Abstract
Glioblastoma (GBM) is the most common intracranial malignancy, but current treatment options are limited. Super-enhancers (SEs) have been found to drive the expression of key oncogenes in GBM. However, the role of SE-associated long non-coding RNAs (lncRNAs) in GBM remains poorly understood. Here, we screened for an up-regulated lncRNA-SLCO4A1-AS1 expressed in GBM by analyzing data from GSE54791, GSE4536 and TCGA. We systematically analyzed its relationship with clinical characteristics, prognosis, epigenetics, tumor microenvironment (TME), biological functions, and transcription factors. We found that SE-driven SLCO4A1-AS1 was significantly upregulated in GBM and correlated with poor prognosis. Knockdown of SLCO4A1-AS1 decreased glioma cell proliferation, invasive ability, self-renewal ability, and increased apoptosis. Epigenetic analysis revealed that SOX2 and SE could drive SLCO4A1-AS1 expression. In vitro experiments further demonstrated that GBM cells with high SLCO4A1-AS1 expression were more sensitive to VX-11e, and overexpression of SLCO4A1-AS1 could reverse the inhibitory effect of VX-11e on GBM cells. In conclusion, this study revealed that SE-driven SLCO4A1-AS1 may be a potential therapeutic target in GBM.
Keywords: GBM, Super-enhancer, SLCO4A1-AS1, Long non-coding RNAs, VX-11e
Subject terms: CNS cancer, Targeted therapies
Introduction
Glioma is one of the most typical intracranial tumors1,2, with the occurrence of more than 10,000 new cases per year and an increasing trend year by year3. Glioblastoma (GBM) is the most malignant subtype of glioma4. The median survival time of patients receiving radiotherapy and chemotherapy is approximately 14.6 months, and more than 80% of GBM patients do not survive longer than 60 months5. Despite extensive research efforts, treatment outcomes for glioma remain largely inadequate6–8.
Super-enhancers (SEs) could bind to a high density of transcription factors and co-activators, providing synergistic binding and synergistic gene activation. In addition, SEs are able to drive high expression of multiple key oncogenes in tumors that have a critical role in determining cell identity and differentiation type. Recent studies have shown that inhibitors targeting SE-related genes have encouraging results in the treatment of a variety of cancers. Thus, exploring the role of SE-related genes in GBM may provide new ideas for the treatment of GBM.
Bioinformatics has gradually been widely used in cancer research9. Although important prognostic biomarkers have gradually been discovered10, the outcomes of glioma treatment still have not been significantly improved. Long non-coding RNAs (lncRNAs) have been found to be associated with the development and progression of cancer11,12. Most of these lncRNAs are endogenous, at least 200 nucleotides in length, and have limited or no ability to encode proteins13. LncRNAs have also been reported to have important regulatory roles in the tumor microenvironment (TME)14,15. However, the potential mechanisms involved in the role of lncRNAs in GBM still need to be further investigated, which is crucial for identifying new targets for GBM therapy.
SLCO4A1-AS1 is a lncRNA that has been found to be associated with a variety of cancers in previous studies. For example, SLCO4A1-AS1 promotes cancer progression by regulating the Hsp90/Cdk2/c-Myc axis in colorectal cancer13. SLCO4A1-AS1 can also act as a molecular sponge for miR-508-3p to regulate autophagy and proliferation in colorectal cancer cells by upregulating partition-defective 3 (PARD3) expression16. SLCO4A1-AS1 can promote the proliferation and metastasis of gastric cancer by interacting with miR-149 and enhancing the expression of the X-linked inhibitor of apoptosis (XIAP)17. It also antagonizes TOX4/NTSR1 signaling to affect lung cancer cell migration and invasion18. However, its role in glioma is unknown.
In this study, we screened SE-driven SLCO4A1-AS1 through multiple databases and found that its expression was upregulated in GBM and associated with a poor prognosis. We further validated the biological function of SLCO4A1-AS1 in vitro experiments and found that VX-11e can play an inhibitory role in GBM by targeting SLCO4A1-AS1, providing a valuable approach for the treatment of GBM.
Results
Identification of highly expressed genes in GBM
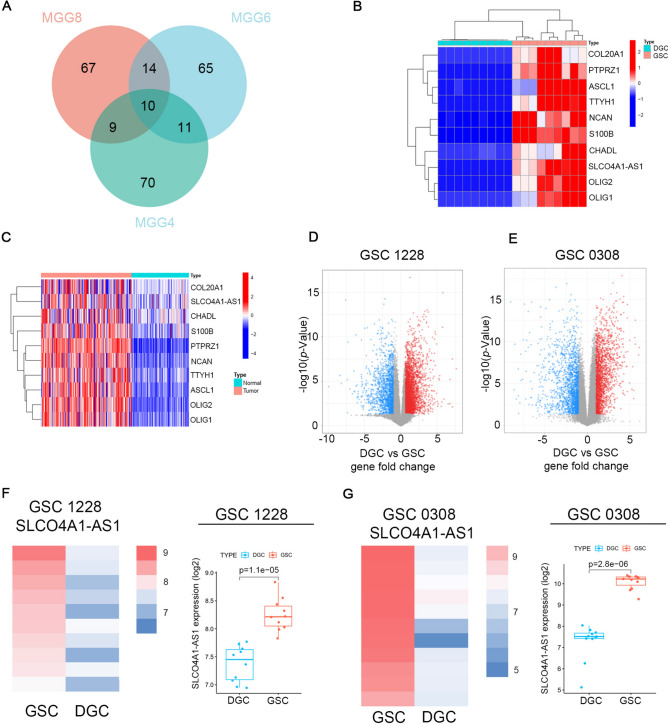
According to previous studies, undifferentiated glioma stem cells (GSCs) cultured in NBE (Neurobasal media with EGF and bFGF) have stronger tumor formation and self-renewal ability compared to differentiated GBM cells (DGCs) cultured in serum conditions19. We first screened 100 genes that were significantly upregulated in MGG4, MGG6, and MGG8 cultured in NBE compared to those cultured in serum conditions through the GSE54791 database and took the intersection to obtain 10 highly expressed genes (Fig. 1A, B). Among them, OLIG2 has been identified as a specific marker for the diagnosis of oligodendroglial tumor cells in previous studies20. ASCL1 is involved in efficient reprogramming of glioma cells into non-proliferating neurons and inhibiting tumor growth21. S100B promotes the infiltration of tumor-associated macrophages by upregulating CCL2, leading to the growth of glioma22. The role of SLCO4A1-AS1 in gliomas has not been reported, and this article solely focuses on SLCO4A1-AS1. SLCO4A1-AS1 expression was also significantly upregulated in GBM by analyzing the TCGA combined GTEx database (Fig. 1C). We validated our findings by analyzing the GSE4536 dataset and found that SLCO4A1-AS1 expression levels were significantly reduced when cells were cultured in serum-containing NBE, implying that SLCO4A1-AS1 is associated with the stemness of GSCs (Fig. 1D–G).
Fig. 1.
SLCO4A1-AS1 expression is upregulated in GBM. (A) The venn diagram shows the intersection of genes up-regulated in MGG4, MGG6 and MGG8 cultured under NBE conditions compared to culturing in serum conditions according to GSE54791 database. (B) Expression of identified genes in GSC and DGC according to GSE54791 database. (C) Expression of identified genes in GBM and normal brain tissue. (D, E) Volcano plot showing DEGs between GSCs and DGCs in 1228 cells (D) and 0308 cells (E) in the GSE4536 database. (F, G) The comparison of SLCO4A1-AS1 expression in GSC 1228 (F) and GSC 0308 (G) cultured under NBE and FBS conditions according to multiple probe sets of GSE4536 dataset. Significant statistical differences between the two groups were assessed using the Mann–Whitney test. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001.
SLCO4A1-AS1 expression was associated with clinical characteristics
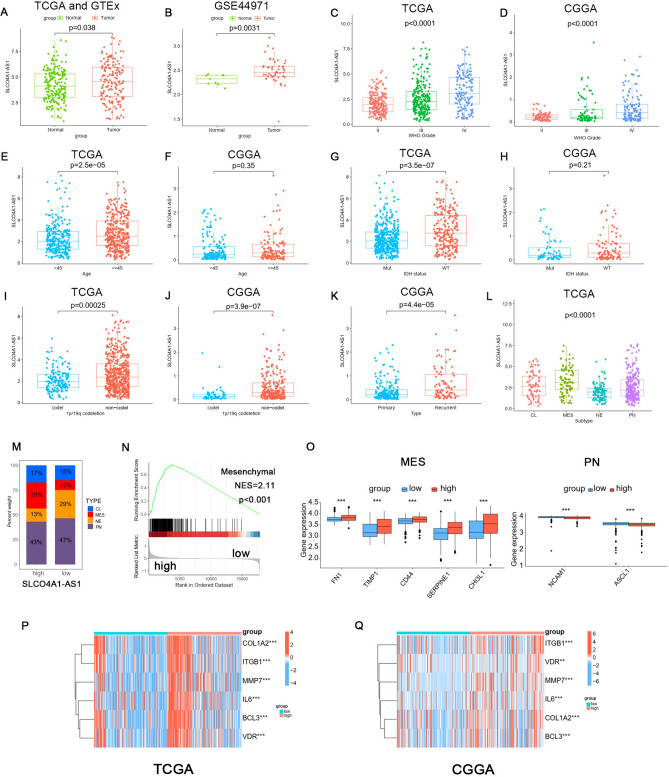
Asymmetric distributions of SLCO4A1-AS1 expression and clinical information were observed. Analyzing the data from TCGA, GTEx and the GSE44971 dataset, we identified significant overexpression of SLCO4A1-AS1 in tumor tissues (P < 0.05, Fig. 2A, B). Furthermore, we investigated its association with grade, age, isocitrate dehydrogenase (IDH) status, 1p/19q co-deletion, and tumor type in TCGA and CGGA datasets (Fig. 2C–K). Our findings indicated that SLCO4A1-AS1 expression is significantly upregulated in higher-grade gliomas and in patients aged 45 years and older. Additionally, IDH-wildtype and 1p/19q non-codeletion are associated with a worse prognosis in gliomas23, our study found that SLCO4A1-AS1 levels were elevated in IDH-wildtype and 1p/19q non-codeletion gliomas. In the CGGA database, there were no statistically significant differences in patient age or IDH status between the high and low expression groups of SLCO4A1-AS1. In contrast, in the TCGA database, we observed statistically significant differences in patient age and IDH status between the high and low expression groups of SLCO4A1-AS1. This discrepancy may be attributable to the relatively smaller sample size in the CGGA dataset. Moreover, SLCO4A1-AS1 expression levels were higher in recurrent gliomas compared to primary gliomas. We next investigated the expression level of SLCO4A1-AS1 in different molecular subtypes and found it to be higher in mesenchymal (MES) and classical (CL) subtypes than in the neural (NE) and proneural (PN) subtypes (Fig. 2L), and the subtype MES is overrepresented in tumors with high expression of SLCO4A1-AS1 (Fig. 2M). Gene Set Enrichment Analysis (GSEA) revealed that glioma samples with high SLCO4A1-AS1 expression were enriched in the MES subtype (Fig. 2N). Correlation analysis showed that MES markers were upregulated in the high expression group, whereas PN markers showed the opposite trend (Fig. 2O), and similar trends were observed for MES signature genes (Fig. 2P, Q). MES GBM is highly aggressive and has a poor prognosis, and the above analysis supported our previous finding that SLCO4A1-AS1 is associated with highly malignant brain tumors.
Fig. 2.
Association between SLCO4A1-AS1 and clinical characteristics. (A) SLCO4A1-AS1 expression levels were elevated in gliomas. The data were obtained from TCGA and GTEx datasets. (B) SLCO4A1-AS1 expression levels in glioma and normal tissues based on GSE44971. (C, D) Expression levels of SLCO4A1-AS1 in different WHO grades in the TCGA and CGGA datasets. (E, F) Distribution of SLCO4A1-AS1 in different ages in TCGA and CGGA datasets. (G, H) Relationship between the SLCO4A1-AS1 expression level and IDH mutation status in the TCGA and CGGA datasets. (I, J) Expression of SLCO4A1-AS1 in the 1p/19q codeletion (codel) and non-codel groups in TCGA and CGGA datasets. (K) SLCO4A1-AS1 is upregulated in recurrent types. The data were obtained from CGGA dataset. (L) SLCO4A1-AS1 expression is increased in MES types in the TCGA dataset. (M) Proportions of glioma samples with various subtypes. The data were obtained from TCGA dataset. (N) MES signature of GSEA. The data were obtained from TCGA dataset. (O) Differential expression analysis of MES markers (FN1, TIMP1, CD44, SERPINE1 and CHI3L1) and PN markers (NCAM1 and ASCL1) in the high SLCO4A1-AS1 and low SLCO4A1-AS1 expression groups. The data were obtained from TCGA dataset. (P, Q) Differential expression analysis of MES signature markers in TCGA (P) and CGGA (Q). Significant statistical differences between the two groups were assessed using the Mann–Whitney test. Significant statistical differences among more than the two groups were assessed using the Kruskal–Wallis test. The data were obtained from TCGA and CGGA datasets. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001.
Impact of SLCO4A1-AS1 and clinical characteristics on glioma prognosis
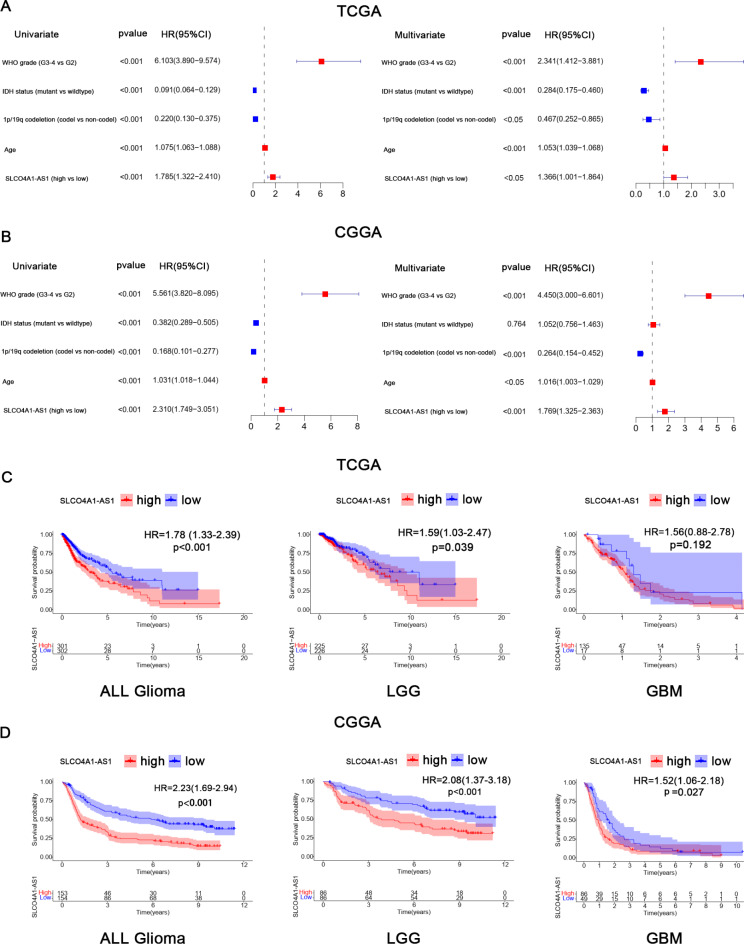
In the TCGA database, univariate Cox regression analysis showed that WHO grade, IDH mutation status, 1p/19q codeletion status, age and SLCO4A1-AS1 expression level were associated with overall survival (OS). Multivariate analysis revealed that WHO grade (HR = 2.341; 95% CI: 1.412–3.881, P < 0.001), IDH mutation status (HR = 0.284; 95% CI: 0.175–0.460, P < 0.001), 1p/19q codeletion status (HR = 0.467; 95% CI: 0.252–0.865, P < 0.05), age (HR = 1.053; 95% CI: 1.039–1.068, P < 0.001) and SLCO4A1-AS1 expression level (HR = 1.366; 95% CI: 1.001–1.864, P < 0.05) were independent prognostic factors for OS in glioma (Fig. 3A). The results of the analyses in the CGGA database were similar to those in the TCGA database (Fig. 3B). Subsequently, Kaplan–Meier survival analysis showed that patients with elevated SLCO4A1-AS1 expression had a worse prognosis in all glioma, lower-grade gliomas (LGG), and GBM groups. However, in the TCGA database, the prognostic impact of SLCO4A1-AS1 in GBM was not significant. This lack of significance may be attributed to the missing survival data for some GBM patients in the TCGA dataset (Fig. 3C). In the CGGA database, survival analysis with the method described above showed similar results (Fig. 3D). In addition, we evaluated the prognosis of patients based on different clinical characteristics (Fig. S1). All of the above results implied that SLCO4A1-AS1 is a potential biomarker for GBM prognosis.
Fig. 3.
Prognostic analysis associated with SLCO4A1-AS1. (A, B) Plots show univariate and multivariate analysis in TCGA (A) and CGGA (B). (C, D) Poor prognosis for patients with high SLCO4A1-AS1 expression in TCGA (C) and CGGA (D).
Enrichment analysis of differentially expressed genes
To investigate the potential mechanisms of SLCO4A1-AS1 affecting glioma progression, glioma samples from the TCGA dataset were divided into SLCO4A1-AS1 high and low expression groups for analysis of differentially expressed genes. A total of 2357 differentially expressed genes (DEGs) were identified. The enriched terms in the biological process (BP), cellular component (CC), and molecular function (MF) and the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were antigen processing and presentation of peptide antigen, regulation of T-cell activation, macrophage migration, synaptic membrane, major histocompatibility complex (MHC) class II protein complex, MHC protein complex, immune receptor activity and ECM-receptor interaction (Fig. S2A–D). The results of CGGA database analysis were similar to those of TCGA database analysis (Fig. S2E–H). To further reveal the key pathways associated with SLCO4A1-AS1, we performed GSEA, and the most important pathways were found to be cell chemokine, regulation of immune effector process, lymphocyte mediated immunity, inflammatory response and activation, and migration of macrophages and T cells (Fig. S2I). We also used the “HALLMARK” gene sets for DEGs enrichment, which showed significant enrichment in cancer activation pathways, inflammatory responses, stromal activation pathways and immune-related pathways (Fig. S2J). These results suggested a possible link between SLCO4A1-AS1 and tumor immune processes and immune cells and may have an impact on the induction and chemotaxis of macrophages and T cells in the TME.
SLCO4A1-AS1 was associated with TME
To investigate the relationship between SLCO4A1-AS1 and TME, we assessed immune cells using the CIBERSORT algorithm and performed correlation analysis (Fig. S3A). The results showed an association with macrophages and T cells and differed between the two groups (Fig. S3B), validating our previous results. We also found that immunosuppressive cells such as Tregs and neutrophils were upregulated, which may promote the formation of an immunosuppressive microenvironment in glioma24. We further quantified the immune cells in the samples using the single-sample GSEA (ssGSEA) algorithm, and the analysis showed that most immune cells were increased in the SLCO4A1-AS1 high expression group, including tumor-associated macrophages (Fig. S3C). We further found that patients with high macrophage infiltration and high SLCO4A1-AS1 expression had the worst prognosis (Fig. S3D).
Stromal cells and immune cells were also found to be present in tumor samples and are involved in cytokine production and signal transmission, potentially influencing tumor immunotherapy and promoting immune escape. We found higher stromal, immune and ESTIMATE scores in the high expression group (Fig. S3E). Immune checkpoints are molecules expressed in immune cells and high expression of immune checkpoints in tumor cells promotes immune escape and greatly increases the difficulty of treatment. We comprehensively evaluated the relationship between SLCO4A1-AS1 and immune checkpoints using two databases, and the results showed correlations except for T-cell immunoreceptor with Ig and ITIM domains (TIGIT) (Fig. S3F), also implying that SLCO4A1-AS1 might become a key lncRNA for immune escape of tumor cells. In addition, we evaluated the correlation with chemokine receptors, chemokines, and human leukocyte antigen (HLA) genes. As shown in Fig. S3G-I, there was an association with all of these genes, further revealing the important role of SLCO4A1-AS1 for the TME.
To further understand the impact of SLCO4A1-AS1 on the biological function of gliomas, we analyzed the glioma samples using gene set variation analysis (GSVA). The results showed that cancer-related pathways in the high expression group were in an activated state, including DNA repair, antigen processing and presentation, and signaling pathways related to immune cell receptors. Some of the classical cancer signaling pathways were also significantly upregulated, such as apoptosis, the JAK-STAT signal pathway and the p53 pathway (Fig. S4A, B). Moreover, we found that signaling pathways of inflammation were also activated (Fig. S4C). The activation of stromal cells has an important impact on the weakening effect of immunotherapy. Therefore, we evaluated the signaling pathways associated with stromal activation (Fig. S4D) and showed a positive correlation with all of the results, suggesting its possible involvement in immunotherapy resistance by influencing stromal activation. The tumor immune cycle is the process by which immune cells recognize and kill tumor cells, and specific targeting of early steps may facilitate the killing of tumor cells by immune cells. We found that most immune cycle steps were upregulated in the SLCO4A1-AS1 high expression group (Fig. S4E). To explore whether SLCO4A1-AS1 is involved in the malignant progression of glioma, we analyzed single-cell sequencing data (GSE103224) and found that the expression of SLCO4A1-AS1 was significantly elevated in malignant glioma, indicating that it may be an important biomarker for the diagnosis of glioma (Fig. S4F). High expression of the stemness index predicts the active biological behavior of tumor cells and is strongly associated with poor prognosis, recurrence after surgery, drug resistance and immunotherapy resistance. The upregulation of the stemness index in the SLCO4A1-AS1 high-expression group also implied that it may play an important role in GSCs (Fig. S4G). Different mutated genes affect the response to immunotherapy differently. We analyzed glioma somatic mutations in the TCGA dataset by grouping them according to SLCO4A1-AS1 expression. The findings showed that SLCO4A1-AS1 was positively correlated with tumor mutation burden (TMB) (Fig. S4H) and that patients in the high SLCO4A1-AS1 expression and high TMB groups had the worst prognosis, while those with low SLCO4A1-AS1 expression and low TMB had a better prognosis (Fig. S4I). The sensitivity of the diagnosis could be improved by a combined analysis of SLCO4A1-AS1 and TMB. Both IDH and capicua (CIC) mutation frequencies were increased in the low expression group, which was also associated with a good prognosis for the patients (Fig. S4J). Mismatch repair genes are capable of repairing faulty nucleotide sequences and are a major regulator of tumor progression. We evaluated the correlation of SLCO4A1-AS1 with mismatch repair genes and showed that the mismatch repair system is also involved in tumor progression in concert with SLCO4A1-AS1 (Fig. S5A). In addition, microsatellite instability (MSI) was also negatively correlated with SLCO4A1-AS1 (Fig. S5B). We repeated the above analysis in CGGA and the results were consistent with the predicted results (Fig. S5C–G).
SLCO4A1-AS1 knockdown inhibited the growth and invasion ability of GBM and enhanced apoptosis
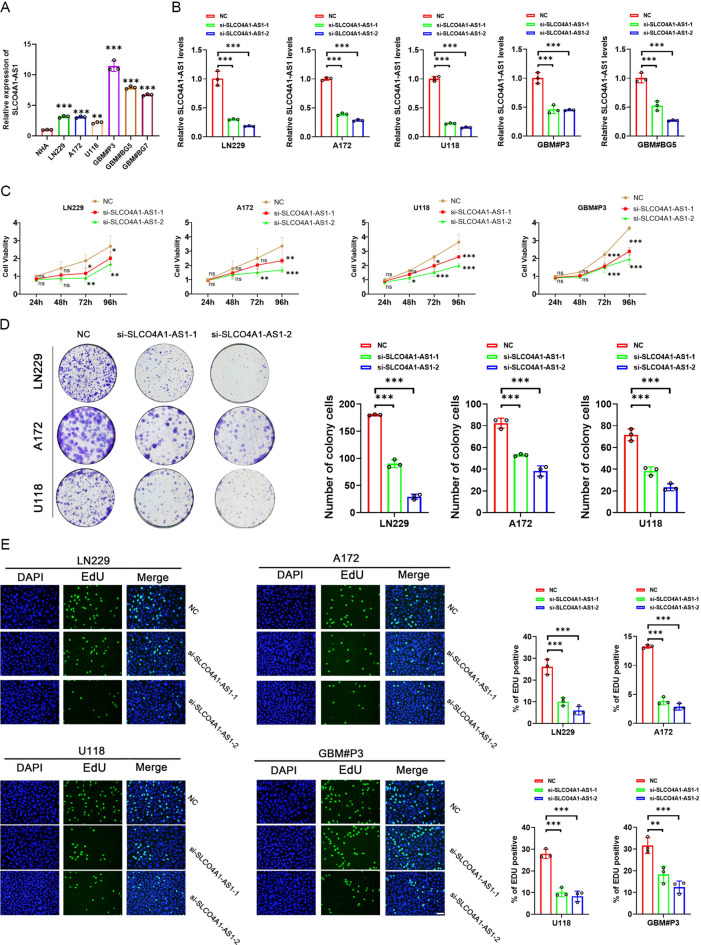
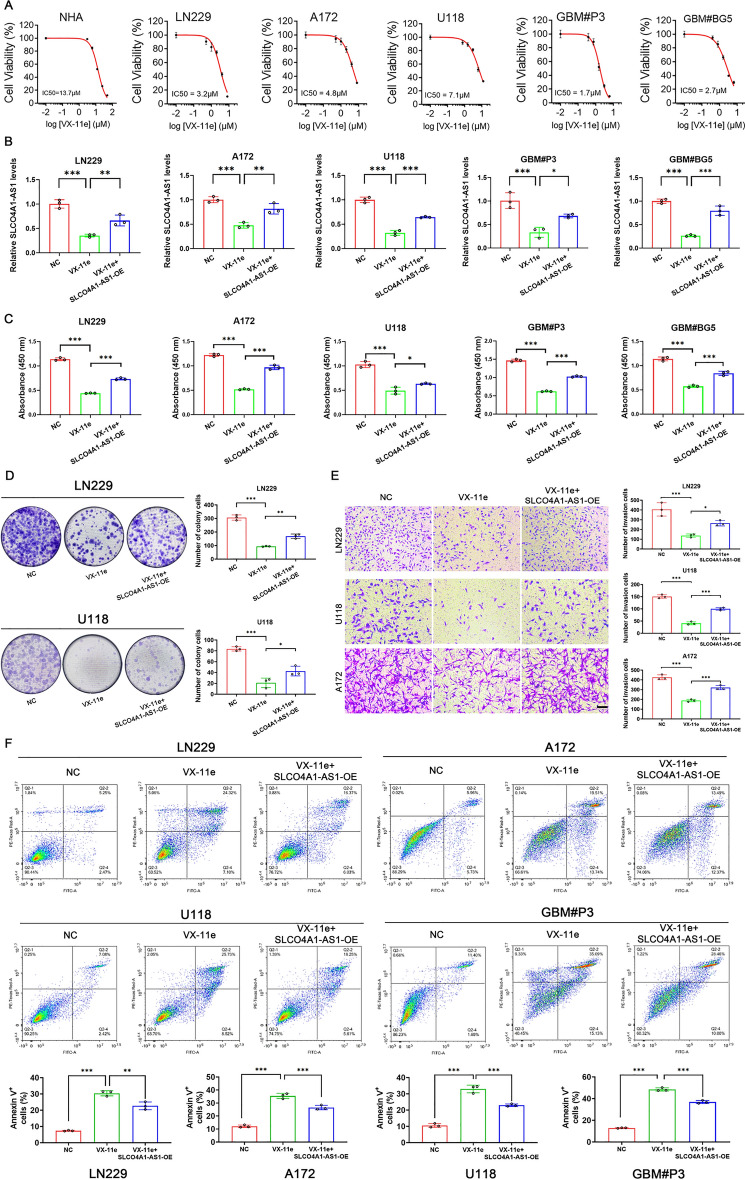
Initially, we analyzed the expression levels of SLCO4A1-AS1 in different glioma cells by qRT-PCR. SLCO4A1-AS1 was expressed at higher levels in GBM cell lines (LN229, A172 and U118) and GSCs (GBM#P3, GBM#BG5 and GBM#BG7) than in normal human astrocytes (NHAs) (Fig. 4A). Two siRNAs were used to knock down SLCO4A1-AS1 in GBM cells and patient-derived primary GSCs. After 48 h of incubation, we detected the knockdown efficiency (Fig. 4B).
Fig. 4.
Knockdown of SLCO4A1-AS1 affects glioma cell proliferation. (A) Based on qRT‒PCR analysis, the SLCO4A1-AS1 expression levels in LN229, A172, U118, GBM#P3, GBM#BG5 and GBM#BG7 cells were higher than those in NHA (n = 3). (B) Knockdown efficiency of siRNAs in LN229, A172, U118, GBM#P3 and GBM#BG5 cells (n = 3). (C) CCK-8 assay results showing that knockdown of SLCO4A1-AS1 inhibits the proliferation of LN229, A172, U118 and GBM#P3 cells (n = 3). (D) Transfection with siRNAs targeting SLCO4A1-AS1 reduces the colony-forming ability of LN229, A172 and U118 cells (n = 3). (E) Proliferative activity of glioma cells after knockdown of SLCO4A1-AS1 analyzed by EdU assay (n = 3). Scale bar = 50 μm. Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. 2-way ANOVA (C) and 1-way ANOVA (A, B, D and E) were used for data analysis.
Cell viability was evaluated between 24 and 96 h in parental and knockdown cells, with the latter showing a lower proliferation rate (Fig. 4C). The colony-forming ability of GBM cell lines (LN229, A172 and U118) was inhibited after knockdown of SLCO4A1-AS1 (Fig. 4D). In the EdU assay, loss of SLCO4A1-AS1 was found to cause inhibition of cell growth in the GBM cell lines and GBM#P3 cells (Fig. 4E).
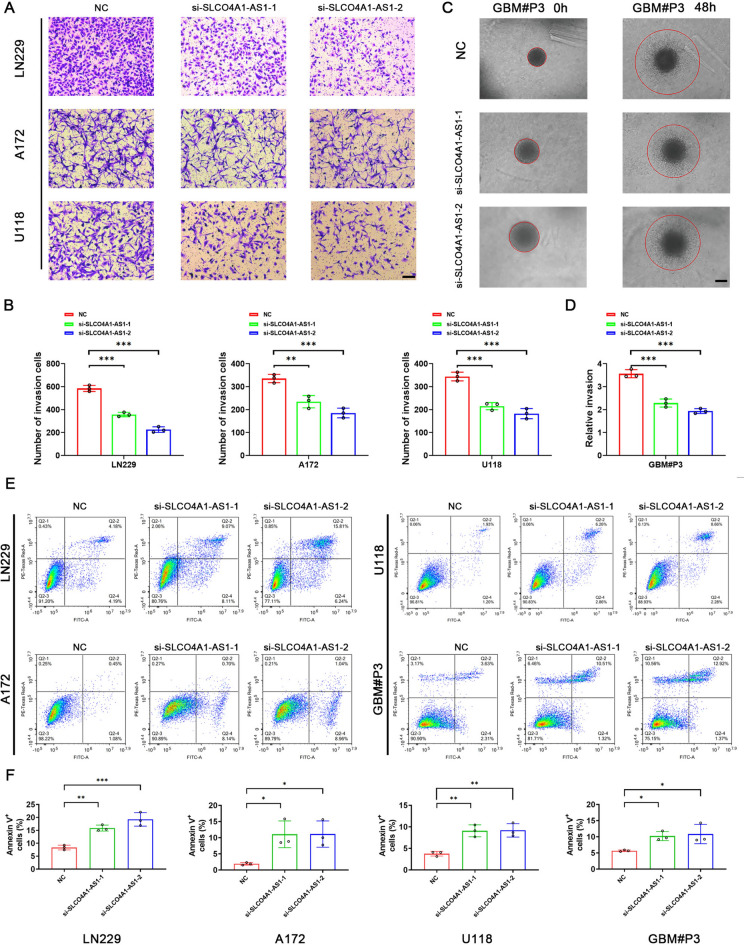
In addition, we continued to investigate the effect of SLCO4A1-AS1 on the invasive ability of GBM cells. The results of the transwell assay showed that SLCO4A1-AS1 knockdown reduced the invasive ability of LN229, A172 and U118 cells (Fig. 5A, B). In the 3D tumor sphere invasion assay, knockdown of SLCO4A1-AS1 led to a decrease in the relative invasion ability of the spheres. (Fig. 5C, D). Next, apoptosis in GBM cell lines and GBM#P3 cells was detected by flow cytometry. The proportion of apoptotic cell death was increased in the SLCO4A1-AS1-deficient LN229, A172, U118, and GBM#P3 cell groups (Fig. 5E, F).
Fig. 5.
Invasion and apoptosis assay. (A) Transwell assay showing that knockdown of SLCO4A1-AS1 reduced the invasive ability of LN229, A172, and U118 cells (n = 3). Scale bar = 100 μm. (B) Quantitative analysis of the transwell assay results to assess the invasion ability of glioma cells following SLCO4A1-AS1 knockdown. (C) 3D tumor sphere invasion assay to assess the invasive capacity of GBM#P3 after knockdown of SLCO4A1-AS1 (n = 3). Scale bar = 100 μm. (D) Quantitative analysis of the 3D tumor sphere invasion assay results to assess the invasive ability of GBM#P3 following SLCO4A1-AS1 knockdown. (E) Annexin V staining of LN229, A172, U118, and GBM#P3 cells following SLCO4A1-AS1 knockdown (n = 3). (F) Quantitative analysis of Annexin V staining in LN229, A172, U118, and GBM#P3 cells following SLCO4A1-AS1 knockdown. Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. 1-way ANOVA was used for data analysis.
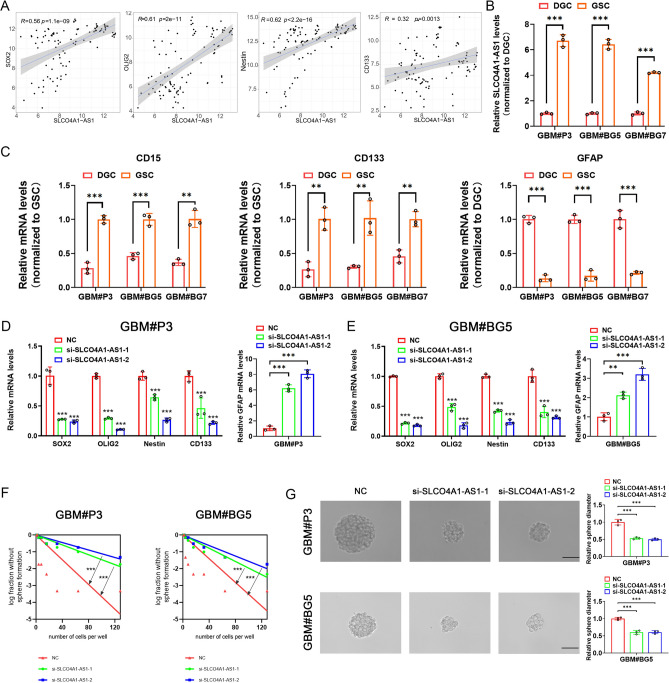
Knockdown of SLCO4A1-AS1 significantly suppressed the stemness of GSCs
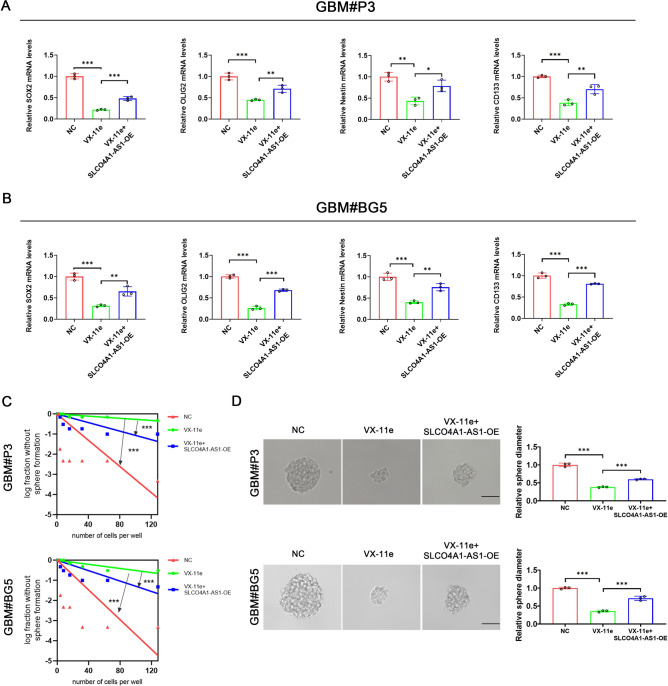
First, we found a significant positive correlation between SLCO4A1-AS1 and stemness markers SOX2, OLIG2, NES and CD133 by analyzing the GSE4536 database (Fig. 6A). We examined that SLCO4A1-AS1 expression was significantly upregulated in GSCs cultured in NBE compared to DGCs cultured in media containing serum (Fig. 6B). CD15, CD133 and GFAP were used to validate the differentiation of GSCs (Fig. 6C). In GBM#P3 and GBM#BG5, we also found that stemness markers were also significantly downregulated after SLCO4A1-AS1 knockdown, while the differentiation marker GFAP was significantly upregulated (Fig. 6D, E). In addition, SLCO4A1-AS1 knockdown significantly reduced sphere formation capacity and tumor sphere expansion capacity (Fig. 6F, G).
Fig. 6.
Knockdown of SLCO4A1-AS1 downregulates stemness markers in GSC. (A) Significant positive correlation between SLCO4A1-AS1 and stemness markers in GSE4536. (B) SLCO4A1-AS1 expression in GSC and matched DGC measured by qRT-PCR (n = 3). (C) CD15, CD133and GFAP expression in GSC and matched DGC measured by qRT-PCR (n = 3). (D, E) Expression of SOX2, OLIG2, NES, CD133 and GFAP after knockdown of SLCO4A1-AS1 in GBM#P3 (D) and GBM#BG5 (E) (n = 3). (F, G) Knockdown of SLCO4A1-AS1 reduced sphere forming ability (F) and tumor sphere expansion capacity (G) in GBM#P3 and GBM#BG5 (n = 3). Scale bar = 50 μm. Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. Student’s t-test (B, C), 1-way ANOVA (D, E, G) and 2-way ANOVA (D, E) were used for data analysis.
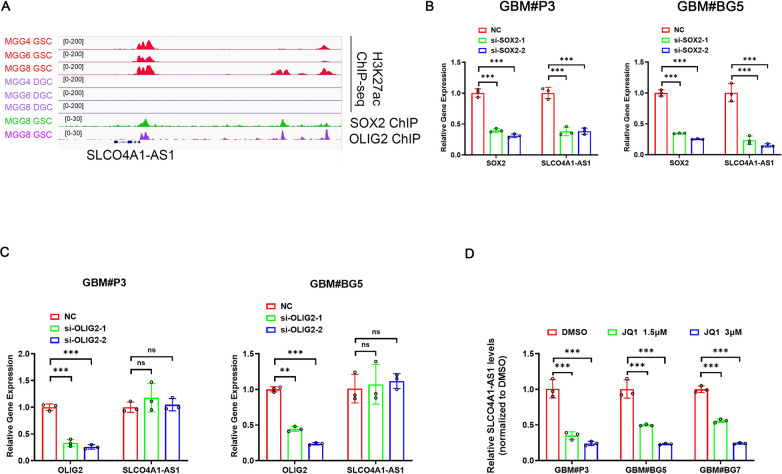
SEs may drive SLCO4A1-AS1 expression
To explore the epigenetic regulation of SLCO4A1-AS1 in GSCs, we analyzed the chromatin landscape of GSCs and matched DGCs and found significantly active SLCO4A1-AS1 SEs in GSCs measured by H3K27ac peak levels, which was in accordance with the previous conclusion that the expression of SLCO4A1-AS1 was upregulated in GSCs (Fig. 1F, G). Next, we used GSC-derived H3K27ac, SOX2, and OLIG2 ChIP-Seq data and found that stemness markers SOX2 and OLIG2 might be underlying drivers of SLCO4A1-AS1 expression (Fig. 7A). Moreover, knockdown of SOX2 also decreased the expression of SLCO4A1-AS1, whereas knockdown of OLIG2 did not have a significant effect on SLCO4A1-AS1 expression, suggesting that SLCO4A1-AS1 expression might be driven by SOX2 (Fig. 7B, C). Transcriptional coactivators can regulate the expression of SE-related genes, prominently bromodomain-containing protein 4 (BRD4). Inhibition of BRD4 could downregulate the expression of SE-driven genes, and the BRD4 inhibitor JQ1 downregulated SLCO4A1-AS1 expression in three GSC lines (Fig. 7D), confirming that SLCO4A1-AS1 expression is driven by a SE.
Fig. 7.
SOX2 and SEs in GSC could drive SLCO4A1-AS1 expression. (A) ChIP-seq profiles of H3K27ac in GSCs and DGCs. (B) Expression levels of SLCO4A1-AS1 and SOX2 after SOX2 knockdown (n = 3). (C) Expression levels of SLCO4A1-AS1 and OLIG2 after OLIG2 knockdown (n = 3). (D) BRD4 inhibitor JQ1 could downregulate SLCO4A1-AS1 expression (n = 3). Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. 2-way ANOVA was used for data analysis.
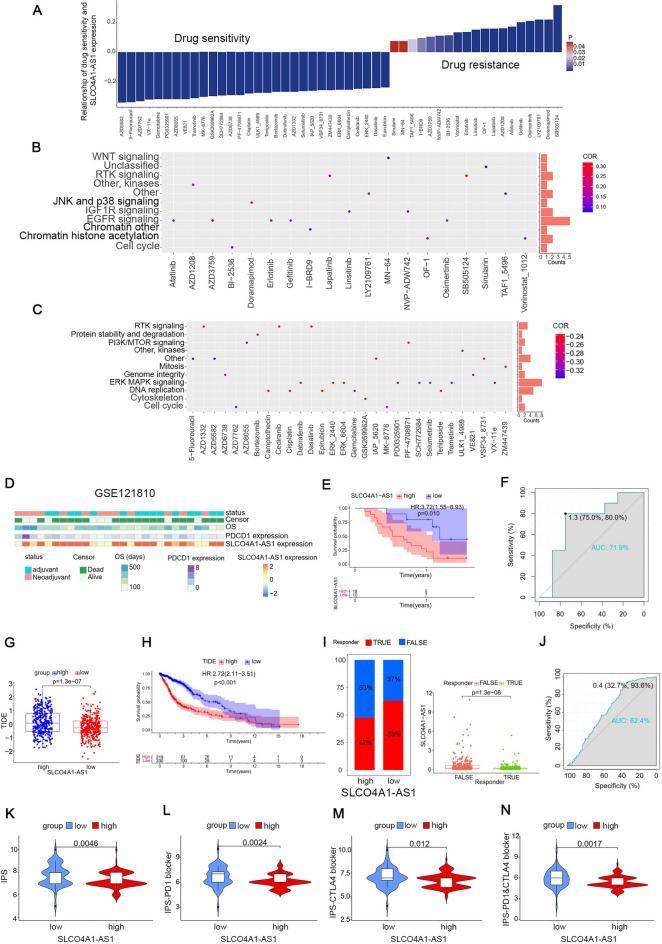
SLCO4A1-AS1 contributed to the selection of chemotherapeutic drugs and the prediction of immunotherapy efficacy
To investigate the predictive role of SLCO4A1-AS1 on drug selection for the treatment of tumors and to help the use of drugs in clinical medicine, we screened 144 drugs associated with SLCO4A1-AS1 in gliomas using the Genomics of Drug Sensitivity in Cancer (GDSC) database, of which 125 were sensitive and 19 were insensitive to patients with high SLCO4A1-AS1 expression. We listed the top 30 most sensitive drugs and 19 insensitive drugs (Fig. 8A). The pathways targeted by these drugs were analyzed and the insensitive drugs were mainly enriched in the EGFR signaling pathway (Fig. 8B), while the sensitive drugs mainly targeted the ERK MAPK signaling pathway (Fig. 8C). We further used the “pRRophetic” package to predict the sensitivity of glioma samples to various drugs and it was found that commonly used chemotherapy drugs such as cisplatin, cytarabine, nilotinib, temozolomide and vinblastine may have better efficacy in patients with high SLCO4A1-AS1 expression (Fig. S6). In conclusion, it showed that SLCO4A1-AS1 was correlated with a variety of chemotherapeutic drugs, which is important for the treatment of cancer patients in the clinic.
Fig. 8.
Drug sensitivity analysis and prediction of immunotherapy effectiveness. (A) List of drugs related to SLCO4A1-AS1. The horizontal axis represents the names of the drugs, while the vertical axis indicates the correlation coefficients between drug sensitivity and SLCO4A1-AS1 expression levels. The colors of the bars represent the statistical P value. (B) Targeted pathways of drug resistance. (C) Targeting pathways for sensitive drugs. The horizontal axis represents the names of the drugs, while the vertical axis indicates the signaling pathways targeted by these drugs. The bar graph on the right side of the plot displays the number of drugs targeting each specific pathway. The colors of the points represent the correlation coefficients between drug sensitivity and SLCO4A1-AS1 expression levels. (D) Distribution of clinical characteristics of patients in the GSE121810 database. (E) GBM patients with high SLCO4A1-AS1 expression had a poorer prognosis after receiving immunotherapy. (F) ROC curves showed that SLCO4A1-AS1 had high accuracy in predicting OS in GBM patients receiving immunotherapy. (G) Higher TIDE scoring in the high expression group. (H) Patients with high TIDE score have a poorer prognosis. (I) Immunotherapy was more effective in the low expression group. (J) ROC curves for predicting the effectiveness of immunotherapy. (K–N) Lower expression group has higher IPS and may have better immunotherapy responsiveness. Significant statistical differences between the two groups were assessed using the Mann–Whitney test. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001.
Immunotherapy is a highly regarded new treatment option that kills cancer cells by activating the immune system. Immunotherapy for gliomas is also one of the most promising treatment modalities available. Thus, we used the GSE121810 database to evaluate the roles of SLCO4A1-AS1 in the efficacy of neoadjuvant anti-PD-1 immunotherapy for recurrent GBM. The clinical status of the patients is shown in Fig. 8D. The analysis showed that patients with low expression of SLCO4A1-AS1 had a better prognosis after immunotherapy (Fig. 8E), and the ROC curve showed that SLCO4A1-AS1 had a better predictive effect on OS (AUC = 0.719; Fig. 8F), which may provide a new strategy for patients with poor treatment outcomes.
We also evaluated the efficacy of immunotherapy in glioma patients using the TIDE tool. The higher TIDE scores in the high SLCO4A1-AS1 expression group implied a greater likelihood of immune escape, a poorer response to immunotherapy (Fig. 8G), and a poorer prognosis for patients in the high TIDE scoring group (Fig. 8H). The prediction of immunotherapy effectiveness in glioma samples was also consistent with our prediction that patients with high SLCO4A1-AS1 expression had poorer and statistically significant immunotherapy effectiveness (Fig. 8I). The ROC curve results showed that SLCO4A1-AS1 had a good ability to predict immunotherapy effectiveness in glioma patients (Fig. 8J). To increase the reliability of the results, we used The Cancer Immunome Atlas (TCIA) online software to make predictions, with a high Immunophenotype Score (IPS) representing a possible better response to anti-PD-1 and anti-CTLA-4. From the results, it is clear that the low expression group may have better responsiveness to immunotherapy, consistent with the results of the previous analysis (Fig. 8K–N).
SLCO4A1-AS1 reversed the effects of VX-11e on proliferation, invasion, apoptosis, and stemness of GBM
AZD558225, 5-Fluorouracil26, and AZD776227 had been validated to have important roles in inhibiting glioma development and progression in previous studies, suggesting that the drug sensitivity analyses we conducted are valuable. VX-11e is an ERK inhibitor that has been reported to have therapeutic effects in multiple diseases28–30; however, the role of VX-11e in GBM has not yet been reported. The CCK-8 assay demonstrated that GBM cells had higher sensitivity to VX-11e compared to NHAs and that GBM cells with high expression of SLCO4A1-AS1 were more sensitive to VX-11e (Fig. 9A). We also found VX-11e treatment reduced the expression of SLCO4A1-AS1 in cells. This effect was reversed by overexpressing the gene through transfection, and functional characteristics in the cells, such as proliferation, colony formation, and apoptosis, were evaluated (Fig. 9B). The results of the CCK-8 and colony formation assays showed that overexpression of SLCO4A1-AS1 reversed the proliferation inhibitory effect of VX-11e on GBM cells (Fig. 9C, D). Overexpression of SLCO4A1-AS1 also rescued the decrease in invasive ability (Fig. 9E) and increase in apoptosis caused by VX-11e (Fig. 9F). Furthermore, overexpression of SLCO4A1-AS1 rescued the downregulation of stemness markers (Fig. 10A, B), sphere formation capacity and tumor sphere expansion capacity caused by VX-11e (Fig. 10C, D).
Fig. 9.
Overexpression of SLCO4A1-AS1 could reverse the effect of VX-11e on GBM. (A) Cell viability of NHA, LN229, A172, U118, GBM#P3, and GBM#BG5 after 48 h of treatment with VX-11e (n = 3). (B) qRT-PCR was performed to examine SLCO4A1-AS1 expression in GBM cells under the conditions indicated (n = 3). (C, D) CCK-8 (C) and colony formation (D) assays examined the proliferative capacity of cells (n = 3). (E) Overexpression of SLCO4A1-AS1 could reverse the inhibitory effect of VX-11e on invasion of GBM cells (n = 3). Scale bar = 100 μm. (F) Apoptosis assay results of GBM cells under the indicated conditions (n = 3). Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. 1-way ANOVA was used for data analysis.
Fig. 10.
Overexpression of SLCO4A1-AS1 could rescue the inhibitory effect of VX-11e on GSC stemness. (A, B) qRT-PCR was performed to examine the expression of stemness markers after treatment under the conditions indicated in GBM#P3 (A) and GBM#BG5 (B) (n = 3). (C, D) Sphere forming ability (C) and tumor sphere expansion capacity (D) were examined after treatment with the indicated conditions in GBM#P3 and GBM#BG5 (n = 3). Scale bar = 50 μm. Data are presented as the mean ± SD. NS, not statistically significant; * P < 0.05; ** P < 0.01; *** P < 0.001. 1-way ANOVA was used for data analysis.
Role of SLCO4A1-AS1 in pan-cancer
We investigated the potential effect of SLCO4A1-AS1 in pan-cancer and found that SLCO4A1-AS1 was differentially expressed in most cancers by comparison with normal samples (Fig. S7A, B). SLCO4A1-AS1 was expressed at higher levels in cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), and stomach adenocarcinoma (STAD) than the corresponding normal tissues. In contrast, the expression levels were lower in breast invasive carcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe carcinoma (KICH), liver hepatocellular carcinoma (LIHC), pheochromocytoma and paraganglioma (PCPG), and thymoma (THYM) samples. In adrenocortical carcinoma (ACC), COAD, esophageal carcinoma (ESCA), kidney renal clear cell carcinoma (KIRC), PAAD, testicular germ cell tumor (TGCT), and thyroid carcinoma (THCA), the expression levels of SLCO4A1-AS1 were associated with stage (Fig. S7C). SLCO4A1-AS1 was related to age in LIHC and THCA and correlated with survival status in HNSC and uveal melanoma (UVM) (Fig. S7D, E). Radar plots revealed that SLCO4A1-AS1 was associated with MSI in 5 cancers and associated with TMB in 7 cancers (Fig. S7F, G). Next, we evaluated TME scoring in other tumors (Fig. S7H-K). Furthermore, we investigated the impact of SLCO4A1-AS1 on prognosis in other cancers (Figs. S8, S9), and the results showed that the upregulation of SLCO4A1-AS1 expression had an unfavorable prognostic impact in most cancers. However, it was favorable for patient prognosis in cancers such as HNSC and UVM. In most cancers, SLCO4A1-AS1 was positively correlated with immune checkpoint expression, immune cell response and HLA gene expression, as well as evaluated for correlation with tumor neoantigens (Figs. S10, S11).
Discussion
Glioma is a common type of intracranial tumor, and treatment with existing methods cannot significantly improve the prognosis31. Therefore, there is a need to find an effective and accurate biomarker to evaluate glioma prognosis. SLCO4A1-AS1 is located on chromosome 20 and previous findings suggested that SLCO4A1-AS1 is elevated in a variety of cancers, including colorectal cancer16,32,33, bladder cancer34, laryngeal squamous cell carcinoma35 and gastric cancer17,36. SLCO4A1-AS1 can also act as a protective factor influencing tumor progression. An analysis of public databases found that high expression of SLCO4A1-AS1 may have a better prognosis for UVM37. In an analysis of data from multiple glioma databases, we found that SLCO4A1-AS1 expression was upregulated in GBM samples and correlated with GBM stemness. By analyzing clinical data from the TCGA and CGGA databases, we also found that SLCO4A1-AS1 was associated with clinical characteristics and prognosis. Molecular subtypes were predictive of glioma prognosis, and SLCO4A1-AS1 was expressed at the highest level in the MES subtype with the worst prognosis38.
According to previous reports showing that lncRNAs also affect the TME, for example, the lncRNA CamK-A leads to cancer microenvironment remodeling by participating in Ca2+ -dependent cell signaling pathways that influence macrophage recruitment, angiogenesis, and tumor progression39. LincRNA-Cox2 regulates epigenetic chromatin remodeling and macrophage inflammatory gene transactivation by acting as a coactivator of NF-κB40. We also focused on the role of SLCO4A1-AS1 in TME and listed the enrichment of DEGs in immune functions such as antigen processing and presentation, synaptic membranes, and ECM-receptor interactions41. GSEA had also found that SLCO4A1-AS1 is associated with immune cell chemotaxis and modulation of immune responses42.This finding suggested that SLCO4A1-AS1 is important in shaping a specific immunosuppressive microenvironment.
In the past decade, immunotherapy has been considered a new strategy for the treatment of glioma43. Immune genes related to glioma have been reported previously44,45. However, the efficacy of immunotherapy in patients with glioma has not met expectations. Therefore, exploring the mechanisms of glioma resistance to immunotherapy may be the current solution46. Based on the ESTIMATE algorithm, samples with low tumor purity showed higher stromal and immune scores47,48. Patients with gliomas of low tumor purity were usually diagnosed with high-grade gliomas and had a worse prognosis14, whereas higher immune scores and stromal scores tend to indicate a higher immune cell infiltration. Our analysis similarly showed increased infiltration of multiple immune cells in the SLCO4A1-AS1 high expression group. Furthermore, ssGSEA showed that macrophages were the immune cells most closely associated with SLCO4A1-AS1 expression. According to previous studies, increased macrophage infiltration may be associated with immunosuppression and affect the immune microenvironment through phenotypic transformation, the release of cytokines and extracellular matrix proteases, suppression of the inflammatory immune response, and promotion of tumor growth, invasion, and angiogenesis, factors that may be associated with the shorter OS times observed in the high expression group49–51. Glioma samples with high SLCO4A1-AS1 expression often had high levels of Th2 cell infiltration, which has previously been studied as a potential link between immunosuppression and low survival rates52, suggesting that SLCO4A1-AS1 may be involved in the immune escape of glioma cells. When further investigating the mechanism behind SLCO4A1-AS1 and immune cell infiltration, we found that multiple chemokines showed a tendency to be highly expressed in the SLCO4A1-AS1 high expression group, and the high expression of chemokines could promote immune cell infiltration. We also found that the expression of almost all immune checkpoints positively correlated with the expression of SLCO4A1-AS1, further revealing its cause of immunosuppression, which may be a new target for ICB therapy53,54. GSVA showed that multiple immune functions, oncogenic pathways and stromal pathways were activated in the SLCO4A1-AS1 high expression group, revealing that SLCO4A1-AS1 is involved in the malignant progression of glioma in multiple ways. Previous studies have shown that activation of stromal cells in the TME can induce T-cell suppression55, which can attenuate tumor responses to immunotherapy56. The SLCO4A1-AS1 high expression group also exhibited higher levels of DNA damage repair activity and DNA damage repair is related to tumor resistance to chemotherapy or radiotherapy, further revealing the reasons for the difficulty in treating gliomas57,58. SLCO4A1-AS1 was positively correlated with TMB, which may influence the production of immunogenic peptides and thus affect the response to immunotherapy59,60. Moreover, patients with high SLCO4A1-AS1 expression had lower IDH and CIC mutation frequencies, which is not beneficial to the prognosis of patients with glioma, consistent with our previous speculation61.
Through in vitro functional experiments, we found that downregulation of SLCO4A1-AS1 expression levels significantly reduced glioma cell proliferation, invasion, self-renewal and tumor sphere formation capacity, and increased apoptosis of glioma cells. Downregulation of SLCO4A1-AS1 also induced a decrease in the expression of multiple stemness markers.
SE had been found to drive high expression of key genes in cancer, and recent studies had identified a role for several genes encoding proteins driven by SE in GBM. But the role of SE-driven lncRNAs in GBM was still rarely reported. Analyzing epigenetics, we found that inhibition of the transcriptional co-activator factor BRD4, which is associated with SE-driven genes, suppressed the expression of SLCO4A1-AS1. Knockdown of SOX2 also reduced SLCO4A1-AS1 expression. These results suggested that the high expression of SLCO4A1-AS1 may be driven by SEs and may be regulated by the stemness transcription factor SOX2, but the underlying mechanisms still need to be further explored. SE-related genes play a key role in the malignant behavior of tumor cells and targeting SE-related genes has become a promising therapeutic strategy in cancer treatment, so we evaluated the therapeutic role played by SLCO4A1-AS1 in GBM and performed sensitivity prediction of commonly used chemotherapeutic drugs by combining the GDSC database and the expression of SLCO4A1-AS1. The results showed that VX-11e might play an inhibitory role in GBM by targeting SLCO4A1-AS1. GBM cells with high expression of SLCO4A1-AS1 were more sensitive to VX-11e. In vitro experiments further showed that VX-11e could decrease the proliferation, invasion and self-renewal ability of GBM and overexpression of SLCO4A1-AS1 could rescue the inhibitory effect of VX-11e on GBM. Thus, SLCO4A1-AS1 could be a potential target for GBM therapy. Although this study revealed the association between SLCO4A1-AS1 and GBM, there were still some limitations. For example, the relationship between the SEs and SLCO4A1-AS1 required more studies to reveal the underlying mechanisms. Secondly, the inhibitory effect of VX-11e on GBM required in vivo functional assays to further validate the findings. Overall, SLCO4A1-AS1 may be a potential marker for GBM treatment and prognosis.
In conclusion, we found that high SLCO4A1-AS1 expression predicted poor prognosis, greater self-renewal and tumor sphere expansion capacity. SE-driven SLCO4A1-AS1 might be a potential marker of GBM prognosis; SLCO4A1-AS1 could serve as a guide for drug selection in GBM patients and assessment of efficacy in patients receiving immunotherapy. VX-11e could play an inhibitory role in GBM by targeting SLCO4A1-AS1 and provide new ideas for further research and treatment of glioma in the future.
Materials and methods
Data sources
TCGA RNA sequencing (RNA-seq) expression data, GTEx RNA-seq expression data and relevant clinical data were downloaded from UCSC XENA (https://xenabrowser.net/datapages/). The RNA-seq data of the GSE44971, GSE54791 and GSE4536 datasets were downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/). ICB data for GBM were obtained from GSE121810. RNA-seq data and clinical information were obtained from CGGA (http://www.cgga.org.cn/) for verification.
Functional enrichment analysis of glioma samples
The R package “limma” was used to identify the DEGs62, and these DEGs were used for GO and KEGG analyses. The cutoff criteria for DEGs were: |log2 (fold change)|> 0.5 and P value < 0.05.
Immune infiltration analysis
The ‘ESTIMATE’ algorithm was used to evaluate the TME in glioma patients47. Analysis of immune cell proportions in gliomas using the CIBERSORT algorithm63. The immune cells were quantified by the ssGSEA algorithm64.
Enrichment analysis of signaling pathways
Use GSVA to calculate scores for immune-related pathways65. ssGSEA was used to score each sample for immune-related functional enrichment.
Drug sensitivity prediction
The GDSC database (https://www.cancerrxgene.org/) and the “pRRophetic” package in R were used to predict the drug sensitivity of glioma samples. The results were presented in a boxplot generated using ggplot2.
Single-Cell RNA-Sequencing
TISCH2 is a scRNA-seq database (http://tisch.comp-genomics.org) to study tumor development and progression by sequencing individual cells to analyze the causes behind cancer development at the cellular level.
Prediction of responsiveness to immunotherapy
Immunotherapy responsiveness was predicted in glioma patients using the TIDE (http://tide.dfci.harvard.edu/), with higher TIDE scores implying a greater likelihood of immune escape and the ineffectiveness of immunotherapy. Download IPS data from TCIA (https://tcia.at/home) for different glioma samples to predict responsiveness to immunotherapy. A high IPS score indicates a likely better responsiveness to immunotherapy.
Correlation analysis of SLCO4A1-AS1 with tumor neoantigens and prognosis in pan-cancer
We used Sangerbox (http://www.sangerbox.com/tool) to calculate the tumor neoantigens in pan-cancer and Spearman correlation analysis to calculate the correlation between tumor neoantigens and SLCO4A1-AS1. We also used Sangerbox to assess the expression of SLCO4A1-AS1 in different tumors and its effect on OS, disease-free survival (DFS), disease-specific survival (DSS) and progression-free survival (PFS) in pan-cancer.
ChIP-seq analysis
BigWig files were obtained from the analysis of GSE54047 and the IGV genome browser was used for visual inspection.
Cell lines and cell cultures
A172, LN229 and U118 cells were purchased from ATCC and cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific; Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) and a 1% penicillin–streptomycin mixture. Patient-derived GSCs P3, BG5 and BG7 were previously isolated and characterized from GBM surgical specimens. GSCs were cultured in neurobasal medium (Gibco/Thermo Fisher Scientific) containing 20 ng/mL epidermal growth factor (EGF; PeproTech; East Windsor, NJ, USA), 10 ng/mL basic fibroblast growth factor (bFGF; PeproTech) and 2% B-27 Neuro Mix (Thermo Fisher Scientific). The medium was changed every 2 to 3 days. NHAs were obtained from Lonza (Walkersville, MD, USA) and cultured in astrocyte medium (ScienCell; Carlsbad, CA, USA) supplemented with the contents of the Astrocyte Growth Medium BulletKit (ScienCell). GBM cells were treated with VX-11e (MCE, HY-14178, USA) for 48 h to verify the role of VX-11e in GBM. The BRD4 inhibitor JQ-1 (MCE, HY-13030, USA) was used to verify that SLCO4A1-AS1 expression is driven by SEs.
Cell transfection
LN229, U118, A172, GBM#P3 and GBM#BG5 cells were uniformly seeded in 6-well plates and divided into NC, si-SLCO4A1-AS1-1 and si-SLCO4A1-AS1-2 groups. In this article, they are labeled as NC, si-1 and si-2, respectively. Transient transfections were performed for siRNAs with 4 µL of Lipofectamine 2000 (Thermo Fisher Scientific) and 5 µL of siRNA, and plasmids were added to 6-well plates using a ratio of 2 µg:5 µL of plasmid to transfection reagent per well. The siRNA sequences targeting SLCO4A1-AS1 were as follows: si-SLCO4A1-AS1-1, 5′-CGUCUGCAGCUUAGUGACATT-3′; si-SLCO4A1-AS1-2, 5′-ACUCUGAAUGCCUGGAACATT-3′. The siRNA sequences targeting SOX2 were as follows: si-SOX2-1, 5′-CCAAGACGCUCAUGAAGAAGG-3′; si-SOX2-2, 5′-GGGACAUGAUCAGCAUGUAUC-3′. The siRNA sequences targeting OLIG2 were as follows: si-OLIG2-1, 5′-CCUUCGGUGCGCAAGCUUUTT-3′; si-OLIG2-2, 5′-CCUACUCAAGUCUCCGUCUTT-3′. si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′.
RNA isolation and qRT-PCR
Total RNA of glioma cells was extracted by an RNA-Quick Purification Kit (ES Science; Shanghai, China), and the RNA was immediately reverse transcribed into cDNA by using Hifair®III 1st Strand cDNA Synthesis SuperMix (Yeasen Biotechnology, #11141ES10). Real-time quantitative PCR amplification was performed using Hieff®qPCR SYBR Green Master Mix (Yeasen Biotechnology, #11201ES03). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize mRNA expression, and the primer sequences used for amplification of SLCO4A1-AS1 lncRNA were as follows: F, TGGGCAGAGTGTCGCTG; R, GGCATTCAGAGTTGCGTTCA; GAPDH: F, GCACCGTCAAGGCTGAGAAC; R, TGGTGAAGACGCCAGTGGA; SOX2: F, ACATGAACGGCTGGAGCAA; R, GTAGGACATGCTGTAGGTGGG; OLIG2: F, TCAAGTCATCCTCGTCCAGC; R, GGCTGTTGATCTTGAGACGC; CD15: F, CAACTGGACGCTCTCCTACC; R, ATGTTGGCTCAGTTGGTGGT; CD133: F, TACAACGCCAAACCACGACT; R, ACCCAGCCACCAGTATGAATC; NES: F, GACCCTGAAGGGCAATCACA; R, GGCCACATCATCTTCCACCA; GFAP: F, GCTTTGCCAGCTACATCGAG; R, TGCCAGATTGTCCCTCTCAAC.
Cell proliferation assay
Cell proliferation assay was measured using CCK-8 following the manufacturer’s protocol. Glioma cells (3 × 103 cells/well) were uniformly seeded into 96-well plates and incubated for 24 h. Then, the glioma cells were treated. 24, 48, 72 and 96 h after treatment, 10 µl CCK-8 solution (Yeasen Biotechnology, #40203ES76) was added to each well. The 96-well plates were incubated at 37 °C for 1 h and the absorbance was measured at 450 nm.
Colony formation assay
Firstly, the glioma cells were treated for 48 h and the treated cells were uniformly seeded into 6-well plates at 800 cells per well. Colony formation experiments were carried out by incubation for approximately 14 days, and the cells were fixed with 4% paraformaldehyde for 20 min and then stained with crystal violet for 15 min, and the results obtained were statistically analyzed.
EdU assay
We used EdU cell proliferation assay kits (Yeasen Biotechnology, #40275ES60) for the EdU assay. Firstly, the glioma cells were treated for 48 h and the treated cells were uniformly seeded into 24-well plates at 20,000 cells per well. After 24 h, the cells were incubated with EdU at 37 °C for 2 h and then fixed with 4% neutral paraformaldehyde fixative solution in the dark for 15 min. The cells were incubated with 0.1 mL of 0.5% Triton X-100 solution for 20 min at room temperature and then incubated with the Click-iT reaction mixture at room temperature for 30 min. The cells were incubated with Hoechst 33342 for 15–30 min to stain nuclei, and imaging and counting were performed under a microscope.
Cell invasion assay
In the transwell assay, after transfection for 48 h, LN229, A172, and U118 cells were placed into the upper chambers in medium without FBS. The lower chambers were filled with medium containing 15% FBS. After 48 h, the cells migrating through the membranes were fixed with 4% paraformaldehyde for 20 min, stained with 0.1% crystal violet for 10 min and counted. In the 3D invasion experiment, GBM#P3 cells (4000 cells/well) transfected for 48 h were uniformly seeded into 96-well plates. After 24 h, invasion gel (R&D Systems; 3500-096-03; Minneapolis, MN, USA) was added into the wells and the diameter of the invasion range of the tumor sphere was measured. 48 h after the invasion gel was added, the invaded spheroids were observed and the diameter of the invasion range of the tumor spheres was measured. Relative invasion was calculated by dividing the 48 h measurements by the initial 0 h measurements and the invasiveness of transfected tumor cells was examined by comparison with the control group. The diameter of the sphere was considered the starting point for quantification.
Apoptosis assay
The apoptosis assay was conducted based on the manufacturer’s protocol (Yeasen Biotechnology, #40302ES50). First, cells were uniformly seeded in 6-well plates and treated for 48 h. After centrifugation and collection, cells were resuspended with binding buffer, and then 5 μl of Annexin V-FITC and 10 μl of PI were added. 15 min later, apoptosis was assayed by flow cytometry.
Extreme limiting dilution assay
GBM#P3 and GBM#BG5 were spread into 96-well plates at a density gradient of 0, 2, 4, 8, 16, 32, 64, and 128 cells to assess self-renewal capacity, and the number of wells with successful tumor sphere formation was counted and analyzed using the ELDA software (http://bioinf.wehi.edu.au/software/elda/) after 10 days.
Neurosphere formation assay
GSCs were seeded at 1000 per well into 6-well plates and treated under certain conditions. 2 weeks later, the diameter of the tumor spheres was measured for statistical analysis.
Statistical analysis
Statistical analyses were completed using R (version 4.2.0) and GraphPad Prism (version 8.0). The Mann–Whitney test was used to compare two groups of data that were not subject to normal distribution and the Student’s t-test was employed to compare two groups of data that were. For comparisons among more than two groups, the Kruskal–Wallis test and one-way ANOVA were used for nonparametric and parametric data. Survival analysis was compared using the Kaplan‒Meier (K-M) method and the log-rank test. Three independent experiments were performed, and results were expressed as the mean ± the standard deviation (SD). P values < 0.05 were considered to indicate statistically significant differences.
Supplementary Information
Author contributions
Y.W. participated in the design of the study, analysis of the data, and writing of the draft. F.L., C.Y., X.Z., Z.X. and Y.S. provided data resources and analysis. X.L., X.L., Z.Z. and B.H. contributed to the draft reviewing and editing. Q.H., X.L. and M.H. participated in the study design and draft editing. All authors contributed to the manuscript and approved the submitted version. All authors read and approved the final draft.
Funding
This work is supported by Clinical Research Center of Shandong University (No. 2020SDUCRCB002), the Natural Science Foundation of China (82103277), Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022003A, JNL-2022041C and JNL-2022042C), the Department of Science and Technology of Shandong Province (ZR2019ZD33, 2021CXGC010903), Shandong Excellent Young Scientists Fund Program (2022HWYQ-035), Shandong Provincial Natural Science Foundation (ZR2021QH030), the Special Foundation for Taishan Young Scholars (tsqn202211041) and the Qilu Young Scholar Program of Shandong University, China.
Data availability
The data utilized in study were downloaded from TCGA (https://xenabrowser.net/datapages/), Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/) and Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/).
Competing interests
The authors declare no competing interests.
Ethics declarations
All the protocols in our study were admitted by the Research Ethics Committee of Shandong University and the Ethics Committee of Qilu Hospital (Shandong, China) (SDULCLL2021-2-26). All experiments and analyses were performed under the guidance of corresponding protocols or guidelines.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qibing Huang, Email: hqibing@163.com.
Xingang Li, Email: lixg@sdu.edu.cn.
Mingzhi Han, Email: mingzhi.han@sdu.edu.cn.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-82109-z.
References
- 1.Jemal, A. et al. Cancer statistics, 2007. CA Cancer J. Clin.57, 43–66. 10.3322/canjclin.57.1.43 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Han, M. et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain J. Neurol.143, 512–530. 10.1093/brain/awz406 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deorah, S., Lynch, C. F., Sibenaller, Z. A. & Ryken, T. C. Trends in brain cancer incidence and survival in the United States: surveillance, epidemiology, and end results program, 1973 to 2001. Neurosurg. Focus20, E1. 10.3171/foc.2006.20.4.E1 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Louis, D. N. et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-oncology23, 1231–1251. 10.1093/neuonc/noab106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, Z., Zhong, H., Liang, X. & Ni, S. Targeting tumor-associated macrophages for the immunotherapy of glioblastoma: navigating the clinical and translational landscape. Front. Immunol.13, 1024921. 10.3389/fimmu.2022.1024921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med.352, 987–996. 10.1056/NEJMoa043330 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Guo, Q. L. et al. Nanosensitizers for sonodynamic therapy for glioblastoma multiforme: current progress and future perspectives. Military Med. Res.9, 26. 10.1186/s40779-022-00386-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, X. et al. TRIM56 promotes malignant progression of glioblastoma by stabilizing cIAP1 protein. J. Exp. Clin. Cancer Res.41, 336. 10.1186/s13046-022-02534-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu, H. et al. TUBA1C is a prognostic marker in low-grade glioma and correlates with immune cell infiltration in the tumor microenvironment. Front. Genet.12, 759953. 10.3389/fgene.2021.759953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding, F., Chen, P., Bie, P., Piao, W. & Cheng, Q. HOXA5 Is recognized as a prognostic-related biomarker and promotes glioma progression through affecting cell cycle. Front. Oncol.11, 633430. 10.3389/fonc.2021.633430 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Augoff, K., McCue, B., Plow, E. F. & Sossey-Alaoui, K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol. Cancer11, 5. 10.1186/1476-4598-11-5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang, J. F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res.24, 513–531. 10.1038/cr.2014.35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, J. et al. SLCO4A1-AS1 promotes colorectal tumourigenesis by regulating Cdk2/c-Myc signalling. J. Biomed. Sci.29, 4. 10.1186/s12929-022-00789-z (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun.6, 8971. 10.1038/ncomms9971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atianand, M. K., Caffrey, D. R. & Fitzgerald, K. A. Immunobiology of long noncoding RNAs. Annu. Rev. Immunol.35, 177–198. 10.1146/annurev-immunol-041015-055459 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Z. & Jin, J. LncRNA SLCO4A1-AS1 promotes colorectal cancer cell proliferation by enhancing autophagy via miR-508-3p/PARD3 axis. Aging11, 4876–4889. 10.18632/aging.102081 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, Y., Sun, B., Gao, J., Huang, Y. & Wang, C. LncRNA SLCO4A1-AS1 accelerates growth and metastasis of gastric cancer via regulation of the miR-149/XIAP axis. Front. Oncol.11, 683256. 10.3389/fonc.2021.683256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, Y. L. et al. LncRNA SLCO4A1-AS1 suppresses lung cancer progression by sequestering the TOX4-NTSR1 signaling axis. J. Biomed. Sci.30, 80. 10.1186/s12929-023-00973-9 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell9, 391–403. 10.1016/j.ccr.2006.03.030 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Marie, Y. et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet (Lond., Engl.)358, 298–300. 10.1016/s0140-6736(01)05499-x (2001). [DOI] [PubMed] [Google Scholar]
- 21.Cheng, X. et al. Inhibition of glioma development by ASCL1-mediated direct neuronal reprogramming. Cells8, 45. 10.3390/cells8060571 (2019). [DOI] [PMC free article] [PubMed]
- 22.Wang, H. et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin. Cancer Res.19, 3764–3775. 10.1158/1078-0432.Ccr-12-3725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, K. et al. High expression of CKS2 predicts adverse outcomes: a potential therapeutic target for glioma. Front. Immunol.13, 881453. 10.3389/fimmu.2022.881453 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, R. et al. The N(6)-methyladenosine-modified pseudogene HSPA7 correlates with the tumor microenvironment and predicts the response to immune checkpoint therapy in glioblastoma. Front. Immunol.12, 653711. 10.3389/fimmu.2021.653711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, Y. C., Yen, M. H., De, S., Rajesh, R. & Tai, C. K. Optimized lipopolymers with curcumin to enhance AZD5582 and GDC0152 activity and downregulate inhibitors of apoptosis proteins in glioblastoma multiforme. Biomater. Adv.154, 213639. 10.1016/j.bioadv.2023.213639 (2023). [DOI] [PubMed] [Google Scholar]
- 26.Lakkadwala, S. & Singh, J. Dual functionalized 5-fluorouracil liposomes as highly efficient nanomedicine for glioblastoma treatment as assessed in an in vitro brain tumor model. J. Pharmaceut. Sci.107, 2902–2913. 10.1016/j.xphs.2018.07.020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozgiray, E., Sogutlu, F. & Biray Avci, C. Chk1/2 inhibitor AZD7762 enhances the susceptibility of IDH-mutant brain cancer cells to temozolomide. Med. Oncol. (Northwood, London, Engl.)39, 166. 10.1007/s12032-022-01769-x (2022). [DOI] [PubMed]
- 28.Jasek-Gajda, E., Jurkowska, H., Jasińska, M., Litwin, J. A. & Lis, G. J. Combination of ERK2 inhibitor VX-11e and voreloxin synergistically enhances anti-proliferative and pro-apoptotic effects in leukemia cells. Apoptosis24, 849–861. 10.1007/s10495-019-01564-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, M. et al. Blockage of extracellular signal-regulated kinase exerts an antitumor effect via regulating energy metabolism and enhances the efficacy of autophagy inhibitors by regulating transcription factor EB nuclear translocation in osteosarcoma. Front. Cell Dev. Biol.9, 650846. 10.3389/fcell.2021.650846 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, W. et al. VX-11e protects articular cartilage and subchondral bone in osteoarthritis by inhibiting the RIP1/RIP3/MLKL and MAPK signaling pathways. Bioorg. Chem.120, 105632. 10.1016/j.bioorg.2022.105632 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Bagley, S. J., Desai, A. S., Linette, G. P., June, C. H. & O’Rourke, D. M. CAR T-cell therapy for glioblastoma: recent clinical advances and future challenges. Neuro-oncology20, 1429–1438. 10.1093/neuonc/noy032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang, R. et al. LncRNA SLCO4A1-AS1 predicts poor prognosis and promotes proliferation and metastasis via the EGFR/MAPK pathway in colorectal cancer. Int. J. Biol. Sci.15, 2885–2896. 10.7150/ijbs.38041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J. et al. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J. Exp. Clin. Cancer Res. CR37, 222. 10.1186/s13046-018-0896-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Y. et al. lncRNA SLCO4A1-AS1 promotes growth and invasion of bladder cancer through sponging miR-335-5p to upregulate OCT4. Onco Targets Ther.12, 1351–1358. 10.2147/ott.S191740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, F. et al. SLCO4A1-AS1 regulates laryngeal squamous cell carcinoma cell phenotypes via the Wnt pathway. Oral Dis.29, 390–401. 10.1111/odi.13893 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Li, Q. et al. SLCO4A1-AS1 facilitates the malignant phenotype via miR-149-5p/STAT3 axis in gastric cancer cells. J. Oncol.2021, 1698771. 10.1155/2021/1698771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, W. & Li, J. Integrated analysis of genes associated with immune microenvironment and distant metastasis in uveal melanoma. Front. Cell Dev. Biol.10, 874839. 10.3389/fcell.2022.874839 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han, M. et al. Comprehensive characterization of TNFSF14/LIGHT with implications in prognosis and immunotherapy of human gliomas. Front. Immunol.13, 1025286. 10.3389/fimmu.2022.1025286 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sang, L. J. et al. LncRNA CamK-A regulates Ca(2+)-signaling-mediated tumor microenvironment remodeling. Mol. Cell72, 71-83.e77. 10.1016/j.molcel.2018.08.014 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Hu, G. et al. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J. Immunol. (Baltim. Md. : 1950)196, 2799–2808. 10.4049/jimmunol.1502146 (2016). [DOI] [PMC free article] [PubMed]
- 41.Zhang, H. J. et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther.9, 1801–1812. 10.2147/ott.S96535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A.102, 15545–15550. 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, S., Tang, L., Li, X., Fan, F. & Liu, Z. Immunotherapy for glioma: current management and future application. Cancer Lett.476, 1–12. 10.1016/j.canlet.2020.02.002 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Zhang, W. et al. GNG5 is a novel oncogene associated with cell migration, proliferation, and poor prognosis in glioma. Cancer Cell Int.21, 297. 10.1186/s12935-021-01935-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong, C. et al. HOXA-AS2 contributes to regulatory T cell proliferation and immune tolerance in glioma through the miR-302a/KDM2A/JAG1 axis. Cell Death Dis.13, 160. 10.1038/s41419-021-04471-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Di, W. et al. Clinical characterization and immunosuppressive regulation of CD161 (KLRB1) in glioma through 916 samples. Cancer Sci.113, 756–769. 10.1111/cas.15236 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun.4, 2612. 10.1038/ncomms3612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai, J. et al. Immunogenic cell death-related risk signature predicts prognosis and characterizes the tumour microenvironment in lower-grade glioma. Front. Immunol.13, 1011757. 10.3389/fimmu.2022.1011757 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gabrusiewicz, K. et al. Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS One6, e23902. 10.1371/journal.pone.0023902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, J. et al. Establishment and validation of a novel prognostic model for lower-grade glioma based on senescence-related genes. Front. Immunol.13, 1018942. 10.3389/fimmu.2022.1018942 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iglesia, M. D. et al. Genomic analysis of immune cell infiltrates across 11 tumor types. J. Natl. Cancer Inst.108, 45. 10.1093/jnci/djw144 (2016). [DOI] [PMC free article] [PubMed]
- 52.De Monte, L. et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med.208, 469–478. 10.1084/jem.20101876 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naranbhai, V. et al. HLA-A*03 and response to immune checkpoint blockade in cancer: an epidemiological biomarker study. The Lancet Oncol.23, 172–184. 10.1016/s1470-2045(21)00582-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jongsma, M. L. M. et al. The SPPL3-defined glycosphingolipid repertoire orchestrates HLA Class I-mediated immune responses. Immunity54, 132-150.e139. 10.1016/j.immuni.2020.11.003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomaszewski, W., Sanchez-Perez, L., Gajewski, T. F. & Sampson, J. H. Brain tumor microenvironment and host state: implications for immunotherapy. Clin. Cancer Res.25, 4202–4210. 10.1158/1078-0432.Ccr-18-1627 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariathasan, S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature554, 544–548. 10.1038/nature25501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashiro, K., Nakao, K., Ohba, S. & Hirose, Y. Human glioma cells acquire temozolomide resistance after repeated drug exposure via DNA mismatch repair dysfunction. Anticancer Res.40, 1315–1323. 10.21873/anticanres.14073 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Núñez, F. J. et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci. Transl. Med.11, 45. 10.1126/scitranslmed.aaq1427 (2019). [DOI] [PMC free article] [PubMed]
- 59.Havel, J. J., Chowell, D. & Chan, T. A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer19, 133–150. 10.1038/s41568-019-0116-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science348, 124–128, 10.1126/science.aaa1348 (2015). [DOI] [PMC free article] [PubMed]
- 61.Zhang, L., Giuste, F., Vizcarra, J. C., Li, X. & Gutman, D. Radiomics features predict CIC mutation status in lower grade glioma. Front. Oncol.10, 937. 10.3389/fonc.2020.00937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, J. et al. 5-Methylcytosine related LncRNAs reveal immune characteristics, predict prognosis and oncology treatment outcome in lower-grade gliomas. Front. Immunol.13, 844778. 10.3389/fimmu.2022.844778 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan, G. et al. Potential impact of ALKBH5 and YTHDF1 on tumor immunity in colon adenocarcinoma. Front. Oncol.11, 670490. 10.3389/fonc.2021.670490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity39, 782–795. 10.1016/j.immuni.2013.10.003 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform.14, 7. 10.1186/1471-2105-14-7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data utilized in study were downloaded from TCGA (https://xenabrowser.net/datapages/), Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/) and Chinese Glioma Genome Atlas (CGGA, http://www.cgga.org.cn/).