Abstract
The mechanism by which DNA-damage affects self-renewal and pluripotency remains unclear. DNA damage and repair mechanisms have been largely elucidated in mutated cancer cells or simple eukaryotes, making valid interpretations on early development difficult. Here we show the impact of ionizing irradiation on the maintenance and early differentiation of mouse embryonic stem cells (ESCs). Our findings demonstrate that irradiation induces the upregulation of the p53 family genes, including p53, p63, and p73, resulting in elevated expression of the E3 ubiquitin ligases Itch and Trim32. Consequently, this impairs ESC maintenance by reducing the protein levels of key pluripotency transcription factors in both mouse ESCs and early embryos. Notably, our study reveals that irradiation-induced DNA damage leads to the recruitment of the BAF complex, causing it to dissociate from its binding sites on the target genes associated with the Yap, Wnt, and TGF-β pathways, thereby increasing signaling and promoting differentiation of ESCs into all three lineages. Importantly, pathway inhibition demonstrates that DNA damage accelerated ESC differentiation relies on Wnt and TGF-β, and is selectively dependent on p53 or p63/ p73 for mesoderm and endoderm respectively. Finally, our study reveals that p53 family proteins form complexes with effector proteins of key signaling pathways which actively contribute to ESC differentiation. In summary, this study uncovered a mechanism by which multiple differentiation signaling pathways converge on the p53 family genes to promote ESC differentiation and are impacted by exposure to ionizing radiation.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-024-05561-0.
Keywords: Chromatin remodeling complex, Pluripotency genes, Embryonic germ layers
Introduction
Ionizing Radiation (IR) is a type of electromagnetic radiation generated by X-ray machines, fluoroscopy, radioisotopes, and nuclear environmental disasters, originating from diverse sources such as cosmic rays, radioactive materials used in medical and industrial processes, and natural radiation from the Earth. The use of radiation-based equipment for diagnosis and disease treatment has raised concerns about its detrimental effects on human health, particularly in relation to fetal malformation, abortion, and childhood cancers [1, 2]. The early stages of human embryonic development are highly sensitive and vulnerable to various teratogenic agents, including ionizing radiation [3]. ESCs are pluripotent cells derived from the inner cell mass of blastocyst-stage embryos [4–6]. ESCs own the capacity to self-renew indefinitely in vitro and to differentiate into every cell type of the body, therefore serve as an in vitro surrogate for the preimplantation embryo in assessing the biological effects of ionizing radiation during early development.
Several studies have examined the DNA damage response of mouse and human embryonic stem cells, noting variation in cell cycle regulation, p53 signaling, and apoptosis compared to cancer cell lines [7–12]. Intriguingly, some investigations have suggested irradiation can affect ESC maintenance and differentiation [2, 12–17]. Despite significant apoptosis observed in γ-irradiated human embryonic stem cells and X-ray-irradiated mouse embryos, the transcript levels of major pluripotency genes Oct4, Sox2, and Nanog did not exhibit significant changes [13, 15]. Surprisingly, surviving γ-irradiated human embryonic stem cells appeared to retain their pluripotency and capacity to form all three embryonic germ layers [13]. Similarly, Hayashi et al. [15] reported that low-dose X-ray-irradiated preimplantation embryos developed into morphologically normal blastocysts that could be successfully implanted and survive in the uterus. However, further studies have demonstrated that irradiation impairs the later stage differentiation of ESCs into cardiomyocytes and neurons [2, 12, 16]. Moreover, Gene ontology analysis has additionally revealed significant alterations in development of cardiovascular, nervous, circulatory, and renal systems in mESCs upon X-irradiation [12, 17], suggesting viable embryo may be significantly impaired from developing or have significant physiological issues. Thus, the level of exposure and the detailed mechanism by which irradiation affects self-renewal and pluripotency remains is vital to understand but remains unclear.
In this study, we investigated the impact of ionizing X-ray irradiation on the maintenance and early differentiation of mouse ESCs. Our findings demonstrate that irradiation induces the upregulation of the p53 transcription factor family, including p53, p63, and p73, resulting in elevated expression of the E3 ubiquitin ligases Itch and Trim32. Consequently, this impairs ESC maintenance by reducing the protein levels of key pluripotency transcription factors in both mouse ESCs and early embryos. Furthermore, we have provided evidence that irradiation treatment promotes the differentiation of ESCs into all three lineages by activating the Wnt, Hippo, and TGF-β signaling pathways. Significantly, our study reveals that irradiation-induced DNA damage leads to the recruitment of the BAF complex, causing it to dissociate from its binding sites on the target genes associated with the Yap, Wnt, and TGF-β pathways. Consequently, this relieves the inhibitory effect of the BAF complex on the activity of these pathways. This accelerated ESC differentiation can be rescued by inhibiting the Wnt or TGF-β pathways or by limiting the expression of p53, p63, and p73. Additionally, we found that p63 and p73 activity appears specifically selective for mesoderm differentiation while p53 activity is associated with endoderm differentiation.
Materials and methods
Animals and mouse embryo collection
Specific pathogen-free (SPF) mice were housed in the animal facility of Tongji University, Shanghai, China, and all animal experiments were carried out in accordance with the Tongji University Guide for the Use of Laboratory Animals.
C57BL/6 (6–8 weeks old) female mice were superovulated by injecting 7 IU of pregnant mare serum gonadotropin (PMSG) and then injecting 7 IU of human chorionic gonadotropin (hCG) 48 h later. Superovulated C57BL/6 female mice were mated with B6D2F1 (C57BL/6 × DBA2) male mice after hCG injection, and then, the zygotes (E0.5) were collected at 22–24 h post-hCG injection and cultured in G1 medium at 37 °C and 5% CO2.
Embryo immunostaining
The embryos were irradiated once with 2 Gy x-ray at E2.5 and E3.5, and cultured in vitro to E4.5 before immunostaining experiments. Upon reaching E4.5, the embryos were fixed in 4% paraformaldehyde for 1 h at room temperature (RT), then washed twice in 0.5% BSA-PBS for 10 min each. The samples were treated with 0.5% Triton X-100 in 3% BSA-PBS for 1 h at RT. After two washes as described above, the samples were incubated in primary antibody at 4 °C overnight in 3% BSA-PBS solution, then washed twice as above. The samples were incubated with secondary conjugated antibody in 3% BSA-PBS at RT for 1 h, then washed twice as above. The nuclei were counterstained with DAPI for 20 min at RT, then washed twice as above, the embryos were observed for fluorescence under a laser-scanning confocal microscope (LSM880, Zeiss).
Cell culture and differentiation
Mouse E14 ESCs were cultured in DMEM (Hyclone) supplemented with 10% FCS (Gibco), 1xNEAA (Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM 2-mercaptoethanol (Sigma), 2 mM L-glutamine (Gibco), and LIF (Millipore) on 0.1% gelatin (Sigma) coated plates. ESCs were frozen in ES medium with 10% of DMSO.
Induction of Embryoid Bodies (EBs): ESCs dissociated with 0.05% of trypsin were centrifuged and re-suspended in ES medium without LIF, and then plated in ultra-low attachment dishes to induce the formation of EBs. Cultures were maintained for indicated days and incubated at 37 °C with 5% CO2. EBs were collected for qPCR analysis.
Plasmid construction and generation of mutant ESCs
1. gRNA Vectors: Single guide RNAs (sgRNAs) were designed using the Benchling website (www.benchling.com/crispr/). To create plasmids with dual sgRNA constructs under the control of two separate U6 promoters, PCR amplicons derived from the pUC57 plasmid were ligated into the pGL3-U6-2sgRNA-CCDB-EF1α-Puro vector, which had been pre-digested with BsaI. Vectors were confirmed by Sanger sequencing.
2. Overexpression Plasmids: The coding regions of the target genes were amplified by PCR using cDNA from mouse or human ESCs as templates. The purified PCR products were ligated into the pPBH-TREtight-MLC-EGFP vector using homologous recombination enzymes (Vazyme).
3. Generation of Knockout ESCs: 2 μg of sgRNA vector(s) targeting specific genes and pCas9 plasmid (Addgene #41815) were co-transfected into ESCs using Lipofectamine 3,000 (Invitrogen). After 7 days’ selection with 1ug/ml of Puro, or 10 μg/ml of BSD, colonies were picked up for genotyping, and finally confirmed by Sanger sequencing and Western blot analysis.
4. Generation of Gene Overexpression ESCs: Overexpression plasmids were transfected into ESCs using Lipofectamine 3,000 (Invitrogen). Following hygromycin selection at a concentration of 100 μg/ml for 5 days, Western blotting was employed for genotyping.
Irradiation of ESCs and embryos
In this study, the Rad Source RS-2000 Pro irradiator was employed. For mouse embryos, irradiation was performed once with 2 Gy x-ray at E2.5 and E3.5, followed by in vitro culture until E4.5 before conducting immunostaining experiments. For ESCs, unless they were intended for the colony formation experiment, approximately 5 × 105 cells were irradiated twice within 48 h with 8 Gy x-ray each. The irradiated cells were then cultured for 24 h before proceeding with further experiments.
Co-Immunoprecipitation (Co-IP) experiments
Approximately 1 × 107 cells were lysed in 600 μl of lysis buffer (1% NP-40, 5% 1 M Tris–HCl pH 8.0, 0.15 M NaCl, and 2% protein inhibitor cocktail) and incubated at 4 °C for 20 min. Following centrifugation at 14,000 rpm for 10 min at 4 °C, the supernatants were incubated with specific primary antibodies on a rocker at 4 °C for 1–2 h. Protein G magnetic beads (20 μL) were added and incubated overnight at 4 °C, followed by three washes with lysis buffer to remove non-specific proteins. The immunoprecipitated complexes were eluted by heating with 100 μl of 2 × loading buffer at 100 °C for 5 min. Eluted proteins were analyzed by Western blotting.
Western blot analysis
Protein samples were fractionated on 10% SDS-PAGE gels, electroblotted onto PVDF membranes (Millipore), and membranes probed sequentially with respective antibodies. Blots were incubated with secondary antibodies and developed with ECL Plus (Bio-Rad) using CLINX chemiluminescence imager (ChemiScope S6). The antibody information was listed in supplementary Table S1.
Quantitative RT-PCR
Quantitative RT-PCR was performed using a ViiA7 real-time PCR system (Applied Biosystems) following a 3-step protocol. Total RNA was isolated using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme), and cDNA was synthesized with the HiScript II Q RT SuperMix (Vazyme). The PCR conditions included an initial denaturation step at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 60 °C for 40 s. Real-time PCR reactions were conducted with Taq Pro Universal SYBR qPCR Master Mix (Vazyme), and gene expression levels were normalized to Gapdh transcript levels. qRT-PCR data were analyzed by GraphPad prism. Error bars represent the standard deviation of three technical replicates from a representative experiment. Primer sequences used for qPCR analysis are detailed in supplementary Table 1.
RNA-seq and ChIP-qPCR experiments
RNA-seq and ChIP-qPCR experiments were conducted as previously described [18]. Mouse ESCs were cultured with and without 8 Gy of X-ray treatment, administered twice over 2 days, for RNA-seq and ChIP experiments.
For RNA-seq experiments, total RNA was extracted using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (Vazyme #RC112-01), and sequencing was performed by GENEWIZ. The RNA Integrity Number (RIN) values for both control and irradiation-treated samples were above 6.8. RNA-seq was conducted using 10 ng of RNA and the VAHTS Universal V10 RNA-seq Library Prep Kit for Illumina (NR606-01) on an Illumina NovaSeq platform.
For ChIP-qPCR experiments, approximately 10^7 cells were crosslinked with a 1% formaldehyde solution. Following cell lysis, chromatin was sheared by sonication using a Bioruptor Plus (Diagenode) and immunoprecipitated with 1 µg of the indicated antibody and Protein G Beads (Thermo Scientific #88847). The DNA, once de-crosslinked, was purified using a DNA Purification Kit (Qiagen #) and subsequently used for qPCR analysis, with bound regions detected using paired primers listed in supplementary Table 1.
Apoptosis assay
A total of 1X106 cells were washed with PBS then labeled with 7-AAD (BD Pharmingen) and Annexin-V (Biolegend) for 15 min at room temperature. Cells were washed with PBS and measured using a FACS Canto II flow cytometer (BD Biosciences, San Jose, CA). Analysis was performed using the Flowjo software (TreeStar, Ashland, OR).
Colony formation assay
For colony formation assays, cells were irradiated once with 4 Gy x-ray irradiation and cultured for 24 h before being dissociated with trypsin and plated in a 10 cm plate. ESCs were cultured for 7 days and stained for alkaline phosphatase using the AP staining kit (Sigma). We scored colonies with ~ 90% AP-positive cells as undifferentiated, colonies with 5% AP-positive staining cells as differentiated, and colonies of intermediate AP-positive cell number as partially differentiated.
Cell cycle assay
Cells were washed with cold PBS, fixed in 70% ethanol for 2 h. Subsequently, cells were stained with propidium iodide (PI) (50 mg/ml, Sigma). PI was visualized using a FACS Canto II flow cytometer (BD Biosciences, San Jose, CA). Analysis was carried out using the Flowjo software (TreeStar, Ashland, OR).
Immunofluorescence staining of ESCs
Cells were fixed in 4% paraformaldehyde for 10 min at room temperature, blocked, and permeabilized with 3% serum in PBS with 0.3% Triton X-100 and then incubated with the indicated antibodies at 4 °C overnight. After washing, cells were incubated with secondary antibodies (1.5 h, room temperature), and counter-stained with DAPI to detect nuclei. The antibody information was listed in supplementary Table 1. Images were captured using an Olympus IX83 microscope with a 10 × objective lens.
RNA seq data analysis
For RNA-seq data analysis, reads were aligned to the mouse genome assembly mm10.GRCm38 using HISAT2 version 2.2.1 using default parameters. Gene expression was quantified by featureCounts version 2.0.3. Differential gene expression analysis was performed via DESeq2 version 1.38.3 in R. The screening criteria for identifying differentially expressed genes were defined as Padj < 0.05 and |log2FoldChange|> = 2. Clustering and visualization of differentially expressed genes were conducted using the R package pheatmap version 1.0.12 and ComplexHeatmap version 2.14.0.
ChIP-seq data analysis
ChIP-seq data analysis was performed as previously described [18]. The published sequencing datasets used in this study were listed in supplementary Table 2.
Quantification and statistical analysis
For all qPCR experiments, one of the three or more biological repeats was presented. All qPCR data represent the mean of three technical replicates. All error bars represent standard deviation (SD). The Student’s t test (unpaired, two-sided) was used to determine the significance of changes in the qPCR using Microsoft Excel. * indicates p < 0.05, ** p < 0.01, *** < 0.001. For RNA-seq experiments, two samples of the control and irradiated cells (ESCs treated with and without irradiation) from independent experiments were selected.
Results
Irradiation impacts ESC self-renewal, resulting in elevated apoptosis rates and cell-cycle abnormalities
To evaluate the impact of X-ray irradiation on the self-renewal capacity, ESCs were first exposed to irradiation and colony formation monitored, decreased numbers of ESC colonies indicated self-renewal ability was significantly impaired (Fig. 1A; Suppl. Fig. S1A). This decline in colony formation coincided with an elevated incidence of apoptosis, as the percentage of apoptotic cells was found to be higher in irradiated ESCs (~ 10.4%) compared to non-irradiated cells (~ 3.2%) (Fig. 1B). Furthermore, the irradiation treatment induced alterations in the cell cycle, characterized by a 51.19% increase in the proportion of cells in the G2-M cell cycle phases and a decrease of 36.12% in the S phase (Fig. 1C). In addition to the diminished colony-forming potential, alkaline phosphatase (AP) staining revealed a reduction in the proportion of homogeneously stained, undifferentiated colonies in irradiated ESCs (Fig. 1D–E). Collectively, our findings underscore the detrimental effects of X-ray irradiation on ESC self-renewal, as evidenced by increased apoptosis, perturbations in the cell cycle, and compromised colony-forming ability.
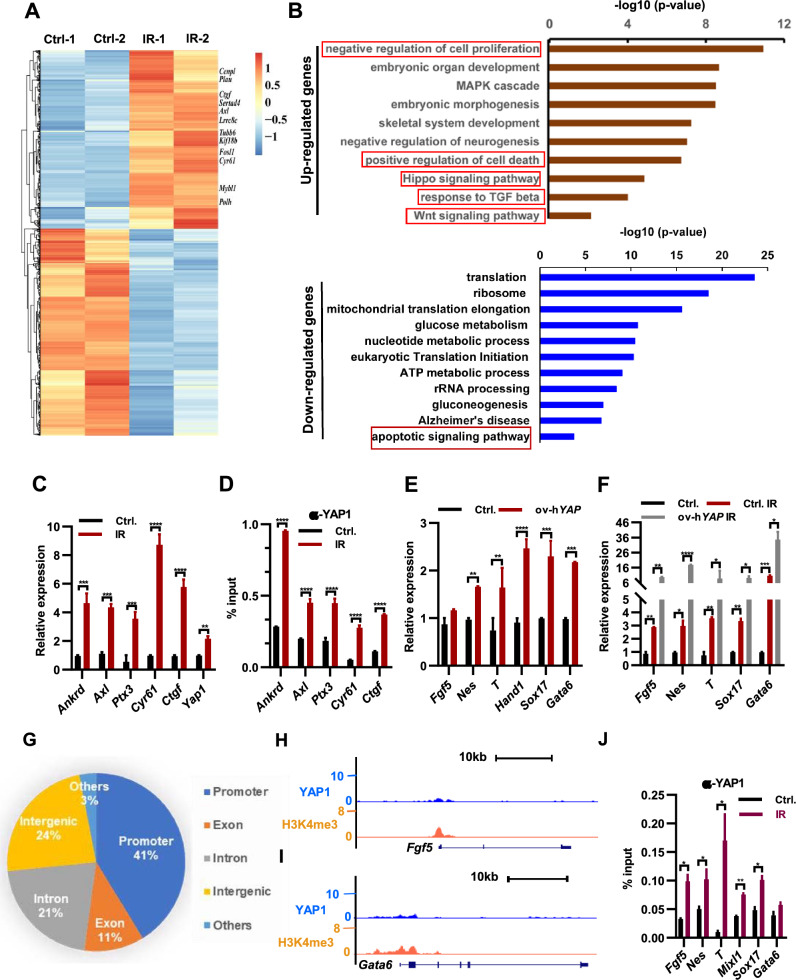
Fig. 1.
Irradiation-induced impairment of ESC self-renewal and promotion of ESC differentiation. A percentage of survived ESC colony formed upon 4 Gy x-ray irradiation treatment. Certain number of ESCs were irradiated with 4 Gy x-ray, and then plated for colony formation assay. B Percentage of ESCs at early and later apoptosis stages after irradiated with 4 Gy x-ray. C cell cycle assay of ESCs upon 4 Gy x-ray irradiation treatment. D AP staining of irradiated ESCs. The ESCs were irradiated with a dose of 4 Gy of x-rays before being plated for the single colony assay. E Percentage of undifferentiated, partially differentiated and fully differentiated colony formed of ESCs upon irradiation with 4 Gy x-ray. F–G qPCR analysis of indicated lineage marker genes (F) and pluripotency genes (G) in ESCs upon irradiated with 8 Gy x-ray twice in 48 h. H Western blot analysis of indicated proteins in ESCs upon irradiated with 8 Gy x-ray twice in 48 h. I Immunostaining analysis of E4.5 embryos with OCT4 antibody following dual 2 Gy x-ray irradiation at E2.5 and E3.5. Scale bar: 50 µm. J–K Western blot analysis of ITCH (J) and TRIM32 (K) in ESCs upon irradiated with 8 Gy x-ray twice in 48 h. L qPCR analysis of indicated lineage marker genes in control (WT) and Oct4−/− ESCs
Irradiation-induced upregulation of Itch and Trim32 promotes the differentiation of ESCs
The observed decrease in the percentage of undifferentiated colonies in irradiated ESCs serves as evidence that irradiation treatment stimulates ESC differentiation (Fig. 1E). To investigate the effect of irradiation on ESC differentiation, we assessed the expression levels of established lineage markers. The transcript levels of the endoderm markers Gata4, Gata6, and Sox17, the mesoderm marker Brachyury (T), and the ectoderm markers Fgf5 and Nes were notably upregulated in ESCs following exposure to X-ray irradiation at the specified dosages when compared to their corresponding control groups (Fig. 1F; Supple. Fig. S1B). These findings demonstrate that irradiation promotes ESC differentiation across all three lineages. Consistent with this, the expression of Rex1, a well-known marker gene for the undifferentiated state of ESCs, notably decreased in irradiated ESCs (Fig. 1G). However, qPCR analysis revealed minimal effects of irradiation on the transcript levels of core pluripotency genes Oct4, Sox2, Nanog, and Klf4 (Fig. 1G). Therefore, we examined the protein levels of OCT4, SOX2, and NANOG. In line with the induced expression of differentiation marker genes in ESCs following X-ray irradiation treatment (Fig. 1F), the protein levels of OCT4, SOX2, and NANOG significantly decreased in irradiated ESCs (Fig. 1H). Furthermore, irradiated E4.5 mouse embryos exhibited reduced OCT4 and NANOG positive cells (Fig. 1I; Supple. Fig. S1C, D). These findings indicate that irradiation promotes ESC differentiation by modulating the stability of key pluripotency proteins.
As an inhibitor of 26S proteasome, MG132 reduces the degradation of ubiquitin-conjugated proteins in ESCs [19]. The addition of MG132 reduced the degradation of OCT4, NANOG and SOX2 (Supple. Fig. S1E), indicating the induced ubiquitination of key pluripotency proteins. WWP2, ITCH, and TRIM32 are E3 ubiquitin ligases known to facilitate the degradation of OCT4 protein [20]. To investigate whether irradiation promotes OCT4 degradation by modulating the expression of Wwp2, Itch, and Trim32, we examined their expression levels in irradiated ESCs. Following irradiation, we observed upregulation of both transcript and protein levels of Itch and Trim32 genes (Supple. Figs. S1F, 1J, K), which was further confirmed by immunostaining experiments (Supple. Fig. S1G, H). However, the transcript level of the Wwp2 gene did not exhibit significant changes (Supple. Fig. S1F). Deletion of the Oct4 gene induced the expression of Gata6, Sox17, FGF5, and Nes in ZHBTc4 ESCs upon Doxycycline treatment (Fig. 1L) [21], suggesting that irradiation-induced ESC differentiation may occur through downregulation of the OCT4 protein. However, the expression of the mesoderm marker gene T, which increased following irradiation treatment (Fig. 1F), was downregulated upon Oct4 gene knockout (Fig. 1L), indicating the involvement of additional mechanisms by which irradiation treatment induces ESC differentiation. In summary, irradiation exerts a promotional effect on the differentiation of embryonic stem cells through the modulation of E3 ubiquitin ligase activity, ultimately resulting in the degradation of pluripotency proteins.
Irradiation promotes the differentiation of ESCs via upregulating the expression of Yap1
To elucidate the underlying mechanism by which irradiation regulates ESC differentiation, we performed RNA sequencing (RNA-seq) to examine global gene expression changes in irradiated ESCs. Our analysis revealed 2627 significantly downregulated genes and 2265 upregulated genes in irradiated ESCs compared to control ESCs (Fig. 2A; Supplementary Table 1). Consistent with the observed induction of apoptosis following irradiation treatment (Fig. 1B), Gene Ontology (GO) analysis demonstrated enrichment of cell death among upregulated genes in irradiated ESCs (Fig. 2B upper panel). Upregulated genes were found to be associated with cell differentiation and cell proliferation (Fig. 2B, upper panel), indicative of an increased propensity for ESC differentiation and alterations in the cell cycle dynamics (Fig. 1C–F). Down-regulated genes were associated with apoptotic signaling pathway (Fig. 2B, lower panel). The expression of genes associated with the Hippo pathway in ESCs was observed to increase under irradiation treatment (Fig. 2A), suggesting a crucial role of this pathway in promoting ESC differentiation under such conditions. Correspondingly, both mRNA and protein levels of Yap1, a transcriptional co-activator of the Hippo pathway [22], were significantly upregulated in irradiated ESCs (Fig. 2C; Supple. Fig. S2A). This upregulation of Yap1 resulted in an enhanced binding of YAP1 to its target genes Ankrd, Axl, Ptx3, Cyr61, and Ctgf (Fig. 2D), thereby leading to their upregulation in irradiated ESCs. Previous studies have demonstrated that overexpression of Yap1 in ESCs disrupts their self-renewal and triggers differentiation by upregulating the expression of lineage-specific genes [22, 23]. Hence, we hypothesized that the regulation of Yap1 expression might be involved in the irradiation-induced changes in the differentiation potential of ESCs. To test this hypothesis, we generated mouse ES cell lines with ectopic expression of either human or mouse Yap1. As expected, we observed an upregulation of the YAP1 target genes in these cells (Supple. Fig. S2B, C). Additionally, in line with a previous report [22], ectopic expression of both mouse and human Yap1 led to differentiation morphological changes in ESCs (Supple. Fig. S2D, E). The expression of core pluripotency regulators, such as Sox2, Nanog, Tbx3, Klf4, Klf5, and Klf2, significantly decreased in both human and mouse Yap1-overexpressing ESCs (Supple. Fig. S2F, G), indicating that ectopic expression of Yap1 impairs self-renewal and promotes ESC differentiation. Consistent with these findings, AP staining revealed a decrease in homogeneously stained, undifferentiated colonies in human YAP1-overexpressing ESCs (Supple. Fig. S2H). Furthermore, both human and mouse Yap1-overexpressing ESCs showed increased expression of established lineage marker genes (Fig. 2E; Supple. Fig. S2I). Interestingly, Yap1-overexpressing ESCs subjected to irradiation treatment exhibited a further decrease in the protein level of pluripotency factors OCT4, SOX2, and NANOG compared to wild-type ESCs (Supple. Fig. S2J). Additionally, the expression of lineage marker genes Fgf5, Nes, T, Sox17, and Gata6 was significantly higher in irradiated ESCs with human or mouse Yap1-overexpression than in wild-type ESCs and irradiated wild-type ESCs (Fig. 2F (human); Supple. Fig. S2K (mouse)). Collectively, these findings suggest that irradiation treatment upregulates Yap1 expression, leading to impaired self-renewal and increased differentiation of ESCs.
Fig. 2.
Irradiation enhances ESC differentiation through upregulation of Hippo signaling pathway. A Heat map depicts the changes in gene expression in mESCs irradiated with 8 Gy x-ray twice in 48 h. The color represents the Z score (row-wise) of the log2 FPKM values of the 4892 genes affected. B GO ontology analysis for biological processes associated with genes whose expression changed in ESCs upon irradiation treatment in A. C qPCR analysis of transcript levels of Yap1 and its target genes in ESCs irradiated with 8 Gy twice in 48 h. D YAP1 levels at indicated target genes in ESCs following dual 8 Gy x-ray irradiation within 48 h determined by ChIP-qPCR. E qPCR analysis of transcript levels of lineage marker genes in ESCs and ESCs overexpressing human YAP1 gene (ov-hYAP1 ESCs). F qPCR analysis of transcript levels of lineage marker genes in ESCs with or without overexpressing human YAP1 gene following dual 8 Gy x-ray irradiation within 48 h determined by ChIP-qPCR. G Binding distribution of YAP1 on the genome based on its ChIP-seq analysis. H–I Genome browser view of YAP1 and H3K4me3 at the Fgf5 (H) and Gata6 (I) loci in ESCs; (J) YAP1 levels at indicated target genes in ESCs following dual 8 Gy x-ray irradiation within 48 h determined by ChIP-qPCR
To investigate the underlying molecular mechanisms of Yap1-mediated ESC differentiation, we analyzed YAP1 chromatin immunoprecipitation sequencing (ChIP-seq) data and found that 41% of YAP binding occurred within the promoter regions of genes (Fig. 2G), indicating that Yap1 primarily regulates gene expression by controlling their promoter activities. Further analysis revealed that YAP1 binds to the promoter regions of lineage marker genes Fgf5, Gata6, T, and Nes in ESCs (Fig. 2H, I; Supple. Fig. S2L–M). ChIP-qPCR experiments revealed that irradiation treatment resulted in an increased YAP binding on the promoters of the lineage markers Fgf5, Nes, T, Mixl1, Sox17, and Gata6 (Fig. 2J), suggesting that irradiation treatment may increase Yap1 expression and upregulate the expression of lineage marker genes through enhanced binding of YAP1 to their promoter regions.
Irradiation lifts the inhibition of BAF complex on Yap and its target genes in mESCs
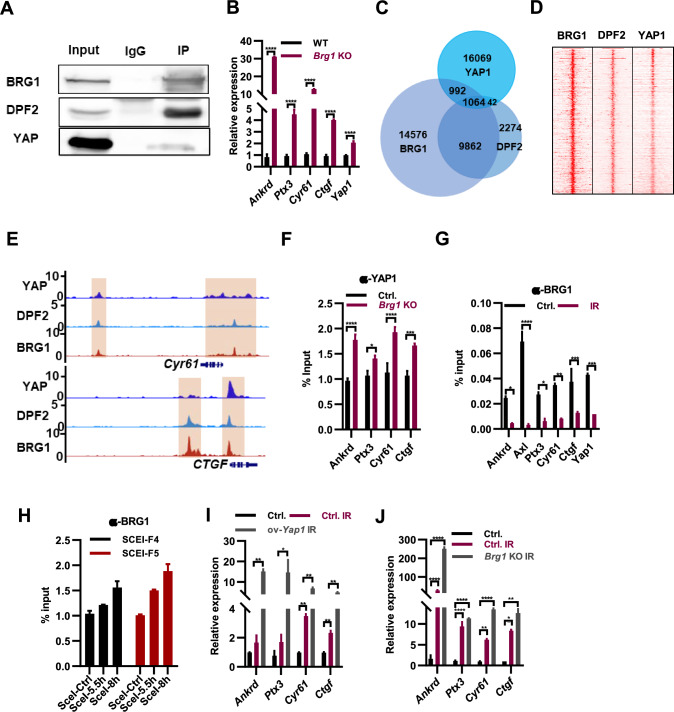
The BAF complex factor ARID1A has been shown to bind to and inhibit the oncogenic transcriptional coactivators YAP and TAZ in cancer cells [24]. We speculated whether BAF complex inhibits the transcription of Yap1 in ESCs. Co-immunoprecipitation (Co-IP) analysis demonstrated the interaction of YAP1 with BAF complex components BRG1 and DPF2 in mESCs (Fig. 3A). Deletion of Brg1, Dpf2, or Arid1a in ESCs resulted in increased expression of Yap1 and its target genes Ankrd, Ptx3, Cyr61, and Ctgf (Fig. 3B; Supple. Fig. S3A, B), indicating that the BAF complex inhibits the expression of Yap1 and its target genes in mESCs. We hypothesized that the BAF complex may prevent the binding of YAP1 to its target genes. To test this idea, we analyzed the ChIP-seq data of BRG1, DPF2, and YAP1 and identified 1,064 co-binding genomic locations (Fig. 3C, D), which included typical Yap1 target genes such as Cyr61 and CTGF (Fig. 3E). The binding of YAP1 to its target genes increased in Dpf2 and Brg1 knockout ESCs (Fig. 3F; Supple. Fig. S3C). Therefore, the BAF complex represses the expression of Yap1 target genes by preventing the binding of YAP1 to them.
Fig. 3.
Irradiation exerts control over Yap target genes by modulating the binding of the BAF complex to them. A Co-immunoprecipitation with BRG1 antibody revealed its interaction with DPF2 and YAP1. B qPCR analysis of Yap1 and its target genes in WT and Brg1−/− ESCs. C Venn diagram depicting the number of peaks that are bound by YAP1, BRG1 and DPF2 from their ChIP-seq analyses. D Heat map of the ChIP-seq signals of BRG1, DPF2 and YAP1 around their common binding sites. The color represents the log2 RPM values. E Genome browser view of YAP1, BRG1 and DPF2 at the Cyr61 and CTGF loci in ESCs. F YAP1 levels at indicated target genes in WT and Brg1.−/− ESCs determined by ChIP-qPCR. G BRG1 levels at indicated target genes in ESCs following dual 8 Gy x-ray irradiation within 48 h determined by ChIP-qPCR. H ESCs were transfected with I- Sce I plasmid. ChIP assay was conducted 5 and 8 h after I-Sce I transfection, and qPCR analyses were used to detect the enrichment of BRG1 relative to the IgG control. I qPCR analysis of YAP1 target genes in ESCs (Ctrl.), irradiated ESCs (Ctrl.IR), and irradiated ESC overexpressing Yap1 (ov-Yap1, IR). J qPCR analysis of Yap1 target genes in ESCs (Ctrl.), irradiated WT ESCs (Ctrl. IR) and Brg1 KO ESCs (Brg1 KO IR)
Irradiation treatment upregulates the expression of Yap1 and its target genes by increasing Yap1 expression (Fig. 2C, D), the molecular mechanism of which remains unknown. Considering the inhibitory role of the BAF complex in the binding of YAP1 to its targets and the regulation of their expression (Fig. 3F; Supple. Fig. S3C), we speculated that irradiation treatment may either decrease the expression of BAF complex components or alleviate the binding of the BAF complex to YAP1 target genes, thereby increasing the binding of YAP1 to its targets. Irradiation did not alter the expression of Brg1, Dpf2, and Arid1a at both mRNA and protein levels (Supple. Fig. S3D, E). Indeed, irradiation treatment reduced the binding of BRG1 to YAP1 and its target genes (Fig. 3G); concurrently, the binding of YAP1 to those locations increased (Fig. 2D). In conclusion, irradiation upregulated the expression of YAP1 target genes by reducing the binding of the BAF complex and increasing the binding of YAP1 to these targets.
Multiple studies have indicated that BAF complexes play crucial roles in facilitating the efficacy of the DNA damage response [25]. The recruitment of ARID1A to DNA double-strand breaks (DSBs) occurs through its interaction with the upstream DNA damage checkpoint kinase ATR [26]. We hypothesized that irradiation-induced DNA damage may recruit the BAF complex to DNA breaks from its binding sites associated with Yap1 and YAP1 target genes, thereby alleviating its inhibition on YAP1 binding. To investigate this, we employed a ChIP assay to examine whether BRG1 was recruited in the vicinity of a site-specific I-SceI-induced DSB [26]. The DR-GFP construct, containing a cutting site for the I-SceI restriction enzyme, was stably integrated into ESCs (Supple. Fig. S3F). ChIP-qPCR analysis revealed enrichment of BRG1 following I-SceI transfection (Fig. 3H). Thus, irradiation-induced DNA damage may recruit BRG1 away from its binding sites on Yap1 and its target genes, leading to increased binding of YAP1 and upregulation of Yap1 and its target genes. Additionally, compared to wild-type ESCs, irradiation treatment of Yap1-overexpressing ESCs further enhanced the expression of Yap1 target genes (Fig. 3I), supporting the notion that IR treatment disrupts the binding of the BAF complex and increases the binding of YAP1 to Yap1 target genes. Furthermore, consistent with these findings, irradiation treatment of Brg1, Dpf2, and Arid1a knockout ESCs resulted in further elevation of the expression of Yap1 target genes, including Ankrd, Ptx3, Cyr61, and Ctgf (Fig. 3J; Supple. Fig. S3G, H), providing further evidence that the deletion of BAF subunits enhances the accessibility of Yap1 to its targets. Therefore, we conclude that irradiation regulates the expression of Yap1 and its target genes by modulating the binding of the BAF complex to these sites.
Irradiation promotes the differentiation ESC via activating Wnt signaling pathway
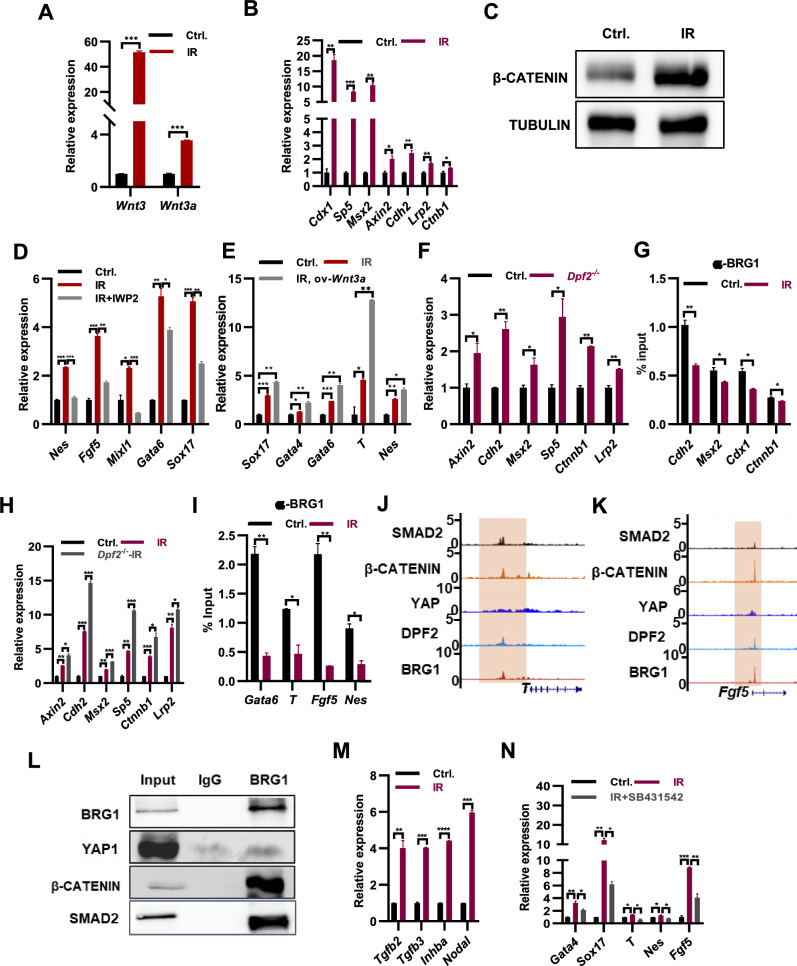
Furthermore, GO analysis of the upregulated genes showed enrichment for the Wnt signaling pathway (Fig. 2B). This observation was corroborated by qPCR analysis, which revealed elevated expression levels of Wnt3, Wnt3a, and several Wnt target genes, including Cdx1, Sp5, Msx2, Axin2, Cdh2, Lrp2, and Ctnb1, in irradiated ESCs (Fig. 4A, B). Consistently, the protein level of β-CATENIN was increased in irradiated ESCs (Fig. 4C). Collectively, these findings indicate that irradiation treatment enhances the activity of Wnt pathway in ESCs.
Fig. 4.
Irradiation treatment induces increased activity of Wnt and TGF-β signaling pathways in ESCs. A qPCR analysis of the expression levels of Wnt3 and Wnt3a transcripts in ESCs (Ctrl.), and ESCs following irradiation with 8 Gy x-ray once in 24 h and twice in 48 h (IR). B qPCR analysis of indicated Wnt target genes in ESCs irradiated with 8 Gy x-ray twice in 48 h. C Western blot revealed the protein levels of β-CATENIN in ESCs (Ctrl.) and irradiated ESCs (IR). D qPCR analysis of the typical lineage genes in ESCs (Ctrl.), irradiated ESCs (IR), and irradiated ESCs with the addition 10 μg/ml of IWP2 (IR + IWP2). E qPCR analysis of transcript levels of the typical lineage genes in WT ESCs (Ctrl.), irradiated WT ESCs (IR), and irradiated ESCs overexpressing Wnt3a (IR, ov-Wnt3a). F qPCR analysis of transcript levels of Wnt target genes in WT (Ctrl.) and Dpf2 KO ESCs. G BRG1 levels at indicated Wnt target genes in WT (Ctrl.) and irradiated ESCs (IR) determined by ChIP-qPCR. H qPCR analysis of transcript levels of Wnt target genes in WT (Ctrl.), irradiated WT (IR) and irradiated Dpf2 KO ESCs (Dpf2.−/− IR). I BRG1 levels at indicated lineage specific genes in irradiated ESCs determined by ChIP-qPCR. J–K Genome browser view of YAP1, β-CATENIN, BRG1 and DPF2 at the T (J) and Fgf5 (K) loci in ESCs. L Co-immunoprecipitation with BRG1 antibody revealed its interaction with β-CATENIN and YAP1. M qPCR analysis of transcript levels of Tgfb2, Tgfb3, Inhba and Nodal genes in WT (Ctrl.), and WT ESCs irradiated with x-ray treatment (IR). N qPCR analysis of transcript levels of lineage marker genes in WT (Ctrl.), irradiated WT ESCs (IR), and irradiated WT ESCs in the presence of the TGF-β receptor inhibitor SB431542 at a concentration of 10 μM (IR + SB431542)
The knockdown of OCT4 in human ESCs has been shown to activate the β-catenin signaling pathway and facilitate ESC differentiation [27]. Conversely, inhibition of the Wnt signaling pathway prevents the activation of mesendoderm differentiation [28]. Based on these findings, we hypothesized that increased Wnt signaling may promote ESC differentiation under irradiation treatment. Indeed, we observed enhanced binding of β-CATENIN to typical lineage marker genes in irradiated ESCs (Supple. Fig. S4A). Furthermore, the addition of the Wnt signaling inhibitor IWP2 during irradiation treatment reduced the expression of lineage marker genes Nes, Fgf5, Mixl1, Gata6, and Sox17 (Fig. 4D), highlighting the crucial role of Wnt signaling in regulating ESC differentiation under irradiation treatment. Consistently, overexpression of Wnt3a further increased the expression of lineage markers such as Gata4, Gata6, Sox17, T, and Nes in ESCs upon irradiation treatment (Fig. 4E). Overexpression of Wnt3 increased the expression of Gata6 and Sox17, but not the mesoderm gene T and ectoderm gene Fgf5 under irradiation treatment conditions (Supple. Fig. S4B). The Wnt signaling pathway has previously been reported to maintain ESC self-renewal [29]. Overexpression of Wnt3a repressed the expression of endoderm marker genes Gata4, Gata6, and Sox17, as well as the mesoderm gene T, in ESCs without irradiation treatment (Supple. Fig. S4C). This suggests that Wnt3a promotes endoderm differentiation of ESCs only after their differentiation has been initiated. To further investigate, we cultured doxycycline-inducible Wnt3a ESCs in non-LIF medium for 24 h to initiate differentiation and then induced Wnt3a overexpression upon the addition of doxycycline for 48 h. qPCR analysis revealed that overexpression of Wnt3a increased the expression of endoderm marker genes Gata4 and Gata6, as well as the mesoderm marker genes T and Mixl1, in differentiating ESCs, while the expression of the neural ectoderm marker gene Nes and the ectoderm gene Fgf5 remained unchanged (Supple. Fig. S4D). Similarly, when inducing EB formation with doxycycline-inducible Wnt3a ESCs in non-LIF medium for 24 h before inducing Wnt3a overexpression for 48 h, qPCR analysis showed that overexpression of Wnt3a increased the expression of endoderm marker genes Gata4 and Gata6, as well as the mesoderm marker gene T, in 3-day EBs, while the expression of the neural ectoderm marker gene Nes decreased and the ectoderm gene Fgf5 did not change (Supple. Fig. S4E). Thus, we conclude that irradiation promotes ESC differentiation by activating the Wnt signaling pathway.
The expression of Wnt3 and Wnt3a, as well as the target genes associated with the Wnt pathway, were found to be upregulated in ESCs following irradiation treatment (Fig. 4A, B). Furthermore, the deletion of Dpf2 or Brg1 in ESCs resulted in an increase in the expression of Wnt pathway target genes (Fig. 4F; Supple. Fig. S4F). ChIP-qPCR analysis using an antibody against BRG1 demonstrated a decrease in the binding of BRG1 to Wnt target genes upon irradiation treatment (Fig. 4G). IR treatment of Dpf2 knockout ESCs resulted in further elevation of the expression of Wnt target genes, including Axin2, Cdh2, Msx2, Sp5, Ctnnb1, and Lrp2 (Fig. 4H), providing further evidence that the deletion of BAF subunits enhances the accessibility of β-CATENIN to its targets. These findings indicate that, similar to the regulation of Yap1 and its target genes, the BAF complex represses the expression of Wnt target genes.
Both the Yap and Wnt pathways play crucial roles in regulating ESC differentiation in response to irradiation treatment (Figs. 2E, F, 4D, E). Following irradiation, there is an increase in the binding of YAP1 and β-CATENIN to typical lineage genes (Fig. 2J; Supple. Fig. S4A), while the binding of BRG1 decreases (Fig. 4I). Notably, the components of the BAF complex, BRG1, and DPF2, co-bind with YAP1 and β-CATENIN at lineage marker genes such as Sox17, T, Fgf5, and Nes (Fig. 4J, K; Supple. Fig. S4G, H). Co-immunoprecipitation experiments using a BRG1 antibody further confirm its interaction with both YAP1 and β-CATENIN (Fig. 4L). Based on these collective findings, we can conclude that irradiation treatment effectively regulates ESC differentiation through several mechanisms. It enhances the binding of YAP1 and β-CATENIN to characteristic lineage marker genes, leading to an increase in their expression. Additionally, irradiation treatment releases the inhibition of the BAF complex, further contributing to the upregulation of gene expression.
Irradiation promotes the differentiation of ESCs via activating TGF-β signaling pathway
The expression of genes associated with the TGF-β pathway in ESCs was observed to increase under irradiation treatment (Fig. 2B), suggesting a crucial role of this pathway in promoting ESC differentiation under such conditions. Correspondingly, the transcript levels of TGF-β pathway associated genes Tgfb2, Tgfb3, Inhba and Nodal were significantly upregulated in irradiated ESCs (Fig. 4M). Irradiation promoted the expression of typical marker genes of all three lineages Gata4, Sox17, T, Nes and Fgf5, which was restored upon the addition of SB431542, the inhibitor of TGF-β pathway (Fig. 4N), suggesting that irradiation promotes the differentiation of ESCs via activating TGF-β signaling pathway. Consistently, ChIP-seq analysis demonstrates the co-localization of SMAD2 with YAP1, β-CATENIN, BRG1 and DPF2 to lineage marker genes of three germ layers T, Fgf5, Sox17 and Nes (Fig. 4J–K; Supple. Fig. S4G, H). Co-IP experiments shown the interaction of SMAD2 with BRG1, YAP1 and β-CATENIN, suggesting the coordination of TGF-β, Hippo and Wnt pathways on the regulation of ESC differentiation. In line with this, the inhibition of TGF-β with SB431542 led to the restoration of irradiation-induced β-CATENIN enrichments at Sox17, Gata6, T, Fgf5, and Nes (Supple. Fig. S4I). Moreover, the elimination of Brg1 resulted in an increase in the expression of Tgfb2, Tgfb3, and Inhba (Supple. Fig. S4J), indicating the suppressive role of the BAF complex in the TGF-β signaling pathway in ESCs. Consistently, the upregulated expression of genes related to the TGF-β pathway such as Tgfb2, Tgfb3, Inhba and Nodal was observed in irradiated ESCs (Fig. 4M), accompanied by a significant reduction in the binding of BRG1 to these genes (Supple. Fig. S4K). Overall, irradiation-induced the alleviation of BAF inhibition on the TGF-β, Hippo, and Wnt signaling pathways, leading to their increased activity. The complex composed of the effector proteins from these three pathways collaboratively regulates the expression of characteristic lineage-specific genes.
Irradiation-induced expression of p53, and p63/p73 drives meso- and endo-dermal differentiation of ESCs, respectively
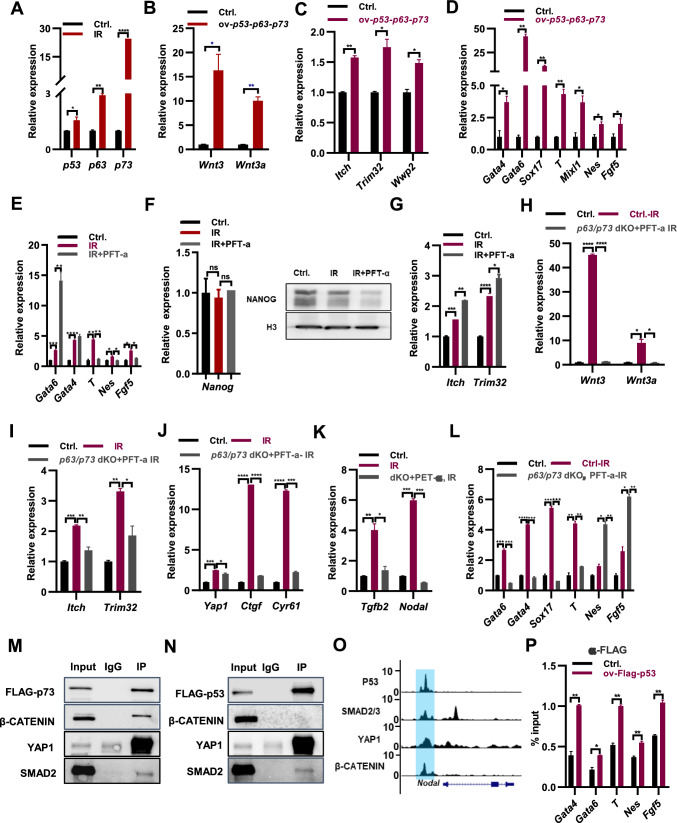
Upon DNA damage, p53, p63 and p73 induce apoptosis and alter the distribution of the cell cycle, leading to a delay in cell growth [30]. In ESCs, irradiation upregulates both the transcript and protein levels of p53, p63, and p73 (Fig. 5A; Supple. Fig. S5A), which in turn may induce apoptosis and perturb the cell cycle (Figs. 1B, C, 2B). Previous studies have identified the Wnt signaling pathway as a target of p53 in mouse ESCs [28, 31]. Furthermore, Wnt-mediated Yap/β-Catenin transcription of Oct4 in ESCs is suppressed by the core hippo scaffold RASSF1A, itself a target of ATM in response to ionizing radiation [32], and switch YAP to support p73 mediated differentiation [23]. Based on this knowledge, we hypothesized that the upregulation of p53, p63, and p73 upon irradiation treatment might similarly switch Wnt signaling pathway away from pluripotency genes and towards differentiation, which was supported by the increased binding of β-CATENIN at lineage marker genes upon the IR treatment (Supple. Fig. S4A). Supporting this notion, overexpression of p53, p63, and p73 increased the transcript levels of Wnt3 and Wnt3a and the protein level of β-CATENIN (Fig. 5B; Supple. Fig. S5B). Irradiation treatment also elevated the expression of Itch and Trim32, leading to increased degradation of OCT4 protein in ESCs (Fig. 1H, J, K; Supple. Fig. S1E). Overexpression of p53, p63, and p73 further upregulated the expression of Itch, Trim32, and Wwp2, and the protein level of ITCH (Fig. 5C; Supple. Fig. S5B). Additionally, the expression of Yap1 and Ctgf, the Tgfb2, Tgfb3, Inbha and Nodal increased in ESCs with overexpressed p53, p63, and p73 (Supple. Fig. S5C-D). Intriguingly, concurrent overexpression of p53, p63, and p73 resulted in elevated expression levels of endoderm marker genes Gata4, Gata6, and Sox17, as well as mesoderm gene T and ectoderm genes Nes and Fgf5 (Fig. 5D). This observation led us to speculate that irradiation treatment might induce ESC differentiation by modulating the expression of Wnt3/3a, Yap1, Trim32, and Itch through the control of p53, p63, and p73 expression. To investigate this, we generated p63/p73 double knockout ESCs using CRISPR/Cas9 technology. Western blot assays confirmed the successful deletion of p63 and p73 in ESCs (Supple. Fig. S5E). Deletion of p63 and p73 restored the upregulated expression of Wnt3, Wnt3a, Itch, Trim32, Yap, Ctgf, and Cyr61 in irradiated ESCs (Supple. Fig. S5F-H). Furthermore, the upregulation of endodermal markers Gata6, Gata4, and Sox17 upon irradiation treatment was restored in p63/p73 double knockout ESCs (Supple. Fig. S5I). However, deletion of p63 and p73 led to further increased expression of mesodermal gene T, as well as ectodermal marker genes Fgf5 and Nes upon irradiation treatment (Supple. Fig. S5I), potentially attributed to p53 expression. Consequently, it can be inferred that p63 and p73 promote endodermal differentiation while repressing mesodermal and ectodermal differentiation in irradiated ESCs.
Fig. 5.
Coordinated regulation of irradiation-induced ESC differentiation by P53, P63, and P73 in conjunction with Hippo, Wnt, and TGF-β signaling pathways. A qPCR analysis of transcript levels of p53, p63 and p73 in ESCs (Ctrl.), and ESCs irradiated with 8 Gy x-ray twice in 48 h (IR). B–D qPCR analysis of transcript levels of Wnt3 and Wnt3a (B), Itch, Trim32 and Wwp2 (C), and typical lineage marker genes (D) in ESCs (Ctrl.), and ESCs overexpressing p53, p63 and p73 (ov-p53/p63/p73). E qPCR analysis of transcript levels of lineage marker genes in ESCs (Ctrl.), irradiated ESCs (IR), and irradiated ESCs in the addition of 30 µM of p53 inhibitor PFT-a (IR, PFT-a). F qPCR analysis of transcript and protein levels of Nanog gene in ESCs (Ctrl.), irradiated ESCs (IR), and irradiated ESCs in the addition of 30 uM of p53 inhibitor PFT-a (IR, PFT-a). G qPCR analysis of transcript levels of Itch and Trim32 in ESCs (Ctrl.), irradiated ESCs (IR), and irradiated ESCs in the addition of 30 uM of p53 inhibitor PFT-a (IR, PFT-a). (H–K) qPCR analysis of transcript levels of Wnt3 and Wnt3a (H), Itch and Trim32 (I), Yap1, Ctgf and Cyr61 (J), TGF-β related genes (K). L qPCR analysis of transcript levels of typical lineage marker genes in ESCs (Ctrl.), irradiated ESCs (IR), and irradiated p63/p73 dKO ESCs supplemented with 30 uM of p53 inhibitor PFT-a (p63/p73 dKO, PFT-a). M Co-immunoprecipitation with FLAG antibody revealed the interaction of FLAG-p73 with β-CATENIN, YAP1 and SMAD2. N Co-immunoprecipitation with FLAG antibody revealed the interaction of FLAG-p53 with YAP1 and SMAD2. O Genome browser view of P53, SMAD2/3, YAP1, and β-CATENIN at the Nodal loci in ESCs. P ChIP-qPCR analysis of FLAG-p53 binding at lineage-specific genes in ESCs overexpressing FLAG-p53 for 48 h
To assess the role of p53 in irradiation-induced ESC differentiation, we employed a p53 inhibitor, Pifithrin-α (PFT-α), and confirmed its inhibitory effect by observing the repression of p53 target genes, including p53, Wnt3, and Wnt3a, which were upregulated in ESCs with exogenous p53 overexpression (Supple. Fig. S5J). Consistently, the inhibition of p53 restored the IR-induced expression of Wnt3, Wnt3a, and YAP1 target genes Ctgf and Cyr61 (Supple. Fig. S5K, L). Therefore, inhibition of either p53 or p63/p73 genes represses the expression of Wnt3, Wnt3a, and YAP1 target genes (Supple. Figs. S5F, H, K, L). The inhibition of p53 with PFT-α restored the upregulated expression of mesodermal markers, including T, and ectodermal markers, such as Nes and Fgf5, in ESCs following irradiation treatment (Fig. 5E). However, the inhibition of p53 further enhanced the upregulated expression of endodermal genes Gata6 and Gata4 in irradiated ESCs (Fig. 5E). Thus, distinct from p63 and p73, p53 represses endodermal differentiation while promoting mesodermal and ectodermal differentiation in irradiated ESCs (Supple. Figs. S5I; Fig. 5E). Irradiating ESCs, either alone or in the presence of the p53 inhibitor PFT-α, did not affect the mRNA levels of Nanog in ESCs (Fig. 5F, left). However, inhibition of p53 further reduced the already downregulated protein levels of NANOG in irradiated ESCs (Fig. 5F, right). Consistently, the upregulated expression of Itch and Trim32 observed upon irradiation treatment was further increased by the inhibition of p53 (Fig. 5G). Considering the repressive role of Nanog on endoderm differentiation [33], the inhibition of p53 further enhanced the upregulated expression of endodermal genes in irradiated ESCs by promoting the expression of Itch and Trim32, thereby impairing the stability of the NANOG protein.
Furthermore, we examined the effect of simultaneous inhibition of p53, p63, and p73 by supplementing PFT-a to p63/p73 double knockout ESCs. This inhibition restored the upregulated expression of Wnt3, Wnt3a, Itch, Trim32, Yap1, Ctgf, Cyr61, Tgfb2, and Nodal in irradiated ESCs (Fig. 5H–K). The deletion of p63 and p73 in ESCs supplemented with PFT-a also restored the upregulation of endodermal markers Gata6, Gata4, and Sox17, along with the mesodermal marker T, in irradiated ESCs (Fig. 5L). This demonstrates that p53 collaboratively regulates mesoendodermal differentiation in irradiated ESCs alongside p63 and p73. Moreover, the inhibition of p53, p63, and p73 in ESCs further increased the expression of ectodermal marker genes Fgf5 and Nes upon irradiation treatment (Fig. 5L), in consistent to the elevated contribution to ectopic neural protrusions observed in p53/p63/p73 triple KO chimeras [28]. The inability to restore the expression of ectodermal markers in irradiated p63/p73 double knockout ESCs with a p53 inhibitor may be attributed to the opposing regulatory effects of p53 and p63/p73 on ectodermal differentiation, with a stronger influence exerted by p63/p73 in this opposing regulation (Fig. 5L). Thus, investigating the detailed mechanisms by which p53 and p63/p73 regulate ectodermal differentiation in ESCs warrants further exploration in future studies.
P53, P63 and P73 coordinately regulate irradiation induced ESC differentiation with Hippo, Wnt and TGF-β signaling pathways
Previous report indicates that p53 and YAP can physically interact in vivo with Smad2, and cooperatively regulates the transcription of TGF-β target genes in Xenopus embryos and human cells [34, 35]. Irradiation induced the significant up-regulation of Wnt3, Wnt3a, p73, Tgfb2, Inhba and Nodal in ESCs in as early as 3 h (Supple. Fig. S5M), suggesting a mechanism by which p53 family proteins, Wnt and TGF-β pathway effector proteins may form a complex to regulate target gene expression and ESC differentiation upon irradiation treatment. To investigate the interaction of P53, P63 and P73 with effector proteins of Hippo, Wnt and TGF-β signaling pathways, we generated ESCs with Flag-tagged p53, p63 and p73 overexpressing ESCs. Co-IP with FLAG antibody demonstrated the interaction between P73 or P63 protein with β-CATENIN, YAP and SMAD2 (Fig. 5M; Supple. Fig. S5N). P53 interacts with YAP and SMAD2, but not β-CATENIN (Fig. 5N). In consistent, ChIP-seq analysis revealed the co-localization of P53, YAP1, SMAD2 and β-CATENIN at Nodal, Win3a and Trim32 loci (Fig. 5O; Supple. Fig. S5O-P). ChIP-qPCR with FLAG antibody revealed the enrichment of FLAG-p53 at Gata4, Gata6, T, Nes and Fgf5 genes in p53 overexpressing or irradiated ESCs (Fig. 5P; Supple. Fig. S5Q), indicating the directing regulation of p53 on lineage marker gene expression. In conclusion, exposure to ionizing radiation upregulates the expression of p53 family proteins, leading to enhanced activation of the Hippo, Wnt, and TGF-β signaling pathways. Consequently, these proteins may interact in a complex manner to cohesively regulate irradiation-induced ESC differentiation.
Discussion
ESCs own the capacity to self-renew indefinitely in vitro and to differentiate into every cell type of the body. Several studies have revealed the seemingly inconsistent results of irradiation on ESC maintenance and differentiation [2, 12–14, 16, 17]. Some reports indicated that irradiation did not change the expression of key pluripotency genes in ESCs and early embryos, and the survived ESCs after irradiation still retained the capacity to form all three embryonic germ layers [13–15]. But others revealed the impaired differentiation of ESCs to cardiomyocytes and neurons et al. [2, 12, 16, 17]. So far, the detailed mechanism by which irradiation affect the maintenance and differentiation is rare. This study establishes the irradiation-induced p53 family gene expression activates the Wnt, TGF-β and Hippo signaling pathways, and form a complex with the effect proteins of these pathways to collaboratively regulate the differentiation of ESCs. Notably, this study demonstrates that irradiation induced the relieves the inhibitory effect of the BAF complex on the activity of these pathways, further promotes the differentiation of ESCs. This study reveals that irradiation affects the maintenance and differentiation by a network comprising p53 family genes and key signaling pathways in ESCs.
Irradiation on pluripotency genes
The influence of irradiation on the expression of critical pluripotency genes has been previously documented [13]. Notably, the microarray analysis of ionizing irradiated hESCs in the study by Wilson et al. [13] revealed no significant alterations in the expression of OCT4, SOX2, and NANOG. Furthermore, transcript levels of Oct4 and Nanog in blastocysts subjected to low-dose X-ray irradiation remained relatively stable compared to non-irradiated embryos [15]. Similarly, the mRNA levels of Oct4, Sox2, and Nanog did not exhibit overt changes in irradiated ESCs exposed to 8 Gy doses (Fig. 1G). However, it is noteworthy that the protein levels of OCT4, SOX2, and NANOG experienced significant reductions in both irradiated mouse ESCs and early embryos (Fig. 1H, I; Supple. Fig. S1D–F).
Previous research has shown that p53 binds to the Nanog promoter and suppresses its expression following DNA damage [36]. In our study, the expression of p53, p63, and p73 was observed to be upregulated in irradiated ESCs (Fig. 5A; Supple. Fig. S5A). Intriguingly, despite these observed changes in p53 family proteins, we did not detect significant alterations in the mRNA levels of Oct4, Sox2, and Nanog in irradiated ESCs (Fig. 1G).
Furthermore, E3 ubiquitin ligases such as WWP2, ITCH, and TRIM32 are known to mediate the degradation of OCT4 protein [20]. In our investigation, both the mRNA and protein levels of Itch and Trim32 displayed upregulation in irradiated ESCs (Supple. Fig. S1H, J), which could account for the reduction in OCT4 protein levels. The precise mechanism responsible for the degradation of NANOG and SOX2 proteins, however, remains unclear. It is worth noting that Trim genes have been shown to regulate the expression of core pluripotency genes in previous studies [37]. Additionally, Mahlokozera et al. [38] reported that TRIM26 competes with WWP2 to stabilize SOX2 protein in glioblastoma stem cells. Interestingly, exposure to ionizing radiation disrupted the expression patterns of multiple Trim genes (Supplementary Table 3). Exploring the systematic role of Trim genes in influencing the stability of major pluripotency proteins presents an intriguing avenue for further investigation.
The shift of BAF complex from its repression on Wnt and Hippo pathway to participation in the irradiation indued DNA damage repair
BAF complexes play crucial roles in facilitating the DNA damage repair [25, 39]. The recruitment of ARID1A to DNA double-strand breaks (DSBs) occurs through its interaction with the upstream DNA damage checkpoint kinase ATR [26]. The phosphorylation of BRG1 is essential for DNA damage repair, as mutation of serine 721 of BRG1 results in defective double-stranded break repair [40]. In this study, we revealed the recruitment of BRG1 to a cutting site with an in vitro system in ESCs (Fig. 3H) [26]. Further, we revealed that irradiation resulted in the decreased enrichment of BRG1 on Yap1 and its target genes, and Wnt target genes (Figs. 3G, 4G), while the binding of YAP1 on the target genes increased (Fig. 2D). We further revealed that IR treatment of Dpf2 knockout ESCs resulted in further elevation of the expression of Wnt target genes (Fig. 4H). BAF complex plays an essential role on the differentiation of ESCs [41]. Co-IP and ChIP-seq analysis indicate that a complex comprising of BAF complex, YAP1 and β-CATENIN bind to the typical lineage marker gene loci (Fig. 4J, K; Supple. Fig. S4G–H). Following irradiation, BRG1 is released from lineage-specific marker genes (Fig. 4I), leading to increased enrichment of YAP1 and β-CATENIN on lineage marker genes (Fig. 2J; Supple. Fig. S4A), consequently promoting the expression of all three typical lineage marker genes (Fig. 1F). Therefore, BAF complex may participate in the regulation of irradiation-induced ESC differentiation in two ways. Frist, irradiation resulted in the released repression of BAF complex on both YAP1 and Wnt pathways, therefore promotes ESC differentiation by increasing their activities. Second, irradiation induced removal of BAF complex on lineage marker genes leads to the access of YAP1 and β-CATENIN, thereby further promotes the expression of lineage marker genes. Consistently, knockdown of Brg1 leads to the increased expression of lineage-specific marker genes in both mouse and human ESCs [42, 43]. Brg1 facilitates PcG function at classical PcG targets, reinforcing their repression in ESCs [44]. The inactivation of PcG complex results in the upregulation of lineage specific-marker genes [19, 45]. Therefore, it is possible that irradiation induced removal of BAF complex at the differentiation lineage marker genes reduces the binding of PcG complex, therefore increases the access of β-CATENIN and YAP1 and the promotes the differentiation of ESCs. It will be of interest to discern how these chromatin remodeling complex, PcG complex and other histone modifiers collaboratively with key signaling pathways to regulate the transition from pluripotency to the initiation of differentiation of ESCs under normal and irradiated conditions.
Integration of p53 family genes with Yap1, Wnt and TGF-β pathways
Wnt signaling activated by p53 family cooperate with Nodal signaling and drives mesendoderm differentiation of ESCs [28]. In consistent, we revealed that IR-induced p53 family gene expression increased the expression of Wnt3, Wnt3a, Tgfb2 and Nodal genes, which was restored upon the deletion of p53, p63 and p73 (Fig. 5H, K). The inhibition of either Wnt or TGF-β pathway with the respective inhibitor restored the promoted mesendoderm differentiation upon irradiation treatment (Fig. 4D, N). The function of Yap1 on the differentiation of ESCs was controversial [46]. Lian et al. reported that ectopic expression of YAP1 prevents ES cell differentiation in vitro and maintains stem cell phenotypes even under differentiation conditions [46]. In contrast, Chung et al. shown that overexpression of Yap1 in ES cells promotes nuclear translocation of YAP1, resulting in disruption of self-renewal and triggering differentiation by up-regulating lineage-specific genes [22]. RASSF1A is a direct target of ATM and ATR upon DNA damage [32, 35]. The activation of Yap is governed by the Hippo pathway kinases MST1/2 which require scaffolding by RASSF1A. RASSF1A-mediated Hippo activation drives pYAP away from TEAD and towards p73, switching YAP from pluripotency to differentiation [23]. It would of interest to study the mechanism by which RASSF1A collaborates with p53 family proteins and key signaling pathway effectors to regulate the self-renewal and differentiation of ESCs upon irradiation.
In this study, overexpression of either human or mouse Yap1 downregulated the expression of pluripotency genes and initiated the differentiation of ESCs (Fig. 2E, F; Supple. Fig. S2F, G, I). Further, irradiation of ESCs with Yap1 overexpression further increased the typical lineage genes (Fig. 2F; Supple. Fig. S2K). Therefore, consistent to Chung et al. [22], overexpression of Yap1 impairs the maintenance of ESCs and promotes their differentiation. Similar to Wnt and TGF-β pathways, ectopic expression of p53 family genes increased the expression of Yap1 and its target genes (Fig. 5B; Supple. Fig. S5C, D). Irradiation induced Yap1 and its target genes were restored upon the inhibition of p53 family genes (Fig. 5J). So, irradiation induced the expression of p53 family genes, thereby increased the activity of Wnt, Yap and TGF-β pathways, therefore promote the differentiation of ESCs.
Wnt activated β-catenin and Nodal activated Smad2/3 are mutually dependent for binding to and activation of key mesendoderm identity genes [28], which was extended in this study by ChIP-seq analysis and Co-IP experiments that a complex comprising of β-CATENIN, YAP1 and SMAD2 proteins collaboratively regulate the expression of typical marker genes of three germ layers (Fig. 4J, K; Supple. Fig. S4G, H). In this study, Wnt3 and Wnt3a, and TGF-β related genes were significantly upregulated as early as 3 h upon irradiation treatment (Supple. Fig. S5M). Wang et al. did not observer the interaction of p53 family members with Smad 2,3 or 4 in both ESCs and EBs, therefor they concluded that p53 acts as a determinant of nodal action in ESCs without physically contacting Smad target loci [28]. In contrast, Co-IP experiments in this study demonstrated that P53 family proteins interact with YAP1, β-CATENIN and SMAD2 (Fig. 5M, N; Supple. Fig. S5N), which might be due to the generation of FLAG-tagged p53 ESCs and the employment of FLAG antibody for Co-IP experiments. But The inhibition of p53 family genes restored the irradiation-induced expression of Wnt and TGF-β genes (Fig. 5H, K), supporting their expression induced by irradiation is p53 family genes dependent. Therefore, this study for the time reveals that p53 family proteins form a complex with effect proteins of key signaling pathway in ESCs and thereby regulate their differentiation. It would be of interest to study their coordination in mouse embryo development in vivo.
P53/p63/p73 family proteins redundantly enable mesendodermal differentiation of ESCs [28]. Consistently, the inhibition of p53 family genes restored the elevated expression of mesendoderm marker genes (Fig. 5L). But the deletion of p63/p73 only restored the irradiation induced expression of endoderm specific genes (Supple. Fig. S5I). Notably, the inhibition of p53 restored the irradiation induced expression of mesoderm gene T (Fig. 5E). A previous study shown the depletion of p53 inhibits mesoderm differentiation during Xenopus embryonic development [34]. In mouse ESCs, knockdown or knockout of p53 leads to a strong inhibition of the mesodermal master gene T [47]. Inhibition of p53 using the p53 inhibitor pifithrin-α or knockdown of p53 expression via p53 RNAi leads to a reduction in Nanog expression and impairs the self-renewal capacity of mouse embryonic stem cells (ESCs) under non-stressful conditions [48, 49]. Wdr5 rescue after its prolonged inhibition targets WDR5 to mesoderm lineage-specifying genes, stimulating differentiation toward mesoderm fates in a p53-dependent fashion [50]. It would be interesting to dissect the mechanism by which p63/p73 and p53 specifically regulate mesoderm and endoderm differentiation of ESCs, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Authors contribution
Y.Y, Z.Z., X. C., X. L., E.O.N., L.C. and W.Z. designed and directed the entire study; Y.Y., W.X., X.W., S.T., and Z.M. performed the cellular experiments and compiled the data, Y.Y., L.Y., and X.L. performed the animal experiments. Y.Y. and X.C. performed the Bioinformatics analysis; Y.Y., Z.Z., X. C., X. L., E.O.N., L.C. and W.Z. participated in writing the manuscript. All authors read and approved the final manuscript.
Funding
Ministry of Science and Technology of the People's Republic of China, 2022YFA1104300, Wensheng Zhang, National Natural Science Foundation of China, 3217060054, Wensheng Zhang, 31970812, Wensheng Zhang.
Data availability
The data supporting the findings of this study are found in the article and the supplementary material. The corresponding author will make all relevant raw data available upon reasonable request.
Declarations
Conflict of interests
There are no competing financial or nonfinancial interests regarding this work.
Ethical approval
The study received approval from the Ethics Committee of Laboratory Animal Welfare in Tongji University (Ethics code: No.TJAB03221111).
Consent for publication
Not applicable.
Consent for publication
All the authors have consented to publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ying Ye, Wenyan Xie, Xuepeng Wang are Co-first authors.
Contributor Information
Eric O’Neill, Email: eric.oneill@oncology.ox.ac.uk.
Lei Chang, Email: changlei@suda.edu.cn.
Wensheng Zhang, Email: zhangwensheng@suda.edu.cn.
References
- 1.McCollough CH, Schueler BA, Atwell TD, Braun NN, Regner DM, Brown DL, LeRoy AJ (2007) Radiation exposure and pregnancy: when should we be concerned? Radiographics 27:909–918. 10.1148/rg.274065149 [DOI] [PubMed] [Google Scholar]
- 2.Helm A, Arrizabalaga O, Pignalosa D, Schroeder IS, Durante M, Ritter S (2016) Ionizing radiation impacts on cardiac differentiation of mouse embryonic stem cells. Stem Cells Dev 25:178–188. 10.1089/scd.2015.0260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stiles J, Jernigan TL (2010) The basics of brain development. Neuropsychol Rev 20:327–348. 10.1007/s11065-010-9148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292:154–156. 10.1038/292154a0 [DOI] [PubMed] [Google Scholar]
- 5.Martin GR (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 78:7634–7638. 10.1073/pnas.78.12.7634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM (1998) Embryonic stem cell lines derived from human blastocysts. Science 282:1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 7.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, Wahl GM (1998) ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol 8:145–155. 10.1016/s0960-9822(98)70061-2 [DOI] [PubMed] [Google Scholar]
- 8.Hong Y, Stambrook PJ (2004) Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci USA 101:14443–14448. 10.1073/pnas.0401346101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J, Li J, Song Z, Qu X, Zhou P, Wu J, Ding M, Deng H (2007) Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem 282:5842–5852. 10.1074/jbc.M610464200 [DOI] [PubMed] [Google Scholar]
- 10.Momcilović O, Choi S, Varum S, Bakkenist C, Schatten G, Navara C (2009) Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells 27:1822–1835. 10.1002/stem.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solozobova V, Rolletschek A, Blattner C (2009) Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol 10:46. 10.1186/1471-2121-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellweg CE, Shinde V, Srinivasan SP, Henry M, Rotshteyn T, Baumstark-Khan C, Schmitz C, Feles S, Spitta LF, Hemmersbach R, Hescheler J, Sachinidis A (2020) Radiation response of murine embryonic stem cells. Cells 9:1650. 10.3390/cells9071650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson KD, Sun N, Huang M, Zhang WY, Lee AS, Li Z, Wang SX, Wu JC (2010) Effects of ionizing radiation on self-renewal and pluripotency of human embryonic stem cells. Cancer Res 70:5539–5548. 10.1158/0008-5472.CAN-09-4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sokolov MV, Panyutin IV, Onyshchenko MI, Panyutin IG, Neumann RD (2010) Expression of pluripotency-associated genes in the surviving fraction of cultured human embryonic stem cells is not significantly affected by ionizing radiation. Gene 455:8–15. 10.1016/j.gene.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi M, Yoshida K, Kitada K, Kizu A, Tachibana D, Fukui M, Morita T, Koyama M (2018) Low-dose irradiation of mouse embryos increases Smad-p21 pathway activity and preserves pluripotency. J Assist Reprod Genet 35:1061–1069. 10.1007/s10815-018-1156-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanu C, Loeliger BW, Panyutin IV, Maass-Moreno R, Wakim P, Pritchard WF, Neumann RD, Panyutin IG (2019) Effect of ionizing radiation from computed tomography on differentiation of human embryonic stem cells into neural precursors. Int J Mol Sci 20:3900. 10.3390/ijms20163900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeliger BW, Hanu C, Panyutin IV, Maass-Moreno R, Wakim P, Pritchard WF, Neumann RD, Panyutin IG (2020) Effect of ionizing radiation on transcriptome during neural differentiation of human embryonic stem cells. Radiat Res 193:460–470. 10.1667/RR15535.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Chronis C, Chen X, Zhang H, Spalinskas R, Pardo M, Chen L, Wu G, Zhu Z, Yu Y, Yu L, Choudhary J, Nichols J, Parast MM, Greber B, Sahlén P, Plath K (2019) The BAF and prc2 complex subunits Dpf2 and Eed antagonistically converge on Tbx3 to control ESC differentiation. Cell Stem Cell 24:138-152.e8. 10.1016/j.stem.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Xiao F, Zhang J, Sun X, Wang H, Zeng Y, Hu J, Tang F, Gu J, Zhao Y, Jin Y, Liao B (2018) Disruption of OCT4 ubiquitination increases OCT4 protein stability and ASH2L-B-Mediated H3K4 methylation promoting pluripotency acquisition. Stem Cell Rep 11:973–987. 10.1016/j.stemcr.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu Y, Zhang W (2021) The Role of E3s in regulating pluripotency of embryonic stem cells and induced pluripotent stem cells. Int J Mol Sci 22:1168. 10.3390/ijms22031168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa H, Miyazaki J, Smith AG (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24:372–376. 10.1038/74199 [DOI] [PubMed] [Google Scholar]
- 22.Chung H, Lee BK, Uprety N, Shen W, Lee J, Kim J (2016) Yap1 is dispensable for self-renewal but required for proper differentiation of mouse embryonic stem (ES) cells. EMBO Rep 17:519–529. 10.15252/embr.201540933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papaspyropoulos A, Bradley L, Thapa A, Leung CY, Toskas K, Koennig D, Pefani DE, Raso C, Grou C, Hamilton G, Vlahov N, Grawenda A, Haider S, Chauhan J, Buti L, Kanapin A, Lu X, Buffa F, Dianov G, von Kriegsheim A, Matallanas D, Samsonova A, Zernicka-Goetz M, O’Neill E (2018) RASSF1A uncouples Wnt from Hippo signalling and promotes YAP mediated differentiation via p73. Nat Commun 9:424. 10.1038/s41467-017-02786-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L, Azzolin L, Di Biagio D, Zanconato F, Battilana G, Lucon Xiccato R, Aragona M, Giulitti S, Panciera T, Gandin A, Sigismondo G, Krijgsveld J, Fassan M, Brusatin G, Cordenonsi M, Piccolo S (2018) The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 563:265–269. 10.1038/s41586-018-0658-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lans H, Marteijn JA, Vermeulen W (2012) ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet Chromatin 5:4. 10.1186/1756-8935-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, Kapoor P, Ju Z, Mo Q, IeM S, Uray IP, Wu X, Brown PH, Shen X, Mills GB, Peng G (2015) ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov 5:752–767. 10.1158/2159-8290.CD-14-0849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson KC, Adams AM, Goodson JM, McDonald CE, Potter JC, Berndt JD, Biechele TL, Taylor RJ, Moon RT (2012) Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct 4. Proc Natl Acad Sci USA 109:4485–4490. 10.1073/pnas.1118777109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Zou Y, Nowotschin S, Kim SY, Li QV, Soh CL, Su J, Zhang C, Shu W, Xi Q, Huangfu D, Hadjantonakis AK, Massagué J (2017) The p53 family coordinates Wnt and nodal inputs in mesendodermal differentiation of embryonic stem cells. Cell Stem Cell 20:70–86. 10.1016/j.stem.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H (2006) Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun 343:159–166. 10.1016/j.bbrc.2006.02.127 [DOI] [PubMed] [Google Scholar]
- 30.Fei P, El-Deiry W (2003) P53 and radiation responses. Oncogene 22:5774–5783. 10.1038/sj.onc.1206677 [DOI] [PubMed] [Google Scholar]
- 31.Lee KH, Li M, Michalowski AM, Zhang X, Liao H, Chen L, Xu Y, Wu X, Huang J (2010) A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci USA 107:69–74. 10.1073/pnas.0909734107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilton G, Yee KS, Scrace S, O’Neill E (2009) ATM regulates a RASSF1A-dependent DNA damage response. Curr Biol 19:2020–2025. 10.1016/j.cub.2009.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643–655. 10.1016/s0092-8674(03)00392-1 [DOI] [PubMed] [Google Scholar]
- 34.Cordenonsi M, Dupont S, Maretto S, Insinga A, Imbriano C, Piccolo S (2003) Links between tumor suppressors: p53 is required for TGF-beta gene responses by cooperating with Smads. Cell 113:301–314. 10.1016/s0092-8674(03)00308-8 [DOI] [PubMed] [Google Scholar]
- 35.Pefani DE, Pankova D, Abraham AG, Grawenda AM, Vlahov N, Scrace S, O’ Neill E, (2016) TGF-β Targets the Hippo pathway scaffold RASSF1A to facilitate YAP/SMAD2 nuclear translocation. Mol Cell 63:156–166 [DOI] [PubMed] [Google Scholar]
- 36.Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y (2005) p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 7:165–171. 10.1038/ncb1211 [DOI] [PubMed] [Google Scholar]
- 37.Jaworska AM, Wlodarczyk NA, Mackiewicz A, Czerwinska P (2020) The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 38:165–173. 10.1002/stem.3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahlokozera T, Patel B, Chen H, Desouza P, Qu X, Mao DD, Hafez D, Yang W, Taiwo R, Paturu M, Salehi A, Gujar AD, Dunn GP, Mosammaparast N, Petti AA, Yano H, Kim AH (2021) Competitive binding of E3 ligases TRIM26 and WWP2 controls SOX2 in glioblastoma. Nat Commun 12:6321. 10.1038/s41467-021-26653-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho PJ, Lloyd SM, Bao X (2019) Unwinding chromatin at the right places: how BAF is targeted to specific genomic locations during development. Development 146:178780. 10.1242/dev.178780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon SJ, Park JH, Park EJ, Lee SA, Lee HS, Kang SW, Kwon J (2015) ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 34:303–313. 10.1038/onc.2013.556 [DOI] [PubMed] [Google Scholar]
- 41.Ye Y, Chen X, Zhang W (2021) Mammalian SWI/SNF chromatin remodeling complexes in embryonic stem cells: regulating the balance between pluripotency and differentiation. Front Cell Dev Biol 8:626383. 10.3389/fcell.2020.626383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kidder BL, Palmer S, Knott JG (2009) SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27:317–328. 10.1634/stemcells.2008-0710 [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Li B, Li W, Ma L, Zheng D, Li L, Yang W, Chu M, Chen W, Mailman RB, Zhu J, Fan G, Archer TK, Wang Y (2014) Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Rep 3:460–474. 10.1016/j.stemcr.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR (2011) esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol 13:903–913. 10.1038/ncb2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- 46.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, Ding S, Guan KL (2010) The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev 24:1106–1118. 10.1101/gad.1903310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hadjal Y, Hadadeh O, Yazidi CE, Barruet E, Binétruy B (2013) A p38MAPK-p53 cascade regulates mesodermal differentiation and neurogenesis of embryonic stem cells. Cell Death Dis 4:e737. 10.1038/cddis.2013.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdelalim EM, Tooyama I (2012) The p53 inhibitor, pifithrin-α, suppresses self-renewal of embryonic stem cells. Biochem Biophys Res Commun 420:605–610. 10.1016/j.bbrc.2012.03.041 [DOI] [PubMed] [Google Scholar]
- 49.Abdelalim EM, Tooyama I (2014) Knockdown of p53 suppresses Nanog expression in embryonic stem cells. Biochem Biophys Res Commun 443:652–657. 10.1016/j.bbrc.2013.12.030 [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Mao F, Zhou B, Huang Y, Zou Z, denDekker AD, Xu J, Hou S, Liu J, Dou Y, Rao RC (2020) p53 integrates temporal WDR5 inputs during neuroectoderm and mesoderm differentiation of mouse embryonic stem cells. Cell Rep 30:465–4806. 10.1016/j.celrep.2019.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are found in the article and the supplementary material. The corresponding author will make all relevant raw data available upon reasonable request.