Abstract
Dissection of the physiological interactomes of histone post-translational modifications (hPTMs) is crucial for understanding epigenetic regulatory pathways. Peptide- or protein-based histone photoaffinity tools expanded the ability to probe the epigenetic interactome, but in situ profiling in native cells remains challenging. Here, we develop a nucleus-targeting histone-tail-based photoaffinity probe capable of profiling the hPTM-mediated interactomes in native cells, by integrating cell-permeable and nuclear localization peptide modules into an hPTM peptide equipped with a photoreactive moiety. These types of probes, such as histone H3 lysine 4 trimethylation and histone H3 Lysine 9 crotonylation probes, enable the probing of epigenetic interactomes both in HeLa cell and hard-to-transfect RAW264.7 cells, resulting in the discovery of distinct interactors in different cell lines. The utility of this probe is further exemplified by characterizing interactome of emerging hPTM, such as AF9 was detected as a binder of histone H3 Lysine 9 lactylation, thus expanding the toolbox for profiling of hPTM-mediated PPIs in live cells.
Subject terms: Chemical tools, Chemical modification, Epigenetics, Proteomics, Histone post-translational modifications
Dissection of the physiological interactomes of histone post-translational modifications is crucial for understanding epigenetic regulatory pathways. Here, authors develop a nucleus-targeting histone-tail-based photoaffinity probe capable of profiling the hPTM-mediated interactomes in native cells.
Introduction
Histone post-translational modifications (hPTMs, e.g., acetylation, methylation, and phosphorylation) play key roles in regulating the diverse protein-protein interactions (PPIs) involved in almost all chromatin-related regulatory events, including transcription, DNA replication, and chromatin remodeling1–3. To fully understand the function of each hPTM in complex cellular events, the set of its physiological binding partners (i.e., interactome) needs to be comprehensively determined, as exemplified by the recent identification of Menin as a key interactor of methylation of histone H3 lysine-79 involved in gene transcriptional regulation4,5. Given that hPTMs are typically highly dynamic and mediate weak or transient PPIs, classical biochemical methods for studying PPIs, such as affinity chromatography and immunoprecipitation, often result in a lack of information on interactors and cannot easily distinguish direct binding partners from indirect ones6–9.
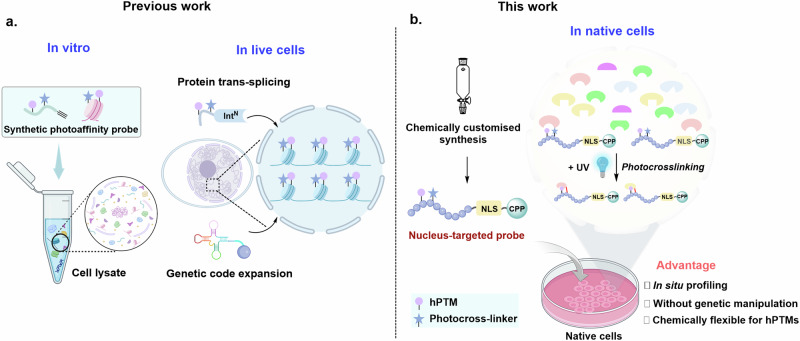
Photo-crosslinking allows the conversion of weak and transient interactions into covalent linkages, providing a powerful platform to examine PTM-mediated PPIs10–13. The recent emergence of peptide- or protein-based histone photoaffinity probes that deployed PTMs of interest at stoichiometric levels with a photo-crosslinking group has greatly expanded the ability to probe the epigenetic interactome14–17. These probes can generally be categorized into those that operate in an in vitro setting and those that manipulate in live cells (Fig. 1a). The former can be constructed through chemical flexibility and atomic customization, but its application requires lysis of cells, which usually leads to dilution of nucleoproteins and potential dissociation of protein complexes18–20; the latter allows in situ profiling of the chromatin interactome, but their assembly in live cells through genetic code expansion or split intein-based technology requires prior genetic manipulation, impeding their application in cells that are not readily amenable to transfection21–24. Thus, chemically tailored tools that enable in situ profiling of hPTM-specific interactomes in the context of live cells without genetic perturbations (native cells) remain to be developed.
Fig. 1. Schematic for profiling the hPTM-mediated interactomes.
a Previous strategies that function in an in vitro setting (a test tube) or rely on prior genetic manipulation. b The nucleus-targeted cell-permeable histone-tail-based photoaffinity probes enable it to penetrate into cells and selectively accumulate in the nucleus. The hPTMs-mediated interactomes could be captured in native cells upon UV irradiation. Some elements of Fig. 1a, b were created in BioRender. Chu, G (2024) https://BioRender.com/u07k630.
Here, we develop a histone-tail-based photoaffinity probe capable of profiling the epigenetic interactomes in native cells, by integrating cell-permeable and nuclear localization peptide modules into an hPTM peptide equipped with a photoreactive moiety via a readily chemically-customized synthesis (Fig. 1b). A simple coincubation of this probe with native cells enables its selective accumulation in the nucleus, after which hPTM-mediated interactomes can be covalently captured in situ upon photo-irradiation. We demonstrate the advantage and practicality of these probes by mapping interacting proteins of hPTMs, such as histone H3 lysine 4 trimethylation (H3K4me3) and histone H3 Lysine 9 crotonylation (H3K9cr) in hard-to-transfect cells (e.g., macrophages). The results further reveal that the interactomes of hPTMs are distinct in different cell lines. Our work also provides a technique platform to characterize the binding partners of emerging hPTM, such as the discovery of AF9 as an interactor of histone lactylation.
Results
Development of nucleus-targeted cell-permeable histone-tail-based photoaffinity probes
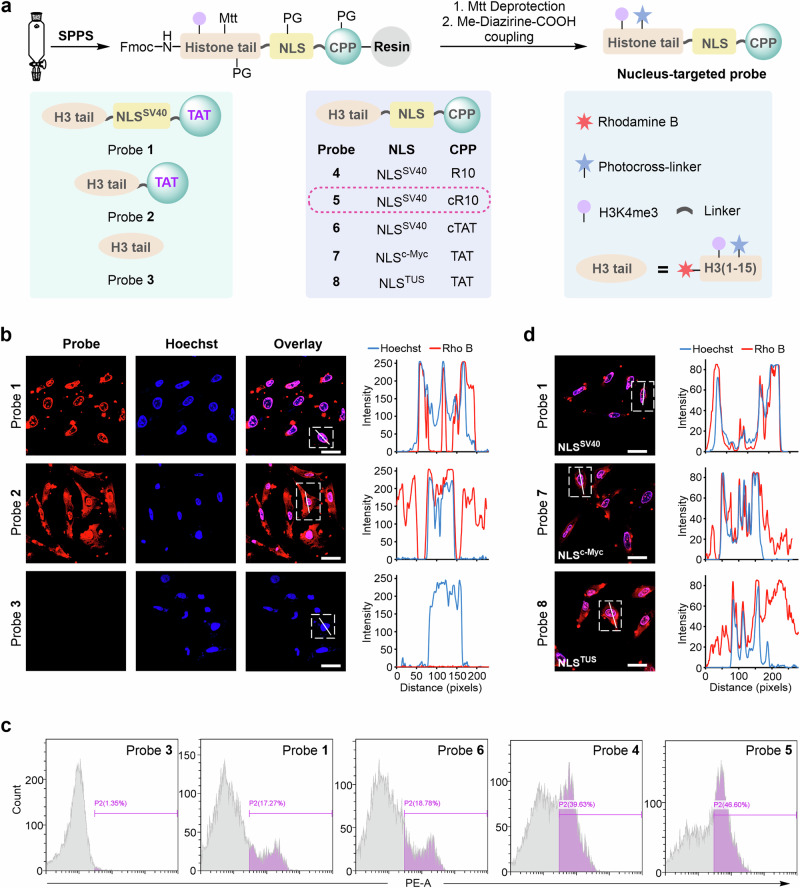
To probe the hPTM-mediated interactomes in live cells, we sought to develop a cell-permeable histone-tail-based photoaffinity probe capable of targeting the nucleus by incorporating a nuclear localization signal (NLS) peptide and a cell-penetrating peptide (CPP) module into the histone-derived peptide carrying a defined hPTM and a proximal photoreactive group (e.g., diazirine). We first synthesized H3K4me3-based probe 1, which carried the SV40-NLS sequence (NLSSV40, I PKKKRKV) and TAT-CPP (KRKKRRQRRRG) at the C-terminus of the H3 peptide (residues 1–15), along with a diazirine-modified lysine at position 7 of H3 (Fig. 2a)25,26. Probe 1 was assembled on Rink amide-AM resin (0.38 mmol/g) using standard Fmoc solid-phase peptide synthesis (SPPS), during which Fmoc-Lys(Me3)-OH and Fmoc-Lys(Mtt)-OH were incorporated at residues 4 and 7 of H31–15, respectively, and rhodamine B (RhoB) was coupled to the N-terminus of probe 1 for imaging (Supplementary Fig. 1). After completion of the chain elongation, the Mtt protecting group on Lys7 was removed by a mixture of HFIP/EDC (v/v = 1:1), resulting in the release of its side chain ε-amine, onto which a diazirine photo-crosslinker (i.e., 3-(3-methyl-3H-diazirine-3-yl)propionic acid) was attached27,28. Following cleavage from the resin and HPLC purification, purified probe 1 was obtained in 35% isolated yield (481 mg from 0.25 mmol resin), and its identity and purity were confirmed by LC-MS analysis (Supplementary Fig. 2, obs. 5465.0 Da, calc. 5465.6 Da). Using a similar approach, we also prepared probe 2 (containing no NLSSV40, Supplementary Fig. 3) and probe 3 (containing no NLSSV40 and TAT, Supplementary Fig. 4) for the subsequent comparison of the cellular permeability and nuclear targeting ability of probe 1.
Fig. 2. Chemical synthesis and visualization of nucleus-targeted cell-permeable histone-tail-based photoaffinity probes.
a Synthetic scheme and structure of probes 1 to 8. b Confocal microscopy images of HeLa cells treated with probes 1–3 for 1 h at 37 °C. Probes 1–3 were visualized using Rho B fluorescence (red channel), and Hoechst 33258 was utilized for nuclear staining (blue channel). 2D intensity profiles corresponding to the lines displayed on the images (line scans) are depicted to the right of the micrographs. Scale bars: 20 μm. Confocal microscopy images shown in (b) are representative of independent biological replicates (n = 3). c Cellular uptake analysis of probes in HeLa cells by flow cytometry. Cells were treated with probes 1 and 3–6 for 30 min at 37 °C. d Confocal microscopy images and line scans of HeLa cells treated with probes 1, 7, and 8. Scale bars: 20 μm. Confocal microscopy images shown in (d) are representative of independent biological replicates (n = 2). Source data are provided as a Source Data file.
Next, HeLa cells were treated with probes 1–3 (15 μM in serum-free DMEM) for 1 h at 37 °C and then washed three times with cold phosphate-buffered saline (PBS) to remove the remaining probe. After nuclear staining with Hoechst, the cellular uptake of the three probes was assessed by confocal laser scanning microscopy (CLSM)29. As shown in Fig. 2b, we observed obvious red fluorescence in cells treated with both probe 1 and probe 2 but not in those treated with probe 3; for probe 1, the red fluorescent signal mainly accumulated in the nucleus, whereas for probe 2, it was dispersed in the cytoplasm. These results suggested that both the NLS and CPP modules were required for the successful nuclear targeting of probe 1 in native cells.
To investigate the impact of different CPPs on the cellular uptake of the histone probes, we prepared probe 4 (containing linear R10, Supplementary Fig. 5), probe 5 (containing cyclic R10, Supplementary Figs. 6 and 7) and probe 6 (containing cyclic TAT, Supplementary Figs. 8 and 9) and evaluated their permeability in HeLa cells30,31. Probes 1, 3 (the control), 4, 5, and 6 (15 μM) were incubated separately with HeLa cells for 30 min under the same conditions. After thorough washing with PBS, the red fluorescence of the cells was quantified by flow cytometry analysis32, which showed that compared with probe 3 (without CPP sequence), treatment with probes 1, 4, 5, and 6 increased the relative fluorescence intensity of the cells from 1.35% to 17.27%, 39.63%, 46.60% and 18.78%, respectively (Fig. 2c and Supplementary Fig. 10). Of these, probe 5 containing the cR10 CPP showed the best cellular uptake, a result that was further confirmed by confocal imaging (Supplementary Fig. 11). In addition, probes with different NLS sequences (i.e., probe 7 with NLSc-Myc and probe 8 with NLSTUS) were produced to test the nuclear targeting ability (Supplementary Fig. 12, 13)33. Through CLSM analysis, we observed that probes 7 and 8 accumulated less in the nucleus than probe 1 (Fig. 2d), suggesting that the NLSSV40 sequence of probe 1 has better nuclear targeting capability. Based on the above results, we selected cyclic R10 and SV40 as the CPP and NLS modules, respectively, to construct histone probes in subsequent studies (Supplementary Fig. 14).
Nucleus-targeted histone-tail-based photoaffinity probes enable the capture of interactors that recognize H3K4me3 markers
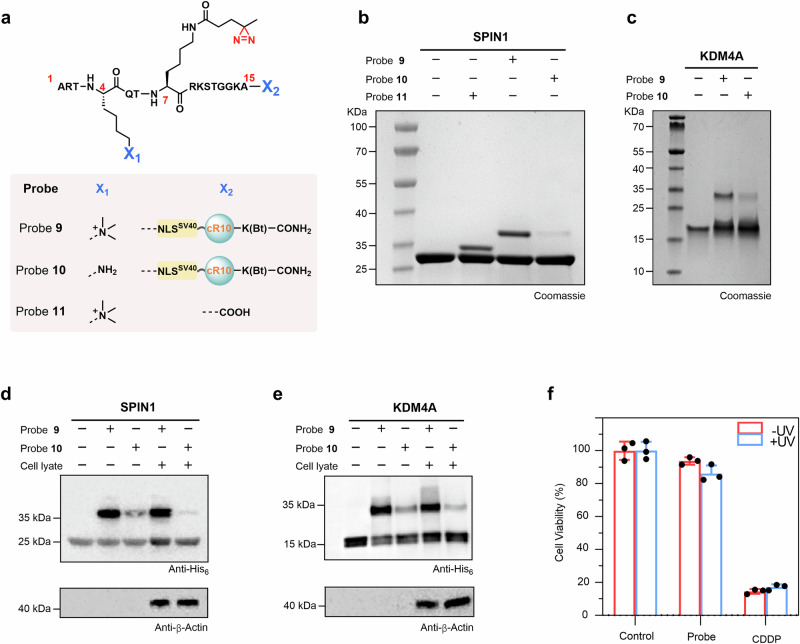
To examine whether our probes could capture hPTM-mediated interacting proteins, probes 9 (with the H3K4me3 marker, Supplementary Figs. 15 and 16) and 10 (without hPTMs, Supplementary Fig. 17), both devoid of the N-terminal rhodamine B, were synthesized for in vitro photo-crosslinking experiments (Fig. 3a). For this purpose, 12 μM 9 or 10 was mixed with a known H3K4me3 reader protein SPIN1 (6 μM) at 4 °C for 2 h, and then the mixture was exposed to 365 nm UV irradiation for 15 min34. SDS-PAGE analysis showed the formation of the expected crosslinked band corresponding to the conjugate of probe 9 and SPIN1 (i.e., ‘9-SPIN1’), whereas no obvious crosslinked band was observed for probe 10 (Fig. 3b). As a control, we tested the crosslinking of SPIN1 with probe 11 (Supplementary Fig. 18), which does not contain NLSSV40 or cR10, in contrast to 9. The expected ‘11-SPIN1’ crosslinked band was detected with an intensity similar to that of ‘9-SPIN1’, indicating that the introduction of NLSSV40 and cR10 does not interfere with the interaction of SPIN1 with H3K4me3 (Fig. 3b). In addition to SPIN1, we tested the crosslinking of probe 9 or 10 with other two known H3K4me3 readers, namely, ING2PHD (PHD domain of ING2) and KDM4ATudor (Tudor domain of KDM4A), both of which were effectively labeled by probe 9 but not by probe 10 after UV irradiation (Fig. 3c and Supplementary Fig. 19)35–37. Furthermore, to test whether probe 9 could capture hPTM-mediated interacting proteins in a complex environment, we performed photo-crosslinking assays in cell lysates using SPIN1 and KDM4ATudor. As we anticipated, the immunoblotting analysis indicated that the expected crosslinked bands (i.e., ‘9-SPIN1’ and ‘9-KDM4ATudor’) were efficiently formed after UV irradiation (Fig. 3d, e). To compare the conjugation efficiency of probe 9 and probe 10, we calculated the (band intensity of 10-SPIN1 conjugate)/(band intensity of 9-SPIN1 conjugate), which showed that the efficiency of probe 10 with SPIN1 was 6% of that of probe 9 (Supplementary Fig. 20). Similarly, the efficiency of probe 10 with KDM4ATudor was 23% of that of probe 9 (Supplementary Fig. 20). Taken together, the above results demonstrated that probe 9 has ability to specifically capture H3K4me3-mediated interacting proteins.
Fig. 3. Nucleus-targeted histone-tail-based photoaffinity probe to capture readers of H3K4me3 in vitro.
a Chemical structure of probes 9 to 11. b, c SDS-PAGE analysis of the labeling of SPIN1 and KDM4A by probe 9. Probes 10 and 11 were used for comparison for SPIN1 (b), and probe 10 was used for comparison for KDM4A (c). Gel images shown in (b, c) are representative of independent biological replicates (n = 2). d, e Cell lysate profiling using probe 9 with probe 10 as a comparison. Labeled proteins were analyzed by immunoblotting using an anti-His6 antibody. Immunoblotting images shown in (d, e) are representative of independent biological replicates (n = 2). f Viability of HeLa cells treated with 60 μM probe 9. Data were plotted as mean ± SEM of three independent biological replicates.
Finally, the effect of probe 9 on cell viability was measured using the MTT assay38, with untreated cells as the negative control and cisplatin treatment as the positive control. As shown in Fig. 3f, the relative cell viability of cells after treatment with 60 μM probe 9 for 12 h was approximately 90%, and UV irradiation for 15 min resulted in a slight decrease in relative cell viability to 87.9%, which was significantly higher than that of cisplatin-treated cells (less than 20%), suggesting that treatment with probe 9, either alone or in combination with UV irradiation, had minimal effect on cell viability. This result was further confirmed via live/dead cell staining analysis (Supplementary Fig. 21).
Chemical proteomics profiling of the H3K4me3 interactome using our probe in HeLa cells
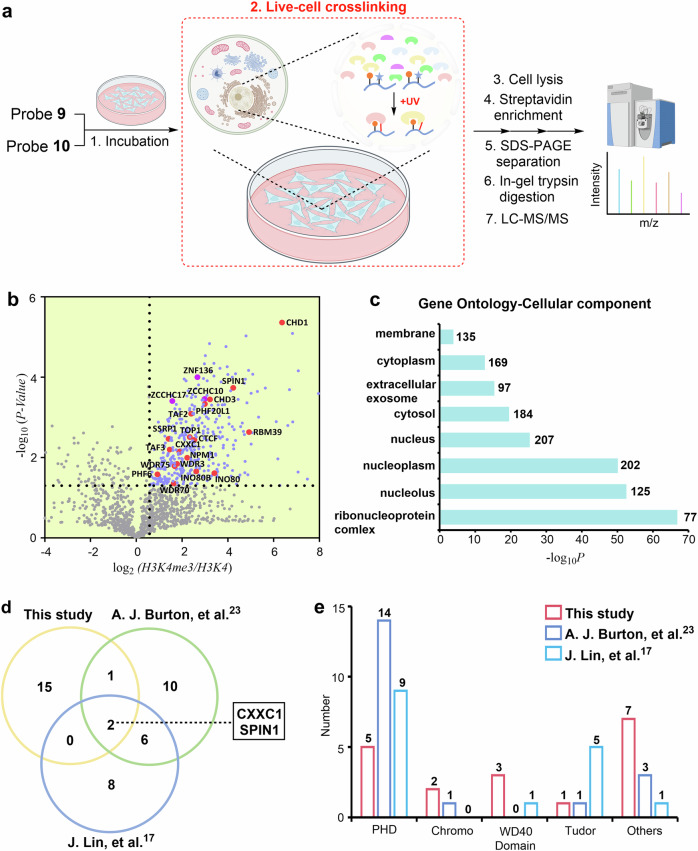
Encouraged by the ability of probe 9 to detect the H3K4Me3 reader, we next moved to a proteome-wide analysis in native cells. We established label-free quantitative (LFQ) mass spectrometry workflow to determine the interactome of H3K4me3 in native HeLa cells using probe 9 and probe 10 was used in parallel as a negative control (Fig. 4a). Briefly, HeLa cells were incubated with 9 or 10 (15 μM) at 37 °C for 12 h and then washed with cold PBS to remove any remaining probe that was not taken up by the cells. Following UV irradiation for 20 min, cell pellets were harvested and lysed by RIPA lysis buffer, and the probe 9- or 10-labeled proteomes were then enriched using streptavidin-coated magnetic beads. After thorough washing to remove nonspecific interactions, the enriched proteins were eluted from beads in the loading buffer. The samples were then separated by SDS-PAGE and analyzed by LFQ mass spectrometry39. Each proteomics assay was performed in triplicate. We analyzed candidates that were significantly enriched by probe 9 compared to probe 10 (fold-change >1.5, false discovery rate <0.05)23.
Fig. 4. Determining the in situ interactome of H3K4me3 in HeLa cells.
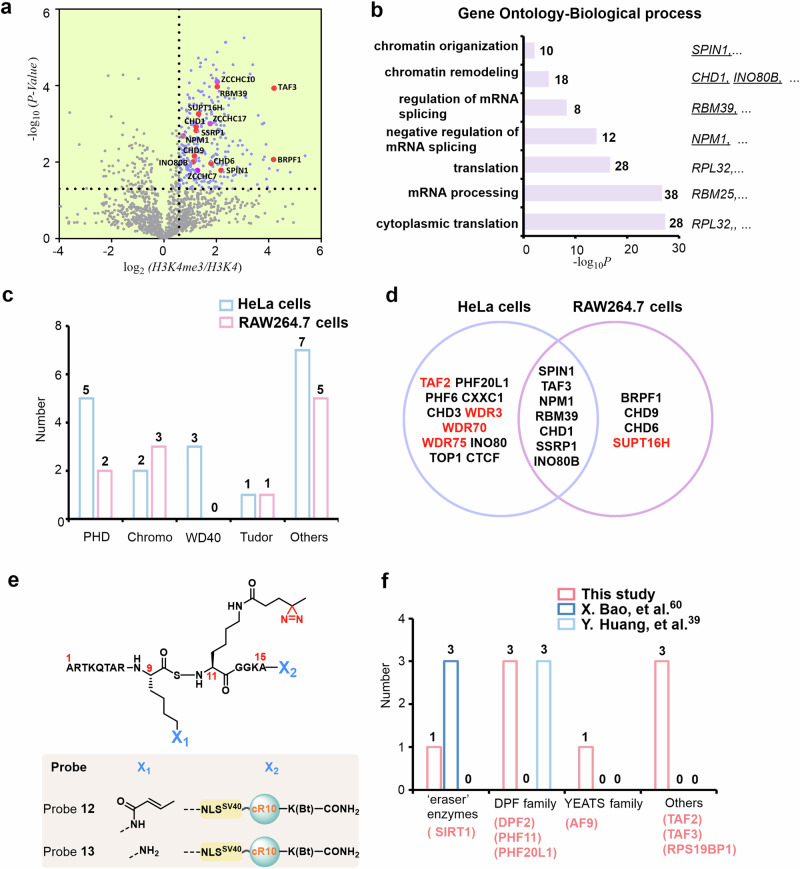
a Schematic for the LFQ proteomics workflow to determine the interactome of H3K4me3 by probe 9. Probe 10 was used for comparison. Some elements of Fig. 4(a) were created in BioRender. Chu, G. (2024) https://BioRender.com/u07k630. b Volcano plots of the quantitative mass spectrometry results. Significantly enriched hits (p < 0.05, >1.5-fold-change) are colored blue. Some established H3K4me3 reader proteins are highlighted and labeled in red. A two-sided test was used. c GO analysis (cellular components) of significantly enriched hits (p < 0.05, >1.5-fold-change) in Fig. 4b. The number of proteins in each GO term is shown. d Venn diagram comparison of identified H3K4me3 reader proteins in the present study and two previous studies, respectively. e Number of identified H3K4me3 reader proteins containing PHD, Chromo, WD40, and Tudor domains in the present study and two previous studies. Source data are provided as a Source Data file.
Analysis of the proteomics data revealed that a total of 381 proteins were significantly enriched by probe 9, 67% of which were distributed in the nucleus according to subcellular location analysis of the UniProtKB database (Fig. 4b and Supplementary Fig. 22), and Gene Ontology cellular component (GOCC) analysis also suggested that most of them were present in the nucleus (Fig. 4c). Among these enriched proteins, 18 previously reported H3K4me3 readers were identified, such as established proteins containing the PHD domain (e.g., CXXC1 and TAF3), chromodomain (e.g., CHD1 and CHD3), WD40 domain (WDR3), Tudor domain (SPIN1) and others (e.g., TOP1 and NPM1) (Fig. 4b and Supplementary Table 1)40–47. Of the 18 proteins, TAF2 (a component of the TFIID complex) and INO80B (a core component of the chromatin-remodeling INO80 complex) indirectly interacted with H3K4me341,48. Moreover, Gene Ontology biological process (GOBP) analysis of these high-confidence probe 9-labeled proteins revealed that the hit proteins were mainly involved in key nuclear functions, such as chromatin organization (e.g., H3K4me3-specific chromatin reader SPIN1), chromatin remodeling (e.g., chromatin-remodeling factors CHD1), and positive regulation of transcription (e.g., transcriptional activator CXXC1) (Supplementary Fig. 23)40,42,45. In contrast, using (FC < 0.67; false discovery rate <0.05) as a cutoff, we did not find any known H3K4me3 interactors.
We also found that our probe-enriched proteins of H3K4me3 in live cells covered four domains, namely, the PHD, chromo, WD40, and Tudor domains (18 in total), which is quite different from previous work (Supplementary Table 2 and Fig. 4d, e)17,23. Moreover, we noted that the subunits of the transcription factors TFIID complex (TAF3, TAF2), FACT complex (SSRP1) and INO80 complex (INO80B, INO80B) were only captured by our probe41,46,48, which may be due to the fact that in situ profiling of our probe is more favorable to capture the subunits of the complexes, whereas lysing the cells may lead to dissociation of the complexes. In addition to the previously known H3K4me3 readers, we also identified several potential interactors in significant hit proteins, which contained the CCHC or C2H2-type zinc finger domain (e.g., ZCCHC10, ZCCHC17, ZNF136) (Table S1). Note that zinc finger CW domain-containing proteins (e.g., ZCWPW1 and ZCWPW2) have been reported as a class of interactors of H3K4me349. Taken together, the above results suggested the capability of our probe for profiling H3K4me3 interactomes in native cells.
The interactome of H3K4me3 and H3K9cr in RAW264.7 cells
hPTM-mediated interactions play an essential role in regulating the fate of macrophages as well as the type and duration of macrophage-mediated inflammatory responses, but the detection of hPTM-specific interactomes in macrophages using genetic code expansion or ultrafast trans-splicing intein-based technology is hampered by the natural defense against gene transfection50,51. To solve this problem, we investigated the interactome of H3K4me3 in macrophage cells (RAW264.7) using probe 9. By taking a similar chemoproteomics workflow as described above (fold-change >1.5, false discovery rate <0.05), a total of 302 candidate proteins were significantly enriched in three biological replicate experiments, of which 201 proteins were characterized as known nuclear proteins (Supplementary Fig. 24). We identified 11 H3K4me3 readers with high confidence, including proteins containing the PHD domain (e.g., TAF3 and BRPF1), Chromodomain (e.g., CHD1, CHD6 and CHD9), Tudor domain (SPIN1), and others (e.g., SSRP1 and SUPT16H) (Fig. 5a)41,42,45,46,52. GOCC analysis showed that the enriched proteins were mainly in the nucleus (e.g., nucleoplasmand, nucleolus) (Supplementary Fig. 25), and GOBP analysis revealed that these proteins were involved in chromatin organization (e.g., SPIN1), chromatin remodeling (e.g., CHD1 and INO80B), translation and mRNA processing (Fig. 5b). These observations demonstrated that our probe enables the profiling of hPTM-mediated interactions in hard-to-transfect cells.
Fig. 5. Determining the interactome of H3K4me3 and H3K9cr in RAW264.7 cells.
a Volcano plots of mass spectrometry results. Hits significantly enriched by probe 9 (p < 0.05, >1.5-fold-change) are colored blue. Some established H3K4me3 reader proteins are highlighted and labeled in red. A two-sided test was used. b GO analysis (biological process) of significantly enriched hits (p < 0.05, >1.5-fold-change) in Fig. 5a. The number of proteins in each GO term is shown. c Number of identified H3K4me3 reader proteins containing PHD, chromo, WD40, and Tudor domains by probe 9 in HeLa and RAW264.7 cells. d Overlap of identified H3K4me3 reader proteins using probe 9 in HeLa and RAW264.7 cells. e Chemical structure of probe 12 and probe 13. f Number of identified H3K9cr reader proteins containing eraser enzymes, DPF, YEATS, and other families in the present study and two previous studies. Red-colored words indicate H3K9cr reader proteins enriched in this study. Source data are provided as a Source Data file.
We then compared the H3K4me3 interactors identified in RAW264.7 cells with those in HeLa cells. We found that the interactors in RAW264.7 cells were also distributed in the PHD, Chromo, or Tudor recognition domains, among which seven of the same reader proteins (SPIN1, TAF3, CHD1, NPM1, SSRP1, INO80B, RBM39) were captured in both cell lines (Fig. 5c, d and Supplementary Table 3), however the proteins belong to WD40 domain were not significantly enriched in RAW264.7 cells. Moreover, probe 9 enriched several proteins involved in chromatin remodeling and chromatin organization in both HeLa cells and RAW264.7 cells, e.g., CHD1 and SPIN1 in HeLa cells and INO80B and SPIN1 in RAW264.7 cells. Notably, we found that WD40 family proteins (e.g., WDR3) and TAF2 were detected only in HeLa cells, which might be due to the expression of WDR3 and TAF2 being upregulated in malignant cells compared to nontumor cells53,54. Moreover, SUPT16H, which exerts key roles in nucleosome reconstruction (component of the FACT complex, histone chaperone), was captured in RAW264.7 cells, possibly because the transcriptional elongation of some inflammation-related genes (e.g., LPS-dependent genes) in macrophages requires destabilization and restoration of nucleosome structure via histone chaperone, whereas the activity of these genes was much lower in HeLa cells (Fig. 5d)55,56.
Moreover, we extended our strategy to determine the interactome of histone crotonylation in RAW264.7 cells. To this end, probes 12 (with the diazirine and crotonylation groups installed at positions 9 and 11 of H3, respectively, Supplementary Figs. 26 and 27) and 13 (without the crotonylation group compared to probe 12, Supplementary Fig. 28) were prepared by modular solid-phase peptide synthesis (Fig. 5e). Subsequent proteomic analysis revealed that a total of 359 candidate proteins were significantly enriched, of which 210 proteins were characterized as known nuclear proteins (Supplementary Fig. 29). We identified 8 proteins that directly or indirectly interact with histone crotonylation with high confidence, including ‘eraser’ enzymes (e.g., Sirt1), DPF family (e.g., DPF2), YEATS family (e.g., AF9) and others (the subunits of the TFIID complex, TAF3 and TAF2) (Fig. 5f and Supplementary Fig. 30)39,57–59. GOCC analysis showed that the enriched proteins were mainly located in the nucleus (e.g., nucleus, nucleoplasm) (Supplementary Fig. 31) and GOBP analysis revealed that these proteins are involved in mRNA processing, RNA splicing, etc (Supplementary Fig. 32). We also note that by using other crosslinking probes, only “eraser” enzymes (e.g., Sirt1, Sirt2, Sirt3) were captured in Hela cell lysates60, whereas only interactors of the DPF family (DPF2, PHF3, and PHF6) were found in 293 T cell39. In contrast, the proteins enriched by our probe cover three families, of which proteins such as AF9, TAF3, and TAF2 were only captured in RAW264.7 cells. These results suggested that the interactomes of hPTMs were quietly distinct in different cell lines, and the development of a generalized method to identify the interactomes in live cells conveniently and rapidly will facilitate mapping a more accurate proteomic landscape.
Proteomic profiling of the H3K9la-specific interactome in RAW264.7 cells
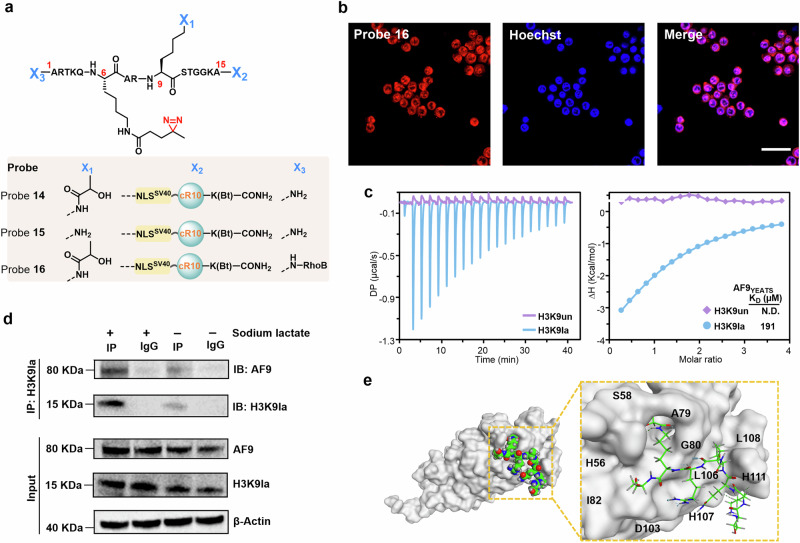
Next, our probes were expanded to characterize the interactome of a recently identified hPTM, i.e., histone lactylation, which serves as an important epigenetic modification regulating gene transcription in RAW264.7 cells during hypoxia and bacterial challenge61–63. To this end, probes 14 (with the diazirine and lactylation groups installed at positions 6 and 9 of H3, respectively, Supplementary Figs. 33 and 34), 15 (without the lactylation group compared to probe 14, Supplementary Fig. 35) and 16 (with the RhoB groups installed at the N-terminal compared to probe 14, Supplementary Figs. 36 and 37) were prepared by modular solid-phase peptide synthesis (Fig. 6a), and CLSM analysis confirmed the successful delivery of these probes into the nucleus (Fig. 6b). Subsequent proteomics analysis revealed that 11 candidate proteins were significantly enriched (Supplementary Fig. 38). Among these, AF9 (a chromatin reader component of super elongation complexes) caught our attention because its YEATS domain has been reported to recognize many other histone lysine acylations (e.g., acetylation and crotonylation)64.
Fig. 6. Determining the in situ interactome of H3K9la.
a Chemical structure of probes 14, 15, and 16. b Confocal microscopy images of RAW264.7 cells treated with probe 14 for 1 h at 37 °C. Probe 14 was visualized using TER fluorescence (red channel), and Hoechst 33258 was utilized for nuclear staining (blue channel). Scale bars: 20 μm. Confocal microscopy images shown in (b) are representative of independent biological replicates (n = 3). c ITC fitting curves of AF9YEATS with H31-10K9la (blue) peptide or H31-10 (red) peptide. d Co-IP assay was conducted to detect the interaction of H3K9la and AF9 in RAW264.7 cells with or without sodium lactate treatment. Immunoblotting images shown in (d) are representative of independent biological replicates (n = 3). e Structural modeling of AF9YEATS and H31-10K9la peptide. The AF9YEATS structure was displayed on a gray surface, the ligand is shown in sphere and sticks, the carbon atoms are green, the oxygen atoms are red, and the nitrogen atoms are blue. Source data are provided as a Source Data file.
To further validate the interaction between AF9 and H3K9la, the binding of AF9 to H3K9la then quantitatively measured by isothermal titration calorimetry (ITC) assays using the recombinant AF9 YEATS domain (residues 2–138, hereafter AF9YEATS) and two synthetic H3 peptides (i.e., H31-10K9la and H31-10, Supplementary Figs. 39 and 40). The dissociation constant (KD value) of AF9YEATS for H31-10K9la was measured to be 191 μM, while the binding affinity between AF9YEATS and H31-10K9 was too weak to determine (Fig. 6c), suggesting that lactylation of H3K9 indeed promotes AF9-YEATS binding. To further investigate the binding of AF9 and H3K9la in vivo, we conducted a co-immunoprecipitation (co-IP) assay in Raw264.7 cells, both with or without sodium lactate treatment. We observed that upon stimulation with sodium lactate (50 mM), the levels of H3K9la and AF9 protein were increased compared to the control group, which did not receive sodium lactate treatment. Moreover, the co-IP results confirmed that sodium lactate treatment significantly enhanced the interaction between H3K9la and AF9 (Fig. 6d and Supplementary Fig. 41). All the results demonstrate that our finding is biologically relevant. Based on the reported cocrystal structure of AF9YEATS with the H33-10K9cr peptide (PDB: 5HJB; ‘cr’ refers to crotonylation), we performed molecular docking of AF9YEATS in complex with H31-10K9la, where the α-hydroxyl group of the lactic acid moiety may form a hydrogen bond with the S58 side chain of AF9YEATS, thereby contributing to AF9YEATS binding (Fig. 6e; Supplementary Figs. 42 and 43)58,65. These results showed that despite the relatively low binding affinity of AF9YEATS for H3K9la, our probe enables us to detect and differentiate such weak protein-protein interactions in the complex cellular environment.
Discussion
In this study, we reported a nucleus-targeted histone-tail-based photoaffinity probe for profiling the hPTM-mediated interactome in native cells. This type of probe, integrating the CPP and NLS modules into the hPTM peptide equipped with a photoreactive moiety, can be easily customized via Fmoc-SPPS-based modular construction. Our studies showed that simple coincubation of this functional probe with cells enables its efficient cellular uptake and selective accumulation in the nucleus, during which hPTM-mediated interacting proteins can be covalently captured in situ upon UV irradiation. By coupling our probe with LFQ-based proteomics analysis, we demonstrated the capability to profile H3K4me3 interactomes in HeLa cells, where 18 established H3K4me3 interactors that cover all four known (PHD, chromo, WD40, Tudor) H3K4me3 recognition domain families were identified with high confidence (Fig. 4b and Supplementary Table 1). Compared with previously reported probes that work in a cell lysate or in the extracted nuclei, our probe exhibited comparable ability to capture the interacting proteins (Fig. 4d, e and Supplementary Table 2). Furthermore, this easy-to-implement chemical probe has a modular and programmable design, that is low technical challenge for the majority of biochemistry laboratories. Notably, the use of our probe capability for noninvasive-treatment of target cells is due to avoiding genetic manipulation or transfection, which is particularly valuable for cells where genetic operation is difficult (e.g., hard-to-transfect cells).
The hPTM-specific interactomes of macrophage cells undergo highly dynamic regulation during immunological processes, and the natural defense of such cells against gene transfection makes it difficult to profile the interactomes of hPTMs using genetic manipulation. With the prepared probe in hand, we performed global profiling of the H3K4me3-specific interactome in RAW264.7 cells in combination with a chemoproteomics workflow and identified 11 known interacting partners (Fig. 5a), similar to the results in HeLa cells, belonging to the three structural domains of PHD, Chromo, and Tudor (Fig. 5c and Supplementary Table 3). We also identified various interactors of H3K4me3 that differed between these two cell types: for instance, WD40 family proteins and TAF2 were significantly enriched only in tumor cells, whereas proteins associated with nucleosome reconstruction, such as SUPT16H, were only captured by the probe from RAW264.7 cells (Fig. 5c, d). We also successfully identified the interactome for histone crotonylation in RAW264.7 cells using this strategy, in which only proteins such as AF9, TAF3, and TAF2 were captured in RAW264.7 cells. These results suggest that diverse cell types may differ in their hPTM interactome landscapes in terms of protein abundance, turnover rates, and location due to various cellular functions and that the development of easily accessible probes is of great importance for the global screening and comparison of hPTM interactomes in different types of native cells. We further probed the interactome of a recently discovered hPTM (lactylation) using our probe and found that AF9 is a potential binding partner of H3K9la (Fig. 6c, d), demonstrating the utility of chemically tailored probes in addressing cutting-edge biology issues. Future studies would extend the utility of probes to profile hPTM-specific interactomes in primary neurons and tissues
Current peptide-based photoaffinity probes lack the context of the chromatin or nucleosome structure, which may not fully recapitulate the native chromatin environment, thus resulting in some failure in identifying chromatin interactors. On the other hand, we were able to identify reliably established binders, suggesting that our nuclear-targeted peptide-based probes are valuable for determining the unstructured regional hPTM-specific interactomes (e.g., H3K4me3). Nevertheless, the extension of the nuclear-targeted approach to nucleosomes, i.e., by preparing cell-penetrating, nuclear-targeted histones (containing CPP and NLS modules) to generate nuclear-targeted nucleosome photoaffinity probes to identify the interactomes of hPTMs in a physiologically relevant nucleosome context, is clearly an exciting direction for future studies. We anticipate that the multifunctional probes described here will help to determine the interactomes of a range of known and emerging hPTMs. We also found that stable isotope labeling by amino acids in cell culture (SILAC) quantification was used to profile H3K4me3 interactors in some previous work17,23, which contributed to a relatively lower false-positive rate compared with LFQ or a tandem mass tag (TMT) quantitative analysis39,66. In the future study, we will utilize SILAC quantitative proteomics combined with our probe to further explore the interactomes of histone PTMs in situ, and we look forward to more in-depth results. Our work showcase here also highlights the advantages of a chemical protein synthesis platform for the construction of tailored-designed probes for biological applications67,68.
Methods
General materials and methods
All reagents have been purchased from commercially available suppliers and are ready to use without needing to be purified. Wang and Rink amide-AM resin were acquired from Tianjin Nankai HECHENG S&T Co., Ltd. in Tianjin, China, while Fmoc-protected amino acids were sourced from GL Biochem in Shanghai, China. Antibodies used include: Anti-His6 antibody (Abcam; ab18184), Anti-β-Actin antibody (Cell Signaling Technology; #4970), anti-AF9 antibody (Cell Signaling Technology; #47577), Anti-H3K9la antibody (PTM BioLab, Inc.; PTM-1419RM). HPLC purification of peptides was conducted using Shimadzu Prominence systems from Shimadzu Corporation, Japan, with each sample monitored at wavelengths of 214 nm and 254 nm. Peptides were analyzed or semi-prepared using Welch Ultimate XB C18 or C4 analytical (flow rate was 1.0 mL/min) or semi-preparative columns (flow rate was 4–5 mL/min). HPLC purification solvent system consisted of buffer A (0.08% TFA in acetonitrile) and buffer B (0.1% TFA in H2O). Fast Protein Liquid Chromatography (FPLC) was carried out using an AKTA system from GE Healthcare Life Sciences, equipped with Superdex 75 or 200 columns. Each sample was detected at wavelengths of 280 nm. Standard ESI mass spectra were acquired using the LC/MS 2020 system from SHIMADZU. For LC-MS/MS, the Orbitrap Exploris 480 mass spectrometer was utilized.
Chemical synthesis of probes
All probes utilized in this study were synthesized using Fmoc-SPPS-based modular synthesis with a scale of 0.25 mmol resin (for probes 1–10, 12–16, Rink amide-AM resin, with a loading of 0.38 mmol/g; for probe 11 and H31-10K9la and H31-10, Wang resin, with a loading: 0.32 mmol/g). In general, the synthesis of peptide was started with swelling of the resin in DMF for 5–8 min. Amino acid coupling was carried out for 30 min at 37 °C, using 4.0 eq. of Fmoc-AA-OH, 8.0 eq. of DIEA, and 4.0 eq. of HCTU in DMF. The Fmoc group was removed by exposing the resin to a 20% (v/v) piperidine solution in DMF for 10 min at 37 °C. Following each coupling reaction and Fmoc-deprotection, the resin was extensively rinsed with DMF and DCM in succession (five times with DMF, five times with DCM, and another five times with DMF). Apart from these, some special operations during peptide synthesis including the Alloc protecting group (0.25 mmol resin) were removed using Pd[P(Ph)3]4 (1.0 eq. 288.9 mg) and PhSiH3 (10.0 eq., 308.5 μL) in 5 mL DCM for 3 h; The Mtt protecting group was removed using HOBT in a mixture of HFIP/EDC in a ratio of 1/1 for 5 min × 8; the coupling of diazirine photo-crosslinker onto the lysine side chain was performed for 30 min at 37 °C, using 4.0 eq. of Me-Diazirine-COOH, 4.0 eq. of HATU, 4.0 eq. of HOAT, and 8.0 eq. of DIEA in DMF; the coupling of crotonic anhydride onto the lysine side chain was performed in the presence of 10 eq. of crotonic anhydride and 5 eq. of DIEA in DMF; the coupling of L-lactate onto the lysine side chain was performed in the presence of 2.0 eq. of L-lactate, 2.0 eq. of HATU, 2.0 eq. of HOAT, and 4.0 eq. of DIEA in DMF; the coupling of Rhodamine B onto the N-terminal amine groups of probes was performed in the presence of 2.0 eq. of Rhodamine B, 2.0 eq. of HATU, 2.0 eq. of HOAT and 4.0 eq. of DIEA in DMF. A configured TFA cocktail (87.5% TFA, 5% phenol, 5% H2O, 2.5% Tips) was added to the dried resin and shaken at room temperature (2 h) to cleave peptides from the resin. Crude peptides were purified by semi-preparative RP-HPLC, and the yields for each probe are listed in Supplementary Table 4.
Cell culture
RAW264.7 was purchased from cellcook (CC9001), and HeLa was a generous gift from Yangzhong Liu (University of Science and Technology of China, Hefei). HeLa cells were cultured in DMEM supplemented with 100 μg/mL streptomycin, 10% FBS, and 100 U/mL penicillin (37 °C, 5% CO2). The cells were exposed to a 0.25% trypsin 0.02% EDTA solution and incubated for 30 s. The supplemented media was added to quench trypsin. The collected cell suspension was subjected to centrifugation (1.0 × g, 5 min). The medium was then aspirated, and the cell was resuspended in a new medium for a new round of adherent growth. RAW264.7 cells were cultured in DMEM containing 10% FBS in an incubator (37 °C, 5% CO2). The cells were scraped off the walls of culture flasks with a cell scraper in PBS. The collected cell suspension was subjected to centrifugation (1.0 × g, 5 min). The PBS was then aspirated, and the cells were placed back into a new medium for a new round of adherent growth.
General procedure for probe delivery
For microscopy experiments, 20,000 cells were seeded into sterile 35 mm glass-bottom dishes. The cells were allowed to attach and proliferate for 24 h in an environment with 5% CO2 at a temperature of 37 °C. Once the cells reached approximately 90% confluency, they were gently rinsed three times with warm PBS. Afterward, they were incubated at 37 °C for 60 min with a serum-free medium that included probes 1–8 (15 or 30 μΜ in final concentration for CLSM analysis, 15 μΜ in final concentration for flow cytometry analysis). Cells were then stained with Hoechst 33258 (2 μg/mL) and then imaged by CLSM analysis.
The dispersion of fluorescent probes within living cells was examined utilizing a confocal laser scanning microscope (Zeiss LSM 880) fitted with a 40x/1.3 Oil DIC M27 objective lens. For the distinct fluorescent labels, dual lasers were employed: a UV laser for Hoechst 33258 at 405 nm and a green laser for Rho B at 543 nm. Throughout the confocal laser scanning microscopy (CLSM) examination, the specimens were maintained at 37 °C within a humidified atmosphere.
Flow cytometry analysis
Cells were incubated with FBS free culture medium containing different probes. After 30 min, cells were washed three times to remove unabsorbed probes. Cells were resuspended in PBS buffer and analyzed by flow cytometer (CytoFLEX, Beckman). The data were analyzed by CytExpert.
Protein expression and purification
The recombinant proteins KDM4ATudor, SPIN1, and AF9YEATS were expressed in the E. coli BL21 cells and triggered for expression by 0.2 mM IPTG at 16 °C for 12 h. The recombinant proteins ING2PHD were expressed in E. coli Rosetta cells and triggered for expression by 0.5 mM IPTG at 18 °C for 12 h. The harvested cells for SPIN1 and KDM4ATudor were suspended in a buffer containing 20 mM Tris and 100 mM NaCl (pH = 7.5), while AF9YEATS was suspended in a buffer with 20 mM Tris and 500 mM NaCl (pH = 7.5). For ING2PHD, the cells were resuspended in a solution of 50 mM HEPES, 150 mM NaCl, and 2 mM DTT (pH = 7.5). Subsequent purification of these proteins was conducted according to established methods32–35.
Briefly, harvested cells were lysed by ultra-sonication in their respective resuspension buffers containing 1 mM fresh PMSF. After centrifugation at 4 °C with a relative centrifugal force of 11.6 × g for 30 min, the supernatant was applied to either a Ni-NTA or GST column, depending on the type of affinity tag present on the protein. Post the initial wash with 5 volumes of the suspension buffer, the proteins were eluted using specific elution buffers. For SPIN1 and KDM4ATudor, an elution buffer consisting of 20 mM Tris, 100 mM NaCl, and 500 mM imidazole (pH = 7.5) was utilized for protein recovery. For AF9YEATS, the column underwent additional extensive washing with resuspension buffers containing high salt concentrations of 750 mM and 1000 mM NaCl. The protein was subsequently eluted using a buffer composed of 20 mM Tris, 500 mM NaCl, and 500 mM imidazole (pH = 7.5). Following elution, the His tag was removed by incubating the protein overnight with thrombin from Solarbio. For the recombinant ING2PHD, elution was achieved using a buffer containing 150 mM NaCl, 50 mM HEPES, 25 mM Glutathione, and 2 mM DTT (pH = 7.5). Following concentration, the purified recombinant proteins underwent additional purification through size exclusion chromatography utilizing a Superdex 75 10/300 column.
Construction of protein expression plasmids
SPIN1
The cDNA encoding the SPIN1 was cloned into the pRSFDuet vector.
Amino acid sequence
R RNIVGCRIQH GWKEGNGPVT QWKGTVLDQV PVNPSLYLIK YDGFDCVYGL ELNKDERVSA LEVLPDRVAT SRISDAHLAD TMIGKAVEHM FETEDGSKDE WRGMVLARAP VMNTWFYITY EKDPVLYMYQ LLDDYKEGDL RIMPDSNDSP PAEREPGEVV DSLVGKQVEY AKEDGSKRTG MVIHQVEAKP SVYFIKFDDD FHIYVYDLVK TS
ING2PHD
The PHD finger domain of human ING2 (residues 208-270) was cloned into a pGEX-6p-1 vector with an N-terminal glutathione S-transferase (GST) tag.
Amino acid sequence
DPN EPTYCLCNQV SYGEMIGCDN EQCPIEWFHF SCVSLTYKPK GKWYCPKCRG DNEKTMDKST
KDM4ATudor
The cDNA encoding the Tudor domain of human KDM4A was cloned into the pRSFDuet vector.
Amino acid sequence
ALQSIT AGQKVISKHK NGRFYQCEVV RLTTETFYEV NFDDGSFSDN LYPEDIVSQD CLQFGPPAEG EVVQVRWTDG QVYGAKFVAS HPIQMYQVEF EDGSQLVVKR DDVYTLDE
AF9 YEATS
Plasmids AF9 (2–138) was cloned into pET28b vector for bacteria expression was a generous gift from Haitao Li (Tsinghua University, Beijing).
In vitro photo-crosslinking experiments
In all, 12 μΜ probe (probe 9, 10, or 11) was mixed with 6 μΜ protein (SPIN1, KDM4ATudor or ING2PHD) in binding buffer (PBS buffer for SPIN1; 50 mM HEPES, 150 mM NaCl, 2 mM MgCl2, pH = 7.5 for KDM4ATudor and ING2PHD) at 4 °C. After incubation for 2 h, the mixtures were exposed to UV light at 365 nm using a SCIENTZ UV lamp (Model: SCIENTZ03-11) for 20 min, and then analyzed by SDS-PAGE and immunoblotting analysis. All photographs of the protein gel were taken on Fusion FX EDGE SPECTRA (Vilber).
Cellular cytotoxicity assays
HeLa cells were inoculated in 96-well plates and incubated at 6000 cells/well for 24 h. After changing the medium containing a different formulation, incubation continued for 12 h, and then PBS washed to remove residual probes. The cells were divided into two groups. One group received 20 min irradiation with 365 nm UV on ice, and the other group did not receive such irradiation. Subsequently, MTT was introduced into the system, and then the cells were placed in an incubator for a further 4 h. After that, 150 μL of DMSO was utilized to dissolve the formazan crystals. The measurement of absorbance at 490 nm was carried out with the employment of a microplate reader (680 Microplate Reader, Bio-Rad). For the live/dead cell staining assay, Hela cells were incubated in six-well culture plates for 24 h, followed by the incubation of different culture mediums for the time range of 0–24 h. The cells were stained with FDA (10 μΜ) and PI (20 μΜ) and then analyzed by fluorescence microscopy (IX71, Olympus).
Photo-crosslinking in live cells
HeLa or RAW264.7 cells underwent treatment with a 60 μΜ probe for 12 h at a temperature of 37 °C (probe 9 or 10 for HeLa cells, probe 9, 10, 12, 13, 14, or 15 for RAW264.7 cells). After PBS washing to eliminate the residual probe, the cells were subjected to 365 nm UV exposure (SCIENTZ; Model: SCIENTZ03-11) for 20 min on ice. The cells were lysed for 30 min in lysis buffer (RIPA buffer containing 1 mM PMSF). The cell lysate was collected and centrifuged at 12 × g for 30 min at low temperature. The resulting supernatant was used for subsequent proteomic analysis.
Mass spectrometry data collection and analysis
After photo-crosslinking and lysis, the cell lysates were incubated with streptavidin agarose beads (Promega) at 4 °C overnight. The beads are then thoroughly washed with solutions C (150 mM NaCl, 1 mM DTT, 25 mM tris, 0.5% v/v NP-40, pH = 7.6), D (PBS solution containing 0.5% w/w SDS, twice), E (PBS solution containing 1 M NaCl, twice), F (tris-buffered, twice) and G (50 mM NH4HCO3, pH = 7.6, 10 times) in this order.
For H3K4me3 (experimental group: probe 9, control group: probe 10, experimental and control groups were each repeated three times, biological replicates) and H3K9cr binding proteins (experimental group: probe 12, control group: probe 13, experimental and control groups were each repeated three times, biological replicates), the identification process was as follows. Enriched proteins on the beads were eluted with loading buffer at 95 °C for 10 min, followed by separation by SDS-PAGE. The protein sample processing protocol was essential, as described69. The proteins in the gel were then sequentially treated with 5 mM DTT and 11 mM IAA. In-gel digestion was performed overnight using trypsin in 50 mM NH4HCO3 solution. Peptides were extracted with solution H (1:1 CH₃CN/H2O (v/v) solution containing 0.1% TFA) and then concentrated to reduce the sample volume.
For H3K9la binding proteins (experimental group: probe 14, control group: probe 15, experimental and control groups were each repeated three times, biological replicates), the identification process was as follows. Beads digestion was performed overnight using trypsin in 50 mM NH4HCO3 solution. Peptides were extracted with solution H and then concentrated to reduce the sample volume. Peptides were dissolved in a solution of 200 mM tetraethylammonium bromide (TEAB) (50 μL), followed by the addition of TMTsixplex labeling reagent (Thermo Fisher) (2 μL), and the reaction was carried out for 1 h at room temperature. Then 5% hydroxylamine (0.5 μL, pH 9–10) was added and reacted for 15 min to quench the reaction. Final desalination with Sep-Pak C18 Vac cartridges (Waters).
A 120 min gradient elution at a flow rate of 0.3 μL/min was performed using the Thermo-Dionex Ultimate 3000 HPLC system, which was directly connected to the Thermo Orbitrap Fusion 480 mass spectrometer. The separation was carried out on a custom-made fused silica capillary column (75 μm ID, 150 mm length; Upchurch, Oak Harbor, WA) loaded with C-18 resin (300 Å, 5 μm; Varian, Lexington, MA). Mobile phase 1 contains an aqueous solution of 0.1% formic acid, and mobile phase 2 is an acetonitrile solution containing 0.1% formic acid.
The Sequest HT algorithm in Proteome Discoverer software (PD, version 1.4) was used to process the data. Hella cells were queried using the human protein database, and RAW264.7 cells were queried using the mouse protein database. Use the following search parameters: fully trypsin specific and allows up to two missed cleavage, oxidation of methionine (M) was considered as a variable modification, while carbamidomethylation of cysteine (C) and TMTsixplex labeling on lysine (K) and the N-terminal were specified as fixed modifications. The precursor ion mass tolerance was set to 20 ppm for MS scans and 20 mmu for MS/MS spectra. The peptide false discovery rate (FDR) was determined by the percolator function within PD software, with a 1% cutoff based on the decoy database. TMT quantification was carried out in PD software, and quantification was restricted to proteins with at least two unique peptide matches. Peptides were considered unique if they were assigned to a specific protein group. Protein ratios were calculated as the median of all peptide identifications for a given protein, and the variability of these ratios was used to assess the precision of the quantification.
ITC measurements
ITC measurement was performed as previously described by using MicroCal PEAQ-ITC (Malvern) at 15 °C33. The recombinant AF9YEAST protein and synthetic histone H3 peptides (H31-10K9la and H31-10) underwent extensive dialysis with ITC buffer: 25 mM Tris, 0.5 M NaCl, 2 mM β-ME, pH = 7.5. Usually, H31–10 peptides at 1.5–3 mM were added dropwise into AF9YEAST protein at 0.1–0.2 mM. There were 20 individual injections of each ITC titration, with 1 μL for the first and 2 μL for the rest. The PEAQ-ITC analysis software was utilized to process the data, with the application of the ‘One Set of Sites’ fitting model.
Co-immunoprecipitation
Raw264.7 cells were lysed in lysis buffer (Western and IP cell lysis buffer with a protease inhibitor mixture), either without or with 50 mM sodium lactate treatment. Samples were incubated overnight at 4 °C with H3K9la antibody and protein A/G magnetic beads for precipitation. The bound proteins were then removed by boiling in SDS buffer and resolved in SDS/polyacrylamide gels, followed by Western blot analysis using H3K9la and AF9 antibodies.
Docking
The protein data bank obtains the three-dimensional structure. (https://www.rcsb.org/structure/5HJB). The crystal model (5HJB), which represents AF9YEATS in complex with the H3K9cr peptide, was protonated and optimized through the “Quickprep” plugin in MOE. The ATP ligand binding site was designated as the docking region. The protein and molecular structures were protonated under the AMBER10: EHT force field. The interaction between the protein and the molecules was analyzed using the “Dock” plugin in MOE. For molecular docking, the induced fit docking protocol was employed, utilizing the triangle match algorithm to generate the docking poses. The London δG scoring function was utilized to calculate the binding energy for each pose, with the top 100 poses being retained. The poses were then optimized using the induced fit algorithm and the GBVI/WAS δG scoring function to assess the binding affinity of optimized poses. The docking poses were aligned with the Histone H3.1 peptide from the crystal structure 5HJB based on the docking regions. Finally, the conformation most similar to the reference was selected as the final result.
Statistics and reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the protein chemistry facility at the Center for Biomedical Analysis of Tsinghua University for sample analysis and technical help, and thank the staff of TOPSCIENCE for the docking and data analysis. This project was supported by the National Key R&D Program of China (No. 2022YFC3401500 for L. Liu), and NSFC (Nos. 22277020, 22227810 for Y. Li, 22137005, 92253302 for L. Liu, 22407121 for G. Chu, 22407040 for Y. Wang), and Anhui Provincial Natural Science Foundation (No. 2208085J42 for Y. Li, 2408085QB057 for G. Chu), and the Fundamental Research Funds for the Central Universities (PA2024GDGP0037, JZ2024YQTD0600 for Y. Li, WK9100000073 for G. Chu, JZ2024HGTA0189 for Y. Wang), and the XPLORER prize (for L. Liu).
Author contributions
Conceptualization and Scientific direction: Y. Li, and L. Liu. Probe synthesis: Y. Wang, Q. Shu, Y. Ye, and R. Ji. Confocal laser scanning microscopy: J. Fan and Q. Shu. Biochemical experiments, cell biological experiments, and MS sample preparation: Y. Wang, J. Fan, R. Ji, Y. Wu, X. Wu, and Q. Shu. MS data collection and analysis: Y. Wang, X. Meng, J. Shi, J. Fan, and H. Deng. Writing (original draft): Y. Li, J. Fan, G. Chu and Y. Wang. Writing (review and editing): Y. Li, Y. Wang, G. Chu, and L. Liu.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange [http://proteomecentral.proteomexchange.org] database under accession code PXD055058, PXD055061, PXD055055 and PXD045620. The crystal structure for AF9 YEATS (PDB: 5HJB) was used in this work. Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yu Wang, Jian Fan, Xianbin Meng, Qingyao Shu.
Contributor Information
Guo-Chao Chu, Email: cgc.2020@tsinghua.org.cn.
Lei Liu, Email: lliu@mail.tsinghua.edu.cn.
Yi-Ming Li, Email: ymli@hfut.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55046-8.
References
- 1.Kouzarides, T. Chromatin modifications and their function. Cell128, 693–705 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res.21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres, I. O. & Fujimori, D. G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol.35, 68–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin, J. et al. Menin “reads” H3K79me2 mark in a nucleosomal context. Science379, 717–723 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Boichenko, I. & Fierz, B. Chemical and biophysical methods to explore dynamic mechanisms of chromatin silencing. Curr. Opin. Chem. Biol.51, 1–10 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Stelzl, U. et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell122, 957–968 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods24, 218–229 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Furey, T. S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet.13, 840–852 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelen, E. et al. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat. Commun.6, 7155 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham, N. D., Parker, R. B. & Kohler, J. J. Photocrosslinking approaches to interactome mapping. Curr. Opin. Chem. Biol.17, 90–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Z. et al. Photo-cross-linking to delineate epigenetic interactome. J. Am. Chem. Soc.144, 20979–20997 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Xie, Y. et al. Fluorescent probes for single-step detection and proteomic profiling of histone deacetylases. J. Am. Chem. Soc.138, 15596–15604 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Xie, Y. et al. Chemical probes reveal Sirt2’s new function as a robust “eraser” of lysine lipoylation. J. Am. Chem. Soc.141, 18428–18436 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Lin, J. & Li, X. D. Peptide-based approaches to identify and characterize proteins that recognize histone post-translational modifications. Chin. Chem. Lett.29, 1051–1057 (2018). [Google Scholar]
- 15.Cuvier, O. & Fierz, B. Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat. Rev. Genet.18, 457–472 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Xie, X. et al. Genetically encoded photoaffinity histone marks. J. Am. Chem. Soc.139, 6522–6525 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Lin, J., Bao, X. & Li, X. D. A tri-functional amino acid enables mapping of binding sites for posttranslational-modification-mediated protein-protein interactions. Mol. Cell81, 2669–2681.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Dao, H. T., Dul, B. E., Dann, G. P., Liszczak, G. P. & Muir, T. W. A basic motif anchoring ISWI to nucleosome acidic patch regulates nucleosome spacing. Nat. Chem. Biol.16, 134–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X. et al. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J. Am. Chem. Soc.134, 1982–1985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai, X. et al. Development of a DNA-templated peptide probe for photoaffinity labeling and enrichment of the histone modification reader. Proteins Angew. Chem. Int. Ed.55, 7993–7997 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zheng, Y., Gilgenast, M. J., Hauc, S. & Chatterjee, A. Capturing post-translational modification-triggered protein-protein interactions using dual noncanonical amino acid mutagenesis. ACS Chem. Biol.13, 1137–1141 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin, F. et al. Linking chromatin acylation mark-defined proteome and genome in living cells. Cell186, 1066–1085.e36 (2023). [DOI] [PubMed] [Google Scholar]
- 23.Burton, A. J. et al. In situ chromatin interactomics using a chemical bait and trap approach. Nat. Chem.12, 520–527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleiner, R. E., Hang, L. E., Molloy, K. R., Chait, B. T. & Kapoor, T. M. A chemical proteomics approach to reveal direct protein-protein interactions in living cells. Cell Chem. Biol.25, 110–120.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niopek, D. et al. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nat. Commun.5, 4404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hameed, D. S., Sapmaz, A., Gjonaj, L., Merkx, R. & Ovaa, H. Enhanced delivery of synthetic labelled ubiquitin into live cells by using next-generation Ub-TAT conjugates. ChemBioChem19, 2553–2557 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Ai, H. et al. Chemical synthesis of post-translationally modified H2AX reveals redundancy in interplay between histone phosphorylation, ubiquitination, and methylation on the binding of 53BP1 with nucleosomes. J. Am. Chem. Soc.144, 18329–18337 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Shi, W. W. et al. Total chemical synthesis of correctly folded disulfide-rich proteins using a removable O-linked β-N-acetylglucosamine strategy. J. Am. Chem. Soc.144, 349–357 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Guidotti, N., Eördögh, Á., Mivelaz, M., Rivera-Fuentes, P. & Fierz, B. Multivalent peptide ligands to probe the chromocenter microenvironment in living cells. ACS Chem. Biol.18, 1066–1075 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herce, H. D. et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem.9, 762–771 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Nischan, N. et al. Covalent attachment of cyclic TAT peptides to GFP results in protein delivery into live cells with immediate bioavailability. Angew. Chem. Int. Ed.54, 1950–1953 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Yu, Y. et al. K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle. Nat. Chem. Biol.17, 896–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ray, M., Tang, R., Jiang, Z. & Rotello, V. M. Quantitative tracking of protein trafficking to the nucleus using cytosolic protein delivery by nanoparticle-stabilized nanocapsules. Bioconjugate Chem.26, 1004–1007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, T., Liu, Z. & Li, X. D. Developing diazirine-based chemical probes to identify histone modification ‘readers’ and ‘erasers’. Chem. Sci.6, 1011–1017 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao, S. et al. Histone H3Q5 serotonylation stabilizes H3K4 methylation and potentiates its readout. Proc. Natl Acad. Sci. USA118, e2016742118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, T., Li, X. M., Bao, X., Fung, Y. M. & Li, X. D. Photo-lysine captures proteins that bind lysine post-translational modifications. Nat. Chem. Biol.12, 70–72 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Peña, P. V. et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature442, 100–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, K. et al. Reversible recognition-based boronic acid probes for glucose detection in live cells and zebrafish. J. Am. Chem. Soc.145, 8408–8416 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang, Y. et al. Deciphering the interactome of histone marks in living cells via genetic code expansion combined with proximity labeling. Anal. Chem.94, 10705–10714 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Lee, J. H., Tate, C. M., You, J. S. & Skalnik, D. G. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J. Biol. Chem.282, 13419–13428 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Lauberth, S. M. et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell152, 1021–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flanagan, J. F. et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature438, 1181–1185 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Hu, Y. et al. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl Acad. Sci. USA109, 5773–5778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wysocka, J. et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell121, 859–872 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Wang, W. et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep.12, 1160–1166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Husain, A. et al. Chromatin remodeller SMARCA4 recruits topoisomerase 1 and suppresses transcription-associated genomic instability. Nat. Commun.7, 10549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui, X. et al. RUNX1/NPM1/H3K4me3 complex contributes to extracellular matrix remodeling via enhancing FOSL2 transcriptional activation in glioblastoma. Cell Death Dis.15, 98 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao, Z., Bai, Y., Huq, E. & Qiao, H. LHP1 and INO80 cooperate with ethylene signaling for warm ambient temperature response by activating specific bivalent genes. bioRxiv. 10.1101/2024.03.01.583049 (2024). [DOI] [PMC free article] [PubMed]
- 49.He, F. et al. Structural insight into the zinc finger CW domain as a histone modification reader. Structure18, 1127–1139 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Qie, J. et al. Integrated proteomic and transcriptomic landscape of macrophages in mouse tissues. Nat. Commun.13, 7389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang, Z. et al. Bioorthogonal photocatalytic decaging-enabled mitochondrial proteomics. J. Am. Chem. Soc.143, 18714–18720 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Zhang, C. et al. BRPF1 bridges H3K4me3 and H3K23ac in human embryonic stem cells and is essential to pluripotency. iScience26, 105939 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, W. et al. WDR3 promotes stem cell-like properties in prostate cancer by inhibiting USF2-mediated transcription of RASSF1A. J. Gene. Med.25, e3498 (2023). [DOI] [PubMed] [Google Scholar]
- 54.Ribeiro, J. R., Lovasco, L. A., Vanderhyden, B. C. & Freiman, R. N. Targeting TBP-associated factors in ovarian cancer. Front. Oncol.4, 45 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostuni, R. & Natoli, G. Transcriptional control of macrophage diversity and specialization. Eur. J. Immunol.41, 2486–2490 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Huang, Z. et al. The corepressors GPS2 and SMRT control enhancer and silencer remodeling via eRNA transcription during inflammatory activation of macrophages. Mol. Cell81, 953–968.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Feldman, J. L., Baeza, J. & Denu, J. M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem.288, 31350–31356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, X. et al. Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking. Nat. Chem. Biol.14, 1140–1149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flynn, E. M. et al. A subset of human bromodomains recognizes butyryllysine and crotonyllysine histone peptide modifications. Structure23, 1801–1814 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Bao, X. et al. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife3, e02999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Izzo, L. T. & Wellen, K. E. Histone lactylation links metabolism and gene regulation. Nature574, 492–493 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature574, 575–580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai, X., Lv, X., Thompson, E. W. & Ostrikov, K. K. Histone lactylation: epigenetic mark of glycolytic switch. Trends Genet.38, 124–127 (2022). [DOI] [PubMed] [Google Scholar]
- 64.Li, Y. et al. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell159, 558–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li, Y. et al. Molecular coupling of histone crotonylation and active transcription by AF9 YEATS domain. Mol. Cell62, 181–193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seath, C. P. et al. Tracking chromatin state changes using nanoscale photo-proximity labelling. Nature616, 574–580 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ai, H., Pan, M. & Liu, L. Chemical synthesis of human proteoforms and application in biomedicine. ACS Cent. Sci.10, 1442–1459 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dong, S. et al. Recent advances in chemical protein synthesis: method developments and biological applications. Sci. China Chem.67, 1060–1096 (2024). [Google Scholar]
- 69.Wang, Y. et al. Photocaging of activity-based ubiquitin probes via a C-terminal backbone modification strategy. Angew. Chem. Int. Ed.61, e202203792 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange [http://proteomecentral.proteomexchange.org] database under accession code PXD055058, PXD055061, PXD055055 and PXD045620. The crystal structure for AF9 YEATS (PDB: 5HJB) was used in this work. Source data are provided in this paper.