Abstract
Radiation therapy is a common treatment modality for lung cancer, and resistance to radiation can significantly affect treatment outcomes. We recently described that lung cancer cells that express more germ cell cancer genes (GC genes, genes that are usually restricted to the germ line) can repair DNA double-strand breaks more rapidly, show higher rates of proliferation and are more resistant to ionizing radiation than cells that express fewer GC genes. The gene encoding TRIP13 appeared to play a large role in this malignant phenotype. However, the molecular regulatory mechanism of TRIP13 in radiation resistance remained largely unknown. Here, we show that TRIP13 is a key contributor to non-small cell lung cancer (NSCLC) treatment resistance, particularly in patients following radiation treatment, for whom levels of TRIP13 expression are correlated with a poor prognosis. Repeated irradiation of led to an increase of basal TRIP13 levels and radioresistance. This effect of radioresistance could be enhanced or abrogated by overexpressing or knocking out TRIP13. Elevated TRIP13 is also correlated with enhanced repair of radiation-induced DNA damage. We further showed the proteins NBS1 and RAD51 (homologous recombination. HR) and XRCC5 (non-homologous end-joining, NHEJ) to act downstream of TRIP13, although inhibition of TRIP13 mostly reduced the HR associated proteins in response to induced resistance to irradiation. This study elucidates a novel mechanism of treatment resistance in NSCLC cells, in which TRIP13 promotes HR mediated DNA repair and resistance to ionizing radiation.
Keywords: Non-small cell lung cancer (NSCLC), Cancer treatment, Irradiation, Homologous recombination, Germ cell cancer genes (GC genes), TRIP13
Subject terms: Radiotherapy, Non-small-cell lung cancer, Double-strand DNA breaks
Introduction
Germ cells, being or giving rise to eggs or sperm, have the ability to pass a relatively intact genome to – in principle – endless future generations, while the somatic cells that make up the rest of the body are subject to limited growth and life span1. As the potential to grow and divide uncontrollably is a key feature of cancer, we hypothesized that partial activation of the “germline program” in somatic cells can contribute to oncogenesis2. We previously described a set of usually germline specific genes that are ectopically expressed in cancer (germ cell cancer genes, GC genes)3,4. Furthermore, expression of these genes appeared to correlate with malignancy in lung cancer cells4. Additionally, we found that higher GC gene expression leads to more rapid repair of DNA double-strand breaks (DSBs), higher rates of proliferation and increased resistance to ionizing radiation (IR), compared to lung cancer cells that express less GC genes5. We identified the gene encoding the thyroid hormone receptor interactor protein 13 (TRIP13) to play an important role in this malignant phenotype.
TRIP13 plays a critical role in cell division and chromosome segregation during meiosis and mitosis6. In addition, during meiosis, TRIP13 acts as a dosage-sensitive regulator, localizing to the synapsed homologous chromosomes at the synaptonemal complex in early pachytene spermatocytes, and to telomeres throughout meiotic prophase I, and its loss leads to meiotic arrest and sterility in both sexes7. In both meiotic and somatic cells, TRIP13 is involved in function of the spindle checkpoint, a mechanism that ensures the accurate segregation of chromosomes during cell divisions8. We previously found that TRIP13 is endogenously expressed in lung cancer cells, and that inhibiting TRIP13 increases the effectiveness of cancer treatment with IR5.
IR induces DNA double-strand breaks (DSBs), a very serious form of DNA damage9. Mammalian cells contain two major pathways for DSB repair, non-homologous end-joining (NHEJ) and homologous recombination (HR)10. In the error-prone NHEJ DNA repair pathway, damaged DNA ends are first recognized and processed by the Ku70/Ku80 heterodimer, followed by recruitment of the core NHEJ complex. This complex includes DNA-PKcs, XLF, and XRCC4/DNA Ligase IV, with DNA-PKcs protecting the DNA ends through auto-phosphorylation, activation of end processing nucleases, and subsequent ligation by the XRCC4-Ligase IV complex and XLF stimulation, involving various additional proteins such as Artemis, PNK and pol λ11. In contrast to NHEJ, HR utilizes the sister chromatid sequence as a template for error-free repair of DSBs12. The HR process initiates with a DNA ends resection reaction, facilitated by CtIP and the MRN-complex, extended by BLM helicase, EXO1, and DNA2 nucleases. This process generates single stranded DNA overhangs (ssDNA), which are essential for the subsequent steps of HR13. Once the ssDNA overhangs are generated, RAD51, a key protein in HR, forms a nucleoprotein filament on the ssDNA. RAD51 and accessory factors catalyse a homology search, strand invasion, and D-loop structure formation to facilitate the restoration of lost information at the DNA ends14. There have been many studies that show TRIP13’s involvement in DNA damage repair15–18. In this study, our aim is to investigate whether TRIP13 mediates resistance to IR. To this end, we generated irradiation-resistant (IRR) cells by subjecting lung cancer cells with various TRIP13 expression levels to repeated radiation exposures to research the role of HR and NHEJ in TRIP13-depend treatment resistance. We utilized TRIP13 inhibitors, knockout and overexpression to further substantiate its role specifically in DSB repair, and observed that inhibiting TRIP13 reduced the expression of the HR marker RAD51. Notably, this inhibition did not affect the levels of the NHEJ proteins KU70 and Ligase IV5.
Materials and methods
Experimental model and subject details
Cells H1703 (ATCC) and BEAS-2B (Sigma)) were cultured in 5% CO2 at 37 °C in RPMI-1640 medium (Thermo Fisher Scientific), supplemented with 10% foetal bovine serum (Thermo Fisher Scientific), 1% HEPES (Gibco), 1% Pen-Strep (Gibco), and 2.2% glucose (Gibco). The cells were refreshed every 3 days and routinely passaged for use. Throughout the entire study, the cells in culture tested negative for mycoplasma contamination.
Generation of TRIP13 over-expression cell line, OE-TRIP13
BEAS-2B cells were transfected (Neon electroporator (Thermo Fisher Scientific) with TRIP13 CRISPR Activation Plasmid (sc-404006-ACT; Santa Cruz Biotechnology) or Control CRISPR plasmid (sc-437275; Santa Cruz Biotechnology), After 48 h, transfected cells were selected using 1ug/mL puromycin (EnzoLife Sciences, ALX-380-028). TRIP13 overexpression was screened by qRT-PCR, fluorescent microscopy and Western blot to analyses.
Irradiation
The cell lines were subjected to 2 Gy of ionizing radiation (IR) in a CellRad system (precision X-Ray, the Netherlands), based on previous results obtained for H1703 cells5. IR resistance cells (-IRR) were irradiated 3 times with a 24 h recovery period between each irradiation, leading to an accumulated dose of ∼sixfold the of the parent cell lines.
Clonogenic assay
Clonogenic assays were performed as described previously19. Cells in both control and treatment groups were plated at an equal density in triplicate in six-well plates. 4 h after plating, the cells were exposed to 2 Gy of IR. Cells were cultured in 3 ml medium, and 2 ml of medium was gently replaced after 7 days. Once the negative control condition (i.e. 0 Gy) showed the formation of colonies of 50 cells, which took approximately 14 days. The medium was removed and cells were gently washed with phosphate-buffered saline (PBS). Subsequently, the cells were fixed and stained with 6% glutaraldehyde + 0,5% crystal violet in PBS. The numbers of colonies with more than 50 cells were electronically counted using Image J (version1.54i) and manually confirmed.
EdU assay
Cells were cultured in 96 well-plates to 60–70% confluency before exposure to IR. After 4 h, 5-ethynyl-2’-deoxyuridine (EdU) was added to the culture and incubated for 2 h. The quantification of EdU-positive cells was performed using the Cell Proliferation Kit (C10337,Thermo Fisher Scientific), as previously described20, and represented by the mean ± SEM of 3 independent experiments. Images were analysed using Leica Application Suite X and counting of nuclei and EdU staining was performed electronically in Image J (version 1.54i).
γ-H2AX and RAD51 staining
Cells were cultured on multi Nunc Lab-Tek II chambered slides (Thermo Fisher Scientific, the Netherlands) for 24 h. Cells were fixed at various time points after irradiation (30 min–24 h) in 4% paraformaldehyde for 10 min and subsequently permeabilized in PBS with 0.1% triton-X for 15 min. Non-specific adhesion sites were blocked for 45 min in 0.25% Tween-20/PBS with 1% bovine serum albumin, followed by the addition of primary antibodies against γ-H2AX (1:10.000, 05–636, Millipore) or RAD51 (1:50, PA5-27,195, Thermo Fisher Scientific), or isotype immunoglobulin G for negative controls. After overnight incubation at 4 °C, the cells were washed and incubated with the corresponding host-specific secondary antibodies, goat-anti-mouse Alexa Fluor 488 (1:1000, A11029, Thermo Fisher Scientific) or goat-anti-rabbit Alexa Fluor 532 (1:1000, A11009, Life Technologies) and counterstained with DAPI. The slides were mounted with Prolong Gold anti-fade (Thermo Fisher Scientific) and visualized using a Leica DM5000B microscope. γ-H2AX and RAD51 foci within the cell nucleus were counted manually. This was repeated 3 times for statistical analysis.
Western blotting
Cells were seeded in 6-well plates. Once 80% confluency was reached, media was discarded and cells were washed twice in PBS. The cells were then treated with 1 ml 0.25% Trypsin in the incubator and, after 5 min, RPMI-1640 medium (with 10%FCS, 1%Hepes, 1% Penstraps, 1.1% glucose) was added to dilute the trypsin. This mixture was pipetted from the plates to tubes and proteins were extracted from the cells used RIPA buffer containing protease- (complete™ Protease Inhibitor Cocktail, Roche) and phosphatase-inhibitors (PhoSTOP™, Roche). Protein concentration was quantified with the Qubit Protein Assay Kit (Thermo Fisher Scientific). Western blot analysis was performed using the LI-COR Odyssey imaging system (LI-COR Biosciences) as previously reported19. The primary antibodies used were against TRIP13 (1:1000, ab128171, Santa Cruz Biotechnology), RAD51 (1:1000, PA5-27,195, Thermo-Fisher), XRCC5(1:1000, ab2172-500, Abcam), NBS1(1:1000, ab32074, Abcam), GAPDH (1:1000; FL-335, Santa Cruz Biotechnology). Secondary antibodies were IRDye 800CW Goat Anti-Rabbit IgG (H + L)(1:1000, 926–32,211, LiCor), IRDye 800CW Goat Anti-Mouse IgG (H + L)(1:1000, 926–32,210, LiCor). Signal was visualized using the Odyssey V3.0 imaging system (LI-COR Biosciences). Band intensities were assessed using Image Studio Lite (Version 5.2). Uncropped full length western blots are included in the Supplementary Information file (Supplementary figure S1).
Quantitative-real time PCR(Q-PCR)
Total RNA was extracted from control and treated cells use RNeasy Mini Kit (Qiagen) and following the protocol provided by the manufacturer. The RNA samples were reversely transcribed using SensiFAST cDNA Synthesis Kit (Bioline). The synthesized cDNA was then used for Q-PCR reactions, using the Roche LightCycler 480 platform in a 384-well plate format. The Q-PCR reaction was performed in a 10uL volume system including 2X LightCycler 480 SYBR Green I Master (Roche). Gapdh, Actb and Tuba1c were used as reference genes, and the data were analyzed using the delta Ct method. The primers for Q-PCR analysis are listed in Table 1.
Table 1.
Primer sequences for Q-PCR analysis.
| Gene | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|
| Gapdh | GTCTCCTCTGACTTCAACAGCG | ACCACCCTGTTGCTGTAGCCAA | 130 |
| Actb | ACCAGAGGCATACAGGGAC | CTAAGGCCAACCGTCAAAAG | 100 |
| Tuba1c | AGCGTGCCTTTGTTCACT | CTCATCCTCTCCGTCAGC | 134 |
| Trip13 | CGGGTCCTGAGAAAACTCCC | CAAACTGCTTGTCCACTGCC | 150 |
Patient survival rates were analysed using Kaplan–Meier survival curves
Lung adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma patients with high TRIP13 expression after radiation treatment had lower survival. This data is from Amsterdam university Medical Center R2 database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). In the R2 platform, choose the Survival (Kaplan–Meier/Cox). For data set choose LUAD/ Lung Squamous Cell Carcinoma, separate by Hazard ratio for a gene (TRIP13), for subset track choose radiation therapy.
Statistical analysis
The data obtained were analysed in Prism Graph Pad 10.1.1 and Excel. For the Figs. 1, 2, 3, 4, two-group comparisons were performed: TRIP13KO cells compared to control (both cell line H1730) and OE-TRIP13 compared to control (both cell line BEAS-2B). In these cases, a Shapiro–Wilk test was used to validate normal distribution of the data and two-sided T-tests were performed. For Fig. 5, one-way ANOVA was performed to compare the means of three groups. Results were corrected for multiple testing using Bonferroni correction where appropriate. p < 0.05 was regarded as statistically significant.
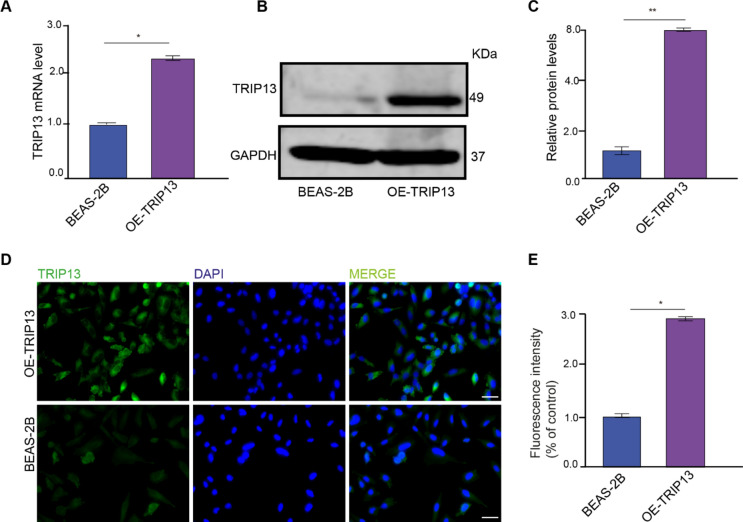
Fig. 1.
Overexpression of TRIP13 in BEAS-2B lung cells. (A) TRIP13 mRNA expression in BEAS-2B (dCas9) and OE-TRIP13 cells (n = 3). (B) lysates from BEAS-2B (dCas9) and OE-TRIP13 were immunoblotted with anti-TRIP13 and anti-GAPDH as loading control. (n = 3). (C) Band intensities were quantified and normalized to corresponding GAPDH. (D) BEAS-2B (dCas9) and OE-TRIP13 cell lines were immunofluorescently labelled with anti-TRIP13 and DAPI nuclear stain. Scale bar = 50 µm. (E) Quantification of the TRIP13 mean fluorescence intensity in the BEAS-2B and OE-TRIP13 cell lines. * = p < 0,05. ** = p < 0,01 by two-sided t test.
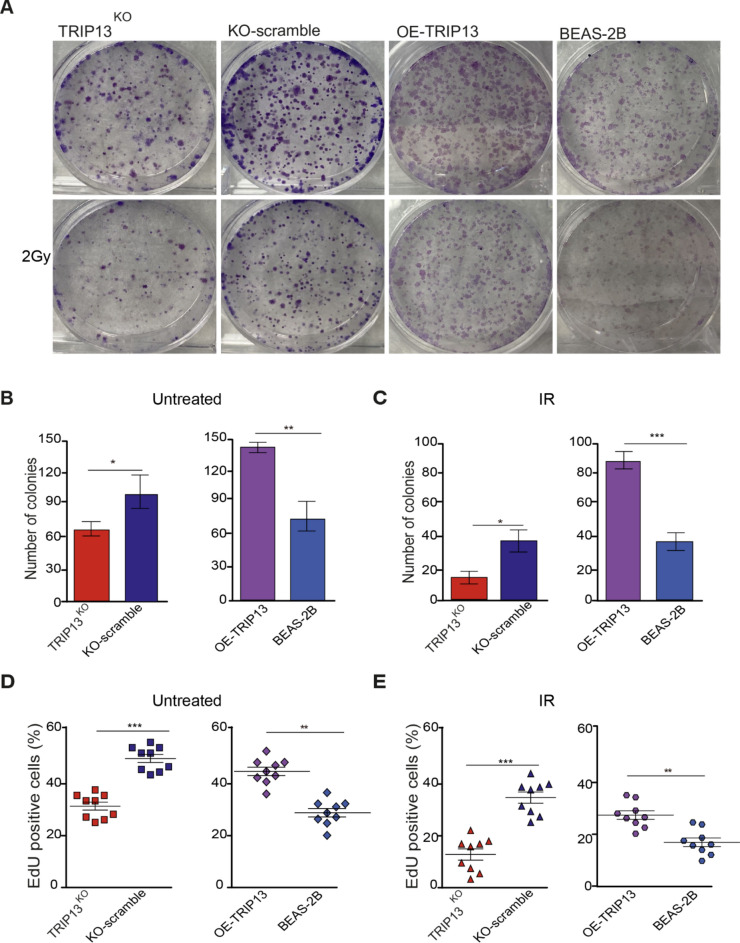
Fig. 2.
TRIP13 promotes radiation resistance and cell proliferation. (A) Clonogenic survival assays were performed with KO-Scramble, TRIP13KO ;BEAS-2B (dCas9) and OE-TRIP13 were irradiated at 2 Gy. (n = 3). (B) Number of colonies in KO-Scramble, TRIP13KO and BEAS-2B (dCas9), OE-TRIP13 cell lines without IR. (C) Number of colonies in KO-Scramble, TRIP13KO and BEAS-2B (dCas9), OE-TRIP13 cell lines following 2 Gy of irradiation. (D) Quantification of EdU positive cells in KO-Scramble, TRIP13KO and BEAS-2B (dCas9), OE-TRIP13 (n = 9) without IR. (E) Quantification of EdU positive cells in KO-Scramble, TRIP13KO and BEAS-2B (dCas9), OE-TRIP13 (n = 9) following 2 Gy of irradiation. * = p < 0,05, ** = p < 0,01, *** = p < 0,001 by two-sided t test.
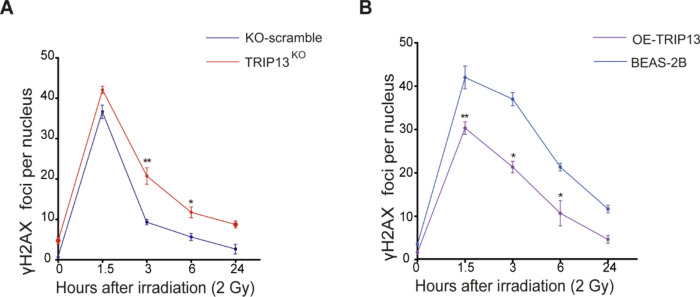
Fig. 3.
TRIP13 promotes DNA double strands break repair. (A) γ-H2AX foci per nucleus after 2 Gy of IR in KO-scramble(blue) and TRIP13KO (red). * = p < 0,05. ** = p < 0,01 (t test comparing KO-scramble and TRIP13KO per single time point). (B) γ-H2AX foci per nucleus after 2 Gy of IR in BEAS-2B (dCas9) (light blue) and OE-TRIP13 (purple). * = p < 0,05. ** = p < 0,01 (t test comparing OE-TRIP13 and BEAS-2B per single time point).
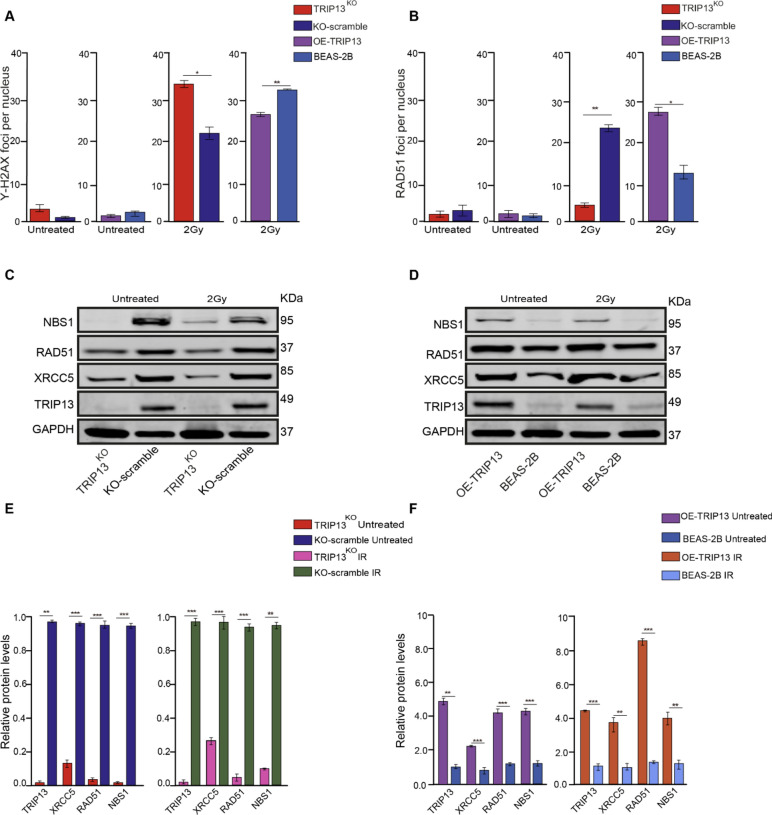
Fig. 4.
TRIP13 promotes DSBs repair through Homologous Recombination and Non- Homologous End Joining. (A) Quantification of γ-H2AX foci per nucleus, with and without 2 Gy of IR, in the four cell lines. (B) Quantification of RAD51 foci per nucleus, with and without 2 Gy of IR, in the four cell lines. (C) NBS1, RAD51, XRCC5 and TRIP13 protein expression in the KO-scramble and TRIP13KO cells, with and without 2 Gy of IR, GAPDH as a loading control (n = 3). (D) NBS1, RAD51, XRCC5 and TRIP13 proteins expression in the BEAS2B (dCas9) and OE-TRIP13 cells, with and without 2 Gy of IR, GAPDH as a loading control (n = 3). (E) Quantification of NBS1, RAD51, XRCC5 and TRIP13 protein levels in the KO-scramble and TRIP13KO cells, with and without 2 Gy of IR, GAPDH as a loading control. (F) Quantification of NBS1, RAD51, XRCC5 and TRIP13 protein levels in the BEAS2B (dCas9) and OE-TRIP13 cells, with and without 2 Gy of IR, GAPDH as a loading control. * = p < 0,05, ** = p < 0,01, *** = p < 0,001 by t test.
Fig. 5.
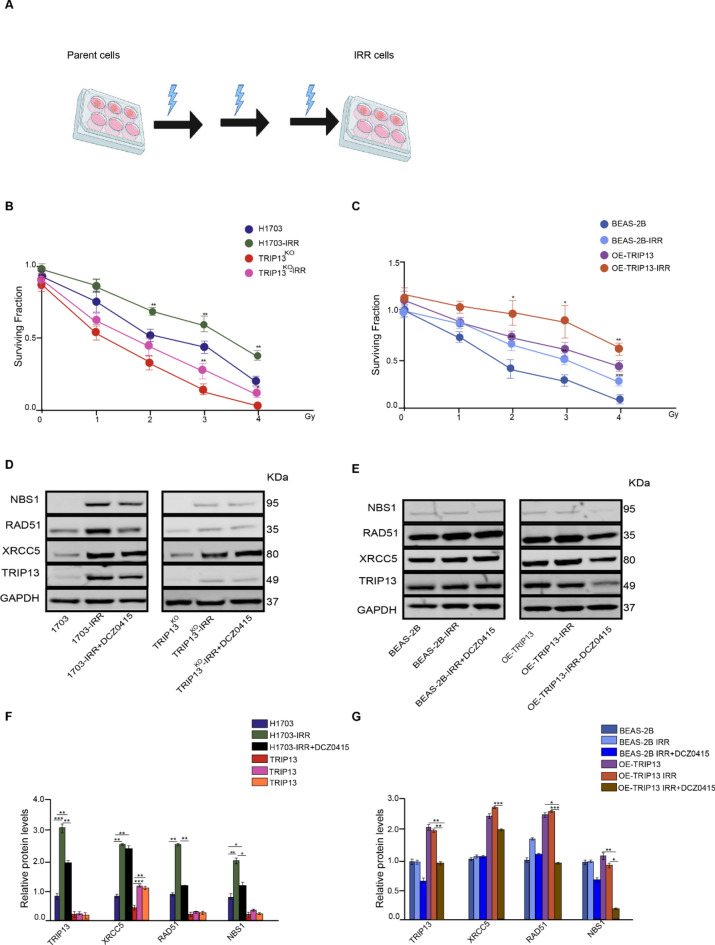
TRIP13 promotes IR resistance through Homologous Recombination. (A) Schematic diagram showing how radioresistant cells were established. (B) Surviving fractions of H1703, H1703-IRR cells and TRIP13KO, TRIP13KO-IRR cells irradiated with increasing of IR. (C) Surviving fractions of BEAS-2B (dCas9), BEAS-2B-IRR cells and OE-TRIP13, OE-TRIP13-IRR cells irradiated with increasing of IR. (D) TRIP13, XRCC5, RAD51 and NBS1 proteins expression in the H1703, H1703-IRR and H1703-IRR with DCZ0415; and TRIP13KO, TRIP13KO-IRR cells and TRIP13KO-IRR with DCZ0415. GAPDH as a loading control (n = 3). (E) TRIP13, XRCC5, RAD51 and NBS1 proteins expression in the BEAS-2B, BEAS-2B-IRR and BEAS-2B-IRR with DCZ0415; and OE-TRIP13, OE-TRIP13-IRR and OE-TRIP13-IRR with DCZ0415 = . GAPDH as a loading control (n = 3). (F) Quantification of TRIP13, XRCC5, RAD51 and NBS1 proteins levels H1703, H1703-IRR and H1703-IRR with DCZ0415 cells relative to GAPDH. (G) Quantification of TRIP13, XRCC5, RAD51 and NBS1 proteins levels, in the BEAS-2B, OE-BEAS-2B and OE-BEAS2B-IRR with DCZ0415 relative to GAPDH. * = p < 0,05, ** = p < 0,01, *** = p < 0,001 by one-way ANOVA.
Results
TRIP13 promotes radiation resistance and cell proliferation
We previously found that suppression of TRIP13 using inhibitor DCZ0415 sensitizes lung cancer cells to IR5. To further investigate the association between TRIP13 and resistance to IR, we used the TRIP13 knockout cell line (TRIP13KO) and control (KO-scramble) we constructed and described previously5. In addition, we used the human lung cell line BEAS-2B to overexpress TRIP13. For this we used the TRIP13 CRISPR activation plasmid, which contains the synergistic activation mediator (SAM) transcription activation system, to overexpress TRIP13 or the CRISPR Activation plasmid encoding the deactivated Cas9 (dCas9) nuclease as control, referred to as BEAS-2B. qRT-PCR, Western blot analysis and immunofluorescence microscopy confirmed that the RNA and protein levels of TRIP13 were significantly higher in cells that overexpress TRIP13 (Fig. 1A, B, C, D and E). Taken together, these results showed that TRIP13 expression was increased in the overexpress TRIP13 cell line compared to BEAS-2B cells.
We subjected all these cell lines, KO-scramble, TRIP13KO, BEAS-2B and OE-TRIP13 cells to 2 Gy of IR and quantified the colony forming rate of cells after exposure (Fig. 2A). We observed that the TRIP13KO cells showed less colony formation, with and without IR, compared to scramble control (Fig. 2B, C). In contrast, TRIP13-overexpressing cells appeared to form more colonies, both with and without IR, compared to control (BEAS-2B, dCas9) (Fig. 2B, C). To investigate the impact of TRIP13 on cell proliferation, we stained four cell lines with DNA synthesis marker 5-ethynyl-2’-deoxyuridine (EdU), which incorporates in the DNA of cells during the S-phase of the cell cycle, with and without IR (Supplementary Fig. 2A, B). This assay showed that, after knockout of TRIP13, cell proliferation significantly decreased in comparison to control with and without IR (Fig. 2D, E). Overexpression of TRIP13 led to significantly increased proliferation compared to control (BEAS-2B, dCas9) with and without IR (Fig. 2D, E). Taken together, higher expression of TRIP13 appeared to promote cell proliferation and resistance to IR.
TRIP13 promotes DNA double strands break repair
To investigate the impact of TRIP13 on IR induced DNA damage, we quantified the number of DSBs in all four cell lines through γ-H2AX staining at 0, 1.5, 3, 6 and 24 h after exposure to 2 Gy of IR (Supplementary Fig. 3). We observed that the TRIP13KO cell line displayed more remaining γ-H2AX foci, compared to its control (Fig. 3A). In contrast, the TRIP13 overexpressing cell line showed less remaining γ-H2AX foci, compared to its control (Fig. 3B). Taken together, these data showed that TRIP13 overexpression enhances DSB repair, while knockout of TRIP13 decreases DSB repair compared to corresponding control cells.
TRIP13 promotes DSBs repair through HR and NHEJ
To investigate whether TRIP13 may mediate DSB-repair via HR, we first co-stained the four cell lines for γ-H2AX and RAD51 after four hours after a dose of 0 or 2 Gy of IR (Supplementary Fig. 4A, B). RAD51 plays a critical role in the search for homologous sequences during DNA strand invasion and is required for HR20. After 2 Gy of irradiation, the number of γ-H2AX and RAD51 foci significantly increased in the four cell lines (Fig. 4A, B). In TRIP13KO cells, more γ-H2AX foci remained after 4 h after IR in comparison to control cells (KO-Scramble); in contrast, compared to BEAS-2B cells (dCas9), more γ-H2AX foci were resolved after 4 h following IR in the TRIP13 overexpressing cells (Fig. 4A, B). Following exposure to 2 Gy of IR, RAD51 foci formation exhibited a substantial reduction in the TRIP13KO cells compared to control cells. Conversely, there was a significant increase in RAD51 foci in TRIP13 overexpressing cells compared to the BEAS-2B(dCas9) control cells (Fig. 4A, B). Combined, it seemed that higher TRIP13 levels lead to faster DSB repair (γ-H2AX foci reduction) via HR (more RAD51 foci).
To further investigate whether TRIP13 induces HR in response to IR, western blot analyses were performed (Fig. 4C, D). We first investigated NBS1, which acts as a scaffold for the formation of the MRN complex (MRE11-RAD50-NBS1), which plays a crucial role in the initial processing of DSBs, including the recruitment of other proteins involved in HR21. In the four cell lines, NBS1 protein levels exhibited no significant changes following IR (Fig. 4C, D, E, F). However, in the TRIP13KO cells, NBS1 exhibited decreased protein levels in comparison to control cells (KO-Scramble), both with and without IR (Fig. 4C, E). The level of NBS1 was elevated in the TRIP13-overexpressing cells in comparison to BEAS-2B (dCas9) cells, both with and without IR (Fig. 4D, F).
RAD51 protein levels also exhibited no significant changes on response to IR (Fig. 4C, D, E, F). Compared to the control cells, RAD51 exhibited lower protein levels in the TRIP13KO cells, both with and without IR (Fig. 4C, E). In the TRIP13-overexpressing cells, the levels of RAD51 were higher in comparison the BEAS-2B (dCas9) cells, both with and without IR (Fig. 4D, F). Taken together, the basal protein levels of TRIP13, NBS1 and RAD51 do not change following 2 Gy of IR. However, the basal protein levels of NBS1 and RAD51 do correlate with TRIP13 levels in these cells. These observations suggest that TRIP13 plays a role in promoting HR through RAD51 and NBS1.
To investigate the Non-Homologous End Joining (NHEJ) pathway, we analysed protein expression of XRCC5 (KU80), a key factor in NHEJ-mediated DSB repair. Along with its heterodimeric partner XRCC6 (Ku70), XRCC5 forms a complex that binds to broken DNA ends, acting as a sensor and facilitating the recruitment other proteins to form a repair complex22,23. XRCC5 protein levels exhibited no significant changes after 4 h following 2 Gy of IR in the four cell lines (Fig. 4C, D, E, F). Compared to the control cells, XRCC5 exhibited lower protein levels in the TRIP13KO cells, both with and without IR (Fig. 4E). In TRIP13-overexpressing cells, the levels of XRCC5 were higher in comparison the BEAS-2B (dCas9) cells, both with and without IR (Fig. 4F). After subjecting the cells to 2 Gy of irradiation, it was observed that also TRIP13 protein levels remained unchanged in response to IR (Fig. 4C, D, E, F). These observations suggest that TRIP13 is also involved in the regulation of NHEJ. TRIP13 thus may increase resistance to IR via multiple DNA repair pathways.
Repeated ionizing radiation exposure induces basal TRIP13 expression, conferring radioresistance
To investigated the mechanism by which TRIP13 mediates resistance to IR, we exposed all cell lines, H1703, TRIP13KO, BEAS-2B (dCas9) and OE-TRIP13 cells, to repeated doses of 2 Gy of IR (Fig. 5A). After each dose we placed the cells back in the incubator for 24 h. After three doses of IR, IR-resistant cell lines were generated (H1703-IRR, TRIP13KO-IRR, BEAS-2B (dCas9) –IRR and OE-TRIP13-IRR). Clonogenic assays verified that IRR cells indeed display higher survival than their parent cells when exposed to the same doses of IR (Fig. 5B, C). Moreover, higher TRIP13 levels in TRIP13-OE and H1703 cells corresponded to a higher resistance to IR compared to their counterparts (BEAS-2B and TRIP13-KO, respectively) (Fig. 5B, C). In line with this result, Kaplan–Meier analysis demonstrated that NSCLC patients with high expression of TRIP13 indeed display a significantly shorter survival time in years following irradiation treatment (Supplementary Fig. 5A, B).
We then performed a western blot analysis to investigate whether TRIP13 levels would be increased in the IRR cells (Fig. 5D, E). The basal levels of TRIP13 in IRR cells appeared higher than those in their parent cells (H1703) (Fig. 5D, F). As observed previously5, also the TRIP13KO cells display very low TRIP13 expression, most likely due to occasional ribosomal frameshifting or transcriptional slippage resulting in restoration of the original reading frame5. As expected, this also led to a light increase of TRIP13 in the TRIP13KO -IRR cells (Fig. 5D, F). We also investigated the TRIP13 expression between IRR and parent cells in the BEAS2B (dCas9) and OE-TRIP13 cells. In the BEAS2B (dCas9)-IRR and OE-TRIP13-IRR cells, no significantly increase in TRIP13 expression was observed (Fig. 5E, G).
As we showed that TRIP13 may promote survival by increasing the protein levels of RAD51, NBS1 and XRCC5 (Fig. 4E, F), we performed western blot analyses to compare the proteins expression levels of these proteins in parent and IRR cells. We found that, in the H1703-IRR cells, XRCC5, RAD51 and NBS1 were increased in comparison to the parent cells. In the TRIP13KO cells, there was only low RAD51 and NBS1 expression, showing only a low increase in TRIP13KO–IRR cells (Fig. 5D, F). However, XRCC5 increased after repeated IR, also in TRIP13KO cells (Fig. 5D, F).
In contrast to the NSCLC-derived H1703 cells, the BEAS-2B (dCas9) and OE-TRIP13 IRR cells showed no significant changes in levels of TRIP13 compared to their parent cells (Fig. 5E, G). Also XRCC5, RAD51 and NBS1 showed no change between the IRR and parent cells (Fig. 5E, G).
To further investigate if the XRCC5, RAD51 and NBS1 increased along with TRIP13, H1703-IRR cells were treated with TRIP13 inhibitor DCZ0415. Following TRIP13 inhibition, RAD51 and NBS1 were decreased in the H1703-IRR cells (Fig. 5D, F). However, XRCC5 showed no significant change in the both cell lines (Fig. 5D, E). The TRIP13-KO cells showed no differences in XRCC5, RAD51 or NBS1 expression in response to TRIP13 inhibition (Fig. 5D, F). Following TRIP13 inhibition, XRCC5, RAD51 and NBS1 were decreased in the OE-TRIP13 IRR cells, but not in the BEAS-2B-IRR cells (Fig. 5E, G).
Taken together, resistance to IR in the IRR cells goes together with elevated TRIP13 levels and HR related proteins.
Discussion
Radiation therapy is a common treatment modality for lung cancer, and resistance to radiation can significantly affect treatment outcomes24. Our group recently described that lung cancer cells that express more genes that are normally specific for germ cells (GC genes) can repair DNA double-strand breaks (DSBs) more rapidly, show higher rates of proliferation and are more resistant to ionizing radiation (IR), compared to lung cancer cells that express fewer GC genes. Specifically, our investigation identified TRIP13 as a significant GC gene associated with enhanced IR resistance. Targeting TRIP13 with the inhibitor DCZ0415 resulted in reduced DNA damage response and heightened sensitivity to IR in high TRIP13-expressing cells5. In line with these results, we here find that TRIP13 plays a key role in the development of resistance to IR. Recurrent irradiation of lung cancer cells led to an increase of basal levels of TRIP13 and radioresistance and elevated TRIP13 induced enhanced repair of IR-induced DNA damage, further increasing cell proliferation and radiation resistance. Indeed, also in NSCLC patients who followed radiation treatment, TRIP13 expression appeared to be correlated with a poor prognosis. We further found the DNA repair proteins NBS1, RAD51 and XRCC5 to act downstream of TRIP13. This study elucidates a novel mechanism of treatment resistance in NSCLC in which TRIP13 promotes DNA repair and radioresistance in NSCLC cells.
For this research, we exposed a lung cancer cell line with high TRIP13 expression (H1703), a TRIP13KO cell line derived from H1703 (TRIP13KO); a “normal” lung epidermal cell line (BEAS-2B), and BEAS-2B cells that overexpress TRIP13 (OE-TRIP13) to repeated doses of IR (-IRR). We found that all IRR cell lines exhibit increased radioresistance upon repeated IR exposure. Interestingly, H1703-IRR cells demonstrated an increase in basal level of TRIP13 expression levels, whereas there was no significant alteration in basal TRIP13 protein levels observed in BEAS-2B cells or in BEAS-2B cells overexpressing TRIP13. This discrepancy between cancer cells and “normal” cells (BEAS2B) suggests a potential distinction in their response to IR between cancer cells and their somatic counterparts. This could also explain why the TRIP13 inhibitor did not influence TRIP13 expression in BEAS-2B cells.
Previously research has been reported that differences in radioresistance are not limited to TRIP13 but have been observed for various other markers and cellular processes. For example, many cancer cells have mutated p53, leading to altered responses to IR compared to normal cells with functional p5325–27. The ATM and ATR kinases, crucial in DNA damage repair signalling, often show dysregulated activity in cancer cells, affecting their IR response28. BRCA1/BRCA2 are critical for homologous recombination repair of DNA double-strand breaks29. Cancers with BRCA mutations, particularly breast and ovarian cancers, show different sensitivities to IR and other DNA-damaging agents30,31. While normal cells can repair DNA damage effectively, BRCA-deficient cancer cells may exhibit increased susceptibility to certain treatments, such as PARP inhibitors, due to their reliance on alternative, less accurate repair pathways32.
Two major DNA repair pathways are involved in repairing IR-induced DSBs: homologous recombination (HR) and non-homologous end joining (NHEJ)10. How TRIP13 mediates DNA damage repair pathway-choice in cancer is still an area of active research. Recent reports indicate that TRIP13 catalyses the conformational change of REV7 from its active ‘closed’ state to an inactive ‘open’ state, dissociating REV7 from the Shieldin complex to promote HR over NHEJ, while also disassembling the REV7-REV3 TLS complex, inhibiting error-prone replicative lesion bypass16. In cancer cells, overexpression of TRIP13 can shift the repair process toward HR or promote improper DNA repair by affecting the balance of these pathways15,33. Inhibiting TRIP13 is a potential therapeutic strategy to sensitize cancer cells to DNA-damaging agents. By blocking TRIP13’s function, cancer cells may be forced to rely on the error-prone NHEJ pathway or fail to repair DNA damage altogether, leading to increased cell death34. In our study, using knockout of TRIP13 in the H1703 cells confirms that decreased TRIP13 levels correspond with reduction of RAD51 and NBS1 (HR), and XRCC5 (NHEJ) expression. Conversely, overexpression of TRIP13 in BEAS-2B cells leads to increased levels of the RAD51, NBS1 and XRCC5 compared to control cells. We further observed that recurrent irradiation of NSCLC cells leads to an increase in basal TRIP13 levels, which is accompanied by elevated levels of the RAD51, NBS1 and XRCC5 in these cells. In the BEAS2B cells and in BEAS-2B overexpressing TRIP13, where basal TRIP13 expression doesn’t change, there were no observable changes in RAD51, NBS1 or XRCC5 levels. Based on these findings, we conclude that TRIP13 is involved in the HR and NHEJ pathways. When IRR-cells were treated with TRIP13 inhibitor DCZ0415, reductions in TRIP13 expression coincided with a decrease in the proteins RAD51 and NBS1, but only in cells with high levels of TRIP13 (H1703-IRR and OE-TRIP13-IRR). Conversely, treatment with the TRIP13 inhibitor DCZ0415 had no discernible effect on TRIP13 expression in BEAS-2B cells. Additionally, there were no significantly changes observed in the NHEJ pathway protein XRCC5 in response to DCZ0415 treatment in IRR-cells, suggesting a more prominent role of TRIP13 in the HR pathway. However, since we only investigated XRCC5, a comprehensive understanding of TRIP13’s role in NHEJ will require further investigation.
Multiple studies have demonstrated TRIP13’s critical role in non-homologous end joining (NHEJ), highlighting its involvement in facilitating the repair of DNA double-strand breaks through this pathway. TRIP13 exhibits a complex, context-dependent function in NHEJ, acting as both a promoter and suppressor of this DNA repair mechanism. Its specific role may vary based on the cellular environment and the particular stage of the DNA damage response.
For instance, in head and neck squamous cell carcinoma (SCCHN), TRIP13 overexpression enhances DNA damage repair by promoting NHEJ through interactions with DNA-PKcs complex proteins, even when homologous recombination (HR) remains intact35. Furthermore, TRIP13 has emerged as a crucial regulator of DNA repair pathway choice, promoting HR while suppressing NHEJ and translesion synthesis. In this context, the suppression of NHEJ by TRIP13 is dependent on the HORMA domain protein REV7. HORMA domain proteins can switch between active and inactive states based on the availability of their C-terminal binding site, suggesting that TRIP13 can also inhibit NHEJ under certain conditions16 Our findings align with this multifaceted role of TRIP13 in DNA repair. We observed lower XRCC5 protein levels in TRIP13 knockout cells compared to control cells, while TRIP13-overexpressing cells exhibited higher XRCC5 levels relative to BEAS-2B (dCas9) cells.
Many studies have demonstrated significant involvement of meiotic genes in cancer. While meiosis is tightly controlled in germ cells, the ectopic expression of such GC genes in cancer may result in the aberrant activation of pseudo-meiotic processes, such as partial meiotic recombination, that can result in chromosomal and genomic instability potentially contributing to tumorigenesis2,36–39. For example, TEX12 is a protein that normally mediates synaptonemal complex assembly, but ectopic expression in somatic cells contributes to oncogenic centrosome amplifications40. Aurora kinase C (AURKC) and MAD2, which are required for the meiotic spindle assembly checkpoint, lead to increased chromosomal abnormalities when ectopically expressed in somatic cells41,42. It has been reported that TRIP13 is aberrantly expressed in various cancers and may be involved in tumorigenesis by promoting chromosome instability and aneuploidy in primary tumors and cancer cell lines43–46. The fact that we find TRIP13 to be involved in HR, in line with its meiotic function recombination, suggests that TRIP13-induced resistance to IR may be associated with pseudo-meiotic activity.
TRIP13 serves as a co-chaperone in various cellular processes47. It exhibits high expression levels during embryogenesis and in meiotic cells, and plays an important role in both mitosis and meiosis48,49. In mitosis, TRIP13 is involved in the Spindle Assembly Checkpoint (SAC) and metaphase to anaphase transition6,50,51. TRIP13 has been identified as playing a crucial role in the regulation of meiosis in both mammalian and budding yeast52. Meiotic DSB formation and repair are a crucial part of meiotic crossover formation and TRIP13 is involved in the resolution of recombination intermediates that arise during meiotic homologous recombination53. Many studies suggest that TRIP13 could contribute to the establishment of inter-homologue biased HR16,54–56. Our study reveals that cells with low TRIP13 expression repair IR-induced DSBs (marked by γ-H2AX) less quickly than high TRIP13 cell lines. We therefore suggest that high TRIP13 expression in cancer cells promotes pseudo-meiotic HR, which leads to resistance to DNA-damaging therapies.
TRIP13 plays a significant role in promoting resistance to various cancer treatments, including radiation and chemotherapy, although whether its expression also increases in patients receiving genotoxic treatment still has to be investigated. Overexpression of TRIP13 in head and neck squamous cell carcinoma (HNSCC) promotes error-prone non-homologous end joining (NHEJ), which can contribute to radiation resistance by allowing cancer cells to survive DNA damage induced by radiation57. TRIP13 overexpression is associated with decreased sensitivity to bortezomib and cisplatin in multiple myeloma58. TRIP13 also facilitates the development of nedaplatin resistance in esophageal squamous cell carcinoma59. In epithelial ovarian cancer, TRIP13 knockdown inhibits cell proliferation and increases sensitivity to treatment60 . Furthermore, TRIP13 amplification confers resistance to PARP inhibitors in homologous recombination (HR)-deficient cells. Overexpression of TRIP13 leads to a striking induction of Olaparib (a PARP inhibitor) resistance in BRCA1-mutant cells34. Additionally, TRIP13-pY56 (phosphorylates TRIP13 on tyrosine 56) could potentially be used to predict the response to radiation or cetuximab and could be explored as an actionable target61.
Our research has unveiled the TRIP13 is a key contributor to treatment resistance, particularly in lung cancer patients following radiation treatment, for whom levels of TRIP13 expression are correlated with a poor prognosis. Repeated irradiation of NSCLC led to an increase of basal TRIP13 levels and radioresistance. Elevated TRIP13 is also correlated with enhanced repair of radiation-induced DNA damage. We further showed the proteins NBS1 and RAD51 and XRCC5 to act downstream of TRIP13. This study elucidates a novel mechanism of treatment resistance in NSCLC cells, in which TRIP13 promotes HR mediated DNA repair and resistance to ionizing radiation.
While specific clinical studies examining changes in TRIP13 levels before and after IR to induces DNA repair are currently lacking. the potential for such research is significant. Previously studies have used mouse models to investigate TRIP13’s role in DNA repair. For example, Trip13 transgenic mice have been used to study the protein’s role in protecting kidney tubular epithelia against cisplatin-induced DNA damage. These mice showed significant protection against cisplatin nephrotoxicity, suggesting TRIP13’s ability to mitigate DNA damage in this context15. Additionally, Mouse models have been crucial for understanding TRIP13’s role in meiotic recombination and chromosome synapsis, with studies demonstrating that TRIP13 is essential for proper meiotic progression and fertility in mice7. Future studies could provide valuable insights into TRIP13’s role in DNA repair treatment response and its prognostic significance in various cancers, potentially leading to new strategies for improving treatment efficacy in cancer patients. However, in our study we chose to focus on studying human lung cancer and patient derived cell lines. We also used TRIP13 knockout (TRIP13-/-) model, provide a direct way to study the consequences of TRIP13 loss on cellular processes, particularly its impact on DNA repair pathways and cell cycle regulation. By observing how cells respond to DNA damage in the absence of TRIP13, we can better understand the specific mechanisms that TRIP13 regulates, such as HR or NHEJ.
Overall, our findings underscore the importance of TRIP13 in influencing treatment outcomes in lung cancer patients, and likely in cancer in general, particularly in the context of IR treatment, highlighting the potential significance of TRIP13 as a prognostic marker and therapeutic target.
Supplementary Information
Acknowledgements
This work has been supported by the Amsterdam Reproduction & Development Research Institute and the China Scholarship Counsel (grant number 202006300005 to Wenqing Liu).
Author contributions
W.L. and G.H. conceived and designed the experiments. W.L. performed the experiments. W.L. and Q.L. conceived and designed the knockout and overexpression cell line experiments. W.L. and G.H. analysed the data. W.L. and G.H. wrote and revised the manuscript. A.v.P. critically read and revised the manuscript.
Data availability
All data and materials generated and analyzed during the present study are available from the corresponding author on reasonable requests.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-84592-w.
References
- 1.Shay, J. W. & Wright, W. E. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell. Biol.1(1), 72–76 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Bruggeman, J. W. et al. How germline genes promote malignancy in cancer cells. Bioessays10.1002/bies.202200112 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Bruggeman, J. W. et al. Massive expression of germ cell-specific genes is a hallmark of cancer and a potential target for novel treatment development. Oncogene37(42), 5694–5700 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruggeman, J. W. et al. Tumors widely express hundreds of embryonic germline genes. Cancers (Basel)12(12), 3812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, W. et al. Germline specific genes increase DNA double-strand break repair and radioresistance in lung adenocarcinoma cells. Cell Death Dis15(1), 38 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vader, G. Pch2(TRIP13): controlling cell division through regulation of HORMA domains. Chromosoma124(3), 333–339 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Chotiner, J. Y. et al. TRIP13 localizes to synapsed chromosomes and functions as a dosage-sensitive regulator of meiosis. Elife10.7554/eLife.92195.3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye, Q. Z. et al. TRIP13 is a protein-remodeling AAA plus ATPase that catalyzes MAD2 conformation switching. Elife10.7554/eLife.07367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan, M. A. & Lawrence, T. S. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin. Cancer Res.21(13), 2898–2904 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vignard, J., Mirey, G. & Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Radiother. Oncol.108(3), 362–369 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Weterings, E. & Chen, D. J. The endless tale of non-homologous end-joining. Cell Res.18(1), 114–124 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Rodgers, K. & McVey, M. Error-prone repair of DNA double-strand breaks. J. Cell Phys.231(1), 15–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nat.450(7169), 509–514 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, L. H. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat. Res.751(2), 158–246 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Hama, T. et al. DNA damage is overcome by TRIP13 overexpression during cisplatin nephrotoxicity. Jci. Insight10.1172/jci.insight.139092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clairmont, C. S. et al. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol.22(1), 87 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Krijger, I. et al. MAD2L2 dimerization and TRIP13 control shieldin activity in DNA repair. Nat. Commun.10.1038/s41467-021-25724-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong, H. et al. TRIP13 participates in immediate-early sensing of DNA strand breaks and ATM signaling amplification through MRE11. Cells11(24), 4095 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franken, N. A. et al. Clonogenic assay of cells in vitro. Nat. Protoc.1(5), 2315–2319 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Park, J. Y. et al. Identification of a novel human Rad51 variant that promotes DNA strand exchange. Nucl. Acids Res.36(10), 3226–3234 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robison, J. G. et al. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J. Biol. Chem.279(33), 34802–34810 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Zahid, S. et al. The multifaceted roles of Ku70/80. Int. J. Mol. Sci.22(8), 4134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fell, V. L. & Schild-Poulter, C. Ku regulates signaling to DNA damage response pathways through the Ku70 von Willebrand a domain. Mol. Cell. Biol.32(1), 76–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baskar, R. et al. Cancer and radiation therapy: Current advances and future directions. Int. J. Med. Sci.9(3), 193–199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abuetabh, Y. et al. DNA damage response revisited: the p53 family and its regulators provide endless cancer therapy opportunities. Exp. Mol. Med.54(10), 1658–1669 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Y., Leslie, P. L. & Zhang, Y. Life and death decision-making by p53 and implications for cancer immunotherapy. Trends Cancer7(3), 226–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlsen, L. et al. The role of p53 in anti-tumor immunity and response to immunotherapy. Front. Mol. Biosci.10, 1148389 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marechal, A. & Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol.5(9), a012716 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setton, J. et al. Long-molecule scars of backup DNA repair in BRCA1- and BRCA2-deficient cancers. Nature621(7977), 129–137 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadeghi, F. et al. Molecular contribution of BRCA1 and BRCA2 to genome instability in breast cancer patients: Review of radiosensitivity assays. Biol. Proced. Online22, 23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrucelli, N., M.B. Daly, and T. Pal, BRCA1- and BRCA2-Associated Hereditary Breast and Ovarian Cancer, in GeneReviews((R)), M.P. Adam, et al., Editors. 1993: Seattle (WA). [PubMed]
- 32.Treszezamsky, A. D. et al. BRCA1- and BRCA2-deficient cells are sensitive to etoposide-induced DNA double-strand breaks via topoisomerase II. Cancer Res.67(15), 7078–7081 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Xue, J. et al. TRIP13 overexpression in hepatocellular carcinoma: Implications for poor prognosis and immune cell infiltration. Discov. Oncol.14(1), 176 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao, L. et al. Targeting TRIP13 for overcoming anticancer drug resistance (Review). Oncol. Rep.10.3892/or.2023.8639 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee, R. et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat. Commun.10.1038/ncomms5527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotillo, R. et al. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature464(7287), 436–440 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bargiela-Iparraguirre, J. et al. Mad2 and BubR1 modulates tumourigenesis and paclitaxel response in MKN45 gastric cancer cells. Cell Cycle13(22), 3590–3601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakhoum, S. F. & Compton, D. A. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J. Clin. Invest.122(4), 1138–1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sou, I. F. et al. Cancer and meiotic gene expression: Two sides of the same coin?. Curr. Top. Dev. Biol.151, 43–68 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Sandhu, S. et al. Centrosome dysfunction associated with somatic expression of the synaptonemal complex protein TEX12. Commun. Biol.4(1), 1371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bejar, J. F., DiSanza, Z. & Quartuccio, S. M. The oncogenic role of meiosis-specific Aurora kinase C in mitotic cells. Exp. Cell. Res.407(2), 112803 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Byrne, T. et al. The association between MAD2 and prognosis in cancer: a systematic review and meta-analyses. Oncotarget8(60), 102223–102234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsueda, R. et al. Oncogenic targets regulated by tumor-suppressive miR-30c-1-3p and miR-30c-2-3p: TRIP13 facilitates cancer cell aggressiveness in breast cancer. Cancers (Basel)15(16), 4189 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen, S. H. et al. Clinical significance and systematic expression analysis of the thyroid receptor interacting protein 13 (TRIP13) as human gliomas biomarker. Cancers13(10), 2338 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agarwal, S. et al. TRIP13 promotes metastasis of colorectal cancer regardless of p53 and microsatellite instability status. Mol Oncol14(12), 3007–3029 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agarwal, S. et al. DCZ0415, a small-molecule inhibitor targeting TRIP13, inhibits EMT and metastasis via inactivation of the FGFR4/STAT3 axis and the Wnt/beta-catenin pathway in colorectal cancer. Mol. Oncol.16(8), 1728–1745 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neuwald, A. F. et al. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res.9(1), 27–43 (1999). [PubMed] [Google Scholar]
- 48.Li, X. C. & Schimenti, J. C. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genet3(8), e130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhlen, M. et al. Tissue-based map of the human proteome. Science347(6220), 1274 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Tipton, A. R. et al. Identification of novel mitosis regulators through data mining with human centromere/kinetochore proteins as group queries. Bmc Cell Biol.10.1186/1471-2121-13-15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Habu, T. et al. Identification of a MAD2-binding protein, CMT2, and its role in mitosis. Embo Journal21(23), 6419–6428 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.San-Segundo, P. A. & Roeder, G. S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell97(3), 313–324 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Roig, I. et al. Mouse TRIP13/PCH2 Is required for recombination and normal higher-order chromosome structure during meiosis. Plos Genetics6(8), e1001062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joshi, N. et al. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Mol. Cell57(5), 797–811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanders, S. et al. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics188(3), 511–521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ho, H. C. & Burgess, S. M. Pch2 Acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. Plos Genet.7(11), e1002351 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banerjee, R. et al. TRIP13 promotes error-prone nonhomologous end joining and induces chemoresistance in head and neck cancer. Nat. Commun.5, 4527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao, Y. et al. TRIP13 impairs mitotic checkpoint surveillance and is associated with poor prognosis in multiple myeloma. Oncotarget8(16), 26718–26731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, L. T. et al. TRIP13 induces nedaplatin resistance in esophageal squamous cell carcinoma by enhancing repair of DNA damage and inhibiting apoptosis. Biomed Res Int2022, 7295458 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, X. Y. & Shu, X. M. TRIP13 promotes proliferation and invasion of epithelial ovarian cancer cells through Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci.23(2), 522–529 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Banerjee, R. et al. Phosphorylation of TRIP13 at Y56 induces radiation resistance but sensitizes head and neck cancer to cetuximab. Mol. Ther.30(1), 468–484 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials generated and analyzed during the present study are available from the corresponding author on reasonable requests.