Abstract
We have investigated the large-scale organization of the human chAB4-related long-range multisequence family, a low copy-number repetitive DNA located in the pericentromeric heterochromatin of several human chromosomes. Analysis of genomic clones revealed large-scale (∼100 kb or more) sequence conservation in the region flanking the prototype chAB4 element. We demonstrated that this low copy-number family is connected to another long-range repeat, the NF1-related (ΨNF1) multisequence. The two DNA types are joined by an ∼2 kb-long tandem repeat of a 48-bp satellite. Although the chAB4- and NF1-like sequences were known to have essentially the same chromosomal localization, their close association is reported here for the first time. It indicates that they are not two independent long-range DNA families, but are parts of a single element spanning ∼200 kb or more. This view is consistent both with their similar chromosomal localizations and the high levels of sequence conservation among copies found on different chromosomes. We suggest that the master copy of the linked chAB4–ΨNF1 DNA segment appeared first on the ancestor of human chromosome 17.
INTRODUCTION
The initial sequencing and analysis of the human genome has been published recently (1,2). The draft genome sequence confirmed that various types of repetitive DNAs classified mainly according to their way of spreading (3) make up nearly one third of chromosomal DNA (1). Over half of the genomic DNA consists of repeated sequences of various types: 45% in four classes of parasitic DNA elements, 3% in repeats of just a few bases, and ∼5% in recent duplications of large segments of DNA (4). Heterochromatic regions including the centromeres, as well as the short arms of acrocentric chromosomes composed primarily of tandem repeats, have been excluded to avoid complications posed by these sequences in the course of contig assembly (1). Although the centromeric region of chromosomes remained largely untouched by the genome project, a number of tandem repeat satellites and complex sequences have been localized to these key functional domains (5). Long-range mapping studies of the centromeric region of chromosomes 7 (6), 13, 14 and 21 (7) and of chromosome Y (8,9) indicated large blocks of satellites joined to non-satellite sequences. Indeed, high-resolution analysis of the centromere of chromosome 10 demonstrated a complex patchwork of satellites and other sequences (10). These complex non-satellite DNA elements were detected at the centromeres of other chromosomes as well (10).
One of these complex sequences is the chAB4 primate-specific long-range multisequence family. The prototype 3.4 kb AB4 sequence was discovered in the nuclear polydisperse circular DNA fraction of an angiofibroma cell line (11). AB4-related sequences were detected in the centromeric region of nine human chromosomes by in situ hybridization, and these chromosomal copies were called chAB4 (12). Comparison of non-human primate chAB4 copies with sequences from different human chromosomes indicated a complex evolutionary pattern (13). The evolution of the family as a unit was attributed to frequent exchanges between copies without noticeable alterations in the structure of chromosomes involved (14). On chromosome 22, chAB4 was found in the inverted arms of a large palindromic structure flanked by centromeric satellite sequences (15). Detailed analysis of Robertsonian translocations revealed that chAB4 copies were in the immediate neighborhood of the centromere on chromosome 22, and perhaps on chromosome 15, while in the case of the remaining three acrocentrics related sequences were detected exclusively on their p arms on both sides of the rDNA cluster (16). Other complex multisequence families have also been described (10,17–25); however, similarly to the chAB4 family, their function, if any, remains unclear. This can be attributed at least in part to the fact that most of these sequences have been localized to the poorly characterized acrocentric short arms and to the centromeric regions of chromosomes.
We describe the large-scale structure of DNA around a chromosomal copy AB4. It is reported here for the first time that the chAB4 multisequence is contiguous with another long-range repeat, the NF1-like (ΨNF1) family, composed of unprocessed pseudogenes of the neurofibromatosis type I gene. The junction sequence is an ∼2 kb tandem array of a 48 bp repeat unit similar to D22Z3. The number of the 48 bp units is variable in the connecting region, but the flanking sequences are highly conserved among different copies. Evidence is provided that the connection of the two long-range families may be a general feature rather than a unique arrangement. We show that the core region of the chAB4 palindrome on chromosome 22 is NF1-like DNA. We propose that the chAB4- and the NF1-related sequences are two different parts of a single low copy-number repeat family. This would explain features of chromosomal distribution and evolution of the two multisequence families previously thought to be independent.
MATERIALS AND METHODS
All restriction enzymes were from New England Biolabs. YAC 857_b_10 was obtained from the CEPH MegaYAC library. YAC handling and DNA manipulation protocols were from Clontech. Human PAC library filters (RPCI1, 3, 4, 5 and 6) and the PAC clones were purchased from Roswell Park Cancer Institute, Buffalo, New York, together with BAC/PAC handling and DNA purification protocols.
Oligonucleotides and sequencing primers
Oligonucleotides were synthesized on a Pharmacia Gene Assembler. Sequence tagged site (STS) primer sequences were described for YAC 857_b_10 as in Assum et al. (14). Primers for chAB4 were CHAB4F primer, 5′-GGGAATTCACCTTTCATCCAAGTAATG-3′ (X57630: 1–26), and CHAB4R primer, 5′-AGGAATTCCCTCCAAATAAAGAGTTTTT-3′ (X57630: 3378–3344). A shorter subfragment chAB4 without the Alu repeat present in X57630 was amplified using primers CHAB4R and an internal primer CHAB4F2, 5′-TTTTGCTCTTATGTTCTATTTGTTCCA-3′ (X57630: 1353–1379). Primers for the vector were designed using the pSacBII sequence: PAC-F primer, 5′-ATATTGCTCTAATAAATTTGCGGC-3′ (U09128: 15 957–159 80), and PAC-R primer, 5′-TCCCGAATTGACTAGTGGGTAGGC-3′ (U09128: 65–42), respectively. PCR primers for amplification of the chAB4–NF1 junction were JCTF, 5′-TTTGTCTGCTTCTGAAGCTTGCTCTGT-3′ (AF401203: 72 593–72 619), and JCTR, 5′-TCGGGGCCTAGGTGGAAAAAGCTTTAA-3′ (AF401203: 76 099–76 073), respectively. Sequencing primers were 20–24mer oligonucleotides, the design based on the sequence to be extended.
STS probe amplification and long-range PCR
STS probes were amplified from human peripheral blood lymphocyte, YAC 857_b_10 and PAC miniprep DNAs using Takara ExTaq DNA polymerase. PCR conditions were: 94°C, 1 min; 35 cycles of 98°C, 20 s; 60°C, 30 s; 68°C, 3 min, followed by cooling to 15°C. Amplifications for generating hybridization probes as well as for STS mapping were done using 100 ng template for human and yeast DNA or 10 ng for the PAC clones, in the presence of 50 pmol of each primer in 75 µl volume. PCR fragments were gel purified and sequenced.
Long-range PCRs to connect and order subclones within PAC 635H14 were done on 10 ng PAC miniprep DNA as above, except that the extension step was 15 min to allow amplification of fragments up to 20 kb.
Amplification of the chAB4–NF1 junction was done under conditions used to obtain the STS probes, but the extension step was for 5 min.
PAC library screening and DNA purification
An approximately equimolar mixture of the STS probes amplified from human lymphocyte DNA was labeled with [α-32P]dCTP, and was used to screen the RPCI4 high-density PAC library filter as recommended by the supplier. Filter coordinates were read and the desired clones were purchased.
Fluorescent in situ hybridization
Fluorescent in situ hybridization (FISH) to metaphases of human peripheral blood lymphocytes were done using biotinylated or DIG-labeled probes using published procedures (26). Images were acquired through a CCD system mounted on an OLYMPUS Vanox microscope.
Subcloning, DNA sequencing and sequence analyses
The PAC 635H14 insert was digested with restriction enzymes SacI, XbaI, BamHI, EcoRI or HindIII and fragments were subcloned in pUC19 using standard techniques. Sequencing of PAC or plasmid miniprep DNA was carried out by primer walking on an ABI model 365 DNA sequencer. Sequence assembly was performed with the DNA Star PC software. GenBank BLAST searches were carried out on the NCBI internet server.
RESULTS
Characterization of chAB4 PAC clones
In order to determine the large-scale organization and the DNA sequences flanking chromosomal copies of AB4, large-insert human genomic clones were analyzed. STS markers were mapped within the chAB4 palindrome on chromosome 22 covered by CEPH YAC 857_b_10 (15). These published primers were used to amplify hybridization probes from human lymphocyte DNA. High-density human PAC library filter RPCI4 was screened with an equimolar mixture of radiolabeled STS fragments N, R2, chAB4ΔAlu, c8E2, 14E2 and 18E2. From over 16 000 clones, 88 showed hybridization to the probe mixture. Ten of the positive clones were chosen at random, PAC miniprep DNAs were purified and tested for the presence of the STS markers mapped in the CEPH YAC by PCR. Human genomic DNA and the YAC were used as positive control templates. The results of STS PCRs are summarized in Table 1. Each of the 10 PACs was found to contain three or more of the markers.
Table 1. STS markers detected in the PAC clones.
| Template | STS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| N | R2 | chAB4 ΔAlu | 1.4 | TE1 | C8E2 | 14E2 | BE1 | 18E2 | |
| HL |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| YAC 857_b_10 |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
| 635H14 | – | – | + | + | + | + | – | – | – |
| 635J14 | – | – | + | + | + | + | – | – | – |
| 654D10 | – | – | – | + | + | + | + | + | + |
| 654O08 |
– |
+ |
+ |
+ |
+ |
+ |
– |
– |
– |
| 655I18 |
– |
– |
+ |
– |
+ |
+ |
– |
– |
– |
| 659M14 |
– |
– |
– |
– |
– |
+ |
+ |
+ |
+ |
| 662K13 |
– |
– |
– |
+ |
– |
+ |
+ |
+ |
+ |
| 663D09 |
– |
– |
– |
+ |
– |
+ |
– |
+ |
+ |
| 668N10 |
– |
– |
– |
– |
– |
– |
+ |
+ |
+ |
| 670G14 | – | – | – | – | + | + | + | + | – |
Human lymphocyte (HL) and YAC 857_b_10 DNA were used as positive controls. PCR amplifications were carried out using STS primers described in Wöhr et al. (15).
PCR products amplified from YAC 857_b_10, corresponding to STS fragments N, R2, chAB4ΔAlu, c8E2, 14E2 and 18E2, were sequenced. These sequences were checked against the GenBank. The 14E2 and 18E2 sequences showed homology to NF1-related sequences. The search results are shown in Table 2.
Table 2. Characterization of STS fragments amplified from CEPH YAC 857_b_10 DNA.
| PCR fragment | Size (kb) | GenBank hits |
|---|---|---|
| chAB4 |
3.38 |
chAB4 varianta, chromosomes 1, 2, 3, 7, 9, 10, 11, 12, 13, 14, 15, 17, 21, 22 |
| N |
0.85 |
Chromosomes 1, 21, 22 |
| R2 |
0.90 |
Chromosomes 1, 2, 5, 10, 11, 13, 17, 21, 22 |
| H1.4 |
0.42 |
chAB4 variant |
| C8E2 |
0.80 |
chAB4 region, chromosomes 1, 2, 3, 8, 10, 11, 12, 13, 15, 17, 21, 22 |
| 14E2 |
0.63 |
NF1-like, chromosomes 1, 2, 3, 8, 9, 10, 11, 12, 13, 15, 17, 18, 21, 22 |
| 18E2 | 0.60 | NF1-like, chromosomes 1, 2, 3, 5, 8, 9, 10, 11, 12, 13, 15, 17,18, 21, 22 |
aWhen we started this work, no other hits were found. More recently, several hits against the draft human genome sequence in the HTGS section of GenBank were identified.
Chromosomal localization of the chAB4 PAC clones
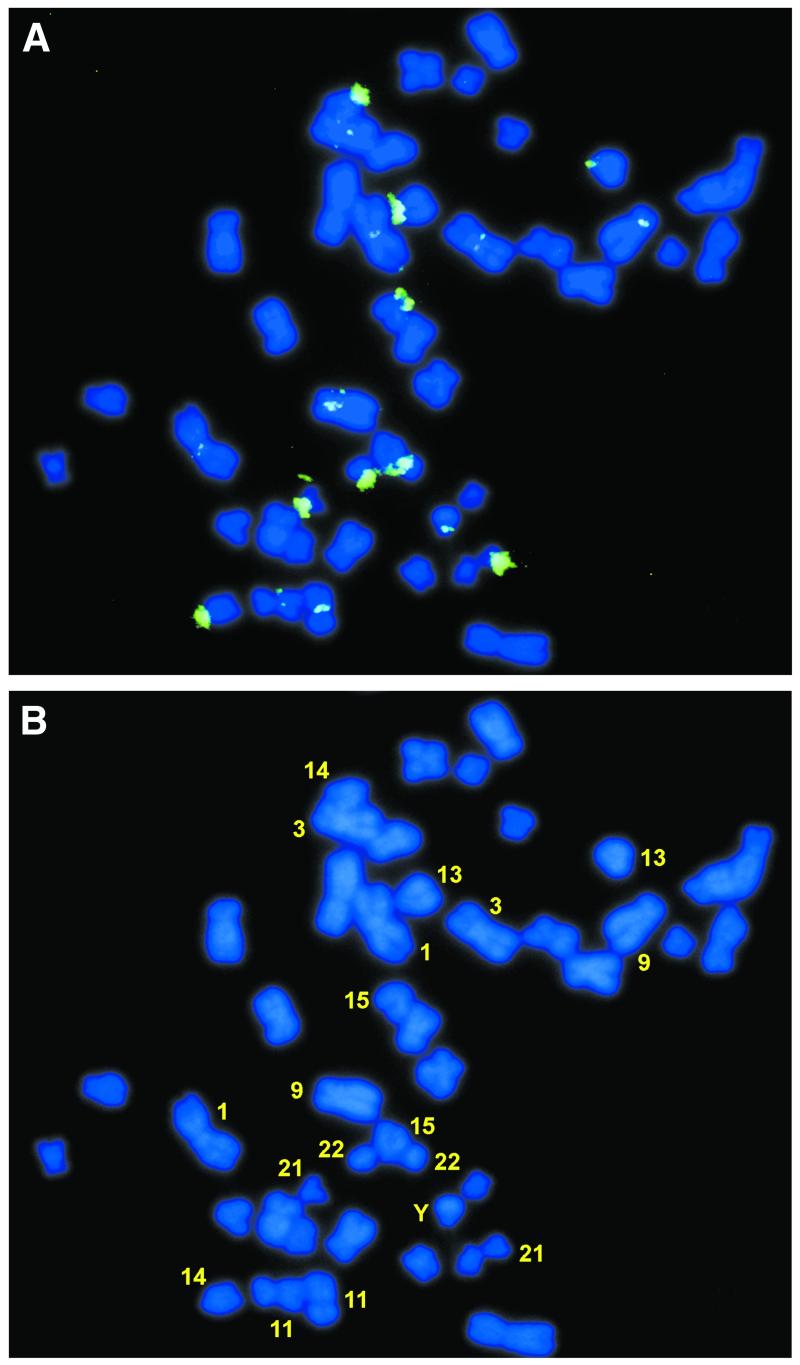
Biotinylated PAC DNA probes were hybridized to human male peripheral blood lymphocyte metaphase spreads. The FISH pattern of PAC 635H14 is shown in Figure 1A, and hybridizing chromosomes, i.e. chromosomes 1, 3, 9, 11, 13, 14, 15, 21, 22 and Y, are indicated in Figure 1B. Chromosomal distributions of the other PACs (data not shown) were found to be very similar to that of clone 635H14: high-intensity signals were detected at the centromeric regions and short arms of the acrocentric chromosomes 13, 14, 15, 21 and 22. Less intense but clear hybridizations were reproducibly observed to the centromeric region of chromosomes 1, 3, 9, and in some cases 2, 10, 11, 12, 17 and Y (summarized in Table 3). All probes detected a 57% overlap in the 14 chromosomes to which hybridization was observed, i.e. chromosomes 1, 3, 9, 13, 14, 15, 21 and 22 were always detected (Table 3). With YAC 857_b_10, derived from chromosome 22 (15), a strong signal was seen at the centromeric region and short arm of chromosome 22 and less intensive ones at chromosomes 1, 3, 9, 13, 14, 21 and Y (data not shown). However, the YAC insert has been shown to carry ∼200 kb of chromosome-specific satellite sequences (15), which may explain the strong chromosome 22 signal. The PAC clones appeared to be devoid of chromosome-specific satellites, rather, the in situ data suggested that their inserts were composed of highly conserved sequences scattered on numerous chromosomes in agreement with the STS data (Table 1). Still, some characteristic differences were observed, e.g. PAC 662K13 reproducibly hybridized to centromere-proximal sites of both arms of chromosome 10 (data not shown).
Figure 1.
FISH of biotinylated chAB4 multisequence family clones to human male peripheral blood lymphocyte metaphase spreads. Shown are (A) FISH pattern of PAC 635H14, and (B) assignment of hybridizations to chromosomes 1, 3, 9, 11, 13, 14, 15, 21, 22 and Y, respectively. This hybridizing chromosome set is identical to that reported for YAC 857_b_10, in agreement with our own data. However, some differences were seen in the hybridization patterns with the other PAC clones; these are summarized in Table 3.
Table 3. Chromosomal distribution of chAB4 family PAC clones.
| Clone | Chromosome | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | X | Y | |
| YAC 857_b_10 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
| 635H14 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
+ |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
| 635J14 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
+ |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
| 654D10 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
+ |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
| 654O8 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
– |
| 655I18 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
| 659M14 |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
+ |
+ |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
– |
| 662K13 |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
+ |
+ |
+ |
+ |
– |
+ |
– |
– |
– |
+ |
+ |
– |
– |
| 663D9 |
+ |
– |
+ |
– |
– |
– |
– |
– |
+ |
– |
– |
– |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
– |
| 668N10 |
+ |
+ |
+ |
– |
– |
– |
– |
– |
+ |
+ |
– |
– |
+ |
+ |
+ |
– |
+ |
|
– |
– |
+ |
+ |
– |
– |
| 670G14 | + | + | + | – | – | – | – | – | + | + | – | – | + | + | + | – | + | – | – | + | + | – | – | |
Chromosomes with FISH signal are indicated (+). YAC 857_b_10 (15) was used as reference. Most probes were found to hybridize to the same set of chromosomes, but clones 662K13 and 670G10 also detected chromosomes 2, 10, 12 and 17 but not 11 and Y, and PACs 668N10 and 670G14 hybridized to chromosomes 2, 10 and 17, but not 11 and Y. There is an 57% overlap in chromosomal localization for all probes used, since chromosomes 1, 3, 9, 13, 14, 15, 21 and 22 make up the chromosome set labeled by all probes.
Sequence analysis of PAC 635H14
When we started to study the chAB4 multisequence family, apart from the prototype sequence (12) only a cosmid (AP000024, withdrawn recently by the submitters) was found in the database that covered chAB4 and flanking DNA, including the STS marker R2. Therefore, it was appropriate to engage in large-scale sequencing. Since the in situ hybridization patterns did not reveal significant differences with the different PAC probes, clone 635H14 was chosen for sequencing from those carrying chAB4 (see Table 2).
Clone 635H14 carried an 86 723 bp human insert (GenBank accession no. AF401203). To begin with, both ends were sequenced using PAC vector primers. The reading with the PAC F primer showed 96% homology to AP000024 from position 24 210 between STS markers R2 and chAB4, while no meaningful similarity was found with the sequence derived from the other end. Subsequently, the PAC was digested with SacI, XbaI, EcoRI, HindIII and BamHI, respectively, and subcloned in pUC19. We sequenced the appropriate subclones by primer walking. Subcontigs without overlaps were connected and ordered using long-range PCR. For two short unclonable parts the overlapping PCR products had to be sequenced. To exclude that PAC 635H14 was a chimera, PCR amplifications were carried out with a selected subset of sequencing primers on human lymphocyte DNA, on the control YAC and the nine remaining PACs (data not shown).
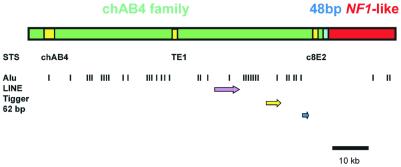
Analysis of the completed sequence was done mainly by searches against the GenBank database. The results of these comparisons are summarized in Figure 2. Thirty-eight Alu repeats (0.4 Alu/kb) were found in clusters. One LINE element and a Tigger transposon (27) were also identified. In addition to simple repeats not indicated in Figure 2, two complex tandem repeat regions were detected. One of them was composed of four copies of a 62 bp repeat unit (5′-TTTGAGATCT TTCTTCTTTT TAATTTAAGC ACCTGCAGCT ATAAGCTTCC CTTTAGCAAG GG-3′). The second repeat block was nearly 2 kb long, and comprised divergent 48 bp repeat units homologous to the D22Z3 satellite sequence (28). The 48-bp satellite array formed the junction region between the chAB4- and ΨNF1-family segments (Fig. 2). The first 72.9 kb of the PAC sequence, containing the chAB4 sequence and STS markers TE1 and c8E2 (called the chAB4 region) was connected by this repeat block to DNA homologous to NF1-related sequences derived from the neurofibromatosis type 1 gene. The NF1-related long-range family has been found using FISH on a number of human chromosomes (17–25).
Figure 2.
Arrangement of the chAB4 and ΨNF1 multisequence families in PAC 634H14 is shown. The chAB4 part (green bar) and the NF1-like region (red bar) are connected through the 48 bp satellite repeat (blue box). Yellow boxes represent locations of STS sequences: the chAB4 copy identified in this PAC clone (nucleotides 5269–8655) showed 94% homology to the prototype AB4 element (12). STS marker TE1 (nucleotides 49 411–49 621) was found to be 93% identical, and the c8E2 marker (nucleotides 71 755–72 615) was 96% homologous to the corresponding chromosome 22-derived sequence. Positions of copies of three different retrotransposon classes are also indicated. Vertical bars show the 38 Alu repeats, the purple arrow denotes a LINE sequence and the yellow arrow represents a Tigger transposon. The blue arrow indicates a short array of 62 bp repeats.
The ends of the remaining nine PAC clones were also sequenced and compared with the 635H14 sequence in order to examine their relationship. The results of these sequence comparisons are summarized in Table 4. PAC 635J14 proved to be identical with 634H14 (their end sequences were identical), whereas one of the end sequences of clones 654D10, 659M14, 662K13 and 670G14 showed 94–98% homology to different regions within the bounds of 635H14, therefore they were certainly independent genomic copies. The AP000024 cosmid sequence had a 15 474 bp overlap with the 5′ end of AF401203, with 95.9% homology. However, one of the ends of clones 654O8 and 655I18 was homologous (94 and 96%, respectively) to sequences around STS marker R2, located in AP000024, and were directed towards 635H14 (Table 4).
Table 4. Locations, lengths and levels of homology of PAC end sequence overlaps within PAC 635H14 (AF401203) and/or AP000024.
| Sequence | Overlap with | Length (bp) | % Homology | |
|---|---|---|---|---|
| AP000024 | AF401203 | |||
|
AF401203 |
24 210> |
1> |
15 474 |
95.9 |
| 655I18 |
7083> |
– |
269 |
94 |
| 654O8 |
10 480> |
– |
545 |
96 |
| 635J14 |
24 210> |
1> |
600 |
96/100a |
| 654D10 |
– |
32 056> |
160 |
98 |
| 670G14 |
– |
39 780> |
530 |
92 |
| 662K13 |
– |
57 261> |
315 |
98 |
| 663D9 |
– |
61 460> |
530 |
97 |
| 668N10 |
– |
65 266> |
500 |
94 |
| 659M14 |
– |
71 531> |
1500 |
96 |
| 635J14 | – | <86 723 | 300 | 1001 |
The AF401203 sequence starts at position 24 210 of AP000024. End sequence readings of the overlapping PAC clones were ordered in increasing position numbers. Arrowheads (< or >) indicate sequence reading orientations with respect to AF401203.
aPAC 635J14 was found to be identical with 635H14 in both of the end sequence readings.
Direct connection of the chAB4- and NF1-related multisequence families
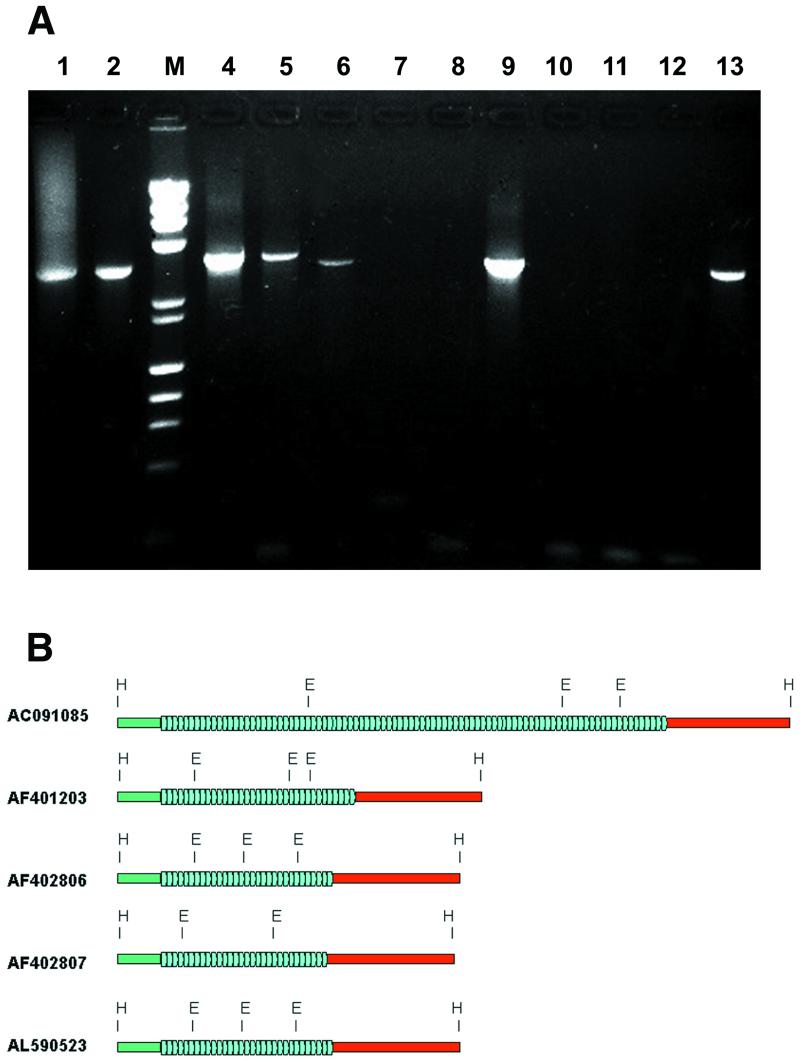
The possibility that these two long-range sequence families could be associated has not been considered previously. To examine whether the chAB4 family—48 bp satellite—NF1-like DNA was a unique feature of PAC 635H14 or whether it could be characteristic of other independent genomic loci as well, PCR amplifications were carried out with primers JCT F and JCT R designed to amplify the junction region. For templates, DNA from human lymphocytes, YAC 857_b_10 and the PAC clones were used. The expected junction fragments of ∼3.2–3.4 kb, based on the 635H14 sequence, were amplified on the lymphocyte, YAC, 635H14, 635J14, 654D10, 659M14 and 670G14 templates (Fig. 3A). The PCR product derived from the YAC was cloned and sequenced (GenBank accession no. AF402807). The corresponding HindIII fragment from PAC 659M14 was subcloned and sequenced (GenBank accession no. AF402806). The YAC-derived junction was 3155 bp, in 659M14, 3189 bp, and in 635H14 it was 3474 bp (nucleotides 72 607–76 081), respectively. These chAB4–NF1 junction regions are shown in Figure 3B. All three sequences were very similar outside the 48 bp repeats: for the 360 bp flanking sequence (green bars in Fig. 3B) on the chAB4 side the homology of AF401203 and AF402806 was found to be 97%, between AF401203 and AF402807, 96%, and it was 97% when AF402906 and AF402807 were compared. The flanking sequences on the ΨNF1 side (red bars in Fig. 3B) showed 94% homology over ∼1400 bp in pairwise alignments. The STS markers 14E2 and 18E2 have been localized to the central part of the large palindrome derived from chromosome 22 (15). We found that these two STSs are NF1-related sequences (Table 1). In addition, the chAB4–NF1 junction amplified from YAC 857_b_10 was placed between sequence-tagged sites c8E2 and 14E2. Therefore, it follows that the non-inverted core of the palindrome on chromosome 22 is composed mainly, if not entirely, of NF1-like DNA, and that the 48-bp satellite chAB4–NF1 junction resides on the chAB4 arm flanked by centromeric α-satellite in YAC 857_b_10.
Figure 3.
(A) PCR amplification of the chAB4–YNF1 junction region. Amplification was done using primers JCT F (chAB4 side) and JCT R (YNF1 side) on human genomic DNA (lane 1), YAC 857_b_10 (lane 2), and PAC clones 635H14, 635J14, 654D10, 654O8, 655I18, 659M14, 662K13, 663D9, 668N10 and 670G14 (lanes 4–13). Extension step during amplification was 10 min. M, size marker lane, mixture of λ HindIII + ΦX174 HaeIII digests. (B) Structure of five independent chAB4–NF1 family junctions. Flanking sequences on the chAB4 side (green bar) and the NF1-related side (red bar), and the tandemly repeated array of degenerate 48 bp satellite units (blue arrowheads) are indicated. The organization of junctions from AC091085 sequence from chromosome 17 (from GenBank), PAC 635H14 (AF401203, this work), PAC 659M14 (AF402806, this work), chromosome 22 YAC 857_b_10 (AF402807, this work), and from the chromosome 22-derived GenBank entry AL590523 are compared. In all cases, these connecting sequences reside in a HindIII restriction fragment. Sequences on either side of the satellite array are highly conserved: 360 bp (green bars) on the chAB4 side was found to be 96% identical in all five junctions. Similarly, the NF1-like regions (red bars) displayed 94% sequence homology. However, substantial heterogeneity was observed in the number of satellite repeats. On the one hand, the copy-number of 48 bp repeats is variable (blue arrowheads); on the other hand, the repeat units show sequence divergence. Polymorphic EcoRI sites in the 48 bp satellite are indicated.
Next, the GenBank was searched for homologies using the conserved sequences flanking the 48 bp repeat array. More than 10 entries were identified in the database, derived from chromosomes 1, 2, 3, 9, 10, 11, 12q, 13, 14, 15, 17, 21 and 22, respectively, and two of them, AL590523 and AC091085, were considered to be of particular significance. GenBank entry AL590523 is from chromosome 22 and it spans the DNA segment defined by STS markers N–R2–chAB4–TE1–c8E2–14E2, corresponding precisely to the arrangement described for the α-satellite arm of the large palindrome (15). The junction of the chAB4 arm to the NF1-like core region in AL590523 is similar to those we sequenced (Fig. 3B). The HindIII fragment spanning the junction region in AL590523 sequence was 3202 bp, whereas in the AC091085 entry from chromosome 17 it was 6102 bp (Fig. 3B). Substantial variation was observed in the number of 48 bp satellite repeats, ranging from 29 units in AF402806 and AF402807 to 92 reiterations in AC091085, while flanking sequences showed 96% homology to each other on the chAB4 side over 360 bp and 94% over 1.4 kb in the NF1-related family region. However, the real significance of AC091085 lies in that it is derived from chromosome 17. This sequence spans ∼65 kb of the chAB4 region, the chAB4–ΨNF1 junction, and ΨNF1 DNA. In addition, another GenBank entry, AF322451, which has been assigned to the 17p13.1 chromosome band, was found to carry a copy of chAB4 together with flanking DNA. Extensive sequence comparisons failed to reveal any connection to the ancestor of the ΨNF1 family, the neurofibromatosis type 1 (NF1) gene (29) in the 17q11.2 region (18). Hence, it was concluded that AC091085 and AF322451 must be derived from a previously unidentified locus on chromosome 17, probably 17p13, where the chAB4 and ΨNF1 families appear to be connected.
DISCUSSION
We describe here the first steps of a systematic large-scale analysis of the chAB4 long-range multisequence family. For this purpose, large-insert PAC clones were isolated and analyzed. One of the clones containing chAB4, the 86 723 bp PAC 635H14, was completely sequenced. This sequence provided a reference to which the structure of other clones could be compared. Apart from an ∼2 kb 48 bp tandem array similar to the D22Z3 sequence (28) no other satellite was detected. However, part of the PAC sequence proved to be NF1-related DNA.
The 48 bp tandem array was found to connect the chAB4 family segment with NF1-like sequence in PAC 635H14. Although these long-range low copy-number families have been studied in detail during the last decade (11–25), they have been thought to be independent mobile DNA elements, and their direct physical connection has not been established previously.
We examined whether direct connection of the two sequence families was a single-copy structure or was it a more or less general structural feature. Therefore, PCRs were carried out with primers designed to amplify junctions similar to that found in the PAC 635H14. Products of ∼3.2–3.5 kb were amplified from the human DNA and YAC template, as well as from PACs 654D10, 659M14 and 670G14. In the three junctions we sequenced, substantial heterogeneity was seen only in the 48 bp satellite repeats. However, the flanking sequences on both sides were found to be 94–97% homologous.
The existence of a conserved connecting structure between the chAB4- and the NF1-like sequence families gained additional support from GenBank searches with sequences from both sides of the junction. From more than ten GenBank entries identified, the junction regions from AL590523 of chromosome 22, and from AC091085 of chromosome 17, were also analyzed. They were shown to be similar to those we sequenced. It will be worthwhile to examine more chAB4–ΨNF1 junctions, since their polymorphism could help in tracing the evolutionary history of the contiguous chAB4–ΨNF1 multisequence.
Detailed examination of the chromosome 17-derived AC091085 sequence indicated that the joint chAB4–ΨNF1 multisequence also existed on this chromosome. GenBank entry AF322451 from 17p13.1 supported this notion. Noteworthy is that neither sequence was found to be associated with the NF1 gene. Therefore, it seemed plausible to assume that this latter region could have been the ancestor of at least some of the related sequences located on other chromosomes, and it resulted from the initial duplication event of the middle part of the NF1 gene. Since no alternative explanation regarding the presence of the chAB4 multisequence family on chromosome 17 has been described so far, the assumption outlined here could provide a working hypothesis to address this question.
We believe that further studies on the large-scale organization of complex long-range sequence families at centromeric regions of the human chromosomes will help to reach a deeper understanding of the mechanisms underlying genome plasticity, and to solve some of the paradoxes (30,31) of the centromeres.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Drs A. Udvardy and I. Raskó for their helpful comments on the manuscript. This work was supported in part by a research fund from Chromos Molecular Systems, Inc., Burnaby, BC, Canada.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +36 62 432 080; Fax: +36 62 433 397; Email: cserpani@nucleus.szbk.u-szeged.hu AF401203, AF402806, AF402807
REFERENCES
- 1. International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Vogt P. (1990) Potential genetic functions of tandem repeated DNA sequence blocks in the human genome are based on a highly conserved ‘chromatin folding code’. Hum. Genet., 84, 301–336. [DOI] [PubMed] [Google Scholar]
- 4.Baltimore D. (2001) Our genome unveiled. Nature, 409, 814–816. [DOI] [PubMed] [Google Scholar]
- 5.Lee C., Wevrick,R., Fisher,R.B., Ferguson-Smith,M.A. and Linc,C.C (1997) Human centromeric DNAs. Hum. Genet., 100, 291–304. [DOI] [PubMed] [Google Scholar]
- 6.Wevrick R. and Willard,H.F. (1991) Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res., 19, 2295–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trowell H.E., Nagy,A., Vissel,B. and Choo,K.H.A. (1993) Long-range analyses of the centromeric regions of human chromosomes 13, 14 and 21: identification of a narrow domain containing two key centromeric DNA elements. Hum. Mol. Genet., 2, 5865–5874. [DOI] [PubMed] [Google Scholar]
- 8.Cooper K.F., Fisher,R.B. and Tyler-Smith,C. (1993) Structure of the sequence adjacent to the centromeric alphoid satellite DNA array on the human Y chromosome. J. Mol. Biol., 230, 787–799. [DOI] [PubMed] [Google Scholar]
- 9.Cooper K.F., Fisher,R.B. and Tyler-Smith,C. (1993) The major centromeric array of alphoid satellite DNA on the human Y chromosome is non-palindromic. Hum. Mol. Genet., 2, 1267–1270. [DOI] [PubMed] [Google Scholar]
- 10.Jackson M.S., Rocchi,M., Thompson,G., Hearn,T., Crosier,M., Guy,J., Kirk,D., Mulligan,L., Ricco,A., Piccininni,S., Marzella,R., Viggano,L. and Archidiacono,N. (1999) Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet., 8, 205–215. [DOI] [PubMed] [Google Scholar]
- 11.Assum G., Böckle,B., Fink,T., Dmochewitz,U. and Krone,W. (1989) Restriction analysis of chromosomal sequences homologous to single-copy fragments cloned from small polydisperse circular DNA (spcDNA). Hum. Genet., 82, 249–254. [DOI] [PubMed] [Google Scholar]
- 12.Assum G., Fink,T., Klett,C., Lengl,B., Schanbacher, M, Uhl,S. and Wöhr,G. (1991) A new multisequence family in human. Genomics, 11, 397–409. [DOI] [PubMed] [Google Scholar]
- 13.Assum G., Gartmann,C., Schempp,W. and Wöhr,G. (1994) Evolution of the chAB4 multisequence family in primates. Genomics, 21, 34–41. [DOI] [PubMed] [Google Scholar]
- 14.Assum G., Pasantes,J., Glaser,B., Schempp,W. and Wöhr,G. (1998) Concerted evolution of members of the multisequence family chAB4 located on various nonhomologous chromosomes. Mamm. Genome, 9, 58–63. [DOI] [PubMed] [Google Scholar]
- 15.Wöhr G., Fink,T. and Assum,G. (1996) A palindromic structure in the pericentromeric region of various human chromosomes. Genome Res., 6, 267–279. [DOI] [PubMed] [Google Scholar]
- 16.Kehrer-Sawatzki H., Wöhr,G., Schempp,W., Eisenbarth,I., Barbi,G. and Assum,G. (1998) Mapping of members of the low-copy-number repetitive DNA sequence family chAB4 within the p arms of human acrocentric chromosomes: characterization of Robertsonian translocations. Chromosome Res., 6, 429–435. [DOI] [PubMed] [Google Scholar]
- 17.Legius E., Marchuk,D.A., Hall,B.K., Andersen,L.B., Wallace,M.R., Collins,F.S. and Glover,T.W. (1992) NF1-related locus on chromosome 15. Genomics, 13, 1816–1818. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki S., Ozawa,N., Taga,C., Kano,T., Hattori,M. and Sakaki,Y. (1994) Genomic analysis of a NF1-related pseudogene on human chromosome 21. Gene, 147, 277–280. [DOI] [PubMed] [Google Scholar]
- 19.Purandare S.M., Breidenbach,H.H., Li,Y., Zhu,X.L., Sawada,S., Neil,S.M., Brothman,A., White,R., Cawthon,R. and Viskochil,D. (1995) Identification of neurofibromatosis 1 (NF1) homologous loci by direct sequencing, fluorescence in situ hybridization and PCR amplification of somatic cell hybrids. Genomics, 30, 476–485. [DOI] [PubMed] [Google Scholar]
- 20.Hulsebos T.J.M., Bijleveld,E.H., Riegman,P.H.J., Smink,L.J. and Dunham,I. (1996) Identification of NF1-related loci on human chromosomes 22, 14 and 2. Hum. Genet. 98, 7–11. [DOI] [PubMed] [Google Scholar]
- 21.Regnier V., Meddeb,M., Lecointre,G., Richard,F., Duverger,A., Nguyen,V.C., Dutrillaux.,B., Bernhem,A. and Danglot,G. (1997) Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum. Mol. Genet., 6, 9–16. [DOI] [PubMed] [Google Scholar]
- 22.Kehrer-Sawatzki H., Schwickardt,T., Assum,G., Rocchi,M. and Krone,W. (1997) A third neurofibromatosis type 1 (NF1) pseudogene at chromosome 15q11.2. Hum. Genet., 100, 595–600. [DOI] [PubMed] [Google Scholar]
- 23.Ritchie R.J., Mattei,M.-G. and Lalande,M. (1998) A large polymorphic repeat in the pericentromeric region of human chromosomes 15q contains three partial gene duplications. Hum. Mol. Genet., 7, 1253–1260. [DOI] [PubMed] [Google Scholar]
- 24.Luijten M., Wang,Y.P., Smith,B.T., Westerveld,A., Smink,L.J., Dunham,I., Roe,B.A. and Hulsebos,T.J.M. (2000) Mechanism of spreading of the highly related neurofibromatosis type 1 (NF1) pseudogenes on chromosomes 2, 14 and 22. Eur. J. Hum. Genet., 8, 209–214. [DOI] [PubMed] [Google Scholar]
- 25.Luijten M., Redeker,S., Minoshima,S., Shimizu,N., Westerveld,A. and Hulsebos,T.J.M. (2001) Duplication and transposition of the NF1 pseudogene regions on chromosomes 2, 14 and 22. Hum. Genet., 109, 109–116. [DOI] [PubMed] [Google Scholar]
- 26.Csonka E., Cserpán,I., Fodor,K., Holló,Gy., Katona,R., Keres,J., Praznovszky,T., Szakál,B., Telenius,A., deJong,G., Udvardy,A. and Hadlaczky,Gy. (2000) Novel generation of human satellite DNA-based artificial chromosomes in mammalian cells. J. Cell Sci., 113, 3207–3216. [DOI] [PubMed] [Google Scholar]
- 27.Smit A.F. and Riggs,A.D. (1995) Tiggers and other DNA transposon fossils in the human genome. Proc. Natl Acad. Sci. USA, 93, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzdorf R., Göttert,E. and Blin,N. (1988) A novel centromeric repetitive DNA from human chromosome 22. Chromosoma, 97, 154–158. [DOI] [PubMed] [Google Scholar]
- 29.Li Y., O’Connell,P., Breidenbach,H.H., Cawthon,R., Stevens,J., Xu,G., Neil,S., Robertson,M., White,R. and Viskochil,D. (1995) Genomic organization of the neurofibromatosis 1 gene (NF1). Genomics, 25, 9–18. [DOI] [PubMed] [Google Scholar]
- 30.Choo K.H.A. (2000) Centromerization. Trends Cell Biol., 10, 182–188. [DOI] [PubMed] [Google Scholar]
- 31.Copenhaver G.P. and Preuss,D. (1999) Centomeres in the genomic era: unraveling paradoxes. Curr. Opin. Plant Biol., 2, 104–108. [DOI] [PubMed] [Google Scholar]