Abstract
Amphioxus (subphylum Cephalochordata) is the closest living relative to vertebrates and widely used for phylogenetic analyses of vertebrate gene evolution. Amphioxus genes are highly polymorphic, but the origin and nature of this variability is unknown. We have analyzed the alcohol dehydrogenase locus (Adh3) in two amphioxus species (Branchiostoma lanceolatum and Branchiostoma floridae) and found that genetic variation is related to repetitive DNA sequences, mainly minisatellites. Small pool-PCR assays indicated that allelic variants are generated by minisatellite instability. We conclude that the generation of new forms was not preferentially linked to germline processes but rather to somatic events leading to mosaic adult animals. Furthermore, most Adh minisatellites belong to a novel class, which we have named mirages. Their distinctive feature is that the repeat subunit spans the exon–intron boundaries and generates potential duplications of the splice sites. However, splicing may not be compromised as no aberrant mRNA variants were detected.
INTRODUCTION
The cephalochordate amphioxus, the closest living relative to vertebrates, is considered the ideal outgroup for phylogenetic reconstructions and developmental analyses of vertebrate gene families (1,2). Several amphioxus genes display inter-individual variability (3–15), which suggests that allelic polymorphism is widespread. However, neither the nature nor the origin of these polymorphisms have been described.
In vertebrates, repeated DNA elements are considered a major source of polymorphisms and one of the main driving forces of genome evolution. Among these, minisatellites are intermediate-size tandem arrays scattered throughout the genome and widely reported in mammals. Although information about the instability processes that involve these repeats is scanty, they have been of special interest for DNA typing, gene mapping and the study of natural variability. New length variants have been characterized in human through pedigree analysis (16) and small pool-PCR (SP-PCR) (17). These studies have shown that germline instability often involves complex inter-allelic conversion-like events at meiosis, whereas the infrequent somatic instability may involve polymerase slippage or, rather, intra-allelic unequal crossover (18).
We analyzed the alcohol dehydrogenase class 3 gene in two amphioxus species, Branchiostoma floridae (BfAdh3) and Branchiostoma lanceolatum (BlAdh3) and found that it is highly polymorphic. Sequence analysis and SP-PCR assays indicated that somatic instability of the repeated elements at the intragenic regions could be the cause of this variability, and would confer a mosaic status to adult organisms. Moreover, most of these repeated sequences belong to a novel class of minisatellites, which we have named mirages, because the repeat subunit spans the exon–intron boundaries. All these features, together with the high polymorphism, make amphioxus a suitable model to monitor repeat dynamics and analyze repeat instability under an evolutionary perspective.
MATERIALS AND METHODS
Genomic DNA, Southern analysis and RT–PCR
Adult specimens of B.lanceolatum were kindly provided by the Laboratoire Arago (Observatoire Océanologique de Banyuls-sur-mer, France) and adult specimens of B.floridae were harvested in Tampa Bay, FL. Animals were kept at –80°C until use. Genomic DNA was isolated by the guanidine thiocyanate method (19) with minor modifications (20). For Southern analysis, 7–10 µg PstI-digested genomic DNA from single animals was resolved in agarose gels, transferred to nylon membranes and hybridized with six distinct probes under high stringency conditions (13).
Branchiostoma lanceolatum RNA was prepared from single animals and cDNA was synthesized using the Marathon cDNA amplification kit (Clontech). PCR assays were performed with the following oligonucleotide pairs: L1S (5′-GCGGCGGTTGCCTGGGA-3′) + 1R (5′-GAACCCGGAGACGCCTTCCC-3′); L6S (5′-CATGGGGGCAAAGGTCGC-3′) + L14A (5′-CTGGATGGGCTTGTCATG-3′); F7S (5′-TTCGGCATGACGGAGTTCGT-3′) + CR2 (5′-TCCCCAGCCCTTGTGGC-3′); L8S (5′-CGAGCAGCCTTGGAGTCCTG-3′) + L10A (5′-CTCCGAAAGCCGTTCCCTT-3′); L6S + L10A. PCR conditions were as follows: an initial denaturation step at 94°C for 2 min was followed by 40 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 30 s and a final extension step at 72°C for 5 min. Amplified products were resolved on agarose gels and stained with ethidium bromide.
Detection of intron length variation by SP-PCR
Somatic regions (head and tail) and gonadal-enriched tissues were dissected from four B.lanceolatum males. Purified genomic DNA from each sample was kept separately. To detect minisatellite variability in each sample, the conventional SP-PCR (17), with minor modifications, was performed. DNA (700 ng) was fully solubilized by digestion with 10 U BamHI (1 h at 37°C in 4 mM spermidine trichloride), centrifuged at 10 000 g for 2 min to remove particulate matter and adjusted to a final concentration of 4 ng/µl in 10 mM Tris–HCl pH 8.0/1 mM EDTA. Approximately 2280 amphioxus haploid genomes, assuming a molecular mass of 1 pg per haploid genome, were diluted into 200 µl of PCR mix: 1 mM MgCl2, 0.2 mM dNTPs, 0.2 µM of each forward (F) and reverse (R) primer (F1, 5′−ACGGCACGCCAAAACCTC-3′ + R2, 5′−GATGCGGACTTCGTGCGC-3′ and F6, 5′−CATGGGGGCAAAGGTCGC-3′ + R7, 5′−CTGGATGGGCTTGTCATG-3′ for introns 1 and 6, respectively), 1× PCR buffer and 0.1 U/µl of Biotherm™ (Genecraft, Germany) Taq polymerase. Multiple 7 µl aliquots (80 haploid genomes) of this solution were SP-PCR amplified on a GeneAmp PCR system 9600 thermal cycler (Perkin Elmer) at 94°C (20 s), 55°C (1 min) and 72°C (1 min) for 26 cycles, followed by a chase at 72°C (5 min). The PCR products were resolved in a 1.7% agarose gel and detected by Southern blot hybridization with a 32P-labeled probe encompassing the analyzed region. Statistical analysis of the homogeneity of the data was assessed with a chi-squared test. Additional experiments were performed to discard PCR artefacts. As mutant-free control DNA, two cloned length variants of intron 6 were amplified with F6 and R7 primers in the SP-PCR conditions described above. DNA from single clones or from a mixture was used as template. To compare the frequency of the new forms that appeared with genomic DNA versus mutant-free DNA in SP-PCR assays, a contrast of proportions was performed with the Fisher’s exact test.
RESULTS
Amphioxus genomic variability
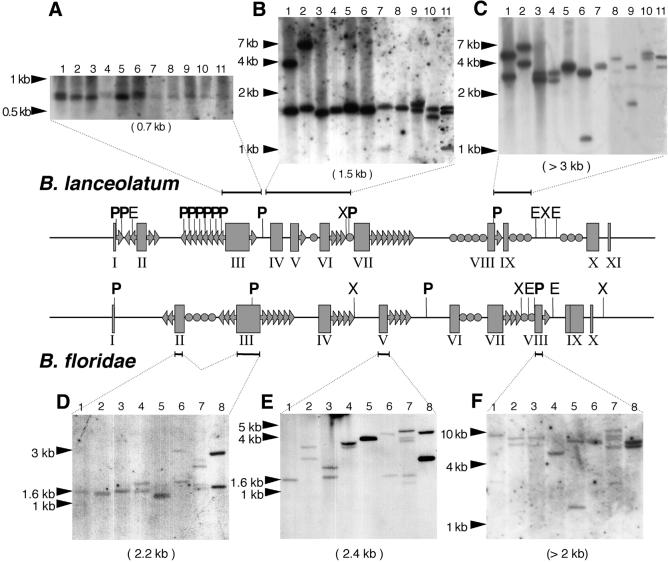
Adh is a single-copy gene, as shown in a previous study on two amphioxus species (B.lanceolatum and B.floridae) (13). However, library screenings (21) yielded recombinant phages encompassing the same genomic region, whose restriction patterns revealed unexpected high variability (data not shown). When Adh probes comprising various segments of the gene were used for Southern analysis on genomic DNA, common hybridization patterns were not identified, as the bands for each of the 19 animals assayed differed in size and number (Fig. 1). Moreover, closer examination of the inter-individual pattern showed an uneven distribution of the variable sites, ranging from a single band of a similar size at exon 3 (Fig. 1A) to several bands of different sizes at exon 9 (Fig. 1C). Remarkably, two individuals displayed more than two bands per lane (Fig. 1B, lane 11 and D–F, lane 7).
Figure 1.
Southern analysis of BlAdh3 (accession nos AF156698, AF156700 and AF156705) and BfAdh3 (AF266713–AF266719). Genomic DNA from single individuals was digested with PstI and probed with BlAdh3 fragments containing exon 3 (A), 4–6 (B) and 8–9 (C), and exons 2–3 (D), 5 (E) and 8 (F) of BfAdh3. The expected band size from the characterized Adh3 genes (under each panel) and the DNA size markers (at the left of each panel) are depicted. Genomic structure of Adh3 (central part): exons (boxes); EcoRI (E) PstI (P, in bold) and XbaI (X) restriction sites, mirages (arrows), other minisatellites (circles) and probes (horizontal lines) are shown. Notice that more than two bands are shown in lane 11 (B), and lane 7 (D–F).
Structural characterization of the Adh repeats: ‘mirages’
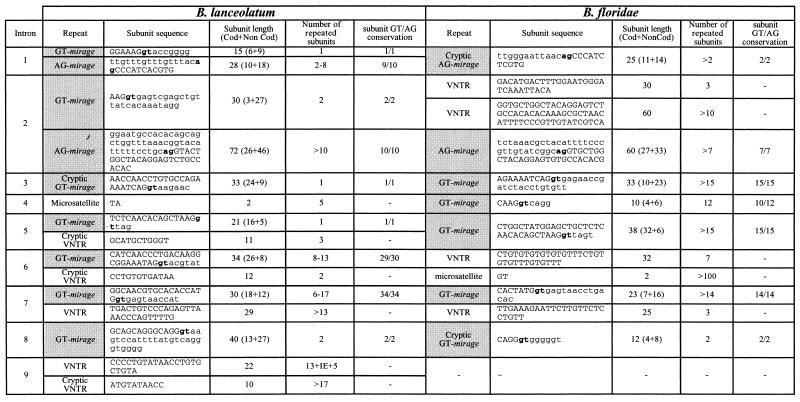
Sequence analysis of the Adh genomic region from previously isolated phages (21) showed that the restriction length polymorphism revealed by Southern analysis was due to repeated DNA sequences in the Adh introns. They were mainly direct tandem repeats [minisatellites, also called variable number tandem repeats (VNTRs)] and also a few internal short tandem repeats (microsatellites) (Table 1).
Table 1. Tandem repeats in BlAdh3 and BfAdh3 genes.
Mirages (gray cells), ag/gt splice sites (bold) and the sequence corresponding to the coding (capital letters) or non-coding (lower-case letters) are indicated. IE, inserted element.
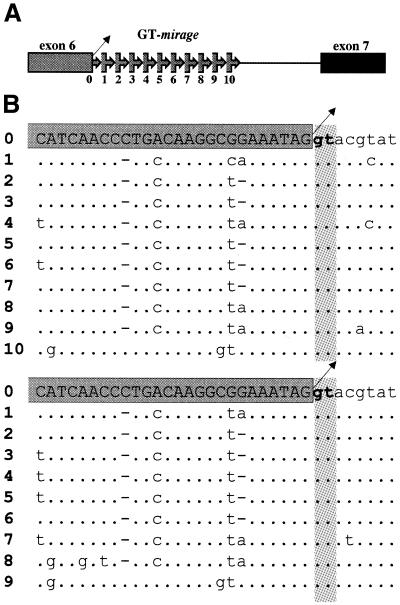
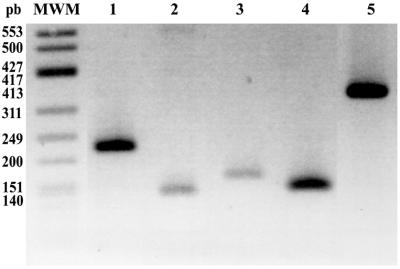
Among VNTRs, we identified a novel type of minisatellite, whose distinct feature was that the repeat subunit spanned the exon–intron boundary (Fig. 2). We have named these novel minisatellites ‘mirages’, since during sequence analysis as soon as a new exon was apparently reached it unexpectedly vanished. Mirages were made of imperfect direct tandem repeats with subunits of 10–72 nt, arranged in stretches of 1 to more than 15 repeats (Table 1). The fragment overlapping the coding region varied from 3 to 32 nt, while the non-coding sequence ranged from 5 to 46 nt. Fourteen out of the 19 BlAdh3 and BfAdh3 introns exhibited mirages. GT-mirages spanned the intron donor splice site and were frequently found, whereas the AG-mirages comprised the acceptor splice site and were found in only introns 1 and 2. Although the consensus splice sites (AG/GT) were conserved in 153 of the 157 (97.5%) repeats analyzed, no aberrant splicing variants were detected by RT–PCR (Fig. 3). Finally, although a few tandem repeat sequences showed poor resemblance to the exon, enough homology had been maintained to classify them as ‘cryptic’ mirages (Table 1).
Figure 2.
(A) Schematic representation of a GT-mirage overlapping the donor splice site at the boundary of exon 6–intron 6 of BlAdh3. Arrow-boxes indicate mirage subunits and the boxed sequence corresponds to the exon. The exon–intron boundary is numbered 0 and each following mirage subunit is consecutively numbered 1–10 (5′–3′). The splice donor site is indicated by an oblique arrow. (B) Sequence alignment of the intron-6 GT-mirage array of two BlAdh3 alleles. The segment corresponding to the coding region (capital letters in the dark gray box) and intron sequences (lower-case letters) are shown. The splice donor site (bold lower case) and its reiteration are depicted on light gray background. Nucleotide substitutions in the repeated sequences are shown, identities are represented by dots and deletions by dashes.
Figure 3.
RT–PCR assays to detect aberrant splicing variants of mirages containing BlAdh3 introns (accession nos AF156698–AF156708) with oligonucleotides located at the flanking exons of intron 2 (lane 1, oligonucleotide pairs L1S–1R), intron 6 (lane 2, L6S–L14A), intron 7 (lane 3, F7S–CR2), intron 8 (lane 4, L8S–L10A) and the segment encompassing intron 6–exon 7–intron 7–exon 8–intron 8 (lane 5, F7S–L10A). Splicing of intron 1 and 9 (Table 1) were also assayed (data not shown). In all samples, no aberrant forms were detected and a single band with the expected size of the properly spliced product was observed. MWM, φX174 DNA HinfI.
Dynamics of the repeats
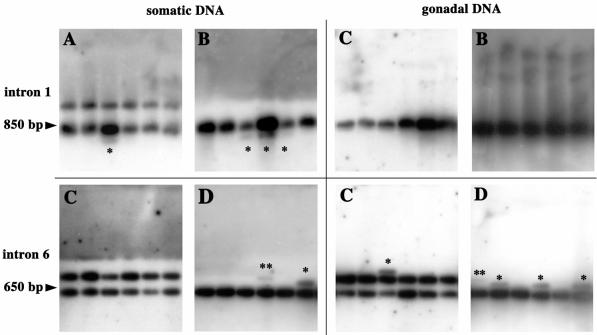
When a particular minisatellite structure was analyzed, we found variation in the number of repeats. To assess whether this variability was of somatic or germline origin, gonadal and somatic DNA was obtained from individual males. Gonadal fractions consisted of meiotic DNA-enriched preparations. SP-PCR assays were performed with primers flanking two BlAdh3 introns containing mirages to detect length variants other than those corresponding to the parental variants (Fig. 4). Four animals, either with one or two parental variants, were analyzed. All the PCRs shared an intense signal, which corresponded to the inherited variants, and occasionally they showed other signals of different length.
Figure 4.
Allelic length variants of BlAdh3 in four single animals (A–D) by SP-PCR. Examples of abnormal length mutants (*) corresponding to somatic (left) and gonadal-enriched DNA fractions (right), of intron 1 (top) and intron 6 (bottom) Adh3. Animals showed one progenitor molecule (B and C for intron 1; D for intron 6) or two progenitor molecules (A for intron 1 and C for intron 6). PCR products were electrophoretically resolved and abnormal-length mutants (*, one-subunit; **, two-subunit variation) were detected by blotting and hybridization.
The 335 PCR amplifications (80 haploid genome/SP-PCR reaction, 26 800 haploid genomes analyzed) yielded a total of 40 new length variants. Assuming that each new variant arose from a single template molecule, this would represent one new variant per 670 haploid genomes (0.15%) (Table 2). The frequency of the new bands was no higher in gonadal-enriched DNA preparations than in the somatic fraction (0.10 and 0.19%, respectively) (χ2 = 6.96; d.f. = 3; P = 0.07), which indicates that repeat instability was not linked to the germline. On the other hand, intron-length variability led to mosaic adults. Moreover, the level of somatic mosaicism in the four individuals differed by up to 10-fold, from 0.02 to 0.27% (χ2 = 16.14; d.f. = 3; P = 0.001). Nevertheless, no significant differences were observed between homozygous and heterozygous animals (0.16 and 0.13%, respectively), nor between the two introns compared (0.13% for intron 1 and 0.16% for intron 6) (χ2 = 4.37; d.f. = 3; P = 0.223). Interestingly, while the new variants of intron 6 increased in size, those from intron 1 decreased.
Table 2. Abnormal length variants detected by SP-PCR assays.
| Abnormal length variants/SP-PCR reactions | Percentage of abnormal length variantsa | ||
|---|---|---|---|
| Introns |
1 |
14/134 |
0.13 |
| |
6 |
23/201 |
0.16 |
| Heterozygosity |
Homozygotes |
27/213 |
0.16 |
| |
Heterozygotes |
13/122 |
0.13 |
| Individuals |
A |
13/115 |
0.10 |
| |
B |
4/67 |
0.07 |
| |
Cb |
22/102 |
0.27 |
| |
D |
1/51 |
0.02 |
| DNAs |
Somatic |
28/189 |
0.19 |
| |
Gonadal |
12/146 |
0.10 |
| Total | 40/335 | 0.15 |
The apparition of new variants is compared between the two introns analyzed (1 and 6), between homozygous and heterozygous animals for parental intron length variants, among the four individuals analyzed (A–D) and between the distinct types of DNA (somatic and gonadal).
aThe percentage has been calculated assuming 80 haploid genomes per SP-PCR.
bTwo of the 22 variants were made of more than one-subunit variation.
SP-PCR control experiments were performed with cloned DNA from two intron 6 alleles as mutant-free DNA (see Materials and Methods) to discard PCR artefacts. No new variant was detected in any of the PCR assays (0/64), indicating that the new length variants were genuine mutated alleles (Fisher’s exact test P = 0.001).
DISCUSSION
Amphioxus intragenic polymorphism
The cephalochordate amphioxus is believed to predate the large-scale gene duplications that marked early vertebrate evolution. To this end, estimates of amphioxus gene number have been crucial. The finding of variable banding patterns by Southern analysis has led to the widespread assumption that the genome of lancelets is highly polymorphic, which may impair gene-copy number assessments. However, no data have been reported on the molecular basis of this polymorphism.
Southern analysis of several individuals corroborates that Adh3 is a single-copy gene in amphioxus (13), although the inter-individual multiple banding pattern allows us to situate this locus among the most polymorphic. The complexity of the Southern patterns was substantially simplified by using reduced-size probes on single animals (Fig. 1). Sequence analysis indicates that most of the differences are due to intragenic repeated DNA, mainly VNTRs and occasionally inserted elements. Additional allelic variation also results from small insertions/deletions and nucleotide substitutions. Moreover, point mutations at the restriction sites could further increase the inter-individual variability observed.
Interestingly, this high polymorphism may be further increased by the VNTR instability that generates somatic mosaic animals. In fact, single individuals showed various degrees of mosaicism (Table 2), probably dependent on the generation rate of the new forms, and the timing and cell target of the mutational event. Hence, if early in development or in a highly proliferative cell lineage, more than two variants of a single copy gene can be isolated from a single animal. Occasionally, the abundance of new variants can allow their visualization as faint bands on Southern blots (Fig. 1), which may have led to the erroneous assumption of another related gene. Certainly, not all amphioxus genes have to behave similarly, because the extent of intragenic variability can be affected by factors such as chromosomal location, methylation, level of transcription, proximity to telomeres, centromeres, as well as to recombination hot-spots, which condition the organization of repetitive DNA in eukaryotic genomes (22–24).
Minisatellite instability
We confirmed that the length variants detected by SP-PCR correspond to genuine mutant alleles rather than PCR artefacts (17): (i) length variants were never detected in the same SP-PCR conditions using cloned alleles as mutant-free DNA controls, and (ii) the frequency of new length variants differed among animals.
SP-PCR analysis on genomic DNA from dissected single animals revealed that the amphioxus repeat instability is related to somatic polymorphism rather than to germline processes. We thus conclude that lancelets are mosaics. The pattern of the length variants pointed to a stepwise mutation fashion, compatible with one subunit variation in each step, in agreement with longer variants only observed at highest mosaicism levels (Fig. 4 and Table 2). Our data also suggest that the repeat dynamics is under a mutational bias (17) because gains or losses did not occur with equal frequency: intron 6 exclusively showed expansions, whereas intron 1 only showed reductions. In human, somatic and germline minisatellite instability leading to new variants have been attributed to separate pathways involving recombination (25). In contrast, although somatic polymorphism in amphioxus is clearly supported by two independent approaches—Southern analysis and SP-PCR experiments—the mechanism underlying the generation of new variants is unknown, since the PCR and cloning strategies performed to characterize recombinant alleles have failed.
‘Mirages’, a novel type of minisatellite
We describe a novel type of minisatellite sequence, termed mirage, whose repeat subunit differs from conventional tandem repeats in that it spans the exon–intron boundaries and duplicates the exon edges. Mirages are abundant in Adh (detected in 75% of the introns). Moreover, in the course of other gene characterizations (15,26) we have gathered evidence that this type of VNTR is not restricted to the Adh locus, but possibly present in other amphioxus genes. Therefore, our data highlight a new minisatellite entity which, to our knowledge, has not been defined before.
Regarding the mirage structure, although none of its subunits is identical to the exon–intron sequence from which it was generated, 97.5% of mirage subunits retain the AG/GT splice site. Redundancy of the consensus signal did not compromise splicing, as no aberrant cDNA variants were detected (Fig. 3). Although the substituted positions within the repeat subunits were frequently the same in a single intron, these were not maintained when different introns were compared. Therefore, functional implications of these positions in the splicing process could not be established.
Concerning its origin, it is reasonable to assume that each mirage arose from a duplication of the adjacent exon and, as the exon–intron boundary is at the core of these structures, the splicing machinery could have been involved. To study the mirage dynamics, we performed a neighbor-joining analysis of the mirage subunits from several alleles (data not shown). When different introns were considered, no common topologies were obtained. Therefore, hints about the history of the repeats could not be inferred.
Mirages preserve sequence similarity in the surrounding regions of exons and, hence, may influence gene evolution by recombination and gene conversion in the course of duplication events.
The presence of three similar minisatellite structures in primates (27–29) allows to speculate on the ancestry of the mirage sequences, which may have predated the cephalochordate–vertebrate split. However, their abundance in the amphioxus genome contrasts with the low number reported in mammals. Indeed, two of the three mirage-type structures in human are associated with aberrant splicing leading to pathological conditions. Unless undetected until present, mirages would have been differentially retained in the different species.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to O. Gonzàlez-Angulo for technical assistance. We thank G. Marfany, A. Jeffreys and A. Carracedo for helpful discussion, C. Arenas for help in the statistical analysis and R. Rycroft for revising the English. We also thank the Serveis Científico-Tècnics (UB) for DNA sequencing. R.G.-D. received a grant from DGICYT (Ministerio de Educación y Cultura, Spain, BMC2000-0536) and C.C. was the recipient of an FPI fellowship from the CIRIT (Generalitat de Catalunya, 1997FI00007).
REFERENCES
- 1.Garcia-Fernàndez J. and Holland,P.W. (1994) Archetypal organization of the amphioxus Hox gene cluster. Nature, 370, 563–566. [DOI] [PubMed] [Google Scholar]
- 2.Holland P.W., Garcia-Fernàndez,J., Williams,N.A. and Sidow,A. (1994) Gene duplications and the origins of vertebrate development. Dev. Suppl., 43, 125–133. [PubMed] [Google Scholar]
- 3.Holland P.W., Koschorz,B., Holland,L.Z. and Herrmann,B.G. (1995) Conservation of Brachyury (T) genes in amphioxus and vertebrates: developmental and evolutionary implications. Development, 121, 4283–4291. [DOI] [PubMed] [Google Scholar]
- 4.Holland L.Z., Kene,M., Williams,N.A. and Holland,N.D. (1997) Sequence and embryonic expression of the amphioxus engrailed gene (AmphiEn): the metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development, 124, 1723–1732. [DOI] [PubMed] [Google Scholar]
- 5.Shimeld S.M. (1997) Characterisation of amphioxus HNF-3 genes: conserved expression in the notochord and floor plate. Dev. Biol., 183, 74–85. [DOI] [PubMed] [Google Scholar]
- 6.Tweedie S., Charlton,J., Clark,V. and Bird,A. (1997) Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol. Cell. Biol., 17, 1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karabinos A. and Riemer,D. (1997) The single calmodulin gene of the cephalochordate Branchiostoma. Gene, 195, 229–233. [DOI] [PubMed] [Google Scholar]
- 8.Kusakabe R., Kusakabe,T., Satoh,N., Holland,N.D. and Holland,L.Z. (1997) Differential gene expression and intracellular mRNA localization of amphioxus actin isoforms throughout development: implications for conserved mechanisms of chordate development. Dev. Genes Evol., 207, 203–215. [DOI] [PubMed] [Google Scholar]
- 9.Williams N.A. and Holland,P.W. (1998) Gene and domain duplication in the chordate Otx gene family: insights from amphioxus Otx. Mol. Biol. Evol., 15, 600–607. [DOI] [PubMed] [Google Scholar]
- 10.Kusakabe R., Satoh,N., Holland,L.Z. and Kusakabe,T. (1999) Genomic organization and evolution of actin genes in the amphioxus Branchiostoma belcheri and Branchiostoma floridae. Gene, 227, 1–10. [DOI] [PubMed] [Google Scholar]
- 11.Shimeld S.M. (1999) The evolution of the hedgehog gene family in chordates: insights from amphioxus hedgehog. Dev. Genes Evol., 209, 40–47. [DOI] [PubMed] [Google Scholar]
- 12.Kozmik Z., Holland,N.D., Kalousova,A., Paces,J., Schubert,M. and Holland,L.Z. (1999) Characterization of an amphioxus paired box gene, AmphiPax2/5/8: developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development, 126, 1295–1304. [DOI] [PubMed] [Google Scholar]
- 13.Cañestro C., Hjelmqvist,L., Albalat,R., Garcia-Fernàndez,J., Gonzàlez-Duarte,R. and Jörnvall,H. (2000) Amphioxus alcohol dehydrogenase is a class 3 form of single type and of structural conservation but with unique developmental expression. Eur. J. Biochem., 267, 6511–6518. [DOI] [PubMed] [Google Scholar]
- 14.Yasui K., Zhang,S., Uemura,M. and Saiga,H. (2000) Left–right asymmetric expression of BbPtx, a Ptx-related gene, in a lancelet species and the developmental left-sidedness in deuterostomes. Development, 127, 187–195. [DOI] [PubMed] [Google Scholar]
- 15.Dalfó D., Cañestro,C., Albalat,R. and Gonzàlez-Duarte,R. (2001) Characterization of a microsomal retinol dehydrogenase gene from amphioxus: retinoid metabolism before vertebrates. Chem. Biol. Interact., 130–132, 359–370. [DOI] [PubMed] [Google Scholar]
- 16.Jeffreys A.J., Royle,N.J., Wilson,V. and Wong,Z. (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature, 332, 278–281. [DOI] [PubMed] [Google Scholar]
- 17.Jeffreys A.J., Tamaki,K., MacLeod,A., Monckton,D.G., Neil,D.L. and Armour,J.A. (1994) Complex gene conversion events in germline mutation at human minisatellites. Nature Genet., 6, 136–145. [DOI] [PubMed] [Google Scholar]
- 18.Jeffreys A.J. and Neumann,R. (1997) Somatic mutation processes at a human minisatellite. Hum. Mol. Genet., 6, 129–136. [DOI] [PubMed] [Google Scholar]
- 19.Chirgwin J.M., Przybyla,A.E., MacDonald,R.J. and Rutter,W.J. (1979) Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry, 18, 5294–5299. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Fernàndez J., Baguñà,J. and Saló,E. (1993) Genomic organization and expression of the planarian homeobox genes Dth-1 and Dth-2. Development, 118, 241–253. [DOI] [PubMed] [Google Scholar]
- 21.Cañestro C., Albalat,R., Hjelmqvist,L., Godoy,L., Jörnvall,H. and Gonzàlez-Duarte,R. (2002) Ascidian and Amphioxus Adh genes correlate functional and molecular features of the ADH family expansion during vertebrate evolution. J. Mol. Evol., 54, 81–89. [DOI] [PubMed] [Google Scholar]
- 22.Hancock J.M. (1996) Simple sequences and the expanding genome. Bioessays, 18, 421–425. [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth B., Sniegowski,P. and Stephan,W. (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature, 371, 215–220. [DOI] [PubMed] [Google Scholar]
- 24.Colot V. and Rossignol,J.L. (1999) Eukaryotic DNA methylation as an evolutionary device. Bioessays, 21, 402–411. [DOI] [PubMed] [Google Scholar]
- 25.Buard J., Collick,A., Brown,J. and Jeffreys,A.J. (2000) Somatic versus germline mutation processes at minisatellite CEB1 (D2S90) in humans and transgenic mice. Genomics, 65, 95–103. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Mir A., Cañestro,C., Gonzàlez-Duarte,R. and Albalat,R. (2001) Characteriztion of the amphioxus presenilin gene in a high gene-density genomic region illustrates duplication during the vertebrate lineage. Gene, 279, 157–164. [DOI] [PubMed] [Google Scholar]
- 27.Bonthron D.T., Smith,S.J. and Campbell,R. (1999) Complex patterns of intragenic polymorphism at the PDGFA locus. Hum. Genet., 105, 452–459. [DOI] [PubMed] [Google Scholar]
- 28.Yang F., Hanson,N.Q., Schwichtenberg,K. and Tsai,M.Y. (2000) Variable number tandem repeat in exon/intron border of the cystathionine beta-synthase gene: a single nucleotide substitution in the second repeat prevents multiple alternate splicing. Am. J. Med. Genet., 95, 385–390. [DOI] [PubMed] [Google Scholar]
- 29.Marchetti G., Patracchini,P., Gemmati,D., DeRosa,V., Pinotti,M., Rodorigo,G., Casonato,A., Girolami,A. and Bernardi,F. (1992) Detection of two missense mutations and characterization of a repeat polymorphism in the factor VII gene (F7). Hum. Genet., 89, 497–502. [DOI] [PubMed] [Google Scholar]