict accordance with the guidelines provided by the herbarium of Ningxia Technical College of Wine and Desertification Prevention and Chinese regulations.
Authors’ contributions
WL and DB planned and designed the research. BZ and ZJ sampled the plant materials, DB and WL analyzed the data. BZ and WL wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the NCBI database at https://www.ncbi.nlm.nih.gov/, reference number OR551485. The associated BioProject, Biosample, and SRA numbers are PRJNA1016474, SAMN37386561, and SRR26058098, respectively.
References
- Amiryousefi A, Hyvönen J, Poczai P.. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34(17):3030–3031. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- Chen T, Jiang X, Zhang Z, Li Q, Lee J.. 2012. A new species and a new variety of Lycium (Solanaceae) from Ningxia, China (in Chinese). Guihaia. 32(1):5–8. [Google Scholar]
- Gao H, Zhang Y-J, Zhao Y-C, Qiu C-G, Liu J-H, Yang J-J, Hua Y-K, Yang S-H, Liu J, Liu Z, et al. 2016. Complete chloroplast genome sequence of Nicotiana tabacum TN90 (Solanaceae). Mitochondrial DNA Part B Resour. 1(1):867–868. doi: 10.1080/23802359.2015.1137808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Tian Y, Yang Y, Shi C.. 2020. Phylogenetic analyses of Lycium chinense (Solanaceae) and its coordinal species applying chloroplast genome. Mitochondrial DNA Part B Resour. 5(3):2201–2202. doi: 10.1080/23802359.2019.1669087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Xin G, Ren X, Du X, Liu H, Hao N, Wang C, Ni X, Liu W.,. 2018. Characterization of the complete chloroplast genome of Lycium barbarum (Solanales: Solanaceae), a unique economic plant to China. Mitochondrial DNA Part B Resour. 3(2):1062–1063. doi: 10.1080/23802359.2018.1509930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley D.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konhar R, Debnath M, Vishwakarma S, Bhattacharjee A, Sundar D, Tandon P, Dash D, Biswal DK.,. 2019. The complete chloroplast genome of Dendrobium nobile, an endangered medicinal orchid from north-east India and its comparison with related Dendrobium species. PeerJ. 7:e7756. doi: 10.7717/peerj.7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao D, Akram MA, Guo C, Jin H, Hu W, Zhang Y, Wang X, Ma A, Xiong J, et al. 2022. Effects of environmental factors on anthocyanin accumulation in the fruits of Lycium ruthenicum Murray across different desert grasslands. J Plant Physiol. 279:153828. doi: 10.1016/j.jplph.2022.153828. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang X, Zhang Q, Yisilam G.. 2020. Complete chloroplast genome of Lycium ferocissimum (Solanaceae), a species native to South Africa. Mitochondrial DNA Part B Resour. 5(1):756–757. doi: 10.1080/23802359.2020.1715301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C.,. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729. [DOI] [PubMed] [Google Scholar]
- Liu W, Rui X, Guo Y, Lu J, Wang Y.. 2016. Nutritional components analysis on two new species of wild Lycium barbarum (in Chinese). JGreen Sci Technol. 15:55–56. [Google Scholar]
- Miller JS. 2002. Phylogenetic relationships and the evolution of gender dimorphism in Lycium (Solanaceae). Syst Botany. 27(2):416–428. [Google Scholar]
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A.. 2020. Using SPAdes De Novo assembler. Curr Protoc Bioinformatics. 70(1):e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- Stefanova P, Taseva M, Georgieva T, Gotcheva V, Angelov A.. 2013. A modified CTAB method for DNA extraction from soybean and meat products. Biotechnol Biotec Eq. 27(3):3803–3810. doi: 10.5504/BBEQ.2013.0026. [DOI] [Google Scholar]
- Tong L, Zhu Y, Lei F, Shen X, Mu X.. 2019. The complete chloroplast genome of Physochlaina physaloides (Solanaceae), an important medicinal plant. Mitochondrial DNA Part B Resour. 4(2):3427–3428. doi: 10.1080/23802359.2019.1674730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Du T, Hu Y, Chen H, Shen E, Fan L, Xiao B., 2022. The complete chloroplast genome of Nicotiana plumbaginifolia. Mitochondrial DNA Part B Resour. 7(1):239–240. doi: 10.1080/23802359.2021.2024772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Jia G, Zhu L, Tian F.. 2019. The complete chloroplast genome sequence of Lycium ruthenicum (Solanaceae), a traditional medicinal plant in China. Mitochondrial DNA Part B Resour. 4(2):2495–2496. doi: 10.1080/23802359.2019.1638324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song H, Jiang D.. 2021. Complete chloroplast genome sequence of Convolvulus arvensis. Mitochondrial DNA Part B Resour. 6(7):1814–1815. doi: 10.1080/23802359.2021.1915202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Huang C, Liu Y, Yuan C, Yu H, Song B.. 2021. The complete chloroplast genome sequence of Nicotiana debneyi (Solanaceae). Mitochondrial DNA Part B Resour. 6(3):1042–1043. doi: 10.1080/23802359.2021.1899074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, He H, Liu W, Ma H.. 2016. Nutrient components and active substances of Lycium barbarum var. implicatum leaves (in Chinese). Northern Horticulture. 23:149–152. [Google Scholar]
- Zou H, Zhang X, Chen J, Wang Z, Huang L, Fang B.. 2021. Complete chloroplast genome of a novel chlorophyll-deficient mutant (clm) in sweet potato (Ipomoea batatas L.). Mitochondrial DNA Part B Resour. 6(3):968–969. doi: 10.1080/23802359.2020.1869616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in the NCBI database at https://www.ncbi.nlm.nih.gov/, reference number OR551485. The associated BioProject, Biosample, and SRA numbers are PRJNA1016474, SAMN37386561, and SRR26058098, respectively.