Abstract
The probiotic gut microbiome and its metabolites are pivotal in regulating host metabolism, inflammation, and immunity. Host genetics, colonization at birth, the host lifestyle, and exposure to diseases and drugs determine microbial composition. Dysbiosis and disruption of homeostasis in the beneficial microbiome have been reported to be involved in the tumorigenesis and progression of colorectal cancer (CRC). However, the influence of bacteria-secreted metabolites on CRC growth is yet to be fully elucidated. In this study, we compared the microbial composition of CRC patients to healthy controls to identify distinct patterns of microbiota-derived metabolites in CRC patients. Metagenomic analysis demonstrated that beneficial bacteria strains; Blautia producta, Bifidobacterium adolescentis, and Bifidobacterium longum decreased, while Parabacteroides distasonis and Bacteroides ovatus were more prevalent in the CRC patient group. Treatment of cancer organoid lines with microbial culture supernatants from Blautia producta, Bifidobacterium adolescentis, and Bifidobacterium longum showed remarkable inhibition of cancer growth. This study demonstrates that the bacterial metabolites depleted in CRC patients may inhibit cancer growth and highlights the effects of microbiome-derived metabolites on CRC growth.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-83048-5.
Keywords: Colorectal cancer, Microbiome, Bifidobacterium, Organoid, Metabolite
Subject terms: Biological techniques, Biotechnology, Cancer, Cell biology, Microbiology, Gastroenterology, Medical research, Oncology
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide, accounting for 10% of all new cancer cases annually1,2. Of all cancer-related deaths, 9% of males and 8% of females died of colorectal cancer. In South Korea, CRC is the third most commonly diagnosed cancer, with 27,877 new cases reported in 2020 and 8,869 deaths attributed to CRC3. In 2020, the number of prevalent CRC cases in Korea was 292,586.
Although the incidence of CRC has tended to decrease due to nationwide screening programs and increased adoption of colonoscopy, the number of patients younger than 50 years presenting with CRC has been steadily increasing in high-income countries since 1988, especially for left-sided colon and rectal cancers4.
Both environmental and hereditary risk factors play a role in the development of CRC2. Around 10–20% of CRC patients have a positive family history, with varying risks depending on the age at CRC diagnosis and the number and degree of affected relatives. Environmental risk factors include smoking, alcohol intake, consumption of red and processed meat, and increased body weight.
Certain microbial species in the colon and rectum, such as Fusobacterium nucleatum and Bacteroides fragilis, may increase the risk of CRC incidence5,6.
Selective activation of STAT3 by Enterotoxigenic Bacteroides fragilis (ETBF) leads to the infiltration of Th17 cells and promotes the development of colorectal cancer (CRC)7. ETBF also contributes to CRC development by releasing Bacteroides fragilis toxin (BFT), which cleaves e-cadherin, weakening the cell-to-cell barrier and potentially leading to chronic colonic inflammation and progression to CRC8–12. Fusobacterium nucleatumalso plays a role in CRC oncogenesis through immune modulation, such as upregulation of myeloid-derived suppressor cells and inhibitory receptors of natural killer cells, as well as the action of specific factors like miR-21, FadA, and Fap213. Additionally, pks + E. coli, which contains a 50 kb polyketide-nonribosomal peptide synthase operon (pks), produces and releases the genotoxin colibactin, causing DNA alkylation on adenine residues14,15. Long-term exposure to colibactin results in T > C transitions and the step-wise accumulation of mutations, contributing to colon carcinogenesis7.
Most cases of early-onset CRC are sporadic, with multiple proposed risk factors, including a western-style diet that can lead to gut dysbiosis, chronic inflammation, and ultimately CRC tumorigenesis4,16.
The gut microbiome, the environment closest to the colon epithelium, reflects the host’s lifestyle and external environments from birth17. Researchers have found that CRC patients’ microbial composition differs from healthy controls. They suggested that an abundant or depleted gut microbiome might play a role in CRC tumorigenesis or progression5,18–21. A meta-analysis of eight metagenomics studies identified a core set of 29 species significantly abundant in the CRC metagenome. These species include Fusobacterium, Porphyromonas, Parvimonas, Peptostreptococcus, Gemella, Prevotella, and Solobacterium22.
Although the role of gut microbiome in the initiation and progression of CRC has been extensively studied, the complete map still needs to be made clear. Certain bacterial species, such as Fusobacterium nucleatum, Peptostreptococcus anaerobius, and enterotoxigenic Bacteroides fragilis, have been reported to contribute to CRC carcinogenesis by accelerating tumor growth23, causing DNA damage24, enhancing inflammation25, and promoting immune evasion of tumors26. Conversely, some species, such as Lactobacillus and Bifidobacterium, are depleted in CRC patients, which may have a protective effect against CRC by regulating colonic inflammation27,28.
The metabolites produced by the microbiome are not only utilized within the gastrointestinal (GI) tract but also contribute to metabolic processes, chronic diseases, and host immunity29–31. In particular, the metabolites created in the colonic lumen are absorbed through the colonic mucosa and enter the bloodstream32.
The metabolome is not a product of a single microorganism but a net metabolic output of the entire gut microbiome, which may elicit tumorigenic or anti-tumorigenic signals in CRC. The major fermentation products are gases and organic acids, including short-chain fatty acids (SCFAs), particularly butyrate, acetate, and propionate.
SCFAs, butyrate, propionate, phenolic acids, and isothiocyanates are known for their anti-carcinogenic properties. The proposed underlying mechanisms involve G protein-coupled receptor-mediated signaling that promotes the differentiation of regulatory T cells and IL-10-producing T cells, blocks the activation of nuclear factor-kB (NF-kB), and induces apoptosis through histone deacetylase inhibition33–36.
In contrast, microbial fermentation products from proteins, including polyamines, hydrogen sulfide, and secondary bile acids, possess pro-carcinogenic properties32. Polyamines are essential for the maintenance of the integrity of membranes and central dogma. However, the synthesis of polyamines in gut bacteria and their accumulation are involved in oxidative stress that could result in cancer37,38. Hydrogen sulfide is produced in the GI tract. Sulfide is known to inhibit butyrate oxidation which results in the breakage of the colonocyte barrier and is genotoxic in the colonic lumen via inducing ROS (reactive oxygen stress)39–41. High-fat diets have been found to be associated with elevated fecal concentrations of secondary bile acids42. Moreover, recent studies have indicated that increased fecal bile acid concentration is observed in patients with colorectal cancer (CRC)43–45. It is suggested that CRC may be linked to alterations in the gut microbiome composition, which can be influenced by secondary bile acids. These acids are known to have significant impacts on antimicrobial functions due to their amphipathic properties, which can cause damage to bacterial cell membranes46.
However, it remains largely unknown how the metabolome of specific species, differing between the guts of CRC patients and healthy controls, affects CRC growth. In this research, we hypothesized that a beneficial microbiome, which is more prevalent in healthy individuals, produces anti-tumor metabolites. This hypothesis is based on previous studies that have shown the anti-cancer effects of cell-free supernatants from Bifidobacterium47–50. However, existing research has focused on 2D cancer cell lines rather than 3D organoid models. As a result, our goal was to demonstrate that bacterial culture supernatants obtained in vitro would not only inhibit tumor growth in 2D models but also in a 3D colorectal cancer organoid model.
Results
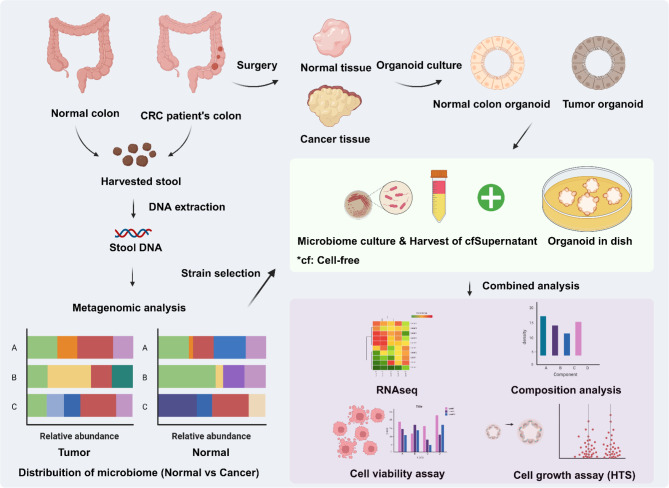
Schematic diagram
The schematic diagram illustrates the overall flow of the study (Fig. 1). We utilized a web-based graphic design platform with BioRender (https://biorender.com/) to visualize the experimental design. Before obtaining CRC tumor and colorectal epithelium biopsies, we secured the patient’s informed consent for their use in this study. The institutional review board of Seoul National University Hospital approved this study (IRB no. H-1701-110-826).
Fig. 1.
Schematic diagram of the study. The comprehensive flow chart provides visual aids of this research.
Demographics
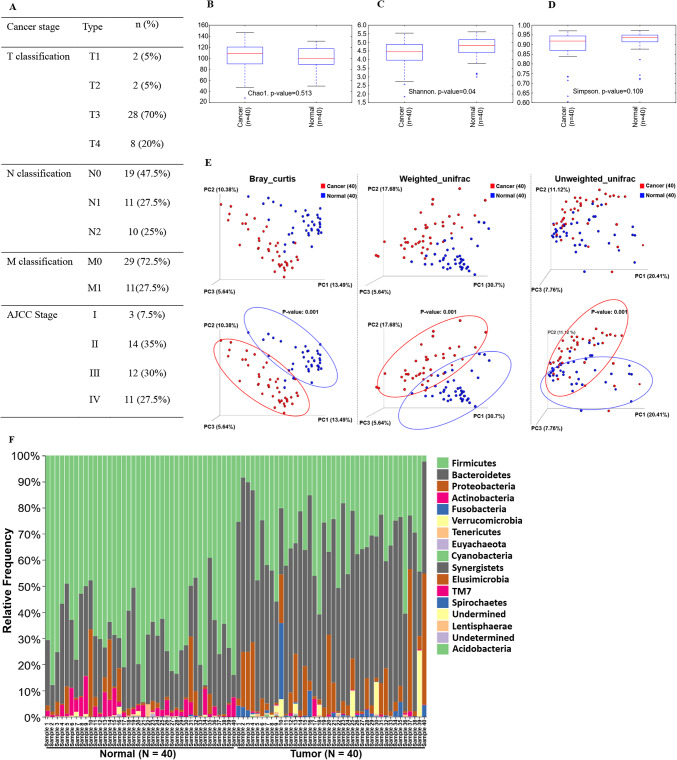
We collected 80 stool samples: 40 from healthy controls and 40 from patients diagnosed with CRC at stages I–IV. The TNM (Tumor, Node, and Metastasis) classifications and AJCC (American Joint Committee on Cancer) stages are summarized in Fig. 2A. In the control group, the average age was 60.6 ± 8.3 years with a male-to-female ratio of 25:15. In the CRC group, the average age was 58.5 ± 10.8 with a male-to-female ratio of 26:14. The most frequently distributed TNM classification was T3 (N = 28, 70%), N0 (N = 19, 47%), and M0 (N = 29, 72.5%). Additionally, 37 of 40 (92.5%) patients were diagnosed at stage 2 or higher. While the sex ratio of the donors was imbalanced, this did not impact the purpose of the study.
Fig. 2.
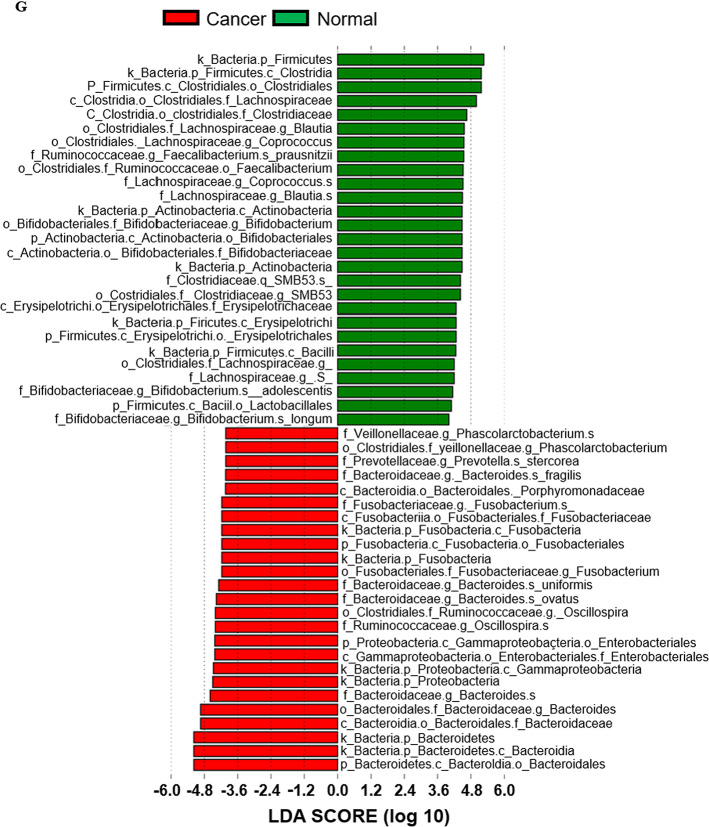
Demographics and metagenomic analysis. TNM classification and AJCC stage of colorectal cancer (n = 40) (A). Alpha (intra-sample) diversity from health controls and CRC patients was analyzed by Chao1 (B), Shannon (C), and Simpson (D) methods. Microbial taxonomic distribution data from healthy control and colorectal cancer patients were patterned by Bray-Curtis, weighted UniFrac, and unweighted UniFrac analysis (E). The microbial distribution of individual samples in the phylum level (F). The linear discriminant analysis (LDA) effect size (LEfSe) analysis. The LDA score was calculated to present the portion differences of cancer-specific strains between samples, compared with healthy control (G). Metagenomic analysis and visualization were conducted using the DADA2 package (version 1.20) in R software (version 4.1.1) and the QIIME2 plugin (version 2021.4, https://qiime2.org). Linear discriminant analysis effect size (LEfSE) (http://huttenhower.sph.harvard.edu/galaxy/) was carried out using a Galaxy computational tool.
Metagenomic analysis presents distinct bacterial distribution between healthy control and CRC patients
We first performed a diversity analysis to compare the beta diversity between healthy control and CRC patient groups. The alpha diversity showed no significant difference between the two groups (Fig. 2B–D). In the beta diversity analysis, the groups were distinctly separated and clustered by Bray Curtis, Weighted Unifrac, and Unweighted Unifrac analysis (Fig. 2E). Showing no significant differences from the alpha diversity test demonstrates that types of species have statistically low relevance between the healthy control and patient’s group. However, the beta diversity test confirmed that the patterns of each strain’s distribution between two groups are showing statistically significant differences. At the phylum level, Phylum Firmicutes was more abundant in the healthy control than in CRC patients, and Phylum Bacteroidetes was more abundant in the CRC patients (Fig. 2F). Using LEfSe (Linear Discriminant Analysis Effect Size), we identified the differential features, revealing distributions in each group by comparing their means for statistical significance (Fig. 2G). The species that were more abundant or depleted in CRC patients were listed in Table 1. Specifically, species such as Blautia obeum, Blautia producta, Faecalibacterium prausnitzii, Bifidobacterium longum, Bifidobacterium adolescentis, Lactobacillus ruminis were more abundant in the fecal samples of healthy subjects. Conversely, Bacteroides ovatus, Parabacteroides distasonis, Bacteroides uniformis, and Parabacteroides gordonii were more abundant in CRC patients.
Table 1.
Highly distributed bacterial strains in healthy controls and CRC patients.
| More abundant in healthy control | More abundant in CRC patients | ||
|---|---|---|---|
| Genus | Species | Genus | Species |
| g__Blautia | s__obeum | g__Bacteroides | s__ovatus |
| g__Blautia | s__producta | g__Parabacteroides | s__distasonis |
| g__Dorea | s__longicatena | g__Bacteroides | s__uniformis |
| g__Faecalibacterium | s__prausnitzii | g__Parabacteroides | s__gordonii |
| g__Clostridium | s__celatum | g__Morganella | s__morganii |
| g__Bifidobacterium | s__longum | g__Clostridium | s__symbiosum |
| g__Bifidobacterium | s__adolescentis | g__Bacteroides | s__fragilis |
| g__Gemmiger | s__formicilis | g__Porphyromonas | s__endodontalis |
| g__Lactobacillus | s__ruminis | g__Prevotella | s__intermedia |
| g__Luminococcus | s__callidus | g__Peptostreptococcus | s__anaerobius |
| g__Roseburia | s__faecis | g__Clostridium | s__hathewayi |
| g__Ruminococcus | s__bromii | g__Veillonella | s__parvula |
| g__Coprococcus | s__eutactus | g__Campylobacter | s__rectus |
The top 13 bacterial strains are listed by their abundance in fecal samples.
Establishment of colorectal normal and tumor organoid lines
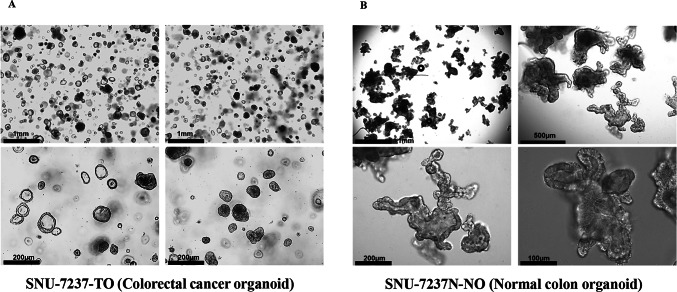
To establish organoid cultures, we isolated crypts from normal tissue and cancer epithelium of tumor tissue, respectively. Each crypt was seeded in Matrigel and passaged up to six times to establish organoid lines. Great care was taken to avoid bacterial or fungal contamination during the culture process. The organoid lines were maintained with periodic feeding and cautious subculture to optimize experimental conditions. We observed morphological differences between CRC organoids (Fig. 3A) and normal colorectal organoids (Fig. 3B); normal organoids exhibited branched crypts absent in tumor organoids. The growth of tumor organoids is not reliant on external niche components, as the Wnt signaling pathway is activated in most CRC tumors51. Related morphological patterns were also observed in a study that presents the growing patterns of established 17 colorectal matched cancer and normal organoids had been daily observed52. However, they also addressed that it is essential to conduct systematic studies that analyze and compare the growth and structure of organoids derived from normal tissue and cancer-adjacent tissue in the same patient or between different patients.
Fig. 3.
Representative bright field images of established organoid lines. Colorectal cancer organoid line (A, SNU-7237-TO) and normal colon epithelium organoid lines (B, SNU-7237N-NO).
Normal organoid lines were passaged every 4–7 days, while tumor organoid lines were passaged every 10–14 days. The identity of all organoid lines was confirmed by matching their short tandem repeat (STR) profiles with those of their parental tissue (Supplementary Table S1).
Bacterial supernatants of Bifidobacterium suppress the tumor growth in colorectal cancer organoid model
Among the differential species identified, Blautia producta, Bifidobacterium adolescentis, Bifidobacterium longum, Parabacteroides distasonis and Bacteroides ovatus were cultured, and their supernatants containing their metabolome were tested to organoids. These supernatants were then used to treat three CRC organoid lines (SNU-7237-TO, SNU-7293-TO, and SNU-7390S3-TO). Blautia producta, Bifidobacterium adolescentis, and Bifidobacterium longum were more abundant in healthy individuals, while Parabacteroides distasonis and Bacteroides ovatus were more prevalent in CRC patients (Supplementary Figure S1).
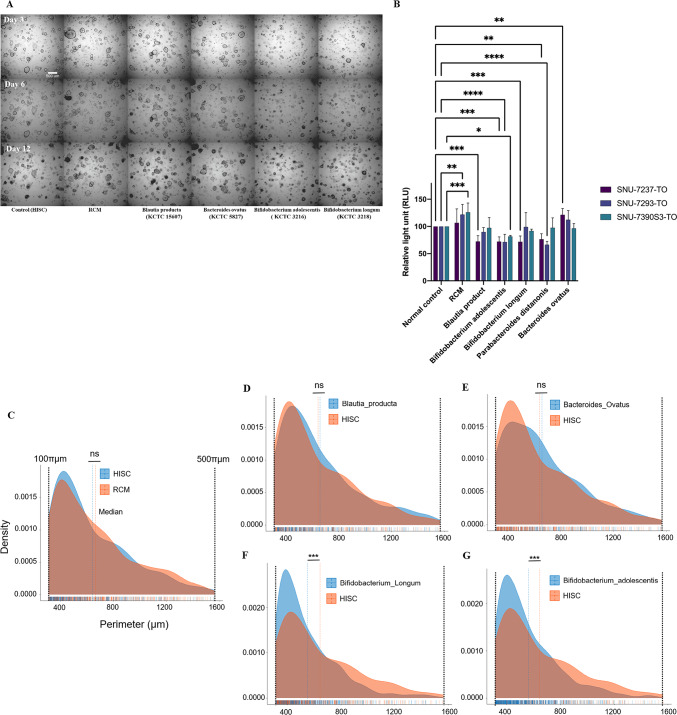
For the assay, 4,000 dissociated cells were seeded onto a 96-well plate and incubated with the organoid culture medium mixed with each of the supernatants at a concentration of 20%. Morphological changes were captured at three different time points (3, 6, and 12 days) after the treatment.
Morphological tracing data does not contain the effects of Parabacteroides distasonis to mainly emphasize the proliferative inhibition by Bifidobacterium longum and Bifidobacterium adolescentis (Fig. 4A). Recent research has confirmed that treating cell-free supernatants or directly injecting Parabacteroides distasoniscan suppress tumor growth and incidence53. It has also been suggested that pentadecanoic acid secreted from Parabacteroides distasonishas the potential to act as an HDAC6 (Histone deacetylase 6) inhibitor and suppress the stemness of stem-like cancer54. Nevertheless, the reasons behind the widespread presence of this tumor-suppressive bacterial strain in the Korean CRC patient group remain unknown.
Fig. 4.
High-throughput screening/analysis and cell viability assay. SNU-7237-TO was treated with four microbial supernatants (three strains abundant in healthy controls and one abundant in tumor patients), organoid plain medium, and bacteria plain medium. The culture state of organoids after 3, 6, and 12 days was captured with optical microscopy (A). Viable tumor (B) organoid cells after treatment of culture metabolites were visualized as a bar plot based on the relative light unit (RLU) parameter. Median distribution plots present perimeters calculated from the captured images of SNU-7237-TO treated with each bacterial supernatant, bacterial culture medium and organoid (C–G). ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
In our data, Parabacteroides distasonis also showed notable tumor-suppressive effects in aspects of cell viability. Quantified data on Parabacteroides distasonis was included in the result of the cell viability assay (Fig. 4B). Experiments were conducted in a triplicated-duplicated manner and the measurements of relative light units (RLU) were taken three days after replacing the medium with that containing microbial supernatants.
Our findings revealed promising patterns in tumor growth and proliferation suppression, noticeable as early as three days after treatment. This effect was observed across all organoids except for those treated with Bacteroides ovatus, a strain relatively enriched in the CRC fecal samples (Fig. 4A).
Each conditioned medium of the bifidobacterial strains demonstrated a distinct decrease in the viability of CRC organoids. Organoid viabilities were confirmed by RLU (Relative Light Unit) quantification, indicating a significant impact on the organoids’ survival (Fig. 4B). This result suggests that metabolites secreted from Bifidobacterium may be involved in anti-tumoric mechanism.
Additional perimeter analysis was performed employing bright field images of organoids treated with bacterial culture supernatants. Samples treated with HISC (organoid medium), RCM (bacterial medium), supernatants of Blautia producta, and Bacteroides ovatus did not show a significant difference. However, both bifidobacterial supernatants significantly decreased cell growth. Supernatants derived from Bifidobacterium adolescentis and Bifidobacterium longum have not shown adverse effects on colon normal organoid (Supplementary Figure S2 and Supplementary Table S2 and S3). The median perimeters of organoids treated with HISC, RCM, B. producta sup., B. ovatus sup., B. adolescentis sup., and B. longum sup. were 644.85 μm, 666.1 μm, 659.86 μm, 663.86 μm, 562.33 μm, and 550.78 μm, respectively (Fig. 4C–G). In four samples, the average perimeter, excluding Bifidobacterium adolescentis and longum, was 658.67 micrometers, while the average in bifidobacterial strains was 556.56 micrometers. According to this data, there was a 15.51% reduction in aspects of organoid perimeter and a 39.67% reduction in aspects of organoid volume, assuming the organoid has a perfect spherical shape. The impact of Bifidobacteriumon perimeter and volume is not as significant as that of anticancer drugs like 5-FU (5-Fluorouracil) and oxaliplatin. Analysis of anticancer treatment image data for one of the colorectal cancer organoids (SNU-4351S2-3-TO) discussed in a previously published study revealed that treatment with 5-FU resulted in a −59.91% change in area and a −41.23% change in perimeter. Similarly, treatment with oxaliplatin led to a −62.17% change in area and a −48.25% change in perimeter (Supplementary Figure S3)55. It is important to note that Bifidobacterium is a probiotic, not an anticancer agent.
Bacterial metabolome induces discriminating transcriptomic impact between colorectal normal and tumor organoids
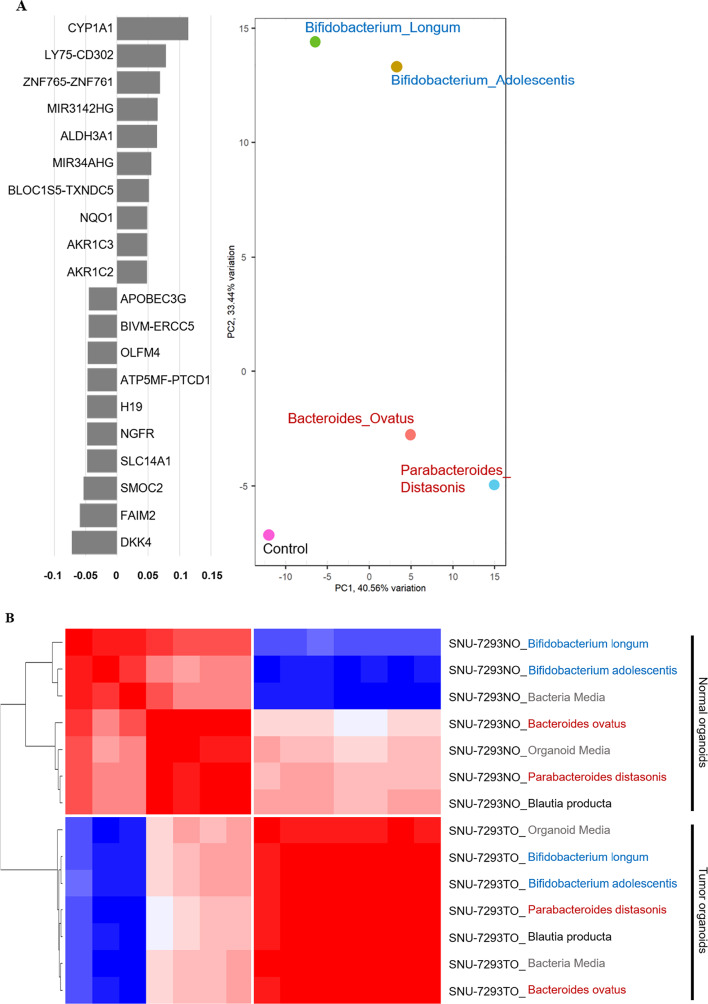
Euclidean distance analysis indicated that CRC-depleted bacteria (Bifidobacterium longum and Bifidobacterium adolescentis) and CRC-abundant bacteria (Bacteroides ovatus and Parabacteroides distasonis) were differently clustered (Fig. 5A). Principal component analysis (PCA) demonstrated that principal component 2 (PC2) primarily separated the CRC-abundant bacteria from the CRC-depleted bacteria group. We selected the top 10 genes accounting for the clear separation. CYP1A1 and DKK4 were the most upregulated genes in the respective bacteria groups (Fig. 5B). CYP1A1, a cytochrome P450 enzyme, was the most upregulated gene following treatment with CRC-depleted microbial supernatants and has been implicated in cancer prevention by detoxifying microenvironmental tumorigenesis factors56. In contrast, Dickkopf protein 4 (DKK4), which was shown at higher levels than other genes from CRC-abundant bacteria supernatants, has been demonstrated to be upregulated in CRC57.
Fig. 5.
RNA sequencing analysis. Altered transcriptomic profiles of 20 candidate genes by mock control or metabolomic supernatants were visualized as a heatmap (A). The separated Bifidobacterium group from the CRC-abundant group is shown on the PCA plot (B). Additional pathway analysis was performed to browse the transcriptomic impact of each supernatant and plotted (C). All visualizations of transcriptomic features were created using R software (version 4.1.1) and the ComplexHeatmap package (version 2.21.0).
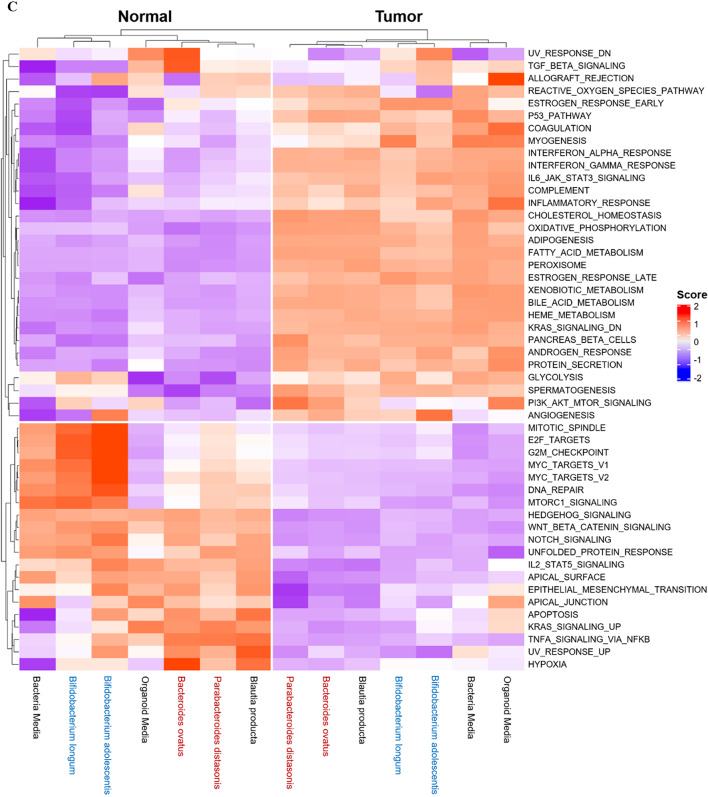
To determine which pathways were mostly affected by the bacterial metabolome, we performed enrichment score analysis (Fig. 5C). Bacteria media was used as a mock control, as it affected the transcriptomic patterns of both normal and tumor organoids. CRC-abundant bacteria group up-regulated multiple pathways, including TGF-β and reactive oxygen species pathways in normal colon organoids. CRC-depleted bacteria group specifically down-regulated reactive oxygen species pathways and up-regulated PIK3_AKT_mTOR signaling pathways among various cancer hallmark pathways.
Component analysis revealed distinct metabolomic variation between cancer and normal subjects
We conducted a component analysis to identify the composition of metabolites in bacterial cell-free supernatant. We divided the supernatant groups into two categories: cancer subjects (Parabacteroides distasonis and Bacteroides ovatus) and normal subjects (Bifidobacterium adolescentis and Bifidobacterium longum). Each group was normalized by referring to the microbial culture medium (RCM).
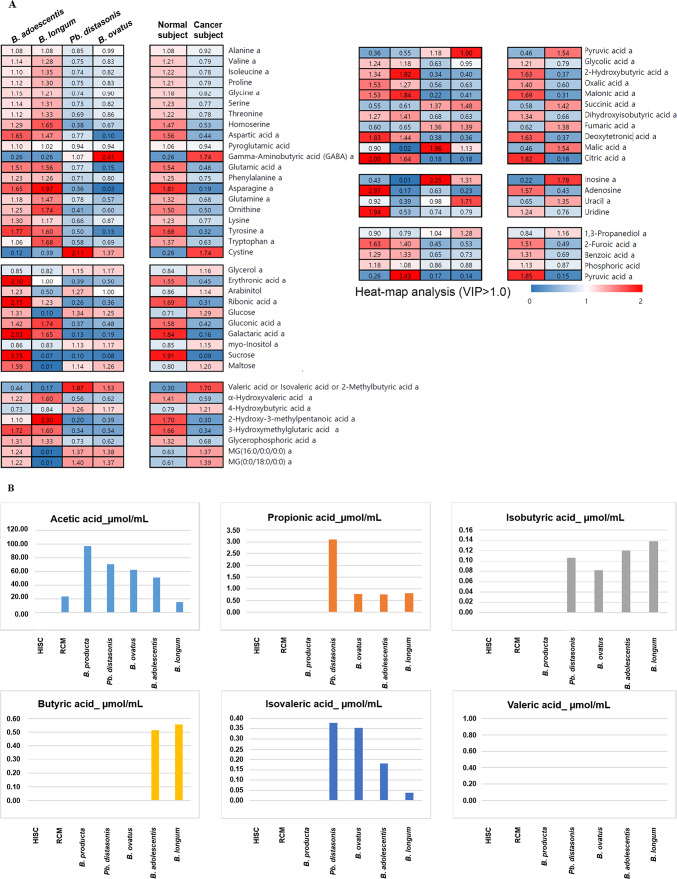
The significance of our findings is underscored by the higher proportion of amino acids such as asparagine, tyrosine, and aspartic acid in the normal subject group and the exclusive abundance of gamma-aminobutyric acid (GABA) and cystine in the cancer subject group compared to the normal group (Fig. 6A).
Fig. 6.
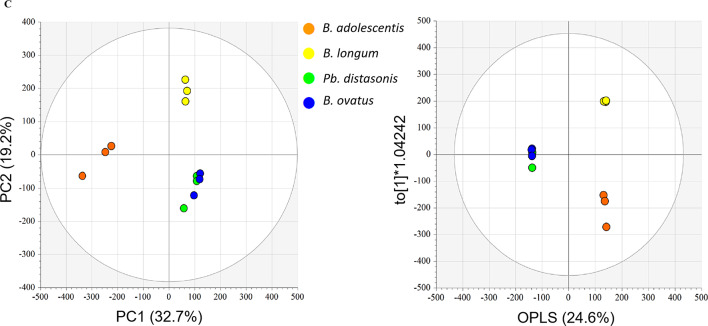
Component analysis. Relative portions of each subtype (‘amino acids,’ ‘carbohydrates,’ ‘fatty acids and lipids,’ ‘organic acids,’ ‘purines, and pyrimidines,’ and ‘etc.’) were calculated through data normalization (A). The concentration of six short-chain fatty acids in each subjected sample (B). Metabolomic distances between normal and cancer subjects were performed for PCA (left) and OPLS-DA (right) PCA; R2X (cum): 0.657, Q2 (cum): 0.301, and OPLS-DA; R2X (cum): 0.608, R2Y (cum): 1, Q2 (cum): 0.938, and ANOVA p-value < 0.05 (C). Charts (A) and (B) were generated and summarized using Microsoft Excel based on raw data. Multivariate statistical analysis (C) was performed using SIMCA P+ (version 16.0, Umetrics, Umea, Sweden).
Our SCFAs analysis revealed some intriguing findings. For instance, butyric acid was detected only in bacterial supernatants from Bifidobacterium adolescentis and Bifidobacterium longum, known for their beneficial function as a metabolic energy source for colon epithelium58 and as HDAC inhibitors linked to tumorigenesis suppression in colorectal cancer59. On the other hand, a higher concentration of acetic acid was observed in cancer subjects, and the level of propionic acid was dramatically higher in Parabacteroides distasonis samples compared to other samples. These results support the notion that Parabacteroides distasonis and Bacteroides ovatusmay produce and secrete tumorigenic metabolites through their physiological activity60.
The culture supernatant from both the cancer and normal groups contains isobutyric acid, which is found in the following microorganisms: Parabacteroides distasonis (0.11 µmol/mL), Bacteroides ovatus (0.08 µmol/mL), Bifidobacterium adolescentis (0.12 µmol/mL), and Bifidobacterium longum (0.14 µmol/mL). While previous studies have suggested that isobutyric acid may have anti-tumor effects61,62, an analysis of the concentrations in the samples collected for this study indicates that there are only slight differences in the absolute concentrations. This makes it difficult to objectively assess the impact of this metabolite based on the available data.
Levels in acetic and propionic acid, cancer subjects showed higher concentration than normal groups. Some studies observed anti-cancer effects when acetic and propionic acid were treated63,64. However, it was also reported that circulating plasma concentration of acetic and propionic acids are associated with an elevated cancer risk in the Czech cohort65. As previously mentioned, metabolites produced in GI tract enters the bloodstream, and they play pivotal roles in homeostasis66. However, Further research is essential to elucidate the specific mechanisms by which microbiome-derived metabolites exert their anticancer effects.
Valeric acid was not detected in our samples while isovaleric acid was detected and higher in the cancer group. Isovaleric acid is known to have anti-tumor potential and is associated with decreased cancer risk but also not known how it is involved in that mechanism65,67.
After conducting GC-TOF-MS analysis, we performed multivariate statistical analysis using raw data (Fig. 6C). PCA distinguished normal subjects from cancer subjects, revealing group variations (Left panel). OPLS-DA (Orthogonal Partial Least Squared Discriminant Analysis) was also conducted to confirm and maximize the difference between the normal and cancer subjects.
Discussion
The intake of beneficial bacteria in the gut has been shown to prevent CRC by improving both the quantitative and qualitative composition of the gut microbiome58–60. Extracts of Bifidobacterium adolescentishave been found to inhibit the growth of CRC cell lines by reducing β-glucuronidase, tryptophanase, and urease activity68. Furthermore, the ingestion of Bifidobacterium longumas probiotics by CRC patients has been observed to alter the composition of the gut microbiome towards increased richness and diversity, resulting in enhanced tight junction integrity and reduced cell permeability, which is crucial in preventing CRC69. In our study, we validated the efficacy of Bifidobacterium supernatant using the 3D CRC organoid model.
The microbiome has garnered significant attention over the last decade due to its roles in developing several in vivo systems, including immunity, gut epithelium, and brain functions70–74. Our data suggest that metabolites secreted by commensal bacteria play a crucial role in the anti-cancer effects, leading to cell death and growth arrest in CRC organoid models. Butyrate, one of the short-chain fatty acids (SCFAs), is a primary energy source for colonocytes and contributes to intestinal robustness through its anti-inflammatory function70. Butyrate is produced in the gut by beneficial bacteria that possess genes coding for butyryl-CoA dehydrogenase, butyryl-CoA transferase, and butyrate kinase75,76.
In our data, we observed high enrichment of butyric acid in both Bifidobacterium adolescentis and Bifidobacterium longum, which are known to be beneficial microbiomes (Fig. 6B). Our principal component analysis (PCA) revealed that the expression of the CYP1A1 gene was the most significant contributing factor in these beneficial microbiomes as well (Fig. 5B). Previous studies have shown that butyrate, the salt and ester of butyric acid, can alter the expression of CYP1A1 through histone deacetylase activity in colon epithelial cell models77. This research confirmed that the presence of butyric acid in Bifidobacterium adolescentis and Bifidobacterium longum is linked to increased expression of CYP1A1, which may have implications for colon cancer proliferation.
This study explores the impact of treating colorectal cancer organoids with individual strains. However, it’s clear that the in vivo system of internal organs is influenced by the combined interactions and synergistic effects of metabolites secreted by different strains. Our internal organs, gut microbiota, and parasites are intricately affected by these secreted metabolites. In addition, further research is needed to comprehend the specific mechanism related to anticancer effects by metabolites derived from microbiome. In future studies, we aim to treat organoids with a combination of supernatants from various strains or to co-culture different microorganisms with organoid models, allowing for exploration from multi-omics and metabolomics perspectives.
Our study confirms that beneficial bacteria can inhibit cancer growth not only in CRC cell lines but also in the 3D organoid model, which is currently emerging as a substitute for mammalian models in the early stages of drug development. Furthermore, with advancements in genomic analysis technology, metagenomic and transcriptional analyses can now be performed on stool samples from healthy controls and actual clinical patients. High-throughput screening systems have also been recently adopted as a platform for various biomedical research efforts, enabling the analysis and documentation of phenotypical changes following treatment with microbial supplements. In summary, our study has demonstrated the applicability of the 3D colorectal organoid model in studying the anti-cancer effects of microbial metabolites, including those produced by bifidobacterial strains, on tumor proliferation and growth in vitro. These findings, along with advancements in genomic analysis and high-throughput screening technologies, offer significant potential for developing effective microbiome-based therapies for colorectal cancer and other inflammatory diseases.
However, even though this study is the first test investigating anti-cancer effects of cell-free supernatants from Bifidobacterium adolescentis and Bifidobacterium longum toward colorectal cancer organoid model, our study has not reached to applying clinical trials that could construct the evidential bridge linking in vitro to in vivo. Furthermore, co-culturing bacterial cells with organoids offers a more physiologically relevant environment for investigating direct interactions than observing changes induced by supernatants. However, since Bifidobacterium is an anaerobic bacterium, it is crucial to advance co-culture methodologies that facilitate selective oxygen supply to normal colon organoids under hypoxic conditions. Addressing this methodological challenge is expected to provide deeper insights into the anticancer mechanisms through which the microbiome plays a role in inhibiting tumor growth. The mentioned above are the main limitations of our study, and further follow-up clinical research and methodological development are still required.
Methods
Collection of fecal samples
Fecal samples from Korean CRC patients and healthy controls were collected and analyzed. Fecal samples from CRC patients were collected at National Cancer Center, Goyang between June 2002 and Apr 2004. The stools from healthy controls were collected in health check-up center at Seoul National University Hospital, Seoul and were selected through age and sex matched selection. The stool samples were collected by subjects in a tube and submitted to the institution and were frozen and stored at − 20 °C. Fecal samples from both CRC patients and healthy control were sequenced in Macrogen Inc, Korea in 2017.
16S bacterial rRNA analysis
Whole DNA was extracted from fecal samples using the Mag-Bind Universal Pathogen 96 Kit (Omega Bio-Tek, Norcross, GA, USA) with a Hamilton Microlab STAR liquid handler (Hamilton Laboratory Solutions, Manitowoc, WI, USA) after bead-beating the samples with TissueLyser (Qiagen, Hilden, Germany), followed by amplicon PCR targeting the V3 to V4 regions of the 16 S bacterial rRNA gene using 341 F and 805R primers (341 F-CCTACGGGNGGCWGCAG, 805R-GACTACHVGGGTATCTAATCC). After DNA library preparation, indexing and quality checks were performed using the Nextera XT index kit (Illumina, San Diego, CA, USA) and Qubit4.0 (Thermofisher, Wilmington, DE, USA), and 300 × 2 paired-end sequencing was performed using the MiSeq system (Illumina, San Diego, CA, USA). Illumina adapter sequences of the paired-end reads were removed using Cutadapt78. Then, the trimmed sequences were processed using QIIME2. Briefly, the reads were assigned to each sample according to a unique index; pairs of reads from the original DNA fragments were merged using an import tool in QIIME279. To remove low-quality bases at the end of the reads, the DADA2 software package80 wrapped in QIIME2 was applied. Alpha and beta diversity were analyzed using core-metrics-phylogenetic in the QIIME2 diversity plugin. Alpha and beta diversities were calculated using alpha- and beta-group significance in the QIIME2 diversity plugin, respectively. Alpha diversity was calculated by observed features, evenness, and Shannon Index. Beta diversity was compared by principal coordinate analysis using Bray–Curtis distances, unweighted Unifrac, and weighted Unifrac. Taxonomic annotation was performed by mapping the training reference set with primers (forward, 5′-CCTACGGGNGGCWGCAG-3′; reverse, 5′-GACTACHVGGGTATCTAATCC-3′) and extracting the V3–V4 region using GreenGenes version 13_881. Linear discriminant effect size analysis (LEfSe) was per- formed to identify differential features at the species level between groups based on linear discriminant analysis (LDA) scores using Galaxy implementation82 Linear discriminant effect size analysis (LEfSe)82 compares abundances of bacterial species levels between normal and cancer samples with a linear discriminant analysis (LDA) score.
Sample preparations of colorectal cancer and colon epithelium for organoid culture
Tissue samples from CRC patients were obtained from surgical specimens and immediately transported to the laboratory in a fresh state. Tissue biopsies were mechanically dissociated using surgical scissors and digested using an enzyme solution containing collagenase II (1.5 mg/ml, Gibco, 17101-015-1G), hyaluronidase (20 µg/ml, Sigma Aldrich, H3506-100MG), and Y-27632 (10 µM, Sigma Aldrich, Y0503-5MG) in DME/F12 medium (supplemented with penicillin and streptomycin). The tissue was incubated at 37 °C with gentle rotation for 1 h, after which the pellets were collected by filtering through a 100 μm-pore cell strainer. The pellets were then resuspended in RGF-BME (Reduced Growth Factor Basement Membrane Extract) Matrigel (Trevigen, 3533-001-02) and seeded onto 24-well plates. For the initial 3 days of culture, the isolated cancer epithelium (tumor) and crypts (normal) were fed with Human Intestinal Stem Cell HISC)( medium supplemented with 10 µM Y-27632 to prevent anoikis.
Organoid culture medium
To produce a complete organoid culture medium, conditioned medium and basal culture medium were mixed in a 1:1 ratio and supplemented with various growth factors, including B27 (1X, Gibco, 17504-044-50X, 10 ml), n-acetyl cysteine (1.25 mM, Sigma Aldrich, A7250-5G), nicotinamide (10 mM, Sigma Aldrich, 72340-100G), hEGF (0.1 mg/ml, Thermo Fisher Scientific, PHG0311L-0.1 mg), hFGF10 (10 ng/ml, Thermo Fisher Scientific, PHG0204-25 µg), A83-01 (500 nM, Sigma Aldrich, SML0788-5MG), SB202190 (3 µM, Sigma Aldrich, S7067-5MG), Prostaglandin E2 (10 nM, Sigma Aldrich, P5640-1MG), and contamination-blocking antibiotics (Primocin, 100 µg/ml, Thermo Fisher Scientific, ant-pm-1–10*1 ml 500 mg). Conditioned medium was prefabricated in advance using a stably transfected cell line (L-WRN, ATCC CRL-3276) purchased from ATCC, which secreted Wnt-3a, R-spondin, and noggin proteins into the culture medium.
To produce conditioned medium, the L-WRN transfected cell line is cultured in T-175 flasks (Nunc, 159910) with DMEM/F-12 medium supplemented with 10% FBS, 1% P/S, and selection antibiotics, Hygromycin B (500 µg/ml) and G418 (2.0 mg/ml). The L-WRN cell line possesses resistance genes for both Hygromycin B and G418, allowing it to survive under dual selection pressure, while non-specific cells lacking resistance genes are eliminated. Once the cells fully occupy the flask, they are passaged into six T-175 flasks. From this stage onward, as the medium will be used as conditioned medium for organoid culture, the cells are cultured in 30 ml of DMEM/F-12 supplemented with 10% FBS and 1% P/S. The medium is replaced daily for 7 days, with the used medium collected and stored at 4 °C. After 7 days, the collected conditioned medium is pooled in an autoclaved 3 L bottle and filtered through a 0.2 μm pore bottle-top filter (Nalgene, 595–4520) for final preparation.
The basal culture medium was composed of GlutaMax (10 mM, Thermo Fisher Scientific, 35050-061 100 ML) and DME/F12 supplemented with 10% FBS and 1% penicillin and streptomycin.
Organoid culture
We established and cultured three sets of organoid lines (SNU-7237-TO, SNU-7293-TO, and SNU-7390S3-TO) derived from CRC patients, as well as their corresponding normal organoid lines (SNU-7237 N-NO, SNU-7239 N-NO, and SNU-7390 N-NO). Tumor organoid lines were passaged using TrypLE express (Gibco, 12604), which was resuspended in DME/F12 supplemented with 5% BCS to dissociate the organoids into single cells. The dissociated cell solution was centrifuged at 1,500 rpm (126×g) for 3 min, and the pellet was mixed with RGF-BME and seeded onto a new 6-well plate. Normal organoids were passaged using Cell Recovery Solution (Corning, 354253 100 ml). To split organoids into single crypts, harvested and collected organoid pellets were gently resuspended in Cell Recovery Solution and incubated at 4℃ for 30 min. Matrigel was liquefied by mechanical pipetting (30–50 times), and the subsequent culture process was identical to that of the tumor organoid lines.
STR profiling
Genomic DNA was extracted from each organoid line using the AmpF1STR PCR amplification kit (Applied Biosystems, Foster City, CA). The amplified DNA was analyzed using a genetic analyzer (Applied Biosystems 3500/3500xL Genetic Analyzer, Foster City, CA) to detect minisatellites and determine their short tandem repeat (STR) profiles. The STR marker libraries used for profiling included D8S1179, D2S441, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, Vwa, TPOX, Penta D, Amelogenin, D1S1656 and D18S51.
Treatment of cell-free microbial supernatant to organoids
A hemocytometer was used to calculate the seeding concentration. 4,000 cells per well were seeded on a white 96-opaque well plate (SPL, 30196), and four days were given for differentiation and organoid formation. Each microbial cell-free supernatant was then diluted to 20% concentration in HISC (Organoid medium, Human Intestinal Stem Cell) medium. The cell-free supernatant was obtained from Five strains: Blautia producta (KCTC 15607), Parabacteroides distasonis (KCTC 5751), Bacteroides ovatus (KCTC 5827), Bifidobacterium adolescentis (KCTC 3216), and Bifidobacterium longum (KCTC 3218). All five strains were cultured according to the depositor’s instructions (https://kctc.kribb.re.kr). The supernatants were harvested when the strains reached an optical density at 0.4 (OD600). Additionally, the pH of all microbial cell-free supernatants was adjusted to 6.8 ≤ pH ≤ 7.2 by adding 1.0 M NaOH and measured using a pH meter. The medium containing the supernatants and organoids were co-cultured for three days to twelve days.
RLU measurement for cell viability assay
Viable cells and cytotoxicity were assessed using a 3D organoid culture model with CellTiter-Glo® 3D, which is used for evaluating cellular ATP levels. The reagent was thawed and equilibrated to room temperature before use, and all experimental procedures were performed at room temperature. The half volume of the culture solution (containing bacterial supernatant) was replaced with CellTiter-Glo® 3D (Promega, G9683) and thoroughly resuspended using a multi-channel pipette. Plates for screening were sealed with aluminum foil and incubated for 30 min. After the light-excluded incubation, the luminescence of intra-organoid ATP content was quantified by Varioskan Lux (Thermo Fisher Scientific, USA).
High-throughput screening
To establish the HTS system, we used the ImageXpress Micro Confocal 4 (Molecular Devices, USA) and corresponding image analysis software (MetaXpress, Molecular Devices, USA). For screening, we selected the µ-Plate Angiogenesis 96-well (Ibidi, 89646, Germany) and calibrated it accordingly. Our method reported previously was partially applied to this study55. We seeded 4,000 cells per well, derived from organoids, onto 10 µl of RGF-BME gel that had been previously dispensed and solidified in each well of the plate. For the side of the organoid that faced the culture medium, 4% of RGF-BME was added to the organoid medium as a feeding ingredient for the ECM. The seeded organoids were allowed to grow for 4 days and then treated with bacterial supernatant in the proportion of 25% culture in the medium. The High-throughput screening (HTS) was performed at 3, 6, and 12 days after treatment using a 4X apochromatic objective lens, z-stacking, and z-projection.
RNA sequencing
Total RNA was isolated from cell lysate using TRIzol (Qiagen, Hilden, Germany) and Qiagen RNeasy Kit (Qiagen, Hilden, Germany). Paired-end sequencing reads from cDNA libraries (101 bp) were generated with an Illumina NovaSeq6000 instrument and the sequence quality was verified with FastQC v.0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). For data preprocessing, low-quality bases and adapter sequences in reads were trimmed using Trimmomatic v 0.3883. The trimmed reads were aligned to the human genome (UCSC hg19) using HISAT v2.1.0, a splice-aware aligner83. Then, transcripts including novel splice variants were assembled with StringTie v1.3.4d84. The abundance of these transcripts in each sample was calculated as read counts or TPM (Transcript per Million mapped reads) values. To perform principal components analysis (PCA) and confirm the similarity distance among the samples, the dist and prcomp functions were used from the ggdendro (v0.1.22) and ggfortify (v0.4.11) R packages respectively. To access the internal data of gene loadings and analyze the contributing variables of PC2 which split the samples by beneficial and harmful intestinal bacteria, the loading components were calculated by advanced features of the pca function from PCAtools (v1.2.0) R package. The 20 highest loading components containing both positive and negative values in PC2 were plotted in a bar graph. Based on the raw read counts of 35,993 transcripts, single sample enrichment level analysis of the cell signaling pathway was conducted by GSEA (v4.1.0) using the hosted MSigDB gene set database of KEGG library (c2.cp.kegg.v7.4.symbols.gmt). The phenotype label was designated as either normal colorectal mucosa versus tumor tissue or corresponding tumor derivatives for the normalized enrichment score (NES) of a single sample. Independent NES of the paired sample (normal and tumor) was calculated on default fields with the setting of permutations as 1,000 and phenotype. The result was annotated by NCBI Gene ID MsigDB.v7.4.chip platform. A total of overlapped pathways (50 gene sets of KEGG pathway) which were significantly different (FDR < 0.25) compared to normal control in both multiple tumors and PDOs (patient-derived organoid) were selected to identify the recapitulations of differentially expressed pathways in derived models. Using the ComplexHeatmap (v2.2.0) R package, the heatmap of NES values was plotted and the mapped color variance was set between minimum and maximum values.
Gas chromatography
Supernatants from bacterial culture were harvested at O.D600 values of 0.35–0.40. Methanol was added to the samples in a 1:1 ratio, and the resulting mixture was vortexed and sonicated for 10 min. After centrifugation at 13,000 rpm (16,609×g) and 4℃ for 10 min, the supernatant was collected and dried using a speed vacuum at 10,000 ppm (1 mg/ml). The dried sample was reconstituted in 150 µl of distilled water and then transferred to a new ep-tube. The sample was dried again with a speed vacuum, and derivatization was performed. Gas chromatography was carried out using a GC-TOF-MS (LECO Corporation, US) with a Rtx-5MS column and helium gas for analysis. The split ratio was set at 20:1, and three analytical replicates were performed.
Ethics approval and consent to participate
Tumor and normal biopsies used in this study were approved for research by patient’s written informed consent and the institutional review board (IRB) of Seoul National University Hospital (IRB index: H-1701-110-826). Also, the study was performed in accordance with the Declaration of Helsinki. Furthermore, all procedures were performed in compliance with relevant laws and institutional guidelines and have been approved by the appropriate institutional committees.
Statistical analysis
In alpha and beta diversity analysis, the significance of similarity among the groups was evaluated using permutational multi-variate analysis of variance (PERMANOVA) with 999 permutations (Fig. 2B). To determine the statistical significance in cell viability of organoid, the RLU values were compared using Mann-Whiteney U test. P-value less than 0.05 were considered as significant (Fig. 4.B). VIP (Variable Importance in Projection) values from the OPLS-DA (Orthogonal Partial Least Squared Discriminant Analysis) model, sorted based on values greater than 1.0, identified metabolites with significant differences between the normal and cancer subjects. This data, normalized with the mean of the total, confirms the quantitative gap between the two groups. The Mann-Whitney U test further validated these differences, as indicated by the ‘a’ notation after the labels of metabolites in a heatmap column when the p-value was below 0.05 (Fig. 6. A).
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was kindly and directly supported by National Research Foundation of Korea (NRF) grant funded by the Korean government (Grant no. 2021M3H9A1030151, 2022M3A9B6018217, and NRF2022R1A5A102641311).
Author contributions
M-J.K. and M-H.S. are equal contributors to this work and designated as co-first authors. Conceptualization, M-J.K., M-H.S., Hs.P., J-L.K. and S-Y.J.; Methodology, M-J.K., M-H.S., Bk-C., Y-S.J., Gh.K., and J-L.K.; Software, M-H.S., S-C.K. and Gh.K.; Validation, M-J.K, M-H.S., and Hs.P., J-L.K., and S-Y.J. ; Formal Analysis, M-J.K., M-H.S., S-C.K., Gh.K., and S-Y.J.; Investigation, M-J.K. and M-H.S.; Resources, J-W.P., Y-K.S., Hs.P., and J-L.K.; Data Curation, M-J.K. and M-H.S. ; Writing – Original Draft, M-J.K. and M-H.S.; Writing – Review and Editing, M-J.K., M-H.S., J-L.K., and S-Y.J.; Visualization, M-J.K., M-H.S., and S-C.K.; Supervision, Hs.P., J-L.K., and S-Y.J.; Project Administration, Hs.P., J-L.K., and S-Y.J.; Funding Acquisition, J-L.K.
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity in patient’s personal information and are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Min Jung Kim and Myoung-Hyun Song contributed equally to this work.
Contributor Information
Ja-Lok Ku, Email: kujalok@snu.ac.kr.
Seung-Yong Jeong, Email: syjeong@snu.ac.kr.
References
- 1.Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin.73, 17–48. 10.3322/caac.21763 (2023). [DOI] [PubMed] [Google Scholar]
- 2.Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M. & Wallace, M. B. Colorectal cancer. Lancet394, 1467–1480. 10.1016/S0140-6736(19)32319-0 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Kang, M. J. et al. Cancer statistics in Korea: Incidence, mortality, survival, and prevalence in 2020. Cancer Res. Treat.55, 385–399. 10.4143/crt.2023.447 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinicrope, F. A. Increasing incidence of early-onset colorectal cancer. N Engl. J. Med.386, 1547–1558. 10.1056/NEJMra2200869 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Nakatsu, G. et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun.6, 8727. 10.1038/ncomms9727 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell. Host Microbe14, 207–215. 10.1016/j.chom.2013.07.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med.15, 1016–1022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu, S., Lim, K. C., Huang, J., SaidiR, F. & SearsC, L. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci.95, 14979–14984 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viljoen, K. S., Dakshinamurthy, A., Goldberg, P. & Blackburn, J. M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PloS ONE10, e0119462 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sears, C. L. Enterotoxigenic Bacteroides fragilis: A rogue among symbiotes. Clin. Microbiol. Rev.22, 349–369 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollander, D. et al. Increased intestinal permeability in patients with Crohn’s disease and their relatives: A possible etiologic factor. Ann. Intern. Med.105, 883–885 (1986). [DOI] [PubMed] [Google Scholar]
- 12.Teahon, K., Smethurst, P., Levi, A., Menzies, I. & Bjarnason, I. Intestinal permeability in patients with Crohn’s disease and their first degree relatives. Gut33, 320–323 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashemi Goradel, N. et al. Fusobacterium nucleatum and colorectal cancer: A mechanistic overview. J. Cell. Physiol.234, 2337–2344 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science363, eaar7785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue, M. et al. Structure elucidation of colibactin and its DNA cross-links. Science365, eaax2685 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta, R. S. et al. Association of dietary patterns with risk of colorectal cancer subtypes classified by fusobacterium nucleatum in tumor tissue. JAMA Oncol.3, 921–927. 10.1001/jamaoncol.2016.6374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature486, 222–227. 10.1038/nature11053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeller, G. et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol.10, 766. 10.15252/msb.20145645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, Q. et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun.6, 6528. 10.1038/ncomms7528 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Vogtmann, E. et al. Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS ONE11, e0155362. 10.1371/journal.pone.0155362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemer, B. et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut66, 633–643. 10.1136/gutjnl-2015-309595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med.25, 679–689. 10.1038/s41591-019-0406-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang, Y. et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology152, 851–866. 10.1053/j.gastro.2016.11.018 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubinstein, M. R. et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell. Host Microbe14, 195–206. 10.1016/j.chom.2013.07.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung, L. et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe23, 203–214. 10.1016/j.chom.2018.01.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long, X. et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol.4, 2319–2330. 10.1038/s41564-019-0541-3 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Khazaie, K. et al. Abating colon cancer polyposis by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. U. S. A.109, 10462–10467. 10.1073/pnas.1207230109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohamadzadeh, M. et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc. Natl. Acad. Sci. U. S. A.108(Suppl 1), 4623–4630. 10.1073/pnas.1005066107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, D. & Medeiros, D. M. The microbiome as a major function of the gastrointestinal tract and its implication in micronutrient metabolism and chronic diseases. Nutr. Res.112, 30–45 (2023). [DOI] [PubMed] [Google Scholar]
- 30.Wu, J. et al. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell12, 360–373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blacher, E., Levy, M., Tatirovsky, E. & Elinav, E. Microbiome-modulated metabolites at the interface of host immunity. J. Immunol.198, 572–580 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol.12, 661–672. 10.1038/nrmicro3344 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Brown, A. J. et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem.278, 11312–11319. 10.1074/jbc.M211609200 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Maslowski, K. M. et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature461, 1282–1286. 10.1038/nature08530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, N. et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity40, 128–139. 10.1016/j.immuni.2013.12.007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thangaraju, M. et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res.69, 2826–2832. 10.1158/0008-5472.CAN-08-4466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Martino, M. L. et al. Polyamines: Emerging players in bacteria–host interactions. Int. J. Med. Microbiol.303, 484–491 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Pegg, A. E. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol.26, 1782–1800 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Roediger, W., Moore, J. & Babidge, W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig. Dis. Sci.42, 1571–1579 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Attene-Ramos, M. S. et al. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen.51, 304–314 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Attene-Ramos, M. S., Wagner, E. D., Gaskins, H. R. & Plewa, M. J. Hydrogen sulfide induces direct radical-associated DNA damage. Mol. Cancer Res.5, 455–459 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Ou, J. et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr.98, 111–120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrasa, J. I., Olmo, N., Lizarbe, M. A. & Turnay, J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol. In Vitro27, 964–977 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Ou, J. et al. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr. Cancer64, 34–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernstein, H., Bernstein, C., Payne, C. M. & Dvorak, K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. WJG15, 3329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louis, P., Hold, G. L. & Flint, H. J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol.12, 661–672 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Faghfoori, Z., Faghfoori, M. H., Saber, A., Izadi, A. & Yari Khosroushahi, A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int.21, 258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahmani, S., Azarpira, N. & Moazamian, E. Anti-colon cancer activity of Bifidobacterium metabolites on colon cancer cell line SW742. Turk. J. Gastroenterol.30, 835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pahumunto, N., Teanpaisan, R. J. P. & Proteins, A. Anti-cancer properties of potential probiotics and their cell-free supernatants for the prevention of colorectal cancer: An in vitro study. Probiotics Antimicrob. Proteins15, 1137–1150 (2023). [DOI] [PubMed] [Google Scholar]
- 50.Lee, D. K. et al. Anti-proliferative effects of Bifidobacterium adolescentis SPM0212 extract on human colon cancer cell lines. BMC Cancer8, 1–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujii, M. et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell. Stem Cell.18, 827–838 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Kashfi, S. M. H., Almozyan, S., Jinks, N., Koo, B. K. & Nateri, A. S. Morphological alterations of cultured human colorectal matched tumour and healthy organoids. Oncotarget9, 10572 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koh, G. Y., Kane, A. V., Wu, X. & Crott, J. W. Parabacteroides distasonis attenuates tumorigenesis, modulates inflammatory markers and promotes intestinal barrier integrity in azoxymethane-treated A/J mice. Carcinogenesis41, 909–917 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Ediriweera, M. K., To, N. B., Lim, Y. & Cho, S. K. Odd-chain fatty acids as novel histone deacetylase 6 (HDAC6) inhibitors. Biochimie186, 147–156 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Song, M. H. et al. Colon cancer organoids using monoclonal organoids established in four different lesions of one cancer patient reveal tumor heterogeneity and different real-time responsiveness to anti-cancer drugs. Biomed. Pharmacother.152, 113260. 10.1016/j.biopha.2022.113260 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Androutsopoulos, V. P., Tsatsakis, A. M. & Spandidos, D. A. Cytochrome P450 CYP1A1: Wider roles in cancer progression and prevention. BMC Cancer9, 187. 10.1186/1471-2407-9-187 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maehata, T. et al. Transcriptional silencing of Dickkopf gene family by CpG island hypermethylation in human gastrointestinal cancer. World J. Gastroenterol.14, 2702–2714. 10.3748/wjg.14.2702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang, J. H., Shim, Y. Y., Cha, S. K., Reaney, M. J. T. & Chee, K. M. Effect of Lactobacillus acidophilus KFRI342 on the development of chemically induced precancerous growths in the rat colon. J. Med. Microbiol.61, 361–368. 10.1099/jmm.0.035154-0 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Zhang, M. et al. Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model. J. Microbiol.53, 398–405. 10.1007/s12275-015-5046-z (2015). [DOI] [PubMed] [Google Scholar]
- 60.Zhu, J. et al. Lactobacillus salivarius Ren prevent the early colorectal carcinogenesis in 1, 2-dimethylhydrazine-induced rat model. J. Appl. Microbiol.117, 208–216. 10.1111/jam.12499 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Nakkarach, A. et al. Anti-cancer and anti-inflammatory effects elicited by short chain fatty acids produced by Escherichia coli isolated from healthy human gut microbiota. Microb. Cell Fact.20, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohara, T. & Mori, T. Antiproliferative effects of short-chain fatty acids on human colorectal cancer cells via gene expression inhibition. Anticancer Res.39, 4659–4666 (2019). [DOI] [PubMed] [Google Scholar]
- 63.Okabe, S. et al. Acetic acid induces cell death: An in vitro study using normal rat gastric mucosal cell line and rat and human gastric cancer and mesothelioma cell lines. J. Gastroenterol. Hepatol.29, 65–69 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Pham, C. H. et al. Anticancer effects of propionic acid inducing cell death in cervical cancer cells. Molecules26, 4951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Genua, F. et al. Association of circulating short chain fatty acid levels with colorectal adenomas and colorectal cancer. Clin. Nutr. ESPEN46, 297–304 (2021). [DOI] [PubMed] [Google Scholar]
- 66.Levy, M., Blacher, E. & Elinav, E. Microbiome, metabolites and host immunity. Curr. Opin. Microbiol.35, 8–15 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Du, X. et al. Alterations of the gut microbiome and fecal metabolome in colorectal cancer: Implication of intestinal metabolism for tumorigenesis. Front. Physiol.13, 854545 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim, Y. et al. Inhibition of proliferation in colon cancer cell lines and harmful enzyme activity of colon bacteria by Bifidobacterium adolescentis SPM0212. Arch. Pharm. Res.31, 468–473. 10.1007/s12272-001-1180-y (2008). [DOI] [PubMed] [Google Scholar]
- 69.Liu, Z. et al. Randomised clinical trial: The effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery - a double-blind study. Aliment. Pharmacol. Ther.33, 50–63. 10.1111/j.1365-2036.2010.04492.x (2011). [DOI] [PubMed] [Google Scholar]
- 70.Arrieta, M. C., Stiemsma, L. T., Amenyogbe, N., Brown, E. M. & Finlay, B. The intestinal microbiome in early life: Health and disease. Front. Immunol.5, 427. 10.3389/fimmu.2014.00427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez, F. D. The human microbiome. Early life determinant of health outcomes. Ann. Am. Thorac. Soc.11(Suppl 1), 7–12. 10.1513/AnnalsATS.201306-186MG (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olszak, T. et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science336, 489–493. 10.1126/science.1219328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sommer, F. & Backhed, F. The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol.11, 227–238. 10.1038/nrmicro2974 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Yang, I. et al. The infant microbiome: Implications for infant health and neurocognitive development. Nurs. Res.65, 76–88. 10.1097/NNR.0000000000000133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vital, M., Howe, A. C. & Tiedje, J. M. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio5, e00889. 10.1128/mBio.00889-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parada Venegas, D. et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol.10, 277. 10.3389/fimmu.2019.00277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zapletal, O. et al. Butyrate alters expression of cytochrome P450 1A1 and metabolism of benzo [a] pyrene via its histone deacetylase activity in colon epithelial cell models. Arch. Toxicol.91, 2135–2150 (2017). [DOI] [PubMed] [Google Scholar]
- 78.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J.17, 10–12 (2011). [Google Scholar]
- 79.Bolyen, E. et al. Author correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol.37, 1091. 10.1038/s41587-019-0252-6 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol.72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol.12, R60. 10.1186/gb-2011-12-6-r60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alli, E. et al. Enhanced sensitivity to cisplatin and gemcitabine in Brca1-deficient murine mammary epithelial cells. BMC Pharmacol.11, 7. 10.1186/1471-2210-11-7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Erratum to. Cancer statistics, 2021. CA Cancer J. Clin.71, 359. 10.3322/caac.21669 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not openly available due to reasons of sensitivity in patient’s personal information and are available from the corresponding author upon reasonable request.