Abstract
The modulation of enzymatic activities of Escherichia coli DnaB helicase by homologous and heterologous single-stranded DNA-binding proteins (SSBs) and its DNA substrates were analyzed. Although DnaB helicase can unwind a variety of DNA substrates possessing different fork-like structures, the rate of DNA unwinding was significantly diminished with substrates lacking a 3′ fork. A 5 nt fork appeared to be adequate to attain the maximum rate of DNA unwinding. Efficient helicase action of DnaB requires the participation of SSBs. Studies involving heterologous SSBs demonstrated that they can stimulate the helicase activity of DnaB protein under certain conditions. However, this stimulation occurs in a manner distinctly different from that observed with cognate E.coli SSB. The E.coli SSB was found to stimulate the helicase activity over a wide range of SSB concentrations and was unique in its strong inhibition of single-stranded DNA-dependent ATPase activity when uncoupled from the DNA helicase activity. In the presence of a helicase substrate, the ATPase activity of DnaB helicase remained uninhibited. Thus, E.coli SSB appears to coordinate and couple the ATPase activity to the DNA helicase activity by suppressing unproductive ATP hydrolysis by DnaB helicase.
INTRODUCTION
In vitro studies with λ phage and Escherichia coli DNA replication have shown that the DnaB protein acts as a helicase in the replication of λ and plasmids containing the E.coli origin of replication, Ori C (1,2). One of the most important discoveries regarding the function of DnaB protein was its DNA helicase activity reported first by LeBowitz and McMacken in 1986 (2). The E.coli DnaB helicase is a hexameric protein comprised of six 52 kDa monomers (3–6). DnaB is a multifunctional enzyme that controls the formation and translocation of the replisome in E.coli and λ phage DNA replication. During replication of DNA, it interacts with a host of other replication proteins including DnaC protein, DnaG protein or primase and λP protein (2–7). Perhaps due to its ability to interact with other replication proteins, DnaB protein plays a pivotal role in the assembly of the primosome and subsequent movement of the replication apparatus (8–10). The N-terminal region of DnaB protein has been shown to be indispensable for the helicase activity of DnaB protein as well as its hexamerization (11). Several laboratories have shown that oligomerization is an important step in the mechanism of action of DNA helicases (9–17). The formation of a hexamer proceeds through leucine zipper-mediated dimerization of monomers, followed by the association of dimers to form the hexamer (12). Although the crystal structure of the protein remains unknown at the present time, protein mapping has delineated the specific domains involved in nucleotide hydrolysis, DNA binding and oligomerization (12,13).
Previously, we have studied the DNA-dependent nucleotidase activity of DnaB and modulation of its ATPase activity by DnaC and λP proteins (7). In addition, detailed mapping has shed light on the specific domains required for its biochemical functions (12,13). However, much remains to be determined regarding modulation of its various enzymatic activities. The helicase activity of DnaB helicase is stimulated by E.coli single-stranded DNA-binding protein (SSB) (2,3), whereas Arai and Kornberg (3) demonstrated that the single-stranded DNA (ssDNA)-dependent ATPase activity is inhibited. Thus, the mechanism of participation of E.coli SSB in helicase action remains unclear. To resolve these questions, we have explored the influence of ssDNA-binding proteins from prokaryotic and eukaryotic sources on DnaB helicase action. The eukaryotic replication protein A (RPA) is homologous in function to E.coli SSB and has been shown to be involved in DNA replication and helicase action (18–22). T4 phage gene 32 protein (gp32 protein), a phage-encoded ssDNA-binding protein, is essential in the replication of T4 bacteriophage in vitro (8,23). These proteins were used to analyze the specific role(s) of ssDNA-binding proteins in the helicase action of DnaB protein.
Early studies of the helicase activity of DnaB protein in vitro indicated that a 99 nt fork was required for processive unwinding of the template DNA containing a large duplex region (2). However, in the in vitro replication of the E.coli origin of replication, OriC, a very small region of duplex DNA is opened that becomes a substrate for DnaB helicase (8,24). The process appears to take place with DnaA protein bound to DNA, rather than the more common SSB-coated DNA, and results in generating a forked structure with a small single-stranded region. This would appear to suggest that the size of the fork required by DnaB to initiate helicase activity is smaller than previously estimated.
Jezewska et al. (25) demonstrated that the optimal size of the DNA for binding to the DnaB hexamer in 20 bp as a single oligonucleotide or as an arm (5′ or 3′) of a fork. However, this study focused on DNA binding only and it was carried out in the absence of SSBs lacking an assessment of the helicase function of DnaB. Kaplan and Steitz (26) analyzed the structural requirement of DnaB helicase from Thermus aquaticus at 55°C, also in the absence of any SSBs. With T.aquaticus DnaB, it appears that 35 bp in the 5′ tail and 10–15 nt in the 3′ end of the duplex fork structure would suffice. However, unlike the original work of LeBowitz and McMacken (2), both of these studies (25,26) were not carried out under physiological conditions of E.coli growth and chromosomal replication. It should be noted that DNA conformation and DNA melting are highly susceptible to SSB and temperature. Consequently, the structural requirements of the DNA substrate for the helicase activity of the DnaB protein in the presence of E.coli SSB and normal growth temperature remain unknown.
In this report, we have investigated the mechanism of modulation of DnaB helicase action by (i) the structure of the DNA substrate, and (ii) the action of SSBs under normal conditions of E.coli growth and chromosomal DNA replication.
MATERIALS AND METHODS
Nucleic acids, enzymes and other reagents
DnaB protein was purified to homogeneity as described by Arai et al. (6) from YS1recA (pKA1), which was received as a kind gift from Dr K. Arai.
Ultrapure deoxy- and ribonucleotides were obtained from AP Biotech (Piscataway, NJ) and were used without further purification. [α-32P]ATP and [γ-32P]ATP were obtained from AP Biotech. Yeast RPA was purified to homogeneity from wild-type yeast (>98% purity in SDS–PAGE) as described (14). Escherichia coli SSB was purchased from Promega Biotech (Milwaukee, WI). gp32 protein of T4 bacteriophage was from U.S. Biochemical (Cleveland, OH). The homogeneity of the commercially obtained SSBs was verified by SDS–PAGE (data not shown). T4 polynucleotide kinase was obtained from New England Biolabs Inc. (Beverly, MA). All chemicals used to prepare buffers and solutions were of reagent grade and were purchased from Fisher Chemical Company (Pittsburgh, PA). Polyethyleneimine–cellulose TLC strips were from J.T. Baker Chemical Co.
Oligonucleotides were synthesized by Oligos Inc. (Corvallis, OR). The oligonucleotides were analyzed and further purified by 20% polyacrylamide–8 M urea gel electrophoresis. The final purity of the oligonucleotides was >99% as judged by autoradiography of the phosphorylated products.
Buffers
1× TBE contained 89 mM Tris–borate (pH 8.3), 2.5 mM EDTA. Buffer A contained 25 mM Tris–HCl (pH 7.5), 10% (v/v) glycerol, 0.1 mg/ml bovine serum albumin and 5 mM DTT.
ATPase assays
The ATPase assays were carried out as previously described (11). The amount of DnaB protein used in the assays was selected such that the rate of hydrolysis would be linear under the reaction conditions. A standard 10 µl reaction mixture contained 10 mM MgCl2, 200 pmol of M13mp18 ssDNA, 100 µM [α-32P]ATP (1000–2000 c.p.m./pmol) and 1.67 nM DnaB protein in buffer A. Reactions were incubated at 37°C for 30 min (unless stated otherwise) and terminated by addition of 2 µl of 200 mM EDTA followed by chilling on ice. Two-microliter aliquots were applied to polyethyleneimine–cellulose strips, which were pre-spotted with ADP–ATP marker. The strips were developed with 1 M formic acid, 0.5 M LiCl and dried. The ADP–ATP spots were located by UV fluorescence. The portions containing ATP and ADP were excised and counted in a liquid scintillation counter using a toluene-based scintillator.
In kinetic analyses a given set of reactions were carried out in a single tube in an appropriate reaction volume and were initiated by addition of DnaB protein. At the indicated time points 10 µl aliquots were removed and transferred to tubes containing 2 µl of 200 mM EDTA at 0°C. The remainder of the assay was carried out as described above.
Preparation of helicase substrates
As shown in Figure 1 and Table 1, two different helicase substrate motifs were utilized in these studies. The method of preparation was the same for each, except that the oligonucleotide hybridized to M13mp19 ssDNA was different in each case. The sequences of the oligonucleotides are given in Table 1. T4 polynucleotide kinase was used to label the 5′ end of the oligomers. Hybridization was carried out as previously described (14). Excess unhybridized labeled oligonucleotide was removed by gel filtration (Sephacryl-S400; Pharmacia, Piscataway, NJ) chromatography. The purified substrate was diluted to a final concentration of 17 fmol/µl (10 000–20 000 c.p.m./µl) with 10 mM Tris–HCl (pH 7.5), 1 mM EDTA.
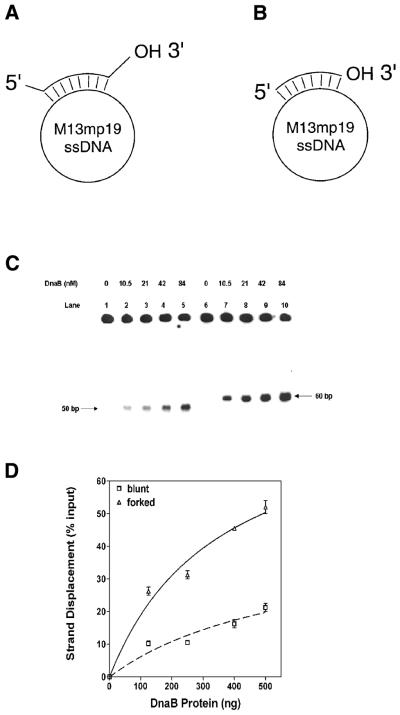
Figure 1.
Analysis of the helicase activity of the DnaB helicase on substrates with forked and recessed termini: (A) structures of the helicase substrates. Substrate A with 5′ and 3′ preformed forks: a synthetic 60 bp oligonucleotide, complementary to a 50 bp sequence between nucleotides 6268 and 6317 of M13mp19 ssDNA, was hybridized to M13mp19. (B) Substrate B without preformed forks: a synthetic 50 bp oligonucleotide, complementary to the sequence between nucleotides 6268 and 6317 of M13mp19 ssDNA was hybridized to M13mp19. The substrates were prepared as described in Materials and Methods. (C) Autoradiogram of the helicase assay comparing activities on forked and recessed substrates. Lanes 1–5, substrate without a preformed fork (substrate 2): lane 1, substrate and SSB in the absence of DnaB protein; lanes 2–5, DnaB protein as indicated. Lanes 6–10, substrate 1 with 5′ and 3′ forks (substrate 1): lane 6, substrate and SSB in the absence of DnaB; lanes 7–10, DnaB protein as indicated. A standard helicase assay was carried out for 30 min in the presence of 0.69 µM SSB and varying amounts of DnaB protein as indicated. (D) Quantification of strand displacement during DnaB protein titration in the DNA helicase assay in (C) above. Quantification was carried out as described in Materials and Methods.
Table 1. Sequences of oligonucleotides used in the preparation of helicase substrates.
| Oligo | Sequence (5′→3′) | ΔG0 (kcal/mol)a |
|---|---|---|
| 1 | GGGTCTCACG ACGTTGTAAA ACGACGGCCA GTGAATTCGA GCTCGGTACC CGGGGTAGGA | –107.14 |
| 2 | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGAGCTCG GTACCCGGGG | –107.14 |
| 3 | GGGTCTCACG ACGTTGTAAA ACGACGGCC GTGAATTCGA | –70.7 |
| 4A | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGA | –70.7 |
| 4B | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGAAT | –70.7 |
| 4C | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGAATCTC | –70.7 |
| 4D | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGAATCTC CATGG | –70.7 |
| 4E | TCACGACGTT GTAAAACGAC GGCCAGTGAA TTCGAATCTC CATGGTCCAC TAGGA | –70.7 |
N, complementary to M13mp19 ssDNA between nucleotides 6268 and 6317.
aBreslauer et al. (28).
Assay conditions
Reaction mixtures were set up on ice as follows. A standard 20 µl reaction volume contained buffer A and 10 mM MgCl2, 3.4 mM ATP, 17 fmol (10 000–20 000 c.p.m./µl) of substrate and the indicated amount of DnaB protein. The mixtures were incubated at 37°C for the times indicated and the reactions were terminated by the addition of 4 µl of 2.5% SDS, 60 mM EDTA and 1% bromophenol blue. A fraction (25%) of each reaction mixture was analyzed on a 8% polyacrylamide gels in 1× TBE and 0.1% SDS. The electrophoresis was carried out in 1× TBE, 0.1% SDS for 1 h at 160 V. Following electrophoresis, the gels were dried and exposed to Fuji RX film at –80°C for 12 h. If needed, helicase activity was quantified by scintillation counting of excised substrate/product bands from the dried gels as described earlier (12,13) and/or by scanning densitometry of the corresponding autoradiogram.
Other methods
Protein concentrations were estimated according to the method of Bradford (27) using bovine serum albumin as standard.
RESULTS
Influence of DNA structure on the kinetics of strand displacement
DnaB protein unwinds the DNA double helix during the initiation and elongation stages of chromosomal DNA replication in E.coli. We have carried out an analysis of the influences of the DNA fork structure on helicase action using defined helicase substrates that mimic a replication fork. These substrates were prepared from M13mp19 ssDNA hybridized to synthetic oligonucleotides creating partial duplexes with or without forked structures at the 5′ and 3′ termini. The sequences of the oligonucleotides used in this study are shown in Table 1 and a schematic diagram of the partial duplex substrates is given in Figure 1A and B. We have utilized thermodynamically stable but defined DNA substrates because such substrates are advantageous for measuring the ability of a DNA helicase to recognize and initiate the helicase action regardless of the processivity under the given assay conditions.
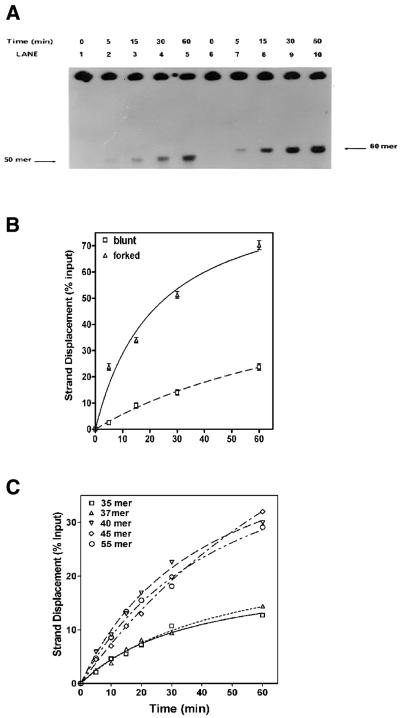
Protein titrations of helicase activity using blunt-ended versus forked substrates are shown in Figure 1C and D. The presence of forked structures in substrate A significantly stimulated the rate of strand displacement when compared with substrate B in a quantitative analysis of strand displacement. Although the size of the fork in substrate A was only 5 bp, it appeared to enhance the rate of strand displacement. These results also demonstrate that DnaB helicase can unwind duplex DNA in the absence of a forked structure, albeit at a substantially reduced rate. With both substrates, DNA unwinding increased with addition of DnaB protein (Fig. 1C and D). However, when a fork was present, there was a significant increase in displacement with increased DnaB protein (Fig. 1D). The results of kinetic studies are shown in Figure 2A. The rate of duplex unwinding of a forked substrate was at least 3-fold higher than that observed with the blunt-ended substrate B. The maximal extent of strand displacement observed with the forked substrate was ∼70% at 60 min whereas the maximal extent of strand displacement observed with the blunt-ended substrate was only 25% during the same period. The requirement of a 5′ or 3′ fork was examined using substrates that had either a 5′ or a 3′ 5 nt preformed fork with a 35 bp duplex region. DnaB protein was able to unwind substrates much more effectively when the substrate had a 3′ fork (data not shown). The influence of a 5′ fork was undetectable in our assay and the amount of displacement was comparable to that observed with recessed substrates.
Figure 2.
Kinetics of DnaB helicase on forked and recessed substrates: a standard helicase assay was carried out with 27 nM DnaB in the presence of 0.69 µM E.coli SSB for the indicated length of time. (A) Autoradiogram: lanes 1–5, helicase activity in the absence of a preformed fork (substrate 2); lanes 6–10, helicase activity in the presence of a preformed fork (substrate 1). (B) Quantification of the kinetics of strand displacement in the helicase assay in (A) above. Quantification was carried out as described in Materials and Methods. (C) Effects of increasing 3′ fork length on the rate of strand displacement: standard helicase assays were carried out over time periods ranging from 0 to 60 min using 160 ng DnaB in the presence of 0.69 µM E.coli SSB. The lengths of the 3′ fork on the substrates were as follows: no fork (squares); 2 nt fork (triangles); 5 nt fork (inverted triangles); 10 nt fork (diamonds); 20 nt fork (circles).
An analysis of 3′ fork length on the rate of displacement is shown in Figure 2C. We have examined substrates with a 35 bp duplex containing 0, 2, 5, 10 and 20 bp 3′ extensions or forked regions. A 5 nt fork length appeared to be adequate for maximal stimulation of DnaB helicase. On the other hand, 3′ forks of lengths >5 nt did not further increase the rate of strand displacement.
DNA helicase activity of DnaB protein is modulated by heterologous and homologous SSBs
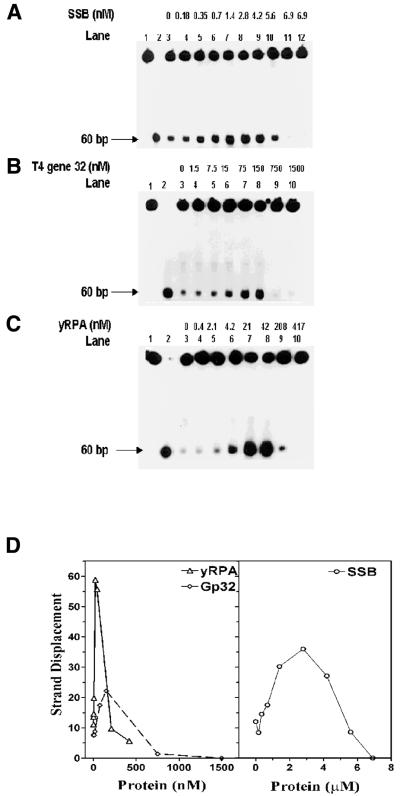
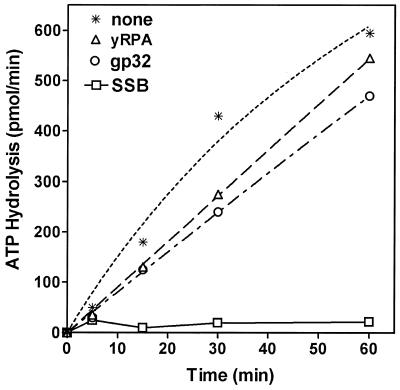
We have examined the effects of E.coli SSB on the DNA helicase activity of DnaB protein. In order to explore the possible functional interaction with its cognate SSB we have examined other SSBs—yeast RPA (yRPA) and gp32. Highly purified SSB, gp32 and yRPA were utilized in these experiments and the possibility of a helicase contaminant was ruled out by helicase assays carried out in the absence of DnaB protein. No detectable helicase activity was observed (data not shown). Escherichia coli SSB stimulated the helicase activity over a broader and higher concentration range (0.01–5 µM), and the extent of stimulation was intermediate between gp32 and yRPA (Fig. 3A–D). gp32 and yRPA affected the DNA helicase activity in comparable ranges (0.01–0.5 µM), but differed in the degree of stimulation. The yRPA stimulated the helicase activity of DnaB protein, by ∼10-fold over the range 4.2–41.7 nM. Beyond 41.7 nM, the stimulation decreased and addition of 210 nM inhibited the activity. Stimulation of DnaB helicase was observed with gp32 protein. At higher concentrations, it inhibited the unwinding similar to that observed with yRPA. Therefore, stimulation of DnaB helicase observed with yRPA and gp32 appeared to be quite different from that observed with cognate SSB.
Figure 3.
Influence of SSBs on the helicase action of DnaB protein: a standard helicase assay was carried out for 30 min utilizing the 5′→3′ forked substrate. The reaction mixtures contained 27 nM DnaB protein and varying amounts of SSBs as indicated. (A) Influence of E.coli SSB: lane 1, substrate alone; lane 2, heat-denatured substrate; lanes 3–12, DnaB protein in the presence of E.coli SSB; lane 12, 6.9 µM E.coli SSB in the absence of DnaB protein. (B) Influence of T4 gp2 protein: lane 1, substrate alone; lane 2, heat-denatured substrate; lanes 3–10, DnaB protein in the presence of T4 gp32 protein. (C) Influence of yRPA: lane 1, substrate alone; lane 2, heat-denatured substrate; lanes 3–10, DnaB protein in the presence of yRPA. (D) Quantification of strand displacement in the helicase assays (A–C) above. Quantification was carried out as described in Materials and Methods.
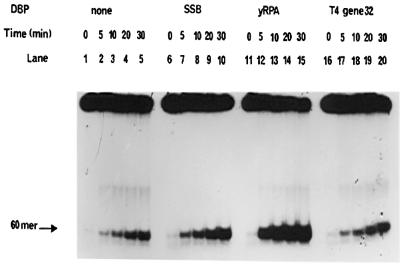
The effects of E.coli SSB, T4 gp32 and yRPA on the kinetics of the DNA unwinding by DnaB helicase are shown in Figure 4. All SSBs increased the rate and extent of unwinding of the substrate, however, they differed uniquely in the concentration ranges they stimulated or inhibited.
Figure 4.
Kinetics of DnaB helicase activity and modulation by SSBs. The standard helicase assay was carried out over time periods ranging from 0 to 30 min utilizing the 5′→3′ forked substrate. The reaction mixtures contained 27 nM DnaB protein and SSBs as indicated. Lanes 1–5, DnaB protein in the absence of SSB proteins. Lanes 6–10, DnaB protein in the presence of 0.69 µM E.coli SSB. Lanes 11–15, DnaB protein in the presence of 21 nM yRPA. Lanes 16–20, DnaB protein in the presence of 75 nM gp32 protein.
Cognate E.coli SSB regulated DNA-dependent ATPase activity of DnaB
The multifunctional DnaB protein has been shown to have a strong DNA-dependent ATPase activity and requires participation of SSB for its DNA helicase function (2). Escherichia coli SSB had a significant effect on the ATPase activity of DnaB protein, which correlates well with the findings of Arai and Kornberg (3). This inhibition appeared to be specific for only E.coli SSB (Fig. 5). Time-course analysis of the inhibition indicated that E.coli SSB inhibited the initial rate of the reaction. Analysis of the ATPase activity of DnaB in the presence of gp32 protein or yRPA further demonstrated that these proteins had no significant effect on the kinetics of ATP hydrolysis. It is important to note that the basal level ssDNA-dependent ATPase activity of DnaB helicase in the absence of SSB is very high and the energy of such ATP hydrolysis is uncoupled to its DNA helicase function and, thus, not used for DNA unwinding. Consequently, the energy released by ATP hydrolysis is wasted. It appears likely that by binding to ssDNA cofactor, E.coli SSB reduces the basal ATPase activity of DnaB protein and blocked nonsense and idle hydrolysis of ATP. Neither yRPA nor gp32 proteins appeared to inhibit the ATP hydrolysis significantly (Fig. 5). Both yRPA and gp32 protein are structurally different from E.coli SSB and, therefore, their mechanism of DNA binding may be different. In addition, DNA helicases are likely evolved to work with the cognate SSBs. This hypothesis could explain the unique inhibition of nonsense ATPase activity of DnaB helicase by its cognate SSB making the ATPase activity specific for the fork structure of the helicase substrate, which was observed here.
Figure 5.
Effects of SSBs on the kinetics of DnaB ATPase activity. Standard ATPase assays were carried out over time periods ranging from 0 to 60 min, using 3.33 nM DnaB protein in the presence of 200 pmol ssDNA and 0.35 µM SSB, 0.75 µM gp32 and 0.21 nM RPA. No SSB (asterisks); yRPA (triangles); gp32 protein (circles); E.coli SSB (squares). Each point represents the average of triplicate determinations.
DNA structure modulates SSB inhibition of DNA-stimulated ATPase
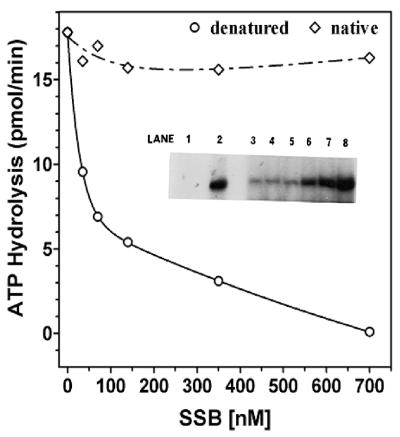
In order to further analyze our hypothesis that E.coli SSB inhibited only the idle ATPase activity, we have carried out identical analysis in which the ssDNA was hybridized with oligonucleotides to generate partial duplex regions terminating in 15 bp 3′ fork structures. Poly(dT) was hybridized with oligo(dA)35–(dC)15 in a 1:0.5 molar ratio (in terms of nucleotide). The ATPase activity with increasing amounts of E.coli SSB in the presence of this DNA substrate was then examined. An analogous series of experiments was carried out in which the poly(dT):oligo(dA)35–(dC)15 substrate had been heat denatured to melt all duplex fork structures. When poly(dT):oligo(dA)35–(dC)15 with duplex regions possessing fork structures (Fig. 6) was used as cofactor for ATPase, no inhibition was observed with E.coli SSB over the range 0–6 µM. When this substrate was converted entirely to ssDNA through heat denaturation, SSB inhibition was observed in a manner similar to that of previous experiments with ssDNA cofactor (Fig. 6); the presence of only 0.7 µM SSB inhibited the ATPase activity by ∼50% and at amounts of >3 µM this inhibition was essentially complete (>90%). The lack of inhibition was sensitive to the molar ratio of duplex regions and, when the ratio of poly(dT):oligo(dA)35–(dC)15 was <1:0.5, the sensitivity to inhibition was the same as that observed with poly(dT) alone (data not shown).
Figure 6.
Influence of DNA structure on SSB inhibition of ssDNA stimulated ATPase. Standard ATPase assays were carried out for a period of 10 min in the presence of 200 pmol (as nucleotide) of poly(dT):oligo(dA)35–(dC)6. The reaction mixtures contained 3.33 nM DnaB protein and varying amounts of SSB as indicated. Heat-denatured DNA co-factor (circles); native DNA co-factor (diamonds). Inset, influence of E.coli SSB on DNA unwinding of poly(dT):oligo(dA)35–(dC)6. A standard helicase assay was carried out in the presence of increasing amounts of E.coli SSB using 16.7 nM DnaB, and poly(dT):oligo(dA)35–(dC)6 as a substrate. Increased DNA unwinding corresponds to increased amounts of liberated [α-32P]oligo(dA)35–(dC)6. Lane 1, native substrate; lane 2, heat-denatured substrate; lane 3, no SSB; lane 4, 35 nM SSB; lane 5, 70 nM SSB; lane 6, 140 nM SSB; lane 7, 0.35 µM SSB; lane 8, 0.7 µM SSB.
Demonstration that poly(dT):oligo(dA)35–(dC)15 is a substrate for the DNA helicase activity of DnaB is presented as an inset to Figure 6. Data indicate that SSB stimulated DNA helicase activity in a dose-dependent manner. Consequently, with this forked substrate, SSB did not inhibit ATPase activity and stimulated the DNA helicase activity in a dose-dependent manner. Inhibition by E.coli SSB of ATPase activity is observed only with non-helicase substrates such as ssDNA and not with forked helicase substrates, which appeared to correlate well with the known function of the DnaB helicase in the replication fork.
DISCUSSION
DnaB helicase plays a pivotal role as a DNA helicase in the initiation of DNA replication, as well as in the organization and translocation of the replisome during replication of the genomes of E.coli and bacteriophage λ (1–2,8). Earlier studies have demonstrated that DnaB helicase requires a preformed fork structure for initiation of DNA unwinding (2). According to the model of Bramhill and Kornberg (24), DnaB protein initiates DNA unwinding in a small region of DNA that is partially melted by DnaA protein at the OriC origin of replication in E.coli. Consequently, we have systematically analyzed the role(s) of E.coli SSB in helicase action and the structural requirement of the forked substrates.
A preformed 3′ fork in the helicase substrate can efficiently stimulate the DNA helicase activity of DnaB protein
The results presented here demonstrate that DnaB helicase can unwind blunt-ended DNA substrates, however, only at a substantially reduced rate and efficiency as compared with an identical substrate with a forked structure at its end (Figs 1 and 2). Substrates containing a much larger duplex region may display different kinetics of strand displacement due to other kinetic factors such as the requirement for high processivity and rapid re-annealing of the unwound DNA, etc. The presence of a small fork, created by a 5 bp mismatch can enhance the rate of unwinding several fold (Fig. 1C). Comparison of two substrates containing a 35 bp identical duplex region with forks at either the 5′ or 3′ end indicated that the 5′ fork did not have measurable effects on the rate of DNA unwinding. This result was likely due to the fact that the polarity of migration of DnaB protein is 5′→3′ (2). Previous studies indicated that a long forked structure is probably a more desirable substrate for DnaB helicase. However, our results show that even a small preformed 3N-fork in the substrate can significantly stimulate helicase activity, and the rate was not observed to increase with increasing fork length beyond 5 nt (Fig. 2C) A preformed fork may enable more rapid complex formation with DnaB helicase and, in turn, lead to more rapid unwinding of the substrate. With a small substrate, a rapid recognition and complex formation would significantly influence the relative rate of unwinding, unlike that observed in much longer duplex substrates. Our results indicated that even a small single-stranded region, which DnaA protein could generate following binding to its recognition sequence, would likely be large enough for the initiation of DnaB helicase action.
Escherichia coli SSB inhibited DNA helicase-uncoupled ssDNA-dependent ATPase activity
In order to further understand modulation of the ssDNA-dependent ATPase activity of DnaB helicase, we have examined the effects of yRPA and gp32 on the ATPase activity of DnaB protein and compared them with E.coli SSB. Protein titrations of SSB, yRPA and gp32 versus DnaB ATPase activity (Fig. 5) showed that E.coli SSB had a significant effect on the ATPase activity of DnaB protein, which correlates well with the findings of Arai and Kornberg (3). Both yRPA and gp32 proteins inhibited the ATP hydrolysis only slightly (Fig. 5). Kinetic analysis of the inhibition indicated that E.coli SSB affects the initial rate of the reaction, and that over time, DnaB is unable to displace the bound SSB. This suggests that the inhibitory effect of SSB on the ssDNA-dependent ATPase activity of DnaB protein is not likely to be due to simple inhibition of DNA binding to DnaB protein by SSB. The inhibitory effect of E.coli SSB on the ATPase appears unique in light of the fact that it stimulates the helicase activity (2). One possible explanation may be that the basal level ATPase activity of DnaB helicase is high and that all of the energy is not needed for DNA unwinding and would thus be wasted. By binding to ssDNA, E.coli SSB reduces the basal ATPase activity of DnaB protein and appears to channel the hydrolysis of ATP.
Specific functional interaction between DnaB helicase and E.coli SSB
To further evaluate the functional interaction between DnaB and its cognate SSB, we have also examined the roles of heterologous SSBs in the helicase activity of DnaB. yRPA and gp32 protein are known to be involved in the replication of their genomic DNA and stimulation of their cognate DNA polymerases and helicases; thus sharing a common role in replication. All of the SSBs examined were capable of stimulating or inhibiting the helicase action depending upon the concentration range at which they were present (Fig. 3). gp32 and yRPA stimulated the helicase activity of DnaB protein several fold at concentrations below 0.56 µM and inhibited the helicase activity at higher concentrations suggesting a common mode of action. On the other hand, the E.coli SSB stimulated the helicase activity over a wider concentration range (0.01–5 µM) and, at higher concentrations (>5 µM), it inhibited the helicase activity. These differences may reflect the number of nucleotides bound per mole of protein as well as the binding affinity of the respective SSB. However, all of these SSBs appear to be able to stimulate DnaB helicase and have a common mechanism of action in altering the DNA structure for DnaB. The mechanism of inhibition of DnaB protein by these SSBs at higher concentrations would likely be due to exclusion of DnaB protein from the fork by an excess of the binding proteins.
Taken together, these results demonstrate that DnaB protein can effectively function with a substrate containing a small fork and with a variety of SSBs other than the endogenous E.coli SSB. Our studies suggest that E.coli SSB may not have any direct and specific interaction with DnaB helicase when it functions as a facilitator of helicase action by binding to the ssDNA region created by DNA unwinding. However, E.coli SSB appears to have a specific role in the modulation of the ssDNA-dependent ATPase activity of DnaB protein that cannot be substituted by other ssDNA-binding proteins such as yRPA or gp32. This attenuation of ATP hydrolysis may be important in channeling the energy of ATP during the replication process and may serve as a mechanism to reduce futile ATP hydrolysis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Ken-ichi Arai of the University of Tokyo for the plasmid pKA1, Dr Robert Nagele of this University and anonymous reviewers for critical review of this manuscript and Mr Stephen Flowers for PowerPoint illustrations. The authors gratefully acknowledge the support of this work by a grant from the National Institute of General Medical Sciences, National Institutes of Health (GM 36002-13).
REFERENCES
- 1.Funnell B., Baker,T.A. and Kornberg,A. (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem., 262, 10327–10334. [PubMed] [Google Scholar]
- 2.LeBowitz J.H. and McMacken,R. (1986) The Escherichia coli dnaB replication protein is a DNA helicase. J. Biol. Chem., 261, 4738–4748. [PubMed] [Google Scholar]
- 3.Arai K. and Kornberg,A. (1981) Mechanism of dnaB protein action. II. ATP hydrolysis by dnaB protein dependent on single- or double-stranded DNA. J. Biol. Chem., 256, 5253–5259. [PubMed] [Google Scholar]
- 4.Arai K. and Kornberg,A. (1981) Mechanism of dnaB protein action. III. Allosteric role of ATP in the alteration of DNA structure by dnaB protein in priming replication. J. Biol. Chem., 256, 5260–5266. [PubMed] [Google Scholar]
- 5.Arai K. and Kornberg,A. (1981) Mechanism of dnaB protein action. IV. General priming of DNA replication by dnaB protein and primase compared with RNA polymerase. J. Biol. Chem., 256, 5267–5272. [PubMed] [Google Scholar]
- 6.Arai K., Yasuda,S. and Kornberg,A. (1981) Mechanism of dnaB protein action. V. Association of dnaB protein, protein n′ and other prepriming proteins in the primosome of DNA replication. J. Biol. Chem., 256, 5273–5280. [PubMed] [Google Scholar]
- 7.Biswas S.B. and Biswas,E.E. (1987) Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lambda P gene products. J. Biol. Chem., 262, 7831–7838. [PubMed] [Google Scholar]
- 8.Kornberg A. and Baker,T.A. (1992) In DNA Replication. Freeman, San Francisco, CA.
- 9.Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- 10.Runyon G.T., Wong,I. and Lohman,T.M. (1993) Overexpression, purification, DNA binding and dimerization of the Escherichia coli uvrD gene product (helicase II). Biochemistry, 32, 602–612. [DOI] [PubMed] [Google Scholar]
- 11.Biswas E.E., Chen,P. and Biswas,S.B. (1994) Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry, 33, 11307–11314. [DOI] [PubMed] [Google Scholar]
- 12.Biswas E.E. and Biswas,S.B. (1999) Mechanism of DNA binding by the DnaB helicase of Escherichia coli: analysis of the roles of domain γ in DNA binding. Biochemistry, 38, 10919–10928. [DOI] [PubMed] [Google Scholar]
- 13.Biswas E.E. and Biswas,S.B. (1999) Mechanism of DnaB helicase of Escherichia coli: structural domains involved in ATP hydrolysis, DNA binding and oligomerization. Biochemistry, 38, 10929–10939. [DOI] [PubMed] [Google Scholar]
- 14.Biswas E.E., Biswas,S.B. and Bishop,J.E. (1986) The dnaB protein of Escherichia coli: mechanism of nucleotide binding, hydrolysis and modulation by dnaC protein. Biochemistry, 25, 7368–7374. [DOI] [PubMed] [Google Scholar]
- 15.Matson S.W. (1991) DNA helicases of Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol., 40, 289–326. [DOI] [PubMed] [Google Scholar]
- 16.Bujalowski W., Klonowska,M.M. and Jezewska,M.J. (l994) Oligomeric structure of Escherichia coli primary replicative helicase DnaB protein. J. Biol. Chem., 50, 31350–31358. [PubMed] [Google Scholar]
- 17.Bujalowski W. and Klonowska,M.M. (1993) Negative cooperativity in the binding of nucleotides to Escherichia coli replicative helicase DnaB protein. Interactions with fluorescent nucleotide analogs. Biochemistry, 22, 5888–5900. [DOI] [PubMed] [Google Scholar]
- 18.Dornreiter I., Erdlie,L.F., Gilberty,I.U., von Winkler,D., Kelly,T.J. and Fanning,E. (1992) Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen.EMBO J., 11, 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erdile L.F., Heyer,W.-D., Kolodner,R.D. and Kelly,T.J. (1991) Characterization of a cDNA encoding the 70 kDa single stranded DNA binding subunit of human replication protein A and the role of the protein in DNA replication. J. Biol. Chem., 266, 12090–12098. [PubMed] [Google Scholar]
- 20.Heyer W.D. and Kolodner,R.D. (1989) Purification and characterization of a protein from Saccharomyces cerevisiae that binds tightly to single-stranded DNA and stimulates a cognate strand exchange protein. Biochemistry, 28, 2856–2862. [DOI] [PubMed] [Google Scholar]
- 21.Heyer W.D., Rao,M., Erdile,L.F., Kelly,T.J. and Kolodner,R.D. (1990) An essential Saccharomyces cerevisiae single stranded DNA binding protein is homologous to the large subunit of human RPA. EMBO J., 9, 2321–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim C., Snyder,R.O. and Wold,M.S. (1992) Binding properties of replication protein A from Human and yeast cells. Mol. Cell. Biol., 12, 3050–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkatesan M., Silver,L.L. and Nossal,N.G. (1982) Bacteriophage T4 gene 41 protein, required for the synthesis of RNA primers, is also a DNA helicase. J. Biol. Chem., 257, 12426–12434. [PubMed] [Google Scholar]
- 24.Bramhill D. and Kornberg,A. (1988) A model for initiation at origins of DNA replication. Cell, 52, 743–755. [DOI] [PubMed] [Google Scholar]
- 25.Jezewska M.J., Rajendran,S. and Bujalowski,W. (1998) Complex of Escherichia coli primary replicative helicase DnaB with a replication fork: recognition and structure. Biochemistry, 37, 3116–3136. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan D.L. and Steitz,T.A. (1999) DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J. Biol. Chem., 274, 6889–6897. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 28.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]