Abstract
Transcription factor IIIA (TFIIIA) is specifically required for transcription of 5S rRNA genes and is the archetypal C2H2 zinc finger protein. All known vertebrate TFIIIAs have a similar organization: nine zinc fingers, followed by a C-terminal domain of unknown structure. The zinc fingers of Saccharomyces cerevisiae TFIIIA are interrupted between fingers eight and nine by an 81-amino acid spacer. Aside from the amino acids required for zinc finger folding, TFIIIAs from different species are remarkably divergent, whereas the natural binding site, the internal control region of the 5S rRNA gene, is well conserved. We now describe the identification and characterization of TFIIIA from Schizosaccharomyces pombe. This protein is organized differently from its known homologs, in that it contains eight closely spaced zinc fingers, a ninth zinc finger missing a C-terminal Zn2+-coordinating histidine, a 53- amino acid spacer, and an unprecedented tenth zinc finger. We have confirmed the identity of this divergent protein as TFIIIA by showing that it binds specifically and with high affinity to the S.pombe 5S rRNA gene. Comparison of DNase I protection patterns produced by TFIIIA from multiple species suggests a novel mode of DNA recognition by the S.pombe protein. Recombinant S.pombe TFIIIA was also shown to support specific transcription of the 5S rRNA gene in vitro.
INTRODUCTION
Transcription of 5S rRNA genes by RNA polymerase III is dependent upon a gene-specific transcription factor that binds to the internal control region (ICR) (1,2) of these genes. Transcription complex assembly is initiated by the binding of this factor, transcription factor IIIA (TFIIIA), to the gene and is completed by the binding of the multi-subunit factors TFIIIC, TFIIIB and RNA polymerase III (3–5). The DNA-binding moiety of TFIIIA consists of multiple consecutive C2H2 zinc fingers, a motif originally defined on the basis of its repeated occurrence in Xenopus laevis TFIIIA (6,7). Although it is the archetypal example of a zinc finger protein, X.laevis TFIIIA is atypical in that it contains an unusually large number of tandemly repeated zinc finger domains (nine), and binds to a particularly large segment of double-stranded DNA. TFIIIA from X.laevis binds not only to the 5S rRNA gene, but also to 5S rRNA, the gene product. Through this mechanism, it participates in feedback inhibition of 5S rRNA synthesis (8,9) and participates in a network of interactions that couple 5S rRNA synthesis to ribosomal protein accumulation (10). The structure, nucleic acid-binding capabilities and transcriptional activity of X.laevis TFIIIA have been studied extensively by our group and others. In addition, Saccharomyces cerevisiae TFIIIA has been analyzed in some detail.
Analysis of TFIIIA sequences from several vertebrate species, including human, catfish, mouse, rat and five species of frogs (11–17), has revealed remarkably poor conservation of primary sequence. In fact, we have estimated a divergence rate of 1.5–3 amino acid changes per residue per billion years for TFIIIA during the evolution and diversification of vertebrates (data not shown). This rate of evolutionary change is high when compared with other zinc finger proteins, other transcription factors or proteins in general (18,19). The rapidity of the rate is particularly surprising given the relatively high degree of sequence conservation observed for the TFIIIA-binding site in 5S rRNA genes (20) (∼58% identity between Schizosaccharomyces pombe and X.laevis), and the fact that dual recognition of 5S rRNA genes and 5S rRNA might have been expected to place additional constraints on TFIIIA’s sequence. Despite variation in primary sequence, however, vertebrate homologs of TFIIIA possess a similar structural organization consisting of nine closely spaced zinc fingers followed by a C-terminal domain of indeterminate structure. A region within the C-terminal domain of X.laevis TFIIIA has been shown to be required for the support of transcription of 5S rRNA genes (21).
The organization of the X.laevis and human TFIIIA genes suggests that the repeated zinc finger motif resulted from duplication of exons encoding individual zinc fingers (11,13,22) (analysis of the human gene performed using BLAT at www.genome.ucsc.edu). In the TFIIIA genes of both species, each of the first six zinc fingers is encoded by an individual exon, while the seventh, eighth and first half of the ninth finger are encoded by a seventh exon. The second half of finger nine and the C-terminal domain are encoded by an eighth exon. Presumably, ancestral introns separating exons encoding individual C-terminal zinc fingers have been lost subsequent to motif duplication. In fact, it is possible that additional zinc fingers present in an ancestral protein have been lost in the vertebrate lineage. Brown et al. (23) have even argued that remnants of three additional zinc fingers can be identified in X.laevis TFIIIA.
TFIIIA from S.cerevisiae has diverged so significantly from the vertebrate proteins that little sequence conservation can be discerned other than at the seven residues in each zinc finger that are important for maintaining the conserved zinc finger fold. The protein contains nine zinc fingers, as do the vertebrate proteins, but also includes a unique 81-amino acid spacer region between fingers eight and nine (24,25). This region includes a leucine-rich oligopeptide that is required for transcription of 5S rRNA genes (26). A 40-amino acid C-terminal region that is dispensable for transcriptional activity follows finger nine (27).
Despite its considerable divergence in sequence and structural organization, TFIIIA appears to function similarly in various vertebrate species and in S.cerevisiae. Therefore, it is of interest to determine if the rapid evolutionary divergence of TFIIIA has resulted in additional categories of structural organization in other distantly related eukaryotic species. To this end, we have identified and characterized TFIIIA from S.pombe, fission yeast, and show that it possesses a number of interesting features that clearly distinguish it from its homologs in vertebrates and S.cerevisiae.
MATERIALS AND METHODS
Isolation of an S.pombe TFIIIA cDNA and purification of recombinant protein
A cDNA encoding the putative S.pombe TFIIIA homolog was cloned for bacterial expression using PCR amplification from an S.pombe cDNA library constructed in plasmid pGAD-GH (provided by G. Hannon, Cold Spring Harbor Laboratory). The oligonucleotide primers were designed based on sequences in the S.pombe genome database. The 5′ primer (TAACCTACATGTGTCATTTCAATG) included an AflIII restriction site (A/CRYGT) surrounding the ATG start codon to permit ligation to a compatible NcoI site in the expression vector. The 3′ primer (TTGCTTGGATCCTTATTATGAAGAGAAGCT) annealed 90 nt downstream of the stop codon and introduced a BamHI site. The complete coding sequence of TFIIIA was obtained in a single PCR and was cloned between the NcoI and BamHI sites of the expression vector pET-11D (Novagen). The insert was sequenced (accession no. AY091590) and the plasmid was named pET-SP3A.
Schizosaccharomyces pombe TFIIIA was subsequently expressed and purified using methods established for production of recombinant TFIIIA from X.laevis (28). In short, pET-SP3A was transformed into Escherichia coli BL21 (DE3) cells, and TFIIIA expression was induced with IPTG. Cells were lysed and soluble material removed by low speed centrifugation. The insoluble recombinant TFIIIA was solubilized from the pellet with 5 M urea, and purified with two sequential ammonium sulfate precipitations followed by BioRex70 (BioRad) ion-exchange column chromatography. TFIIIA was eluted in a 1 M NaCl step following an initial wash in a 0.25 M NaCl-containing buffer.
Cloning of an S.pombe 5S rRNA gene
A clone of the S.pombe 5S rRNA gene, including 130 nt of sequence upstream of +1 and 140 nt downstream of the 5S rRNA coding sequence, was constructed using PCR amplification from genomic DNA. Primers were designed to anneal to sequences flanking the known 5S rRNA gene (29), adding a HindIII site at the 5′ end (TCAATAAAGCTTCATCTC) and including a naturally occurring EcoRI site at the 3′ end (GTTAATAGAATTCGGGCTAA). The resulting fragment was cloned between the HindIII and EcoRI sites of plasmid pGP12 (30). The insert was sequenced and the plasmid was named pGP-SP5S.
Gel mobility shift assay
The 390-bp HindIII–EcoRI fragment of plasmid pGP-SP5S was gel purified and labeled with [α-32P]dATP using the Klenow fragment of DNA polymerase I. DNA-binding reactions were carried out as described for X.laevis TFIIIA (31). Reactions contained fixed concentrations of recombinant TFIIIA and labeled probe and variable concentrations of unlabeled DNA in 20 mM Tris–HCl (pH 7.5), 70 mM KCl, 7 mM MgCl2, 10 µM ZnCl2, 1 mM dithiotheitol, 10% glycerol, 100 µg/ml bovine serum albumin and 10 µg/ml poly(dI–dC). Unlabeled DNA referred to as ‘specific’ (see Fig. 2) is the HindIII–EcoRI fragment of pGP-SP5S, while ‘non-specific’ refers to a 393-bp NlaIII fragment from pGP-SP5S that does not contain 5S rDNA sequence. Reaction mixtures were incubated for 30 min at 30°C and loaded on a pre-run (30 min, 400 V) non-denaturing polyacrylamide gel, while running, at 4°C. Electrophoresis was for 3 h at 400 V, and gels were dried and scanned on an Ambis Radioanalytic Imaging System for quantification.
Figure 2.
Sequence-specific DNA binding by putative S.pombe TFIIIA. A gel mobility shift assay was used to show specificity of DNA binding. Increasing concentrations of unlabeled 5S rDNA (A) or non-specific competitor (B) were added to binding reactions including labeled 5S rDNA and TFIIIA. (C) Graphical representation of the binding data. The curves shown were generated by fitting parameters for specific and non-specific equilibrium binding constants to the data.
Determination of equilibrium binding constant
A series of reaction mixtures containing a constant concentration of TFIIIA and variable concentrations of a 390-bp DNA fragment containing the S.pombe 5S rRNA gene were incubated for 30 min at 30°C and analyzed by gel mobility shift assay as described above. Concentrations of bound and free DNA in each lane were quantified and a Kd determined using Scatchard analysis (32). Data from multiple determinations were combined and used to determine a best estimate of the Kd and an associated standard error using analysis of covariance. Details of the method of analysis are described elsewhere (31).
Determination of rate constant for dissociation
A single large-scale binding reaction was prepared as above and incubated for 30 min at 30°C. This equilibrated binding reaction was then mixed with pre-incubated trap DNA, consisting of double-stranded oligonucleotide containing the S.pombe 5S rRNA gene ICR (SPICR-T, non-template strand from +38 to +95, TCCGATCACTGCAGTTAAGCGTCTGAGGGCCTCGTTAGTACTATGGTTGGAGACAACA annealed to SPICR-B, template strand from +44 to +100, TCCCATGTTGTCTCCAACCATAGTACTAACGAGGCCCTCAGACGCTTAACT-GCAGTG). The time at which the equilibrated binding reaction mixture and trap were combined was defined as t = 0. A single control reaction, including trap, was pre-incubated to ensure that sufficient trap was included in the reaction to prevent re-binding of TFIIIA upon dissociation from the labeled probe. At various time intervals, aliquots of the large-scale reaction were loaded on to a pre-run, non-denaturing polyacrylamide gel, while running, at 4°C. The gel was run, dried and quantified as described above. The fraction of DNA bound at various time points (Bt) was determined. The rate constant (koff) for complex dissociation was obtained by fitting the data to the equation Bt = Ae–kt + C, where Bt is the fraction of DNA bound at time t, and A, C and k are fitted parameters. k is the dissociation rate constant (koff). The reported koff for the S.pombe TFIIIA/5S rRNA gene complex is the average of four independent determinations. Half-life was calculated using the equation t1/2 = (ln 2)/koff. Details of the method are described elsewhere (31).
DNase I protection assay (footprint)
A 5′ end-labeled PCR fragment, including 5S rDNA sequence from –63 to +185, was generated using pGP-SP5S as template. The 5′ oligonucleotide (ATAACATTAGAGTATCTC) included in the reaction mixture was labeled on the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase for footprinting of the non-template strand, while the 3′ primer (CACAAACCCAATGAGTGC) was labeled similarly for footprinting of the template strand. Saturating amounts of TFIIIA were added to a fixed amount of probe under conditions described above. After 30 min, an aliquot of the reaction mixture was loaded on a non-denaturing gel at 4°C to confirm the absence of unbound probe in the reaction mixture. The remainder of the reaction was treated with 1.5 U DNase I (Gibco/BRL) for 5–10 s. Reactions were stopped using 150 mM NaCl, 5 mM EDTA, 0.5% SDS, 50 mM Tris pH 8.0, with 50 ng/µl tRNA as carrier (SETS/tRNA). Nucleic acids were extracted, precipitated with ethanol, and redissolved in 90% formamide, 20 mM EDTA, 0.05% bromphenol blue and 0.05% xylene cyanol. Samples were analyzed on an 8% polyacrylamide, 7 M urea gel run at 1300 V for 4 h. The gel was dried and exposed to a phosphor storage screen overnight. The screen was scanned and the data analyzed using Image Quant software (Molecular Dynamics).
Schizosaccharomyces pombe extract preparation
Extract was prepared using a modification of a protocol developed by Schultz et al. (33) and described previously (36). In short, wild-type (strain 972h–) S.pombe cells were grown to mid-log phase in 2 l of YEL media (5 g yeast extract and 30 g dextrose per liter), pelleted, washed twice (once without and once with protease inhibitors) in 9 ml extraction buffer [100 mM HEPES pH 7.9, 245 mM KCl, 5 mM EGTA, 1 mM EDTA, 2.5 mM DTT (added fresh), and 30 µl/ml Sigma protease inhibitor cocktail (mammalian cell P8340)]. The cells were extruded from a syringe into liquid nitrogen and then ground extensively under liquid nitrogen with a mortar and pestle. These broken cells were then spun at 4°C at 36 000 r.p.m. in a Ti70 ultracentrifuge rotor for 2 h (100 000 g). The supernatant was removed, carefully avoiding the lipid layer floating on the surface, and was diluted with 2 vol of 60% glycerol, 5 mM EGTA, 0.05 mM EDTA, 2.5 mM DTT and brought to a final KCl concentration of 100 mM. For preparation of whole cell extract, the supernatant was dialyzed rather than diluted at this point (see below).
To remove endogenous TFIIIA activity, the whole cell extract was fractionated on a phosphocellulose (Whatman P11) column. The phosphocellulose resin was equilibrated in 100 mM KCl, 20 mM HEPES pH 7.9, 5 mM EGTA, 0.05 mM EDTA, 2.5 mM DTT. The sample was loaded, the column washed with the same buffer, and fractions of a high salt elution (0.6 M KCl) were collected. Peak fractions were combined and dialyzed in a 3500 MWCO slide-a-lyzer (Pierce) against 20 mM HEPES pH 7.9, 100 mM KCl, 5 mM EGTA, 0.05 mM EDTA, 20% glycerol, 2.5 mM DTT and 0.2 mM PMSF.
In vitro transcription assay
Transcription reactions were performed at 30°C in a total volume of 20 µl with 200 ng (4.1 nM) pGP-SP5S as template, with 0.6 mM CTP, 0.6 mM UTP, 0.6 mM ATP, 0.05 mM GTP, 2 µCi [α-32P]GTP (3000 Ci/mmol), in 20 mM HEPES pH 7.9, 10% glycerol, 10 µM ZnCl2, 0.1 mM BSA, 2.5 mM MgCl2, 0.4 mM DTT, and 3 µl of the 0.6 M P11 fraction (amount of P11 fraction used was preparation specific). The monovalent salt concentration (NaCl/KCl) was held constant in each reaction at 100 mM (significant salt is carried over from the recombinant TFIIIA preparation). TFIIIA was added at variable concentrations. Reactions were stopped in SETS/tRNA (see above), extracted, precipitated, and run on a denaturing gel as above. The dried gel was analyzed on a PhosphorImager using Image Quant software (Molecular Dynamics).
RESULTS
Periodic searching of the S.pombe genome sequence database (37), using X.laevis or S.cerevisiae TFIIIA as a query sequence in a TBLASTN search, resulted in the identification of an open reading frame (ORF) encoding a multi-zinc finger protein (accession no. AL13265). The overall sequence identity of the protein encoded by this ORF to known TFIIIAs from vertebrate species or S.cerevisiae was low. Nonetheless, this ORF was the closest match for TFIIIA in the S.pombe genome. Because of the large evolutionary distance between S.pombe and either S.cerevisiae or vertebrates, we expected S.pombe TFIIIA to be significantly diverged from other known TFIIIAs. Therefore, we considered this divergent ORF a likely candidate to encode S.pombe TFIIIA.
Sequence similarity between putative S.pombe TFIIIA and TFIIIA from either X.laevis or S.cerevisiae is dominated by the seven residues found in each zinc finger that are required for proper folding (Fig. 1). These include three conserved hydrophobic residues, two Zn2+-coordinating cysteines and two Zn2+-coordinating histidines. Spacing between these residues is also conserved [consensus sequence, (F/Y-X-C-X(1–5)-C-X3-F/Y-X5-L-X2-H-X(3–4)-H, X = any residue)]. Excluding consensus Zn2+-coordinating and hydrophobic residues present in all zinc fingers, however, there is only 15% sequence identity between putative S.pombe TFIIIA and either X.laevis or S.cerevisiae TFIIIA.
Figure 1.
Sequence alignment of TFIIIA from X.laevis, S.pombe and S.cerevisiae. Sequences were aligned manually to show the conservation of the structural residues in each zinc finger and to illuminate the differences in sequence and spacing in each case. Note that alignment of the S.cerevisiae sequence with finger nine from X.laevis and S.pombe is an artificial designation, shown only for illustration. We have defined the C-terminus of a zinc finger as the second Zn2+-coordinating histidine residue or, in the case S.pombe finger nine, the glycine residue found at the equivalent position. This differs from the convention used by Archambault et al. (24) and results in the S.cerevisiae and S.pombe spacers being six amino acids longer here than as described previously and in the text. Residue number is shown to the right of each line of sequence. Conserved residues that constitute the zinc finger consensus motif (see Results) are highlighted in yellow, and residues shaded in gray are conserved in at least two of the three proteins shown here.
The TFIIIA homolog from S.pombe includes eight closely spaced C2H2 zinc fingers, followed by a ninth, probable zinc finger motif that lacks the conserved C-terminal histidine. A cysteine is found six residues downstream of the first Zn2+-coordinating histidine, however. This motif variation has been noted in a small number of putative zinc fingers in the S.cerevisiae genome (38), but has yet to be demonstrated to mediate proper folding. A zinc finger missing the C-terminal histidine has not been observed in any other TFIIIA for which sequence information is available, although a likely candidate for Drosophila TFIIIA found in the genome sequence database appears to have a similar variant finger (gene CG9609) (39) (data not shown).
Following the unusual ninth finger, the S.pombe protein contains a 53-amino acid spacer region lacking known sequence motifs, followed by a tenth consensus zinc finger. All other characterized TFIIIAs contain just nine zinc fingers. Vertebrate TFIIIAs contain a variable length (49–68 amino acid) non-zinc finger region at the C-terminus, and an 81-amino acid spacer is located between fingers eight and nine of S.cerevisiae TFIIIA (24,25). These regions have no primary sequence similarity to one another, or to the spacer region in the S.pombe protein. Sequences within these non-zinc finger regions, however, have been shown to be necessary for support of 5S rRNA synthesis in both the X.laevis (21) and S.cerevisiae (26) systems. Interestingly, the size of the S.pombe spacer plus the non-canonical finger nine that precedes it is approximately equal to the size of the spacer region in the S.cerevisiae protein.
Cloning and expression of S.pombe TFIIIA
A cDNA encoding putative S.pombe TFIIIA was obtained from a cDNA library using PCR amplification, sequenced, and used for expression of the protein in E.coli. Annotations in the S.pombe genome database indicated that the TFIIIA gene includes a 42-bp intron between the sequences encoding zinc fingers one and two. The sequence of our newly obtained cDNA clone (accession no. AY091590) confirmed that the predicted intron sequence was absent from the encoded mRNA; the remainder of the coding sequence was identical to that in the genome database. Recombinant protein was successfully purified using methods previously developed for the purification of X.laevis TFIIIA (see Materials and Methods) and has an electrophoretic mobility consistent with the 43 kDa molecular weight predicted from the primary sequence (data not shown).
Sequence-specific DNA-binding activity
The activities of TFIIIA include specific binding to the 5S ribosomal RNA gene (5S rDNA) and support of transcription of the gene. Gel mobility shift assays were used to demonstrate that the recombinant protein does indeed possess specific 5S rDNA-binding activity (Fig. 2). Binding of the protein to the probe can be competed by the addition of unlabeled 5S rDNA, but not by addition of non-specific DNA (see Materials and Methods), confirming that the DNA binding observed is sequence specific.
DNase I protection
In order to investigate the specific manner in which S.pombe TFIIIA interacts with its binding site, a DNase I protection assay was used (Fig. 3). Saturating amounts of protein were added to a radioactively end-labeled DNA fragment containing the 5S rRNA gene and the complexes were treated with DNase I. We confirmed that virtually all of the probe molecules were bound by protein prior to DNase I digestion by running an aliquot of the reaction mixture on a non-denaturing gel and observing little or no unbound probe in the mixture (data not shown). Protection patterns on both the template and non-template strands were assessed.
Figure 3.
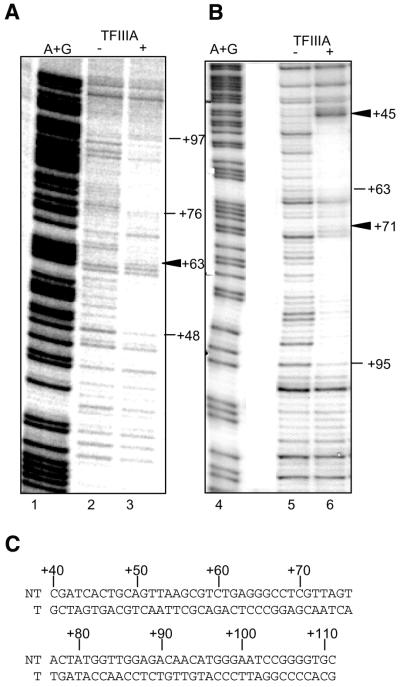
DNase I protection analysis of the S.pombe 5S rRNA gene. Saturating concentrations of S.pombe TFIIIA were used. Lanes 1 and 4 contain A + G Maxam–Gilbert sequencing reactions using the labeled DNA fragment. The DNase I digestion products in the absence of protein are in lanes 2 and 5 and the digestion products in the presence of TFIIIA are in lanes 3 and 6. Numbers to the right of each gel designate the position of DNase I cleavage [relative to the start of transcription (+1)] necessary to generate the indicated band. Triangles designate positions of hypersensitivity to DNase I cleavage. (A) Non-template strand was labeled. (B) Template strand was labeled. (C) Sequence of the S.pombe 5S rRNA gene from +40 to +111. NT, non-template strand; T, template strand.
The protein protects a region of the DNA from position +45 to +95, relative to the start site for transcription (+1), on the template strand (Fig. 3B) and from +48 to +97 on the non-template strand (Fig. 3A). There is an unprotected region in the center of the footprint from +63 to +73 on the template strand, and +61, +62 and +70 to +76 are unprotected on the non-template strand. DNase I hypersensitive sites are induced at position +63 on the non-template strand and at +45 and +71 on the template strand upon TFIIIA binding.
Further characterization of the protein–DNA interaction
The affinity of TFIIIA for its binding site, the 5S rRNA gene, was assessed quantitatively. Increasing amounts of unlabeled competitor 5S rDNA were added to reaction mixtures containing constant amounts of TFIIIA and labeled DNA. The amounts of bound and free DNA were quantified using a gel mobility shift assay and the equilibrium binding constant (Kd) for the interaction was determined as described in Materials and Methods and elsewhere (31,40). We measured a Kd of 0.19 nM (SE ±0.03) for the interaction between S.pombe TFIIIA and 5S rDNA. This value is roughly comparable with those reported previously for the binding of X.laevis or S.cerevisiae TFIIIA to 5S rDNA (40,41). Precise values for each of these systems are presented in Table 1.
Table 1. Equilibrium and kinetic constants for DNA binding by TFIIIA.
| Species | Kd (nM) | ΔG (kcal/mol) | Temperature (°C) | koff (s–1) | t1/2 (min) |
|---|---|---|---|---|---|
| Xenopus laevis | 0.42 (±0.03)a,b | –12.8 | 25 | 0.00197 (±0.00018)c,d | 5.9 |
| Schizosaccharomyces pombe | 0.19 (±0.03)a | –13.5 | 30 | 0.00148 (±0.00021)c | 7.8 |
| Saccharomyces cerevisiae | 0.11 (±0.03)e | –13.6 | 25 | ND | ND |
The kinetics of the dissociation of the TFIIIA–5S rDNA complex was also analyzed. The rate of dissociation of the complex was determined to be 0.00148 (±0.00021) s–1 with a corresponding half-life of 7.8 min, compared with a half-life of 5.9 min measured for the X.laevis TFIIIA–DNA complex (42; M.Bumbulis, S.Ponnampalam, K.Brady and D.R.Setzer, manuscript in preparation).
Transcriptional activity of putative S.pombe TFIIIA
To test the ability of putative S.pombe TFIIIA to support transcription of the 5S rRNA gene, an in vitro transcription system from S.pombe was developed. A whole cell extract was prepared from S.pombe, based upon a method developed for the analysis of the S.pombe RNA polymerase I machinery (33–35) and used previously to study the transcription of the S.pombe 7S L gene (36). This extract is capable of transcribing a plasmid-borne 5S ribosomal RNA gene in vitro, with or without added recombinant TFIIIA (Fig. 4), at an efficiency of approximately 0.5 transcripts per template per 2 h. The transcript generated migrates close to the position of authentic X.laevis 5S rRNA, which is 120 nt long. The additional band generated in these reactions is likely to be a post-transcriptionally labeled tRNA molecule (R. Maraia, personal communication).
Figure 4.
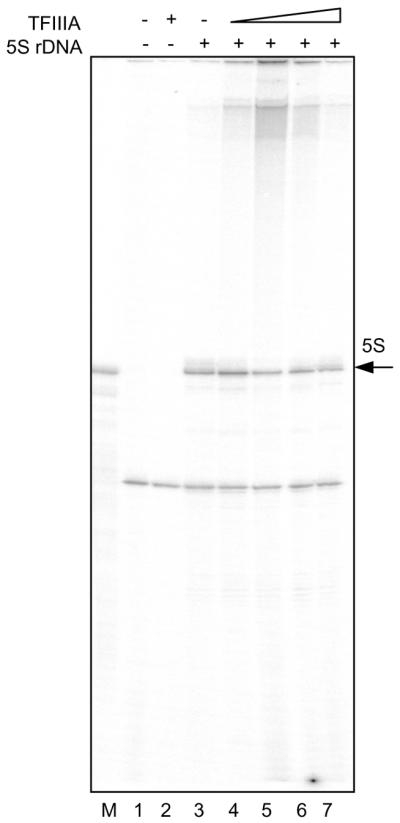
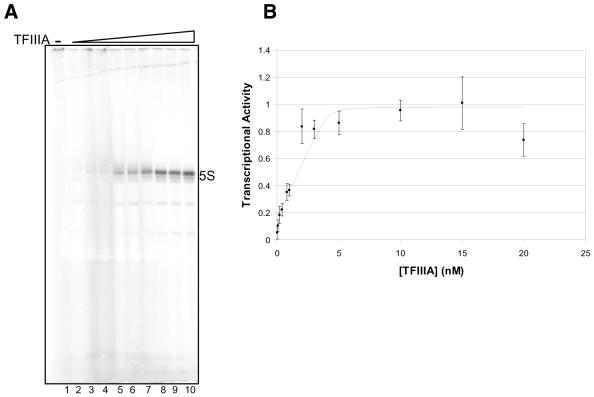
Schizosaccharomyces pombe 5S rRNA gene transcription in whole cell extract. Reactions were performed without (lanes 1 and 2) or with (lanes 3–7) added plasmid-borne 5S rDNA template and without (lanes 1 and 3) or with (lanes 2, 4–7) added recombinant TFIIIA. The reactions in lanes 4–7 included increasing concentrations (5.6, 11.2, 22.3 and 33.5 nM respectively) of TFIIIA. Lane M contains Xenopus 5S rRNA as a marker.
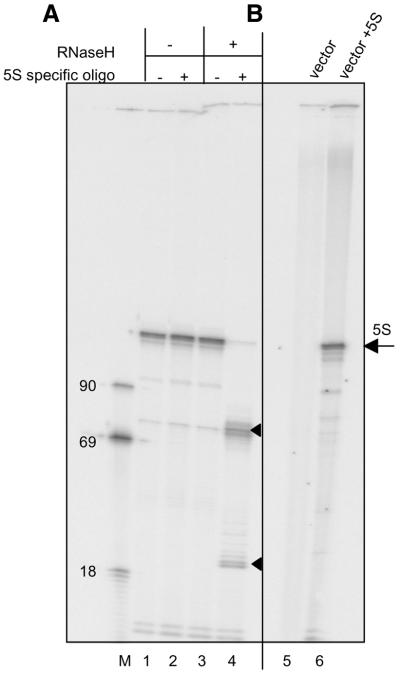
We further confirmed that the 120-nt transcript is in fact 5S rRNA using oligonucleotide-directed RNase H cleavage (Fig. 5A). An oligonucleotide was designed to anneal to 5S rRNA within structurally conserved loop C (positions +33 to +44) and was expected to direct cleavage of the transcript by RNase H into fragments of ∼30 and ∼70 nt. The RNA was specifically cleaved to generate the expected two fragments only in the presence of the oligonucleotide and RNase H. The transcript was also shown to be generated only when a 5S rRNA gene sequence was present in the plasmid template (Fig. 5B). In addition, we demonstrated that generation of the transcript is sensitive to the transcriptional inhibitor α-amanitin at concentrations previously observed to inhibit synthesis of S.pombe tRNA in a differently prepared in vitro transcription system (50% inhibition at ∼0.5 mg/ml) (43) (data not shown).
Figure 5.
Authenticity of the in vitro S.pombe 5S rRNA gene transcription product. (A) Lane M, size markers. Lanes 1–4, 5S rRNA was synthesized in TFIIIA-supplemented 0.6 M P11 fraction and incubated without (lanes 1 and 3), or with (lanes 2 and 4) a 5S rRNA-specific oligonucleotide and without (lanes 1 and 2), or with (lanes 3 an 4) RNAse H treatment. Triangles designate positions of RNAse H cleavage products. (B) Transcription reactions performed in TFIIIA-supplemented extract with vector only (lane 5) or with vector including the 5S rRNA gene (lane 6).
Analysis of 5S rRNA synthesis in the extract as a function of time revealed the existence of a lag of ∼1 h before any appreciable product accumulation could be detected. Similar, though not as lengthy, lags in activity have been described for in vitro transcription by RNA polymerase III in crude extracts from X.laevis (4,44). Products accumulated almost linearly for at least 3 h after the initial lag, from approximately 0.03 transcripts per template at 1 h to 1.5 transcripts per template at 4 h (data not shown). Further experiments were done at a fixed incubation time of 2 h.
In order to assay the activity of recombinant TFIIIA in this system, it was necessary to remove endogenous TFIIIA activity from the whole cell extract. To this end, the extract was fractionated on phosphocellulose, using conditions similar to those used previously to fractionate factors involved in RNA polymerase III-dependent transcription in other systems (45–47). The transcriptional activity of the 0.6 M P11 fraction was tested for 5S rDNA- and TFIIIA-dependence. We showed that the 120 nt RNA was produced only in the presence of recombinant TFIIIA (Fig. 6) and of a plasmid containing an S.pombe 5S rRNA gene (data not shown).
Figure 6.
Activity of recombinant S.pombe TFIIIA in in vitro transcription assay. (A) Transcription reactions were performed with the 0.6 M P11 fraction, supplemented with recombinant TFIIIA. All reactions included plasmid-borne 5S rDNA template. The reaction in lane 1 contained no added TFIIIA and those in lanes 2–10 contained increasing amounts of TFIIIA. (B) Quantification and graphical presentation of the data from several experiments similar to that shown in (A). Error bars present the standard error of the mean for measurements of transcriptional activity made in multiple indepndent experiments. The curve presented was generated using non-linear curve-fitting methods that will be described in detail elsewhere (D.R.Setzer and D.B.Schulman, manuscript in preparation.
Conditions for transcription were optimized with respect to monovalent salt and magnesium concentration. Optimal activity was observed at 2.5 mM MgCl2 and 100 mM monovalent salt. Activity is quite sensitive to deviations from these optimal salt concentrations. At 5 mM MgCl2, activity was reduced by ∼30% and by ∼85% at 7.5 mM. Similarly, transcriptional activity exhibits a surprisingly narrow monovalent salt optimum, with a decrease in activity of ∼90% at salt concentrations of either 50 or 150 mM (data not shown).
Reactions were performed at a range of TFIIIA concentrations with a fixed amount of 0.6 M P11 fraction to determine if transcriptional activity is sensitive to increasing amounts of added TFIIIA. The results were quantified and the number of transcripts per input template molecule was determined. At high concentrations of recombinant protein, transcriptional activity reached a plateau, corresponding to an efficiency of approximately 0.2 transcripts per template per 2 h. Thus, in these reactions, a component of the transcriptional apparatus other than TFIIIA is limiting. This component is unlikely to be the template, as adding additional template beyond the 4.1 nM concentration used in these experiments did not increase the transcriptional activity (data not shown). Rather, it is likely that one of the protein components of the transcription complex or RNA polymerase III, included in the extract, is entirely sequestered in complexes in the presence of high concentrations of TFIIIA.
DISCUSSION
Despite considerable sequence divergence when compared with TFIIIAs from vertebrates or S.cerevisiae, it is clear from the results presented here that the protein we have characterized is indeed S.pombe TFIIIA. We have shown that the protein has specific 5S rDNA-binding activity, with an affinity and DNase I-protection pattern similar to those exhibited by previously characterized TFIIIAs from distantly related species. Additionally, we have shown that the recombinant form of the protein is capable of supporting 5S rDNA transcription in vitro.
Several novel features of TFIIIA from S.pombe add additional complexity to considerations of TFIIIA’s evolutionary ancestry. Most notably, S.pombe TFIIIA is unique among characterized homologs in that it contains 10, rather than nine, zinc finger motifs. Putative TFIIIA homologs have also been identified from genome sequencing efforts in the fungi Neurospora crassa [Neurospora Sequencing Project, Whitehead Institute/MIT Center for Genome Research (www-genome.wi.mit.edu)] and Aspergillus fumigatus (The Institute for Genomic Research website at http://www.tigr.org). Interestingly, a potential tenth zinc finger can be identified in both cases, although there are multiple deviations from consensus in several fingers. Thus, S.pombe TFIIIA may be representative of a larger sub-family of 10-finger-containing TFIIIA homologs. An interesting possibility is that the common ancestor of vertebrates, S.cerevisiae and S.pombe possessed a 10-finger version of TFIIIA similar in overall organization to that now observed in S.pombe. In this scenario, ancestral finger nine would have diverged extensively in the S.cerevisiae lineage, resulting in an extended spacer between ancestral fingers eight and 10, now considered to be fingers eight and nine. Similarly, finger 10 would have diverged in the vertebrate lineage to become part of the C-terminal non-finger region; alternatively, it may have been deleted entirely. Thus, S.pombe finger nine would be orthologous to vertebrate finger nine and to the first 30 or so amino acids of the S.cerevisiae spacer, although no vestige of sequence similarity remains to validate this hypothesis. Similarly, S.cerevisiae finger nine would be the ortholog of S.pombe finger 10 and the last part of the C-terminal tail of the vertebrate protein. Again, however, no remaining sequence similarity between the vertebrate C-terminal tail and any zinc finger can be discerned.
In principle, the evolutionary relationships between the zinc fingers in vertebrate, S.pombe and S.cerevisiae TFIIIAs might be revealed by inter-finger sequence comparisons. Unfortunately, the sequence divergence is so great that we have been unable to detect any convincing patterns of similarity when all possible pair-wise comparisons among the zinc fingers of the three proteins are made. Thus, the evolutionary scenario presented remains speculative, and establishing which of the 10 S.pombe zinc fingers is the ‘extra’ one in comparison with the nine fingers found in other species is likely to be difficult.
Schizosaccharomyces pombe TFIIIA includes several other deviations from the typical zinc finger consensus. Most significant among these is the absence of a C-terminal Zn2+-coordinating histidine residue in finger nine. We have shown previously that replacement of the C-terminal histidine with other residues in at least some zinc fingers of X.laevis TFIIIA produces adverse effects on DNA binding (48). Furthermore, one might expect that the absence of a C-terminal histidine would interfere with stable folding of the putative zinc finger domain, and that S.pombe finger nine would therefore not form a functional zinc finger. Although this may very well be the case, other considerations suggest that the C-terminal histidine might not be essential. Merkle et al. (49) have shown that model zinc finger peptides can fold even in the absence of a C-terminal histidine, presumably using a water molecule as the fourth Zn2+ ligand. It is also noteworthy that a cysteine residue is found six positions C-terminal to the first putative Zn2+-coordinating histidine, and Bohm et al. (38) have described the existence of a small number of such variant putative zinc finger motifs in the genome of S.cerevisiae. Although no variant zinc finger of this type has yet been studied biochemically, it is certainly possible that zinc finger nine of S.pombe TFIIIA is a functional member of this zinc finger domain subfamily.
Other unusual features of the zinc fingers of S.pombe TFIIIA include the absence of a highly conserved hydrophobic residue two residues before the first Zn2+-coordinating cysteine of finger seven, non-consensus spacing between the Zn2+-coordinating cysteines and histidines in finger six, and a very short spacer between fingers six and seven. Interestingly, the sixth zinc finger in S.cerevisiae and S.pombe share an identical heptapeptide (WSQLQ) preceding the first Zn2+-coordinating histidine. In fact, the initial tryptophan residue of this sequence is conserved in all characterized TFIIIAs and has been implicated in 5S rRNA recognition (50,51).
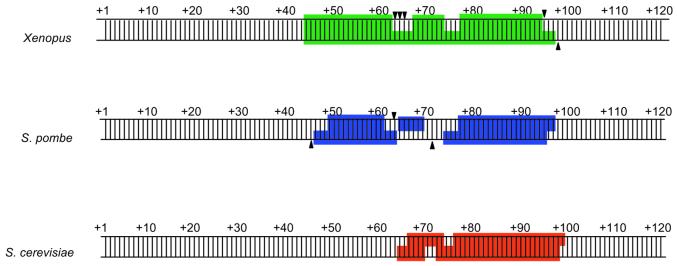
Comparison of the DNase I protection patterns produced on 5S rRNA genes by TFIIIAs from X.laevis, S.cerevisiae and S.pombe reveals another distinctive feature of S.pombe TFIIIA (Fig. 7). The overall size of the region protected by the S.pombe protein is similar to that protected by X.laevis TFIIIA (40,46), but larger than that observed in S.cerevisiae (41,52). There is, however, a striking difference in the pattern of protection within the putative ICRs of the X.laevis and S.pombe 5S rRNA genes. Whereas the X.laevis protein completely protects the template strand from residues +45 to +96, a region of ∼11 bp, from +63 to +73, remains unprotected by S.pombe TFIIIA. The differences in the protection patterns observed with these three proteins suggest that DNA recognition occurs differently in each case. In S.cerevisiae, the lack of protection at the 5′ end of the binding site is consistent with results from Rowland and Segall (41) showing that the four C-terminal zinc fingers of TFIIIA can be deleted without affecting the pattern of DNase protection. Thus, the C-terminal zinc fingers of S.cerevisiae TFIIIA are unlikely to be intimately associated with the 5S rRNA gene, even though cross-linking results suggest that some extended association of TFIIIA with the 5′ end of the ICR does occur (53). In X.laevis, individual DNA subsites across the length of the control region can be associated with almost every zinc finger in TFIIIA (40,54–56), even though fingers four and six have been proposed to span successive minor grooves with little or no direct DNA contact (40,56–58). The extended region of non-protection in the middle of the S.pombe TFIIIA footprint suggests that contacts made by X.laevis TFIIIA to the intermediate element of the 5S rRNA gene ICR (58) may not occur in S.pombe, and that DNA contacts are restricted to the 5′ and 3′ ends of the control region. It seems likely, therefore, that binding of S.pombe TFIIIA to the 5S rRNA gene is mediated by some combination of N- and C-terminal zinc fingers only. Additional investigation of this issue is clearly merited.
Figure 7.
Schematic diagram of the DNase I protection patterns generated on 5S rDNA by X.laevis (40,46), S.pombe and S.cerevisiae (41,52) TFIIIA proteins. Boxed regions represent regions of protection from DNase I cleavage. Triangles designate positions rendered hypersensitive to DNase I cleavage upon binding of the protein.
Others have identified short oligopeptide sequences in both S.cerevisiae (26) and X.laevis TFIIIA (21) that are important for transcription of 5S rRNA genes but that play no role in DNA recognition. Presumably, structures involving these sequences play a role in higher order interactions that are important for transcription complex assembly and/or function. Interestingly, the relevant sequences from the two species exhibit no similarity to each other, although the X.laevis sequence is conserved among tetrapod vertebrates. We can find no peptide sequence in S.pombe TFIIIA that resembles either of the sequences specifically important for transcriptional activity in S.cerevisiae or X.laevis. It will be important to determine just what region of the S.pombe protein, if any, is dispensable for DNA binding but important for transcriptional activity, and that therefore may be functionally analogous to the characterized sequences in S.cerevisiae and X.laevis TFIIIA. It should be noted that there is also striking sequence diversity among the homologous polypeptide subunits of TFIIIC and TFIIIB from humans, S.cerevisiae and S.pombe (reviewed in 5,59). It is reasonable to hypothesize that the divergent regions of TFIIIA found to be specifically important for transcription may interact with similarly divergent surfaces on one or more subunits of TFIIIC or TFIIIB.
It is intriguing that different structural organizations characterize TFIIIAs in the three evolutionary lineages represented by X.laevis, S.cerevisiae and S.pombe, even though the same ultimate function, recognition of 5S rRNA genes and nucleation of transcription complex assembly, is served in each case. The use of different mechanisms to satisfy the same biological requirement—synthesis of 5S rRNA—affords an opportunity to gain insight into fundamental biochemical processes through comparative analysis of the 5S rRNA synthetic machinery in these three species.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Rich Maraia, Ying Huang, Jo Ann Wise, Sujata Lakhe-Reddy and Chris Webb for many helpful discussions and for providing S.pombe strains and reagents. We would also like to thank Greg Hannon for providing an S.pombe cDNA library prepared in vector pGAD-GH. Sequencing of A.fumigatus was funded by the National Institute of Allergy and Infectious Disease U01 AI 48830 to David Denning and William Nierman. This work was supported by grant number GM48035 from the National Institute of General Medical Sciences.
DDBJ/EMBL/GenBank accession no. AY091590
REFERENCES
- 1.Bogenhagen D.F., Sakonju,S. and Brown,D.D. (1980) A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3′ border of the region. Cell, 19, 27–35. [DOI] [PubMed] [Google Scholar]
- 2.Sakonju S., Bogenhagen,D.F. and Brown,D.D. (1980) A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5′ border of the region. Cell, 19, 13–25. [DOI] [PubMed] [Google Scholar]
- 3.Lassar A.B., Martin,P.L. and Roeder,R.G. (1983) Transcription of class III genes: formation of preinitiation complexes. Science, 222, 740–748. [DOI] [PubMed] [Google Scholar]
- 4.Setzer D.R. and Brown,D.D. (1985) Formation and stability of the 5 S RNA transcription complex. J. Biol. Chem., 260, 2483–2492. [PubMed] [Google Scholar]
- 5.Geiduschek E.P. and Kassavetis,G.A. (2001) The RNA polymerase III transcription apparatus. J. Mol. Biol., 310, 1–26. [DOI] [PubMed] [Google Scholar]
- 6.Miller J., McLachlan,A.D. and Klug,A. (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J., 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg J.M. (1988) Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc. Natl Acad. Sci. USA, 85, 99–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pelham H.R. and Brown,D.D. (1980) A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc. Natl Acad. Sci. USA, 77, 4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rollins M.B., Del Rio,S., Galey,A.L., Setzer,D.R. and Andrews,M.T. (1993) Role of TFIIIA zinc fingers in vivo: analysis of single-finger function in developing Xenopus embryos. Mol. Cell. Biol., 13, 4776–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittman R.H., Andrews,M.T. and Setzer,D.R. (1999) A feedback loop coupling 5 S rRNA synthesis to accumulation of a ribosomal protein. J. Biol. Chem., 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 11.Drew P.D., Nagle,J.W., Canning,R.D., Ozato,K., Biddison,W.E. and Becker,K.G. (1995) Cloning and expression analysis of a human cDNA homologous to Xenopus TFIIIA. Gene, 159, 215–218. [DOI] [PubMed] [Google Scholar]
- 12.Hanas J.S., Hocker,J.R., Cheng,Y.G., Lerner,M.R., Brackett,D.J., Lightfoot,S.A., Hanas,R.J., Madhusudhan,K.T. and Moreland,R.J. (2002) cDNA cloning, DNA binding and evolution of mammalian transcription factor IIIA. Gene, 282, 43–52. [DOI] [PubMed] [Google Scholar]
- 13.Arakawa H., Nagase,H., Hayashi,N., Ogawa,M., Nagata,M., Fujiwara,T., Takahashi,E., Shin,S. and Nakamura,Y. (1995) Molecular cloning, characterization and chromosomal mapping of a novel human gene (GTF3A) that is highly homologous to Xenopus transcription factor IIIA. Cytogenet. Cell Genet., 70, 235–238. [DOI] [PubMed] [Google Scholar]
- 14.Ogilvie M.K. and Hanas,J.S. (1997) Molecular biology of vertebrate transcription factor IIIA: cloning and characterization of TFIIIA from channel catfish oocytes. Gene, 203, 103–112. [DOI] [PubMed] [Google Scholar]
- 15.Gaskins C.J., Fiser-Littell,R.M., Duke,A.L. and Hanas,J.S. (1989) Species variation in transcription factor IIIA. Nucleic Acids Res., 17, 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaskins C.J. and Hanas,J.S. (1990) Sequence variation in transcription factor IIIA. Nucleic Acids Res., 18, 2117–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaskins C.J., Smith,J.F., Ogilvie,M.K. and Hanas,J.S. (1992) Comparison of the sequence and structure of transcription factor IIIA from Bufo americanus and Rana pipiens. Gene, 120, 197–206. [DOI] [PubMed] [Google Scholar]
- 18.Dayhoff M.O. (1978) Atlas of Protein Sequence and Structure. Vol. 5. National Biomedical Research Foundation.
- 19.Page R.H. and Holmes,E. (1998) Molecular Evolution: A Phylogenetic Approach. Blackwell Science, Oxford; Malden, MA.
- 20.Szymanski M., Barciszewska,M.Z., Erdmann,V.A. and Barciszewski,J. (2002) 5S ribosomal RNA database. Nucleic Acids Res., 30, 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao X. and Darby,M.K. (1993) A position-dependent transcription-activating domain in TFIIIA. Mol. Cell. Biol., 13, 7496–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tso J.Y., Van Den Berg,D.J. and Korn,L.J. (1986) Structure of the gene for Xenopus transcription factor TFIIIA. Nucleic Acids Res., 14, 2187–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown R.S., Sander,C. and Argos,P. (1985) The primary structure of transcription factor TFIIIA has 12 consecutive repeats. FEBS Lett., 186, 271–274. [DOI] [PubMed] [Google Scholar]
- 24.Archambault J., Milne,C.A., Schappert,K.T., Baum,B., Friesen,J.D. and Segall,J. (1992) The deduced sequence of the transcription factor TFIIIA from Saccharomyces cerevisiae reveals extensive divergence from Xenopus TFIIIA. J. Biol. Chem., 267, 3282–3288. [PubMed] [Google Scholar]
- 25.Woychik N.A. and Young,R.A. (1992) Genes encoding transcription factor IIIA and the RNA polymerase common subunit RPB6 are divergently transcribed in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 89, 3999–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowland O. and Segall,J. (1998) A hydrophobic segment within the 81-amino-acid domain of TFIIIA from Saccharomyces cerevisiae is essential for its transcription factor activity. Mol. Cell. Biol., 18, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milne C.A. and Segall,J. (1993) Mapping regions of yeast transcription factor IIIA required for DNA binding, interaction with transcription factor IIIC and transcription activity. J. Biol. Chem., 268, 11364–11371. [PubMed] [Google Scholar]
- 28.Del Rio S. and Setzer,D.R. (1991) High yield purification of active transcription factor IIIA expressed in E. coli. Nucleic Acids Res., 19, 6197–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao J., Appel,B., Schaack,J., Sharp,S., Yamada,H. and Soll,D. (1982) The 5S RNA genes of Schizosaccharomyces pombe. Nucleic Acids Res., 10, 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Setzer D.R., Hmiel,R.M. and Liao,S.Y. (1990) A simple vector modification to facilitate oligonucleotide-directed mutagenesis. Nucleic Acids Res., 18, 4175–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Setzer D.R. (1999) Measuring equilibrium and kinetic constants using gel retardation assays. Methods Mol. Biol., 118, 115–128. [DOI] [PubMed] [Google Scholar]
- 32.Scatchard G. (1949) The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci., 51, 660–672. [Google Scholar]
- 33.Schultz M.C., Choe,S.Y. and Reeder,R.H. (1991) Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc. Natl Acad. Sci. USA, 88, 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols M., Willis,I. and Soll,D. (1990) Yeast suppressor mutations and transfer RNA processing. Methods Enzymol., 181, 377–394. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., Zhao,A., Chen,L. and Pape,L. (1997) Activated levels of rRNA synthesis in fission yeast are driven by an intergenic rDNA region positioned over 2500 nucleotides upstream of the initiation site. Nucleic Acids Res., 25, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodicker F., Ossenbuhl,F., Michels,D. and Benecke,B.J. (1999) Faithful in vitro transcription by fission yeast RNA polymerase III reveals unique alpha-amanitin sensitivity. Gene Expr., 8, 165–174. [PMC free article] [PubMed] [Google Scholar]
- 37.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 38.Bohm S., Frishman,D. and Mewes,H.W. (1997) Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res., 25, 2464–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 40.Del Rio S., Menezes,S.R. and Setzer,D.R. (1993) The function of individual zinc fingers in sequence-specific DNA recognition by transcription factor IIIA. J. Mol. Biol., 233, 567–579. [DOI] [PubMed] [Google Scholar]
- 41.Rowland O. and Segall,J. (1996) Interaction of wild-type and truncated forms of transcription factor IIIA from Saccharomyces cerevisiae with the 5 S RNA gene. J. Biol. Chem., 271, 12103–12110. [DOI] [PubMed] [Google Scholar]
- 42.Del Rio S. (1992) Structural and functional studies of Xenopus laevis transcription factor IIIA zinc finger mutants. Doctoral Dissertation, Molecular Biology and Microbiology, Case Western Reserve University.
- 43.Hamada M., Sakulich,A.L., Koduru,S.B. and Maraia,R.J. (2000) Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J. Biol. Chem., 275, 29076–29081. [DOI] [PubMed] [Google Scholar]
- 44.Bieker J.J., Martin,P.L. and Roeder,R.G. (1985) Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell, 40, 119–127. [DOI] [PubMed] [Google Scholar]
- 45.Segall J., Matsui,T. and Roeder,R.G. (1980) Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J. Biol. Chem., 255, 11986–11991. [PubMed] [Google Scholar]
- 46.Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- 47.Shastry B.S., Ng,S.Y. and Roeder,R.G. (1982) Multiple factors involved in the transcription of class III genes in Xenopus laevis. J. Biol. Chem., 257, 12979–12986. [PubMed] [Google Scholar]
- 48.Bumbulis M.J., Wroblewski,G., McKean,D. and Setzer,D.R. (1998) Genetic analysis of Xenopus transcription factor IIIA. J. Mol. Biol., 284, 1307–1322. [DOI] [PubMed] [Google Scholar]
- 49.Merkle D.L., Schmidt,M.H. and Berg,J.M. (1991) Design and characterization of a ligand-binding metallopeptide. J. Am. Chem. Soc., 113, 5450–5451. [Google Scholar]
- 50.Clemens K.R., Wolf,V., McBryant,S.J., Zhang,P., Liao,X., Wright,P.E. and Gottesfeld,J.M. (1993) Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science, 260, 530–533. [DOI] [PubMed] [Google Scholar]
- 51.Hamilton T.B., Turner,J., Barilla,K. and Romaniuk,P.J. (2001) Contribution of individual amino acids to the nucleic acid binding activities of the Xenopus zinc finger proteins TFIIIIA and p43. Biochemistry, 40, 6093–6101. [DOI] [PubMed] [Google Scholar]
- 52.Braun B.R., Riggs,D.L., Kassavetis,G.A. and Geiduschek,E.P. (1989) Multiple states of protein–DNA interaction in the assembly of transcription complexes on Saccharomyces cerevisiae 5S ribosomal RNA genes. Proc. Natl Acad. Sci. USA, 86, 2530–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- 54.Hayes J.J. and Clemens,K.R. (1992) Locations of contacts between individual zinc fingers of Xenopus laevis transcription factor IIIA and the internal control region of a 5S RNA gene. Biochemistry, 31, 11600–11605. [DOI] [PubMed] [Google Scholar]
- 55.Clemens K.R., Liao,X., Wolf,V., Wright,P.E. and Gottesfeld,J.M. (1992) Definition of the binding sites of individual zinc fingers in the transcription factor IIIA–5S RNA gene complex. Proc. Natl Acad. Sci. USA, 89, 10822–10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clemens K.R., Zhang,P., Liao,X., McBryant,S.J., Wright,P.E. and Gottesfeld,J.M. (1994) Relative contributions of the zinc fingers of transcription factor IIIA to the energetics of DNA binding. J. Mol. Biol., 244, 23–35. [DOI] [PubMed] [Google Scholar]
- 57.Hayes J.J. and Tullius,T.D. (1992) Structure of the TFIIIA–5 S DNA complex. J. Mol. Biol., 227, 407–417. [DOI] [PubMed] [Google Scholar]
- 58.Nolte R.T., Conlin,R.M., Harrison,S.C. and Brown,R.S. (1998) Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex. Proc. Natl Acad. Sci. USA, 95, 2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang Y. and Maraia,R.J. (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res., 29, 2675–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]