Abstract
Stevens-Johnson syndrome (SJS) and toxic epidermal necrolysis (TEN) are severe mucocutaneous disorders characterized by extensive tissue necrosis; they are often accompanied by severe ocular complications (SOC). The regulatory role of microRNAs (miRNAs) in modulating immune responses in SJS/TEN is not fully understood, particularly in relation to chronic SOC. We explored the expression profiles of specific miRNAs and their potential impact on the regulation of key innate immune genes in patients with SJS/TEN with SOC. We analyzed plasma samples from 100 patients with chronic stage SJS/TEN with SOC and 92 healthy controls to examine the expression levels of eight specific miRNAs (let-7a-5p, let-7d-3p, let-7e-5p, miR-146a-5p, miR-130a-3p, miR-151a-3p, miR-151a-5p, miR-27b-3p) using quantitative RT-PCR (RT-qPCR). In addition, we subjected mononuclear cells from 12 SJS/TEN patients and 9 controls to RT-qPCR to assess the expression of the innate immune-related genes IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2. Significant upregulation of 4 miRNAs (let-7a-5p, let-7e-5p, miR-146a-5p, and miR-27b-3p) was observed in the plasma of SJS/TEN patients; this correlated with the increased expression of TLR3, RIG-I, and MDA5. Furthermore, MDA5, IFI44L, RSAD2, CXCL10, and IFIT2 were also significantly up-regulated in the mononuclear cells from these patients, indicating a systemic modulation of immune response genes. Our findings demonstrate that specific miRNAs are up-regulated in SJS/TEN with SOC and associated with the upregulation of critical immune response genes, suggesting their involvement in the pathogenesis and persistence of SOC. These miRNAs and their target genes may serve as potential biomarkers or therapeutic targets in managing SJS/TEN with SOC.
Keywords: Stevens-Johnson syndrome, miRNAs, Plasma, Innate immunity, Severe ocular complications
Subject terms: Biomarkers, Diseases, Pathogenesis, Risk factors
Introduction
Stevens-Johnson syndrome (SJS)/toxic epidermal necrolysis (TEN) is an acute inflammatory vesiculobullous reaction affecting the mucosa of the ocular surface, oral cavity, genitals, and the skin. Severe ocular complications (SOC) are observed in about half of the SJS/TEN patients diagnosed by dermatologists and in burn units. Ophthalmologists often treat SOC not only in the acute- but also in the chronic stage1–4.
Cold medicines, such as NSAIDs and multi-ingredient cold medications, were identified as the main causative drugs for SJS/TEN with SOC1,5–7. We also reported that single nucleotide polymorphisms (SNPs) in Toll-like receptor 3 (TLR3)8–10, prostaglandin-E receptor 3 (PTGER3)11,12, and the IKZF1 gene13,14 are significantly associated with cold medicine related SJS/TEN with SOC.
Interestingly, these genes have been reported to regulate mucocutaneous inflammation, including inflammation of the ocular surface. Allergic dermatitis and conjunctivitis were significantly down-regulated in TLR3 KO mice15,16, while they were significantly up-regulated in EP3 KO mice17,18. Ikzf1 Tg mice, in which the Ik1 isoform was introduced into cells expressing keratin 5, not only spontaneously developed dermatitis, blepharoconjunctivitis, and inflammation of the paronychia and oral cavity, allergic dermatitis was also significantly up-regulated in the Ikzf1 Tg mice19. These findings suggest the role of common mechanisms in the pathogenesis of SJS/TEN with SOC and of allergic diseases such as allergic dermatitis and conjunctivitis.
Thus, abnormal innate mucosal immunity may contribute to the ocular surface inflammation seen in SJS/TEN with SOC3,7,20.
Elsewhere we reported that miR-628-3p, up-regulated in the plasma of patients in the chronic stage of SJS/TEN with SOC, negatively regulates innate immunity by suppressing pattern recognition receptors (PRRs) such as TLR3, RIG-I, and MDA-521. We also documented that let-7a-5p was significantly up-regulated in atopic dermatitis with or without severe allergic conjunctivitis, and that this miRNA positively regulates important innate immune-related genes including TLR3, RIG-I, and MDA522. It also positively regulates STAP1, IFI44L, CXCL11, TNFSF10, AIM2, RSAD2, IFITM1, CXCL10, CCL8, TRIM22, HERC5, IFI27, IFIT2, GBP4, IFIT1, TNFSF13B, and USP4122, which are reportedly down-regulated by the miR-628-3p mimic21.
Thus, because innate immune-related genes were positively regulated by let-7a-5p and negatively regulated by miR-628-3p, we suspect that the balance between let-7a-5p and miR-628-3p contributes to the development of ocular surface inflammation21,22.
MicroRNAs (miRNAs), small noncoding RNAs consisting of about 22 nucleotides in their mature form, function as endogenous regulators of many genes. They typically bind to complementary sequences in the untranslated regions of target mRNAs, inhibiting their expression. miRNAs play roles in cell and tissue functions, including immune response, apoptosis, cell differentiation, and they are involved in the pathogenesis of various human diseases23.
Compared to the controls, not only miRNA-let-7a-5p but also let-7d-3p, let-7e-5p, miR-146a-5p, miR-130a-3p, miR-151a-3p, miR-151a-5p, and miR-27b-3p are significantly upregulated in patients with severe atopic keratoconjunctivitis22.
Based on our hypotheses that abnormal innate mucosal immunity contributes to the ocular surface inflammation seen in SJS/TEN with SOC, and that common mechanisms play a role in the pathogenesis of SJS/TEN with SOC and allergic diseases, we investigated miRNA let-7a-5p, because we suspected that it positively regulates important innate immune-related genes, e.g. TLR3, RIG-I, and MDA522. We also examined let-7d-3p, let-7e-5p, miR-146a-5p, miR-130a-3p, miR-151a-3p, miR-151a-5p, and miR-27b-3p which, compared to the controls, were significantly upregulated in severe atopic keratoconjunctivitis patients22.
To investigate the function of the miRNAs significantly up-regulated in the plasma of SJS/TEN with SOC patients, we used their mimic-transfected THP-1 cells and subjected the dsRNA receptors TLR3, RIG-I, and MDA5 and innate immune-related genes, e.g. IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2 to quantitative RT-PCR.
Materials and methods
Human plasma
This study was approved by the institutional review board of Kyoto Prefectural University of Medicine. All experimental procedures were conducted in accordance with the tenets of the Declaration of Helsinki. Written informed consent was obtained from all participants after they were given a detailed explanation of the research purpose and experimental protocols.
Quantitative miRNA PCR
As previously reported21,22,24, we utilized TaqMan MicroRNA Reverse Transcription Kits (Applied Biosystems, Vilnius, Lithuania) for the reverse transcription (RT) reaction. Quantitative miRNA PCR assays were performed on a StepOne Plus instrument (Applied Biosystems) following the manufacturer’s instructions. The specific primer and probe mix included in predesigned Taqman microRNA assays were let-7a-5p, ID: 000377; let-7d-3p, ID: 001178; let-7e-5p, ID: 002406; miR-146a-5p, ID: 000468; miR-130a-3p, ID: 000454; miR-151a-3p, ID: 002254; miR-151a-5p, ID: 002642; and miR-27b-3p, ID: 000409 (Applied Biosystems). We normalized the expression of miRNA using spike-in cel-miR-39 (miR-39, ID: 000200, P/N: 4440887; Applied Biosystems).
Quantitative RT-PCR of THP-1 cells transfected with miRNA mimics
THP-1 cells were sourced from the JCRB Cell Bank (Osaka, Japan). As previously reported21,22, for transfection and RT-qPCR, THP-1 cells were cultured as recommended by the manufacturer; 2-day stimulation was with 100 ng/ml PMA (Sigma-Aldrich, Saint Louis, MO). The miRNA mimics and controls for miRNA-let-7d-3p, let-7e-5p, 146a-5p, 130a-3p, 151a-3p, 151a-5p, and 27b-3p were from Applied Biosystems. The mimics and their negative controls were mixed with Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) and added to the THP-1 cells for 24 h (80% confluence). Total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. For the RT reaction, ReverTra Ace (TOYOBO, Japan) was used. RT-qPCR assays were performed on a StepOne Plus instrument (Applied Biosystems). Primers and probes for TLR3 (Hs00152933), IFIH1 (MDA5) (Hs00223420), DDX58 (RIG-I) (Hs00204833), IFI44L (Hs00915292), TNFSF10 (Hs00921974), AIM2 (Hs00915710), RSAD2 (Hs00369813), CXCL10 (Hs00171042), TRIM22 (Hs01001179), IFI27 (Hs01086373), and IFIT2 (Hs01922738) were purchased from Applied Biosystems. Quantification data were normalized to the expression of the housekeeping gene GAPDH.
Quantitative RT-PCR of mononuclear cells
Mononuclear cells were isolated using the Lymphoprep™ (Veritas, Japan) according to the manufacturer’s instructions. Total RNA was isolated using the TRIzol™ Reagent (ThermoFisher, USA) according to the manufacturer’s instructions. For the RT reaction, ReverTra Ace (TOYOBO, Japan) was used. RT-qPCR assays were performed on a StepOne Plus instrument (Applied Biosystems).
Data analysis
Data from quantitative miRNA PCR and RT-qPCR assays were expressed as mean ± standard error (SE). The assays were evaluated using Student’s t-test performed with Microsoft Excel. We also examined the interactions between different miRNAs and their target mRNAs using miRNeta (https://www.mirnet.ca/).
Results
Quantitative miRNA PCR analysis
We investigated miRNA-let-7a-5p; it positively regulate key innate immune-related genes such as TLR3, RIG-I, and MDA5, and 7 other miRNAs, i.e. let-7d-3p, let-7e-5p, 146a-5p, 130a-3p, 151a-3p, 151a-5p, and 27b-3p that, compared to the controls, were significantly upregulated in the plasma of patients with severe atopic keratoconjunctivitis22. We performed quantitative miRNA PCR assays using plasma samples from SJS/TEN patients with SOC in the chronic stage (n = 100) and healthy controls (n = 92). All 8 miRNAs were significantly up-regulated in the plasma of SJS/TEN patients compared to the controls (Fig. 1).
Fig. 1.
Comparative analysis of miRNA expression levels between SJS/TEN with SOC patients and healthy controls. This figure displays the quantitative miRNA PCR analysis results for specific miRNAs (let-7a-5p, let-7d-3p, let-7e-5p, miR-151a-3p, miR-151a-5p, miR-146a-5p, miR-130a-3p and miR-27b-3p) in plasma samples from two distinct groups: SJS: SJS/TEN with SOC patients, control: healthy controls. Quantification was performed using miR-39 as an internal control to normalize the data. The Y-axis indicates the fold increase in specific miRNA expression levels over the control samples. Data points represent the mean ± standard error of the mean (SEM) for patients with SJS/TEN with SOC in the chronic stage (n = 100) compared to the healthy controls (n = 92). Statistical significance between the groups is denoted by asterisks: *p < 0.001, **p < 0.0001, and ***p < 0.00001, suggesting significant upregulation of miRNAs in the SJS/TEN with SOC patient group relative to the controls.
SJS/TEN with SOC: Stevens-Johnson syndrome/Toxic Epidermal Necrolysis with Severe Ocular Complications.
RT-qPCR analysis of THP-1 cells transfected with miRNA mimics
We focused on blood cells because some miRNAs were significantly up-regulated in the plasma of our patients. Our earlier RT-qPCR analysis of THP-1 cells (a monocyte cell line) transfected with the let-7a-5p miRNA mimic showed significant up-regulation of TLR3, RIG-I, and MDA5, receptors of dsRNA and critical innate immune-related genes22.
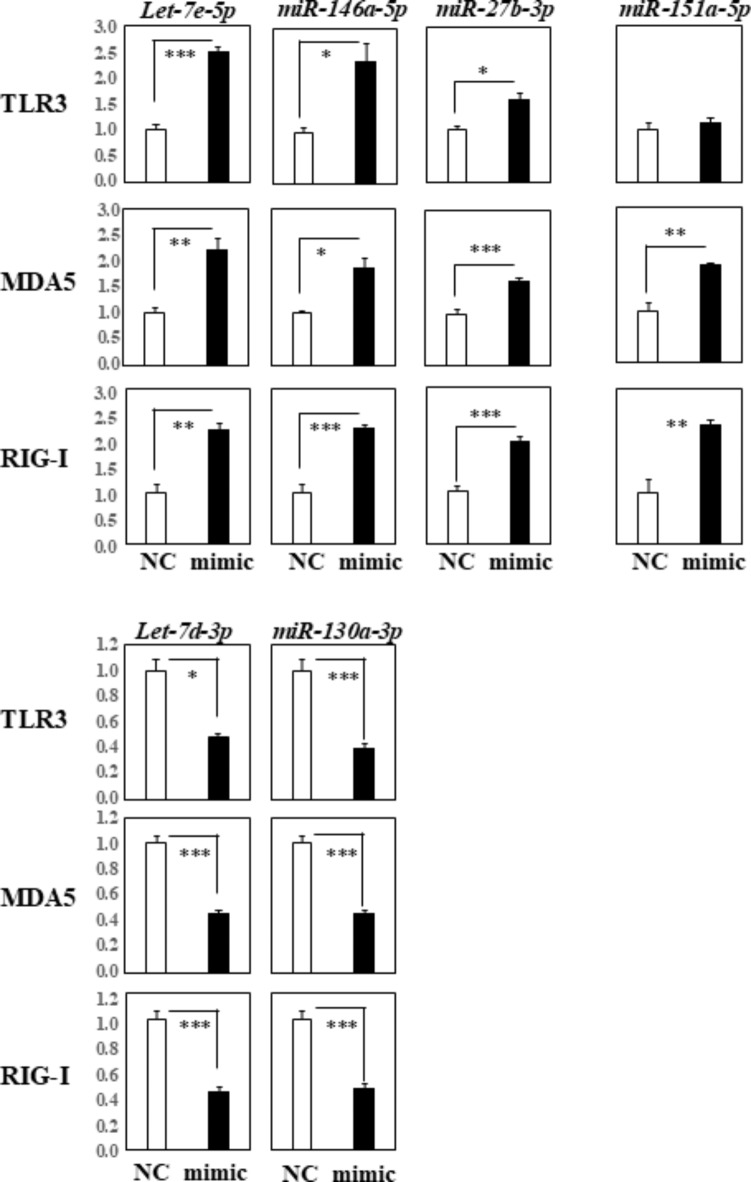
We conducted RT-qPCR analyses using THP-1 cells transfected with 7 miRNAs, i.e. let-7d-3p, let-7e-5p, 146a-5p, 130a-3p, 151a-3p, 151a-5p, and 27b-3p, to further confirm their regulatory impact on TLR3, RIG-I, and MDA5. Compared to the negative control, the expression of TLR3, MDA5, and RIG-I was significantly up-regulated in cells transfected with the let-7e-5p, miR-146a-5p, and miR-27b-3p mimics (Fig. 2, top row). The expression of MDA5 and RIG-I, but not of TLR3, was significantly increased in cells transfected with the miR-151a-5p mimic. Conversely, the expression of TLR3, MDA5, and RIG-I was significantly down-regulated in cells transfected with the let-7d-3p and miR-130a-3p mimics (Fig. 2, bottom row).
Fig. 2.
RT-qPCR Analysis of TLR3, MDA5, and RIG-I Gene Expression in THP-1 Cells Transfected with Specific miRNAs. This figure shows the results of our RT-qPCR analysis measuring the expression levels of TLR3, MDA5, and RIG-I genes in THP-1 cells following transfection with different miRNA mimics: let-7e-5p, miR-146a-5p, miR-27b-3p, miR-151a-5p, let-7d-3p, and miR-130a-3p. Quantification data were normalized to the expression of the housekeeping gene GAPDH. The Y-axis represents the fold increase in specific mRNA levels over the control samples (NC: non-targeting mimic control, mimic; each mimic of let-7e-5p, 146a-5p, 27b-3p, 151a-5p, let-7d-3p, 130a-3p miRNA). Data are presented as the mean ± standard error of the mean (SEM) from three independent experiments, with each group consisting of four replicates. Statistical significance is indicated by asterisks: *p < 0.05, **p < 0.005, ***p < 0.0005, highlighting differences in gene expression induced by each miRNA mimic compared to the control.
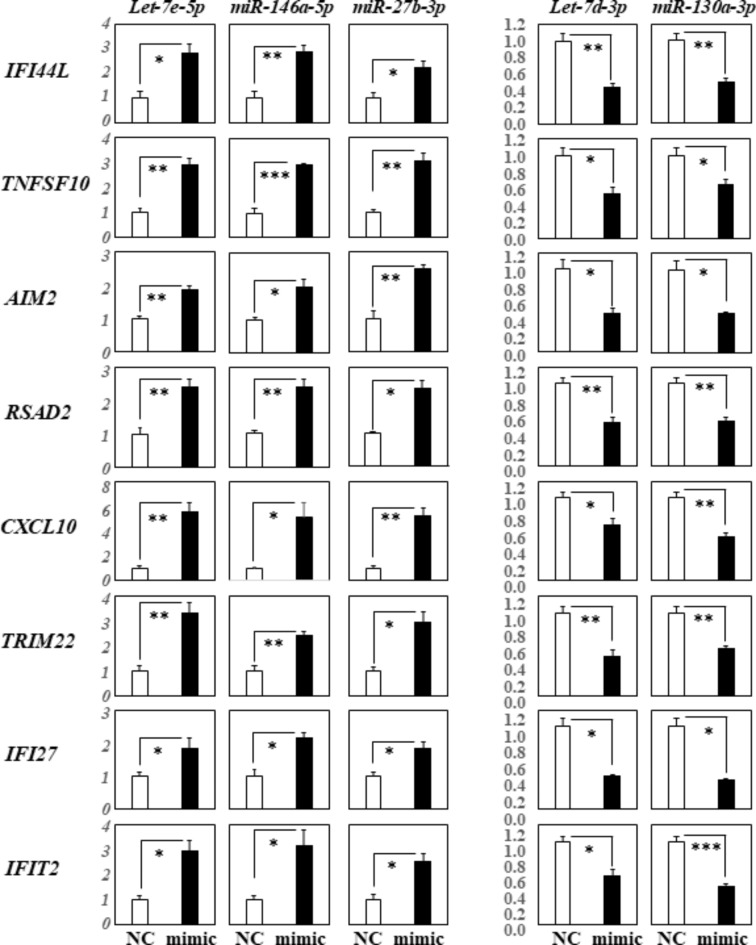
Elsewhere22, we reported that let-7a-5p could positively regulate STAP1, IFI44L, CXCL11, TNFSF10, AIM2, RSAD2, IFITM1, CXCL10, CCL8, TRIM22, HERC5, IFI27, IFIT2, GBP4, IFIT1, TNFSF13B, and USP41; genes that were negatively regulated by the miR-628-3p mimic. To study these effects further, we performed RT-qPCR analysis of the innate immune-related genes; IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2; using THP-1 cells transfected with the three miRNAs (let-7e-5p, 146a-5p, 27b-3p) that significantly up-regulated the expression of TLR3, MDA5, and of RIG-I, and THP-1 cells transfected with two miRNAs (let-7d-3p, 130a-3p) that down-regulated these receptors, respectively. We found that the expression of IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2 was significantly up-regulated in cells transfected with the let-7e-5p, miR-146a-5p, and miR-27b-3p mimics (Fig. 3, left row). Conversely, their expression was significantly down-regulated in cells transfected with the let-7d-3p and 130a-3p mimics (Fig. 3, right row).
Fig. 3.
RT-qPCR analysis of innate immune-related gene expression in THP-1 cells transfected with specific miRNA mimics. This figure displays the results of RT-qPCR analysis assessing the expression of innate immune-related genes in THP-1 cells transfected with different miRNA mimics: hsa-let-7e-5p, miR-146a-5p, miR-27b-3p, let-7d-3p, and miR-130a-3p. The quantification data were normalized to the expression of the housekeeping gene GAPDH. The Y-axis indicates the relative increase in specific mRNA levels compared to the control samples (NC: non-targeting mimic control, mimic; each mimic of let-7e-5p, 146a-5p, 27b-3p, let-7d-3p, 130a-3p miRNA). Data are presented as the mean ± standard error of the mean (SEM) for each group (n = 4), derived from three representative experiments. Statistical significance is indicated by asterisks: *p < 0.05, **p < 0.005, ***p < 0.0005, it demonstrates significant changes in gene expression induced by each miRNA mimic relative to the control.
Figure 4 summarizes the potential interactions or competitions among different miRNAs and their target mRNAs. The data shown were confirmed by quantitative RT-PCR; they are based on our experimental data using comprehensive gene expression analysis of each miRNA mimic. We also examined the interactions between different miRNAs and their target mRNAs using miRNeta (https://www.mirnet.ca/) and found interactions between mir-146a-5p and its target mRNAs, such as RSAD2, IFI27, TRIM22, IFIT1, and IFITM1, and between mir-130a-3p and IFITM1. Since the database used in miRNeta may include data from various cells and tissues, we found only a few interactions that matched the experimental data shown in Fig. 4.
Fig. 4.
Interaction and competition among miRNAs and their target mRNAs in SJS/TEN with SOC. This figure illustrates the potential interactions and competitions between various miRNAs and their target mRNAs, as identified in plasma samples from patients with SJS/TEN with SOC. The study revealed that 8 miRNAs were up-regulated in these plasma samples, in addition to the previously identified miR-628-3p. Target mRNAs were confirmed by RT-qPCR, utilizing comprehensive gene expression analysis of mimics for each of the 9 miRNAs involved. Hexagons represent the eight up-regulated miRNAs in SJS/TEN with SOC. MiRNA-151a-3p was excluded because it showed no interaction with the examined target mRNAs. Green circles indicate the 11 target mRNAs examined in the context of all 9 miRNA mimics used in this study. Gray circles represent target mRNAs identified in preliminary experiments using mimics of 5 or 6 miRNAs.
Comparison of the expression of the innate immune-related genes in mononuclear cells
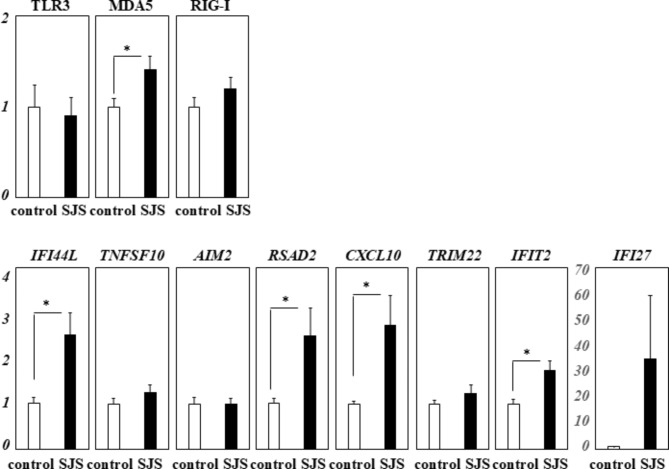
We performed RT-qPCR analysis of the innate immune-related genes; IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2; using mononuclear cells from 12 SJS/TEN patients with SOC in the chronic stage and 9 healthy controls. We found that MDA5, IFI44L, RSAD2, CXCL10, and IFIT2 were significantly up-regulated (Fig. 5).
Fig. 5.
RT-qPCR analysis of innate immune-related gene expression in mononuclear cells derived from SJS/TEN with SOC patients and controls. This figure presents the results of RT-qPCR analysis measuring the expression of innate immune-related genes in mononuclear cells from SJS/TEN patients with SOC in the chronic stage (n = 12) compared to the healthy controls (n = 9). The expression data were normalized to the housekeeping gene GAPDH. The Y-axis indicates the relative increase in specific mRNA levels over control samples. Data are expressed as the mean ± standard error of the mean (SEM). Statistical significance is denoted by an asterisk (*p < 0.05), highlighting differences in gene expression between the two groups.
Discussion
We examined 8 miRNAs, i.e. miRNA-let-7a-5p, let-7d-3p, let-7e-5p, 146a-5p, 130a-3p, 151a-3p, 151a-5p, and 27b-3p, that were significantly upregulated in patients with severe atopic keratoconjunctivitis compared to controls. These miRNAs were also found to be elevated in the plasma of patients with SJS/TEN with SOC, suggesting a common pathway of immune modulation across these inflammatory conditions.
As miRNA-let-7a-5p positively regulates key innate immune-related genes such as TLR3, RIG-I, and MDA522, we explored the potential regulatory effects of the other 7 miRNAs on these genes. RT-qPCR analysis of THP-1 cells transfected with each respective miRNA mimic revealed that let-7e-5p, miR-146a-5p, and miR-27b-3p mimics could positively regulate TLR3, RIG-I, and MDA5. Conversely, let-7d-3p and miR-130a-3p mimics appeared to negatively regulate these receptors. The miRNAs that positively affected TLR3, RIG-I, and MDA5 also enhanced the expression of other innate immune-related genes such as IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2. In contrast, let-7d-3p and miR-130a-3p, which negatively regulated TLR3, RIG-I, and MDA5, down-regulated these immune-related genes.
As shown in Fig. 4, let-7a-5p, let-7e-5p, miR-146a-5p, and miR-27b-3p up-regulated target mRNAs related to the innate immune system, e.g., TLR3, MDA5, RIG-I, IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2. On the other hand, let-7d-3p, miR-130a-3p, and miR-628-3p down-regulated these mRNAs.
The innate immune related genes MDA5, IFI44L, RSAD2, CXCL10, and IFIT2 were significantly up-regulated in the mononuclear cells from patients with SJS/TEN with SOC.
Our findings suggest that these miRNAs up-regulated in SJS/TEN with SOC, and the innate immune-related genes MDA5, IFI44L, RSAD2, CXCL10, and IFIT2 contribute to the pathogenesis of SJS/TEN with SOC.
Members of the let-7 miRNA family, including let-7a-5p, let-7d-3p, and let-7e-5p, have roles in regulating immune responses and inflammation25. Interestingly, while let-7a-5p and let-7e-5p positively regulate key innate immune receptors, let-7d-3p exhibits an opposite effect. This suggests that these miRNAs family members might contribute to the heightened immune reactivity observed in SJS/TEN with SOC.
MiR-146a-5p has been associated with the negative regulation of the NF-κB pathway and with inflammation26. MiR-27b-3p plays a significant role in the regulation of inflammation27. Both revealed a complex role in our study by upregulating immune responses, suggesting that depending on the cellular context and environmental cues, their functions include pro- and anti-inflammatory actions.
MiR-130a-3p plays a significant role in inflammation, particularly in the context of metabolism-related inflammation28. MiR-130a-3p and let-7d-3p down-regulated the expression of TLR3, RIG-I, and MDA5, while let-7a-5p and let-7e-5p up-regulated their expression. This suggests that their regulatory mechanism or mechanisms temper excessive immune responses in SJS/TEN with SOC.
The differential regulation of innate immunity-related genes by miRNAs, e.g. let-7a-5p, hsa-let-7e-5p, miR-146a-5p, and miR-27b-3p, which up-regulate TLR3, RIG-I, and MDA5, and by miR-130a-3p and let-7d-3p, which down-regulate these genes, illustrates the complexity of immune responses in SJS/TEN with SOC (Fig. 4). This intricate regulation underscores the delicate balance necessary for maintaining immune homeostasis. Disruption of this balance can lead to severe clinical manifestations, such as SJS/TEN with SOC.
Analysis of an association between the levels of each miRNA and clinical data such as the patients’ age of onset, the interval between onset and sample collection, and the grade of conjunctival invasion into the cornea revealed no significant differences. We cannot rule out the possibility that yet unknown factors play a complex role in the levels of each miRNA.
Genes such as IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2 are crucial components of the innate immune system, and their dysregulation might contribute to the severe inflammatory responses observed in SJS/TEN with SOC. The regulation of these genes by miRNAs highlights the complex genetic landscape of SJS/TEN and underscores their importance in mediating immune responses.
IFI44L (Interferon-Induced Protein 44-Like) and IFIT2 (Interferon-Induced Protein with Tetratricopeptide Repeats 2) are induced by interferons and play crucial roles in the antiviral response29,30. IFIT2 enhances the type I interferon signaling pathway, presenting an antiviral response29, while IFI44L negatively modulates innate immune responses following viral infections30. IFIT2 mediates apoptosis31. RSAD2 (Radical S-adenosyl Methionine Domain Containing 2), also known as viperin, is a key enzyme in innate immune responses, it is highly expressed in many cell types in response to viral infection and inflammatory stimuli31. The upregulation of these genes in SJS/TEN with SOC might reflect an exacerbated immune response to viral or drug-induced stress, potentially leading to widespread tissue damage.
CXCL10, a chemokine induced by interferons, plays a crucial role in the innate immune response by enhancing defence mechanisms through the recruitment and activation of natural killer cells in the central nervous system during viral infections32. Its upregulation in SJS/TEN with SOC might be essential for controlling the migration of immune cells to sites of inflammation, which can either exacerbate or mitigate the disease process.
TNFSF10 (Tumor Necrosis Factor Superfamily Member 10), also known as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), plays a significant role in the innate immune response. It involves the non-secretory apoptotic cytotoxic mechanism of natural killer- and dendritic cells33. The regulation of TRAIL suggests a mechanism by which cellular apoptosis is either promoted or inhibited.
AIM2 (Absent in Melanoma 2) is crucial in the innate immune response, particularly in recognizing cytoplasmic DNA and initiating the inflammasome complex34. It may play a vital role in triggering inflammatory responses in SJS/TEN with SOC.
TRIM22 (Tripartite Motif Containing 22) is significant in the context of viral infections35. IFI27, a type I interferon-stimulated gene, counteracted innate immune responses by interfering with RIG-I signaling; it is crucial for the antiviral defence36.
The modulation of these genes by the miRNAs we identified hints at a complex interplay between viral defence, cell survival, and apoptosis pathways that may become dysregulated in SJS/TEN with SOC.
The regulation of genes such as IFI44L, TNFSF10, AIM2, RSAD2, CXCL10, TRIM22, IFI27, and IFIT2 by miRNAs highlights the complex genetic landscape of SJS/TEN with SOC. Our findings underscore the critical role these genes play in mediating the immune responses characteristic of SJS/TEN with SOC.
Because some innate immune related genes such as MDA5, IFI44L, RSAD2, CXCL10, and IFIT2, were significantly up-regulated in the mononuclear cells of patients with SJS/TEN with SOC, the up-regulated miRNAs and those genes might strongly contribute to the pathogenesis of SJS/TEN with SOC.
An understanding of the regulation of these genes by miRNAs may yield therapeutic targets. Modulating the expression of these genes through miRNA-based therapies might help control the immune response and prevent the progression of ocular and skin complications in SJS/TEN with SOC. However, there are limitations in the sample size, particularly of mononuclear cells from these patients, and the cross-sectional nature of our study limits our ability to draw causal inferences about the role of these miRNAs in disease progression. Further studies are needed to explore the direct impact of these genes on the clinical outcomes of SJS/TEN and of their interactions with other signaling pathways.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the author(s) used ChatGPT4 in order to edit our English. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Abbreviations
- RT-qPCR
Quantitative RT-PCR
- TLR3
Toll like receptor 3
- RIG-I
Retinoic acid-inducible gene-I
- MDA5
Melanoma differentiation-associated gene 5
- IFI44L
Interferon-induced protein 44-like
- TNFSF10
Tumor necrosis factor (ligand) superfamily, member 10
- AIM2
Absent in melanoma 2
- RSAD2
Radical S-adenosyl methionine domain containing 2
- CXCL10
Chemokine (C-X-C motif) ligand 10
- TRIM22
Tripartite motif containing 22
- IFI27
Interferon, alpha-inducible protein 27
- IFIT2
Interferon-induced protein with tetratricopeptide repeats 2
Author contributions
M.U. plan this research and M.U, H.N., and H.Y. performed the experiments.M.U. wrote the main manuscript text and H.N. prepared Figs. 1, 2, 3 and 4. All authors reviewed the manuscript.
Funding
Our work was partly supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of the Japanese government, and by grants-in-aid for scientific research form the Japanese Ministry of Health, Labor, and Welfare.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sotozono, C. et al. Predictive factors associated with acute ocular involvement in Stevens-Johnson syndrome and toxic epidermal necrolysis. Am. J. Ophthalmol.160 (2), 228–237 (2015). e222. [DOI] [PubMed] [Google Scholar]
- 2.Ueta, M. Stevens-Johnson syndrome/toxic epidermal necrolysis with severe ocular complications. Expert Rev. Clin. Immunol.16 (3), 285–291 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ueta, M. Pathogenesis of Stevens-Johnson Syndrome/Toxic epidermal necrolysis with severe ocular complications. Front. Med. (Lausanne). 8, 651247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueta, M. et al. Severe ocular complications of SJS/TEN and associations among pre-onset, acute, and chronic factors: a report from the international ophthalmology collaborative group. Front. Med. (Lausanne). 10, 1189140 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ueta, M. et al. Independent strong association of HLA-A*02:06 and HLA-B*44:03 with cold medicine-related Stevens-Johnson syndrome with severe mucosal involvement. Sci. Rep.4, 4862 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueta, M. et al. Trans-ethnic study confirmed independent associations of HLA-A*02:06 and HLA-B*44:03 with cold medicine-related Stevens-Johnson syndrome with severe ocular surface complications. Sci. Rep.4, 5981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ueta, M. & Kinoshita, S. Ocular surface inflammation is regulated by innate immunity. Prog Retin Eye Res.31 (6), 551–575 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Ueta, M. et al. Toll-like receptor 3 gene polymorphisms in Japanese patients with Stevens-Johnson syndrome. Br. J. Ophthalmol.91 (7), 962–965 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueta, M. et al. Epistatic interaction between toll-like receptor 3 (TLR3) and prostaglandin E receptor 3 (PTGER3) genes. J. Allergy Clin. Immunol.129 (5), 1413–1416e1411 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Ueta, M. et al. HLA-A*0206 with TLR3 polymorphisms exerts more than additive effects in Stevens-Johnson syndrome with severe ocular surface complications. PLoS One. 7 (8), e43650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueta, M. et al. Association between prostaglandin E receptor 3 polymorphisms and Stevens-Johnson syndrome identified by means of a genome-wide association study. J. Allergy Clin. Immunol.126 (6), 1218–1225e1210 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Ueta, M. et al. HLA-A*02:06 and PTGER3 polymorphism exert additive effects in cold medicine-related Stevens-Johnson syndrome with severe ocular complications. Hum. Genome Var.2, 15023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chantaren, P. et al. Association of IKZF1 SNPs in cold medicine-related Stevens-Johnson syndrome in Thailand. Clin. Transl. Allergy. 9, 61 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueta, M. et al. IKZF1, a new susceptibility gene for cold medicine-related Stevens-Johnson syndrome/toxic epidermal necrolysis with severe mucosal involvement. J. Allergy Clin. Immunol.135 (6), 1538–1545e1517 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Nakamura, N., Tamagawa-Mineoka, R., Ueta, M., Kinoshita, S. & Katoh, N. Toll-like receptor 3 increases allergic and irritant contact dermatitis. J. Invest. Dermatol.135 (2), 411–417 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Ueta, M., Uematsu, S., Akira, S. & Kinoshita, S. Toll-like receptor 3 enhances late-phase reaction of experimental allergic conjunctivitis. J. Allergy Clin. Immunol.123 (5), 1187–1189 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Honda, T. et al. Prostaglandin E(2)-EP(3) signaling suppresses skin inflammation in murine contact hypersensitivity. J. Allergy Clin. Immunol.124 (4), 809–818e802 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Ueta, M., Matsuoka, T., Narumiya, S. & Kinoshita, S. Prostaglandin E receptor subtype EP3 in conjunctival epithelium regulates late-phase reaction of experimental allergic conjunctivitis. J. Allergy Clin. Immunol.123 (2), 466–471 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Ueta, M. et al. Mucocutaneous inflammation in the Ikaros Family Zinc Finger 1-keratin 5-specific transgenic mice. Allergy73 (2), 395–404 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Ueta, M. & Kinoshita, S. Innate immunity of the ocular surface. Brain Res. Bull.81 (2–3), 219–228 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Ueta, M. et al. Regulation of innate immune response by mir-628-3p upregulated in the plasma of Stevens-Johnson syndrome patients. Ocul Surf.21, 174–177 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Ueta, M. et al. Positive regulation of innate immune response by miRNA-let-7a-5p. Front. Genet.13, 1025539 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ardekani, A. M. & Naeini, M. M. The role of MicroRNAs in human diseases. Avicenna J. Med. Biotechnol.2 (4), 161–179 (2010). [PMC free article] [PubMed] [Google Scholar]
- 24.Ueta, M. et al. Regulation of gene expression by miRNA-455-3p, upregulated in the conjunctival epithelium of patients with Stevens-Johnson syndrome in the chronic stage. Sci. Rep.10 (1), 17239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilles, M. E. & Slack, F. J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin. Ther. Targets. 22 (11), 929–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su, Y. L. et al. Myeloid cell-targeted miR-146a mimic inhibits NF-kappaB-driven inflammation and leukemia progression in vivo. Blood135 (3), 167–180 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, J. et al. microRNA-27b shuttled by mesenchymal stem cell-derived exosomes prevents sepsis by targeting JMJD3 and downregulating NF-kappaB signaling pathway. Stem Cell. Res. Ther.12 (1), 14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng, H. et al. Regulation and mechanism of mouse miR-130a/b in metabolism-related inflammation. Int. J. Biochem. Cell. Biol.74, 72–83 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Chen, L. et al. IFITM2 presents antiviral response through enhancing type I IFN signaling pathway. Viruses 15 (4) (2023). [DOI] [PMC free article] [PubMed]
- 30.DeDiego, M. L., Martinez-Sobrido, L. & Topham, D. J. Novel functions of IFI44L as a feedback regulator of host antiviral responses. J. Virol.93 (21) (2019). [DOI] [PMC free article] [PubMed]
- 31.Stawowczyk, M., Van Scoy, S., Kumar, K. P. & Reich, N. C. The interferon stimulated gene 54 promotes apoptosis. J. Biol. Chem.286 (9), 7257–7266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trifilo, M. J. et al. CXC chemokine ligand 10 controls viral infection in the central nervous system: evidence for a role in innate immune response through recruitment and activation of natural killer cells. J. Virol.78 (2), 585–594 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vujanovic, N. L. Role of TNF superfamily ligands in innate immunity. Immunol. Res.50 (2–3), 159–174 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Man, S. M., Karki, R. & Kanneganti, T. D. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol.46 (2), 269–280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Pietro, A. et al. TRIM22 inhibits influenza a virus infection by targeting the viral nucleoprotein for degradation. J. Virol.87 (8), 4523–4533 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villamayor, L. et al. The IFN-stimulated gene IFI27 counteracts innate immune responses after viral infections by interfering with RIG-I signaling. Front. Microbiol.14, 1176177 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.